Abstract
MicroRNAs (miRNAs), such as miR-192, mediate the actions of transforming growth factor β1 (TGF-β) related to the pathogenesis of diabetic kidney diseases. We found that the biphasic induction of miR-192 expression by TGF-β in mouse renal glomerular mesangial cells initially involved the Smad transcription factors, followed by sustained expression that was promoted by acetylation of the transcription factor Ets-1 and histone H3 by the acetyltransferase p300, which was activated by the serine and threonine kinase Akt. In mesangial cells from Ets-1–deficient mice or in cells in which Ets-1 was knocked down, basal amounts of miR-192 were higher than those in control cells, but sustained induction of miR-192 by TGF-β was attenuated. Furthermore, inhibition of Akt or ectopic expression of dominant-negative histone acetyltransferases decreased p300-mediated acetylation and Ets-1 dissociation from the miR-192 promoter, and prevented miR-192 expression in response to TGF-β. Activation of Akt and p300 and acetylation of Ets-1 and histone H3 were increased in glomeruli from diabetic db/db mice compared to non-diabetic db/+ mice, suggesting that this pathway may contribute to diabetic nephropathy. These findings provide insight into the regulation of miRNAs through signaling-mediated changes in transcription factor activity and in epigenetic histone acetylation under normal and disease states.
Introduction
Increasing evidence shows that microRNAs (miRNAs), including miR-192, play major roles in kidney development and the pathogenesis of diabetic kidney diseases, such as diabetic nephropathy (1, 2); however, the mechanisms of miRNA regulation and biogenesis in renal cells are not well understood. The abundance of miR-192 is increased in renal glomeruli of mouse models of diabetes (2–7), in kidneys of patients with certain renal diseases (8, 9), and in sera of patients with primary focal segmental glomerulosclerosis (in which miR-192 abundance correlated with proteinuria and interstitial fibrosis) (10). It is also increased in cultured human or mouse renal glomerular mesangial cells (MCs), podocytes, endothelial cells, or tubular cells treated with high concentrations of glucose or with transforming growth factor β1 (herein referred to simply as TGF-β) (3, 5, 7, 11). In diabetic nephropathy, TGF-β has been shown to be the major profibrotic molecule that increases the expression of extracellular matrix (ECM) genes, such as those that encode collagens. It is also a major mediator of the effects of high glucose concentration in renal cells, and the abundance of TGF-β is increased by high glucose in MCs through the actions of upstream stimulatory factors (12, 13). miR-192 targets Zeb2, an E-box repressor (14), which inhibits expression of the gene encoding type 1 collagen α2 (Col1a2) in mouse renal MCs through E-boxes in the far-upstream region of Col1a2 (3). miR-192 also induces increased abundance of other renal miRNAs, such as miR-200b, miR-200c, miR-216a, and miR-217 in MCs through de-repression at E-boxes in their promoter regions (13, 15). In addition, miR-200b and miR-200c increase the expression of Col1a2, Col4a1, and TGF-β1 by targeting E-box repressors (Zeb1 and Zeb2) (14, 16) in MCs, which amplifies the fibrotic response (13). miR-216a can also promote Col1a2 expression by yet another mechanism that involves inhibition of its target Ybx1 and enhanced action of the E-box activator Tfe3 (17). miR-216a and miR-217 activate the serine and threonine kinase Akt by targeting the phosphatase PTEN (phosphatase and tensin homolog deleted from chromosome 10) in mouse MCs and in glomeruli from a mouse model of type 2 diabetes (db/db) (15). These miRNA cascades initiated by miR-192 under diabetic conditions have key roles in fibrosis (the production of increased amounts of ECM proteins, such as collagens) and hypertrophy (Akt activation and enhanced protein synthesis) related to the pathogenesis of diabetic nephropathy (3, 13, 15). Furthermore, treatment with a locked nucleic acid–modified miR-192 inhibitor or with low-dose paclitaxel ameliorated renal fibrosis in animal models of diabetes or kidney injury by decreasing the expression of miR-192 (4, 15, 18).
Studies have identified a role for the transcription factor Smad3 in TGF-β–induced miR-192 expression in tubular cells (11). In addition, the region upstream of miR-192 is regulated by p53 in other cells types (19–21). However, some reports showed that TGF-β inhibits miR-192 expression in cancer cell lines, tubular epithelial cells, and immortalized kidney cell lines (22–24) and the hepatocyte nuclear factor1 (HNF1) has also been implicated in miR-192 expression (25, 26). Because this could be a result of cell-specific signaling elicited by TGF-β, further details of the regulation of miR-192 expression by TGF-β need to be clarified. We examined the molecular mechanisms underlying the early and sustained expression of miR-192 in MCs treated with TGF-β. We observed a quick, relatively transient recruitment of Smad2 and Smad3 (within 1 hour) to a Smad-binding site in the upstream region of miR-192 after the treatment of MCs with TGF-β. In addition, we observed the presence of multiple potential Ets-1–binding sites in the upstream regions of the genes encoding human and mouse miR-192, which was suggested previously (3, 27). However, the relevance of these Ets-1 sites has not been reported. Ets-1 was cloned as a proto-oncogene (28). Ets-1 protein functions as a sequence-specific (GGAA/T) DNA-binding transcription factor, and there are several Ets-1 family members that can bind to this consensus sequence (29, 30). With respect to renal fibrosis, Ets-1 promotes matrix remodeling through the regulation of matrix metalloproteinases and ECM proteins, such as collagens (31–33). Therefore, we focused primarily on Ets-1 to investigate the regulation of miR-192 expression in response to TGF-β.
We found that Ets-1 is a negative regulator of basal miR-192 expression, and that removal of Ets-1 from the miR-192 promoter region promotes miR-192 expression in response to TGF-β. Ets-1 and histone lysine acetylation in the miR-192 promoter region were mediated by p300, which was phosphorylated by Akt in response to treatment with TGF-β. Together, these events correlated with the sustained expression of miR-192 in response to TGF-β in MCs.
Results
TGF-β increases miR-192 expression through acetylation of Ets-1 and activation of Akt and p300
Both high glucose (25 mM) and TGF-β induce the expression of ECM genes related to the pathogenesis of diabetic nephropathy, and TGF-β mediates the effects of high glucose concentrations in renal MCs through upstream stimulatory factors (12, 13). We first confirmed that TGF-β is the critical factor for the downstream effects of high glucose conditions on miR-192 expression by examining the effects of a TGF-β neutralizing antibody in mouse MCs treated with 25 mM glucose (fig. S1). The expression of miR-192 (as well as other downstream miRNAs, such as miR-200b, and fibrotic genes such as Col1a2) was increased 72 hours after treatment with high glucose, and this increase was abolished by pre-treatment with the TGF-β antibody, suggesting that TGF-β is the key mediator of miR-192 expression in high glucose or diabetic conditions. We also confirmed that the abundance of miR-192 was increased in MCs treated with TGF-β alone from 1 to 24 hours compared to untreated MCs (Fig. 1A). This increase in miR-192 abundance in MCs was sustained even at 48 and 72 hrs after treatment with TGF-β (Fig. 1A). The initial early induction of miR-192 expression by TGF-β in other cell types can be mediated by Smad3, p53, or both (11, 19–21). However, the mechanisms underlying the sustained expression of miR-192 in response to TGF-β in MCs are unclear and thus warranted further examination.
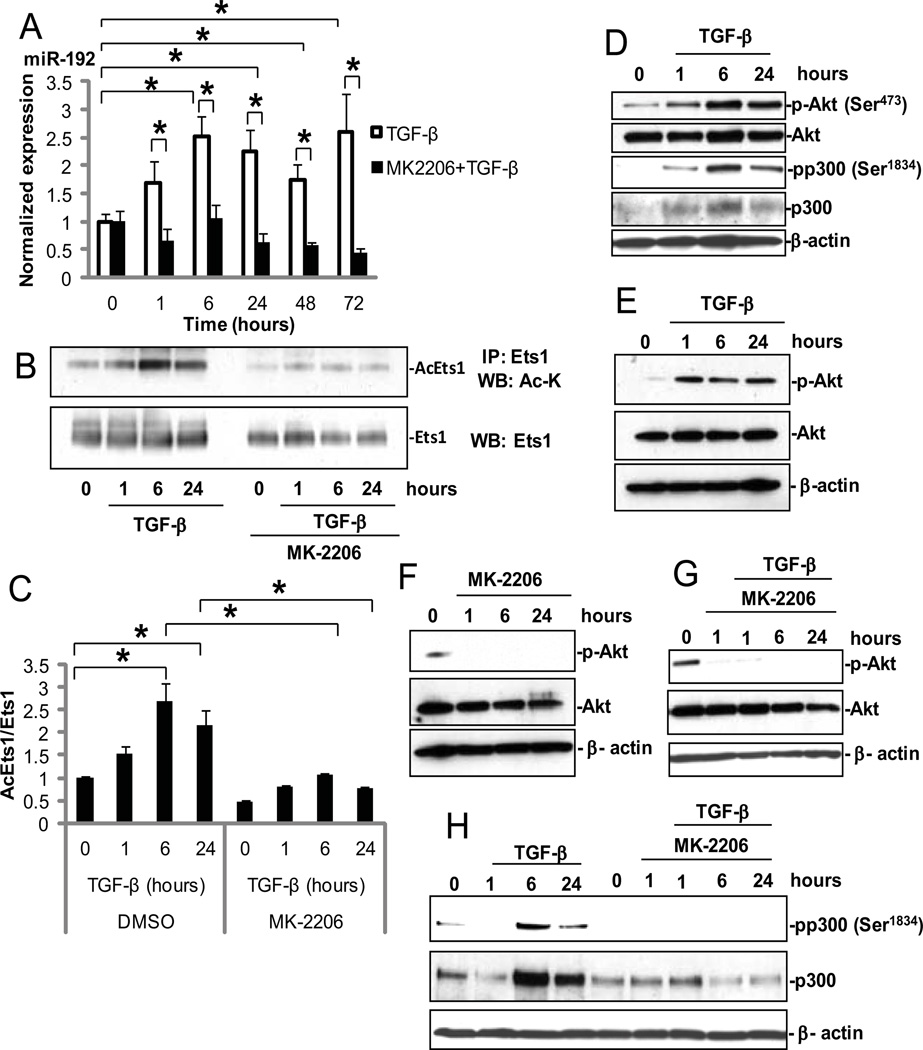
Fig. 1. TGF-β increases miR-192 expression through acetylation of Ets-1 and activation of Akt and p300.
(A) Time-course of miR-192 expression in MCs treated from 1 to 72 hours with 10 ng/ml TGF-β (white bars) or with TGF-β and 1µM MK-2206 (black bars). Culture medium was changed before these treatments with no more changes during the experiments. MK-2206 was treated at 1 hour before TGF-β treatment. Data are means ± SEM from three experiments. All comparisons were between 0 hour and each time point. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis. (B) MC lysates were immunoprecipitated (IP) with an anti-Ets-1 antibody and analyzed by Western blotting for acetylated Ets-1 (Ac-Ets-1) with an antibody against acetyl lysine (Ac-K). (C) Normalized ratio of Ac-Ets-1 over total Ets-1. Data are means ± SEM from three experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis. (D) Amounts of p-Akt and p-p300 in MCs treated with TGF-β. (E to H) MCs were pre-treated with 1 µM MK-2206 or DMSO for 1 hour and then treated with TGF-β for the indicated times before being analyzed by Western blotting. β-actin and total Akt served as loading controls. (E) Amount of p-Akt at Ser473 by TGF-β and (F) its inhibition by MK-2206 in MCs. (G and H) Effect of MK-2206 on TGF-β–induced phosphorylation of p300 at Ser1834 in MCs.
Because several potential Ets-1–binding sites were identified in the upstream region of miR-192, (3, 27) and because Ets-1 is implicated in ECM remodelling that is related to diabetic nephropathy (31–33), we focused on Ets-1 in this study. We examined the abundance of total and phosphorylated Ets-1 (pEts-1) protein by Western blotting analysis and detected no significant changes in either Ets-1 protein abundance or the extent of its phosphorylation in response to TGF-β (fig. S2, A to C). On the other hand, we detected an increase in the extent of Ets-1 acetylation 6 hours after treatment with TGF-β, and this was sustained for up to 72 hours (Fig. 1, B and C, and fig. S3, A and B). We next examined the phosphorylation of the histone acetyltransferase p300 at Ser1834, an Akt target site, to determine whether increased activation of p300 was responsible for the increased acetylation of Ets-1 (Fig. 1D). The amount of total and phosphorylated p300 was increased after treatment with TGF-β, maximally at 6 hours (Fig. 1D). Concurrently, Akt activation increased, indicated by an increase in the phosphorylation of Akt at Ser473 (Fig. 1D).
To confirm whether Akt was the critical kinase for the phosphorylation of p300 in MCs treated with TGF-β, we pre-treated MCs with the Akt-specific inhibitor MK-2206. TGF-β activated Akt over 1 to 24 hours (Fig. 1E), and MK-2206 markedly inhibited both basal Akt phosphorylation (Fig. 1F) and TGF-β–induced Akt activation in MCs (Fig. 1G). In the same experiment, MK-2206 also abrogated the TGF-β–induced increase in phosphorylated p300 relative to those in cells treated with vehicle alone (Fig. 1H). MK-2206 also inhibited the TGF-β–induced increase in Ets-1 acetylation (Fig. 1, B and C, fig. S3, A and B). Moreover, induction of miR-192 expression by TGF-β in MCs was abolished by MK-2206 (Fig. 1A). These results suggested that Akt, activated by TGF-β, phosphorylated and activated p300, which in turn increased the acetylation of Ets-1 and induced miR-192 expression in MCs treated with TGF-β.
It is known that mTORC2 (mammalian target of rapamycin complex 2) is the major kinase targeting Akt at Ser473 (34). MK-2206 is, however, a specific inhibitor of Akt and blocks Akt phosphorylation at Ser473 (35–37). We also tested a dominant negative (DN) Akt mutant (kinase-dead) which inhibits wild-type Akt function(38) and verified that this DN-Akt also inhibited phosphorylation of Akt at Ser473 in MCs treated with TGF-β (fig. S4, A and B). These results confirmed that MK-2206 inhibits the phosphorylation of Akt at Ser473 by specifically and directly inhibiting Akt, not indirectly through mTORC2 for example.
Ets-1 is a negative regulator of basal miR-192 expression, and a decision-maker in the TGF-β response
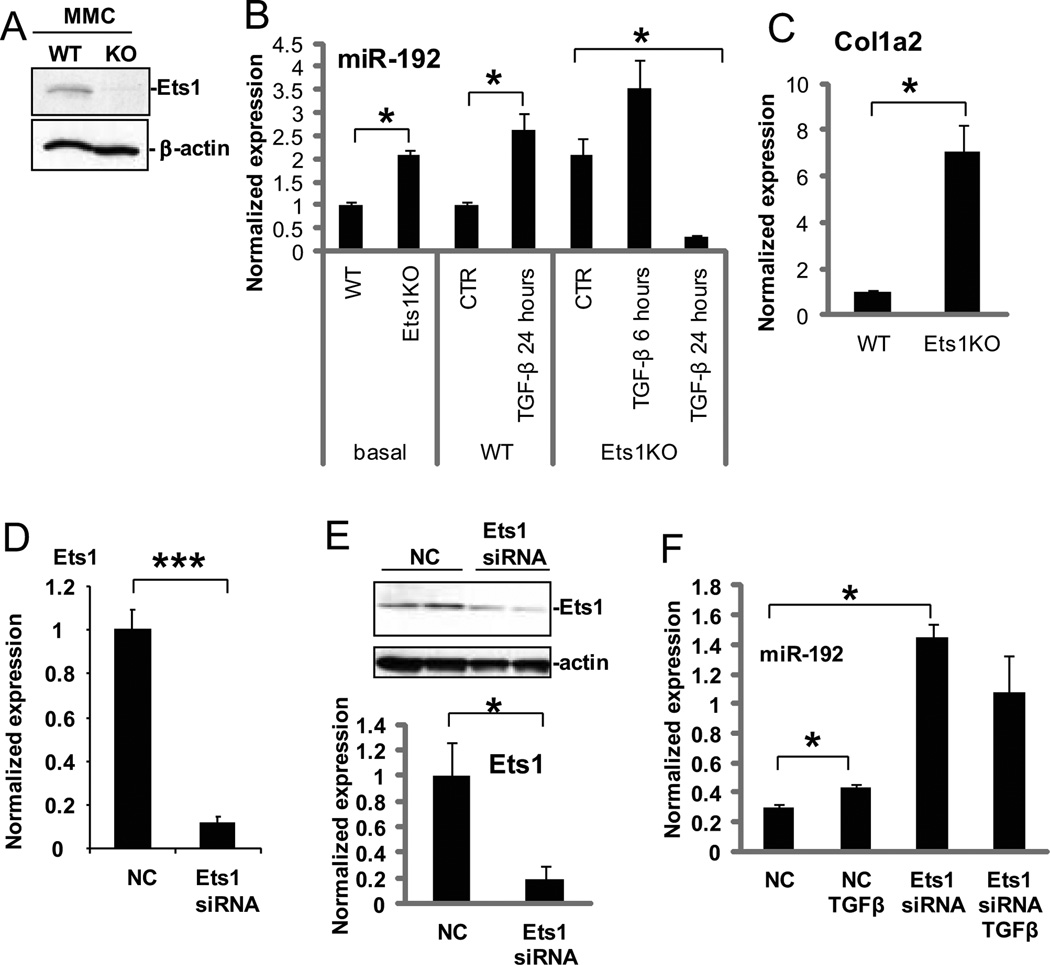
Because Ets-1 appeared to be an important transcriptional regulator of miR-192 expression in MCs, we next tested the expression of miR-192 in MCs derived from Ets-1-deficient mice (39). Absence of Ets-1 protein in these murine MCs was confirmed by Western blotting (Fig. 2A). Basal expression of miR-192 was two-fold higher in Ets-1–deficient MCs compared with wild-type MCs (Fig. 2B), suggesting Ets-1 may repress its basal expression. Basal Col1a2 expression was also significantly higher in Ets-1–deficient murine MCs compared with wild-type MCs (Fig. 2C). Treatment with TGF-β increased miR-192 expression by 2.5 fold over control in wild-type MCs as anticipated (Fig. 2B). However, in Ets-1–deficient MCs, expression of miR-192 slightly increased with TGF-β at 6 hours, but its expression was even lower than basal expression at 24 hours (Fig. 2B). These results demonstrate that MCs from Ets-1–deficient mice have increased miR-192 expression in the basal state but have lost the persistent induction by TGF-β.
Fig. 2. Expression of miR-192 is increased in Ets-1–deficient MCs and MCs treated with Ets-1 siRNA.
(A) Confirmation of Ets-1 absence in murine MCs from Ets-1–deficient (KO) mice compared with wild-type mice (WT) by Western blotting. (B) Basal or induced (TGF-β or control, CTR) miR-192 expression in Ets-1–deficient MCs compared to wild-type MCs. Data are means ± SEM from 3 experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis. (C) Col1a2 expression in Ets-1-deficient MCs compared twith wild-type MCs. Data are means ± SEM from 3 experiments. *P <0.05 by Student’s t-tests. (D and E) Abundance of Ets-1 mRNA (by PCR, D), and protein (by Western blotting, E) in MCs treated with Ets-1 siRNA compared to MCs treated with negative control oligonucleotides (NC). Data are means ± SEM from 3 experiments.*P <0.05; *** P <0.001 by Student’s t-tests. (F) Basal and TGF-β-induced miR-192 expression in MCs treated with Ets-1 siRNA. Data are means ± SEM from three experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis.
Next, we tested the effects of Ets-1 knockdown using siRNAs in wild-type MCs. Efficient reduction of Ets-1 mRNA and protein amounts with the siRNA pool treatment was confirmed (Fig. 2, D and E). Similar to the results observed in Ets-1–deficient MCs, Ets-1 siRNA treatment significantly increased basal miR-192 expression compared to treatment with a negative control siRNA pool (Fig. 2F). Whereas treatment with TGF-β increased miR-192 expression in the negative controls as expected, it did not further increase miR-192 expression in Ets-1 siRNA treated cells. However, the expression of miR-192 in MCs treated with Ets-1 siRNA and TGF-β was still higher than that observed in those treated with control siRNA and TGF-β (Fig. 2F), suggesting that knockdown of Ets-1 may increase basal miR-192 expression but may also saturate miR-192 expression, and therefore simultaneous TGF-β treatment may be unable to further increase miR-192 expression. Taken together, these data suggested that Ets-1 is a negative regulator of basal miR-192 expression but that its removal is critical for enhanced miR-192 expression in the response to TGF-β.
Smad and Ets-1 sites in the upstream region of murine miR-192 are responsive to TGF-β
A non-coding RNA CJ241444 (CJ24) is transcribed from the genomic sequence just upstream of miR-192, and the Atg2a gene is further upstream (Fig. 3A). Because the expression patterns of CJ24 were very similar to those of miR-192 in MCs treated with TGF-β or MK-2206 (fig. S5A and Fig. 1A, respectively), it is likely that miR-192 expression is also controlled by the region upstream of CJ24. Inhibition of Akt by MK-2206 also attenuated TGF-β-induced expression of Col1a2 and Col4a1, genes involved in the fibrotic response, but their expression was at least partially restored by co-treatment with a miR-192 mimic in MCs (fig. S5, B and C), further supporting the notion that miR-192 is an important mediator of TGF-β signaling to downstream fibrotic genes.
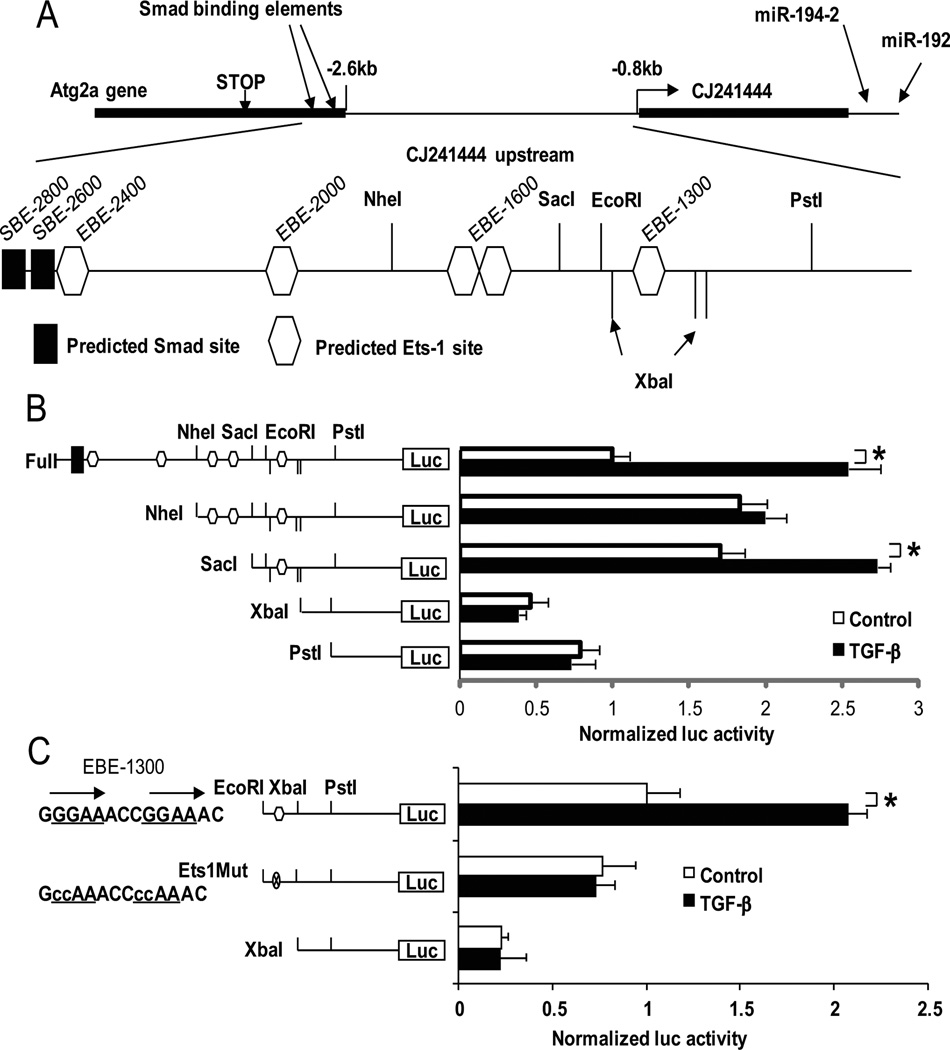
Fig. 3. Smad and Ets-1 sites in the upstream region of murine miR-192 are responsive to TGF-β.
(A)The region upstream of CJ24 (downstream of poly (A) signal of Atg2a gene including SBE-2600 site and just upstream of CJ24 transcription start site) was cloned into pGL3P. Recognition sites for restriction enzymes, NheI, SacI, XbaI, EcoRI and PstI were used to create deletion mutants. (B) TGF-β treatment increased the luciferase activity of full-length (with SBE-2600) and SacI constructs. (C) Mutant constructs of the EBE-1300. Tandem repeats of GGAA (Ets binding consensus) (29) were found at the site. These repeats were replaced with CCAA. Luciferase activity of the SacI construct was abolished in the construct with the Ets-1 site mutation or deletion. Ratios of firefly to renilla luc activities were obtained and results expressed as Normalized luc activity after normalization to activities in control untreated cells (as described in the Methods). Data are means ± SEM from four experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis.
Taking a closer look at the region upstream of miR-192 (those of Atg2a, CJ24 and in between), we identified Smad binding elements (SBE) in the 3’UTR of Atg2a and two potential Smad sites and five potential Ets-1 binding elements (EBE) (four EBE clusters) in the upstream region of CJ24 (Fig. 3A). We hypothesized that these Smad and Ets-1 sites are potential enhancer elements of miR-192 and CJ24. To test this, we cloned full-length or fragments of the region upstream of CJ24 into the pGL3P vector which has a promoter. Deletion constructs containing none or one to five of the Ets-1 sites were then cloned. Luciferase assays showed that the full-length (including the Smad binding element SBE-2600) and SacI constructs responded to TGF-β treatment in MCs (Fig. 3B). It may be interesting that SBE-2600 in the neighbouring Atg2a gene was responsive to the TGF-β treatment, because usually important regulatory elements in one gene are separated from neighbouring genes. The full-length construct showed lower basal reporter activity relative to the NheI or SacI constructs, suggesting that the upstream region might have some repressive elements (Ets-1 sites) for basal promoter activity. XbaI and PstI constructs (without EBE-1300) showed even lower basal activity and did not respond to TGF-β, further supporting the notion that the upstream Ets-1 binding sites are negative regulators of the CJ24 upstream region, whereas the EBE-1300 site appears to be critical for the TGF-β response.
To further confirm this, nucleotide sequence substitutions were introduced into the EBE-1300 site by replacing the tandem repeats of GGAA (Ets binding consensus) (29) with CCAA. The resulting Ets-1 site mutant construct lost the TGF-β response (Fig. 3C), demonstrating that the EBE-1300 site plays a major role in TGF-β induced CJ24 or miR-192 expression in MCs. The EBE-1300 Ets-1 site mutant did not show significant decrease in basal activity compared with that of wild-type EBE-1300, while the XbaI construct (without EBE-1300) showed significantly decreased basal activity, suggesting that the region between EcoRI to XbaI sites may have positive regulatory elements, although we could not identify any characteristic motifs by in silico transcription factor binding site searches. Because the NheI construct has three Ets-1sites (three negative elements), it may be resistant to transcriptional activation by TGF-β in the absence of the Smad sites. On the other hand, the Sac I construct responds to TGF-β most likely because it has only one Ets-1 site and thus may be transcriptionally activated relatively more easily through dissociation of Ets-1 from EBE-1300. Moreover, TGF-β did not have any significant effects on reporter activity in MCs derived from Ets-1– deficient mice (fig. S6), further supporting a critical function for Ets-1 in the TGF-β response.
TGF-β changes the occupancies of Smad2 and Smad3, Ets-1 and p300 as well as histone acetylation at the miR-192 upstream region
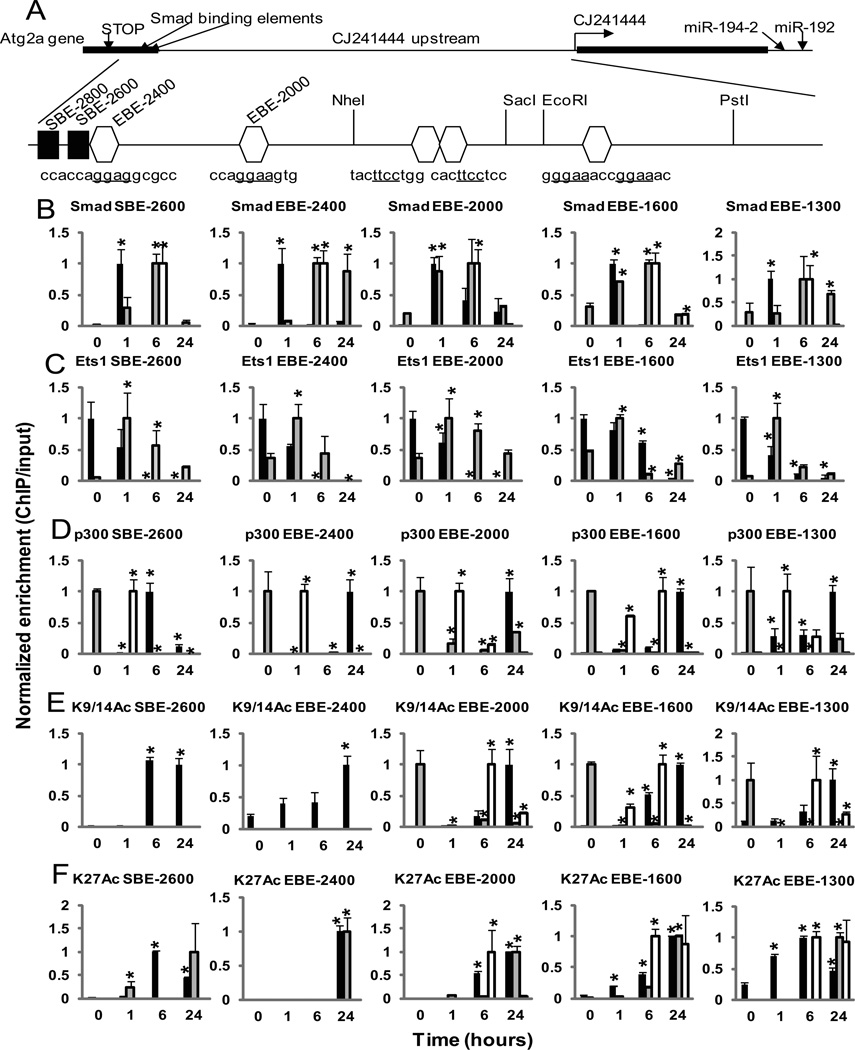
Because our data indicated a key role for Ets-1 sites during the transcriptional activation of miR-192 by TGF-β, we used ChIP assays to evaluate Ets-1 occupancy as well as other chromatin events at all these sites (Fig. 4A). We first performed ChIP experiments at the two Smad sites (SBE-2800 and SBE-2600). Since we could not detect any signal at SBE-2800 with Smad2 and Smad3 ChIP even after TGF-β treatment, the remaining experiments were done only at the SBE-2600 site. Smad2 and Smad3 occupancy at SBE 2600 was increased at 1 hour after treatment with TGF-β and this quickly decreased soon after, within 6 to 24 hours (Fig. 4B, black bars). Smad2 and Smad3 also bound to all the Ets-1 sites, EBE-2400, EBE-2000, EBE-1600 and EBE-1300 within 1 hour of TGF-β treatment in a pattern very similar to the Smad site, suggesting that Smad and Ets-1 may form a complex which interacts with Ets1 sites. Furthermore, we found that Ets-1 also occupied the Smad site SBE-2600 and this was reduced after TGF-β treatment similar to the reduction seen at the four Ets-1 binding sites (Fig. 4C). Six hours after treatment with TGF-β, p300 was recruited at SBE-2600, and in parallel, the acetylation of histone H3 at lysines 9, 14, and 27 (H3K9, H3K14, and H3K27, respectively) were also increased at this site (Fig, 4, D to F, black bars). From the timings of their activation (Fig. 1, D and H, and Fig 4, D to F), these ChIP results suggest that in response to TGF-β, Smad binds to the Smad binding site first (at 1 hour) and then Akt-activated p300 binds to the Smad binding site later (at 6 hours) to promote acetylation of H3K9, 14, and 27, thereby enabling chromatin remodeling associated with histone acetylation (40).
Fig. 4. Chromatin Immunoprecipitation (ChIP) assays reveal TGF-β induced changes in the occupancies of Smads and Ets-1 and histone acetylation in the upstream region of miR-192.
(A) Schematic representation of the upstream region of miR-192. A transcript (CJ241444, CJ24) as well as the Atg2a gene are located in the upstream region of the miR-192 gene. Smad binding elements (SBE-2800 and SBE-2600) are found in the 3’UTR of Atg2a. Four clusters of potential Ets-1 binding sites (EBE-2400, EBE-2000, EBE-1600, and EBE-1300) are present in the upstream region of CJ24. (B to F) Cross-linked chromatin samples from MCs treated with TGF-β for 0 to 24 hours were immunoprecipitated with indicated antibodies: Smad2 and Smad3 (Smad) (B), Ets-1 (Ets1) (C), p300 (D), acetylated histone H3K9 and H3K14 (K9/14Ac) (E), and H3K27Ac (K27Ac) (F). ChIP enriched DNA samples were amplified by qPCR using primers spanning each of these sites. Black bars, MCs treated with TGF-β alone; Gray bars, MCs treated with TGF-β and MK-2206; White bars, Ets-1-deficient MCs treated with TGF-β. Data are means ± SEM from three experiments. *P < 0.05 compared to the 0 time point by one-way ANOVA followed by Tukey’s post-hoc test analysis.
Next Ets-1 binding was examined by ChIP analysis. All four potential Ets-1 sites evaluated (Fig. 4A) were occupied by Ets-1 under control conditions, but Ets-1 dissociated from these sites after TGF-β treatment over 6 to 24 hours (Fig. 4C, black bars). Because increased phosphorylation of p300 at Ser1834 by TGF-β was also observed (Fig. 1, D and H), we next examined p300 occupancy as well as histone acetylation at these Ets-1 sites. ChIP assays showed significantly increased p300 occupancy and increased histone H3K9, 14, and 27 acetylation at all of the Ets-1 sites in the miR-192 upstream region at 6 to 24 hours in MCs after TGF-β treatment (Fig. 4, D to F, black bars). To determine if acetylated Ets-1 (AcEts-1) dissociates from these Smad and Ets-1 sites, Ets-1-ChIPed chromatin was re-ChIPed with antibody against AcLys (fig. S7). Enrichment of AcEts-1 was significantly lower than Ets-1 at all of these Smad and Ets-1 sites. These results suggested that p300 is recruited to the Ets-1 site by TGF-β treatment, and its activation by phosphorylation most likely leads to the observed increase in acetylation of histone H3K9, 14 and 27, and Ets-1, and the subsequent dissociation of acetylated Ets-1 from the Ets-1 sites of the CJ24 and miR-192 upstream regions.
The Akt inhibitor, MK-2206, affects the occupancies of Smad2 and Smad3, Ets-1, and p300 as well as histone acetylation at the miR-192 upstream region
Next, we investigated the role of Akt in these TGF-β-induced changes at the miR-192 upstream region. ChIP assays performed in the presence of Akt inhibitor MK-2206 showed a delay of 6 hours in Smad recruitment to SBE-2600 after treatment with TGF-β (Fig. 4B, gray bars) compared to experiments without Akt inhibitor, which showed Smad recruitment within 1 hour (Fig. 4B),suggesting that Akt may assist in the recruitment of Smad to the Smad binding site upstream of miR-192 in response to TGF-β. Ets-1 occupancy was enhanced by treatment with MK-2206, increasing at 1 hour and then steadily dropping over 6 to 24 hours at SBE-2600 and at all of the Ets-1 sites (Fig. 4C, gray bars). This Akt inhibitor also decreased p300 occupancy at the Smad site even by the early time point of 1 hour relative to time 0 (Fig. 4D, gray bars), suggesting that Akt inhibition decreases p300 phosphorylation which leads to a concomitant increase in Ets-1 occupancy and decrease in the actual expression of miR-192 (Fig. 1, A and H, and Fig. 4C). TGF-β treatment partially overcame the effects of MK-2206 and led to a subsequent decrease in Ets-1 occupancy at SBE-2600 and all the Ets-1 sites (Fig. 4C). However, the Ets-1 occupancy was still high enough to suppress the expression of miR-192 (Fig.1A). In MK-2206 treated cells, H3K9 and H3K14 acetylation was undetected at the Smad site, whereas H3K27 acetylation was slightly increased (Fig. 4, E and F, gray bars), suggesting that H3K9 and H3K14 acetylation by p300 (but not H3K27 acetylation) is regulated by Akt and is essential for induction of miR-192 expression (Fig. 1A). In addition, p300 occupancy and H3K9 and H3K14 acetylation were also clearly reduced at EBE-2000, −1600 and −1300 by combined MK-2206 and TGF-β treatment (Fig. 4, D and E, gray bars), suggesting that p300 may be the critical histone acetyltransferase of H3K9 and H3K14 related to miR-192 induction. Overall, this altered profile of Smad and p300 recruitment and histone acetylation in the presence of the Akt inhibitor may contribute to the suppression of TGF-β-induced miR-192 expression seen in the presence of the Akt inhibitor (Fig. 1A).
Because we previously showed that a cascade from miR-192 to miR-216a and miR-217 activates Akt by inhibiting PTEN, and that a miR-192 inhibitor inhibits Akt activation through the same cascade in MCs (15), we also tested whether the miR-192 inhibitor increases Ets-1 occupancy at the Smad or Ets-1 sites in the upstream region of miR-192 (fig. S8). Ets-1 enrichment at the Smad site (SBE-2600) (fig. S8A) and Ets-1 sites (EBE-2400 and EBE-1300) (fig. S8, B and C) was higher in MCs treated with miR-192 inhibitor compared with control MCs. These results suggest that miR-192 inhibition leads to Ets-1 enrichment at the upstream region of miR-192 because of decreased Ets-1 acetylation as a result of reduced Akt activation (15) and subsequently decreased p300 phosphorylation.
Altered occupancies of Smad2 and Smad3 and p300 as well as histone acetylation at the miR-192 upstream in Ets-1–deficient MCs
We next performed ChIP assays with antibodies against Smad2 and Smad3, p300, and acetylated histone H3K9, 14, and 27 in MCs derived from Ets-1–deficient mice. Smad2 and Smad3 had delayed recruitment to the Smad site (SBE-2600) and all of the Ets-1 binding sites in Ets-1-deficient MCs (recruited at 6 hours) compared with wild-type MCs (recruited at 1 hour) (Fig. 4B, white and black bars, respectively), suggesting that Ets-1 may assist in recruiting Smad proteins to DNA elements. The recruitment of p300 was observed within 1 hour at the Smad and all of the Ets-1 binding sites (Fig. 4D, white bars), although Smad recruitment and histone acetylation was not detected until 6 hours after TGF-β treatment in the Ets-1–deficient cells, and this acetylation at EBE-2000, −1600 and −1300 was reduced by 24 hours (Fig. 4, D and E, white bars). These results were consistent with the expression pattern of miR-192 in Ets-1–deficient MCs treated with TGF-β (Fig. 2D), suggesting that Smad recruitment to these sites may not be very efficient in the absence of Ets-1, even though Akt is active and p300 is phosphorylated (Fig. 1, D and H). Interestingly, at 6 hours, both p300 and Smad were also recruited at EBE-2000, −1600 and −1300, but not at SBE-2600 and EBE-2400 (Fig. 4, B and D, white bars). This suggests that increased histone H3 acetylation may be detected because both Smad and p300 were present together at EBE-2000, −1600 and −1300 at 6 hours after TGF-β treatment, and that such “coexistence” of Smad and p300 might be necessary for p300 to acetylate histone H3, at least in Ets-1 deficient conditions. Delayed recruitment of Smad2, Smad3, and p300 and delayed acetylatoin of H3K9, 14, and 27 in the absence of Ets-1 (Fig. 4, B to E, white bars) might contribute to the slight induction of miR-192 in these Ets-1–deficient cells, which was observed at about 6 hours (Fig. 2B). The reason for this relatively weak induction of miR-192 by TGF-β at 6 hours in the Ets-1–deficient cells could be because miR-192 expression is already saturated due to the absence of repression from Ets-1. In addition, Smad2, Smad3, and p300 occupancies and H3K9, 14, and 27 acetylation were attenuated at later time points (at about 24 hours) (Fig. 4, B to E), and this could be the reason for loss of persistently high expression of miR-192 in Ets-1–deficient cells (Fig. 2B).
TGF-β-induced miR-192 expression is dependent on acetyltransferase and Akt activity
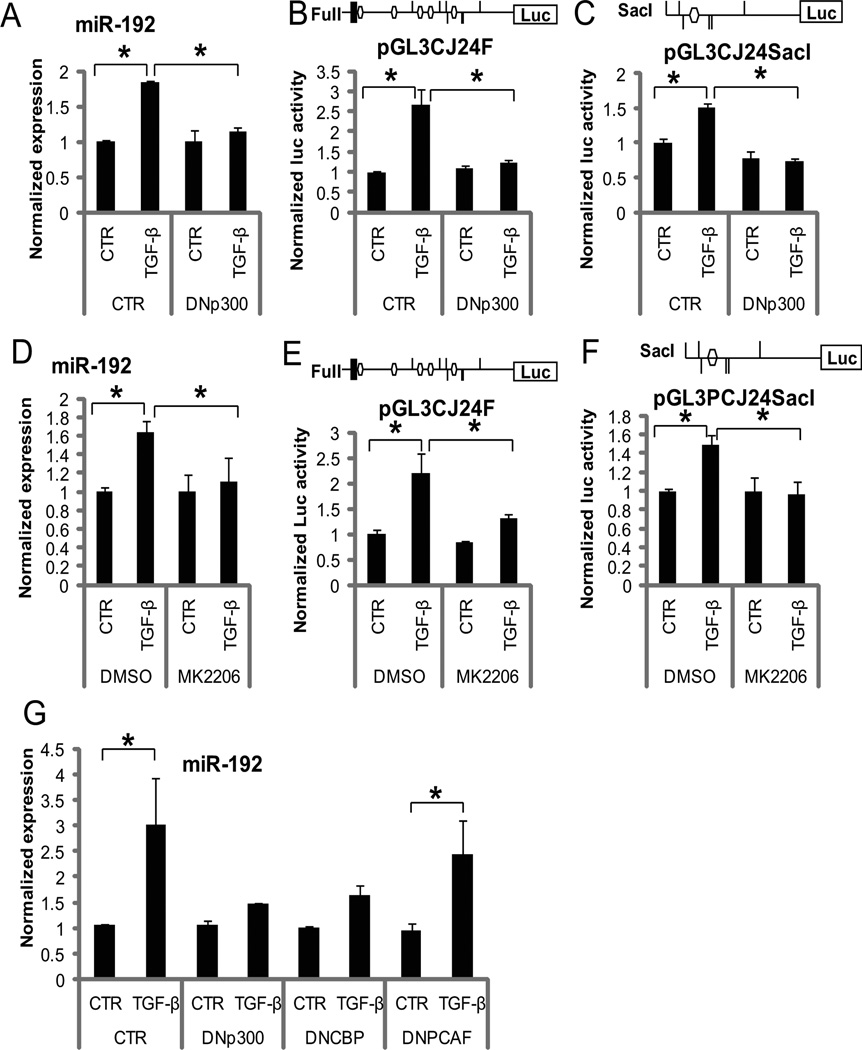
As mentioned above, acetylation of Ets-1 and chromatin histones by p300 (phosphorylated and activated by Akt) appear to drive the transcription of miR-192 (and also CJ24). To determine the functional relevance, we tested the effects of dominant negative p300 (DNp300) or the Akt inhibitor (MK-2206) on miR-192 expression and its upstream enhancer activity (Full-length and SacI luciferase reporter constructs depicted in Fig. 3). DNp300 inhibited TGF-β-induced miR-192 expression (Fig. 5A) and miR-192 upstream enhancer activity (Fig. 5, B and C) relative to that seen in controls, demonstrating that the acetyltransferase activity of p300 is essential for TGF-β-mediated induction of miR-192 in MCs. The Akt inhibitor MK-2206 also inhibited TGF-β-induced miR-192 expression (Fig. 5, D) and upstream enhancer activity (Fig. 5, E and F) relative to that observed in cells treated with vehicle (DMSO). These results strongly suggest that Akt-activated p300 is involved in increased expression of miR-192 in MCs in response to TGF-β.
Figure 5. TGF-β induced miR-192 expression is dependent on acetyltransferase activity and Akt activity.
(A to C) Effects of DNp300 on TGF-β induced miR-192 expression (A) and miR-192 promoter activity (Full-length and SacI constructs) (B and C) in MCs. MCs were transfected with pCMV or pCMV-p300DN (DNp300) and treated without (CTR) or with TGF-β for 24 hours. Data are means ± SEM from 3 experiments. (D to F) Effects of MK-2206 on TGF-β-induced miR-192 expression (D) and on miR-192 promoter activity (Full-length and SacI constructs) (E and F) in MCs. MCs were pre-treated with MK-2206 or vehicle DMSO and then treated with TGF-β. Data are means ± SEM from four experiments. (G) Effects of other histone acetyltransferases (CBP and PCAF) compared with p300 on miR-192 expression. MCs were transfected with DNp300, DNCBP, or DNPCAF, and treated with TGF-β (10 ng/ml) for 24 hours. Data are means ± SEM from three experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis.
To further investigate the contribution of other histone acetyltransferases, we assessed the expression of miR-192 in MCs transfected with dominant negative cyclic AMP response element binding protein (CREB)-binding protein (CBP) (DNCBP) or dominant negative p300/CBP-associated factor(PCAF) (DNPCAF), compared with cells transfected with DNp300 (Fig. 5G). Similar to DNp300 (also in Fig. 5A), DNCBP inhibited TGF-β-induced miR-192 expression, whereas DNPCAF did not have a significant effect, suggesting that CBP (but not PCAF) may have a similar function as p300 in promoting miR-192 expression in response to TGF-β in MCs.
DNp300 and DNCBP, but not DNPCAF, inhibit histone acetylation at the Smad and Ets-1 sites in the miR-192 upstream region
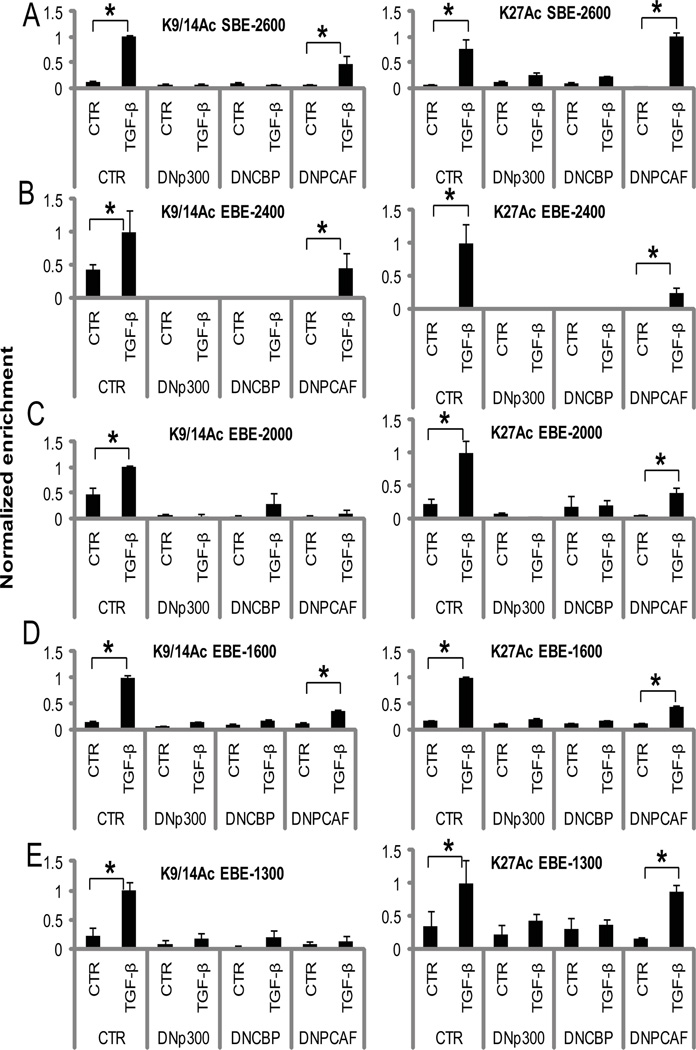
In order to further delineate causal relationships between chromatin modifying enzymes and the chromatin changes at the SBE and EBE sites (Fig. 4A), we performed ChIP assays for acetylated H3K9, H3K14, and H3K27 in MCs transfected with DNp300, DNCBP, or DNPCAF with or without TGF-β. DNp300 and DNCBP similarly decreased TGF-β-induced H3K9, H3K14, and H3K27 acetylation at the Smad and all Ets-1 binding sites (Fig. 6, A to E), although DNCBP had slightly less inhibitory effects on H3K9 and H3K14 acetylation at EBE-2000 (Fig. 6C). However, DNPCAF had either a small or no effect at most of these sites, with only some inhibition of H3K9 and H3K14 acetylation at EBE-2000 and EBE-1300 (Fig. 6, A to E). These results correlate with the trend of miR-192 expression in MCs transfected with DNp300, DNCBP, or DNPCAF (Fig. 5G), and suggest that p300 and CBP are the key histone acetyltransferases involved in H3K9, H3K14, and H3K27 acetylation at the upstream region of miR-192, and that H3K9 and H3K14 acetylation at all the sites might be necessary for maximum expression of miR-192.
Fig. 6. DNp300 and DNCBP, but not DNPCAF inhibit chromatin histone acetylation at the upstream region of miR-192 in MCs treated with TGF-β.
(A to E) Cross-linked chromatin samples from DNp300, DNCBP, or DNPCAF-transfected MCs treated with or without (CTR) 10 ng/ml TGF-β for 24 hours were immunoprecipitated with indicated antibodies: acetylated histone H3K9 and H3K14 (H3K9/14Ac) and H3K27 (H3K27Ac). ChIP-enriched DNA was amplified by qPCR using primers spanning each site: SBE-2600 (A), EBE-2400 (B), EBE-2000 (C), EBE-1600 (D), and EBE-1300 (E). Data are means ± SEM from three experiments. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis.
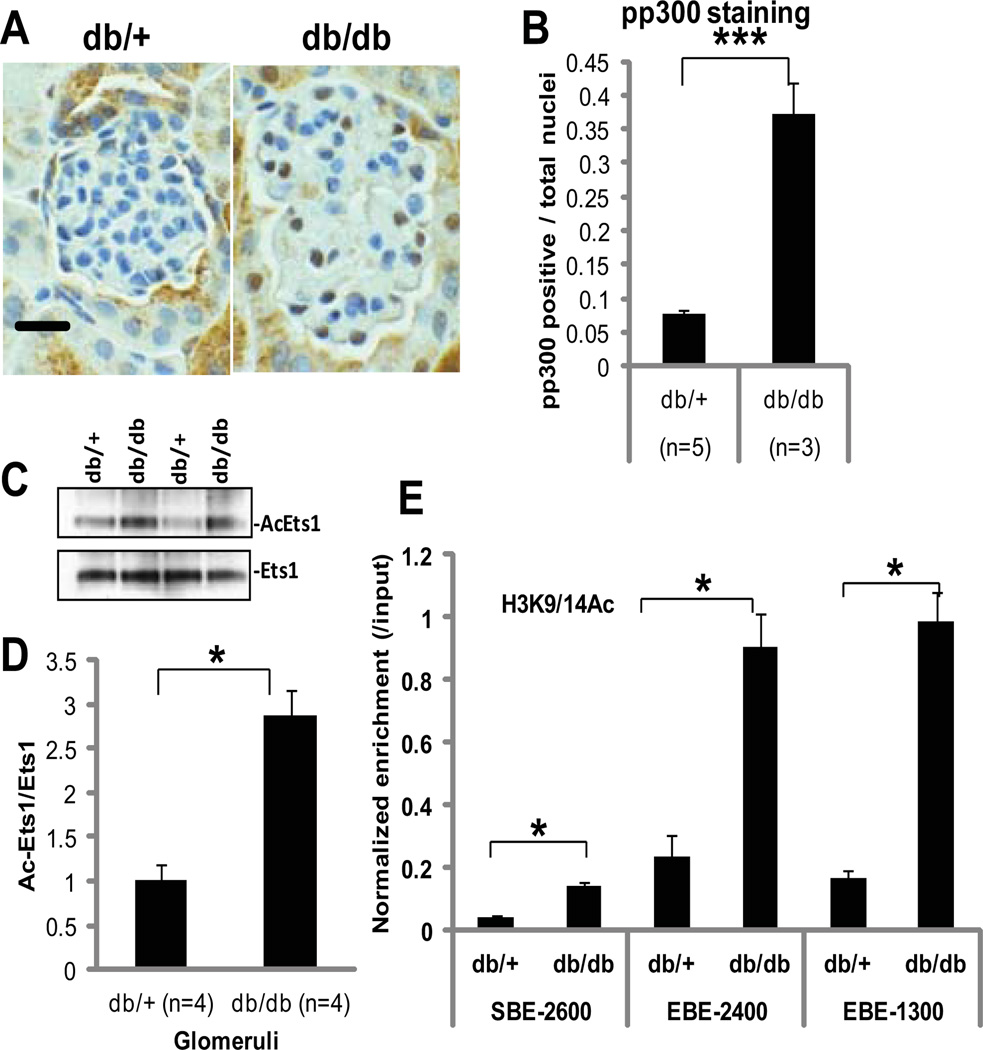
The phosphorylation of p300 and acetylation of Ets-1 and the miR-192 promoter are augmented in glomeruli from diabetic db/db mice
The amount of phosphorylated p300 was increased in vivo in the glomeruli from type 2 diabetic mice (db/db) compared with that in control mice (db/+) (Figure 7, A and B). Ets-1 acetylation in the glomeruli from db/db mice was also significantly higher than those from control db/+ mice (Fig. 7, C and D). Furthermore, histone H3K9 and H3K14 acetylation at the Smad and Ets-1 sites in the promoter region of miR-192 was increased in glomeruli of db/db mice compared with db/+ mice (Fig. 7E). Together, these results provide in vivo relevance and suggest that diabetic conditions can also activate Akt to phosphorylate p300, which in turn acetylates Ets-1 and histone H3 in the glomeruli of diabetic mice.
Fig. 7. Increased p300 phosphorylation, Ets-1 acetylation, and histone acetylation in glomeruli from diabetic mice.
(A) Immunohistochemical staining of phosphorylated p300 in glomeruli from a genetic control (non-diabetic) db/+ mouse compared with a diabetic db/db mouse. Images are representative of sections from 5 db/+ and 3 db/db mice. Scale bar, 20µm. (B) Ratio of the number of phospho-p300-containing nuclei to that of total nuclei. Data are means + SD from glomeruli from five db/+ mice and glomeruli from three db/db mice. ***P < 0.001 by Student’s t-tests. (C and D) Acetylation of Ets-1 in glomeruli from diabetic db/db mice compared to control db/+ mice. Data are means + SD from glomeruli from four db/+ mice and four db/db mice. *P < 0.05 by Student’s t-tests. (E) ChIP-qPCR detecting the acetylation of histone H3K9 and H3K14 (H3K9/14Ac) at Smad binding site (SBE-2600) and Ets-1 binding sites (EBE-2400 and EBE-1300) in the upstream region of miR-192 in glomeruli from db/db mice compared to db/+ mice. Data are means + SD from glomeruli from four db/+ mice and four db/db mice. *P < 0.05 by one-way ANOVA followed by Tukey’s post-hoc test analysis.
Discussion
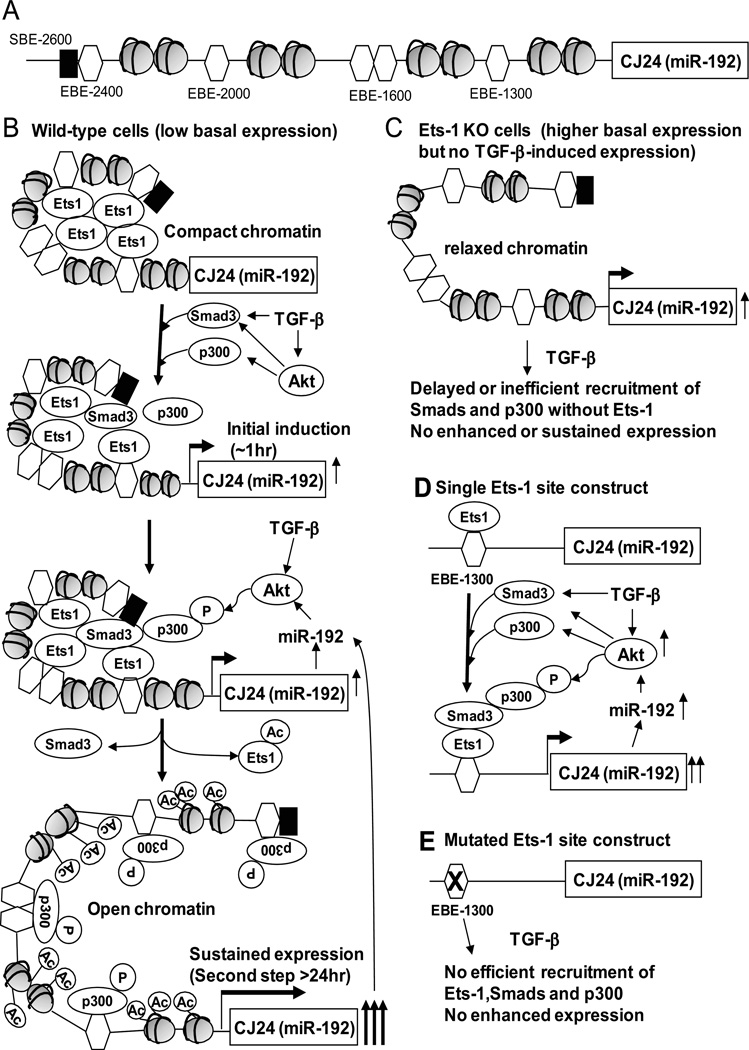
The expression of miR-192 is increased by TGF-β in several human and mouse renal cell types, such as mesangial, tubular, and endothelial cells, and podocytes (2–11), suggesting that the regulation of miR-192 in these cells under diabetic conditions (high glucose or increased activity of TGF-β) may be conserved. In some of these studies, the regulation of miR-192 by TGF-β was shown to be mediated through Smad3 at putative Smad binding sites in the miR-192 upstream region (11). This mechanism is responsible for the initial induction of miR-192 expression by TGF-β (Fig. 8). However, in the current study, we also found a sustained expression of miR-192 (even up to 72 hours) in response to TGF-β in MCs (Fig. 1A), suggesting that other non-Smad dependent mechanisms may also be involved. Using multiple approaches, we uncovered a new role for Ets-1 and epigenetic chromatin remodeling by enhancer histone acetylation in promoting sustained expression of miR-192. We identified a Smad site (SBE-2600) and Ets-1 binding site (EBE-1300) in the miR-192 upstream region important for the TGF-β induced transcriptional regulation of miR-192 in MCs (Fig. 3). Smad2 and Smad3 were recruited to SBE-2600 at 1 hour and dissociated over 6 to 24 hours (Fig. 4), suggesting that the initial, quick induction of miR-192 may be mediated by Smad. All Ets-1 sites, and even the SBE-2600 Smad site, in the miR-192 upstream region were occupied by Ets-1 in the basal state, but TGF-β induced the dissociation of Ets-1 from these sites (Fig. 4). Basal miR-192 expression was increased in Ets-1–deficient or knockdown MCs (Fig. 2), suggesting that Ets-1 is a negative regulator of miR-192 expression. Further, the presence of Ets-1 at the upstream region seems to also be required for the recruitment and activation of transcriptional co-activators such as p300, the subsequent acetylation of Ets-1 and upstream histone regions, and as a result the dissociation of Ets-1 in response to TGF-β, thereby promoting the sustained phase of miR-192 expression (Fig. 8, A and B).
Fig. 8. Proposed model for the step-wise regulation of miR-192 in MCs treated with TGF-β.
(A)Schematic structure of the upstream region of miR-192. (B to E) A proposed model for the regulation of miR-192 in response to TGF-β. (B) In the normal state, unacetylated Ets-1 at the Ets-1 sites in the miR-192 upstream region maintains a compact state of chromatin and represses miR-192 expression. Initially TGF-β induces activation of Smad3 to increase miR-192 expression at about 1 hour. The increased expression of miR-192 activates Akt which phosphorylates p300 that acetylates Ets-1 and dissociates Ets-1 from the upstream region. Phosphorylated p300 also acetylates H3K9 and H3K14 to relax the chromatin structure at the miR-192 upstream region, and this leads to sustained miR-192 expression. (C) In Ets-1–deficient cells, chromatin structure of upstream region of miR-192 is relatively relaxed which results in higher basal expression of miR-192. Since Ets-1-deficiency leads to inefficient recruitment of Smads and p300, miR-192 induction is not enhanced even after TGF-β treatment. (D) Based on our luciferase reporter assays, one Ets-1 element cannot repress miR-192 expression. However, Ets-1 binding to this Ets-1 element helps recruit Smads and p300 in response to TGF-β and thus increases miR-192 expression. (E) The reporter construct with a mutation in the Ets-1 element cannot recruit Smads and p300 even after TGF-β treatment because Ets-1 cannot be recruited to the upstream region of miR-192.
On the other hand, because the upstream region of miR-192 may be relatively relaxed and accessible without the repressive effects of Ets-1, the basal expression was higher in Ets-1-deficient cells than that seen in wild-type cells (Fig. 2 and Fig. 8C). However, due to the lack of Ets-1 in Ets-1– deficient cells, Smads or p300 may not be efficiently recruited to the upstream region after TGF-β treatment even with active Akt and phosphorylated p300 (Fig.1, D and H), and hence the sustained expression of miR-192 may not be observed in Ets-1–deficient cells (Fig. 8C). Some reporter constructs with one Ets-1 binding element (SacI and EcoRI in Fig. 3) responded to TGF-β treatment, but this was lost in the mutant EBE-1300. A construct with three Ets-1 binding elements (NheI in Fig. 3) did not respond to TGF-β treatment (Fig. 3B). Although plasmid reporters are artificial and do not have the true cellular chromatin environment, these results demonstrate that multiple Ets-1 elements together can be repressive, whereas a single Ets-1 element is enhancive. This suggests that Ets-1 interaction with multiple upstream Ets-1 elements may promote chromatin condensation (Fig. 8B), whereas a single Ets-1 site is unable to make repressive structures and even enhances transcription by recruiting Smads and p300 which acetylates and dissociates Ets-1 (Fig. 8D). This may be the reason why the mutant construct of EBE-1300 (Ets-1 mutant in Fig. 3) had similar basal activity but lost the TGF-β response (Fig. 8E), compared with the wild-type construct (EcoRI in Fig. 3 and Fig. 8D).
A significant increase in Ets-1 acetylation was observed in MCs treated with TGF-β, accompanied by increased Akt and p300 phosphorylation (Fig. 1, D and H). TGF-β activates Akt kinase in MCs through miRNA circuits from miR-192 to miR-216a and miR-217 cluster targeting PTEN (15) and to miR-200b and miR-200c (13) targeting Fog2, an inhibitor of phosphatidylinositol-3-kinase which activates Akt (41). Several other reports also show that Akt is activated by TGF-β in MCs and other cell types (15, 17, 42–46). Akt phosphorylates and activates p300 (47). In this study, we detected a significant increase in p300 phosphorylation at the Akt target site (Ser1834) after TGF-β treatment with a concomitant increase of Akt activation (phosphorylation at Ser473) (Fig. 1). The Akt-specific inhibitor MK-2206 also inhibited Akt and p300 phosphorylation and Ets-1 acetylation as well as the increased expression of miR-192 even in MCs treated with TGF-β (Fig. 1), strongly suggesting that Akt mediated p300 phosphorylation and Ets-1 acetylation in MCs are involved in the increased expression of miR-192. Acetylated Ets-1 (by p300) dissociates from the COL1A2 promoter in human dermal fibroblast cells treated with TGF-β (31), and TGF-β decreases Ets-1 DNA binding activity and increases type I collagen in MCs (32). Likewise our data suggests that acetylated Ets-1 dissociates from the miR-192 upstream region, an event that contributes to sustained miR-192 expression in MCs treated with TGF-β (Fig. 8B).
We also detected increased p300 occupancy, as well as increased H3K9, H3K14 and H3K27 acetylation at the miR-192 upstream region in response to TGF-β (Fig. 4), suggesting that p300 may be recruited to the Ets-1 site by TGF-β, and that Akt-activated p300 acetylates Ets-1 as well as histone H3. The subsequent release of Ets-1 led to sustained expression of miR-192 (Fig. 8B).The Akt inhibitor MK-2206 decreased p300 occupancy and acetylation of H3K9 and H3K14 but not of H3K27 at Smad and Ets-1 sites (Fig. 4), suggesting that p300 preferentially acetylates H3K9 and H3K14 when activated by Akt. A parallel inhibition in TGF-β-induced miR-192 expression by MK-2206 (Fig. 1A) suggests that acetylation of H3K9 and H3K14 (but not H3K27) was critical for TGF-β-induced miR-192 expression. DNp300 and DNCBP (but not DNPCAF) inhibited TGF-β–induced increase of miR-192 and histone acetylation at the miR-192 upstream region, confirming that acetylation of Ets-1 and histone H3 is essential (Fig. 5G and Fig. 6). Based on our data, we conclude that Akt, activated by TGF-β, phosphorylates p300 which preferentially acetylates histone H3K9 and H3K14 as well as Ets-1, enabling nucleosome relaxation and thereby allowing prolonged expression of miR-192 (Fig. 8B), although we cannot totally exclude CBP as a mediator in this signaling.
Phosphorylation of p300 and acetylation of Ets-1 and promoter histones H3K9 and H3K14 were increased in glomeruli from type 2 diabetic mice (db/db) compared to control mice (db/+) (Fig. 7). Enhanced histone acetylation, Akt activation, and phosphorylation of Akt downstream molecules, such as mTOR and FoxO3a, have been observed in the kidneys of animal models of diabetic nephropathy (15, 42, 46, 48, 49), suggesting that Akt activation and increased phosphorylation of Akt downstream targets – including p300 – may also be functionally active in the diabetic kidney in vivo. The signaling mechanism (involving TGF-β, Akt, p300, Ets-1, and miR-192) indicated by our in vitro experiments were supported by in vivo analysis of kidney cells in db/db mice, a model of diabetic nephropathy (50).
Because genome-wide ChIP on microarray analysis of Smad2 and Smad3 identified numerous genomic regions which included Ets-1 binding sites (51) and our data suggests that Ets-1 is involved in TGF-β signaling, Smad proteins may interact with Ets-1 and regulate downstream gene expression (including miR-192 and collagens) through Ets-1 binding sites (apart from SBEs) in response to TGF-β. In fact, we detected Smad2 and Smad3 at all potential Ets-1 binding sites as well as a Smad site (SBE-2600) in the miR-192 upstream region, and also Ets-1 at the same Smad site (Fig. 4). Therefore, the Smad binding site-independent TGF-β response (but still Smad-dependent) may be mediated by Ets-1 to enhance miR-192 expression. Because TGF-β induces PAI-1 expression through association of Smad3 with CBP as well as Akt-activated p300 in rat MCs (52), TGF-β signaling may generally involve these histone modifying proteins (p300 and CBP) that are activated by Akt to enhance chromatin remodeling and target gene expression.
A recent study showed that co-occupation of Smad3 with cell-type specific master transcription factors, such as Oct4, Myod1 or PU.1, determines cell-type specific gene response to TGF-β (53). TGF-β leads to a decrease in miR-192 expression in some other cell types (22–24), which also suggests a cell-type specific and context dependent TGF-β response (2, 54, 55). Because the expression of miR-192 is stimulated by Smad3 and p53 (11, 19–21), p53 and Smad may affect the miR-192 response to TGF-β (11, 56, 57). Our observations showing initial induction of miR-192 expression over 6 hours but loss of its sustained expression in Ets-1–deficient or knockdown MCs (Fig. 2) also implicate Ets-1 as an additional important cell-type-specific decision-maker of TGF-β response.
Taken together, along with its role in kidney fibrosis and hypertrophy(2–4, 32, 42, 48), Ets-1 is also a strong candidate target for controlling the expression of miRNAs and downstream genes involved in fibrosis and renal cell hypertrophy related to diabetic nephropathy. From a translational standpoint, Akt inhibitors (such as MK-2206 or related derivatives) might be evaluated for the treatment of diabetic nephropathy by inhibiting glomerular fibrosis and hypertrophy promoted by key miRNAs such as miR-192. Other possibilities may include approaches for overexpressing Ets-1 or certain histone deacetylases. Overall, these findings provide new insights into the regulation of miRNAs through signaling-mediated changes in transcription factor activity and in epigenetic histone acetylation in MCs under normal and disease states.
Materials and Methods
Mice
All animal studies were conducted according to a protocol approved by the Institutional Animal Care and Use Committee of the Beckman Research Institute of City of Hope. Obese type 2 diabetic db/db mice and genetic control db/+ mice (10–12 weeks old) were obtained from Jackson Laboratories. Ets-1–deficient mice have been described earlier (39). Glomeruli were sieved from renal cortical tissue as described (3).
Cell culture and Materials
Primary mouse MCs from wild type and from Ets-1–deficient mice (39) were isolated and cultured as described (3). Recombinant human TGF-β1 and pan-specific TGF-β1 antibody (MAB1835) were from R&D Systems, Inc. MK-2206 was purchased from Selleck Chemicals, dissolved in DMSO and used at a final concentration of 1µM. To knockdown Ets-1 in MCs, Ets-1 ON-TARGETplus SMART pool siRNA (Thermo Scientific) was transfected using Amaxa Nucleofector and Basic Nucleofector Kit (Lonza) as described (15). Target sequences are 5’-CAGCUACGGUAUCGAGCAU-3’, 5’-UCAAGUAUGAGAACGACUA-3’, 5’-GCUUAGAGAUGUAGCGAUG-3’ and 5’-CCUGUUACACCUCGGAUUA-3’. ON-TARGETplus Non-targeting Pool (Thermo Scientific) was used as control.
Real time quantitative PCR (qPCR)
Real-time qPCRs were performed using SYBR Green PCR Master Mix and 7300 or 7500 Realtime PCR System (Applied Biosystems, Foster City, CA USA), as reported (15). miRNA expression levels were also analyzed by the miScript System (Qiagen). PCR primer sequences are: mouse Ets-1 5’-TAGTTGTGACCGCCTCACCC-3’; 5’-TCGGCCCACTTCCTGTGTAG-3’; mouse CJ24 5’-GCTCTGTTTAGAAGGGAAGGAGGAA-3’; 5’-ATTCATCCAAGGCCCACACC-3’.
Western blot analyses and Immunoprecipitation
Western blotting was performed as described (3). Antibodies against Ets-1 (Santa Cruz Biotechnology, Inc), phospho-Ets-1(MBL International Corp.), acetyl lysine, Akt, phospho-Akt, and beta-actin (Cell Signaling Technology), p300 (Millipore), and phospho-p300 (Ser1834) (Thermo Scientific) were used at 1:1000 dilution. To detect acetylated Ets-1, cell lysates (in SDS Lysis Buffer, Millipore) were precleared with Protein-A beads for 1 hour, immunoprecipitated with Ets-1 antibody overnight, incubated with Protein-A beads for 1 hour, washed 4 times with TNE buffer, 10 mM Tris-HCl (pH 7.4), 1% Nonidet P-40, 1 mM EDTA, and eluted with SDS sample buffer. Immunoprecipitates were analyzed by Western blotting using Acetyl lysine antibody (AcEts1), followed by Western blotting with Ets-1 antibody to confirm that the bands represent Ets-1 (total Ets1).
Plasmids and luciferase reporters
The mouse CJ241444 (an EST present upstream of miR-192) upstream region was amplified using primers, miR192-2.6F 5’-actAGATCTCTGAGCGTGTGGGTCAGT-3’ miR192-0.8R 5’-tgaaagcttGATAGGTGGGCAGGGACTCTCA-3’ and cloned into BglII and HindIII sites of pGL3P (Promega). Lower case letters indicate extra sequences for cloning of PCR fragments. The Ets-1 site mutant was made by site directed mutagenesis as described (15) using primers, 5’-GCTGTGGccAAACCccAAACCATCTAGTCT-3’; 5’-AGACTAGATGGTTTggGGTTTggCCACAGC-3’. Lower case letters indicate sequence substitution for mutagenesis of Ets-1 binding sites (GGAA to CCAA). Dominant negative p300 and PCAF expression plasmids (pCMV-p300DN/PCAF) were from Dr. Michael Stallcup (University of Southern California, LA) (58). Dominant negative CBP plasmid was from Dr. Christopher Glass (University of California, San Diego) (59).
Luciferase assays
MCs were transfected with plasmids using a Nucleofector and Basic Nucleofector Kit (Lonza). After 48 h, Luciferase activities were measured as described (15). Briefly, Fire-fly luciferase reporter plasmids (pGL3P with miR-192 upstream region) were cotransfected with internal control vector, pRL-TK (Renilla luciferase, Promega) and the ratio of Fire-fly/Renilla was calculated. The values were normalized to control conditions (normalized luc activity). For TGF-β treatment, transfected cells were treated with 10 ng/ml TGF-β for 6~24hr.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as reported (15). Briefly, MCs were treated with10 ng/ml TGF-β and then cross-linked with formaldehyde. Isolated glomeruli from mice were cross-linked with formaldehyde (final concentration 1% in PBS for 20 min at room temperature), and then quenched with 125 mM glycine (5 min at RT). The cross-linked chromatin was sheared and immunoprecipitated using antibodies against Ets-1 (Santa Cruz Biotechnology), Smad2 and Smad3 (Cell Signaling), p300 (Millipore), H3K9Ac and H3K14Ac (Millipore), or H3K27Ac (Abcam). To detect enrichment of acetylated Ets-1, chromatin immunoprecipitates obtained with Ets-1 antibody were reimmunoprecipitated with acetyl lysine antibody. ChIP-enriched DNA was purified and used as template for real time qPCR and genomic region spanning the Ets-1 sites in the upstream region of miR-192 were amplified using primers, SBE −2800, 5’-CACTGTCCCAACATGCTCAC-3’, 5’-GGTAGCAGGGAGAAGCAATG-3’; SBE-2600, 5’-AGCCAAGTGTCTCTCCGTGT-3’, 5’-GGTGGCAGGAGGTGTGTACT-3’; EBE-2400, 5’-actAGATCTCTGAGCGTGTGGGTCAGT-3’, 5’-ATGACAGGCACCAGGATGGTAAGTAG-3’; EBE-2000, 5’-GCTGGGACAAAGCTGTGTTGTCCAGA-3’, 5’-CCAGGGATGCCCAACAAGTGATCC-3’; EBE-1600, 5’-AGGGTCAGTATTGATGGCAGTGGGCT-3’, 5’-GGAGGACAGCCTGGGAGCTCTTA-3’; EBE-1300, 5’-cagaGAATTCTTAAGCAGCCATGTCTG-3’, 5’-gcatTCTAGAGTATGCTTGGATCATGC-3’. ChIP-QPCR results (Normalized enrichment) were calculated by 2-delta delta Ct method and normalized to input DNA (purified from 5% of the same crosslinked chromatin).
Data Analysis
Statistical analyses were performed by Student’s t-tests to compare two groups (planned comparison) or one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test analysis to compare multiple groups. P<0.05 was considered as statistically significant.
Supplementary Material
Acknowledgements
We are grateful to all members of the Natarajan laboratory for helpful discussions. Funding: This work was supported by grants from the National Institutes of Health, R01 DK081705, R01 DK058191 (to RN) and the American Heart Association, Scientist Development Grant (to YZ).
Footnotes
Author contributions: M.K., V.D., M.W., J.T.P, S.D., S.K., A.M., S.P., H.Y. and L.L. performed and analyzed experiments. Y.Z. and P.O. provided Ets-1-deficient mice. M.K. and R.N. designed experiments and wrote the paper.
Competing interests: The authors have no competing interests.
References and Notes
- 1.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 2011;7:286. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 2.Kato M, Park JT, Natarajan R. MicroRNAs and the glomerulus. Experimental cell research. 2012;318:993. doi: 10.1016/j.yexcr.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A. 2007;104:3432. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting MicroRNA-192 Ameliorates Renal Fibrosis in Diabetic Nephropathy. J Am Soc Nephrol. 2012;23:458. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XX, Jiang T, Shen Y, Caldas Y, Miyazaki-Anzai S, Santamaria H, Urbanek C, Solis N, Scherzer P, Lewis L, Gonzalez FJ, Adorini L, Pruzanski M, Kopp JB, Verlander JW, Levi M. Diabetic Nephropathy is Accelerated by Farnesoid X Receptor Deficiency and Inhibited by Farnesoid X Receptor Activation in a Type 1 Diabetes Model. Diabetes. 2010;59:2916. doi: 10.2337/db10-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is upregulated and can lead to increased fibronectin production in diabetic nephropathy. Faseb J. 2008;22:4126. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of miRNAs in patients with hypertensive nephrosclerosis. American journal of hypertension. 2010;23:78. doi: 10.1038/ajh.2009.208. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Kwan BC, Lai FM, Choi PC, Chow KM, Li PK, Szeto CC. Intrarenal expression of microRNAs in patients with IgA nephropathy. Laboratory investigation; a journal of technical methods and pathology. 2010;90:98. doi: 10.1038/labinvest.2009.118. [DOI] [PubMed] [Google Scholar]
- 10.Cai X, Xia Z, Zhang C, Luo Y, Gao Y, Fan Z, Liu M, Zhang Y. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatric nephrology (Berlin, Germany) 2013 doi: 10.1007/s00467-013-2434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol. 2010;21:1317. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Casado M, Vaulont S, Sharma K. Role of upstream stimulatory factors in regulation of renal transforming growth factor-beta1. Diabetes. 2005;54:1976. doi: 10.2337/diabetes.54.7.1976. [DOI] [PubMed] [Google Scholar]
- 13.Kato M, Arce L, Wang M, Putta S, Lanting L, Natarajan R. A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011;80:358. doi: 10.1038/ki.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 15.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, Rossi JJ, Natarajan R. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol. 2009;11:881. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, Lanting L, Todorov I, Rossi JJ, Natarajan R. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem. 2010;285:34004. doi: 10.1074/jbc.M110.165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun L, Zhang D, Liu F, Xiang X, Ling G, Xiao L, Liu Y, Zhu X, Zhan M, Yang Y, Kondeti VK, Kanwar YS. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by downregulating miR-192. The Journal of pathology. 2011;225:364. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer cell. 2010;18:367. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive MicroRNAs 192 and 215 Are Capable of Inducing Cell Cycle Arrest. Cancer research. 2008;68:10094. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer research. 2008;68:10105. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 22.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 Promotes Fibrogenesis in Diabetic Nephropathy. J Am Soc Nephrol. 2010;21:438. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, Saleem M, Goodall GJ, Twigg SM, Cooper ME, Kantharidis P. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins RH, Martin J, Phillips AO, Bowen T, Fraser DJ. Transforming growth factor beta1 represses proximal tubular cell microRNA-192 expression through decreased hepatocyte nuclear factor DNA binding. The Biochemical journal. 2012;443:407. doi: 10.1042/BJ20111861. [DOI] [PubMed] [Google Scholar]
- 26.Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. Rna. 2008;14:1433. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson DK, McWilliams-Smith MJ, Nunn MF, Duesberg PH, O’Brien SJ, Papas TS. The ets sequence from the transforming gene of avian erythroblastosis virus, E26, has unique domains on human chromosomes 11 and 21: both loci are transcriptionally active. Proc Natl Acad Sci U S A. 1985;82:7294. doi: 10.1073/pnas.82.21.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 30.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. The EMBO journal. 2010;29:2147. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czuwara-Ladykowska J, Sementchenko VI, Watson DK, Trojanowska M. Ets1 is an effector of the transforming growth factor beta (TGF-beta ) signaling pathway and an antagonist of the profibrotic effects of TGF-beta. J Biol Chem. 2002;277:20399. doi: 10.1074/jbc.M200206200. [DOI] [PubMed] [Google Scholar]
- 32.Mizui M, Isaka Y, Takabatake Y, Sato Y, Kawachi H, Shimizu F, Takahara S, Ito T, Imai E. Transcription factor Ets-1 is essential for mesangial matrix remodeling. Kidney Int. 2006;70:298. doi: 10.1038/sj.ki.5001541. [DOI] [PubMed] [Google Scholar]
- 33.Razzaque MS, Naito T, Taguchi T. Proto-oncogene Ets-1 and the kidney. Nephron. 2001;89:1. doi: 10.1159/000046034. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 35.Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan BS, Kotani H. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Molecular cancer therapeutics. 2010;9:1956. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 36.Tan S, Ng Y, James DE. Next-generation Akt inhibitors provide greater specificity: effects on glucose metabolism in adipocytes. The Biochemical journal. 2011;435:539. doi: 10.1042/BJ20110040. [DOI] [PubMed] [Google Scholar]
- 37.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Aktmediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou BP, Hu MC, Miller SA, Yu Z, Xia W, Lin SY, Hung MC. HER-2/neu blocks tumor necrosis factor-induced apoptosis via the Akt/NF-kappaB pathway. J Biol Chem. 2000;275:8027. doi: 10.1074/jbc.275.11.8027. [DOI] [PubMed] [Google Scholar]
- 39.Zhan Y, Brown C, Maynard E, Anshelevich A, Ni W, Ho IC, Oettgen P. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. The Journal of clinical investigation. 2005;115:2508. doi: 10.1172/JCI24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annual review of biochemistry. 2001;70:81. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 41.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 42.Mahimainathan L, Das F, Venkatesan B, Choudhury GG. Mesangial cell hypertrophy by high glucose is mediated by downregulation of the tumor suppressor PTEN. Diabetes. 2006;55:2115. doi: 10.2337/db05-1326. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh Choudhury G, Abboud HE. Tyrosine phosphorylation-dependent PI 3 kinase/Akt signal transduction regulates TGFbeta-induced fibronectin expression in mesangial cells. Cellular signalling. 2004;16:31. doi: 10.1016/s0898-6568(03)00094-9. [DOI] [PubMed] [Google Scholar]
- 44.Runyan CE, Schnaper HW, Poncelet AC. The phosphatidylinositol 3-kinase/Akt pathway enhances Smad3-stimulated mesangial cell collagen I expression in response to transforming growth factor-beta1. J Biol Chem. 2004;279:2632. doi: 10.1074/jbc.M310412200. [DOI] [PubMed] [Google Scholar]
- 45.Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem. 2005;280:10870. doi: 10.1074/jbc.M413223200. [DOI] [PubMed] [Google Scholar]
- 46.Kato M, Yuan H, Xu ZG, Lanting L, Li SL, Wang M, Hu MC, Reddy MA, Natarajan R. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol. 2006;17:3325. doi: 10.1681/ASN.2006070754. [DOI] [PubMed] [Google Scholar]
- 47.Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol Cell Biol. 2005;25:6592. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagai K, Matsubara T, Mima A, Sumi E, Kanamori H, Iehara N, Fukatsu A, Yanagita M, Nakano T, Ishimoto Y, Kita T, Doi T, Arai H. Gas6 induces Akt/mTOR-mediated mesangial hypertrophy in diabetic nephropathy. Kidney Int. 2005;68:552. doi: 10.1111/j.1523-1755.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 49.Xin X, Chen S, Khan ZA, Chakrabarti S. Akt activation and augmented fibronectin production in hyperhexosemia. American journal of physiology. 2007;293:E1036. doi: 10.1152/ajpendo.00271.2007. [DOI] [PubMed] [Google Scholar]
- 50.Sharma K, McCue P, Dunn SR. Diabetic kidney disease in the db/db mouse. Am J Physiol Renal Physiol. 2003;284:F1138. doi: 10.1152/ajprenal.00315.2002. [DOI] [PubMed] [Google Scholar]
- 51.Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol. 2009;29:172. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das F, Ghosh-Choudhury N, Venkatesan B, Li X, Mahimainathan L, Choudhury GG. Akt kinase targets association of CBP with SMAD 3 to regulate TGFbeta-induced expression of plasminogen activator inhibitor-1. J Cell Physiol. 2008;214:513. doi: 10.1002/jcp.21236. [DOI] [PubMed] [Google Scholar]
- 53.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, Dekoter RP, Young RA. Master Transcription Factors Determine Cell-Type-Specific Responses to TGF-beta Signaling. Cell. 2011;147:565. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikushima H, Miyazono K. Cellular context-dependent "colors" of transforming growth factor-beta signaling. Cancer Sci. 2010;101:306. doi: 10.1111/j.1349-7006.2009.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasinath BS, Feliers D. The complex world of kidney microRNAs. Kidney Int. 2011;80:334. doi: 10.1038/ki.2011.165. [DOI] [PubMed] [Google Scholar]
- 56.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 57.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelialmesenchymal transition through microRNAs targeting ZEB1 and ZEB2. The Journal of experimental medicine. 2011;208:875. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurokawa R, Kalafus D, Ogliastro MH, Kioussi C, Xu L, Torchia J, Rosenfeld MG, Glass CK. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.