Morphological novelties arise through changes in development. The origin of the Chinese lantern phenotype or inflated calyx syndrome (ICS), a postfloral novelty in Physalis, was previously shown to be associated with the MADS box gene MPF2. This study reveals that molecular interactions between MPF3 (a euAP1 MADS box gene) and MPF2 and their products played crucial roles in the evolution of ICS.
Abstract
The Chinese lantern phenotype or inflated calyx syndrome (ICS) is a postfloral morphological novelty in Physalis. Its origin is associated with the heterotopic expression of the MADS box gene 2 from Physalis floridana (MPF2) in floral organs, yet the process underlying its identity remains elusive. Here, we show that MPF3, which is expressed specifically in floral tissues, encodes a core eudicot APETALA1-like (euAP1) MADS-domain protein. MPF3 was primarily localized to the nucleus, and it interacted with MPF2 and some floral MADS-domain proteins to selectively bind the CC-A-rich-GG (CArG) boxes in the MPF2 promoter. Downregulating MPF3 resulted in a dramatic elevation in MPF2 in the calyces and androecium, leading to enlarged and leaf-like floral calyces; however, the postfloral lantern was smaller and deformed. Starch accumulation in pollen was blocked. MPF3 MPF2 double knockdowns showed normal floral calyces and more mature pollen than those found in plants in which either MPF3 or MPF2 was downregulated. Therefore, MPF3 specifies calyx identity and regulates ICS formation and male fertility through interactions with MPF2/MPF2. Furthermore, both genes were found to activate Physalis floridana invertase gene 4 homolog, which encodes an invertase cleaving Suc, a putative key gene in sugar partitioning. The novel role of the MPF3-MPF2 regulatory circuit in male fertility is integral to the origin of ICS. Our results shed light on the evolution and development of ICS in Physalis and on the functional evolution of euAP1s in angiosperms.
INTRODUCTION
MADS box genes, as core transcriptional regulators, are associated with the evolution and development of the flower (Theissen and Melzer, 2007), which is the central identifying feature of angiosperms. Alterations in either spatial or temporal expression are involved in floral structural diversification and the origin of floral morphological novelties (Kanno et al., 2003; He et al., 2004; He and Saedler, 2005). Within Solanaceae, the genus Physalis features the Chinese lantern as a postfloral morphological novelty, while its close relative Solanum does not develop any such traits. The lantern is a derivative of the calyx, which inflates to a balloon-like structure encapsulating the berry after fertilization; thus, the Chinese lantern phenotype is also termed inflated calyx syndrome (ICS). Hormonal signals that appear to be released at fertilization trigger calyx inflation (He and Saedler, 2007). Further molecular genetic analyses revealed that the MADS box gene MPF2, an ortholog of AGAMOUS-LIKE24 (AGL24) in Arabidopsis thaliana, is heterotopically expressed in floral organs by the alteration of its own promoter, a key step associated with the evolutionary origin of ICS in Physalis (He and Saedler, 2005). The integration of MPF2 into floral development partially occurs via protein interactions with other floral MADS-domain proteins, such as MPF3 (encoded by MADS box gene 3 from Physalis floridana [MPF3]) and PFAG (encoded by the Arabidopsis AGAMOUS ortholog from P. floridana [PFAG]) (He et al., 2007). MPF2-specific downregulation via RNA interference (RNAi) revealed that MPF2 has two functions during floral development of Physalis: (1) expression of postfloral calyx inflation by regulating cell division and cell expansion and (2) regulation of male fertility (pollen development). Since unsuccessful fertilization usually leads to the abolition of ICS development, male fertility appears to be integral to the origin of ICS (He and Saedler, 2005). ICS is hypothesized to be the result of postfloral calyx inflation. However, the processes underlying the identity of the floral calyx and postfloral ICS are not yet understood.
These processes have been elucidated by investigations using model plant systems. Genetic analyses of floral mutants in Arabidopsis identified the underlying genetic regulation of floral organ identity as formulated in the ABC model (Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994): A-function alone specifies sepal identity; A- and B-functions together control petal identity; B- and C-functions together control stamen identity; and C-function alone specifies carpel identity. The SEPALLATA (SEP) genes, SEP1/2/3/4, termed E-function genes, are also required for the specification of organ identity (Pelaz et al., 2000; Theissen and Saedler, 2001; Ditta et al., 2004). These MADS-domain proteins regulate each other in a reciprocal fashion and form complexes so as to exert their regulatory roles (Honma and Goto, 2001; Ng and Yanofsky, 2001; Theissen and Saedler, 2001; Theissen and Melzer, 2007; Gregis et al., 2009). The first and only identified A-function MADS box gene is APETALA1 (AP1) in Arabidopsis (Mandel et al., 1992); however, the concept of A-function was challenged by the role of SQUAMOSA (SQUA) in Antirrhinum majus, which does not determine the sepal and petal organ identity (Schwarz-Sommer et al., 1990; Huijser et al., 1992). Accumulated evidence now shows that the A-function is not conserved in eudicots (Litt and Kramer, 2010), but that several AP1/SQUA homologs are involved in determining sepal identity within the core eudicots (Berbel et al., 2001; Taylor et al., 2002; Vrebalov et al., 2002; Benlloch et al., 2006). Furthermore, this function requires participation of other genes. In Arabidopsis, sepal organ identity has been well documented as being regulated by AP1, AGL24, and SEP (Mandel et al., 1992; Bowman et al., 1993; Ditta et al., 2004; Yu et al., 2004). In addition, the direct repression of AGL24 by AP1 is a crucial step in flower development (Yu et al., 2004). Thus, both AGL24 overexpressors and ap1 mutants feature an identical phenotypic variation with leaf-like sepals and indeterminate inflorescences (He et al., 2004; Yu et al., 2004). AGL24 in Arabidopsis is the ortholog of MPF2 in Physalis and of Solanum tuberosum MADS-box gene 16 (STMADS16) in Solanum (He and Saedler, 2005). Unlike STMADS16, MPF2 mRNA is heterotopically expressed in floral organs (∼10 to 20% of the leaf level). Heterotopic expression is crucial to the origin of ICS in Physalis (He and Saedler, 2005) and a hallmark for plants of Solanaceae family that feature ICS (Hu and Saedler, 2007). Further overexpression of either MPF2 or STMADS16 can lead to leaf-like sepals in transgenic Solanum plants (He and Saedler, 2007). Similarly, an AP1/SQUA-like transcription factor is assumed to repress STMADS16 or MPF2 in floral organs. As observed in Solanum, STMADS16 was expressed in vegetative tissues only; however, alterations in the MPF2 promoter itself led to leakage of its expression to floral organs in Physalis (He and Saedler, 2005).
Multiple copies of AP1/SQUA-like genes are known to have evolved in plant genomes, and they fall into the core eudicot APETALA1-like (euAP1), core eudicot FRUITFULL-like (euFUL), and FRUITFULL-like (FL) subclades (Ferrándiz et al., 2000; Litt and Irish, 2003; Preston and Kellogg, 2006; Litt, 2007; Shan et al., 2007). Only the euAP1 members have been shown to be involved in specifying sepal identity within the core eudicots (Mandel et al., 1992; Gustafson-Brown et al., 1994; Berbel et al., 2001; Taylor et al., 2002; Vrebalov et al., 2002; Benlloch et al., 2006). Since in Solanum, Le-MADS-MC regulates sepal organ identity (Vrebalov et al., 2002), we assumed that its putative ortholog MPF3 in Physalis described by He et al. (2004) might also specify calyx identity. MPF2 was shown to interact with MPF3 in yeast (He et al., 2007); therefore, we hypothesized that MPF3 interacts with MPF2/MPF2 genetically and physically to specify calyx identity and regulate ICS development. We conducted this study with three main objectives: namely, to determine (1) whether a regulatory circuit exists in Physalis to control floral calyx identity; (2) whether the postfloral calyx inflation, driven by the heterotopic expression of MPF2, results from the alteration of floral calyx identity; and (3) the role of MPF3 in the development of ICS.
We isolated four AP1/SQUA-like genes from Physalis. Phylogenetic and microsynteny analyses demonstrated that MPF3 is one of the euAP1 members that shares a similar expression pattern in floral development. Using two RNA silencing approaches, RNAi and virus-induced gene silencing (VIGS), we discovered the role of MPF3 in determining calyx identity and the size of the Chinese lantern structure. In addition, we showed that MPF3 plays a vital role in male fertility. These findings are supported by observations regarding MPF3, including its mRNA accumulation pattern, interacting protein spectrum, and its genetic regulation of MPF2. PFINV4 is a putative ortholog of INV4, which encodes an invertase that cleaves Suc, and is the key gene that regulates pollen maturation in rice (Oryza sativa) and lily (Lilium longiflorum) (Ranwala and Miller, 1998; Oliver et al., 2005). Genetic regulation of PFINV4 by MPF3 indicates a novel way for MADS box transcription factors to influence the development of male fertility. We showed that the development of ICS after fertilization in Physalis was not a result of altered floral calyx organ identity. Furthermore, we discuss neofunctionalization or functional diversification that accompanies the evolution of novel genetic and physical interactions within euAP1 genes of angiosperms.
RESULTS
Molecular Characterization of AP1/SQUA-Like Genes from Physalis
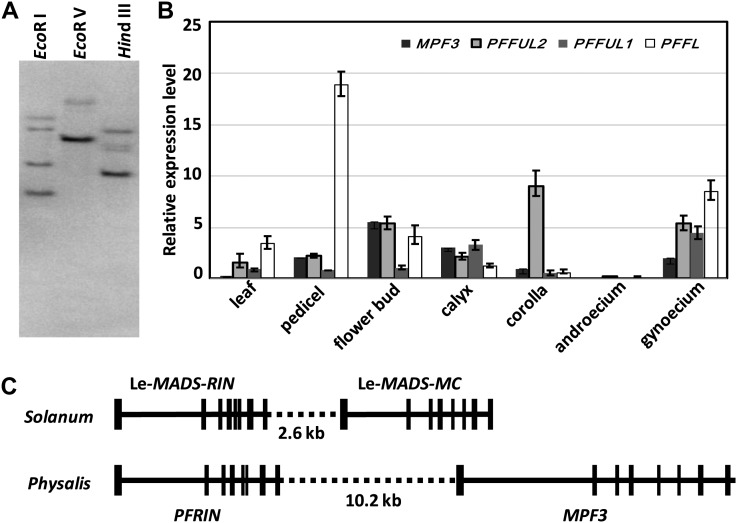
Multiple copies of AP1/SQUA-like genes exist in the genomes of all angiosperm species examined, and they share high sequence identities (Litt and Irish, 2003; Preston and Kellogg, 2006; Litt, 2007; Shan et al., 2007). An AP1/SQUA-like cDNA, named MPF3, was previously isolated from P. floridana (He et al., 2004). We performed a DNA gel blot analysis using an MPF3 cDNA fragment encoding the IKC domain as a probe to estimate the copy numbers of MPF3-like genes in P. floridana. The results suggest that four copies may exist in the genome of P. floridana (Figure 1A). In line with this, we obtained four AP1/SQUA-like cDNAs. Bayesian and maximum likelihood phylogenetic trees were reconstructed to show that the four genes belong to three subclades, and these genes were named MPF3 (euAP1), PFFUL1, PFFUL2 (euFUL), and PFFL (FL) (see Supplemental Figure 1 and Supplemental Data Set 1 online). Since the coding region of these paralogs had changed to the extent that the four copies were evolutionarily distinct, their expression patterns also might have become more diversified.
Figure 1.
Characterization of AP1/SQUA-Like Genes in Physalis.
(A) DNA gel blotting using the MPF3-specific cDNA fragment as a probe. Genomic DNA was digested using the restriction enzymes indicated.
(B) Expression of AP1/SQUA-like genes. For each sample, the genes from left to right are MPF3, PFFUL2, PFFUL1, and PFFL. Actin was used as an internal control. Experiments were performed on three independent biological samples. The average expression level and sd are presented.
(C) Local microsynteny of MPF3 and its ortholog Le-MADS-MC. Vertical lines indicate exons, and horizontal lines indicate promoter, intron, and intergenic sequences.
The mRNA expression patterns of AP1/SQUA-like genes in P. floridana were therefore measured using real-time RT-PCR. Total RNA was extracted from leaves, pedicels, flower buds, and the four whorls of mature flowers. As shown in Figure 1B, among these AP1/SQUA-like paralogs (MPF3, PFFUL1/2, and PFFL), only MPF3 was found not to be expressed in vegetative organs (leaves). Their transcripts were detected at different levels, predominantly in flower buds, pedicels, calyces, corolla, and gynoecia, of mature flowers. All transcripts were expressed in the androecium at extremely low levels or not at all. Consistent with the diverse expression patterns of AP1/SQUA-like genes (Litt, 2007), the four paralogs in P. floridana also evolved distinct expression patterns after duplication.
Local Synteny Provides Further Support That MPF3 Is an Ortholog of Le-MADS-MC
The phylogeny and expression analyses suggest that MPF3, as a euAP1, is a putative ortholog of Le-MADS-MC. However, additional evidence for its orthology comes from microsynteny analysis. In tomato (Solanum lycopersicum), Le-MADS-MC is physically linked to another MADS box gene, Le-MADS-RIN, in the genome (Vrebalov et al., 2002). If MPF3 is the true ortholog of Le-MADS-MC, MPF3 would also be linked to the Physalis ortholog of Le-MADS-RIN. In the Physalis floral transcriptome, the putative ortholog of Le-MADS-RIN was found and named PFRIN (Unigene119993_P106; see Supplemental Data Set 2 online), and the two shared 86.4% amino acid sequence identity. Next, we analyzed the linkage of MPF3 and PFRIN using the long-template PCR approach. First, we retrieved a 9763-bp fragment harboring the MPF3 locus and a 5725-bp fragment harboring the PFRIN locus. Both gene pairs share a conserved intron-exon structure between Solanum and Physalis (Figure 1C). We then amplified the linkage region (∼10.2 kb) between MPF3 and PFRIN. Overall, we constructed a 25.69-kb DNA fragment from the Physalis genome (Figure 1C). These data indicate that local microsynteny has been well maintained between the two genomes during evolution, which further substantiates the orthology of MPF3 and Le-MADS-MC, which share 91.5% amino acid sequence identity, suggesting that the role of MPF3 in flowers might be similar to that of Le-MADS-MC.
To elucidate the function of MPF3, we characterized the fine floral expression of the gene, the subcellular localization of the protein, and the interacting proteins in Physalis.
Fine Floral Expression Pattern of MPF3 in Physalis
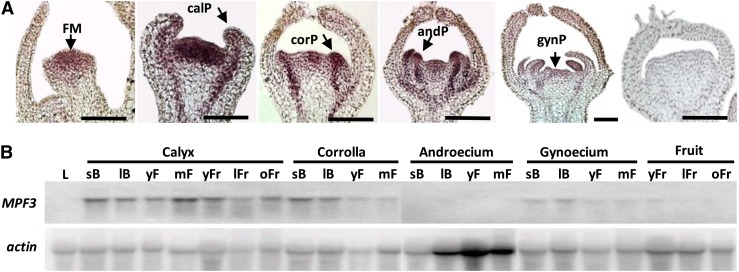
Real-time RT-PCR showed that MPF3 expression was restricted to floral organs (Figure 1B). To characterize MPF3 expression during floral development, we performed RNA gel blotting and mRNA in situ hybridization. To avoid possible cross-hybridization with other paralogs, a fragment spanning the most variable C-domain and the 3′-untranslated region of MPF3 was used as a MPF3-specific probe. Using mRNA in situ hybridization (Figure 2A), MPF3 transcripts were detected throughout the floral meristem. During floral organ initiation, MPF3 was found to be expressed in the primordia of the two outer whorls (calyx and corolla) and in the inner whorls (androecium and gynoecium). At the completion of flower bud formation, MPF3 expression was mainly observed in the outer whorl regions. The expression signal was also detected in vascular tissues of the pedicel during flower formation. These findings suggest that MPF3 may initiate floral meristems and specify floral organ identity. RNA gel blots showed that MPF3 mRNA was abundant in the calyx, corolla, and gynoecium from the time that floral buds appeared to anthesis, while no expression was detected in the androecium (Figure 2B). After fertilization, MPF3 was also detectable in developing fruits, albeit at a relatively low level. Consistent with the results obtained from real-time RT-PCR (Figure 1B), no MPF3 mRNA was detected in leaves (Figure 2B), thereby verifying the specificity of the MPF3 probe. This expression pattern strongly suggests a role for MPF3 throughout floral development, including the initiation of floral meristems, specification of floral organ identity, and growth of floral and postfloral organs.
Figure 2.
Expression of MPF3 during Flower Development in Physalis.
(A) MPF3 mRNA in situ hybridization in early floral development. The floral meristem (FM), primordia of calyx (calP), corolla (corP), androecium (andP), and gynoecium (gynP) are indicated by arrows. From left to right, MPF3-antisense was probed in the first five images, and MPF3-sense was probed in the last image. Bars = 50 μm.
(B) RNA gel blot analysis of MPF3 after flower formation. Total RNAs were isolated from the leaf (L), calyx, corolla, androecium, gynoecium, and fruit. The developmental stages were roughly determined as small buds (sB), large buds (lB), young flowers (yF), mature flowers (mF), young fruits (yFr), large fruits (lFr), and old fruits (oFr). Actin was reprobed after stripping off MPF3 and was used as a loading control.
MPF3 Is Primarily Localized to the Nucleus
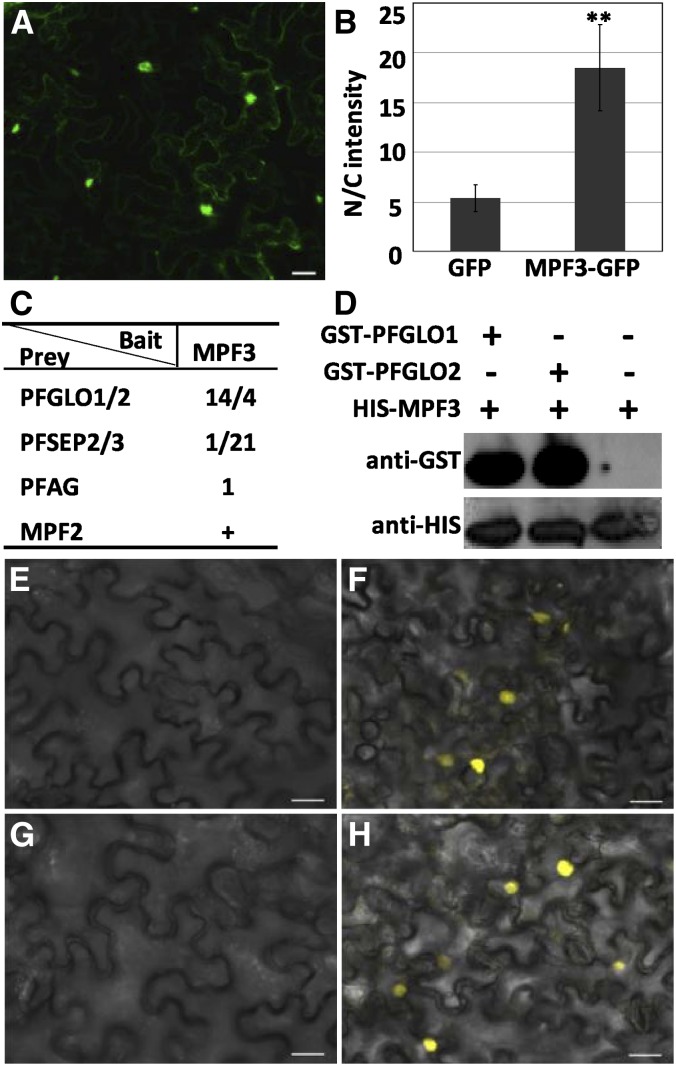
In addition to mRNA accumulation and distribution, the localization of a protein is another important aspect of gene activity. The precise subcellular localization of MPF3 would indicate whether or not this protein acts as a transcription factor. To test this, we linked the completed MPF3 open reading frame (ORF) with GFP (for green fluorescent protein) and infiltrated this construct via Agrobacterium tumefaciens into plant cells to synthesize a MPF3-GFP fusion protein. Cells transfected with GFP alone were used as a control. In our analyses, the GFP signal was detected in whole cells in both the MPF3-GFP fusion protein and control (Figures 3A and 3B). However, for the MPF3 fusion protein, the signal was primarily detected in the nucleus and only slightly in the cytoplasm (Figure 3A). We verified this by quantifying the relative GFP fluorescence intensity. In comparison to the GFP control, the nuclear-to-cytoplasmic ratio of the GFP signal in cells harboring the MPF3-GFP fusion increased significantly (P = 2.45 × 10−21; Figure 3B), indicating that, after translation, MPF3 was primarily transported to the nucleus. This localization suggests that MPF3, alone or via an interaction with other proteins, has a regulatory role in the nucleus.
Figure 3.
Subcellular Localization of MPF3 and Its Interacting Proteins.
(A) Subcellular localization of MPF3.
(B) The relative proportional fluorescence of the nucleus to the cytoplasm (N/C) between the GFP control and MPF3-GFP fusion. Thirty-two cells were analyzed in the GFP control and 20 in the MPF3-GFP fusion. The mean and sd are presented. Asterisks designate significance relative to the control (two-tailed Student’s t test, P = 2.45 × 10−21).
(C) Results of library screen using MPF3 as bait. The number stands for the independent colony that was obtained and sequenced. The previous observation of MPF3 interacting with MPF2 (He et al., 2007) is included and indicated by a plus symbol.
(D) Pull-down assay to confirm that MPF3 interacts with PFGLO1/2. The first two lanes are the results of heterodimer formation of the two proteins as indicated (see Methods). The last lane denotes His-MPF3 proteins loaded as control. Plus and minus symbols stand for inclusion or exclusion of the corresponding proteins, respectively.
(E) to (H) Results of BiFC analyses. The YFPn and YFPc empty vectors were coexpressed as a negative control (E). MPF3-MPF2 (F), MPF3-PFGLO1 (G), and MPF3-PFAG (H). A YFP signal suggests that the corresponding interaction occurs. As in (G), no YFP signal was detected in MPF3-PFGLO2. The results ([F] to [H]) are from the combination of MPF3-YFPn, and the indicated proteins were fused with YFPc at their C termini. Bars = 20 μm.
Characterization of MPF3′ MADS-Domain Interacting Proteins
MADS-domain proteins often form dimers and complexes that regulate transcription (Davies et al., 1996; Egea-Cortines et al., 1999; Honma and Goto, 2001). To identify which other MADS-domain proteins interact with MPF3, yeast two-hybrid analysis was performed. The Physalis expression library (He et al., 2007) was directly screened using MPF3 as bait, since it did not self-activate in yeast. Besides MPF2 (He et al., 2007), MPF3 was observed to interact with a series of floral MADS-domain proteins, including PFGLO1, PFGLO2, PFSEP2, PFSEP3, and PFAG (Figure 3C). PFGLO1 and PFGLO2 are homologous to GLOBOSA (GLO) (Tröbner et al., 1992); thus, they might serve as B-function MADS-domain proteins. PFSEP2 and PFSEP3 are E-function proteins and PFAG is a C-function protein (He et al., 2007). We verified the specific interactions between MPF3 (a euAP1 protein) and PFGLO1/2 (B-function proteins) using pull-down assays. Our results suggest that both PFGLO1 and PFGLO2 bind to MPF3 (Figure 3D), thus confirming the observations made in the yeast two-hybrid assays.
To verify these interactions in plant cells, bimolecular fluorescence complementation (BiFC) analyses using yellow fluorescent protein (YFP)-split technology were performed. The N-terminal (YFPn) and C-terminal (YFPc) halves of YFP were fused in frame to the C termini of the proteins examined, and all combinations of the two fusion proteins with either YFPn or YFPc were coexpressed in plant cells to assess whether or not they underwent heterodimerization. The YFPn and YFPc halves without the MADS-domain protein fusion were coexpressed as a negative control. Coexpression of MPF3-YFPn with MPF2-YFPc and with PFAG-YFPc yielded clear YFP signals exclusively in the nucleus (Figures 3F and 3H). This confirmed that MPF3 stably interacted with MPF2 and PFAG in the nucleus. Surprisingly, no visible YFP signal was detected using any combinations of MPF3 and PFGLO1/2 fusion protein with either YFPn or YFPc (Figure 3G). As expected, no signal was observed in the control (Figure 3E). The failure to detect YFP signals in the MPF3 and PFGLO1/2 interactions suggests that these interactions are weak or unstable in plant leaf cells. Accordingly, we quantified their interacting strengths in yeast and found that MPF3 interacted less strongly with PFGLO1 and PFGLO2 than with MPF2 and PFSEP3, as revealed by relatively low β-galactosidase activity (see Supplemental Figure 2A online). Nonetheless, MPF3 and PFAG had equivalent weak interacting strengths. In addition, all of these proteins formed homodimers (see Supplemental Figure 2B online).
The stable and transient interactions of MPF3 with these floral MADS-domain transcription factors further support the possibility that these proteins have roles in floral pathways in Physalis.
Transgenic Analyses Reveal Crucial Roles for MPF3 in Floral Development
As there are currently no mpf3 mutants available, we inferred the role of MPF3 using reverse genetic approaches in Physalis. Overexpression and RNAi strategies were used to infer the roles of MPF3 in P. floridana. Three MPF3 overexpression lines (OE1, OE2, and OE3) were generated (see Supplemental Table 1 online). Overexpression of MPF3 did not promote any visible morphological variations compared with wild-type Physalis. However, the timing of the floral phase transition was found to be altered. Wild-type Physalis flowered ∼30 d after germination, when the node number reached 8 to 9 in a growth chamber under long-day conditions, while the transgenic plants flowered ∼28 d after germination, with approximately six nodes under the same conditions (see Supplemental Table 1 online). This result deviated significantly from the wild type (P<4.2 × 10−5 for the time of first flower opening and P<1.59 × 10−10 for node number at flowering time), suggesting that MPF3 promoted flowering in Physalis.
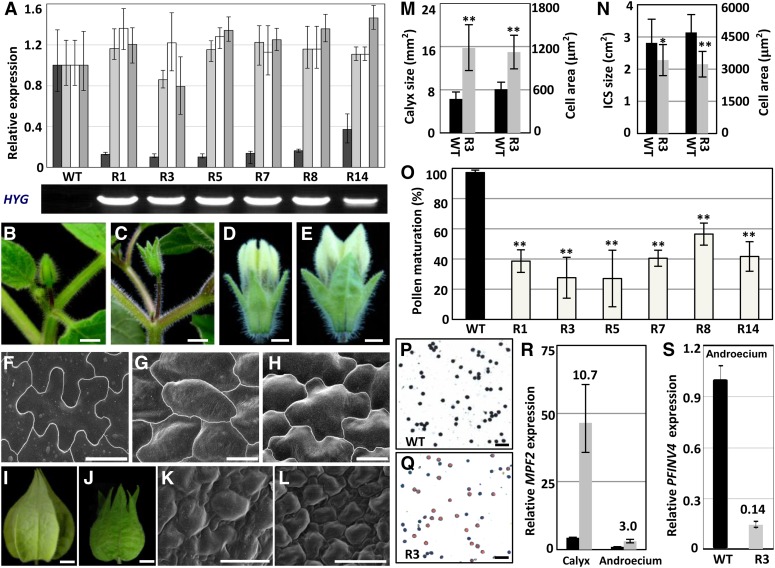
Six independent lines (R1, R3, R5, R7, R8, and R14) of MPF3-RNAi transgenic Physalis plants were generated and analyzed. Real-time RT-PCR analyses indicated that MPF3 mRNA was reduced by 60 to 90% in comparison to the wild type, while the other three homologs were not downregulated; yet, the level of PFFL mRNA appeared to be slightly elevated, indicating that MPF3 was specifically knocked down in these transgenic lines (Figure 4A). The phase transition of the transgenic plants was not affected (see Supplemental Table 1 online). In comparison with the wild type (Figure 4B), the solitary flowers with elongated pedicels were uniformly seen (Figure 4C), suggesting that floral meristem identity was not affected by the downregulation of MPF3. Nevertheless, dramatic floral phenotypic variations were observed (e.g., the morphology of the floral calyx was affected).
Figure 4.
Generation and Analyses of 35S:MPF3-RNAi Transgenic Physalis Plants.
(A) Genotyping analysis of 35S:MPF3-RNAi lines. The expression of MPF3 and its paralogs (PFFUL1/2 and PFFL) were compared between 35S:MPF3-RNAi lines (R1, R3, R5, R7, R8, and R14) and wild-type (WT) Physalis via real-time RT-PCR analysis. Expression of the genetic resistance to hygromycin (HYG) was monitored to confirm the identity of transgenic lines. Actin gene was used as an internal control. The experiments were repeated using three independent biological samples. Mean expression and sd are presented.
(B) and (C) Flower buds of the wild type (B) and R3 35S:MPF3-RNAi transgenic line (C). Bars = 5 mm.
(D) and (E) Floral calyx of the wild type (D) and R3 35S:MPF3-RNAi transgenic line (E). Bars = 1 mm.
(F) Epidermal cells of the leaf. Bar = 20 μm.
(G) and (H) Calyx epidermal cells of the wild type (G) and line R3 (H). Bars = 20 μm.
(I) and (J) Mature ICS of the wild type (I) and line R3 (J). Bar = 5 mm.
(K) and (L) Epidermal cells of the ICS from the wild type (K) and line R3 (L). Bars = 20 μm.
(M) Size of the calyx surface and epidermal cells of the floral calyx in the wild type and line R3. The number of cells analyzed was 20 and 18 in the wild type and R3, respectively. The number of calyces analyzed was 50 and 25 in the wild type and R3, respectively. Error bar is sd.
(N) Surface area of the ICS and its epidermal cell size in the wild type and line R3. The number of cells analyzed was 20 for both the wild type and R3. The number of calyces analyzed was 46 and 60 in the wild type and R3, respectively. Error bar is sd.
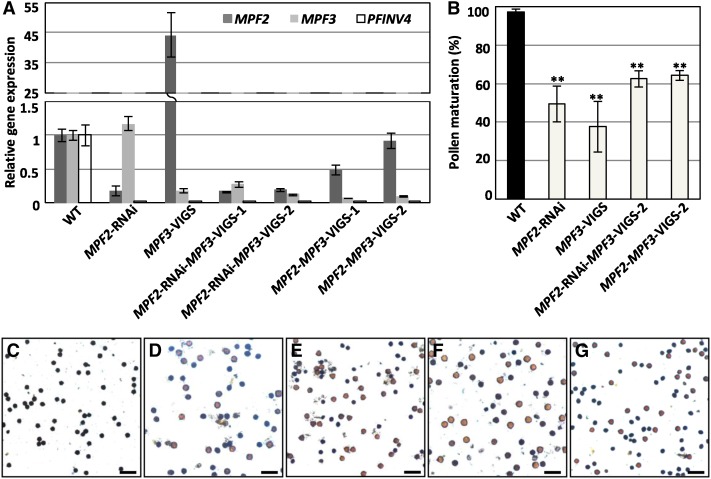
(O) Reduction of pollen maturation of 35S:MPF3-RNAi transgenic lines. For each line, 10 flowers were evaluated. The mean and sd are presented.
(P) and (Q) I2-KI stained pollen from the wild type (P) and 35S:MPF3-RNAi transgenic line R3 (Q). Active pollen is blue, and sterile pollen is tawny. Bars = 100 μm.
(R) and (S) Expression of MPF2 in the calyx and androecium was elevated in the MPF3-RNAi line (R), and the expression of PFINV4 in the androecium was repressed in the MPF3-RNAi line (S). Expression of these genes in 35S:MPF3-RNAi-R3 (gray column) was compared with those in the wild type (black column). The gene expression in the androecium of the wild type was taken as 1, and Actin was used as an internal control. The experiments were repeated using three independent biological samples. Mean expression and sd are presented.
Compared with the calyces of the wild type (Figures 4B and 4D), leaf-like calyces were observed throughout the entire spectrum from young flower buds to mature flowers in the MPF3-RNAi transgenic Physalis lines (i.e., R3; Figures 4C and 4E). The epidermal cells of the floral calyx were subjected to scanning electron microscopy analysis. Prior to anthesis, the calyx cells of wild-type Physalis were usually small and undifferentiated (Figure 4G; Hu and Saedler, 2007). The cells were enlarged, and cellular morphology was transformed into shape typical of leaf cells (strongly lobate) in the floral calyx of MPF3-RNAi mutants (Figure 4H), resembling leaf cells (Figure 4F). Therefore, knocking down of MPF3 caused a homeotic transformation of the floral calyx into a leaf-like structure by altering cell identity, and this was accompanied by a significant increase in cell size (P = 4.26 × 10−10; Figure 4M). As a result, a significant enlargement of the floral calyx (about twice the size of the wild-type calyx, P = 1.63 × 10−14) was seen in mature flowers (Figures 4E and 4M). However, the final size of the transformed ICS was significantly (P = 0.04) reduced and the morphology deviated from the wild-type ICS (Figures 4I, 4J, and 4N). Surprisingly, in comparison with the wild-type ICS (Figures 4K and 4N), the cell size was significantly (P = 1.32 × 10−7) reduced in the deformed ICS of MPF3-RNAi line R3 (Figures 4L and 4N). In both developmental stages, variation in cell size could not completely account for the variation in organ size (see Supplemental Table 2 online), suggesting that cell number also increased. These findings indicate that MPF3 regulates cell identity and, thus, the calyx identity prior to anthesis. Moreover, the manner in which MPF3 modulates cell division and expansion was found to be dependent on the organ contexts. MPF3 is mainly a negative regulator of cell division in both floral and postfloral calyx development. Cell expansion is also negatively regulated in the floral calyx, while it is positively regulated in the fruiting calyx.
The reproductive organs appeared to be normal; however, the fertility of the transgenic plants was poor, as indicated by a low number of berries produced in the absence of artificial pollination. However, pollination with wild-type pollen led to normal fruit development, indicating that the impaired fertility was primarily due to defects in the male organs. Pollen development was first evaluated by iodine-potassium iodide (I2-KI) staining. Normally, developed pollen stains blue, but we observed that more than 40% of pollen grains did not stain at all in these transgenic plants, a significant deviation from the wild type (P < 2.20 × 10−6; Figure 4O). One representative sample depicts pollen I2-KI staining of R3 (27.6% ± 13.5%; Figure 4Q) compared with that of the wild type (97.3% ± 2.8%, Figure 4P). In the MPF3-RNAi line, the basic structure of the stamen was evaluated further; however, we did not find any abnormality in stamen development (see Supplemental Figure 3 online). Therefore, downregulation of MPF3 may directly or indirectly affect sugar translocation to the pollen instead of affecting the stamen structure, thus leading to immature pollen grains.
These transgenic analyses suggest that MPF3 is a multifunctional protein in Physalis. It may either play a redundant role or a small role in specifying floral meristem identity so as to promote flowering, and it regulates calyx identity and ICS size. Moreover, our findings suggest that MPF3 has acquired a novel role in male fertility. As a transcription factor, MPF3 might repress or activate downstream genes so as to fulfill its biological functions. We studied this by comparing gene expression between wild-type and MPF3-knockdown lines.
Dramatic Repression of MPF2 and Marked Activation of PFINV4 by MPF3
MPF2 encodes the key regulator of ICS and male fertility. Knockdown of MPF2 leads to a small ICS (in extreme cases no ICS develops) and lower pollen yields (He and Saedler, 2005), and overexpression of MPF2 causes leaf-like sepals in transgenic Solanum (He and Saedler, 2007). These aspects were also affected in MPF3 knockdown lines. Therefore, the expression of MPF2 was first investigated in the 35S:MPF3-RNAi transgenic plants. The calyx and androecium of mature flowers were collected, and the RNA was subjected to real-time RT-PCR. MPF2 mRNA accumulation was elevated ∼11-fold in the transformed calyx and threefold in the androecium of R3, in comparison to each respective organ of the wild type (Figure 4R). These data indicate that MPF3, through direct or indirect genetic interaction with MPF2, play an important role in the development of the calyx and, subsequently, in male fertility.
To characterize the pollen defect in MPF3 knockdowns, we also evaluated the genes involved in sugar-partitioning pathways. The Suc transporter genes (SUT1 and SUT2), monosaccharide transporter gene (MST8), the carbon starved anther gene (CSA) for a MYB domain protein, and the anther cell wall invertase gene (INV4) all play essential roles in sugar-partitioning pathways in Oryza, Solanum, and Lilium (Ranwala and Miller, 1998; Leggewie et al., 2003; Oliver et al., 2005; Büttner, 2007; Zhang et al., 2010). We first searched for closely related homologs in the Physalis floral transcriptome (see Supplemental Data Set 2 online) and retrieved PFSUT1 (Unigene54711_P106), PFSUT2 (Unigene13767_P106), PFMST8 (Unigene17675_P106), PFCSA (Unigene38938_P106), and PFINV4 (Unigene70960_P106). These genes are thought to exert similar roles in Physalis, since pollen maturation was inhibited in MPF3 knockdowns. RT-PCR demonstrated that wild-type PFINV4 was mainly expressed in the androecium and PFCSA was specifically expressed in the androecium, while other genes were expressed in both leaves and the androecium (see Supplemental Figure 4 online). Furthermore, we found that PFINV4 was specifically repressed in the MPF3-RNAi lines; however, the other genes were not altered. INV4, a key gene regulating sugar partitioning in the Suc sequestration phloem-unloading pathway, encodes an anther-specific invertase for cleaving sucrose (Ranwala and Miller, 1998). Its repression is correlated with Suc accumulation and pollen sterility in rice (Oliver et al., 2005). The expression of PFINV4 was further monitored in the androecium of the MPF3-RNAi line R3. Our results showed that PFINV4 mRNA accumulation was significantly reduced (to only 14.0% of wild-type levels) in the organs of R3 (Figure 4S). These data suggest that MPF3 regulates pollen maturation, either directly or indirectly, by activating PFINV4, and this may regulate sugar partitioning in Physalis.
Knockdown of MPF3 via a VIGS Approach
There was pronounced transgene silencing in the case of 35S:MPF3-RNAi. In the T3 generation, the effect of MPF3 silencing was completely ablated. Generation of new double-gene RNAi plants was not feasible, since the procedure is time-consuming. To unravel the role of MPF3 in flower development through genetic interaction with MPF2, which is a key regulator of ICS development and pollen maturation (He and Saedler, 2005), we exploited the VIGS approach. The MPF3-specific fragment used in our RNAi analysis was introduced into the tobacco rattle virus (TRV) system (Liu et al., 2002) to generate a MPF3-VIGS construct. Infection of Physalis seedlings resulted in MPF3 downregulated mutated flowers whose phenotypic variation resembled that of the 35S:MPF3-RNAi lines. In comparison with the wild type (see Supplemental Figures 5A to 5C online), the floral calyx became leaf like, a smaller deformed ICS developed, and the pedicel was elongated (see Supplemental Figures 5D to 5F online). These alterations in flower morphology and size phenocopied those observed in 35S:MPF3-RNAi transgenic plants, suggesting that TRV-mediated VIGS is applicable for gene silencing in Physalis. Total mRNA from three flowers with dramatic phenotypic deviation from the wild type was subjected to real-time RT-PCR. Although an elevation in PFFL was observed (similar to that in MPF3-RNAi lines), transcript abundance of MPF3 in these mutated flowers was efficiently knocked down, and the lowest residual level was 4.1% that of the wild type (see Supplemental Figure 5G online), corroborating the observed phenotypic variations. In these MPF3-VIGS flowers, the expression of MPF2 in the calyx and androecium was significantly increased, particularly in the calyx (44 times that of the wild type; see Supplemental Figure 5H online). PFINV4 expression in the androecium was dramatically decreased to 0.5% of that of the wild type (see Supplemental Figure 5I online); this knockdown approached that of a PFINV4 null-expression mutant, reflecting the existence of genetic interactions among MPF3 and MPF2/PFINV4. Concomitantly, we downregulated the Physalis phytoene desaturase gene (PFPDS) in an independent VIGS experiment as a control. We observed specific organ bleaching effects as observed in tomato when a phytoene desaturase gene was silenced (Liu et al., 2002), but we did not observe pollen defects and alterations in PFINV4 expression (see Supplemental Figure 6 online), indicating the specificity of pollen defects in the manipulated genes. We therefore used a VIGS approach to confirm the genetic interactions between MPF3 and MPF2.
Genetic Interactions of MPF3 and MPF2 in Flower Development
To elucidate the genetic interactions between MPF3 and MPF2, double gene-silencing mutants of the two genes were created. We took advantage of the TRV-VIGS approach to silence MPF3 in the KO38 line of MPF2-RNAi (He and Saedler, 2005) to generate MPF2-RNAi-MPF3-VIGS flowers. Concomitantly, we silenced both MPF2 and MPF3 in the wild type through VIGS to produce MPF2-MPF3-VIGS flowers. We genotyped the mutated flowers using real-time RT-PCR (Figure 5A). In MPF3 single downregulated mutants, the MPF2 levels were dramatically elevated, while the expression of MPF3 was not affected in flowers carrying a single MPF2 mutation, indicating that MPF2 is a downstream target of MPF3. Unlike their single silencing mutants, MPF3 and MPF2 expression levels were dramatically downregulated in the double mutants (Figure 5A). We then thoroughly characterized the silencing effects of the two genes on floral phenotypic variations.
Figure 5.
The Role of Genetic Interactions between MPF3 and MPF2 in Determining Male Fertility.
(A) Genotyping analysis. Expression of MPF3 and MPF2 in calyces and PFINV4 in the androecium was revealed via real-time RT-PCR. The experiments were performed in biological triplicate. Actin was used as an internal control. Mean expression and sd are presented. WT, the wild type.
(B) Pollen maturation rate. For each genotype, 10 flowers were evaluated. The mean and sd are presented. Double asterisks indicate P < 0.01 in two-tailed Student’s t test.
(C) to (G) I2-KI–stained pollen from the wild type (C) and MPF2 and/or MPF3 downregulated flowers, as indicated ([D] to [G]). Active pollen is blue, and sterile pollen is yellowish-brown. Bars = 100 μm.
Genetic Interaction between MPF3 and MPF2 Is Essential to Male Fertility
Male fertility is a primary role of a flower. A dramatic downregulation of MPF2 blocks pollen production (He and Saedler, 2005). In the progeny of the KO38 line, some pollen grains were produced due to transgene silencing. However, in comparison with pollen maturation in the wild type (97.3% ± 2.8%; Figures 5B and 5C), only 49.4% ± 9.3% of pollen grains were found to be mature as revealed by I2-KI staining (Figures 5B and 5D), suggesting that pollen maturation was significantly inhibited (P = 6.65 × 10−12 ) by MPF2 downregulation.
Once MPF3 was silenced via the VIGS approach, 37.5% ± 13.2% of pollen reached maturity (Figures 5B and 5E), which is similar to the effect observed in 35S:MPF3-RNAi lines (Figure 4O), indicating that pollen maturation processes were also significantly blocked (P = 4.04 × 10−11) by the downregulation of MPF3. Nonetheless, in MPF3 single downregulated mutants, MPF2 was upregulated, hinting that poor male fertility might also result from a higher dosage of MPF2 in the androecium. To verify the cause of immature pollen in MPF3 single downregulated mutants, we assessed pollen maturation in MPF2 and MPF3 double downregulated flowers. We found that although the abnormality was alleviated (with ∼65% of pollen reaching maturity; Figures 5B, 5F, and 5G), it was still significantly lower than in the wild type (P<2.57 × 10−14). These findings suggest that both MPF3 and MPF2 directly regulate male fertility.
In all of these cases, stamen development was normal (see Supplemental Figure 3 online). However, PFINV4 was markedly downregulated when either MPF2 or MPF3 was silenced (Figure 5A; see Supplemental Figure 4 online). Thus, abnormalities in male fertility in these mutant flowers may have resulted from a dramatic suppression of PFINV4. These findings suggest that activation of PFINV4 by interactions between MPF3 and MPF2/MPF2 are essential for pollen maturation.
MPF3 Interacts Genetically with MPF2 to Specify the Identity of the Floral Calyx
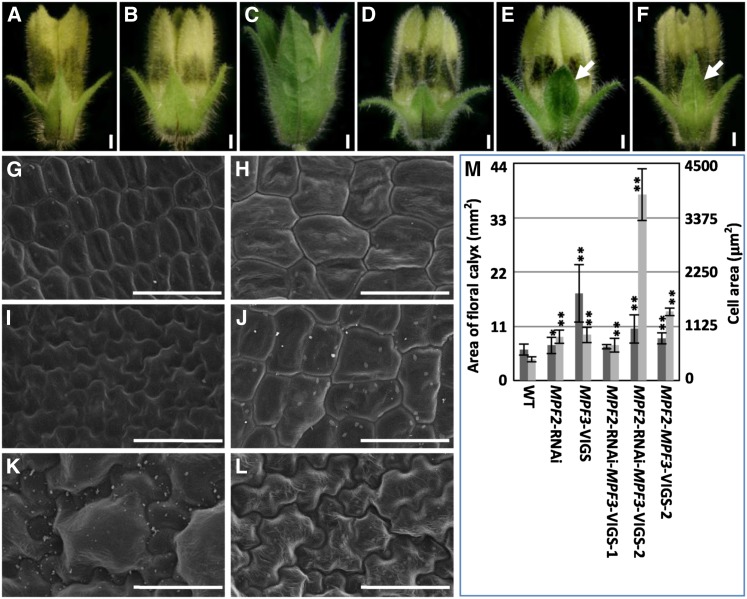
Compared with the wild-type floral calyx (Figure 6A), knocking down of MPF2 did not alter calyx identity (Figure 6B; He and Saedler, 2005). A dramatic downregulation of MPF3 significantly altered the morphology of the calyx, which became more leaf-like (Figures 4 and 6C; see Supplemental Figure 5 online). In these cases, MPF2 was upregulated in the floral calyx (Figures 4R and 5A; see Supplemental Figure 5H online). In our previous work, we discovered that overexpression of MPF2 led to leaf-like calyces in Solanum (He and Saedler, 2005); therefore, we assumed that MPF2 was a key regulator of calyx organ identity. This assumption was validated in our double gene silencing analysis.
Figure 6.
Both MPF3 and MPF2 Regulate Floral Calyx Identity.
(A) to (F) Phenotypic variation of the floral calyx. The white arrows indicate the leaf-like tip of calyces. Bars = 1 mm.
(A) The wild type.
(B) MPF2-RNAi.
(C) MPF3-VIGS.
(D) MPF2-RNAi-MPF3-VIGS-1.
(E) MPF2-RNAi-MPF3-VIGS-2.
(F) MPF2-MPF3-VIGS-2.
(G) to (L) Cell morphology of the floral calyx of the aforementioned phenotypes in (A) to (F), respectively. Bars = 50 µm.
(M) Quantification of calyx size and the size of its respective cells (for details, see Supplemental Table 2 online). The genotype of each sample is referred to in Figure 5A.
Unlike the mutated flowers seen upon silencing of just MPF3 (Figures 4 and 6C; see Supplemental Figure 5 online), we observed either no obvious alterations in calyx organ identity in the double gene downregulated flowers (i.e., MPF2-RNAi-MPF3-VIGS-1 flowers; Figure 6D) or only slight leaf-like structures in the tips of the floral calyces (as seen in MPF2-RNAi-MPF3-VIGS-2 and MPF2-MPF3-VIGS flowers; indicated by arrows, Figures 6E and 6F). In these cases, both MPF3 and MPF2 were knocked down (Figure 5A), implying genetic interactions between MPF3 and MPF2 in the determination of calyx organ identity.
This observation was also supported by variations in epidermal cell morphology in the floral calyx (Figures 6G to 6L). Prior to anthesis, cells of the wild-type floral calyx were small and undifferentiated, with regular shapes (Figure 6G), while cells in the MPF2-RNAi floral calyx were significantly larger (P = 8.53 × 10−17; Figure 6H). Cells also became significantly larger (P = 2.77 × 10−16), irregular in shape typical of leaf cells on the floral calyx once MPF3 was strongly downregulated (Figure 6I). This was accompanied by a dramatic elevation of MPF2 transcript (Figures 4R and 5A; see Supplemental Figure 5H online). In MPF2 and MPF3 double knockdowns, consistent with variations in the morphology of the floral calyx (Figures 6D to 6F), two types of cell morphologies were observed. Cells on the MPF2-RNAi-MPF3-VIGS-1 flowers (Figure 6D) were even larger than those on the wild type, with a regular shape (Figure 6J), while cells of MPF2-RNAi-MPF3-VIGS-2 and MPF2-MPF3-VIGS flowers (Figures 6E and 6F) became larger than these of the wild type and irregular before anthesis (Figures 6K and 6L). These findings coupled cell identity with floral calyx identity, which are both governed by the repression of MPF2 by MPF3. Interestingly, we discovered that the MPF2/MPF3 transcript ratio in the floral calyx was important for cell identity (shape). When the ratio of MPF2/MPF3 was greater than 1, cells were the shape typical of leaf cells, in comparison to the wild type.
The cell size within the floral calyx increased in all cases; however, the increase in cell size did not fully explain the variation in organ size (Figure 6M; see Supplemental Table 2 online). MPF2 promotes cell division but inhibits cell expansion in the floral calyx (He and Saedler, 2005), but MPF3 negatively regulates the two processes. However, once both genes were downregulated, cells in the floral calyx became significantly larger (P<1.29 × 10−10), while organ size was not altered (Figure 6M; see Supplemental Table 2 online); this suggested that cell division was basically inhibited. Therefore, genetic and physical interactions between MPF3 and MPF2 may promote cell division and inhibit cell expansion in the floral calyx.
These findings suggest that the repression of MPF2 by MPF3 is a crucial step in specifying calyx identity and that their interactions also regulate cell division and expansion during the development of the floral calyx.
Both MPF3 and MPF2 Regulate Cell Division and Cell Expansion and, Thus, PostFloral ICS Size
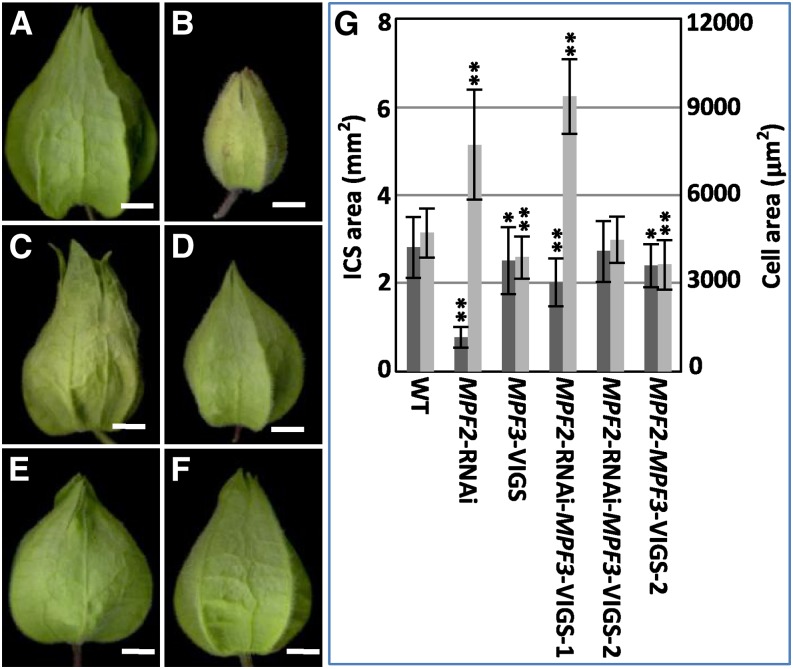
Once cell identity is determined at the preanthesis stage, the ICS results from cell division and cell expansion. Cells in the mature ICS become irregular and dramatically expand. In the wild type, the ICS area increased ∼46.1 ± 16.9 times, while cells in the ICS increased 10.9- ± 1.8-fold with respect to cells of the floral calyx. In contrast with the wild type (Figures 7A and 7G), with either MPF2 or/and MPF3 downregulated mutants, a smaller ICS was created after fertilization (Figures 7B to 7G); however, variation in cell size was quite complicated (Figure 7G; see Supplemental Figure 7 online). The variation in cell size did not completely explain the variation in organ size (see Supplemental Table 2 online). When the MPF2/MPF3 ratio was smaller than one in comparison with the wild type (see Supplemental Figure 7A and Supplemental Table 2 online), significantly larger cells were found in the ICS, as in MPF2-RNAi (P = 3.81 × 10−7; see Supplemental Figure 7B online) and MPF2-RNAi-MPF3-VIGS-1 (P = 3.29 × 10−14; see Supplemental Figure 7D online), and cell division was thus suppressed. However, when the ratio was greater than 1, the cells of the ICS became significantly smaller (in the MPF3-VIGS and the MPF2-MPF3-VIGS-2; P = 0.00; Figure 7G; see Supplemental Figures 7C and 7F online) or were unaffected (in the MPF2-RNAi-MPF3-VIGS-2; P = 0.44; see Supplemental Figure 7E online); cell division was therefore enhanced. Hence, interactions between MPF3 and MPF2 may elegantly regulate cell division and cell expansion and regulate postfloral calyx growth, modulating the ultimate size of the ICS.
Figure 7.
MPF3 and MPF2 Regulate Postfloral Calyx Size.
(A) to (F) Phenotypic variation of various ICSs. Bars = 5 mm.
(A) The wild type.
(B) MPF2-RNAi.
(C) MPF3-VIGS.
(D) MPF2-RNAi-MPF3-VIGS-1.
(E) MPF2-RNAi-MPF3-VIGS-2.
(F) MPF2-MPF3-VIGS-2.
(G) Quantification of ICS size and of its respective cells. Error bars are sd. Single and double asterisks stands for P < 0.05 and P < 0.01 in two-tailed Student’s t test, respectively. (For details, such as sample sizes, see Supplemental Table 2 and Supplemental Figure 7 online.) The genotype of each sample is referred to in Figure 5A.
These results confirm that genetic interaction between MPF3 and MPF2 is essential for male fertility, calyx identity, and ICS size in Physalis and suggest that protein–protein interactions between these two proteins affect these developmental processes.
Binding Capacity of MPF3-Associated Heterodimers to the CArG-Boxes in the MPF2 Promoter
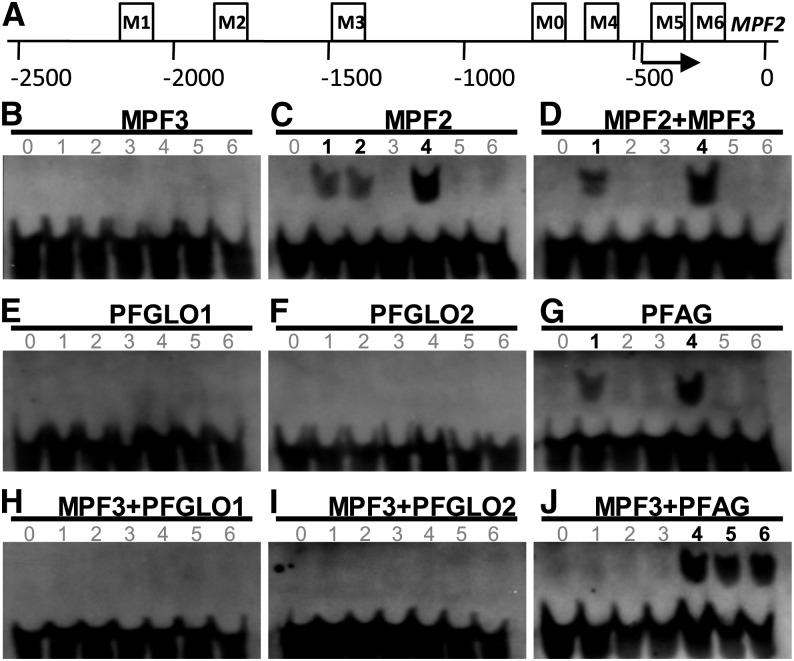
The CArG-box constitutes major binding sites for the MADS-domain transcription factors (West et al., 1997). However, no classical CArG motif, such as CC(A/T)6GG and C(A/T)8G, is present in the 1.4-kb MPF2 promoter region (He and Saedler, 2005). The promoter was then extended to 2.5 kb upstream of the translation initiation site (Riß, 2009; Khan et al., 2012), and sequence analysis revealed different types of CArG-boxes, including two CC(A/T)6GG, one C(A/T)8G, and one CC(A/T)7G (Figure 8A; see Supplemental Table 3 online). In addition, two variants C(A/T)7GG were found in the 5′-untranslated region. Altogether six motifs were designated as M1 to M6 in ordered sequence (Figure 8A; see Supplemental Table 3 online). Therefore, we assumed that MPF3 and its associated heterodimers fulfilled their biological functions by selectively binding to some of these motifs. To test this hypothesis, we conducted electrophoretic mobility shift assays (EMSAs).
Figure 8.
Ability of the MPF3 Interacting Partners and the Associated Heterodimers to Bind to a CArG-Box in the MPF2 Promoter.
(A) Distribution of CArG-box variants in the MPF2 promoter. M1 to M6 designate the CArG-boxes and M0 the random non-CArG-box as a negative control in the EMSA. The arrow indicates the putative transcriptional initiation site (He and Saedler, 2005). Position 0 indicates the translational start site. The boxes stand for M0 to M6 motifs and their relative positions on MPF2. For sequence details, see Supplemental Table 3 online.
(B) to (J) EMSA results of the MADS-domain protein binding to the CArG-boxes in the MPF2 promoter. The numbers from 0 to 6 indicate M0 to M6, respectively. Numbers in black and bold stand for the CArG-box bound by the indicated MADS-dimers, and the numbers in gray represent the motif that cannot be bound by the indicated MADS-dimers.
(B) MPF3.
(C) MPF2.
(D) MPF3 and MPF2.
(E) PFGLO1.
(F) PFGLO2.
(G) PFAG.
(H) MPF3 and PFGLO1.
(I) MPF3 and PFGLO2.
(J) MPF3 and PFAG.
Dimerization of MADS-domain proteins is required for DNA binding (Immink et al., 2010). Since these proteins can homodimerize, we first evaluated the binding ability of the homodimers to these CArG-boxes and found that MPF2 homodimers bound to the first three CArG-boxes (i.e., M1, M2, and M4) in the MPF2 promoter (Figure 8C) and that PFAG homodimers could bind to M1 and M4 (Figure 8G). By contrast, homodimers of MPF3, PFGLO1, and PFGLO2 were not able to bind to any of the CArG-boxes (Figures 8B, 8E, and 8F). Next, we monitored ability of MPF3-associated heterodimers to bind to these CArG-boxes. We demonstrated that MPF3-MPF2 heterodimers selectively bound to M1 and M4 (Figure 8D); MPF3-PFAG bound to M4, M5, and M6 (Figure 8J); and MPF3 and PFGLO1/PFGLO2 heterodimers did not bind with any of these motifs (Figures 8H and 8I). Thus, these DNA binding activities provide a basis for the regulation of MPF2 by the related protein dimers and specifically for the genetic interaction between MPF3 and MPF2 in floral organs.
MPF3 appears to have secured novel floral roles through the evolution of novel physical and genetic interactions, which largely result from alterations in its coding regions, as it features an expression pattern similar to that of the other euAP1 genes. This was further supported by selection analysis and a functional proof using a transgenic heterologous host.
Positive Selection of MPF3 Supports Its Functional Diversification
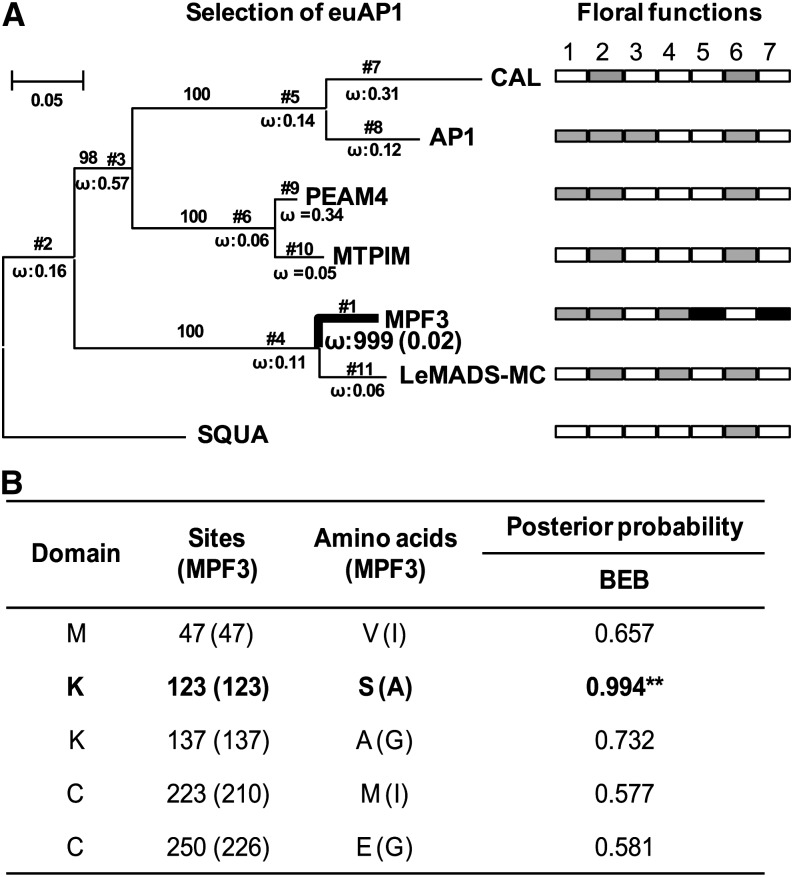
The sequence diversification of euAP1 was evaluated via selection analysis. The sequences with known functions from Brassicaceae, Fabaceae, Plantaginaceae, and Solanaceae were included for selection tests using two types of models (branch and branch-site models). In the free ratios branch model test (model = 1, giving separate ω values for each branch), the MPF3 branch was detected as having significant positive selection (ω = 999; highlighted with bold lines, Figure 9A). When all branches were set as foreground branches, the branch model test (model = 2, giving different ω values between foreground and background branches) further confirmed this (see Supplemental Table 4 online). Furthermore, to uncover the positive sites in MPF3, the branch-site model test was executed; this allows positive selection at a small number of sites along a specific lineage and is the most powerful model to detect positive selection. In this test, the MPF3 branch in euAP1 was set as the foreground branch, and five potential sites were found to be under positive selection. Notably, these sites were all distributed in the M, K, and C domains; in particular, Ala-123 in the K domain had a high posterior probability (Figure 9B). This evidence suggests that functional differentiation in these sites may be associated with alterations of MPF3 in DNA binding ability and protein–protein interactions during evolution.
Figure 9.
Positive selection of MPF3 supports diversification of its floral functions.
(A) The bold line indicates that positive selection was detected in the branches or the proteins. The proportion of sites with ω > 1 for the foreground branches is indicated under the branches in the bracket. SQUA was used as outgroup. Floral functions include (1) flowering time, (2) sepal identity, (3) petal identity, (4) calyx size, (5) ICS formation, (6) inflorescence or floral meristem, and (7) male fertility. As indicated, the gray box designates the genes that possess the role, while the empty box depicts the absence of such a role. The novel role for MPF3 is highlighted by the black box.
(B) Selectively relaxed constrained sites in MPF3. The sites and amino acids in parentheses are relative to those in MPF3. Bold font and asterisks indicate a positively selected codon site having P > 95% in Bayes Empirical Bayes (BEB) analysis.
[See online article for color version of this figure.]
Our selection results support functional differentiation of MPF3 in comparison with other euAP1 members, and we show that euAP1 genes from various genera encode MADS-box proteins with multiple functions. The floral roles for each gene in its own native host vary and include flowering time, sepal identity, petal identity, calyx size, ICS formation, floral/inflorescence meristem, and male fertility (Figure 9A). MPF3 appears to have secured a novel role in male fertility and ICS formation in Physalis.
Functional Divergence Is More Pronounced in a Transgenic Heterologous Host
We pose the following questions. How does sequence divergence under positive selection affect the functional divergence of euAP1 between Solanaceae and Brassicaceae? As orthologs of AP1, do MPF3 from P. floridana and its ortholog St-MC from Solanum tuberosum really function as AP1 does in Arabidopsis? To answer these questions, we overexpressed the two genes in Arabidopsis and its ap1 mutants, respectively. At least 15 independent transgenic lines were obtained for each construct and showed similar phenotypic variations. Single transgenic lines for St-MC and for MPF3 were characterized in a wild-type Arabidopsis background via DNA gel blot analysis. The bolting time and rosette leaf number at the time of bolting under either long-day or short-day conditions were recorded as the flowering time. In both conditions, the plants harboring St-MC or MPF3 had a flowering time comparable to that of wild-type Arabidopsis, while 35S:AP1 transgenic plants apparently flowered earlier (see Supplemental Figure 8 online), indicating that MPF3 and St-MC did not promote earlier flowering in Arabidopsis. In an ap1 background (three different alleles ap1-1, ap1-10, and ap1-12 were used), neither protein complemented the floral phenotype of these ap1 mutants. These results suggest that orthologs of euAP1 from Solanaceae and Brassicaceae, which are separated by a great evolutionary distance, underwent functional divergence. We therefore reiterate that using a heterologous system, such as Arabidopsis, is not the best method to evaluate gene function and the evolution of gene function.
DISCUSSION
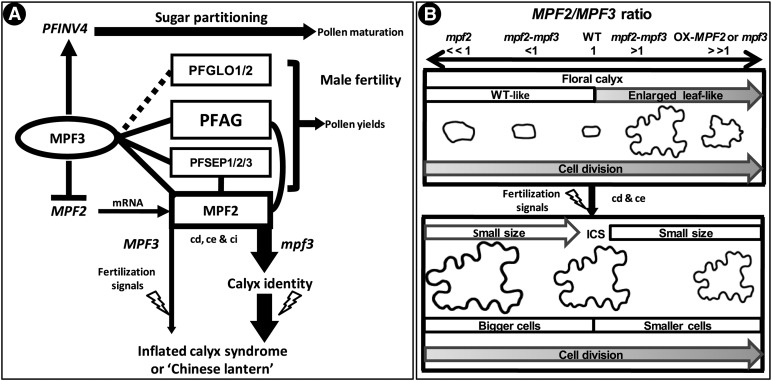
Gene duplication events have occurred at least twice during the evolution of the AP1/SQUA-like genes, resulting in the euFUL, FL, and euAP1 subclades in eudicots (Becker and Theissen, 2003; Litt and Irish, 2003; Shan et al., 2007). Within Solanaceae, an additional duplication event resulted in two copies (PFFUL1/2) of euFUL. Although these paralogs became diversified in sequence and expression, they are expressed in floral organs and possess diverse functions (Litt, 2007). The euAP1 have drawn more attention because of their role in the specification of sepal organ identity in flower development (i.e. AP1 in Arabidopsis [Mandel et al., 1992] and Le-MADS-MC in Solanum [Vrebalov et al., 2002]); therefore, their orthologs might have played a role in the evolution of the Chinese lantern or ICS. Four AP1/SQUA-like homologs were isolated in Physalis, and we demonstrated an essential role for MPF3 in flower development. As a euAP1 member, the prevalent expression of MPF3 in the floral meristem appears to support the hypothesis that MPF3 specifies floral meristem identity and regulates flowering time. Constitutive expression of MPF3 indeed promoted flowering in transgenic Physalis, while specific silencing analyses revealed that these roles were likely redundant or insignificant. MPF3 regulates floral calyx identity and growth and has novel functions in flower development through genetic and physical interactions with the key components of both postfloral ICS and male fertility (Figure 10A). This protein thus couples the two developmental processes of both ICS and male fertility during the evolution of Physalis.
Figure 10.
Summary of Floral Calyx and Postfloral ICS Development in Physalis.
(A) Current integrated MPF3-associated regulatory and interacting networks for ICS development in Physalis. The networks summarize knowledge gained from previous studies (He and Saedler, 2005; He and Saedler, 2007; He et al., 2007; Zhang et al., 2012) and this article. Proteins are presented in vertical circles or boxes, and genes or mRNAs are in italics. The solid lines indicate protein–protein interactions, while the dashed line indicates that the interactions need to be verified. Arrows stand for activation or promotion, and blocked lines for repression. cd, cell division; ce, cell expansion; ci, cell identity. The lightning symbol represents fertilization signals. For details, see text.
(B) The MPF2/MPF3 ratio model in the development and growth of the floral calyx and ICS in Physalis. Calyx development after primordia initiation consists of (top portion) the stage that organ identity is determined, organ growth before flowering, and (bottom portion) postfloral calyx outgrowth, which is triggered by fertilization signals (lightning symbol). The MPF2/MPF3 ratio is proposed to play an important role in these processes, particularly in establishing the identity of the floral calyx. MPF2 and MPF3 also regulate cell division and cell expansion in floral and postfloral calyx growth. Cell size and cell morphology are given proportionally for each context. Cell size for the fruiting calyx is ∼11 times that in the floral calyx, while the ICS area is ∼46 times that of the floral calyx area, indicating that ICS is a result of cell division (cd) and cell expansion (ce). WT, the wild type; OX-MPF2, MPF2 overexpression line; mpf2, mpf3, and mpf2-mpf3, downregulation mutants. The MPF2/MPF3 ratio in the wild type is set to 1; in the mutants, the ratio is then either >1 (>>1) or <1 (<<1). Shaded arrows in the floral calyx context represent a gradual promotion of the corresponding trait. Empty arrows in the ICS context depict a severe downregulation of either MPF2 or MPF3, which results in flowers with a smaller ICS than in those of the wild type. When the ratio is <1, larger cells are produced, whereas a ratio >1 results in smaller cells. This model is based on the quantification of cell and organ size of the transgenic mutants in comparison with the wild type (summarized in Supplemental Table 2 online).
Genetic Regulation of MPF3 and MPF2 Primarily Specifies Floral Calyx Identity
MPF3 primarily determines calyx organ identity, since its knockdown leads to a leaf-like calyx and its orthologs from various plant species regulate sepal organ identity (Mandel et al., 1992; Gustafson-Brown et al., 1994; Berbel et al., 2001; Taylor et al., 2002; Vrebalov et al., 2002; Benlloch et al., 2006). MPF3 is distinct from AP1 in Arabidopsis, but similar to other euAP1 genes, and does not control petal organ identity; however, it is expressed in corolla and corolla primordia during floral development. The role of MPF3 in corolla development might be redundant or partitioned by its paralogs, as was recently observed in poppy (Papaver somniferum, Pabón-Mora et al., 2012). In addition, MPF3 may not repress the expression of a C-function gene (PFAG) since PFAG is expressed in the gynoecium and its primordia, where the C-function gene exerts its role. Thus, MPF3 partially serves as an A-function gene to regulate calyx organ identity. The concept of the floral A-function has been problematic since its inception (Schwarz-Sommer et al., 1990; Litt and Irish, 2003; Litt, 2007). Our findings provide additional evidence to challenge an evolutionary conservation of the A-function gene as defined in Arabidopsis (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). The controversy regarding the A-function appears to resolve when subfunctionalization after gene duplications is considered (Litt and Kramer, 2010; Pabón-Mora et al., 2012). Regardless, the core role of the A-function is to specify sepal identity.
Alteration of sepal identity often accompanies alteration of cell morphology or identity; for example, trichome type is a typical morphological marker used to distinguish the sepal and leaf in Arabidopsis (Mandel et al., 1992; Gustafson-Brown et al., 1994). However, cell morphology is not necessarily considered to be a marker that distinguishes the calyx and leaf in Solanaceae (Hu and Saedler, 2007). The epidermal cells on the floral calyx stay undifferentiated and are smaller prior to anthesis in Physalis and then differentiate to be lobate in the postfloral calyx. However, in the MPF3-silenced mutants, the cell morphology of the transformed calyx before anthesis was already strongly lobate (as with cells making leaves), indicating that MPF3 specifies calyx identity by regulating cell identity. According to the floral quartet model (Theissen and Saedler, 2001), interaction with SEPALLATA-like proteins in Physalis (PFSEPs) (e.g., PFSEP1, PFSEP2, and PFSEP3; He et al., 2007) appears to be sufficient for MPF3 to specify calyx identity (Figure 10A).
However, the precise specification of floral calyx identity might also require the activity of MPF2, which has a dose-dependent effect (Figure 10B). Overexpression of MPF2 in Solanum (He and Saedler, 2007), similar to the overexpression of its ortholog, AGL24, in Arabidopsis (Yu et al., 2004), superimposed a vegetative developmental program in flower development and generated leaf-like sepals, phenocopying ap1 mutants and indicating that the repression of AGL24 is a crucial step in promoting flower development (He et al., 2004; Yu et al., 2004). By analogy, MPF3 (the ortholog of AP1) was thought to repress MPF2, while variation in the promoter of MPF2 led to heterotopic expression of the gene itself in floral organs (He and Saedler, 2005); however, the native expression of MPF2 in the calyx may be too low to alter the identity of the floral calyx in Physalis. In MPF3-silenced flowers, however, MPF2 expression was more than 10 times greater than in the wild-type calyx, as in lines that overexpress MPF2, which led to a change in the identity of the calyx and its cells (Figure 10). When both MPF3 and MPF2 were silenced, no obvious leaf-like calyx was seen or only a weak homeotic transformation in the calyx tips was observed. Thus, the MPF2/MPF3 ratio was critical for specifying the floral calyx identity (Figure 10B). Once the ratio was greater than 1, the vegetative growth in calyx development was superimposed and a leaf-like floral calyx was created; otherwise, the calyx was determined. These findings confirm the role of genetic control of MPF2 by MPF3 during floral development.
There is no classical CArG-box present in the 1.4-kb MPF2 promoter (He and Saedler, 2005), but some variants exist near the transcriptional initiation site, and three classical CArG-boxes occur in a remote region (Riß, 2009; Khan et al., 2012), providing the structural basis for a direct genetic interaction between MPF3 and MPF2. Our EMSA revealed that MPF2 bound to M1, M2, and M4 and that MPF3 itself cannot bind to any CArG-box of the MPF2 promoter. However, MPF3-MPF2 selectively bound to M1 and M4, indicating that expression of MPF2 was at least self-regulated and that the floral organs were more tightly regulated by MPF3-MPF2 together than by MPF3 alone. M1 bound by MPF3 seems to repress MPF2, since the CC(A/T)7G CArG-box (designated as M4) in the vicinity of the transcriptional initiation site might contribute to the heterotopic expression of MPF2 in floral organs, which was also proposed for MPF2-like expression in sepals (Khan et al., 2012). The M4 site may result from a point mutation in the classical CArG boxes [either CC(A/T)6GG or C(A/T)8G] or from a unique origin in Physalis. In Solanum, the M4 motif is missing and the activation of STMADS16 by St-MC is abolished in the floral organs (He and Saedler, 2005; Riß, 2009; Khan et al., 2012). By contrast, in Physalis (very likely due to presence of the M4 motif), MPF2 is activated by MPF3; hence, MPF2 is heterotopically expressed in floral organs.
Thus, the complex of MPF3 and PFSEPs regulates calyx identity by interacting directly with MPF2/MPF2 either genetically or physically in Physalis (Figure 10A), similar to the situation suggested for the orthologous pair AP1-AGL24 in Arabidopsis (Yu et al., 2004; de Folter et al., 2005). Our results indicate that the key regulatory networks involved in sepal organ identity are evolutionarily conserved across plant families and further suggest that a mutation or evolution of a key regulatory element in a key gene alters gene expression to dramatically shape the morphology in Solanaceae.
MPF3 and MPF2 Regulate Cell Cycle Processes in Calyx Growth, Thus Determining ICS Size
Organ growth is the result of cell division and cell expansion. In Physalis, calyx growth can be divided into floral and postfloral processes. In both cases, the MPF2/MPF3 ratio is associated with cell cycle processes (Figure 10B). In our study, cell division was promoted as the ratio increased; thus, MPF2 is the major determinant promoting cell division, while MPF3 inhibits cell division during both developmental processes in the calyx of Physalis. However, the cell size changes in the downregulated mutants proved to be quite interesting.
During floral development, cell size in the calyx increased if either MPF3 or MPF2 was downregulated (Figure 10). Thus, both MPF3 and MPF2 act as repressors of cell expansion in the growth of the floral calyx. Although downregulation of MPF2 increased cell size, the unaffected MPF3 inhibited cell division; thus, floral calyx size was largely unaffected. Once MPF3 was knocked down, the concentration of MPF2 mRNA was significantly elevated. Thus, the rate of cell division and cell size both increased, and the floral calyx was transformed into an enlarged leaf-like organ. In the double downregulated mutants, the ratio of MPF2/MPF3 varied. Once again, when the ratio was under 1, the cells were enlarged, while when the ratio was greater than 1, the cell size fluctuated, but was still much larger than in the wild type. This implies a physical interaction between MPF2 and MPF3 (in addition to their genetic interaction) in the growth of the floral calyx.
In contrast with the floral calyx, once MPF3 was downregulated, the size of the mature ICS and its cells was reduced, indicating that MPF3 promoted cell expansion in the fruiting calyx. Opposite effects on cells were observed once MPF2 was knocked down (He and Saedler, 2005), but this also resulted in a smaller lantern. In the absence of MPF3 and with a dramatic increase in MPF2 mRNA, cell size was reduced and this resulted in a smaller ICS. Moreover, cell expansion in the fruiting calyx was also regulated by the MPF2/MPF3 ratio in the double-silenced mutants (Figure 10B). Cells were larger than wild-type cells when the ratio was less than one and smaller when the ratio was greater than one. However, the ICS became smaller once either of the two genes was downregulated. Thus, both MPF3 and MPF2 and their interactions are essential to the final size of the Chinese lantern in Physalis.
The molecular details regarding how MPF3 and MPF2 regulate the cell cycle in the development and growth of both the floral calyx and postfloral calyx are not yet understood; however, floral transcriptomic comparisons between the wild type and various mutants could improve our understanding of these processes. However, abundance and dosage balance of MPF2 and MPF3, and their genetic and physical interactions, regulate cell division, cell expansion, and cell differentiation. Interactions with fertilization or hormonal signals ultimately trigger postfloral calyx outgrowth and the development of ICS (He and Saedler, 2005; He and Saedler, 2007) and might relate to male fertility.
Both MPF3 and MPF2 Are Integral to Male Fertility
Stamen identity is regulated by B-, C-, and E- function proteins (Theissen and Saedler, 2001). The euAP1 genes are not involved in this developmental process, since mutations of these genes do not affect male fertility. Although silencing of MPF3 did not affect anther development, pollen maturation was severely impaired, which affected male fertility. MPF3 is expressed in androecium primordia during early floral development; however, its expression drops once the androecium is formed. One possibility is that the earlier expression might impact pollen development. Alternatively, the low level could still be enough to direct pollen maturation. Furthermore, a discrepancy between mRNA expression and protein expression might also play a role (Immink et al., 2010). Finally, as reported previously (Wu et al., 2003; Urbanus et al., 2009, 2010), the possibility of both intercellular transport of MPF3 and non-cell-autonomous function cannot be excluded. Nevertheless, all of the above could essentially alter the effect of MPF3 on pollen development.
MPF2 knockdown results in male infertility primarily due to a dramatic reduction in pollen yield (He and Saedler, 2005). The defects in pollen maturation in successive generations resemble those in MPF3-silenced lines. However, downregulation of MPF3 resulted in increased expression of MPF2 in the androecium, indicating that MPF2 overexpression could also damage pollen maturation. A similar defect in pollen development was seen in MPF2 MPF3 double knockdowns, although more pollen grains appeared to be normal. Therefore, the activity of both MPF3 and MPF2 is required for normal pollen development (Figure 10A).
MPF2 may affect male fertility by interacting with florally expressed MADS-domain proteins in Physalis (He et al., 2007). Thus, both interactions with other MADS-domain proteins and the regulation of their target genes are important but not essential for MPF3 to affect male fertility. GLO in Antirrhinum (a B-function protein; Tröbner et al., 1992), TM29 (a SEP-like protein, E-function) in Solanum (Ampomah-Dwamena et al., 2002), and FARINELLI (C-function) in Antirrhinum (Davies et al., 1999) are all involved in male fertility. Since MPF3 and MPF2 interact with their orthologs, including PFGLO1, PFGLO2, PFSEP2/3, and PFAG (this study; He et al., 2007), the roles of these orthologs in male fertility should also be considered. The interactions between MPF3 and PFAG and MPF2 seem to be weak but stable, since they were detected in vitro and in vivo. Direct interactions between euAP1 proteins and B-function proteins have not yet been reported in angiosperms. Even Le-MADS-MC from Solanum, a close relative of Physalis, does not show this capability (Leseberg et al., 2008). We demonstrated that MPF3 interacts weakly with PFGLO1 and PFGLO2 in vitro, but no YFP signal was detected in our BiFC analyses using plant leaf cells. Thus, such interactions might also be transient in Physalis. Coexpressing these proteins in a plant cellular environment allows for various putative partners with stronger attractions for these MADS-domain proteins to become available and compete with each other for dimerization and complex formation (Immink et al., 2010), thus abolishing any YFP signal. The molecular details regarding this hypothesis need to be extensively investigated. Nonetheless, the stable and transient protein–protein interactions of both MPF3 and MPF2 with these male fertility–associated MADS-domain proteins could together affect the dynamics of the quartet determining the stamen (Theissen and Saedler, 2001), and this might make it essential for MPF3 and MPF2 to influence pollen yields and maturation in Physalis (Figure 10A), as observed in the previous work (He and Saedler, 2005) and in this work.
In a parallel fashion, Suc accumulation in pollen grains is essential for pollen maturity (Ranwala and Miller, 1998; Oliver et al., 2005; Zhang et al., 2010). In our study, insufficient sugar accumulated in the pollen grains once either MPF3 or MPF2 or both were knocked down. Expression of several related genes was monitored and PFINV4 was concomitantly, specifically, and strongly repressed in the androecium. Its ortholog INV4 in Oryza and Lilium was demonstrated to be a key component in sugar partitioning during anther development (Ranwala and Miller, 1998; Oliver et al., 2005). Thus, it is likely that the marked downregulation of PFINV4 causes an abnormal photosynthetic carbon allocation, leading to immature pollen grains. Thus, MPF3, by interacting with MPF2, may activate the PFINV4 gene to regulate male fertility by affecting sugar partitioning (Figure 10A). In particular, PFAG could bind to the CArG-boxes, M1 and M4, and MPF3-PFAG could bind to the CArG-boxes, M4, M5, and M6, in the upstream regions of MPF2. It is interesting that PFAG and MPF3-PFAG share a CArG-box M4 with MPF2-MPF3 and MPF2. Moreover, MPF3-PFAG could bind to variants of CArG-boxes in the 5′-untranslated region. Although the biological relevance of these interactions needs to be substantiated, these findings indicate that PFAG and MPF3-PFAG also fulfill a role in this pathway that may be mediated by MPF2. However, PFGLO1, PFGLO2, and their MPF3-associated heterodimers did not bind to any CArG-boxes of the MPF2 promoter; thus, they might not affect male fertility via direct genetic interaction with MPF2. Whether they also regulate PFINV4 in this novel pathway by MADS-domain transcription factors needs to be further investigated.
Therefore, MPF3 has an integral role in male fertility without affecting stamen organ identity and development. The MPF3-MPF2/MPF2–regulating and –interacting complex, as a primary regulator of cell division and cell expansion, is integrated into the male fertility–determining networks through novel genetic and physical interactions. This integration further corroborates the previous assumption of the role of male fertility in the evolution and development of the ICS (He and Saedler, 2005; He et al., 2007).
Therefore, MPF3 may orchestrate floral initiation and ICS formation by integrating identity, growth, patterning, and hormonal pathways in Physalis, as suggested for AP1 in Arabidopsis (Kaufmann et al., 2010), but it has acquired novel and essential functions in ICS formation and male fertility. The latter findings are different from data obtained for other euAP1 genes (Figure 9), and functional divergence within the euAP1s in floral development is also discussed below.
Positive Selections Underlie Functional Diversification of euAP1 Orthologs
Several euAP1 family members from various genera have been functionally characterized thus far, such as AP1 and CAL in Arabidopsis (Mandel et al., 1992; Bowman et al., 1993), SQUA in Antirrhinum (Huijser et al., 1992), Le-MADS-MC in Solanum (Vrebalov et al., 2002), PEAM4 in Pisum (Berbel et al., 2001; Taylor et al., 2002), MTPIM in Medicago (Benlloch et al., 2006), and MPF3 in Physalis (this study). In addition to SQUA, other molecules play a role in sepal identity, and the divergences between MPF3 and other euAP1 genes are apparent due to the functional pleiotropic processes of the involved genes (Figure 9).
Particular functions have been ascribed to some orthologs; for example, the role in petal development seems to be unique for AP1 in Arabidopsis (Mandel et al., 1992; Bowman et al., 1993). The functional divergence is more pronounced when euAP1s from different plant families are heterologously tested in the same system (e.g., Arabidopsis from Brassicaceae). In addition to its role in floral organ identity, AP1 plays an important role in the fate of the floral meristem, as revealed by loss-of-function studies (Mandel et al., 1992; Bowman et al., 1993; Yu et al., 2004), and a gain-of-function mutation of AP1 promotes flowering in Arabidopsis (Mandel and Yanofsky, 1995) and in a heterologous host (Peña et al., 2001). However, both MPF3 and St-MC from Solanaceae failed to complement ap1 mutants, and their overexpression in wild-type Arabidopsis did not lead to earlier flowering. Therefore, we emphasize that the function of a gene should be inferred in its native host.
Nearly all euAP1s reported affect floral or inflorescence meristem identity in their own hosts (Figure 9; Huijser et al., 1992; Mandel et al., 1992; Taylor et al., 2002; Vrebalov et al., 2002; Berbel et al., 2001; Benlloch et al., 2006). However, solitary flowers with a leaf-like calyx are seen in 35S:MPF3-RNAi Physalis, indicating that floral meristem identity is not altered by knocking down MPF3. The low abundance of MPF3 transcript in the MPF3 knockdowns may be sufficient to promote flower development; conversely, the role of MPF3 in floral meristem identity may be shared with its paralogs. Studies of mpf3 null mutants and genetic analyses of MPF3 and its paralogs in Physalis could resolve this.
Even within the family Solanaceae, euAP1 from different genera behave differently. Solanum is a close relative of Physalis, since the microsynteny is well maintained for the MPF3 orthologs in Physalis and Le-MADS-MC orthologs in Solanum. Loss of Le-MADS-MC in tomato led to a leaf-like macrocalyx and altered the determinacy of the inflorescence (Vrebalov et al., 2002); however, this gene is not involved in male fertility as MPF3 is in Physalis.
MPF3 shares a similar expression domain with other euAP1s (Huijser et al., 1992; Mandel et al., 1992; Carr and Irish, 1997; Vrebalov et al., 2002) that are exclusively expressed in floral development. Thus, their functional divergence mainly results from variations in their coding regions. MPF3 underwent strong Darwinian selection, and the five selected sites located in the M, K, and C domains (Figure 9) might alter the spectrum and strength of the target genes and protein–protein interactions, thus resulting in functional divergence. In particular, site 123 (with Bayes Empirical Bayes > 0.95) in the K domain, which is known to regulate protein–protein interactions (reviewed in Immink et al., 2010), seems to be critical for the divergence of protein–protein interactions. The lower probability of the other sites undergoing selections indicates that they might play a lesser role in affecting the protein–protein interactions. In accordance with this, we observed that the interacting protein spectrum differed in different species. The euAP1 orthologs, as observed for both AP1 in Arabidopsis (de Folter et al., 2005) and Le-MADS-MC in Solanum (Leseberg et al., 2008), did not interact with any B- and C-function proteins, while SQUA in Antirrhinum (Davies et al., 1996) and MPF3 in Physalis interacted with C-function proteins. MPF3 interacted with B-function proteins only in Physalis. Positive selection in protein sequences might also have led to the evolution of novel regulatory pathways. As we have demonstrated, MPF3 evolved its regulatory role in sugar partitioning pathways most likely by activating PFINV4, and MPF3 was involved in the origin of the ICS by interacting with MPF2/MPF2 (Figure 10A). Such a specific interaction and its effects on pollen maturation have not been observed in other euAP1 mutants.
Thus, changes in the molecular interaction networks of MPF3 in comparison to the molecular interaction networks of its orthologs may form the basis for the diverse phenotypic variations observed in the MPF3-silenced mutants. The roles of euAP1 proteins have diversified, and this diversification is undoubtedly driven by the action of different selective pressures during evolution.
Conclusions
Although the expression domains of euAP1 proteins are basically identical, these proteins, under positive selection, underwent functional diversification during the course of evolution. Nonetheless, most of the euAP1s are determinants of sepal identity. The formation of the novel trait, the Chinese lantern, in Physalis is not a result of a change in organ identity. The MPF3-MPF2 regulatory circuit is associated with cell division, cell expansion, and cell identity, and this circuit thus regulates floral calyx development and postfloral outgrowth to an ICS (Figure 10B); however, the evolution of the circuit’s interactions with PFAG, PFGLO1/2, and PFSEP1/2/3 implicates it in male fertility (Figure 10A). The evolution of a role in regulating the sugar partitioning pathway through the activation of PFINV4, which encodes an invertase, suggests that MPF3, a euAP1 MADS box gene, is associated with stamen functionality. The involvement of MPF3 and MPF2, as key regulators in calyx development, in male fertility seems to be integral to the origin of the Chinese lantern, since a fertilization signal is required for the development of this novel trait (He and Saedler, 2005; He and Saedler, 2007). Thus, the regulation of these identified MADS box genes and interactions among their proteins as determined by this study and others (He and Saedler, 2005; He et al., 2007), are involved in the evolution and development of the postfloral novelty, the Chinese lantern or ICS, in Physalis.
METHODS
Plant Materials
Solanum macrocarpon, potato (Solanum tuberosum), and Arabidopsis thaliana were grown in greenhouses at the Max-Planck Institute for Plant Breeding Research (Cologne, Germany) under long-day or short-day conditions. Physalis floridana P106 (He and Saedler, 2005) was grown in a growth chamber at the State Key Laboratory of Systematic and Evolutionary Botany of the Chinese Academy of Sciences (Beijing, China) under long-day conditions. The lines of three different alleles of ap1 mutants (ap1-1, ap1-10, and ap1-12) and a 35S:AP1 transgenic Arabidopsis were purchased from The Arabidopsis Information Resource (http://www.Arabidopsis.org/). Leaves and floral materials were harvested for genomic DNA and total RNA isolation.
Gel Blot Analyses
RNA and DNA gel blot analyses were performed as previously described (He et al., 2002). For RNA hybridization, the Actin gene was reprobed as a loading control after the filters were stripped. MPF3 cDNA fragments encoding the C-domain and the 3′-untranslated region were used as probes for both RNA and DNA gel blot hybridizations in Physalis. A MPF3 cDNA fragment or a corresponding fragment of St-MC cDNA was also used as a probe in DNA gel blots to identify transgenic Arabidopsis lines harboring single copies of MPF3 or St-MC, respectively.
mRNA in Situ Hybridization
A 275-bp cDNA fragment spanning the C-domain and 3′-untranslated region of the MPF3 cDNA was designed as probe template. Probes were synthesized using the T7 RNA polymerase driven by a T7 promoter and labeled with digoxigenin using the DIG-RNA labeling kit (Roche). Hybridization was performed as described by Carr and Irish (1997), except that the temperature for washing slides was 50°C in our experiments.
RT-PCR Analyses
Routine RT-PCR analysis was performed and PCR products were separated on a 1.2% agarose gel. Real-time RT-PCR was performed using the SYBR Premix Ex Taq (Perfect Real Time) kit (TaKaRa) on an Mx3000p real-time system (Stratagene) at an amplification cycle consisting of 30 s at 95°C for 1 cycle, followed by 40 cycles of 5 s at 95°C and 40 s at 60°C, which were followed by a dissociation curve. The Actin gene was used as the internal control.
Yeast Two-Hybrid Assays
The open reading frame (ORF) of MPF3 was inserted into pGBKT7 and transformed into the yeast strain AH109 to check self-activation. MPF3 cannot self-activate, and the MPF3 construct was transformed into Y187 as bait to screen the Physalis library. The medium, library information, and the procedures used were the same as described by He et al. (2007). The ORF of the MPF3-interacting partners was cloned into pGADT7. The cotransformed yeast cells with MPF3 bait were grown on the selected plates SD/-His-Leu-Trp at 28°C for 5 d. Their interaction strength was determined by a galactosidase activity assay using o-nitrophenyl-β-d-galactoside as substrate according to the protocol of Clontech.
Pull-Down Assays
The ORF of MPF3 was cloned into a pET-30a(+) vector containing a 6×His tag, expressed in Escherichia coli BL21 (DE3), and then purified on Ni Sepharose columns (GE Healthcare). ORFs of PFGLO1 and PFGLO2 were cloned into a pGEX-4T-1 vector with a glutathione S-transferase (GST) tag, expressed in E. coli BL21, and then loaded onto Glutathione Sepharose columns (GE Healthcare). The purified recombinant HIS-MPF3 proteins were loaded into the Glutathione Sepharose columns with GST-PFGLO1- and GST-PFGLO2-recombinant proteins, respectively, and incubated overnight at 4°C. The columns were then washed four times with binding buffer, and the bound proteins were eluted with washing buffer. The eluted proteins were heated and then separated on 12% SDS polyacrylamide gels. Proteins were blotted on nitrocellulose filter membranes by the semidry electrophoretic transfer method, and the membranes were blocked with 5% BSA-Tris-buffered saline containing 0.05% Tween-20 buffer for 1 h. Membranes were incubated with anti-GST or anti-HIS antibodies for 2 h and then with anti-horseradish peroxidase antibodies for 1 h. The membranes were washed with Tris-buffered saline containing 0.05% Tween-20 buffer three times after each incubation. Radioautographs of the filters were obtained using Kodak film at room temperature for 1 min and fixed according to standard procedures.
Subcellular Localization and BiFC
For subcellular localization, the ORF of MPF3 was cloned into the Super1300 expression vector (Chen et al., 2009) using the XbaI-KpnI cutting sites and fused to a GFP gene. For BiFC assays, the ORFs of MPF3, MPF2, PFGLO1, PFGLO2, and PFAG were cloned into a pair of vectors, pSPYNE-35S and pSPYCE-35S (Walter et al., 2004) through the XbaI and BamHI cutting sites, respectively. The two vectors were designed to express either N- or C-terminal halves of a YFP. The recombinant MPF3-GFP construct or the construct combination of two proteins fused with the N- or C-terminal halves of YFP was injected into leaf epidermal cells of Nicotiana benthamiana via Agrobacterium tumefaciens. Forty-eight hours after injection, the GFP or YFP fluorescence signal was detected using confocal laser scanning microscopy (Olympus FV1000 MPE). The GFP signal intensity was quantified using ImageJ (http://rsb.info.nih.gov/nih-image; Abràmoff et al., 2004).
EMSA
The recombinant proteins HIS-MPF2, HIS-PFAG, GST-MPF3, GST-PFGLO1, and GST-PFGLO2 were expressed in E. coli and purified using Ni and Glutathione Sepharose columns (GE Healthcare). The EMSA was performed using the DIG gel-shift kit, 2nd generation (Roche). The 3′end labeling with digoxigenin-11-ddUTP double-stranded DNA probes was generated by annealing complementary oligonucleotides. The probe sequences are shown in Supplemental Table 3 online. The protein-DNA binding mixture was incubated at room temperature for 15 min and separated on a 6% native polyacrylamide gel. After blotting and cross-linking, the DIG-labeled probes were detected by chemiluminescence and then the membranes were exposed to x-ray film.
Plant Transformation and Genotyping Analysis
A 389-bp fragment of MPF3 cDNA encoding partial K and full C domains and a partial 3′-untranslated region were inserted into the binary vector pFGC1008 (ChromDB; http://www.chromdb.org), resulting in a MPF3-RNAi construct. MPF3 ORF cDNA was cloned into the plant binary vector pBin19 and overexpressed in Physalis. For transformation of Arabidopsis, the full-length cDNAs for MPF3 and St-MC were inserted into pBAR. For Physalis transformation, the Agrobacterium strain LBA4404 was used. The medium and transformation procedures were identical to those described previously (He and Saedler, 2005). The GV3101 strain was used for transformation of Arabidopsis using the floral dip method (Clough and Bent, 1998). The transgenic Physalis plants were confirmed by real-time RT-PCR, and the transgenic Arabidopsis plants were confirmed by DNA gel blots.
VIGS Analyses
Three gene-silencing constructs were created to specifically downregulate MPF2, MPF3, or both. The fragments for MPF2-RNAi (He and Saedler, 2005) and for MPF3-RNAi were introduced either solely or tandemly into a TRV-mediated gene silencing system (Liu et al., 2002) and then applied to the leaves of 2-week-old Physalis seedlings. A 300-bp PFPDS-specific cDNA fragment was introduced into the TRV system as a control.
Morphological Analyses
For scanning electronic microscopy, fresh material was frozen in liquid nitrogen, sputter-coated with gold, and examined with a digital scanning microscope (Hitachi S-800). Cell and calyx size were quantified using the AxioVision LE image-processing program (http://www.zeiss.de). Calyces were taken from preanthesis flowers of the wild type and MPF3-RNAi lines indicated. Stamen sectioning was performed. Pollen maturation was detected using I2-KI staining. The main stem node number was recorded for flowering time in Physalis. The bolting time and rosette leaf number at the time of bolting were recorded to determine flowering time in Arabidopsis. For histological analyses, mature stamens were fixed in formalin-acetic acid-alcohol solution and dehydrated in a graded ethanol series. After replacing the ethanol with xylene, the stamens were embedded in paraffin wax (Sigma-Aldrich). Tissue sections were cut at 10 μm on a microtome and stained with safranine and fast green. All P values are based on two-tailed t tests.
Phylogenetic Reconstruction and Selection Evaluation
BLAST searches were performed in GenBank (http://www.ncbi.nlm.nih.gov/) using four P. floridana AP1/SQUA-like protein sequences as queries. The coding sequences were aligned and manually adjusted in BioEdit version 7.0.9 (Hall, 1999). The phylogeny was constructed based on the amino acid sequences, excluding the unalignable regions in the I and C domains, with MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001) (prset aamodelpr = mixed; ngen =1000000), using two sequences from Euptelea pleiosperma as the outgroup. The maximum likelihood tree was also constructed using the PhyML v3.0 program (Guindon and Gascuel, 2003).
To determine whether there were changes in selection pressure among members of the euAP1 clade, we used codon-based substitution models as implemented in PAML (Yang, 2007). The branch (model = 1; NSsites = 0; or model = 2; NSsites = 0) and branch-site models (model = 2; NSsites = 2) were performed (Yang, 1998; Zhang et al., 2005). Likelihood ratio tests were conducted using χ2 tests to detect positive selection between each of the pairs of models (Nielsen and Yang, 1998).
Sequence Isolation
Total RNA was extracted from young floral buds with Plant RNA reagent (Invitrogen). First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) with the poly(T) primer 5′-CCGGATCCTCTAGAGCGGCCG C(T)17-30. The 3′ and 5′ ends of the AP1/SQUA-like cDNAs from Physalis were amplified according to the standard manual of the 3′/5′ RACE kit (Roche). The degenerate primers were 5′-AGNCAAGTNCANTTYTCAAAGAG-3′ and 5′-AARGGNAARATCTTTGARTAYTC-3′, and the adapter primer was 5′-CCGGATCCTCTAGAGCGGCCGC-3′. Full-length cDNA sequences were then obtained using gene-specific primers (see Supplemental Table 5 online). The orthologs of MPF3 were previously isolated from S. macrocarpon (MSM3) and S. tuberosum (St-MC) with a routine RT-PCR (He et al., 2004).
All sequences were amplified using TaKaRa LA Taq polymerase. The amplified fragments were cloned into the pGEM-T Easy Vector (Promega), and the various expression constructs were commercially sequenced by Beijing Genomics Institute. The primer sequences used in this work are presented in Supplemental Table 5 online.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL databases under the following accession numbers: KC414859 (St-MC), KC414860 (MSM3), KC414861 (PFFUL1), KC414862 (PFFUL2), KC414863 (PFFL), KC414864 (MPF3), JX467691 (PFGLO1), KC174706 (PFGLO2) and KC794937 (PFAG).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Bayesian Interference of AP1/SQUA-Like Genes.
Supplemental Figure 2. Protein–Protein Dimerization among MPF3-Interacting Partners within the Yeast Two-Hybrid System.
Supplemental Figure 3. Anther Transverse Section Analyses.
Supplemental Figure 4. Expression of the Candidate Genes Involved in Sugar Partitioning Pathways.
Supplemental Figure 5. VIGS-Mediated MPF3 Silencing Phenocopies MPF3-RNAi.
Supplemental Figure 6. Normal Pollen Development in the 35S:PDS-VIGS Flowers.
Supplemental Figure 7. Cell Morphology in the Various ICSs.
Supplemental Figure 8. Functional Analyses in Transgenic Arabidopsis.
Supplemental Table 1. Variation in Flowering Time in Transgenic Physalis Plants.
Supplemental Table 2. MPF3 and MPF2 Modulate Cell Division and Cell Expansion during Calyx Development in Physalis.
Supplemental Table 3. Probe Sequences for EMSA.
Supplemental Table 4. Parameter Estimates under Branch and Branch-Site Models.
Supplemental Table 5. List of Primers Used in This Work.
Supplemental Data Set 1. Sequence Alignment of AP1/SQUA-Like Genes.
Supplemental Data Set 2. Sequence Information Used from the Physalis Floral Transcriptome.
Acknowledgments
We thank Heinz Saedler and Claus-Peter Stelzer for their valuable suggestions to improve the article. The assistance of Yinhou Xiao in scanning electron microscopy analyses, Miss Xiang Ao in mRNA in situ hybridization, and Jingquan Li in operation of the confocal laser scanning microscope are acknowledged. This work was supported by Hundred Talents Project of the Chinese Academy of Sciences to C.H., by grants (31070203 and 30870175) from the National Natural Science Foundation of China (to C.H.), and by the 46th Postdoctoral Science Foundation of China to Y.T.
AUTHOR CONTRIBUTIONS
C.H. designed the research project and conceived the experiments. J.Z., Y.T., S.R., and C.H. isolated sequences. Y.T. and J.Z. characterized expressions with real-time RT-PCR. Y.T. and J.Z. performed overexpression and RNAi analyses. J.S.Z. conducted mRNA in situ hybridization. J.Z. and P.G. searched Physalis genes involved in sugar partitioning pathways. J.S.Z., P.G., and J.Z. performed relative quantification of protein–protein interaction strength in yeast. J.S.Z. performed pulldowns. R.S. performed yeast library screens. C.H. and S.R. performed transgenic Arabidopsis analyses. J.Z. conducted other experiments. M.Z. and Y.T. performed evolution analyses. C.H. and J.Z. analyzed the data. C.H. wrote the article.
Glossary
- ICS
inflated calyx syndrome
- RNAi
RNA interference
- VIGS
virus-induced gene silencing
- BiFC
bimolecular fluorescence complementation
- TRV
tobacco rattle virus
- EMSA
electrophoretic mobility shift assay
- ORF
open reading frame
- I2-KI
iodine-potassium iodide
References
- Abràmoff M.D., Magalhães P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- Ampomah-Dwamena C., Morris B.A., Sutherland P., Veit B., Yao J.L. (2002). Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Benlloch R., d’Erfurth I., Ferrandiz C., Cosson V., Beltrán J.P., Cañas L.A., Kondorosi A., Madueño F., Ratet P. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A., Navarro C., Ferrándiz C., Cañas L.A., Madueño F., Beltrán J.P. (2001). Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J. 25: 441–451 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Alvarez J., Weigel D., Meyerowitz E.M., Smyth D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Büttner M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581: 2318–2324 [DOI] [PubMed] [Google Scholar]
- Carr S.M., Irish V.F. (1997). Floral homeotic gene expression defines developmental arrest stages in Brassica oleracea L. vars. botrytis and italica. Planta 201: 179–188 [DOI] [PubMed] [Google Scholar]
- Chen Y.F., Li L.Q., Xu Q., Kong Y.H., Wang H., Wu W.H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Davies B., Egea-Cortines M., de Andrade Silva E., Saedler H., Sommer H. (1996). Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15: 4330–4343 [PMC free article] [PubMed] [Google Scholar]
- Davies B., Motte P., Keck E., Saedler H., Sommer H., Schwarz-Sommer Z. (1999). PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18: 4023–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Immink R.G.H., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M.M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60: 626–637 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C., Savidge B., Yanofsky M.F. (1994). Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76: 131–143 [DOI] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98 [Google Scholar]
- He C.Y., Münster T., Saedler H. (2004). On the origin of floral morphological novelties. FEBS Lett. 567: 147–151 [DOI] [PubMed] [Google Scholar]
- He C.Y., Saedler H. (2005). Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc. Natl. Acad. Sci. USA 102: 5779–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C.Y., Saedler H. (2007). Hormonal control of the inflated calyx syndrome, a morphological novelty, in Physalis. Plant J. 49: 935–946 [DOI] [PubMed] [Google Scholar]
- He C.Y., Sommer H., Grosardt B., Huijser P., Saedler H. (2007). PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-domain protein MPF2 from Physalis floridana. Mol. Biol. Evol. 24: 1229–1241 [DOI] [PubMed] [Google Scholar]
- He C.Y., Zhang J.S., Chen S.Y. (2002). A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor. Appl. Genet. 104: 1125–1131 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hu J.Y., Saedler H. (2007). Evolution of the inflated calyx syndrome in Solanaceae. Mol. Biol. Evol. 24: 2443–2453 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Huijser P., Klein J., Lönnig W.E., Meijer H., Saedler H., Sommer H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink R.G.H., Kaufmann K., Angenent G.C. (2010). The ‘ABC’ of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21: 87–93 [DOI] [PubMed] [Google Scholar]
- Kanno A., Saeki H., Kameya T., Saedler H., Theissen G. (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52: 831–841 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Khan M.R., Hu J., Ali G.M. (2012). Reciprocal loss of CArG-boxes and auxin response elements drives expression divergence of MPF2-Like MADS-box genes controlling calyx inflation. PLoS ONE 7: e42781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie G., Kolbe A., Lemoine R., Roessner U., Lytovchenko A., Zuther E., Kehr J., Frommer W.B., Riesmeier J.W., Willmitzer L., Fernie A.R. (2003). Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology. Planta 217: 158–167 [DOI] [PubMed] [Google Scholar]
- Leseberg C.H., Eissler C.L., Wang X., Johns M.A., Duvall M.R., Mao L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Litt A. (2007). An evaluation of A-function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168: 73–91 [Google Scholar]
- Litt A., Irish V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A., Kramer E.M. (2010). The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 21: 129–137 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Mandel M.A., Gustafson-Brown C., Savidge B., Yanofsky M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mandel M.A., Yanofsky M.F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377: 522–524 [DOI] [PubMed] [Google Scholar]
- Ng M., Yanofsky M.F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13: 739–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z. (1998). Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S.N., Van Dongen J.T., Alfred S.C., Mamun E.A., Zhao X., Saini H.S., Fernandes S.F., Blanchard C.L., Sutton B.G., Geigenberger P., Dennis E.S., Dolferus R. (2005). Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 28: 1534–1551 [Google Scholar]
- Pabón-Mora N., Ambrose B.A., Litt A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158: 1685–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Peña L., Martín-Trillo M., Juárez J., Pina J.A., Navarro L., Martínez-Zapater J.M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19: 263–267 [DOI] [PubMed] [Google Scholar]
- Preston J.C., Kellogg E.A. (2006). Reconstructing the evolutionary history of paralogous APETALA1/FRUITFULL-like genes in grasses (Poaceae). Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwala A.P., Miller W.B. (1998). Sucrose-cleaving enzymes and carbohydrate pools in Lilium longiflorum floral organs. Physiol. Plant. 103: 541–550 [Google Scholar]
- Riß, S. (2009). Isolation and Analysis of MPF2-Like MADS-Box Genes from Physaleae and Characterization of Their cis-Regulatory Regions. PhD dissertation (Koeln, Germany: Universität zu Köln). [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936 [DOI] [PubMed] [Google Scholar]
- Shan H.Y., Zhang N., Liu C.J., Xu G.X., Zhang J., Chen Z.D., Kong H.Z. (2007). Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol. Phylogenet. Evol. 44: 26–41 [DOI] [PubMed] [Google Scholar]
- Taylor S.A., Hofer J.M.I., Murfet I.C., Sollinger J.D., Singer S.R., Knox M.R., Ellis T.H. (2002). PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol. 129: 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Melzer R. (2007). Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. (Lond.) 100: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W.E., Saedler H., Sommer H., Schwarz-Sommer Z. (1992). GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus S.L., de Folter S., Shchennikova A.V., Kaufmann K., Immink R.G.H., Angenent G.C. (2009). In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus S.L., Martinelli A.P., Dinh Q.D., Aizza L.C., Dornelas M.C., Angenent G.C., Immink R.G.H. (2010). Intercellular transport of epidermis-expressed MADS domain transcription factors and their effect on plant morphology and floral transition. Plant J. 63: 60–72 [DOI] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Weigel D., Meyerowitz E.M. (1994). The ABCs of floral homeotic genes. Cell 78: 203–209 [DOI] [PubMed] [Google Scholar]
- West A.G., Shore P., Sharrocks A.D. (1997). DNA binding by MADS-box transcription factors: A molecular mechanism for differential DNA bending. Mol. Cell. Biol. 17: 2876–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dinneny J.R., Crawford K.M., Rhee Y., Citovsky V., Zambryski P.C., Weigel D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Yang Z.H. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573 [DOI] [PubMed] [Google Scholar]
- Yang Z.H. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yu H., Ito T., Wellmer F., Meyerowitz E.M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Zhang H., Liang W.Q., Yang X.J., Luo X., Jiang N., Ma H., Zhang D.B. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.S., Khan M.R., Tian Y., Li Z.C., Riss S., He C.Y. (2012). Divergences of MPF2-like MADS-domain proteins have an association with the evolution of the inflated Calyx syndrome within Solanaceae. Planta 236: 1247–1260 [DOI] [PubMed] [Google Scholar]
- Zhang J.Z., Nielsen R., Yang Z.H. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22: 2472–2479 [DOI] [PubMed] [Google Scholar]