Abstract
Marijuana is the most widely abused illegal drug and its spectrum of effects suggests that several receptors are responsible for the activity. Two cannabinoid receptor subtypes, CB1 and CB2, have been identified, but the complex pharmacological properties of exogenous cannabinoids and endocannabinoids are not fully explained by their signaling. The orphan receptor GPR55 binds a subset of CB1/CB2 ligands and has been proposed as a cannabinoid receptor. This designation, however, is controversial as a result of recent studies where lysophosphatidylinositol (LPI) is identified as a GPR55 agonist. To define a biological role for GPR55 requires GPR55-selective ligands that have been unavailable. From a β-arrestin, high-throughput, high-content screen of 300,000 compounds run in collaboration with the Molecular Libraries Probe Production Centers Network initiative (PubChem AID1965), we identified potent GPR55 selective agonists. By modeling of the GPR55 activated state, we compared the GPR55 binding conformations of three of the novel agonists obtained from the screen CID1792197, CID1172084 and CID2440433 [PubChem Compound IDs] with that of LPI. Our modeling indicates the molecular shapes and electrostatic potential distributions of these agonists mimic that of LPI; the GPR55 binding site accommodates ligands that have inverted-L or T shapes with long, thin profiles that can fit vertically deep in the receptor binding pocket while their broad head regions occupy a horizontal binding pocket near the GPR55 extracellular loops. Our results will enable the optimization and design of second generation GPR55 ligands and provide a means for distinguishing GPR55 selective ligands from those interacting with cannabinoid receptors.
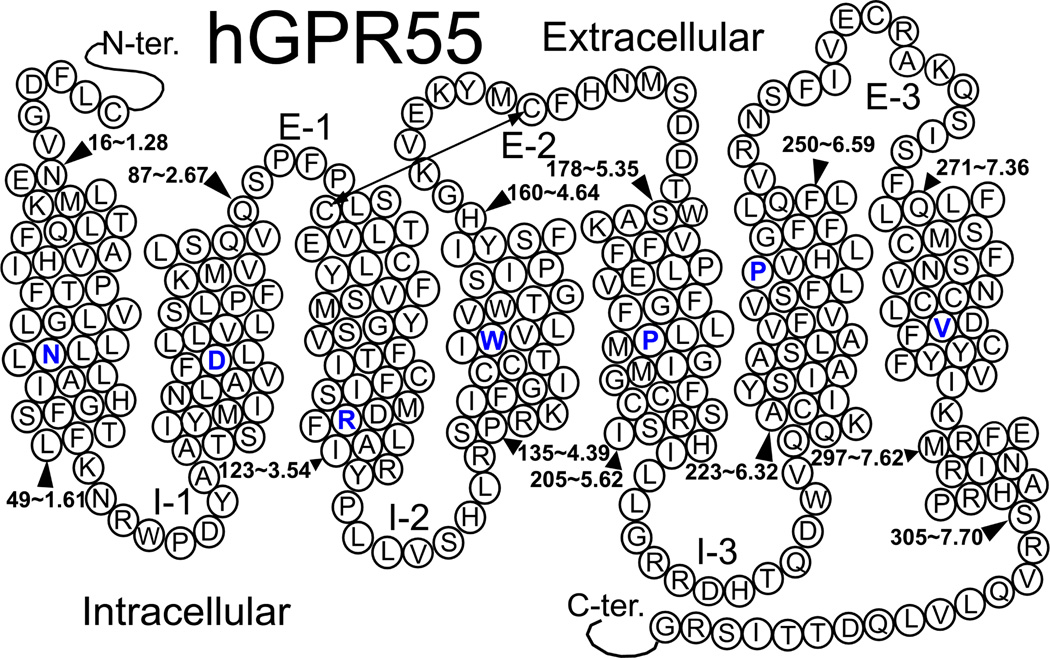
GPR55 is a rhodopsin-like (Class A) G protein-coupled receptor (GPCR), highly expressed in human striatum (1), that has the potential to become an important therapeutic target (Genbank accession # NM-005683; see Figure 1). Characterizations of GPR55(−/−) knock-out mice (2, 3) reveal a role for GPR55 in inflammatory pain, neuropathic pain, and bone development, while other studies indicate that GPR55 activation is pro-carcinogenic(4–6). The GPCR proteins most homologous to GPR55 are GPR35 (27%), P2Y (29%), GPR23 (30%), and CCR4 (23%) (1), but GPR55 also exhibits low amino acid identity to cannabinoid CB1 (13.5%) and CB2 (14.4%) receptors. This homology to cannabinoid receptors and the potential utility of marijuana derivatives in therapy prompted a search for GPR55 ligands among known cannabinoid receptor compounds. Thus, some synthetic and natural cannabinoids were among the first chemicals recognized to bind GPR55 (7).
Figure 1.
Helix net representation of the human GPR55 sequence.
Subsequent studies of GPR55 pharmacology suggested that lysophosphatidylinositol (LPI; 1) compounds are endogenous GPR55 agonists (8), with 2-AGPI, a 2-arachidonoyl containing LPI species, possessing significantly higher potency and maximal efficacy of the LPI species observed to date (9). 2-AGPI may represent the true natural ligand of GPR55. In this manuscript we reference efficacy relative to the commercially available LPI (1). Using a β-arrestin green fluorescent protein biosensor to assess a cohort of CB1/CB2 ligands for GPR55 activity, Kapur et al. (10) confirmed LPI as a GPR55 agonist, while observing that the cannabinoid antagonists AM251 and SR141716A were also GPR55 agonists. These GPR55 ligands possess comparable efficacy in inducing β-arrestin trafficking, and moreover, activate the G-protein dependent signaling of PKCβII. Conversely, the potent synthetic cannabinoid agonist CP55940 acts as a GPR55 antagonist/partial agonist, inhibiting GPR55 internalization, the formation of β-arrestin GPR55 complexes, and the phosphorylation of ERK1/2 (10).
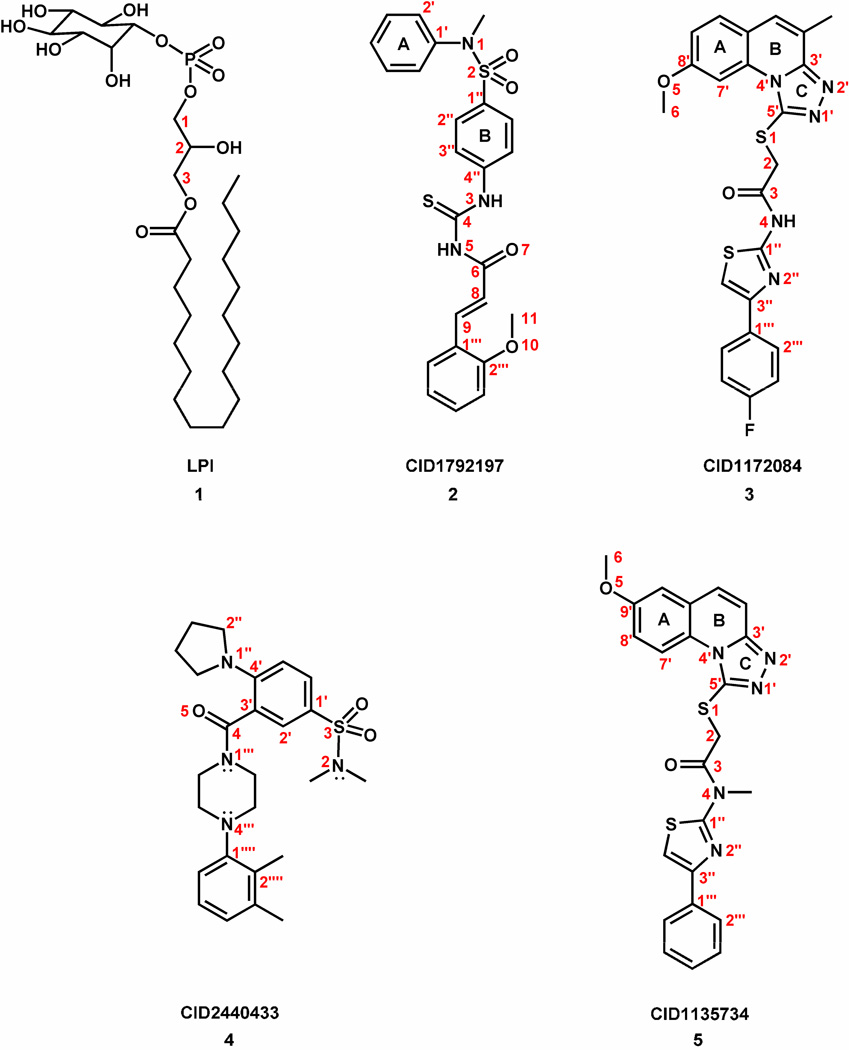
More potent and selective GPR55 ligands have been unavailable predominantly because many of the known GPR55 ligands were identified from cannabinoid receptor/lipid biased compound libraries (11, 12). The discovery and molecular characterization of better GPR55 chemotypes is an important next step towards the design of higher quality GPR55 ligands that can serve either as research tools or as a basis for designing novel drugs. During a collaborative project between our individual laboratories and the Sanford-Burnham screening center of the Molecular Libraries Probe Production Centers Network (MLPCN), we identified a series of GPR55 agonists that belong to novel, unreported GPR55 agonist chemotypes. These were discovered by a high content, high throughput β-arrestin screen (see http://mli.nih.gov/mli/mlp-probes/). In this manuscript we focus on three GPR55 agonists identified in this screen: CID1792197 (2), CID1172084 (ML185, 3), and (CID2440433; ML184, 4) (see Figure 2 for compound drawings).
Figure 2.
The structures of GPR55 agonists (1–4) and control compound (5) are illustrated here.
Only a few Class A GPCRs have been crystallized to date. These include Rhodopsin (Rho) (13–15), the β2-adrenergic receptor (β2-AR) (16–18), β1-adrenergic receptor (β1-AR) (19), adenosine A2A receptor (20) and most recently the CXCR4 (21) and dopamine D3 (22) receptors. These crystal structures reveal a common topology that includes: (1) an extracellular N terminus; (2) seven transmembrane alpha helices (TMHs) arranged to form a closed bundle; (3) loops connecting TMHs that extend intra- and extracellularly; and (4) an intracellular C terminus that begins with a short helical segment (Helix 8) oriented parallel to the membrane surface. Although no crystal structure is available for GPR55, it is possible to build a homology model of GPR55 based on available crystal structures and refine this model based on sequence dictated differences between GPR55 and the template crystal structure. This refined model can then be used to explore the GPR55 ligand binding pocket and key amino acid interactions for GPR55 ligands. We report here the development of a refined homology model of the GPR55 activated state (GPR55 R*) and its subsequent use to explore the interaction between GPR55 and the novel CID agonists (2–4) identified in our screen. Our modeling data indicate that the similarity between the CID compounds and LPI enables them all to be recognized by a single GPR55 binding pocket. The chemical diversity provided by these three lead compounds, CID (2–4), combined with the identification of key receptor interaction sites should provide a basis for the design of more potent and efficacious second generation GPR55 ligands that retain GPR55 selectivity.
METHODS
Chemical Library Screening
Potent GPR55 selective agonists were identified using a β-arrestin (see methods below), high-throughput, high-content screen of 300,000 compounds run in collaboration with the Molecular Libraries Probe Production Centers Network program. For more details about this library of compounds, see http://mli.nih.gov/mli/compound-repository/mlsmr-compounds/. Compounds were screened for agonism (PubChem AIDs 2347) at GPR55, as well as, for both agonism and antagonism at GPR35 (PubChem AIDs 2397, 2338), CB1 (PubChem AIDs 2341, 2339) and CB2 (PubChem AIDs 434928, 434922). The high content imaging assay utilized in each case employs a cell line permanently expressing a beta-arrestin GFP biosensor and an enhanced receptor of interest (i.e., GPR55, GPR35, CB1 or CB2). A full description of each of these assay protocols (according to AID number) is accessible at the PubChem website (http://pubchem.ncbi.nlm.nih.gov/). Potent GPR55 agonists lacking agonism or antagonism at GPR35, CB1 or CB2, were further evaluated for pERK activation and PKCβII translocation (see methods below). A set of novel GPR55 agonist molecular scaffolds were selected from the screen (CID1792197 (2); CID1172084 (3); CID2440433 (4), including a chemically related inactive compound (CID1135734 (5)) and the binding of each compound was explored using a computer model of the GPR55 activated state (see methods below).
Biochemical Studies
β-Arrestin Assessment of GPR55 Activation
U2OS cells permanently expressing HA-GPR55E and βarr2-GFP have previously been described (10). Cells were plated onto 384-well assay plates (Greiner) at 80–85% confluency and maintained at 37 °C in 5% CO2 overnight. Prior to drug application, serum-containing media was removed and replaced with serum-free MEM. Agonist stimulated redistribution of βarr2-GFP was assessed following drug treatment for 75 minutes at 37 °C in 5% CO2. Cells were then fixed with 4% paraformaldehyde for 40 minutes at room temperature followed by two washes with PBS. The nuclear stain, DAPI (Invitrogen) was added and plates were sealed for imaging. Image acquisition was performed on an Opera™ QEHS (PerkinElmer) using the following settings: 20 × 0.45 NA air objective, camera set to 2 by 2 binning for an image size of 688 by 512 pixels, 2 channels acquired sequentially- 488 nm excitation for β-arrestin-GFP and 365 nm excitation for DAPI (details in PubChem AID1965).
Data Analysis
βarr2-GFP aggregates were identified using the Acapella spot detection algorithm (Perkin Elmer). Briefly, the analysis protocol extracts from two dimensional images those pixels that generate objects of interest that fall within a predetermined range of sizes and intensities and that are embedded among widely varying local backgrounds. The primary assay read-out was based on the number of detected fluorescent spots representing pits or vesicles per cell. Concentration-effect curves for agonist-mediated receptor activation were analyzed by nonlinear regression techniques and data were fitted to sigmoidal dose-response curves to obtain EC50 values. Activity values were normalized to the positive controls (LPI (1) as 100% at 10 µM) and negative controls (vehicle only as 0%) for each well plate. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Dunnett’s post-test or two-tailed unpaired Student’s t-test and p values of <0.05 were considered significant.
Western Blot Analysis of ERK Activity
GPR55E-expressing U2OS cells were grown to sub-confluence in 60-mm plates and serum-starved overnight before assay. After drug treatment the cells were disrupted in a lysis buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 10 µM MgCl2, 20mM p-nitrophenyl phosphate, 1mM Na3VO4, 25mM NaF, and a protease inhibitor mixture (1:25, pH 7.5). Lysates were immediately placed on ice for 10 min and then centrifuged at 16,000 χ g for 30 min at 4°C. Supernatants, corresponding to the cytosolic fraction, were collected, and protein concentrations were determined by the Bradford assay (Bio-Rad) using bovine serum albumin as a standard. Cytosolic fractions (20 µg) were separated on a 10% gel by SDS-PAGE followed by immunoblotting (23). Antibodies against double-phosphorylated ERK1/2 (1:5000) were detected using a Fuji imager LAS-1000 (Fujifilm Life Science, Woodbridge, CT). A monoclonal antibody against actin (1:10,000) was used to confirm equal protein loading. Densitometric analysis was performed using ImageJ software (rsb.nih.gov/ij). The value obtained for both ERK1 and ERK2 was normalized to anti-actin levels. The data were normalized to control and presented as percentage stimulation.
PKCβII Assay of GPR55 Activation
HEK 293 cells plated in 35-mm glass well Matek plastic dishes were transiently transfected with 175 µl of solution containing 1.5 µg/ml PKCβII-GFP cDNA or the PKC plasmid and 5 µg/ml human GPR55 cDNA in pCMV-Sport6 (Open Biosystems, Huntsville, Al) using a standard calcium phosphate protocol. Cells expressing GPR55 and PKCβII-GFP were utilized 24 h after transfection. Cells were washed with warm MEM and maintained at 37°C in 5% CO2 for 30–45 min after drug application. Agonist stimulated redistribution of PKCβII-GFP was assessed after drug treatment at room temperature.
Receptor Model Development
Modeling of GPR55 Using GPCR Crystallization Data
A model of the activated form of GPR55 was created using the 2.4Å crystal structure of β2-AR (16). First, the GPR55 sequence was aligned with the sequences of β2-AR, Rho, A2-adenosine receptor, CB1 and CB2 receptors. The same highly conserved residues that have been used in the past to align CB1 (24–29) and CB2 (30, 31) receptors to the sequence of bovine Rho were used as alignment guides for the GPR55 sequence. GPR55 possesses most of the conserved Class A patterns in TMHs 1, 2, 4 and 5 (N1.50, D2.50, W4.50 and P5.50). There are a few notable conserved motif differences between the GPR55 sequence and the sequence of not only the cannabinoid receptors but also the sequence of the crystallized GPCRs: 1) GPR55 has a conservative substitution (DRF) for the TMH3 E/DRY motif, 2) it has a conservative substitution (SFLP) for the TMH6 CWXP motif, and 3) it has a non-conservative substitution (DVFCY) for the THM7 NPXXY motif. Finally, the GPR55 extracellular-1 (EC-1) loop is shorter than most (3 amino acids (aa) vs. 6 aa in β2-AR and Rho) and the GPR55 EC-3 loop is noticeably longer than most (14 aa long vs. 5 aa in β2-AR, 6 aa in Rho and CB1/CB2).
Conformational Memories (CM) Technique for Calculating GPCR Structure
We explored the possible sequence-dictated conformational deviations of GPR55 TMHs 1, 2, 5, 6, and 7 from the β2-AR template using the CM technique (32–34). The CM method uses multiple Monte Carlo/simulated annealing random walks employing the CHARMM force field. Backbone φ and ψ torsions in regions of interest were allowed to vary ±50°, while all other backbone torsion angles were allowed to vary ± 10°. Side chain torsions were allowed to vary ±180°. All bond angles were allowed to vary ± 8° except from the C-S-C angles which were allowed to vary ± 15°. A minimum of 105 conformers were generated for each GPR55 helix independently in a distance dependent dielectric at 310K. Output helices were analyzed using the program ProKink (35). This program, which is embedded in the Simulaid Conversion program (36) was used to calculate the face shift, wobble angle, and bend angle of each helix.
TMH1 in GPR55 has a Pro at 1.41 not found in other sequences. An ideal helix (φ=−62.9° and ψ=−41.6°) with the GPR55 sequence was built and the region around the Pro (A1.37 to P1.41) was varied using CM.
TMH2 in GPR55 has a Pro at 2.58 instead of at 2.59 as in the β2-AR. The β2-AR TMH2 was mutated to the GPR55 sequence and the residues V2.54 to P2.58 were considered the variable region. S2.56 near P2.58 was set to be in g minus χ1.
TMH5 in GPR55 has two prolines, the conserved P5.50 that both β2-AR and Rho have and an additional one at 5.41. The β2-AR TMH5 was mutated to the GPR55 sequence. Only the region from K5.37 to P5.41 was varied because the β2-AR TMH5 already has a proline kink at P5.50.
TMH6 in GPR55 does not have the highly conserved Class A CWXP motif. Instead, it has the conservative substitution SFXP. Since TMH6 is implicated in conformational changes during receptor activation (37–42) it was very important to explore the conformational space of GPR55 TMH6. X-ray crystal structures of Rho and others have shown that an “ionic lock” salt bridge is formed between R3.50 near the intracellular (IC) end of TMH3 and D/E6.30 at the IC end of TMH6. This salt bridge is facilitated by the proline kink in TMH6 which bends the IC end of TMH6 towards TMH3. Biophysical studies have indicated that this salt bridge breaks upon activation and TMH6 straightens (40). An ideal helix (φ=−62.9° and ψ=−41.6°) with the GPR55 TMH6 sequence was built and the region around the Pro (V6.46 to P6.50) was varied using CM, while the S6.47 χ1 was held at trans position (see Supplemental Information for further details).
THM7 in GPR55 lacks the highly conserved NPXXY motif of Class A GPCRs and instead has a DVXXY sequence. The NPXXY motif not only influences the conformation of TMH7, but also places Y7.53 in the correct position to interact with F7.60 on the intracellular extension of TMH7, Helix 8 (Hx8). For receptors that possess an NPXXY motif, we typically designate the 7.46 to 7.50 region as variable. For GPR55 TMH7, an ideal helix (φ=−62.9° and ψ=−41.6°) was built and the region around the Pro (C7.46 to V7.50) was varied using CM.
Construction of the TMH region of GPR55
The outputs of the CM runs were used to build the TMH region of the receptor. The TMHs 1, 2, 5 and 7 were aligned with the intracellular part of the corresponding helices of the β2-AR, TMH 6 was aligned with the intracellular end of the β2-AR TMH6 for the inactive form of the receptor and with the extracellular end of the β2-AR TMH6 for the activated form of the receptor. The GPR55 inactive and activated bundles were then energy minimized to relieve any sidechain clashes and to pack the TMHs. The OPLS2005 all atom force field in Macromodel 9.1 (Schrodinger Inc., Portland, OR) was used employing a distance dependent dielectric with extended cutoffs (8.0 Å for nonbonded, 20.0 Å for electrostatic, and 4.0 Å for hydrogen bonding). A harmonic constraint was placed on all the TMH backbone torsions (φ, ψ, and ω) to preserve the general shape of the helices and let the sidechains adjust. The constraint on the TMH backbone torsions was gradually reduced to zero in the course of the minimization.
Modeling of the Intra/Extracellular Loops and Receptor Termini
Modeler 8.2 (43, 44) was used to add the N- and C-termini (Met1-Asn16 and Ser305-Gly319 respectively) and the intra-and extracellular loops (IC1: Leu49-Trp53, IC2: Leu127-Arg136, IC3: Gly210-Gln220, EC1: Ser88-Phe90, EC2: Phe159-Ser178, and EC3: Asp253-Ile266). The final loops were energy minimized to a gradient of 0.01 kcal/mol in a high dielectric using the Generalized Born/Surface Area (GB/SA) continuum solvation model for water, while the TMHs were frozen. The same force field, cutoffs and conjugate gradient method mentioned above were used.
MD Simulation of GPR55 Activated State (R*) in POPC Bilayer
The MD simulation cell for the GPR55 R* model embedded in a bilayer containing 151 palmitoyl-oleoyl-phosphatidylcholine (POPC) lipid molecules was built as described in Hurst et al. (45). The membrane was generated de novo using a library of lipid conformations from previous neat lipid simulations, as described in Grossfield et al (46). The system was neutralized with 10Cl− ions and NaCl was added to achieve 0.15M ionic strength. Three internal waters were included, two waters flanking Asp2.50 as seen in other crystal structures of GPCRs (13–20) and one water close to Asp7.49 (usually Asn in other GPCRs) which is surrounded by hydrophobic residues and has no partner. The system was then solvated with 13454 bulk waters and was run in the NPAT ensemble (using experimental area per lipid value for POPC of 68.3Å2)(47) for a total of 70ns. The NAMD2 simulation package (48) was used to run the dynamics employing the CHARMM27 force field for proteins and lipids (49–51). A plot of the RMSD of the TMH bundle versus time was prepared to identify the range of time points for which the TMH bundle geometry has stabilized in the lipid bilayer (plateau region in plot).
Ligand Conformational Analysis of LPI
LPI is a very flexible molecule and can assume a very large number of conformations. CM (32–34) was used to explore the conformational space of LPI because CM has been shown to achieve complete sampling of the conformational space of very flexible ligands, it overcomes energy barriers efficiently and it converges in a reasonable number of steps (32). The CM output was then analyzed and separated into groups (clusters) of similar conformations using X-Cluster as implemented in Macromodel (52). Increasing rms deviation from the first structure of the CM output is used as a criterion for partitioning the output into clusters.
Conformational Analysis of CID Compounds
Complete conformational analyses of the CID compounds were performed using ab initio Hartree-Fock calculations at the 6–31G* level as encoded in Jaguar (version 9.0, Schrodinger, LLC, New York, NY). HF 6–31G* 6-fold conformer searches were performed for the rotatable bonds (CID1792197 (2) bonds: N1-S2, S2-C1", C4"-N3, C6-C8, C9-C1"' and C2"'-O10, CID1172084 (3) bonds: C8'-O5, C5'-S1, S1-C2, C2-C3, N4-C1"', and C3"-C1"'; CID2440433 (4) bonds: N2-S3, S3-C1', C4'-N1"', C3'-C4, C4-N1"', N4"'-C1"" and CID1135734 (5) bonds: C9'-O5, C5'-S1, S1-C2, C2-C3, N4-C1"', and C3"-C1"'). In each conformer search, local energy minima were identified by rotation of a subject torsion angle through 360° in 60° increments (6-fold search), followed by HF 6–31G* energy minimization of each rotamer generated. To calculate the energy difference between the global minimum energy conformer of each compound and its final docked conformation, rotatable bonds in the global minimum energy conformer were driven to their corresponding value in the final docked conformation and the single point energy of the resultant structure was calculated at the HF 6–31G* level.
Electrostatic Potential Map Calculation
The electrostatic potential density surface was calculated using Spartan '08 (Wavefunction, Inc., 18401 Von Karman Ave., Suite 370, Irvine, CA 92612). The electrostatic potential energy was calculated using the Hatree-Fock method at 6–31G* level of theory and was mapped on the 0.002 isodensity surface of each molecule. The surface was color coded according to the potential, with electron rich regions colored red and electron poor regions colored blue.
Docking of Ligands
A low free energy conformer of LPI generated by CM, and the lowest energy conformation of each CID compound were used as inputs for receptor docking studies. Initially, each compound was manually docked in the binding site of GPR55 R* model that had been equilibrated in POPC (see above). The automatic docking program, Glide (Schrodinger Inc., Portland, OR) was then used to explore other possible binding conformations and receptor site interactions. Glide was used to generate a grid based on the centroid of select residues in the binding site (from the manual dock). Extra precision (XP) and flexible docking with ring sampling were selected for the docking setup. K2.60(80), the only positively charged residue in the transmembrane region, was defined as a hydrogen bond donor and was used as the only constraint for the automatic docking of all the compounds.
Ligand/Receptor Minimization
The receptor/ligand complexes were minimized using the OPLS2005 all atom force field in Macromodel 9.1 (Schrodinger Inc., Portland, OR). An 8.0-Å nonbonded cutoff (updated every 10 steps), a 20.0-Å electrostatic cutoff, and a 4.0-Å hydrogen bond cutoff were used in each stage of the calculation. The first stage consisted of 20000 steps of conjugate gradient minimization in 1000-step increments using a distance dependent dielectric. No harmonic constraints were placed on the ligand and the receptor sidechains in order to optimize the ligand/receptor interaction. A harmonic constraint was placed on all the TMH backbone torsions (φ, ψ, and ω) to preserve the general shape of the helices, with this gradually reduced to zero in the course of the first stage of the minimization. The loops and termini backbone torsions were also constrained with a 250 kcal/mol force during the first stage. In the second stage of the minimization, the Generalized Born/Surface Area (GB/SA) continuum solvation model for water was used as implemented in Macromodel. This stage of the calculation consisted of 20000 steps of Polak-Ribier conjugate gradient minimization in 1000 step increments or until the bundle reached the 0.05 kJ/mol gradient, in which the ligand and TMH bundle were frozen, but the loops and termini were allowed to relax.
RESULTS
Identification and Biochemical Characterization of GPR55 agonists
The three CID agonists (2–4) (Figure 2) were identified on the basis of the ability of a β-arrestin-GFP reporter to distinguish the active versus inactive conformation of GPCRs in general and GPR55 in particular (53). These ligands,
(CID1792197; 2) (E)-3-(2-methoxyphenyl)-N-[[4[methyl(phenyl)sulfamoyl]phenyl]-carbamothioyl]-prop-2-enamide,
(CID1172084; 3) N-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]-2-[(8-methoxy-4-methyl-[1,2,4]triazolo-[4,3-a]quinolin-1-yl)sulfanyl]acetamide, and
(CID2440433; 4) 3-[4-(2,3-dimethylphenyl)piperazine-1-carbonyl]-N,N-dimethyl-4-pyrrolidin-1-ylbenzenesulfonamide,
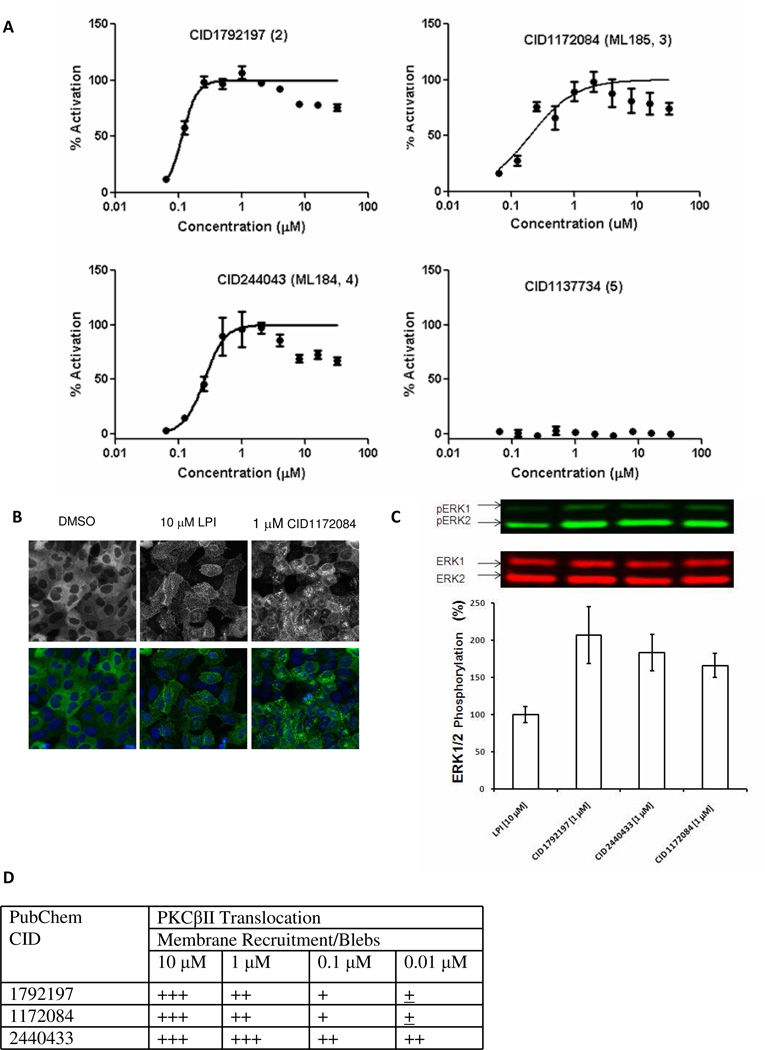
were further chosen for computer modeling of the GPR55 binding pocket due to their potencies, 110–250 nM EC50s, and greater than 100-fold selectivity for GPR55 over GPR35, CB1 and CB2 (see Table 1) in the β-arrestin translocation assay in U2OS cells permanently expressing HA-GPR55E and βarr2-GFP (10). Figure 3A illustrates concentration response curves for agonist activity in the β-arrestin trafficking assay. CID1792197 (2), CID1172084 (ML185, 3) and CID244043 (ML184, 4) induced trafficking with EC50’s of 0.11 ± 0.01, 0.16 ± 0.02 and 0.26 ± 0.01 µM, respectively (mean ± S.E.M., n = 2). Activity values were normalized to LPI (1) as 100% at 10 µM. No trafficking was observed with CID1135735 (5). The dose response curves in Figure 3A reach 100% activation as compared to LPI and then drop back down to a slightly lower plateau. This is likely due to a combination of the biology of the pit/vesicle formation with the image analysis assay read-out. The primary assay read-out is based on the well average of the “Number of Spots Detected per Cell” parameter. As higher concentrations of the compounds are added, the pits either cannot be resolved properly and multiple pits may be detected as one larger pit or the pits merge into larger vesicles, resulting in an apparent drop of activity at higher concentrations. See Figure S-1 in Supplemental Information for an illustration of this point ((zooming in to >250% is necessary to see the smaller dimmer spots well). In addition, it is possble that compound solubility problems at higher concentrations may also be contributing to this drop to a lower plateau.
Table 1.
High Throughput β-arrestin Screen Results
| PubChem CID |
Compound Number |
Agonist | Agonist Assay | Antagonist Assay | ||||
|---|---|---|---|---|---|---|---|---|
| GPR55 | GPR35 | CB1 | CB2 | GPR35 | CB1 | CB2 | ||
| EC50 (µM) | EC50 (µM) |
EC50 (µM) |
EC50 (µM) |
IC50 (µM) |
IC50 (µM) |
IC50 (µM) |
||
| 1792197 | 2 | 0.11/0.12 Avg 0.11 |
> 32 | > 32 | > 32 | > 32 | > 32 | > 32 |
| 1172084 | 3 | 0.15/0.18 Avg 0.16 |
> 32 | > 32 | > 32 | > 32 | > 32 | > 32 |
| 2440433 | 4 | 0.25/0.27 Avg 0.26 |
> 32 | > 32 | > 32 | > 32 | 21.8 | 15.1 |
| 1135734 | 5 | > 32 | > 32 | > 32 | > 32 | > 32 | > 32 | > 32 |
Figure 3.
Agonist activity produced by CID compounds. A. Concentration-response curves for agonist activity in the β-arrestin trafficking assay. CID1792197 (2), CID1172084 (ML185, 3) and CID244043 (ML184, 4) induced trafficking with EC50’s of 0.11 ± 0.01, 0.16 ± 0.02 and 0.26 ± 0.01 µM, respectively (mean ± S.E.M., n = 2). Activity values were normalized to LPI (1) as 100% at 10 µM. No trafficking was observed with CID1135735 (5). B. Representative images showing β-arrestin translocation of LPI vs. CID1172084 (see Table S-1 in the Supplemental information for images for all compounds (1–5)). C. CID compounds elicit ERK1/2 phosphorylation. U2OS cells expressing GPR55E cells were incubated with 1 µM CID2440433, 1 µM CID1792197, 1 µM 1172084 and 10 µM LPI. ERK1/2 was monitored and normalized to LPI (10 µM as 100%). Notably treatment with either of the CID compounds at 1 µM exhibited ERK1/2 phosphorylation that was at least as high as the previously reported activation upon LPI (10 µM) treatment. A representative gel is shown and mean ± SEM from three different experiments is shown. D. PKCβII translocation produced by CID compounds. ‘+’ indicates ~1% GFP membrane recruitment or blebs formation, ‘++’ indicates ~10% GFP membrane recruitment or blebs formation, ‘+++’ indicates ~30% GFP membrane recruitment or blebs formation.
Figure 3B presents representative images showing β-arrestin translocation in GPR55 (seen as punctate pattern in cell interiors) produced by application of LPI (10 µM) vs. CID1172084 (3) (1 µM) compared to the lack of translocation seen for the DMSO control. It is clear here that both LPI and CID1172084 (3) induce translocation in HA-GPR55E expressing U2OS cells compared to the control (HA-GPR55E expressing U2OS cells treated with DMSO) (see Table S-1 in Supplemental Information for images for all compounds (1–5)).
Treatment of HEK or U2OS cells expressing GPR55 with the CID(2–4) compounds or LPI(1) also activates at least two other intracellular signaling pathways common to agonists of many GPCRs, pERK activation and translocation of PKCβII to the plasma membrane. Figure 3C illustrates that CID compounds elicit ERK1/2 phosphorylation. U2OS cells expressing GPR55E cells were incubated with 1 µM CID2440433, 1 µM CID1792197, 1 µM 1172084 and 10 µM LPI. ERK1/2 was monitored and normalized to LPI (10 µM as 100%). Notably, treatment with either of the CID compounds at 1 µM exhibited ERK1/2 phosphorylation that was at least as high as the previously reported activation upon LPI (10 µM) treatment. A representative gel is shown and mean ± SEM from three different experiments is shown. Figure 3D presents a table summarizing PKCβII translocation results for application of CID compounds (2–4) to GPR55 expressing cells. Here ‘+’ indicates ~1% GFP membrane recruitment or blebs formation, ‘++’ indicates ~10% GFP membrane recruitment or blebs formation, ‘+++’ indicates ~30% GFP membrane recruitment or blebs formation.
GPR55 Receptor Model Development
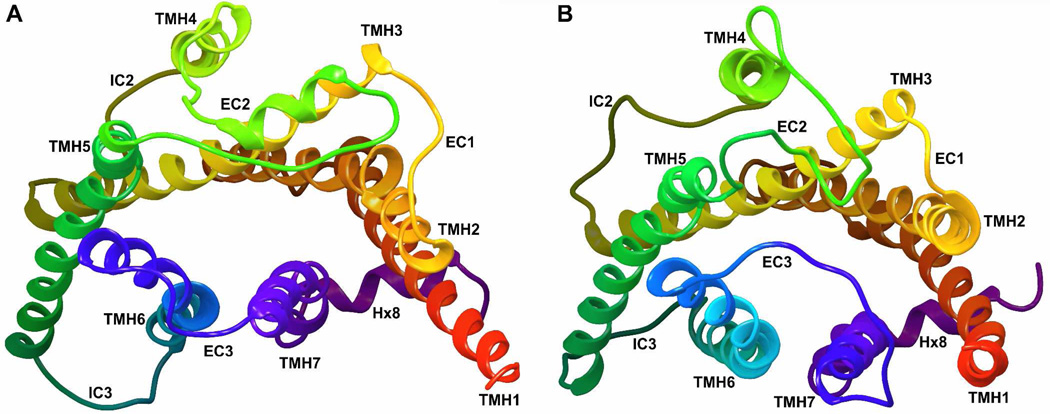
A homology model of the GPR55 inactive state was developed using the β2-AR crystal structure (PDB Name: 2RH1 (16)) as the template. This initial model was refined based on sequence dictated differences between GPR55 and the β2-AR crystal structure. Figure 4 illustrates a comparison of the β2-AR crystal structure (Figure 4A) and the GPR55 inactive state model (Figure 4B). Here the view is from the extracellular side of each receptor. The key differences between these two bundles in the transmembrane helices are described below.
Figure 4.
A comparison of (A) the β2-AR crystal structure to (B) the GPR55 inactive state model is illustrated here. The view is from the extracellular side of each receptor.
TMH1: TMH1 in the β2-AR pulls away from the TMH bundle on the EC side of the receptor and hydrophobic residues on TMH2 and TMH7 form a wall that excludes the EC portion of TMH1 from the binding site crevice. CM results show that the lipid-facing residue P1.41 in GPR55 bends the EC end of TMH1 instead toward the binding crevice, filling the space between TMH1/2 and TMH1/7. See Table 2 for helix geometry details..
Table 2.
ProKink Analysis Results for the Chosen TMH Conformers
| Helix | Hinge Residue | Bend Angle | Wobble Angle | Face Shift |
|---|---|---|---|---|
| TMH1 | P1.41 | 21.1° | 163.7° | 43.2° |
| TMH2 | P2.58 | 28.4° | −100.1° | 67.3° |
| TMH5 | P5.41 | 14.6° | −160.6° | 20.9° |
| TMH6 R state | P6.50 | 32.2° | −97.8° | 54.0° |
| TMH6 R* state | P6.50 | 25.7° | −98.3° | 68.4° |
| TMH7 | V7.50 | 31.9° | 173.0° | −0.9° |
TMH2: Since TMH1 in GPR55 bends toward the binding pocket on the EC end, TMH2 must take a different conformation as well to avoid steric overlaps with the EC end of TMH1. CM results show that GPR55 has this built into its sequence because GPR55 has a proline at 2.58 instead of 2.59 as in β2-AR, which bends the extracellular portion of TMH2 away from TMH1 and toward TMH3. See Table 2 for helix geometry details. This TMH2 helix exposes residues S2.64(84), L2.63(83) and K2.60(80) to the binding site crevice. It is important to note that had TMH2 retained the β2-AR TMH2 conformation, the EC-1 loop would not have been able to join the EC ends of TMH2 and TMH3 since the GPR55 EC-1 loop is only three amino acids long (SPF). In contrast, the EC-1 loop of the β2-AR is six amino acids in length (KMWTFG).
TMH5: Since the β2-AR TMH5 has the conserved P5.50 as does GPR55 TMH5, the backbone torsions for that Pro kink were preserved and only the region around P5.41 in GPR55 TMH5 was explored via CM. P5.41 bends the extracellular end of TMH5 inward toward the binding crevice and toward the extracellular end of TMH6. See Table 2 for helix geometry details.. The TMH5 was chosen from the CM output to avoid a clash with the EC end of TMH6.
TMH6: The CM output gives helices that bend inward toward the binding crevice and also helices that bend toward TMH7. From the helices generated by CM, we picked two helices, the more bent one was chosen for the inactive GPR55 bundle. This conformer allowed a hydrogen bond to form between R3.50 and Q6.30 at the IC end of the TMH bundle. A second TMH6 conformer was chosen that was straighter that we incorporated in the activated GPR55 model. See Table 2 for helix geometry details. This conformer was chosen so that the R3.50/Q6.30 hydrogen bond was broken. In the β2-AR, aromatic residues at 6.44 and 6.52 flank the toggle switch residue, W6.48. These residues, along with C6.47 have been proposed to be part of a larger rotamer toggle switch within the binding site crevice of the β2-AR (54). GPR55 also has aromatic residues flanking F6.48, F6.44 and H6.52, while the residue at 6.47 is a serine. The CM results showed that there is a correlation between the conformation of F6.48(239) and H6.52(243) (see Table S-2 in Supplemental information).
TMH7: GPR55 TMH7 lacks the highly conserved Class A GPCR NPXXY motif, having instead DVFCY. The lack of a proline residue in TMH7 resulted in CM output that did not have the usual proline induced bend and deformation seen in crystal structures of other Class A GPCRs. See Table 2 for helix geometry details.
GPR55 Model Activated Form (R*)
The hallmark of GPCR activation is the breaking of the ionic lock between R3.50 and E/D6.30 which allows TMH6 to straighten. The result is an intracellular opening of the receptor, exposing residues that can interact with the C-terminus of Gα. The active form of the GPR55 receptor model differs from the inactive form mainly in changes at the intracellular side of the receptor. The intracellular end of TMH6 has straightened and moved away from the TMH bundle, breaking the hydrogen bond between R3.50 and Q6.30 . The distance between the Cα carbons of R3.50 and Q6.30 is 13.3Å in the GPR55 R* model, indicating loss of interaction between the IC ends of TMH3 and 6. This distance is comparable to the opening formed in the very recent rhodopsin meta II (activated state) crystal structure (R3.50 -E6.30 Cα distance = 14.7Å)(55) in which the C-terminal fragment of transducin’s G-alpha is inserted. The size of this same opening in another recent activated GPCR crystal structure, the nanobody stabilized β2-AR crystal structure, is larger (R3.50 and E6.30 Cα distance=17.2Å)(56), due to the physical insertion of the nanobody higher than the C-terminal fragment of transducin’s G-alpha which causes the IC opening to enlarge.
Molecular Dynamics, GPR55 R* in a POPC Simulation
In order to equilibrate our GPR55 R* model in a realistic environment, a 70 ns MD simulation was performed in a POPC bilayer environment. The most noticeable change that took place during the MD simulation involved the shape of TMH7 and location of Hx8. Due to the lack of NPXXY motif (GPR55 has DVXXY instead), CM produced TMH7 conformations that lacked the pronounced bend and discontinuity seen in the β2-AR template structure. Energy minimization of the GPR55 bundle produced a distortion in TMH7 as described above. Further changes in TMH7 occurred during the MD simulation of GPR55 R*. Both the wobble angle and face shift angles for TMH7 changed (bend angle = 31.2°; wobble angle=-127.3°, face shift= 17.9°) and its intracellular end along with Hx8 moved away from the bundle increasing the IC opening of the receptor. This change is not unexpected since the R3.50/Q6.30 interaction is broken in R* and the IC ends of the TMH domain must open up during activation to receive the G protein Gα C-terminus. A plot of RMSD versus time (data not shown here) revealed that the RMSD of the GPR55 R* model stabilized by 35 ns. A frame from the 36th ns of this simulation was selected for initial ligand docking.
Ligand Conformational Analysis
Binding Conformation of LPI from Conformational Memories (CM) Calculations
For LPI (1), the CM distance dependent dielectric calculation produced conformers with bent head groups containing two hydrogen bonds between the inositol C5 hydroxyl (equatorial) and a phosphate oxygen and also between the glycerol hydroxyl and a phosphate oxygen. An LPI conformer with this bent head group orientation and an all-trans fatty acid chain (lowest energy conformation for saturated fatty acid chain) was used for initial docking studies in the GPR55 R* model.
Global Minimum Energy Conformations of Compounds 2–5
The key dihedral angles defining the minimum energy conformations of compounds 2–5 are listed in Table 3. See Supplemental Information for a more detailed description of the minimum energy conformations.
Table 3.
Dihedral Angle Definition of the Minimum Energy Conformers of the CID Compounds
| Dihedral Definition | CID1792197 | CID1172084 | CID2440433 | CID1135734 |
|---|---|---|---|---|
| C4"-N3-C4-N5 | −176.6° | |||
| N3-C4-N5-C6 | 4.2° | |||
| N5-C6-C8-C9 | 180.0° | |||
| C8-C9-C1"'-C2"' | 161.4° | |||
| C3"-C4"-N3-C4 | −66.5° | |||
| N1-S2-C1"-C2" | 86.2° | |||
| C1'-N1-S2-C1" | 70.2° | |||
| C2'-C1'-N1-S2 | 81.2° | |||
| C5'-S1-C2-C3 | −179.8° | |||
| S1-C2-C3-N4 | −178.4° | |||
| N4'-C5'-S1-C2 | −178.8° | |||
| C3-N4-C1"-N2" | −179.6° | |||
| C7'-C8'-O5-C6 | 179.9° | |||
| N2"-C3"-C1"'-C2"' | −18.8° | |||
| C2'-C1'-S3-N2 | −90.0° | |||
| C1'-S3-N2-LP | −179.7° | |||
| C3'-C4'-N1"-C2" | 173.7° | |||
| C2'-C3'-C4-O5 | 114.1° | |||
| O5-C4-N1"'-LP | 87.4° | |||
| LP-N4"'-C1""-C2"" | 28.0° | |||
| C5'-S1-C2-C3 | −179.3° | |||
| S1-C2-C3-N4 | 178.2° | |||
| N4'-C5'-S1-C2 | 179.4° | |||
| C3-N4-C1"-N2" | 178.1° | |||
| C8'-C9'-O5-C6 | −0.1° | |||
| N2"-C3"-C1"'-C2"' | −19.2° |
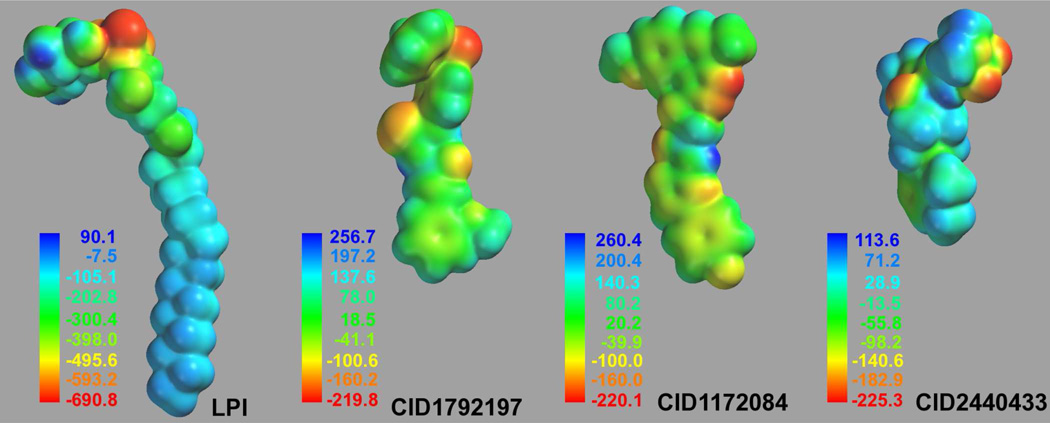
GPR55 Agonist Molecular Electrostatic Potential Maps
Figure 5 illustrates the molecular electrostatic potential maps (ranges in kJ/mol given next to each ligand) of the docked conformations of LPI (1) and CID compounds (2–4) at GPR55 R*. These ligands have inverted-L or T shapes with the most electronegative atom (or group of atoms) in each compound located in the “elbow” of the L or at one end of the T cross bar, with the rest of the molecule assuming a nearly straight conformation. The phosphate oxygens of LPI, the adjacent nitrogens of the triazole ring of CID1172084 (3) and the sulfonamide oxygens of CID1792197 (2) and CID2440433 (4) are the most electron rich regions of the ligands and were used as the interaction point with K2.60(80). LPI’s phosphate group has the most negative electrostatic potential region as would be expected for a negatively charged group.
Figure 5.
The molecular electrostatic potential maps of the docked conformations of compounds 1–4 are illustrated here. The range of the electrostatic potential (in kJ/mol) is given next to the map of each compound.
Docking of LPI and CID Compounds in GPR55 R* Model
K2.60(80), the only positively charged TMH residue in the putative binding site was used as the primary interaction site for all the ligands docked in GPR55 R* here. The putative binding site for LPI (1) and CID compounds 2–4 was identified to be the TMH 2–3–5–6–7 region of GPR55 R*, with only the CID compounds reaching to TMH 5.
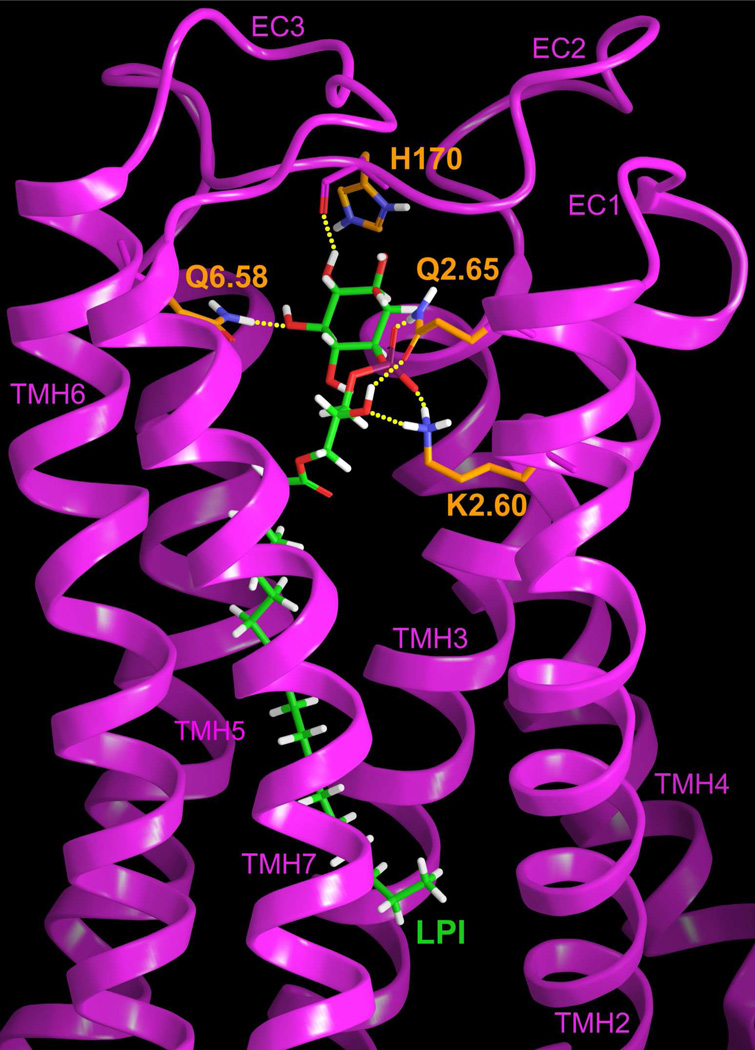
Figure 6 illustrates the final LPI (1)/GPR55 R* complex obtained using Glide. The binding site for LPI (1) is located between transmembrane helices (TMHs) 2, 3, 6 and 7. The primary interaction site for LPI (1) involves a salt bridge between the exposed, charged phosphate oxygen with K2.60(80). The hydrogen bond (N–O) distance and (N-H—O) angle are 2.45Å and 168°. K2.60(80) also hydrogen bonds to the glycerol hydroxyl of LPI. The hydrogen bond (N–O) distance and (N-H—O) angle are 2.68Å and 157°. Additionally, Q2.65(85) can hydrogen bond to the other exposed oxygen of the LPI (1) phosphate and to the glycerol hydroxyl that is involved with the K2.60(80). The hydrogen bond (N–O) and (O–O) distances and (N-H—O) and (O-H—O) angles are 2.58Å and 2.67Å, and 175° and 141 ° respectively. The inositol hydroxyls participate in hydrogen bonding with the backbone carbonyl of the extracellular EC2 loop residue H170 (hydrogen bond (O–O) distance and (O-H—O) angle: 3.07Å and 129°) and also Q6.58(249) at the top of TMH6 (hydrogen bond (N–O) distance and (N-H—O) angle: 2.82Å and 159°). The total pairwise interaction energy for LPI (1) at this docking site in GPR55 is −89.31 kcal/mol, making this the ligand with the best interaction energy within the set studied here (see Table S-3 in the Supplemental Information). The charge-charge interaction of the LPI (1) phosphate group with K2.60(80) is a major contributor to the interaction energy, however, hydrogen bonding interactions with H170 and Q6.58(249), as well as Van der Waals interactions with F3.33(102), M3.36(105), F169, L7.35(270) and M7.39(274) (residues not shown in Figure 6) also contributed significantly to the overall interaction energy.
Figure 6.
The LPI/GPR55 R* complex is illustrated here. The view is from the lipid bilayer with TMH1 removed to simplify the view. TMHs 2 and 7 are closest to the viewer here. K2.60 has been used as the primary interaction site for the most electronegative region of LPI.
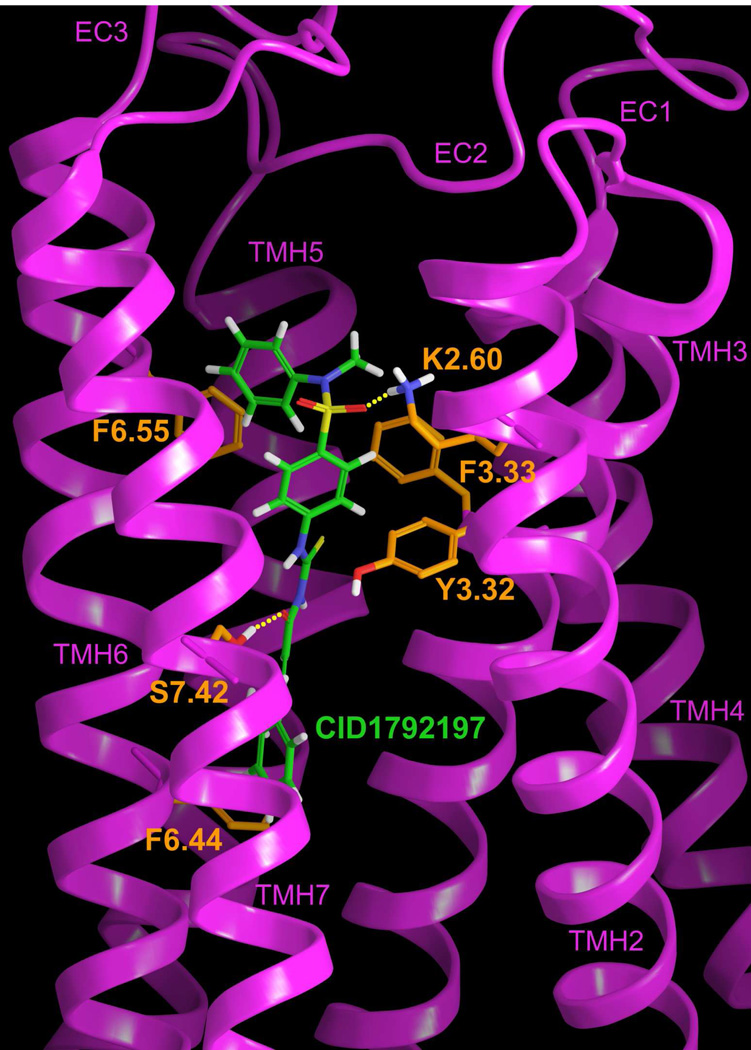
Figure 7 illustrates the final CID1792197 (2)/GPR55 R* complex obtained using Glide. The CID1792197 (2) sulfonamide oxygen hydrogen bonds with K2.60(80) and S7.42(277) hydrogen bonds with the carbonyl oxygen adjacent to the thiourea group. The hydrogen bond (N–O) and (O–O) distances and (N-H—O) and (O-H—O) angles are 2.66Å and 2.60Å and 166° and 164° respectively. CID1792197 (2), just like the other two CID compounds, forms a number of aromatic stacks. The pendant phenyl ring next to the sulfonamide group forms an offset parallel stack with F6.55(246) (ring centroid to ring centroid distance 5.66Å), the phenyl group between the sulfonurea and sulfonamide stacks with Y3.32(101) and F3.33(102). The ring centroid to ring centroid distances are 5.43Å and 4.87Å and the angles between ring planes are 74° and 30° respectively. Finally, the methoxy phenyl group stacks with F6.44(235) with a ring centroid to centroid distance of 5.22Å and an angle between the ring planes of 87°. The total pairwise interaction energy for CID1792197 (2) with GPR55 is −57.84 kcal/mol (see Table S-4 in the Supplemental Information). The major interaction energy contributions for this compound are the hydrogen bonding interactions with K2.60(80), the aromatic stacking interactions with F3.33(102) and the Van der Waals interactions with M3.36(105), F5.47(190)and L7.35(270) (residues not shown in Figure 7). Aromatic stacking interactions with F6.44(235) and Van der Waals interactions with S6.47(238) and V6.51(242) (not pictured in Figure 7) also contribute a small amount to the interaction energy. The remainder of the hydrogen bonding and aromatic stacking interactions identified contribute much less to the overall interaction energy.
Figure 7.
The CID1792197 (2)/GPR55 R* complex is illustrated here. The view is from the lipid bilayer with TMH1 removed to simplify the view. TMHs 2 and 7 are closest to the viewer here. K2.60 has been used as the primary interaction site for the most electronegative region of CID1792197 (2).
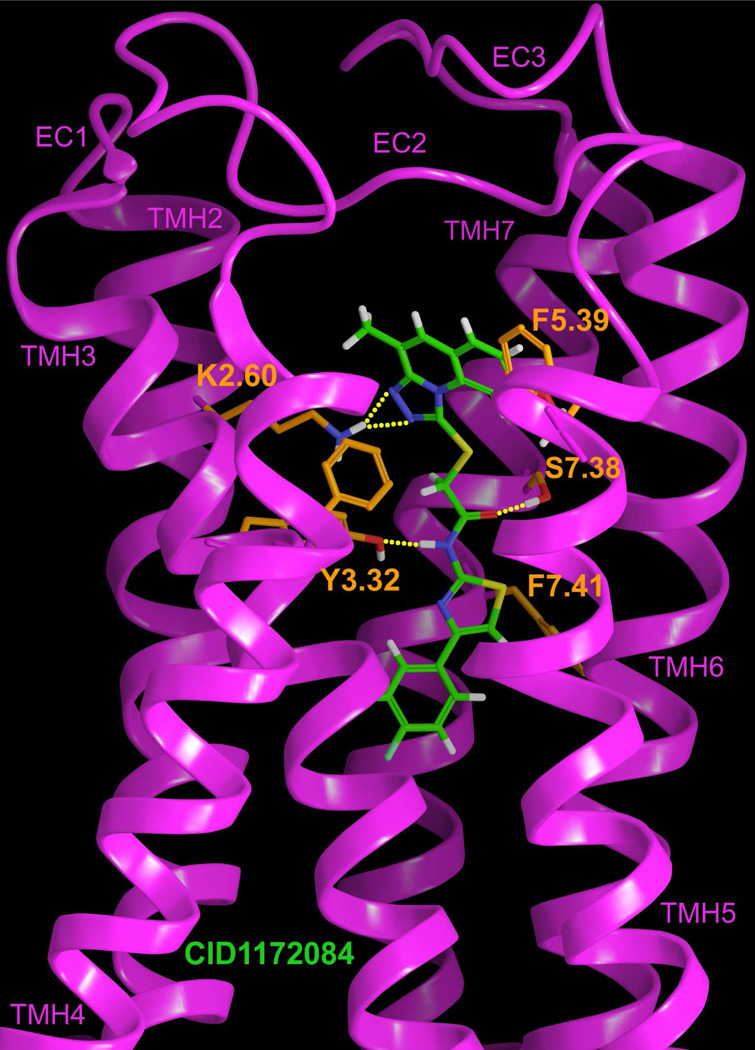
Figure 8 illustrates the final CID1172084 (3)/GPR55 R* complex obtained using Glide. The CID1172084 (3) 1,2,4-triazolo[4,3-a]quinoline ring nitrogens (positions 1′ and 2′) form hydrogen bonds with K2.60(80). The hydrogen bond (N-N1′) and (N-N2′) distances and (N-H—N1′) and (N-H—N2′) angles are 2.92Å and 2.82Å and 155° and 159 ° respectively. Y3.32(101) hydrogen bonds to the amide nitrogen (N4) and S7.38(273) hydrogen bonds to the carboxamide oxygen. The hydrogen bond (O–N) and (O–O) distances and (O-H—N) and (O-H—O) angles are 2.77Å and 2.57 Å and 158° and 175° respectively. Additionally, three phenylalanines form aromatic stacking interactions with the ligand. F3.33(102) and F7.41(276) form a T-stack with the 1,2,4-triazole and the thiazole rings of the compound respectively (ring centroid to ring centroid distances: 4.96Å and 5.94Å and angles between ring planes: 76° and 25° respectively) and F5.39(182) forms an offset parallel stack with the first ring of the 1,2,4-triazolo[4,3-a]quinoline (distance 6.21Å). The total interaction energy for CID1172084 (3) with GPR55 was found to be −65.19 kcal/mol (see Table-S-5 in the Supplemental Information). While the hydrogen bonding interactions with K2.60(80) and Y3.32(101) and the aromatic stacking interactions with F3.33(102) and F7.41(276) contribute to the interaction energy, the Van der Waals interactions with the hydrophobic residues M3.36(105), L7.35(270), M7.39(274) and S7.42(277) (residues not shown in Figure 8) have a major contribution to the total interaction energy of this complex.
Figure 8.
The CID1172084 (3)/GPR55 R* complex is illustrated here. The view is from the lipid bilayer with TMH1 removed to simplify the view. TMHs 4 and 5 are closest to the viewer here. K2.60 has been used as the primary interaction site for the most electronegative region of CID1172084 (3).
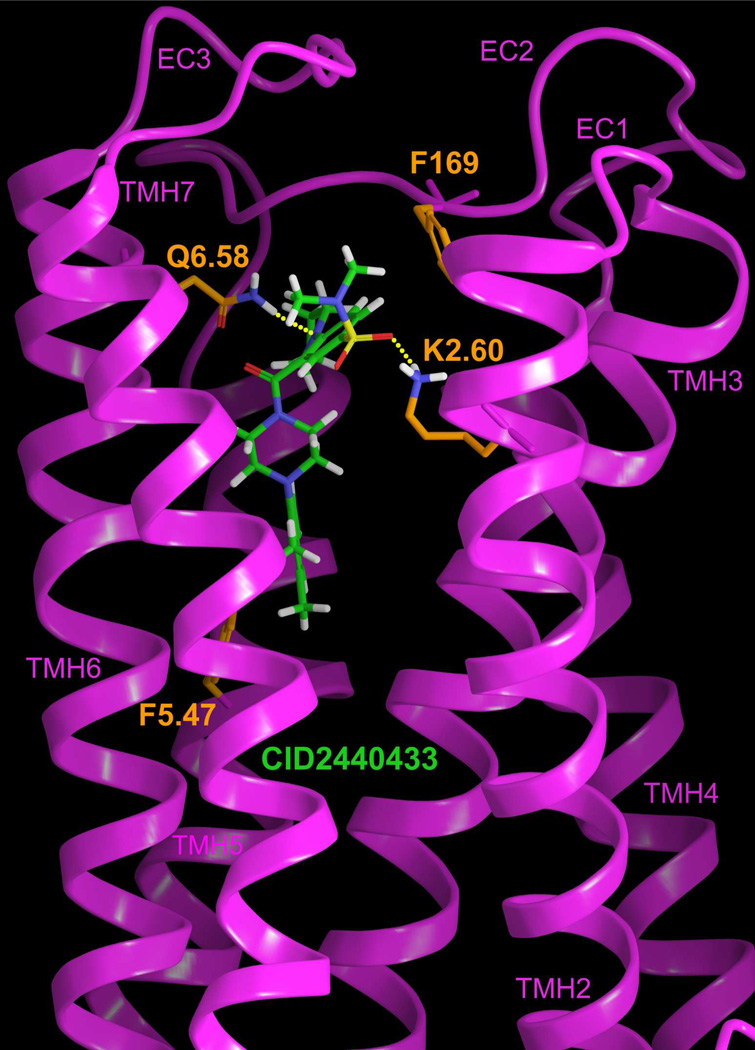
Figure 9 illustrates the final CID2440433 (4) /GPR55 R* complex obtained using Glide. The CID2440433 (4) sulfonamide oxygen forms a hydrogen bond with K2.60(80) and Q6.58(249) hydrogen bonds with the pyrrolidine nitrogen. The hydrogen bond (N–O) and (N–N) distances and (N-H—O) and (N-H—N) angles are 2.70Å and 2.96Å and 147° and 172° respectively. CID2440433 (4) also forms π-π stacking interactions with the receptor. The benzene ring adjacent to the sulfonamide forms an aromatic T-stack with F169 of the EC-2 loop (ring centroid to ring centroid distance 6.24Å and angle between ring planes 58°) and the dimethyl benzyl ring adjacent to the piperazine forms multiple aromatic stacks with the residue F5.47(190) (ring centroid to ring centroid distance 5.46Å and angle between ring plane 23° ). The total interaction energy for CID2440433 (4) at this docking site in GPR55 is −52.88 kcal/mol (see Table S-6 in the Supplemental Information). The hydrogen bonding interactions with K2.60(80) and Q6.58(2.49) are major contributors to the interaction energy, as well as the aromatic stacking interactions with H169. However, the Van der Waals interactions with the hydrophobic residues F3.33(102), V6.51(242) and L7.35(270) (residues not shown in Figure 9) make a substantial contribution to the overall interaction energy.
Figure 9.
The CID2440433 (4)/GPR55 R* complex is illustrated here. The view is from the lipid bilayer with TMH1 removed to simplify the view. TMHs 2 and 7 are closest to the viewer here. K2.60 has been used as the primary interaction site for the most electronegative region of CID2440433 (4).
Assessment of GPR55 R* Binding with the Negative Control CID1135734 (5)
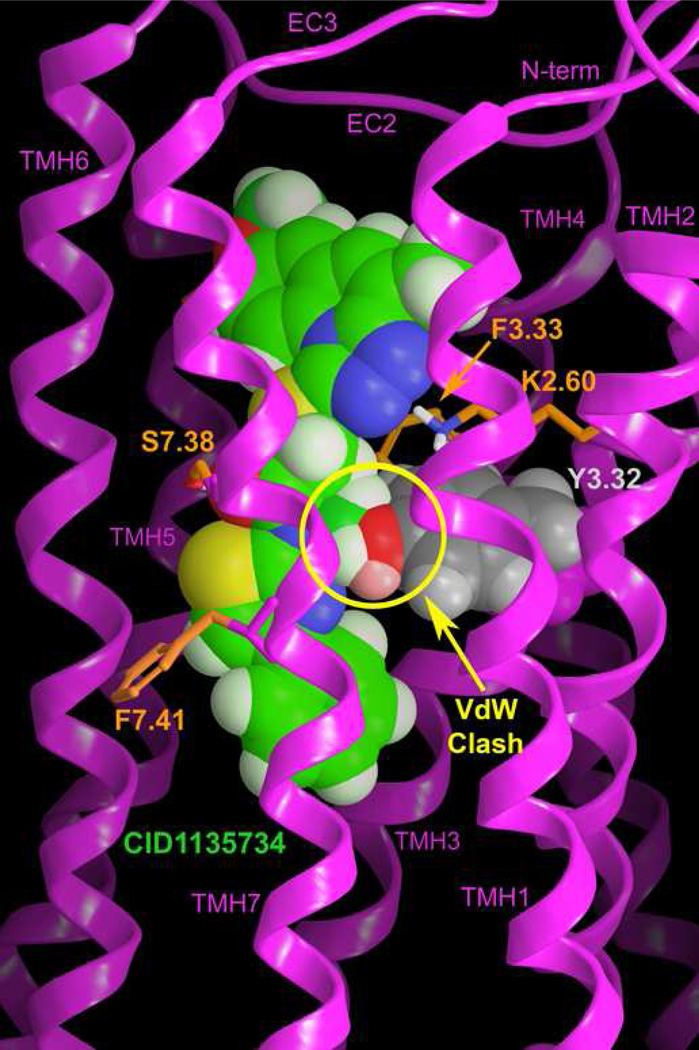
CID1135734 (5) is structurally related to CID1172084 (3), differing only in the absence of a fluorine, the presence of an N-methyl in the amide group and the position of the methoxy group on Ring A (see Figure 2). Despite the fact that the differences are small, CID1135734 (5) is inactive at GPR55 whereas CID1172084 (3) is an agonist. CID1135734 (5) was used here as a negative control to test the GPR55 R* model. Figure 10 illustrates the structure of CID1135734 (5) superimposed on the CID1172084 (3)/GPR55 R* complex. Of the structural differences between these closely related compounds, only one appears to interfere with CID1135734 (5) binding. The methoxy attached on the 1,2,4-triazolo[4,3-a]quinoline ring does not impact the N1-N2 ring nitrogens interaction with K2.60. This methoxy group is located in the spacious extracellular loop region where it does not have any steric clashes with the receptor. Additionally, the replacement of the 4-fluoro phenyl ring with the smaller, phenyl ring does not affect interactions with GPR55 R*. However, the CID1135734 (5) N-methyl group has major steric overlap with Y3.32(102), a residue that hydrogen bonds with the CID1172084 (3) amide moiety. The yellow circle in Figure 10 indicates the region of major steric overlap with Y3.32 of the receptor. This overlap likely explains why CID1135734 (5) does not activate GPR55 (see Table 1).
Figure 10.
The structure of CID1135734 (5) superimposed on the CID1172084 (3)/GPR55 R* complex is illustrated here. The view is from the lipid bilayer with TMHs 1 and 7 closest to the viewer here. The yellow circle indicates a major steric overlap with Y3.32 of the receptor. This overlap likely explains why CID1135734 (5) does not activate GPR55 (see Table 1).
DISCUSSION
High-Throughput Screening
High-throughput screening remains a powerful tool for the identification of new chemotypes acting at GPCRs. This is especially true in the GPR55 field where many of the known GPR55 ligands were identified from cannabinoid receptor/lipid biased compound libraries, leading to low chemical diversity and lack of receptor selectivity. Here we have identified three novel chemotypic GPR55 ligands (2–4) that lack agonism or antagonism at GPR35, CB1 or CB2. Despite their unique structures, we show that all of these ligands adopt a similar binding mode at GPR55 that is related to the binding mode of the endogenous GPR55 ligand, LPI (1). Our modeling indicates that the molecular shapes and electrostatic potential distributions of these CID agonists mimic that of LPI (1).
Receptor TMH Bundle Shape
It is clear in Figure 4 that while the GPR55 model preserves the general topology of a Class A GPCR, the TMH bundle is not identical in shape to the template structure, the β2-AR. The differences are dictated here by sequence divergences between GPR55 and β2-AR that our model development protocol (see Methods section) is designed to elucidate. There are numerous examples in the literature demonstrating how divergences in a single transmembrane helix, such as TMH1, affect the overall structure of the TMH bundle. For example, Rho TMH1 (13–15) contains a proline at position 1.48, while the β2-AR TMH1 (16–18) lacks a proline. The proline in TMH1 of Rho bends the EC end of TMH1 back towards the TMH bundle. In contrast, β2-AR TMH1 is straight and tilts away from the TMH bundle on the EC side; and this extreme tilt away from the binding pocket is accommodated by the presence of a “hydrophobic wall” of residues on β2-AR TMHs 2 and 7 that shields the binding pocket.
GPR55 is similar to Rho in that it also contains a proline in TMH1, P1.41, and that TMH1 leans into the TMH bundle as a consequence. Likewise, sequence differences in other GPR55 helices and loop segments relative to the β2-AR template also permit TMH bundle formation by accommodating the difference between GPR55 and β2-AR TMH1. Whereas β2-AR (16–18) contains a TMH2 proline at position 2.59 that causes the TMH2 EC end to lean towards TMH1, GPR55 TMH2 displays a sequence difference from β2-AR (16–18) that allows it to lean away from TMH1. For GPR55, this is a proline at position 2.58 placed such that TMH2 leans toward TMH3. This is a critical sequence change due to the very short length of the EC-1 loop in GPR55. The new CXCR4 crystal structure (21) provides evidence of the consequence of the shift in proline position in TMH2 to P2.58. Here it is clear that this change causes TMH2 to lean toward TMH3. The conformation of TMH2 is also critical for the GPR55 ligand binding pocket. The only positively charged TMH residue in GPR55 is K2.60. This residue faces into the binding pocket in our model, as does residue 2.60 in the CXCR4 crystal structure (21). It is important to note that residue 2.60 in the β2-AR structure (16) is in the TMH2–3 interface and not available from within the binding pocket.
The large majority of Class A GPCRs contain an NPXXY motif with Pro at sequence position 7.50. However, GPR55, GPR35 and GPR18 are rare exceptions and instead have a DXXXY motif that lacks proline. This small change in sequence, nonetheless, has an appreciable consequence on hydration and local transmembrane flexibility. For GPR55, although the TMH7 conformation determined from CM is relatively straight, bundle energy minimization induces a bend in TMH7 (see Figure 4) due to N7.43 interacting with the backbone of TMH7. The proximity of two methionine residues from TMH7 (M7.39) and TMH2 (M2.61) causes the EC end of TMH7 to bend away from the bundle as TMH2 and TMH7 pack during the minimization. When the GPR55 receptor model was then placed in the fully hydrated lipid bilayer environment, D7.49 (a residue that is normally N7.49 in other Class A GPCRs), along with D2.50, recruits water and this water further perturbs the backbone structure on TMH7 by hydrogen bonding to backbone carbonyls in the S7.42 to D7.49 region of TMH7. The net result is an increase in flexibility allowing TMH7 to bend at S7.42 and compensate for the lack of a proline at position 7.50.
Receptor Binding Pocket Shape
In most Class A GPCR crystal structures, the co-crystallized ligand is oriented horizontally in the binding pocket. This is true for the initial template crystal structure for the GPR55 homology model, the β2-AR structure which has the partial inverse agonist carazolol oriented horizontally (16). In contrast, ZM241385 is oriented nearly vertically in the adenosine A2A crystal structure (20). Because the structure of β2-AR was used as the template to build the model of the activated GPR55, the model initially contained a “horizontal” space for binding which did not collapse during minimization of the TMH bundle. However, as the GPR55 R* model equilibrated in the hydrated lipid bilayer, the shape of the binding pocket changed. In addition to the retained horizontal opening, the GPR55 R* model developed a rather straight and deep vertical channel reaching as far as N7.46, the residue that forms the floor of the binding pocket. In the MD simulations, the GPR55 R* binding pocket is highly hydrated, with a string of waters that extends to the IC side as would be expected for an R* state of the receptor (45, 57).
The GPR55 R* model agonist binding pocket is lined with hydrophilic residues almost to the intracellular side of the receptor, (S3.39(108), S3.47(116), S6.36(227) and S6.40(238)), in contrast to the highly hydrophobic binding pockets of the cannabinoid CB1 and CB2 receptors. The horizontal part of the GPR55 binding pocket underneath the EC loops is lined by K2.60(80), S2.64(84), Q2.65(85), E3.29(98), Q6.58(249), S7.32(267) and Q7.36(271) and the deep vertical channel is lined by Y3.32(101), S6.47(238), S7.42(277), N7.43(278) and D7.49(284).
The resultant shape of the GPR55 R* binding site accommodates ligands that are inverted-L shapes or T shapes with long, thin profiles that can fit vertically deep in the receptor binding pocket while their broad head regions occupy the horizontal binding pocket opening near the EC loops. The vertical pore is narrow enough such that it cannot accommodate the N-methyl group of CID1135735 (5). For GPR55 agonist ligands (1–4), the highest negative electrostatic potential region is exposed either at the “elbow” of the L or at one end of the T cross bar (see Figure 5, red regions are most negative regions). It is this region that interacts with K2.60 in each of the docks pictured in Figures 6–9.
For LPI (1), the highly negative electrostatic potential is formed by the phosphate group (see Figures 2 and 6). In the LPI (1) docked conformation pictured in Figure 6, the phosphatidylinositol (bulky head group) is bent at the phosphate and is perpendicular to the 18-carbon saturated fatty acid tail. The “elbow” of CID1792197 (2) is formed by a sulfonamide group. The two oxygens of the sulfonyl are exposed and the pendant, N-methyl-N-benzylamino group, from the sulfur is positioned almost perpendicular to the long part of the molecule, placing the sulfur in the “elbow” (see Figures 2 and 7). The exposed sulfonyl oxygens are the most electronegative region of CID1792197 (2) that mimic the exposed phosphate oxygens of LPI. CID1172084 (3) does not have a sulfonamide in the “elbow” but rather a 1,2,4-triazolo[4,3-a]quinoline group that is attached on the 5 position to the long segment of the molecule placing the fused ring system perpendicular to the long segment of the molecule and exposing the N-N of the 1,2,4-triazole. The adjacent nitrogens of the 1,2,4-triazole of CID1172084 (3) form the electronegative region that mimics the exposed phosphate oxygens of LPI (see Figures 2 and 8). CID2440433 (4) is T-shaped and has an N,N-dimethyl sulfonamide group at the end of the cross bar of the T (see Figures 2 and 9) which is the most electronegative region in CID2440433 (4).
Similarity to Other Reported GPR55 Ligands
The cannabinoid CB1 inverse agonist, N-(piperidin-1-yl)-1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-1H–pyrazole-3-carboxamide (AM251) has been reported to act as an agonist at GPR55 (10). Glide docking studies of this compound show that this compound also can adopt a T-shape in the GPR55 binding pocket. The 2,4-dichlorophenyl and pyrazole rings form the cross bar of this T-shape, with the 4-iodophenyl ring penetrating into the vertical section of the binding pocket (see Figure S-2 in Supplemental Information). The energy of interaction (EOI) of AM251 with GPR55 is also comparable to the EOI of CID2440433 (CID2440433 (4)/GPR55 EOI=-52.88 kcal/mol;AM251/GPR55= −53.64 kcal/mole). Further details concerning the modeled AM251/GPR55 R* complex are provided in Supplemental Information.
The structure of CID2440433 (4) is similar to the benzoylpiperazine GPR55 agonist, (1-{2-fluoro-4-[1-(methyloxy)ethyl]phenyl}-4-{[4-fluoro-4-(methylsulfonyl)-2-biphenylyl]carbonyl} piperazine; GSK494581A) that was reported by Brown and co-workers while this paper was under review (12). Our GPR55 R* docking study shows that GSK494581A adopts a T-shape at GPR55 R*, binding in the same receptor region and in a similar orientation as CID2440433 (4) (see Figure 9 and Figure S-3 in Supplemental Information), with comparable energy of interaction (EOI) (CID2440433 (4)/GPR55 EOI=-52.88 kcal/mol; GSK494581A/GPR55 EOI=-52.74 kcal/mol. Further details concerning the modeled GSK494581A /GPR55 R* complex are provided in Supplemental Information.
Future Directions
The chemical diversity provided by the three lead compounds, CID (2–4), combined with the identification of key receptor interaction sites should provide a basis for the design of more potent and efficacious second generation GPR55 ligands that retain GPR55 selectivity. Such studies are underway in our laboratories. In addition, mutation studies currently underway include mutations to test the effects of key amino acids in the ligand pocket, including S7.42 S7.38/Y3.32 and Q6.58 which should affect the binding of CID1792197, CID1172084 and CID2440433/LPI respectively. Preliminary results for K2.60 (K2.60A) suggest that this residue is essential for ligand activation at GPR55 (unpublished observations).
CONCLUSIONS
As a result of a high throughput β-arrestin screen, we have identified three potent novel chemotypic GPR55 ligands (2–4) that lack agonism or antagonism at GPR35, CB1 or CB2. Their shapes mirror the bioactive conformation of the endogenous GPR55 ligand LPI (1), and docking experiments indicate that CID compounds (2–4), as well as LPI (1), bind GPR55 in the TMH 2, 3, 5, 6 and 7 region. The primary interaction for each of these agonists with GPR55 occurs between K2.60 and an exposed highly electronegative region on each ligand. The chemical diversity of the three agonist lead compounds not only provides a basis for designing improved second generation GPR55 ligands, but also provides the tools for separating the biological activity of GPR55 from that of cannabinoids.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge Drs. Russell Dahl and Shenghua Shi for selecting compounds from the primary screen for further testing.
This research was supported by grants from the National Institutes of Health [DA023204, DA05274, DA029432, DA022950 and DA021358(PHR)]. The HCS screening portion of this work was supported by National Institutes of Health Roadmap Program grant [U54HG005033] and performed at Sanford-Burnham Medical Research Institute’s Conrad Prebys Center for Chemical Genomics (CPCCG), a comprehensive screening center with an X01 DA026205 grant to MEA.
Abbreviations
- GPCR
G protein coupled receptor
- CB1/CB2
Cannabinoid type 1 receptor/Cannabinoid type 2 receptor
- LPI
lysophosphatidylinositol
- 2-AGPI
2-arachidonoyl-sn-glycero-3-phosphoinositol
- MLPCN
Molecular Libraries Probe Production Centers Network
- TMH
transmembrane helix
- EC
extracellular
- IC
intracellular
- β2-AR
β2 Adrenergic receptor
- GPF
Green Fluorescent Protein
- CM
Conformational Memories
Footnotes
Supplemental Information: The Supplemental material includes a table (Table S-1) with example β-arrestin images for Compounds 1–5 and DMSO control; an example (Figure S-1) of the functional readout results that corresponds to a dose response curve in Figure 3A; a description of the conformations of the selected TMHs that were incorporated in the GPR55 model; a table (Table S-2) that shows the correlation of χ1 F6.48 (239) with F6.44 (235) and H6.52 (243); a description of the minimum energy geometries of the CID compounds; a set of tables that include all the pairwise interactions of the Compounds 1–4 at the GPR55 R* binding site (Tables S-3 to S-6); and, a description with image of the GPR55 R* binding site and resultant energies of interaction for two other known GPR55 agonists, AM251(Figure S-2 and Table S-7) and GSK494581A (Figure S-3 and Table S-8). The Supplemental Information may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Sawzdargo M, Nguyen T, Lee DK, Lynch KR, Cheng R, Heng HH, George SR, O'Dowd BF. Identification and cloning of three novel human G protein-coupled receptor genes GPR52, PsiGPR53 and GPR55: GPR55 is extensively expressed in human brain. Brain Res Mol Brain Res. 1999;64:193–198. doi: 10.1016/s0169-328x(98)00277-0. [DOI] [PubMed] [Google Scholar]
- 2.Staton PC, Hatcher JP, Walker DJ, Morrison AD, Shapland EM, Hughes JP, Chong E, Mander PK, Green PJ, Billinton A, Fulleylove M, Lancaster HC, Smith JC, Bailey LT, Wise A, Brown AJ, Richardson JC, Chessell IP. The putative cannabinoid receptor GPR55 plays a role in mechanical hyperalgesia associated with inflammatory and neuropathic pain. Pain. 2008;139:225–236. doi: 10.1016/j.pain.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Whyte LS, Ryberg E, Sims NA, Ridge SA, Mackie K, Greasley PJ, Ross RA, Rogers MJ. The putative cannabinoid receptor GPR55 affects osteoclast function in vitro and bone mass in vivo. Proc Natl Acad Sci U S A. 2009;106:16511–16516. doi: 10.1073/pnas.0902743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford LA, Roelofs AJ, Anavi-Goffer S, Mowat L, Simpson DG, Irving AJ, Rogers MJ, Rajnicek AM, Ross RA. A role for L-alpha-lysophosphatidylinositol and GPR55 in the modulation of migration, orientation and polarization of human breast cancer cells. Br J Pharmacol. 2010;160:762–771. doi: 10.1111/j.1476-5381.2010.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andradas C, Caffarel MM, Perez-Gomez E, Salazar M, Lorente M, Velasco G, Guzman M, Sanchez C. The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene. 2010;30:245–252. doi: 10.1038/onc.2010.402. [DOI] [PubMed] [Google Scholar]
- 6.Pineiro R, Maffucci T, Falasca M. The putative cannabinoid receptor GPR55 defines a novel autocrine loop in cancer cell proliferation. Oncogene. 2010;30:142–152. doi: 10.1038/onc.2010.417. [DOI] [PubMed] [Google Scholar]
- 7.Henstridge CM, Balenga NA, Ford LA, Ross RA, Waldhoer M, Irving AJ. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. FASEB J. 2009;23:183–193. doi: 10.1096/fj.08-108670. [DOI] [PubMed] [Google Scholar]
- 8.Oka S, Nakajima K, Yamashita A, Kishimoto S, Sugiura T. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem Biophys Res Commun. 2007;362:928–934. doi: 10.1016/j.bbrc.2007.08.078. [DOI] [PubMed] [Google Scholar]
- 9.Oka S, Toshida T, Maruyama K, Nakajima K, Yamashita A, Sugiura T. 2-Arachidonoyl-sn-glycero-3-phosphoinositol: A Possible Natural Ligand for GPR55. J Biochem. 2009;145:13–20. doi: 10.1093/jb/mvn136. [DOI] [PubMed] [Google Scholar]
- 10.Kapur A, Zhao P, Sharir H, Bai Y, Caron MG, Barak LS, Abood ME. Atypical responsiveness of the orphan receptor GPR55 to cannabinoid ligands. The Journal of biological chemistry. 2009;284:29817–29827. doi: 10.1074/jbc.M109.050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin H, Chu A, Li W, Wang B, Shelton F, Otero F, Nguyen DG, Caldwell JS, Chen YA. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. J Biol Chem. 2009;284:12328–12338. doi: 10.1074/jbc.M806516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AJ, Daniels DA, Kassim M, Brown S, Haslam CP, Terrell VR, Brown J, Nichols PL, Staton PC, Wise A, Dowell SJ. Pharmacology of GPR55 in Yeast and Identification of GSK494581A as a Mixed-Activity Glycine Transporter Subtype 1 Inhibitor and GPR55 Agonist. J Pharmacol Exp Ther. 2011;337:236–246. doi: 10.1124/jpet.110.172650. [DOI] [PubMed] [Google Scholar]
- 13.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 14.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 16.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 19.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science (New York, N.Y. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher S, Winston SE, Fuller SA, Hurrell JG. Current Protocols in Immunology. New York, NY: Wiley; 2008. Immunoblotting and Immunodetection. [DOI] [PubMed] [Google Scholar]
- 24.Bramblett RD, Panu AM, Ballesteros JA, Reggio PH. Construction of a 3D model of the cannabinoid CB1 receptor: determination of helix ends and helix orientation. Life Sci. 1995;56:1971–1982. doi: 10.1016/0024-3205(95)00178-9. [DOI] [PubMed] [Google Scholar]
- 25.Song ZH, Slowey CA, Hurst DP, Reggio PH. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2) Molecular pharmacology. 1999;56:834–840. [PubMed] [Google Scholar]
- 26.Hurst DP, Lynch DL, Barnett-Norris J, Hyatt SM, Seltzman HH, Zhong M, Song ZH, Nie J, Lewis D, Reggio PH. N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H–p yrazole-3-carboxamide (SR141716A) interaction with LYS 3.28(192) is crucial for its inverse agonism at the cannabinoid CB1 receptor. Molecular pharmacology. 2002;62:1274–1287. doi: 10.1124/mol.62.6.1274. [DOI] [PubMed] [Google Scholar]
- 27.Barnett-Norris J, Hurst DP, Lynch DL, Guarnieri F, Makriyannis A, Reggio PH. Conformational memories and the endocannabinoid binding site at the cannabinoid CB1 receptor. J Med Chem. 2002;45:3649–3659. doi: 10.1021/jm0200761. [DOI] [PubMed] [Google Scholar]
- 28.McAllister SD, Hurst DP, Barnett-Norris J, Lynch D, Reggio PH, Abood ME. Structural mimicry in class A G protein-coupled receptor rotamer toggle switches: the importance of the F3.36(201)/W6.48(357) interaction in cannabinoid CB1 receptor activation. The Journal of biological chemistry. 2004;279:48024–48037. doi: 10.1074/jbc.M406648200. [DOI] [PubMed] [Google Scholar]
- 29.McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Abood ME. An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152. doi: 10.1021/jm0302647. [DOI] [PubMed] [Google Scholar]
- 30.Nebane NM, Hurst DP, Carrasquer CA, Qiao Z, Reggio PH, Song ZH. Residues accessible in the binding-site crevice of transmembrane helix 6 of the CB2 cannabinoid receptor. Biochemistry. 2008;47:13811–13821. doi: 10.1021/bi8007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, Hurst DP, Barnett-Norris J, Reggio PH, Song ZH. Cysteine 2.59(89) in the second transmembrane domain of human CB2 receptor is accessible within the ligand binding crevice: evidence for possible CB2 deviation from a rhodopsin template. Molecular pharmacology. 2005;68:69–83. doi: 10.1124/mol.104.007823. [DOI] [PubMed] [Google Scholar]
- 32.Guarnieri F, Weinstein H. Conformational Memories and the Exploration of Biologically Relevant Peptide Conformations:u An Illustration for the Gonadotropin-Releasing Hormone. J Am Chem Soc. 1996;118:5580–5589. [Google Scholar]
- 33.Guarnieri F, Wilson SR. Conformational memories and a simulated annealing program that learns: Application to LTB4. J Comput Chem. 1995;16:654–658. [Google Scholar]
- 34.Whitnell RM, Hurst DP, Reggio PH, Guarnieri F. Conformational memories with variable bond angles. J Comput Chem. 2008;29:741–752. doi: 10.1002/jcc.20822. [DOI] [PubMed] [Google Scholar]
- 35.Visiers I, Braunheim BB, Weinstein H. Prokink: a protocol for numerical evaluation of helix distortions by proline. Protein engineering. 2000;13:603–606. doi: 10.1093/protein/13.9.603. [DOI] [PubMed] [Google Scholar]
- 36.Mezei M. SIMULAID: a simulation facilitator and analysis program. J Comp Chem. 2010;31:2658–2668. doi: 10.1002/jcc.21551. [DOI] [PubMed] [Google Scholar]
- 37.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science (New York N.Y. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 38.Ghanouni P, Steenhuis JJ, Farrens DL, Kobilka BK. Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc Natl Acad Sci U S A. 2001;98:5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javitch JA, Fu D, Liapakis G, Chen J. Constitutive activation of the beta2 adrenergic receptor alters the orientation of its sixth membrane-spanning segment. The Journal of biological chemistry. 1997;272:18546–18549. doi: 10.1074/jbc.272.30.18546. [DOI] [PubMed] [Google Scholar]
- 40.Jensen AD, Guarnieri F, Rasmussen SG, Asmar F, Ballesteros JA, Gether U. Agonist-induced conformational changes at the cytoplasmic side of transmembrane segment 6 in the beta 2 adrenergic receptor mapped by site-selective fluorescent labeling. The Journal of biological chemistry. 2001;276:9279–9290. doi: 10.1074/jbc.M004871200. [DOI] [PubMed] [Google Scholar]
- 41.Lin SW, Sakmar TP. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 42.Nakanishi J, Takarada T, Yunoki S, Kikuchi Y, Maeda M. FRET-based monitoring of conformational change of the beta2 adrenergic receptor in living cells. Biochem Biophys Res Commun. 2006;343:1191–1196. doi: 10.1016/j.bbrc.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 43.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurst DP, Grossfield A, Lynch DL, Feller S, Romo TD, Gawrisch K, Pitman MC, Reggio PH. A lipid pathway for ligand binding is necessary for a cannabinoid G protein-coupled receptor. J Biol Chem. 2010;285:17954–17964. doi: 10.1074/jbc.M109.041590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc Natl Acad Sci U S A. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 48.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feller SE, MacKerell JAD. An Improved Empirical Potential Energy Function for Molecular Simulations of Phospholipids. Journal of Physical Chemistry B. 2000;104:7510–7515. [Google Scholar]
- 50.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of protein. Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 51.Pitman MC, Suits F, Mackerell AD, Feller SE., Jr Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry. 2004;43:15318–15328. doi: 10.1021/bi048231w. [DOI] [PubMed] [Google Scholar]
- 52.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. Macromodel - an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–467. [Google Scholar]
- 53.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. The Journal of biological chemistry. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 54.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. The Journal of biological chemistry. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 55.Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011 doi: 10.1038/nature09789. EPub March 9. [DOI] [PubMed] [Google Scholar]
- 56.Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a Nanobody-Stabilized Active State of the Beta-2-Adrenoreceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. Journal of molecular biology. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.