Abstract
Fat is stored or mobilized according to food availability. Malfunction of the mechanisms that ensure this coordination underlie metabolic diseases in humans. In mammals, lysosomal and autophagic function is required for normal fat storage and mobilization in the presence or absence of food. Autophagy is tightly linked to nutrients. However, if and how lysosomal lipolysis is coupled to nutritional status remains to be determined. Here we identify MXL-3 and HLH-30 as transcriptional switches coupling lysosomal lipolysis and autophagy to nutrient availability and controlling fat storage and ageing in Caenorhabditis elegans. Transcriptional coupling of lysosomal lipolysis and autophagy to nutrients is also observed in mammals. Thus, MXL-3 and HLH-30 orchestrate an adaptive and conserved cellular response to nutritional status and regulate lifespan.
The importance of lipolysis to general metabolism became apparent when it was discovered that fat can only be mobilized in its hydrolysed form1. Classically, lipolysis has been associated with cytosolic or ER-associated neutral lipases. However, cytosolic lipids are also catabolized through autophagy-mediated lipolysis, also termed lipophagy2. Both lysosomal acid lipases3 (LALs) and Atg15 (ref. 4), an autophagy protein with predicted triglyceride-lipase activity, have been proposed to catabolize lipid-droplet fat stores through lipophagy. Although lysosomal inhibitors induce increased levels of lipid-droplet fats, lysosomal inhibition impairs the function of the endocytic pathway and autophagy, making this approach insufficient to define which lipases actually break down lipids through lipophagy. Similar to general macroautophagy, lipophagy is activated by fasting. However, it is unknown if the assembly of autophagosomes is the only regulated step or if the lipases breaking down lipids in the autophagolysosome would be separately regulated by changes in nutrient availability. If the lipases were independently regulated, then the molecular players that link their activity to nutrient availability would remain to be uncovered. Finally, it has not been established if lipophagy is confined to mammals or if it is an ancient mechanism of energy homeostasis.
Here we present the lysosomal lipases LIPL-1 and LIPL-3 as key enzymes breaking down lipid-droplet fats through lipophagy in C. elegans. In addition, we show that the transcription factors MXL-3 and HLH-30 link nutrient availability to lysosomal lipolysis, uncovering two fat regulators. We also show that lysosomal lipolysis and autophagy are similarly linked to nutrient availability in mammalian cells in culture. Finally, we show that lysosomal lipolysis and its regulators MXL-3 and HLH-30 influence C. elegans ageing.
RESULTS
lipl-1, 2, 3, 4 and 5 respond to fasting
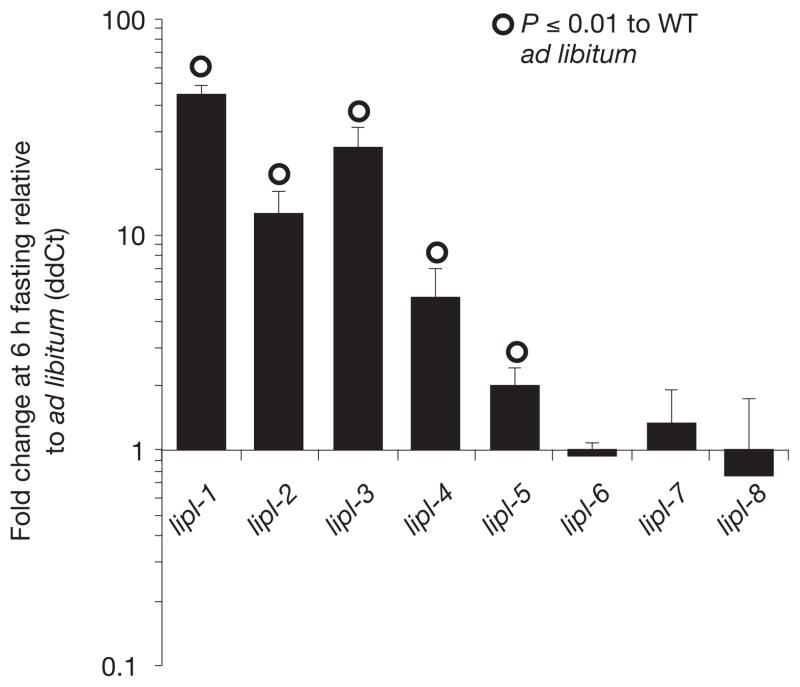
A list of 84 conserved C. elegans genes likely to be regulated by nutritional status was generated through comparative analyses of the transcriptional response to starvation of C. elegans5, Drosophila melanogaster6, and mice7 (Supplementary Tables S1 and S2). We tested these genes for differential expression in 6 h-fasted versus well-fed fertile adult worms. The most upregulated genes of the set belonged to a family of predicted triglyceride lipases. lipl-1, 2, 3, 4 and 5 were upregulated 44-, 12-, 25-, 7- and 2-fold, respectively (Fig. 1). lipl-6, 7 and 8 showed no change in expression (Fig. 1). Transcriptional reporters for lipl-1, 2 and 3 are observed in the intestine of fasted, but not well-fed, larvae and adults (Supplementary Fig. S1). The lipl-4 P::GFP (green fluorescent protein) fusion gene is expressed in the intestine of starvation-induced dauer larvae, and in the pharynx of both well-fed and fasted animals (Supplementary Fig. S1). lipl-5 P::GFP is expressed in the intestine8.
Figure 1.
lipl-1 to 5 are upregulated following fasting. RNA was extracted from young fertile adults fed ad libitum or fasted for 6 h. ddCts (delta delta cycle thresholds) were calculated normalizing to ama-1 and the efficiency of the primer sets as previously described28. Means+ s.e.m. are depicted. lipl-1 to lipl-5, n = 6 independent experiments. lipl-6 to lipl-8, n = 3 independent experiments. Significant t-test derived P -values are indicated. WT, wild type.
LIPL-1 and LIPL-3 are lysosomal lipases controlling lipid-droplet fats
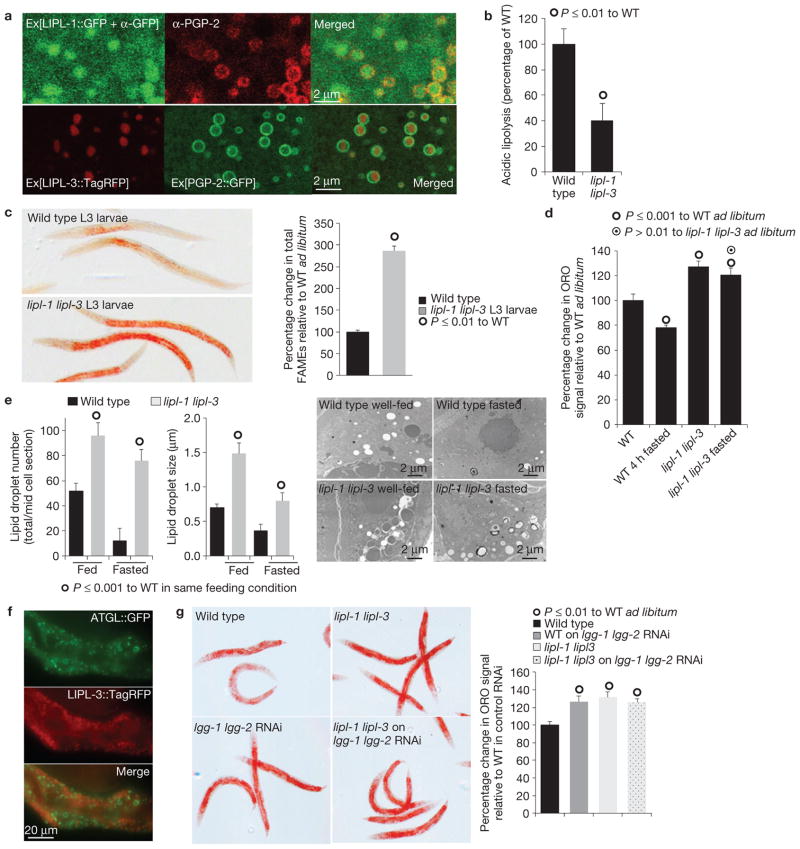
The lipl genes encode for uncharacterized triglyceride lipases with extensive sequence similarity to human lysosomal acid lipase (BLAST scores 9e-78 and 3e-75 for lipl-1 and lipl-3, respectively). The primary sequence of the fasting-responsive LIPL proteins contains predicted signals for lysosomal localization (http://golgi.unmc.edu/ptarget/), with confidence scores of 87%, 75%, 62% and 56% for LIPL-1, 3, 2 and 5, respectively. LIPL-4 was not predicted to be lysosomal9,10. We characterized the body distribution and subcellular localization of LIPL-1, 2 and 3. Translational fusions of lipl-1, 2 and 3 to either GFP or TagRFP (red fluorescent protein) reveal expression of these proteins in the lumen of the gut and/or vesicles within the intestine (Supplementary Fig. S2a). Confocal microscopy shows that LIPL-1 and 3 co-localize with the lysosomal marker PGP-2 (ref. 11; Fig. 2a). Furthermore, fractionation of whole lysates of worms expressing LIPL-3::TagRFP shows that both the acid lipase activity and LIPL-3::TagRFP co-fractionate with the canonical lysosomal enzyme acid phosphatase (Supplementary Fig. S2b), confirming that LIPL-1 and 3 localize to the lysosomal-related organelle of C. elegans. Moreover, lipl-1(tm1954) lipl-3(tm4498) double mutant worm lysates have reduced acid lipase activity (pH 4.5; Fig. 2b), but normal neutral lipase activity (Supplementary Fig. S2c), suggesting that LIPL-1 and 3 are only active in the acidic conditions that characterize the lysosomal lumen.
Figure 2.
LIPL-mediated lysosomal lipolysis controls lipid-droplet fat stores. (a) Representative confocal images of LIPL-1::GFP and LIPL-3::TagRFP localization relative to the lysosomal marker PGP-2 show that LIPL-1 and LIPL-3 are localized to the lysosomal-related organelle. (b) Acid lipase activity, measured in 1-day adult whole lysates, shows that lipl-1 lipl-3 double mutant animals have reduced acidic lipolytic capacity. Mean± s.d. are presented relative to wild type (WT); significant differences are indicated; n = 4 independent experiments. (c) Oil Red O (ORO) staining and fatty acid methyl ester (FAME) analyses of wild-type and lipl-1(tm1954) lipl-3(tm4498) double mutant L3 larvae show that lysosomal lipases regulate cytosolic fat stores (see Fig. 2d for adult measurements). Means ± s.e.m. are presented relative to wild type; significant differences are indicated; n = 3 independent experiments. (d) ORO quantification of wild-type and lipl-1(tm1954) lipl-3(tm4498) double mutant young adults fasted for 4 h shows that LIPL-1 and LIPL-3 contribute to fat mobilization following fasting. (Mean percentage ± s.e.m. relative to fed wild type). Wild-type fasted worms show 20% less ORO signal than animals fed ad libitum (P ≤ 0.001), whereas lipl-1(tm1954) lipl-3(tm4498) double mutant worms show 7% reduction in ORO signal (a difference that is not significant to well-fed fat levels at P ≤ 0.01 but it is significant at P ≤ 0.05). n = 3 independent experiments. (e) Representative transmission electron microscopy images of well-fed and 6 h-fasted wild-type and lipl-1 lipl-3 mutant animals (11,500×) show that the lipl mutants have more lipid-droplet stores in ad libitum fed and fasted conditions. Quantification of vesicle number and size is depicted as mean+ s.e.m., n = 4 independent experiments. (f) Representative images of well-fed transgenic animals expressing LIPL-3::TagRFP and ATGL::GFP shows that LIPL-3 does not localize to lipid droplets. (g) ORO staining and quantification of wild-type and lipl-1(tm1954) lipl-3(tm4498) double mutant young adults treated post-developmentally (from L4) with RNAi against the essential autophagy genes lgg-1 and lgg-2 or vector control show that lipl-1 lipl-3 and the autophagy genes lgg-1 lgg-2 are in the same fat regulatory pathway. (Mean percentage+ s.e.m. relative to wild type on vector control is depicted.) n = 4 independent experiments.
Lysosomal lipases have been proposed to break down lipid-droplet fats3. We investigated the impact of inactivating lipl-1 and lipl-3 on the accumulation of cytoplasmic fats. lipl-1 and lipl-3 double mutant larvae show threefold greater fat stores than wild-type larvae (Fig. 2c). A significant increase in fat signal in adult animals is also observed (Supplementary Fig. S3a). Furthermore, dual inactivation of lipl-1 and lipl-3 impaired fat utilization following fasting (Fig. 2d). Transmission electron microscopy confirms that lipl-1 lipl-3 double mutant animals contain while feeding, and retain during fasting, more and larger lipid droplets than wild-type animals (Fig. 2e and Supplementary Fig. S3b), further suggesting that lipl-1 and 3 break down lipids contained in lipid droplets. However, the LIPL lipases do not co-localize with lipid droplets (Fig. 2f).
In mammals, in basal and fasting conditions, autophagy delivers lipid-droplet lipids to the lysosome: a process termed lipophagy2. In C. elegans, post-developmental dual inactivation of the essential autophagy genes lgg-1 and lgg-2 (LC3 homologues) leads to increased fat accumulation (Fig. 2g), suggesting that lipophagy is conserved across metazoans. Lysosomal3 and autophagic4 lipases were proposed to break down lipid-droplet fats through lipophagy. Inhibition of autophagy by RNA interference (RNAi) of lgg-1 and lgg-2 in the lipl-1 lipl-3 double mutant animals does not lead to further increases in lipid-droplet fat stores (Fig. 2g), confirming that these lysosomal lipases and autophagy are part of the same fat regulatory pathway and suggesting that LIPL-1 and LIPL-3 are the enzymes breaking down lipids through lipophagy in C. elegans. By contrast, no changes in the levels of expression of lipid synthesis or β-oxidation genes were observed in lipl-1 lipl-3 double mutant animals (Supplementary Fig. S3c).
MXL-3 represses the lipl genes when food is available
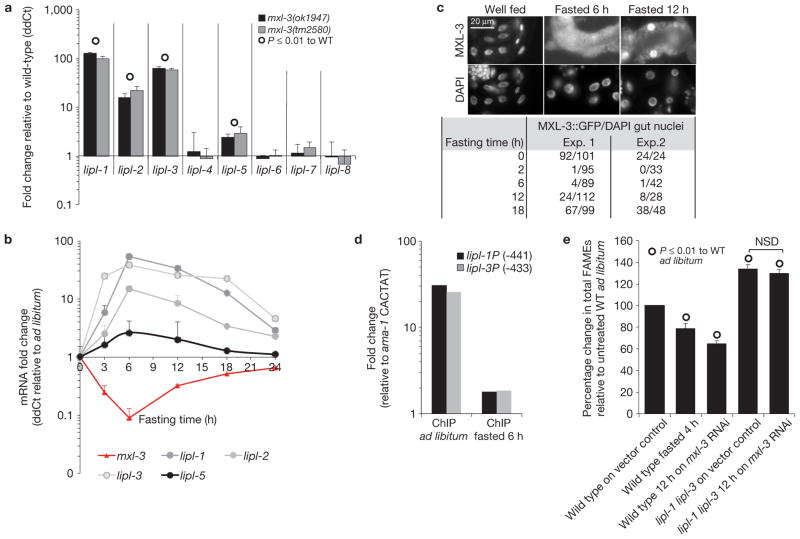
To identify the transcription factors that link lysosomal lipase messenger RNA levels to nutrient availability, a GFP transcriptional fusion to the lipl-1 promoter (lipl-1P::GFP) was used to screen an RNAi sublibrary containing 403 predicted C. elegans transcriptional regulators and 193 nuclear hormone receptors (Supplementary Table S3). Inactivation of the basic-helix–loop–helix transcription factor mxl-3 (Max-like 3) enabled lipl-1P::GFP expression under well-fed conditions. Gene expression analyses of two mxl-3 null mutants, mxl-3(ok1947) and mxl-3(tm2580), confirmed that MXL-3 represses lipl-1, 2, 3 and lipl-5, but not lipl-4, in well-fed animals (Fig. 3a). mxl-3 mRNA levels are also responsive to fasting, dropping by up to 15-fold after 5 h of fasting (Fig. 3b), suggesting that mxl-3 inactivation is part of a physiological response to fasting. Levels of mxl-3 transcripts return to 30% of those observed in well-fed animals after 12 h of fasting. Similarly, the lipl genes return to near basal levels after 18 h of fasting, suggesting that MXL-3 orchestrates a transient response to fasting. An MXL-3::GFP fusion protein, which rescues the mxl-3(ok1947) transcriptional phenotype, localizes to the nuclei of intestinal cells and sensory neurons (Supplementary Fig. S4a–c). Tissue-specific inactivation of mxl-3 in the gut, rather than neurons, triggered a fasting-like transcriptional response (Supplementary Fig. S4d). MXL-3::GFP expressed from a low-copy array localized to the nucleus of well-fed animals, but the signal disappeared from the nuclei after 2 h of fasting and remained undetectable during 6 h of fasting (Fig. 3c and Supplementary Fig. S4e). Twenty-two and 68% of intestinal cells showed nuclear-localized MXL-3::GFP after 12 and 18 h of fasting, respectively (Fig. 3c and Supplementary Fig. S4e). The transient regulation of MXL-3 suggests that the transcriptional changes coupled to inactivation of MXL-3 are part of an acute, rather than prolonged, response to nutritional deprivation. The observation that the lipl genes and mxl-3 have opposite patterns of expression and that they are both expressed in the intestinal cells supports the hypothesis that MXL-3 directly represses the lipl genes. Furthermore, MXL-3 binds in vitro to CACGTG (ref. 12), and this target motif is present within 500 base pairs (bp) of the transcriptional start sites of lipl-1, 2, 3 and 5, but not in the promoter region of the fasting-responsive gene lipl-4, whose transcription is mxl-3 independent. Also, MXL-3 binds and drives expression from the promoter regions of lipl-1, 2 and 3, but not lipl-4 in yeast one-hybrid experiments (Supplementary Fig. S5a). Chromatin immunoprecipitation (ChIP) of MXL-3::GFP shows that MXL-3 binds in vivo to the promoters of lipl-1 and 3 in well-fed animals and that MXL-3 occupancy of these promoters drops by between 5- and 30-fold in fasting worms (Fig. 3d and SI_ChromatinIP). In summary, MXL-3 acts as a nutritionally regulated transcriptional repressor of an acute response to fasting that includes induction of the lysosomal lipase genes lipl-1 and 3.
Figure 3.
MXL-3 represses lysosomal lipolysis in ad libitum-fed conditions. (a) Expression of lipl genes in well-fed mxl-3(ok1947) and mxl-3(tm2580) young adults normalized to same age well-fed wild-type (WT) worms shows that mxl-3 loss of function is sufficient to induce lipl-1 to 3 and lipl-5. Mean ddCts+ s.e.m. are depicted. n = 6 independent experiments for lipl-1 to lipl-5, and n = 4 independent experiments for lipl-6 to lipl-8. (b) L4 larvae were fasted and RNA was extracted at the indicated times. Mean ddCts+ s.e.m. show that the response mxl-3 orchestrates is transient. Time 0, n = 6 independent experiments; time 3–12 h, n = 4 independent experiments; time 18–24 h, n = 2 independent experiments. (c) Immunostainings of well-fed or 6 or 12 h fasted MXL-3::GFP young adults are presented. Quantification of GFP-positive nuclei relative to total intestinal nuclei (4,6-diamidino-2-phenylindole, DAPI) of two independent experiments shows that MXL-3 transiently delocalizes from the intestinal nuclei during fasting. (d) ChIP–quantitative PCR (ChIP-qPCR) analysis of well-fed and 6 h fasted mixed-stage worms expressing MXL-3::GFP presented as Ct in αGFP immunoprecipitated DNA normalized to input DNA and relative to a mock promoter region (CACTAT site −88 of ama-1 gene) shows that MXL-3 vacates the lipl promoters during early fasting. Three sets of primers surrounding CACGTG target sites found up to 500 bp of the ATG of the lipl-1 and lipl-3 genes were used. A representative experiment is presented; see raw data of two independent experiments in Supplementary Table S9. (e) Wild-type or lipl-1(tm1954) lipl-3(tm4498) double mutant animals grown on control RNAi plates were transferred as L4 larvae to control or mxl-3 RNAi plates, incubated for 12 h at 20 °C, and processed for total FAMEs; as a reference, an aliquot of wild-type young adults on control RNAi bacteria was fasted for 6 h. FAME quantification (mean percentage of fed wild type ±s.e.m.) shows that acute mxl-3 inactivation, as fasting, reduces fat stores in a lipl-dependent manner. n = 3 independent experiments. NSD, P-value > 0.05.
Acute inactivation of mxl-3 through RNAi leads to a depletion of fat stores that is comparable to the levels of fat consumption induced by fasting (Fig. 3e). Although it is possible that mxl-3 inactivation also affects β-oxidation, mxl-3 mutant animals show wild-type levels of oxygen consumption (Supplementary Fig. S5c). By contrast, the fast fat depletion induced by acute mxl-3 inactivation depends on the activity of the lipases LIPL-1 and 3 (Fig. 3e). Taken together, the data support the model that MXL-3 is a key molecular switch, whose inactivation turns on a fat-mobilizing program in response to fasting, and that the LIPL lipases are major players executing this metabolic program.
We then asked if certain well-characterized nutrient sensors of C. elegans regulated the expression of mxl-3. Inactivation of CeTOR (target of rapamycin) through RNAi or transforming growth factor (TGF-) β(daf-7(e1372)) did not affect mxl-3 expression (Supplementary Fig. S5d,e). On the other hand, impaired insulin signalling (daf-2(e1368)) leads to modest repression of mxl-3 transcription (50%) and induction of lipl-1 and lipl-3 (6 and 2-fold, respectively; Supplementary Fig. S5f), suggesting that reduced insulin signalling is only part of the regulatory cascade that links MXL-3 function to nutrients.
HLH-30 induces the lipl genes following fasting
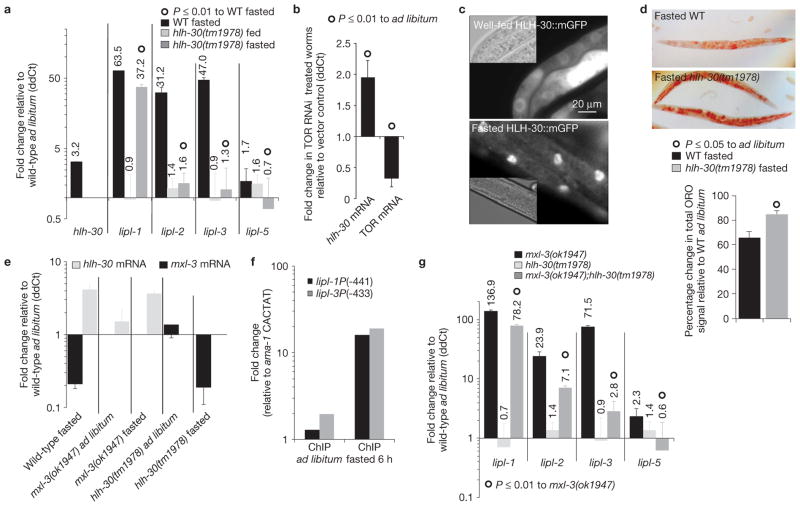
A candidate gene approach was used to identify transcription factors that activate the lipl genes in response to fasting. MXL-3 has the same binding site and shares target genes with HLH-30 (ref. 12), thus we investigated the role of hlh-30 in the activation of lysosomal lipolysis in fasting worms. In contrast to mxl-3, hlh-30 transcription is induced in fasting worms (Fig. 4a), and repressed following refeeding (Supplementary Fig. S6a). Inactivation of hlh-30 by a chromosomal mutation, allele tm1978, delays the activation of an hlh-30 transcriptional fusion to GFP (Supplementary Fig. S6b), suggesting that HLH-30 positively regulates its own transcription. Inactivation of CeTOR induces the expression of hlh-30 (Fig. 4b), revealing that mTOR controls hlh-30/CeTfeb at the transcriptional level. Following fasting, HLH-30 is enriched in the nuclei (Fig. 4c). In turn, hlh-30 induces lipl-2, 3 and 5 expression during fasting, and contributes to the induction of lipl-1 (Fig. 4a), suggesting that hlh-30 would control lipid mobilization following fasting. As expected, hlh-30 mutants show a reduced capacity to mobilize cytosolic lipids after food withdrawal (Fig. 4d). Mutational inactivation of hlh-30 does not affect either the basal transcription or the fasting-induced repression of mxl-3. Conversely, mutation of mxl-3 does not affect either the basal transcription or the fasting-triggered induction of hlh-30 (Fig. 4e). These findings show that mxl-3 and hlh-30 transcripts are independently regulated. Nevertheless, as analysis by ChIP shows that HLH-30 is rarely associated with the promoters of the lipase genes in well-fed animals (Fig. 4f and SI_ChromatinIP), even when the hlh-30 gene is expressed from a high copy number array at higher levels than those observed in fasting animals (Supplementary Fig. S6c), the coordinated relay from repression by MXL-3 to activation by HLH-30 seems critical for the proper expression of the lipase genes in response to nutrient availability. Consistent with this hypothesis, lipl-1 and 3 expression is only slightly increased in well-fed animals overproducing HLH-30 (Supplementary Fig. S6c). In contrast, during fasting, when MXL-3 is not bound to the lipase promoters, HLH-30 occupies the promoters of the lipase genes and induces expression of these targets (Fig. 4f and Supplementary Fig. S6c). Moreover, loss of hlh-30 function suppresses the constitutive induction of the lipase genes observed in mxl-3 mutants (Fig. 4f). Taken together, these findings show that mxl-3 and hlh-30 antagonistically regulate lysosomal lipase gene expression in response to nutrient availability. Finally, adaptive induction of the lipl genes in fasting animals is independent of the metabolic regulators NHR-49 and SIR-2.1 (Supplementary Fig. S6d,e).
Figure 4.
HLH-30 induces lysosomal lipolysis following fasting. (a) Transcriptional levels of hlh-30 and the mxl-3-dependent lipl genes measured in 5 h fasted wild-type (WT) or hlh-30(tm1978) young-adult worms depicted as mean ddCt + s.e.m. relative to wild-type ad libitum-fed worms show that hlh-30 is induced following fasting and HLH-30 induces lysosomal lipolysis. N = 4 independent experiments for lipl data, n = 2 independent experiments for hlh-30 mRNA. HLH-30 deficiency fully abrogates lipl-2, 3 and 5 (P < 0.0001), and impairs lipl-1 (P < 0.01) transcriptional activation following fasting. (b) Animals treated from late L3 stage with RNAi against TOR or vector control were harvested as young adults. Mean + s.e.m. of three independent experiments shows that inhibition of TOR is sufficient to induce hlh-30 transcription. (c) L3 animals carrying the rescuing construct hlh-30P::HLH-30::mGFP::hlh-30 3′UTR (mGFP, monomeric GFP, and UTR, untranslated region), well fed or fasted for 8 h show that HLH-30 is enriched in intestinal nuclei of fasted worms (exposure time: well fed, 500 ms; fasted, 100 ms). (d) ORO staining of wild-type or hlh-30(tm1978) young adults fasted for 8 h reveals that hlh-30 is required for optimal lipid mobilization on fasting. Mean percentages+s.e.m. are shown below relative to well-fed worms treated in parallel, n = 3 independent experiments. (e) mxl-3 and hlh-30 transcriptional levels in wild-type, hlh-30(tm1978) and mxl-3(ok1947) mutants in the basal and fasted states show that mxl-3 transcription is hlh-30 independent, and hlh-30 transcription is mxl-3 independent. Expression is presented relative to wild-type fed animals as mean ddCt + s.e.m. No significant differences relative to wild type in the same feeding state were observed, n = 3 independent experiments. (f) Representative ChIP-qPCR analysis of well-fed and 6 h fasted mixed stage worms expressing HLH-30::GFP presented as in Fig. 3d shows that HLH-30 occupies the lipl promoters during early fasting. See raw data of two independent experiments in Supplementary Table S9. (g) Transcriptional levels of the mxl-3-dependent lipl genes in wild-type, mxl-3(ok1947), hlh-30(tm1978) or mxl-3(ok1947);hlh-30(tm1978) double mutant young-adult worms show that hlh-30 suppresses the constitutive-induction-of-the-lipl-genes phenotype of mxl-3 mutant animals (mean ddCt + s.e.m. relative to wild-type ad libitum-fed worms). n = 3 independent experiments.
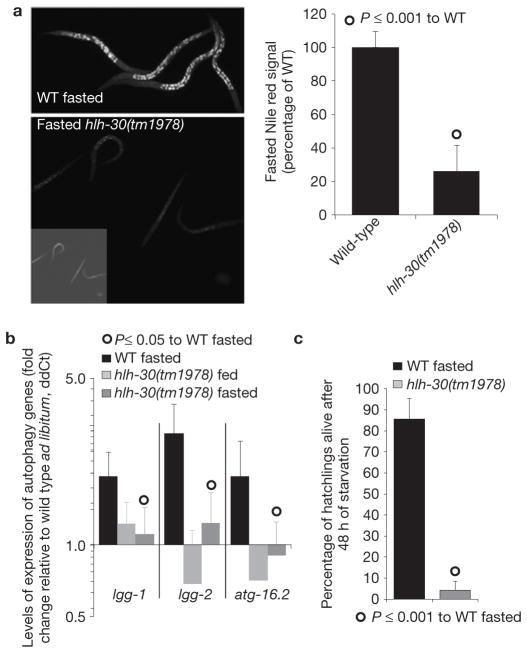
During fasting the C. elegans lysosomal compartment expands13. Inactivation of hlh-30 causes a reduction of the live Nile red-labelled lysosomal compartment in well-fed (ref. 14 and Supplementary Fig. S6f) and fasting conditions (Fig. 5a), suggesting that, as does its mammalian counterpart15,16, hlh-30 links lysosomal biogenesis to nutrient availability in C. elegans.
Figure 5.
HLH-30 activates vital cellular responses to starvation. (a) LRO (lysosome-related organelle) live Nile red-stained images of hlh-30(tm1978) and wild-type (WT) L3 larvae starved for 60 h show that hlh-30 is required for the expansion of the lysosomal compartment following fasting. Quantification as mean± s.e.m. relative to fasted wild-type worms is also presented; n = 3 independent experiments. (b) Transcriptional levels of the autophagy genes lgg-1, lgg-2 and atg-16.2 in fasted wild-type and hlh-30(tm1978) young-adult worms show that transcriptional activation of autophagy following fasting is hlh-30 dependent (mean ddCt + s.e.m. relative to wild-type ad libitum-fed worms). n = 3 independent experiments. (c) The mean percentage (+s.d.) of L1 larvae alive after 48 h fasting in minimum media shows that hlh-30 (tm1978) mutant animals are sensitive to starvation. n = 3 independent experiments. For L4 survival see Supplementary Fig. S6g.
Autophagy is also activated following fasting17, and we had observed that autophagy genes are transcriptionally upregulated in fasted animals, so we tested the role of hlh-30 in the induction of autophagy genes. Fasting hlh-30 mutants fail to activate the transcription of essential autophagy genes (Fig. 5b and Supplementary Table S4). The results support the model that HLH-30 coordinates the activation of lysosomal lipolysis and autophagy to meet the nutritional needs of the cells. Notably, hlh-30 mutant animals die prematurely in starvation conditions (Fig. 5c and Supplementary Fig. S6g), demonstrating that hlh-30 is required to mount an essential metabolic response to starvation.
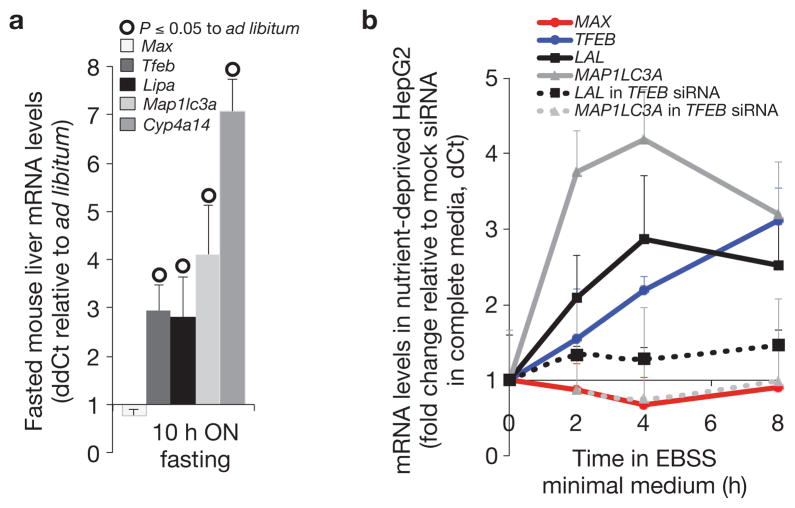
Essential metabolic responses to food deprivation are expected to be under strong selective pressure and consequently conserved. Supporting the view that lysosomal lipolysis is also regulated by nutrients in mammals, increased acid lipase activity was observed in liver lysates from fasted, compared with well-fed, C57L/B6 mice (Supplementary Fig. S7a). Furthermore, TFEB, LAL and the essential autophagy gene product LC3 were induced both in the liver of fasted C57L/B6 mice and in serum-deprived HepG2 cells (Fig. 6a,b), suggesting that lysosomal lipolysis and autophagy may be controlled by similar transcriptional mechanisms across metazoans. Confirming this hypothesis, siRNA inactivation of TFEB impairs the transcriptional activation of LAL and LC3 in nutrient-deprived HepG2 cells (Fig. 6b). In contrast, expression of the closest mammalian homologue of mxl-3, MAX, was not repressed in either fasted mice or serum-deprived hepatocytes (Fig. 6,b). Nonetheless, expression of mouse MAX in C. elegans rescued the constitutive induction of lipase genes in mxl-3 mutants (Supplementary Fig. S7b), suggesting that MAX, or a close member of the MAX family, could regulate energy homeostasis in higher organisms.
Figure 6.
Lysosomal lipolysis and autophagy are transcriptionally linked to nutrients in mammals. (a) Expression analyses of the liver of C57BL/6J 9-week females fasted overnight (ON) for 10 h relative to siblings feeding ad libitum show that LipA (mouse lysosomal acid lipase), Map1lc3a (mammalian lgg-1/lgg-2) and Tfeb (mammalian hlh-30) but not Max (mammalian mxl-3) are transcriptionally linked to nutrients in the mouse liver (median + s.e.m. fold change; ddCt). Levels of expression were normalized to ActB and Cog2 as internal controls; all but Max differences are significant (P < 0.05), n = 3 independent experiments. Cyp4a14 is a positive control. (b) Expression analyses of control or hTFEB siRNA treated HepG2 cells incubated in EBSS minimal medium for 2, 4 or 8 h, compared with the expression of control or hTFEB siRNA treated hepatocytes in complete media, show that LAL (human lysosomal acid lipase), MAP1LC3A and TFEB but not MAX are transcriptionally linked to nutrients in human hepatocytes, and that LAL and MAP1LC3A induction under nutrient deprivation are TFEB dependent (median dCt+ s.e.m.). Levels of expression were normalized to ACTB as internal control; all butMAX differences are significant (P < 0.05), n = 3 independent experiments. IGFBP is a positive control. EBSS, Earles’s balanced salt solution.
MXL-3 and HLH-30 regulate C. elegans ageing
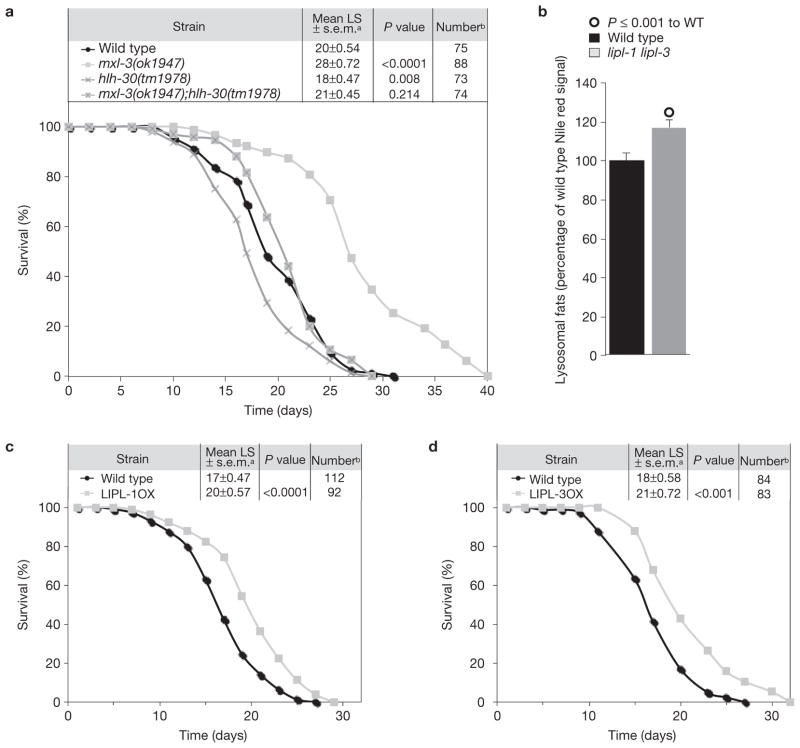
mxl-3 mutant animals show a constitutive fasting-like transcriptional profile, specifically, the constitutive activation of the lysosomal lipases. Similar to animals subjected to low nutrient intake, mxl-3 mutant worms are long lived in abundant food conditions (Fig. 7a). The mxl-3 gene is expressed in the intestine and AWC-sensory neurons (Supplementary Fig. S4). However, mxl-3 mutants undergo chemotaxis normally towards the AWC-specific odorant isoamyl alcohol (Supplementary Fig. S8a), suggesting that this mutant has an intact chemosensory system and that its extended lifespan phenotype is not due to impaired sensory abilities. By contrast, intestinal-specific expression of mxl-3 rescues the mxl-3 extended lifespan phenotype (Supplementary Table S5), suggesting a focus of action of MXL-3 for lifespan in this fat processing/storage organ. The mxl-3 mutant long-lived phenotype is additive to the lifespan extension derived from deficient germline stem cells (glp-1(e2141)), mitochondrial dysfunction (clk-1 RNAi) and a hypomorphic mutation in the insulin receptor (daf-2(e1368)), and it was not suppressed by inactivation of the transcription factors skn-1(zu67) or daf-16(mu86), which are required for lifespan extension due to insulin deficiency (Supplementary Table S6). Although skn-1 and daf-16 inactivation partially suppressed the extended lifespan phenotype of the mxl-3 mutant animals, they affect wild-type longevity to the same extent that they affect the lifespan of the mxl-3 mutant (20–40%), suggesting they are not required for the mxl-3 extended lifespan phenotype. The mxl-3 ageing phenotype was also additive to the eat-2 model of caloric restriction (eat-2(ad465)), and it was not suppressed by inactivation of the transcription factor pha-4 (RNAi), or rheb-1, a GTPase required for lifespan extension by intermittent fasting (Supplementary Table S7). Finally, the mxl-3 extended lifespan phenotype was additive to starvation-induced lifespan extension (Supplementary Table S7), suggesting that mxl-3 does not represent a genetic model of classical caloric-restriction or starvation-induced longevity.
Figure 7.
Lysosomal lipolysis delays C. elegans ageing. (a) Wild-type (WT), mxl-3(ok1947), hlh-30(tm1978) or mxl-3(ok1947);hlh-30(tm1978) double mutant were used in longevity epistasis analyses, revealing that hlh-30 suppresses the mxl-3 extended lifespan phenotype. Animals were incubated at 20 °C and transferred every other day until cessation of reproduction. The cumulative survival curve is depicted. Kaplan–Meier statistics, calculated using SSPS 17, are also shown. (b) Live Nile red quantification of wild-type and lipl-1 lipl-3 double mutant worms shows that lipl-1 and lipl-3 contribute to the clearance of lipids from the endocytic pathway. Mean + s.e.m. signal intensity is shown as percentage relative to wild type; significant differences are indicated; n = 4 independent experiments. (c,d) Animals overexpressing LIPL-1 or LIPL-3 were incubated at 25 °C and transferred every other day until cessation of reproduction. Data presented as in Fig. 7a show that activated lysosomal lipolysis extends C. elegans lifespan. a Survival presented as mean lifespan (LS) ± s.e.m. b Number of uncensored animals (animals that crawled off the plate, bagged, exploded or became contaminated were censored).
Conversely, hlh-30 mutants were modestly short lived (Fig. 7a) and loss of hlh-30 function suppressed the longevity phenotype of mxl-3 mutants (Fig. 7a). Targets of hlh-30 affect both lysosomal function and autophagy, two processes implicated in ageing18–20. The mxl-3 mutants did not show increased autophagy (Supplementary Fig. S8b). Consequently, it is unlikely that hlh-30 suppresses the mxl-3 extended lifespan phenotype by impairing autophagic function. In support of this idea, inactivation of the autophagy genes bec-1 (beclin1 orthologue) or atg-16.2, or the transcriptional regulator of autophagy pha-4, did not suppress the mxl-3 longevity phenotype (Supplementary Table S8). The mxl-3 mutant does not show activated autophagy, but exhibits activated lysosomal lipolysis, and inactivation of hlh-30 suppressed the induction of lysosomal lipases in mxl-3 mutants. These findings support the model that increased lysosomal lipolysis extends C. elegans lifespan. Efficient clearance of extra- and intracellular components delivered to the lysosome or passing through the endocytic pathway is required for normal ageing18. In addition to controlling cytosolic fat stores through lipophagy, the lysosomal lipases LIPL-1 and 3 promote fat mobilization through the endocytic pathway, as made evident by the increased vital Nile red staining observed in lipl-1 lipl-3 double mutants (Fig. 7b). In agreement with the hypothesis that efficient clearance of lipid moieties from the endosomal/lysosomal compartment slows ageing, overexpression of the lipl genes modestly increased C. elegans lifespan (Fig. 7c,d). The longevity data therefore suggest that increased lysosomal lipolysis enhances somatic endurance, possibly by improving cellular clearance, re-routing energy reserves for somatic maintenance, or some combination thereof.
DISCUSSION
Our data support the hypothesis that the C. elegans homologues of human LAL, the LIPL lipases, mediate the mobilization of cytosolic fats through lipophagy, and that active regulation of lipophagy is an ancient mechanism of energy homeostasis selected to adapt to fluctuations in food availability. Furthermore, we find that lysosomal lipolysis is tightly linked to nutrient availability through two metabolic regulators, MXL-3 and HLH-30 (Fig. 8). When food is available, MXL-3 represses the transcription of the lysosomal lipase genes. Following fasting, mxl-3 mRNA levels drop and the protein quickly disappears from the intestinal nuclei. Concomitantly, HLH-30 is translocated to the nucleus and the hlh-30 gene is transcriptionally upregulated, at least in part through an autoregulatory mechanism. This reinforces the activation of a fasting transcriptional program that leads to the transient induction of lysosomal lipolysis and autophagy, which, in conjunction with the canonical neutral lipases21,22, are required to use internal reserves of energy and survive starvation. Lipid consumption is most important and active in the initial phase of the metabolic response to food deprivation of C. elegans males23. Here we show that fertile hermaphrodite C. elegans transiently activate lysosomal lipolysis, providing a contributing mechanism to the rapid consumption of fat stores in adult worms.
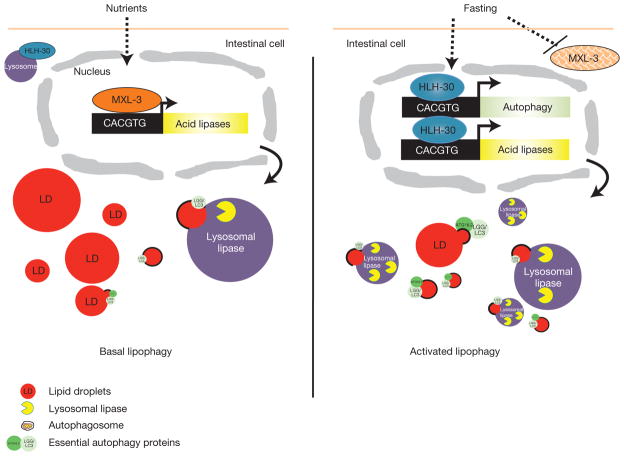
Figure 8.
MXL-3 and HLH-30 model of action. In the presence of nutrients, MXL-3 represses the expression of the lysosomal lipase genes, which contribute to breaking down fats through lipophagy. Upon fasting, MXL-3 is transcriptionally repressed and delocalizes from the nuclei, releasing the repression of the lysosomal lipases. Concomitantly, HLH-30 translocates from the cytoplasm to the nucleus and hlh-30 expression is induced, in part by an autoregulatory mechanism. In the nucleus, HLH-30 enables the activation of lipophagy through the induction of lysosomal lipase and autophagy genes. The activation of this transcriptional program enables utilization of internal reserves of energy, and promotes survival of starvation.
The transcriptional regulation mediated by MXL-3 and HLH-30 functions independently of the metabolic regulators NHR-49 (ref. 24) and SIR-2.1 (ref. 25). Also, although TOR regulates mammalian TFEB protein function26,27 and C. elegans hlh-30/CeTfeb transcription, the inactivation of CeTOR is insufficient to repress mxl-3. We show evidence suggesting that the repression of mxl-3 is required for the activation of its target genes, supporting the hypothesis that in C. elegans CeTOR inhibition is insufficient to fully activate this metabolic programme. CeTOR-independent insulin signalling seems to partially control mxl-3 expression. However, inhibition of insulin signalling is also insufficient to fully activate lysosomal lipolysis, supporting a model in which multiple nutrient-sensing pathways act upstream of lysosomal lipolysis.
The MXL-3-HLH-30 transcriptional circuit is required not only for proper nutrient mobilization in well-fed and fasted conditions but also for normal ageing. MXL-3 promotes ageing and, at least in the context of the mxl-3 mutant model, HLH-30 prevents ageing, suggesting that a transcriptional program selected to adapt to fluctuations in food availability regulates ageing when nutrients are available. We show that the mxl-3 longevity phenotype is additive to caloric restriction and starvation. Although at first surprising, this observation is in agreement with the fact that the response that mxl-3 orchestrates is transient. Fasting leads to the repression of mxl-3 and the activation of lysosomal lipolysis, but this response lasts only hours. In contrast, the mean lifespan of post-reproductively starved animals is 29 days. Consequently, it is unlikely that the transient activation of lipophagy would be a major contributor to starvation-induced lifespan extension. Alternatively, we propose that active clearance of lipid moieties from the endosomal/lysosomal compartment and/or signalling molecules derived from this transient lipid breakdown slow ageing in C. elegans.
In summary, we show that lysosomal lipolysis is an ancient mechanism of energy homeostasis and we present MXL-3 and HLH-30 as metabolic regulators that orchestrate a conserved and adaptive response to food deprivation, in addition to regulating ageing in conditions of food abundance. Finally, the conservation of the presented response suggests that malfunction of the mechanisms that link nutrients to lysosomal function may underlie metabolic disorders of as yet unknown aetiology and age-related disorders in higher organisms.
METHODS
Strains
N2 Bristol was used as the wild-type strain. The following mutant strains were used: rrf-3(pk1426), daf-2(e1368), daf-16(mu86), eat-2(ad465), skn-1(zu67), glp-1(e2141), lipl-1(tm1954), lipl-3(tm4498), mxl-3(ok1947), mxl-3(tm2580) and hlh-30(tm1978). For lipl transcriptional fusions, Is[lipl-1p::GFP-pest], Ex[lipl-2p::RFP-pest;myo-2p::GFP], Ex[lipl-3p::RFP-pest;myo-2p::GFP] and Ex[lipl-4p::GFP;myo-2p::RFP] were used. For LIPL overexpression, Ex[ges-1p::LIPL-1cDNA::SL2::GFP;myo-2p::RFP], Ex[ges-1p::LIPL-2 cDNA::SL2::GFP; myo-2p::RFP] and Ex[ges-1p::LIPL-3 cDNA::SL2::GFP; myo-2p::RFP] were used. ges-1 P driving lipl-2 and lipl-3 complementary DNAs were fused to TagRFP, and LIPL-1 to eGFP for subcellular localization experiments. LIPL-1::TagRFP is toxic, and LIPL-1::mGFP shows very dim signal. For tissue distribution, rescue analysis and ChIP of MXL-3, mxl-3p::MXL-3::mGFP;myo-2p::RFP was used. The vha-6 promoter was used to drive intestinal tissue-specific rescue of mxl-3. For mxl-3 tissue-specific knockdown analyses, lines were created following the strategy of ref. 29. Briefly, double stranded RNAi against mxl-3 was specifically generated in the gut or AWC neurons of the non-spreading sid-1(qt9) strain expressing the lipl-1 P::GFP transcriptional reporter. Is[lipl-1p::GFP-pest] was crossed into sid-1(qt9). Ex[ges-1p::Mxl-3IR::SL2::GFP;myo-2p::RFP], Ex[vha-6p::Mxl-3IR::SL2::GFP;myo-2p::RFP], Ex[odr-1p::Mxl-3IR::SL2::GFP;myo- 2p::RFP], Ex[odr-3p::Mxl-3IR::SL2::GFP;myo-2p::RFP] and Ex[mxl-3p::Mxl-3IR::SL2::GFP;myo-2p::RFP] were created by cloning the desired promoters in the AscI/Pme sites of pWormgatePro plasmids and then using the LR/BP recombinase Gateway system to introduce inverted repeat sequences of the mxl-3 open reading frame. The constructs were injected into sid-1(qt9) animals carrying lipl-1 P::GFP. For tissue distribution, rescue and ChIP analysis of HLH-30, hlh-30p::HLH-30cDNA::mGFP::hlh-30 3′UTR; myo-2p::RFP was used. DA2123 was used to assess autophagy levels; the transgene was crossed into mxl-3(ok1947) and hlh-30(tm1978). Strain VS20 was used in ATGL-1 localization analyses.
C. elegans fasting assay
Transcriptional analyses
Young adults (adult vulva with fewer than five eggs) were harvested, washed in a 35 μm nylon mesh, and seeded in empty NGM plates (fasting) or back onto OP50 plates (well-fed control). After the fasting period, treated and control worms were harvested, washed in a 35 μm nylon mesh, and quickly frozen in liquid nitrogen. qRT–PCR data presented are from at least three independent experiments and all values are normalized to ama-1 as internal control as well as to transcript levels in the untreated or wild-type animals.
Fat content assessment
Except for hlh-30(tm1978) experiments, hatchlings were synchronized for 48 h on S-basal, then seeded on OP50 plates, and incubated at 20 °C, unless otherwise stated. Animals were fasted for the indicated amounts of time and immediately treated for ORO staining13, fixed for electron microscopy, or frozen for later processing. FAMEs were extracted and measured as previously reported30. Equal numbers of worms were compared.
Starvation survival
Approximately 10,000 synchronized hatchlings obtained by egg preparation and incubation in minimal medium, or synchronized young L4s extensively washed through a 20 μm mesh, were followed for each strain. No difference in survival was observed between hlh-30(tm1978) and wild-type worms if eggs were seeded immediately after bleaching on food-containing plates, suggesting that hlh-30(tm1978) mutant worms are not hypersensitive to bleach. Hatchlings or larvae were resuspended in 10 ml S-basal to which 2.5 μl 5 mM Sytox Green (Molecular Probes) and 1 μl 10% Triton X-100 were added. The worm suspensions were maintained rocking at 20 °C. A minimum of 500 worms were run through a COPAS Biosort (Union Biometrica) at the indicated times. Animals with green fluorescent signal of 50 or over were dead, as previously established by lack of movement in a liquid drop. Percentage survival = (1 –(worms with green signal ≥ 50/total worms)) × 100.
Mammalian fasting assays
Mouse experiments
Five 9-week-old females were fasted for 10 h (8 pm–6 am), and five were fed ad libitum. Liver samples were extracted and immediately frozen in liquid nitrogen. RNA was extracted according to manufacturer’s recommendations (TriReagent, MBP). Cyp4a14 was used as positive control and actin B to normalize cDNA. Each experiment was repeated twice. Lipase activity from whole lysates was measured in a reaction mix as described below.
HepG2 experiments
HepG2 human hepatocytes were grown in IMDM complete medium supplemented with 10% heat-inactivated fetal calf serum. At 50% confluence, three plates were washed with PBS and the cells were incubated in serum-free Earle’s balanced salt solution, whereas another three plates were left in complete media as controls. At the indicated times cells from one complete medium and one salt solution plate were harvested and frozen in liquid nitrogen. RNA was extracted according to manufacturer ’s recommendations (TriReagent, MBP). Specific TaqMan probes were purchased from Applied Biosystems. insulin-like growth factor-binding protein (IGFBP) was used as a positive control, and actin B to normalize cDNA. Each experiment was repeated three times. TFEB was knocked down in HepG2 cells using human TFEB Stealth RNAi siRNAs (HSS111870, Invitrogen). Twenty-four hours after transfection using Lipofectamine RNAiMAX transfection reagent (Invitrogen), two plates of TFEB knocked-down cells and two plates of negative-control transfected cells (Stealth RNAi siRNA Negative Control) were harvested and split into a total of seven plates. Forty-eight hours after transfection, half the TFEB knocked-down and negative control cells were resuspended in EBSS. At the indicated times, cells from one TFEB knocked-down complete medium and one TFEB knocked-down EBSS, plus one negative control complete medium and one negative control EBSS plate, were harvested and processed for transcriptional analysis as described above. TFEB expression was tested by TaqMan PCR.
Lipase assay
Animals were resuspended in 300 μl of 200 mM sodium acetate buffer pH 4.5, and immediately frozen in liquid nitrogen. To assay, samples were sonicated and protein concentrations were measured. Reaction mix: 250 μg of total protein (up to 100 μl of lysate), 20 μl 2 M sodium acetate buffer at pH 4.5, 10 μl 4-methylumbelliferyl palmitate (4 mg ml−1), bring reaction volume to 200 μl with water. Fluorescence was read after 2 h at room temperature in a plate reader, excitation 355 nm–emission 460 nm.
Immunostaining
GFP was revealed with anti-GFP antibody (Roche, catalogue no 11814460001) and PGP-2 was revealed with anti-PGP-2 antibody kindly provided by G. Hermann11. TagRFP antibody was raised in house.
RNAi screen
An RNAi sublibrary containing 403 predicted transcriptional regulators and co-regulators and 193 nuclear hormone receptors (Supplementary Table S2) was built by cherry-picking clones from the Ahringer genome-wide and the Vidal RNAi library. The strain carrying the GFP transcriptional fusion to lipl-1 was used to screen for transcriptional regulators that affect transcriptional activation of lipolysis. RNAi bacteria were cultured for 12 h in Luria–Bertani medium with 100 μg ml−1 ampicillin, and then washed with S-basal and seeded onto RNAi agar plates containing 5 mM isopropyl-β-D-thiogalactoside (IPTG). The plates were left to dry in a laminar-flow hood and incubated at room temperature overnight to induce dsRNA expression. Synchronized Is[lipl-1p::GFP-pest; myo-2p::RFP] hatchlings were seeded on the plates. After 3 days of incubation at 20 °C, young adults were scored for GFP signal (animals fed empty vector control showed no signal or a dim signal limited to the tail in these conditions).
Yeast one-hybrid analyses
The ability of MXL-3 to bind to the promoters of the lipases was tested in the yeast one-hybrid system (Y1H), according to ref. 31 with some modifications. Briefly, lipl-1 to 4 promoters (3 kb upstream up to the ATG) were fused to β-galactosidase or HIS3 and integrated in YM4271. Two clones of each promoter that did not grow in 25 mM 3-aminotriazole (3AT) and showed a pale blue colour in the β-gal assay were transformed with only the Gal-4 activation domain or the activation domain fused to the MXL-3 open reading frame. Twenty-four random colonies from each transformation were picked and tested for survival in 3AT or β-gal signal. Expression of a transcriptional regulator able to bind to the tested promoters renders yeast resistant to 3AT and β-gal positive.
ChIP-qPCR
qPCR of DNA immunoprecipitated from well-fed or 6 h-fasted transgenic animals expressing MXL-3::GFP or HLH-30::GFP was carried out as previously described32. The MXL-3::GFP line used expresses 4.2± 0.38 times more mxl-3 than wild-type worms. The HLH-30::GFP line expresses 6.75 ± 0.91 times more hlh-30 than wild-type worms. Monoclonal mouse αGFP antibody from Roche (catalogue no 11814460001) was used for immunoprecipitation. Three sets of primers surrounding the CACGTG target sites up to 500 bp from the start site of lipl-1 or lipl-3 genes were used in qPCRs to compare the immunoprecipitated to input DNA. Fold change was calculated relative to the amplification obtained using two sets of primers surrounding the CACTAT sequence at −88 bp of the ama-1 start site from immunoprecipitated and input DNA. Primer sequences can be found in Supplementary Table S10.
Electron microscopy
Transmitted electron microscopy of osmium-, propylene oxide- and EPON-treated wild-type or mutant young adults was carried out in the Microscopy Core of the Center for Systems Biology/Program in Membrane Biology, which is partially supported by Inflammatory Bowel Disease Grant DK43351 and Boston Area Diabetes and Endocrinology Research Center (BADERC) Award DK57521.
LRO (lysosome-related organelle) content assessment
Animals were synchronized through a 2 h synchronous egg lay or by 24 h hatching at 20 °C in S-basal minimal media. Worms were seeded on NGM plates containing E. coli OP50 supplemented with 1 μM of the fluorescent fatty acid analogue C1-BODIPY 500/510-C12 (Invitrogen) or 25 ng ml−1 of Nile red. Imaging and quantification was conducted using an Axioplan microscope and Axiovision software (Zeiss). At least 25 animals were imaged in at least two independent experiments.
Lysosomal fractionation
Lysosomes were isolated from mixed stage LIPL-3::TagRFP animals using a LYSISO1 kit (Sigma) as previously described33. The lysosome enrichment was determined by measuring acid phosphatase activity (BioVision) normalized to protein concentration. LIPL-3::TagRFP was detected using α-TagRFP antibodies from Evrogen. Acid lipase activity was measured as described above.
Longevity assays
Synchronized day-1 adults were transferred to fresh NGM OP50 or RNAi plates every two days until no progeny was observed and then once a week for RNAi to keep the strength of the treatment. Two to six independent assays were carried out. Lifespan experiments were conducted at 20 °C, unless otherwise stated. Kaplan–Meier survival analysis was done using SSPS 17 software; significance was determined using log-rank statistics.
Statistical analyses
t-test analyses were carried out in all experiments with three or more biological replicates (independent experiments). For experiments with n = 2, representative data are presented. Log-rank statistics was used for the analysis of the longevity experiments as explained above.
Supplementary Material
Acknowledgments
We thank members of the Ruvkun, Ausubel and Kaplan laboratories for helpful comments, especially S. Curran, A. Lee-Conery and M. Wang for help with longevity experiments, J. Larkins-Ford for carrying out Biosort analyses, J. Xu for help with qRT–PCR experiments, J. Bai for acquiring confocal microscopy images, and J. Melo, C. Danna and A. Frand for helpful reading of the manuscript. We are grateful to R. Niedra, B. Seed, Y. Namiki and M. Oettinger for sharing expertise and reagents for mammalian experiments, and thanks H. Y. Mak, A. Soukas, M. Van Gilst, M. Freeman, A. Saghatelian and A. Tyler for protocols, access to equipment and discussions on lipid measurements. We are also grateful to A. Mah and D. Baillie for generating some transgenic strains used early in this project but not presented here, and thank N. Martinez and M. Walhout for sharing reagents and expertise on yeast one-hybrid experiments. We would like to thank the National Bioresource Project, the C. elegans Genetics Center, C. Bargmann and G. Hermann for strains. E.J.O’R. was a recipient of a Human Frontiers Science Program Postdoctoral fellowship. This work was supported by grants R01DK070147 to G.R. and K99DK087928 to E.J.O’R.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
E.J.O’R. designed the overall studies, carried out the experiments and wrote the manuscript. G.R. discussed results and revised the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at www.nature.com/reprints
References
- 1.Whitehead RH. A note on the absorption of fat. Am J Physiol. 1909;24:294–296. [Google Scholar]
- 2.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czaja MJ, Cuervo AM. Lipases in lysosomes, what for? Autophagy. 2009;5:866–867. doi: 10.4161/auto.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovsan J, Bashan N, Greenberg AS, Rudich A. Potential role of autophagy in modulation of lipid metabolism. Am J Physiol Endocrinol Metab. 2010;298:E1–E7. doi: 10.1152/ajpendo.00562.2009. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 6.Zinke I, Schutz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer M, et al. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol Genom. 2004;17:230–244. doi: 10.1152/physiolgenomics.00203.2003. [DOI] [PubMed] [Google Scholar]
- 8.Mallo GV, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 9.Guda C. pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 2006;34:W210–W213. doi: 10.1093/nar/gkl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guda C, Subramaniam S. pTARGET [corrected] a new method for predicting protein subcellular localization in eukaryotes. Bioinformatics. 2005;21:3963–3969. doi: 10.1093/bioinformatics/bti650. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder LK, et al. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol Biol Cell. 2007;18:995–1008. doi: 10.1091/mbc.E06-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grove CA, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009;10:430–435. doi: 10.1016/j.cmet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashrafi K, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 15.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 16.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 20.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- 22.Jo H, Shim J, Lee JH, Lee J, Kim JB. IRE-1 and HSP-4 contribute to energy homeostasis via fasting-induced lipases in C. elegans. Cell Metab. 2009;9:440–448. doi: 10.1016/j.cmet.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Tan KT, Luo SC, Ho WZ, Lee YH. Insulin/IGF-1 receptor signaling enhances biosynthetic activity and fat mobilization in the initial phase of starvation in adult male C. elegans. Cell Metab. 2011;14:390–402. doi: 10.1016/j.cmet.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci USA. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briese M, Esmaeili B, Johnson NM, Sattelle DB. pWormgatePro enables promoter-driven knockdown by hairpin RNA interference of muscle and neuronal gene products in Caenorhabditis elegans. Invert Neurosci. 2006;6:5–12. doi: 10.1007/s10158-005-0011-x. [DOI] [PubMed] [Google Scholar]
- 30.Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deplancke B, Dupuy D, Vidal M, Walhout AJ. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Du H, Rutkowski R, Gartner A, Wang X. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337:351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.