A region on chromosome 8 of several Solanum species contains genes for terpene synthases and cis-prenyl transferases, the latter encoding the enzymes that catalyze the formation of the substrates used by enzymes encoded by the former. Detailed sequence and biochemical analyses identify molecular events that gave rise to distinct gene composition and function in the different Solanum species.
Abstract
Functional gene clusters, containing two or more genes encoding different enzymes for the same pathway, are sometimes observed in plant genomes, most often when the genes specify the synthesis of specialized defensive metabolites. Here, we show that a cluster of genes in tomato (Solanum lycopersicum; Solanaceae) contains genes for terpene synthases (TPSs) that specify the synthesis of monoterpenes and diterpenes from cis-prenyl diphosphates, substrates that are synthesized by enzymes encoded by cis-prenyl transferase (CPT) genes also located within the same cluster. The monoterpene synthase genes in the cluster likely evolved from a diterpene synthase gene in the cluster by duplication and divergence. In the orthologous cluster in Solanum habrochaites, a new sesquiterpene synthase gene was created by a duplication event of a monoterpene synthase followed by a localized gene conversion event directed by a diterpene synthase gene. The TPS genes in the Solanum cluster encoding cis-prenyl diphosphate–utilizing enzymes are closely related to a tobacco (Nicotiana tabacum; Solanaceae) diterpene synthase encoding Z-abienol synthase (Nt-ABS). Nt-ABS uses the substrate copal-8-ol diphosphate, which is made from the all-trans geranylgeranyl diphosphate by copal-8-ol diphosphate synthase (Nt-CPS2). The Solanum gene cluster also contains an ortholog of Nt-CPS2, but it appears to encode a nonfunctional protein. Thus, the Solanum functional gene cluster evolved by duplication and divergence of TPS genes, together with alterations in substrate specificity to utilize cis-prenyl diphosphates and through the acquisition of CPT genes.
INTRODUCTION
Gene duplications provide the raw material for the evolution of genes with new functions by allowing selection to act on divergent sequences while maintaining the original function of the gene (Ohno, 1970). Various mechanisms in eukaryotes lead to gene duplications, and many gene duplication events result in a pair of genes that are initially linked in tandem (Pichersky, 1990). The process can repeat itself; thus, clusters of genes that are homologous are often found on eukaryotic chromosomes. These homologous genes may have the same expression characteristics and encode proteins with the same biochemical function, particularly if the duplication events occurred recently, but over time they often diverge in both aspects.
In bacteria, genes encoding nonhomologous proteins catalyzing different steps in the same biochemical pathway are often physically located next to each other in units called operons and may be transcribed as one polycistronic RNA (Osbourn and Field, 2009; Koonin, 2009). In eukaryotes, polycistronic mRNAs are rare, and physical clusters of nonhomologous genes encoding proteins that participate in the same pathway (referred to here as “functional gene clusters”) are also uncommon, although such clusters are observed in fungal genomes (Cho et al., 1998; Kruglyak and Tang, 2000; Hurst et al., 2002, 2004; Wong and Wolfe, 2005) and recently have been identified in plant genomes. Plant functional gene clusters typically encode enzymes for specialized metabolism, such as the cluster of genes for the synthesis of cyclic hydroxamic acid in maize (Zea mays; Frey et al., 1997), for the triterpenes avenacin in Avena strigosa (Qi et al., 2004) and thalianol and modified marmeral in Arabidopsis thaliana (Field and Osbourn, 2008; Field et al., 2011), for cyanogenic glucosides in Lotus japonica, cassava (Manihot esculenta), and Sorghum bicolor (Takos et al., 2011), for the diterpenes momilactone and phytocassane in rice (Oryza sativa; Wilderman et al., 2004; Shimura et al., 2007), and for the alkaloid noscapine in poppy (Papaver somniferum; Winzer et al., 2012). It is noteworthy that such functional clusters also often contain duplicated genes that may (e.g., the cyclic hydroxamic acid cluster) or may not have evolved different functions (Osbourn, 2010).
Because of limited research into genomic organization of metabolic genes in plants, it is not yet clear whether functional clusters are completely or mostly restricted to specialized metabolism and how common they are within specialized metabolism (Field et al., 2011; Field and Osbourn, 2012). However, some areas of plant specialized metabolism have received more attention in this respect. The terpenoid pathway leading to the synthesis of the specialized metabolites of the monoterpene, sesquiterpene, and diterpene classes (with skeletons of 10, 15, and 20 carbons, respectively; Figure 1A) is widespread in plants, and the organization of genes encoding terpene synthases (TPSs; a term designating the enzymes responsible for the synthesis of the basic skeleton structures of mono-, sesqui-, and diterpenes; Figure 1A) has been examined in several plant genomes (Chen et al., 2011; Zhuang et al., 2012). In Arabidopsis, a number of TPS genes are found in close association with other confirmed or putative terpenoid metabolism-related genes, for example, genes encoding geranylgeranyl diphosphate synthases (the enzyme that synthesizes the substrate for diterpene synthases; Figure 1A), cytochrome P450s (which often hydroxylate terpenes), and glycosyl transferases (Aubourg et al., 2002). However, evidence is currently lacking for the role of any of these genes in terpenoid biosynthesis, and a similar association of such genes with TPS genes is not evident in the grape (Vitis vinifera) genome (Martin et al., 2010; Chen et al., 2011).
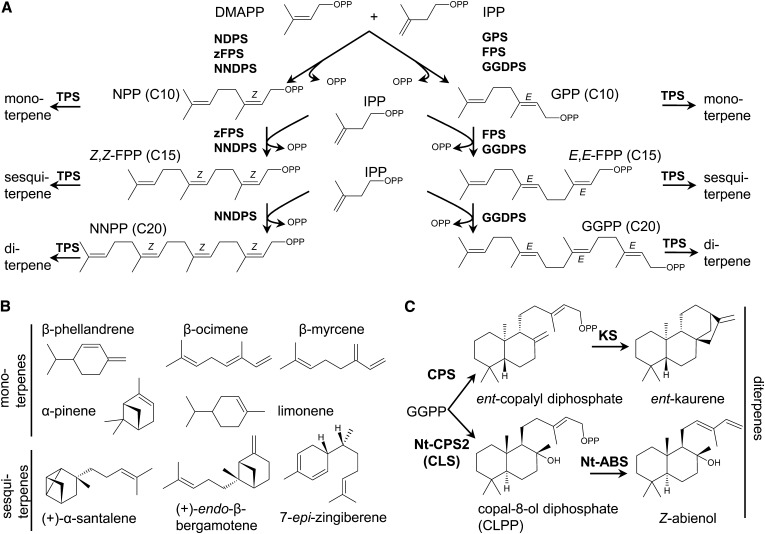
Figure 1.
Terpene Biosynthesis in Solanum.
(A) Biosynthesis of cis- and trans-prenyldiphosphates.
(B) Chemical structures of some monoterpenes and sesquiterpenes synthesized by tomato TPSs.
(C) The general pathway in plants to the synthesis of ent-kaurene (a precursor of gibberellins) from GGPP and the synthesis of Z-abienol from GGPP in N. tabacum.
E,E-FPP, 2E,6E-farnesyl diphosphate; zFPS, cis-farnesyl diphosphate synthase; NNDPS, nerylneryl diphosphate synthase; GPS, geranyl diphosphate synthase; FPS, trans-farnesyl diphosphate synthase; GGDPS, geranylgeranyl diphosphate synthase; Nt-CPS2, N. tabacum ent-copalyl diphosphate synthase 2 (=copal-8-ol diphosphate synthase); Nt-ABS, N. tabacum Z-abienol synthase.
We recently reported a functional cluster of genes for terpene biosynthesis on chromosome 8 of cultivated tomato (Solanum lycopersicum; Falara et al., 2011). The cluster contains five TPS genes (TPS18, TPS19, TPS20, TPS21, and TPS41). In addition, it contains two complete cis-prenyl transferase (CPT) genes, one of which, NERYL DIPHOSPHATE SYNTHASE1 (NDPS1), is expressed mainly in trichomes and encodes an enzyme catalyzing the formation of neryl diphosphate (NPP) that is used by TPS20 (also known as PHELLANDRENE SYNTHASE1 [PHS1]), a member of the TPS-e/f subfamily, or clade, to synthesize β-phellandrene and several other monoterpenes (Schilmiller et al., 2009; Falara et al., 2011; Figures 1A and 1B). Similar enzymes are also expressed in the trichomes of a wild tomato species Solanum pennellii, although they predominantly synthesize α-phellandrene (Falara et al., 2011). Interestingly, in a second wild tomato species, Solanum habrochaites accession LA1777, two homologous trichome-expressed genes on the same region of chromosome 8 were identified that encode proteins with different activities (Sallaud et al., 2009). One of these genes, the CPT gene CIS-FARNESYL DIPHOSPHATE SYNTHASE (zFPS), shows high identity to Sl-NDPS1 and encodes an enzyme that catalyzes the synthesis of the atypical TPS substrate 2Z,6Z-farnesyl diphosphate (Z,Z-FPP) (Figure 1A). The second gene, a TPS gene, encodes an enzyme designated SANTALENE AND BERGAMOTENE SYNTHASE (Sh-SBS) that utilizes Z,Z-FPP to synthesize a mixture of sesquiterpenes in which santalene and bergamotene predominate (Figure 1B).
The use of the cisoid substrates NPP and Z,Z-FPP for mono- and sesquiterpene synthesis is atypical as plants typically use the trans substrates geranyl diphosphate (GPP) and 2E,6E-FPP (E,E-FPP) to synthesize mono- and sesquiterpenes (Figure 1A). In addition, whereas monoterpene biosynthesis using GPP occurs in the chloroplast, sesquiterpene biosynthesis derived from E,E-FPP generally occurs in the cytosol (Chen et al., 2011). However, in the trichomes of tomato and its wild relatives, biosynthesis of both mono- and sesquiterpenes from NPP and Z,Z-FPP, respectively, occurs in the chloroplast (Sallaud et al., 2009; Schilmiller et al., 2009). Furthermore, we recently reported considerable variation in terpene biosynthesis in the trichomes of diverse S. habrochaites accessions collected from the geographic range of the species (Gonzales-Vigil et al., 2012). Four additional TPS transcripts were identified, including those encoding a sesquiterpene synthase, Sh-ZIS, which utilizes Z,Z-FPP to synthesize 7-epi-zingiberene, and three monoterpene synthases, Sh-PHS1, Sh-PIS, and Sh-LMS, which utilize NPP to synthesize β-phellandrene, α-pinene, and limonene, respectively (Figure 1B). Overall, these data suggest a diverse pattern of terpene biosynthesis in the trichomes of Solanum species. However, although these compounds are all synthesized by CPTs and TPSs that are quite similar to each other (within each type), the absence of genomic sequences of the wild tomato species made it difficult to accurately determine the evolutionary changes that gave rise to this great diversity and, in particular, made discussions of orthology between genes impossible.
With the exception of Sl-TPS20, the functions of the enzymes encoded by the other TPS genes in the chromosome 8 cluster of cultivated tomato have not been reported. The sequence of the Sl-TPS41 protein, a member of the TPS-c clade (Chen et al., 2011), is most similar to copal-8-ol diphosphate synthases (CLS) from several species, including tobacco (Nicotiana tabacum; Sallaud et al., 2012). Copal-8-ol diphosphate (CLPP; Figure 1C) is a substrate in the synthesis of the specialized labdane-type diterpenes (Peters, 2006; Falara et al., 2010; Sallaud et al., 2012). Sl-TPS18, Sl-TPS19, and Sl-TPS21 are members of the highly diverse TPS-e/f clade, which in all plant species contains ent-kaurene synthase (KS), the enzyme involved in gibberellin biosynthesis, and in select species also contains mono-, sesqui-, or diterpene synthases (Chen et al., 2011).
Tomato is a member of the Solanaceae family, and the availability of genomic sequences (Tomato Genome Consortium, 2012) and additional genomic and EST databases for several species in this family makes it possible to examine and compare the composition of genes in this cluster in relatively closely related species. Here, we report the results of such an analysis, followed by biochemical analysis to determine the function of the enzymes these genes encode. The combined data were used to determine the evolutionary trajectories of the genes in this cluster in Solanaceae and to further understand how functional clusters in specialized metabolism are formed and evolve over time.
RESULTS
The TPS and CPT Gene Clusters at the Tip of Chromosome 8 in S. lycopersicum and Solanum pimpinellifolium Share the Same Gene Complement and Organization
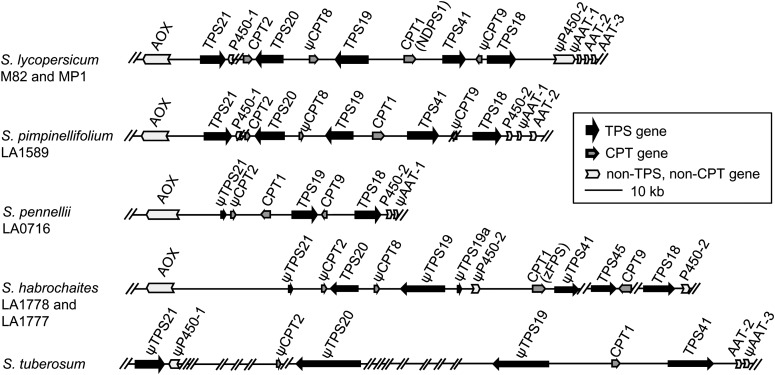
While the released tomato genome sequence suggested that S. lycopersicum TPS18, TPS19, TPS20, TPS21, TPS41, and NDPS1 are present in the chromosome 8 cluster, the actual sequence assembly was poor, with many gaps and incomplete gene sequences. In a subsequent study (Falara et al., 2011), we obtained complete genomic sequences for most of these genes by genomic PCR and reported the presence of a second gene related to NDPS1. The cause of the incomplete and erroneous assembly of this region is likely the presence of multiple copies of highly related sequences, both coding and noncoding. In this study, to solve this problem and to obtain a complete and correct assembly of this region showing the arrangement of the genes, we obtained a BAC of tomato DNA covering this area and physically isolated separate fragments and determined their sequence (Figure 2; see Supplemental Figure 1 online).
Figure 2.
The Terpene Metabolic Gene Cluster at the Tip of Chromosome 8 in Solanaceae.
The genes that are not functional because of deletions or insertions are shown with a “ψ” symbol. The sequences of S. tuberosum were obtained from the database of SOL Genomics Network (http://solgenomics.net/). Scaffold numbers are PGSC0003DMB000000773 and PGSC0003DMS000001497. AOX, alcohol oxidase; NDPS1, neryl diphosphate synthase 1; P450, cytochrome P450; AAT, alcohol acyltransferase; zFPS, cis-farnesyl diphosphate synthase.
The complete sequence of this region (with one gap, flanked by AT repeats) now shows that the cluster contains five complete TPS genes, TPS18, TPS19, TPS20, TPS21, and TPS41, as well as NDPS1 (also referred to as CPT1 in this study and in Akhtar et al., 2013), a second CPT (CPT2; Akhtar et al., 2013), and two partially deleted CPTs (ψCPT8 and ψCPT9). An apparently functional cytochrome P450 gene is situated between TPS21 and TPS20, and the cluster is flanked on one side by a gene for aldehyde oxidase and on the other side by a second cytochrome P450 gene that contains an insertion, followed by three alcohol acyl transferase genes, with the first having multiple deletions and mutations and the other two potentially functional (Figure 2; see Supplemental Figure 1 online). We note that the annotation of TPS41 (see Supplemental Figure 1 online) now shows a gene with 15 exons rather than only 12 as previously described (Falara et al., 2011). The additional three exons (exons 1, 2, and 3) were discovered by comparison with the recently published CLS gene from tobacco, CPS2 (Sallaud et al., 2012; Figure 1C). The 3′ splice site of intron 3 of the S. lycopersicum cultivar Heinz TPS41 gene, as reported in the released genome sequence, is AT rather than the consensus AG, which is found in the tobacco gene (Sallaud et al., 2012). We sequenced the regions in cultivars M82 and MP1, and this splice site is also AT in both. Sequencing of cDNAs from the M82 and MP1 tomato cultivars showed proper splicing of exons 3 and 4 despite this aberrant 3′ splice sequence.
Among the wild relatives of the cultivated tomato, S. pimpinellifolium is phylogenetically the most closely related species to S. lycopersicum (Olmstead et al., 2008), and a draft genome sequence of S. pimpinellifolium accession LA1589 obtained using short sequence reads was recently published (Tomato Genome Consortium, 2012). BLASTN searches utilizing sequences from the S. lycopersicum chromosome 8 cluster identified several homologous contigs of high nucleotide sequence identity from S. pimpinellifolium. These contigs were utilized as a framework for primer design, and gaps between the scaffolds were closed using PCR and sequencing, leading to a final assembly of 107 kb. Analysis of the S. pimpinellifolium (LA1589) chromosome 8 cluster revealed that the gene complement, order, and orientation was completely conserved with S. lycopersicum with the only observed differences being single nucleotide polymorphisms within the predicted protein coding regions of each gene and single nucleotide polymorphisms and insertion/deletions within intron and other noncoding regions. Furthermore, each of the genes in the S. pimpinellifolium cluster are highly similar to their S. lycopersicum counterpart and do not contain obvious deletions or mutations that could render them nonfunctional (Figure 2; see Supplemental Figure 1 online).
Rearrangements and Deletions Define the Chromosome 8 TPS and CPT Gene Clusters of S. pennellii and S. habrochaites
Previous research has documented several distinct TPS-e/f genes located on the top of chromosome 8 in the wild tomato species S. pennellii and S. habrochaites together with variation in the terpene products of the enzymes that they encode (Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011; Gonzales-Vigil et al., 2012). However, genomic sequence of the chromosome 8 cluster from these wild tomato relatives was unavailable, rendering assignment of orthology and paralogy impractical. To address the evolutionary relationships of these genes, genomic sequences of the chromosome 8 TPS/CPT cluster of S. pennellii and S. habrochaites were obtained. Similar to the approach utilized for generating sequence of the S. pimpinellifolium cluster, a draft genome sequence assembly of S. pennellii LA0716 (http://www.usadellab.org/cms/index.php?page=projects-some) was queried using sequences derived from the S. lycopersicum cluster and scaffolds were obtained that were utilized as a framework for completing the remaining sequence of the cluster using sequencing of PCR fragments, leading to the assembly of a 76-kb region (Figure 2). While the gene cluster in this region is marked by Spe-TPS21 and Spe-TPS18 on either end, as in S. lycopersicum and S. pimpinellifolium, Spe-TPS21 is likely nonfunctional as deletions removed exon 6 and all exons after exon 7, Spe-TPS19 and Spe-CPT1 are inverted in their arrangement and orientation compared with their position in the S. lycopersicum genome, there is no Spe-TPS20, and a 13-kb deletion removed the entire Spe-TPS41 gene (Figure 2; see Supplemental Figure 1 online). The lack of Spe-TPS41 in LA0716 and additional S. pennellii accessions was confirmed using PCR-based markers at this locus (see Supplemental Figure 2 and Supplemental Table 1 online). Note that the gene now designated as Spe-TPS19 (accession number KC807997) was originally designated as Spe-TPS20 (Falara et al., 2011; cDNA accession number JN412071) because its exact position was not known at the time, but our analysis indicates that it is most likely orthologous to Sl-TPS19 since it is located immediately adjacent to Spe-CPT1 (Figure 2), and its sequence overall is more similar to other TPS19 sequences than to TPS20 sequences in Solanum (Figure 3; see Supplemental Figure 3 online).
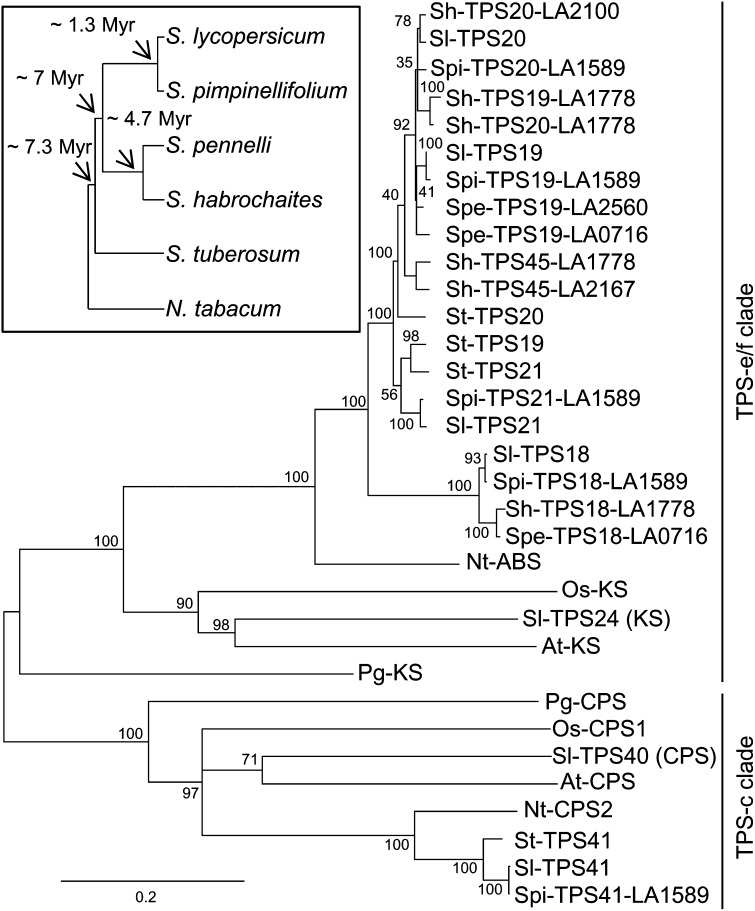
Figure 3.
Phylogenetic Analysis of Solanaceae TPS Genes.
A maximum likelihood phylogenetic tree obtained as described in Methods. Inset, phylogenetic relationships in the genus Solanum, with estimated time of divergence in millions of years, based on Tomato Genome Consortium (2012) and Nakazato et al. (2010). Sl, S. lycopersicum; Spe, S. pennellii; Spi, S. pimpinellifolium; Sh, S. habrochaites; St, S. tuberosum; Nt, Nicotiana tabacum; Os, Oryza sativa; At, Arabidopsis thaliana; Pg, Picea glauca. Myr, million years ago.
To examine the composition and arrangement of this functional gene cluster in S. habrochaites, an ordered BAC library from S. habrochaites LA1778, an accession that possesses a trichome-derived terpene profile identical to that previously described for LA1777 (Sallaud et al., 2009; see Supplemental Figure 4 online), was screened with cDNA probes derived from Sl-TPS20 and Sh-CPT9, identifying a single BAC clone of 119 kb that was subcloned and sequenced. The BAC clone covered the region from the AOX gene to the end of Sh-TPS41. Additional sequences covering the region downstream from Sh-TPS41 to Sh-P450-2 were obtained by PCR. Sequence analysis of the reassembled region (Figure 2) revealed substantial rearrangement of the cluster together with several deletions that removed all or part of Sh-CPT2, Sh-TPS19, Sh-TPS21, Sh-TPS41, and Sh-P450-1. Next to the mutated Sh-TPS19, there are two exons that resemble exons 12 and 13 of Sh-TPS19, and this truncated gene was labeled Sh-ψTPS19a. However, Sh-CPT9, similarly to S. pennellii but unlike in S. lycopersicum, has a complete open reading frame. Furthermore, an additional gene, designated Sh-TPS45, which is identical to the previously published Sh-SBS sequence (Sallaud et al., 2009), is located between Sh-TPS41 and Sh-CPT9 (Figure 2; see Supplemental Figure 1 online). Sh-SBS is highly similar to Sh-ZIS, and recent screening of diverse S. habrochaites accessions failed to identify individual accessions that contained both of these genes (Gonzales-Vigil et al., 2012), suggesting the possibility that they may be different alleles of the same locus. To test this hypothesis, we sequenced the genomic region encompassing Sh-ZIS and Sh-CPT9 from three zingiberene-producing accessions, LA1696, LA2104, and LA2167, with data indicating that the connection and orientation between Sh-ZIS and Sh-CPT9 is the same as that observed between Sh-SBS and Sh-CPT9 (see Supplemental Figure 5 online).
Organization of the TPS and CPT Gene Cluster in S. tuberosum and Identification of the Potential N. tabacum Orthologs
In potato (S. tuberosum), we were able to identify scaffolds that map to chromosome 8 and appear to contain the orthologous TPS and CPT genes found in the chromosome 8 functional cluster of the other Solanum species. While the quality of the sequence is poor and there are many gaps (see Supplemental Figure 1 online), the overall arrangement of the genes is similar to that seen in the other Solanum species (Figure 2). However, while St-TPS41, St-CPT1, St-TPS19, St-CPT2, St-TPS20, and St-TPS21 are recognizable by their position, there appears to be no St-TPS18 gene (and no EST of a hypothetical St-TPS18 was found in any database). Furthermore, with the exception of St-TPS41, all the other genes present contain multiple deletions that shift the open reading frames.
In tobacco, cDNAs of Nt-CPS2, the gene most closely related to Solanum TPS41-type genes outside Solanum, and Nt-ABS (N. tabacum Z-abienol synthase; Figure 1C), the non-Solanum TPS gene most similar to the Solanum TPS-e/f genes in the chromosome 8 cluster (see below), have been reported (Sallaud et al., 2012). A search for genomic sequences in the databases allowed assembling parts of Nt-ABS gene including exons 1 to 9 and part of exon 12 to exon 14. A complete sequence of Nt-ABS (except for one small gap remaining in intron 5) was obtained by bridging the gaps by PCR and determining the sequence of the spanning fragments (see Supplemental Figure 1 online).
Phylogenetic Analysis of TPS and CPT Genes from the Functional Clusters
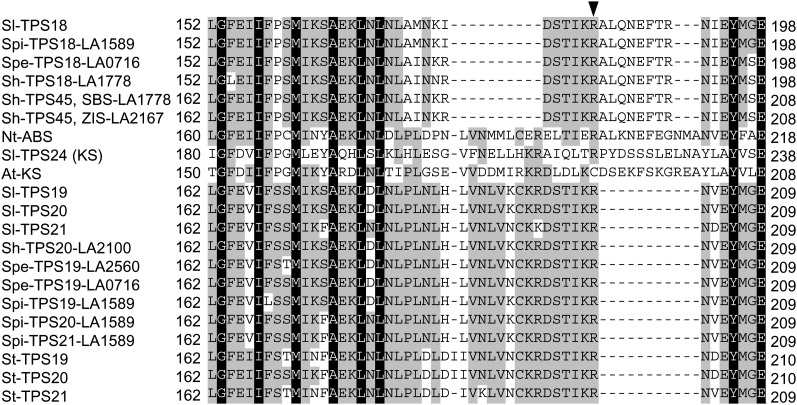
Phylogenetic analysis of the coding nucleotide sequences of the Solanaceae TPS genes (except those with major deletions) was performed using the maximum likelihood method (Figure 3). Because this method gave low bootstrap values to some branch points in the clade that included TPS18, TPS19, TPS20, TPS21, and TPS45 genes, a second analyses of these genes based on Bayesian inference was conducted, and it resulted in essentially the same branch topography, but with much higher support (see Supplemental Figure 3 online). Analysis of all the TPS-e/f genes in the cluster indicated that they all form a distinct clade that diverged from KS genes before the divergence of monocots and dicots but after the divergence of angiosperms and gymnosperms (Figure 3), a similar observation to that made for Nt-ABS (Sallaud et al., 2012). The phylogenetic tree makes clear that all the TPS-e/f genes in the Solanum cluster evolved from a single gene after the split of the Solanum-Nicotiana lineages. All the Solanum TPS18 genes form a separate branch, and all TPS21 genes and the S. tuberosum TPS19 and TPS20 genes are located on a parallel branch to that of the other Solanum TPS20 and TPS19 genes. Sh-TPS45 (Sh-SBS) appears to be distinct but closely related to TPS19 and TPS20 genes (Figure 3). It was previously shown, based on protein sequence comparisons (derived from cDNA sequences), that the Sh-SBS and Sh-ZIS proteins are most similar to TPS20/19 proteins across most of their sequence but have an internal segment of ∼35 amino acids that is more similar to the corresponding region in Sl-TPS18 (Gonzales-Vigil et al., 2012). Our analysis of the genomic sequences strengthen this observation, since the genomic sequences of TPS18 genes as well as of Sh-SBS and Sh-ZIS show that they all contain a nine-codon deletion in exon 4 relative to TPS19, TPS20, and TPS21 genes, while the genes in the latter group have a deletion of 8 codons in exon 5 relative to TPS18 genes, Sh-SBS and Sh-ZIS (Figure 4).
Figure 4.
Alignment of Protein Sequences Encoded by the Region Flanking Intron 4 in the TPS Genes in the Functional Gene Cluster.
The arrow shows the position of the intron, which splits an Arg codon. Numbers indicate the amino acid positions in each protein. The alignment with full-length cDNAs is shown in Supplemental Data Set 4 online.
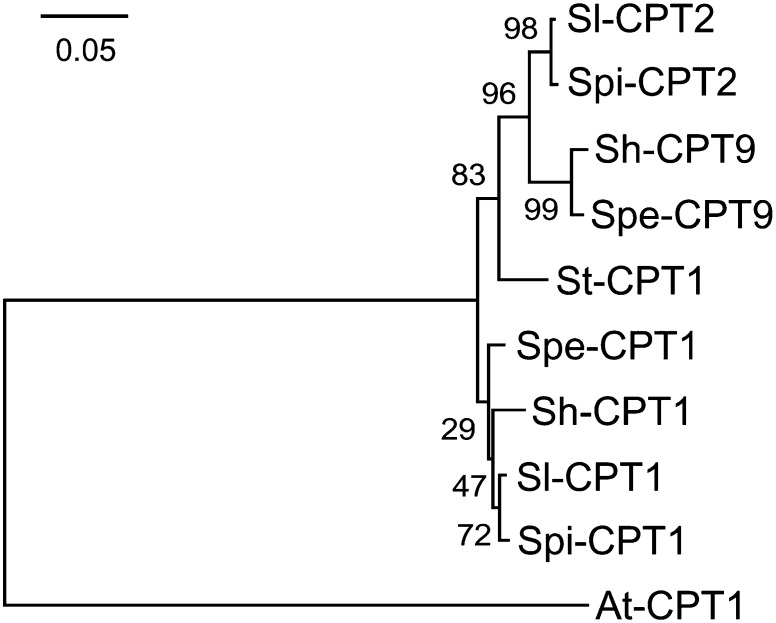
The phylogenetic analysis also shows that while TPS41-type genes from tobacco and the Solanum species are related to genuine copalyl diphosphate synthase (CPS) genes, belonging to the TPS-c clade of the TPS family, they form a distinct cluster that separated from CPS genes around the split between monocots and dicots, and after the split of the angiosperms from gymnosperms (Figure 3). A maximum likelihood phylogenetic analysis of CPT genes indicated that CPT1 genes from S. lycopersicum, S. pimpinellifolium, S. pennellii, and S. habrochaites cluster together, and CPT2 and CPT9 genes form another cluster (Figure 5).
Figure 5.
Phylogenetic Analysis of Solanaceae CPT Genes.
A maximum likelihood phylogenetic tree obtained as described in Methods. Sl, S. lycopersicum; Spe, S. pennellii; Spi, S. pimpinellifolium; Sh, S. habrochaites; St, S. tuberosum; At, Arabidopsis thaliana.
Enzymatic Activities of Proteins Encoded by the TPS and CPT Genes within the Functional Cluster
To attempt to deduce the direction in the evolution of the enzymatic capacities of the proteins encoded by the genes in this functional cluster, extensive in vitro enzymatic assays utilizing various substrates were performed (Table 1). Some previously characterized enzymes were included in these analyses to serve as reference points or for direct comparisons.
Table 1. The Preferred Substrates of the Enzymes Encoded by the Solanum TPS Genes of the Functional Cluster.
| Substrate | S. lycopersicum | S. habrochaites | |||||
|---|---|---|---|---|---|---|---|
| TPS18 | TPS19 | TPS20 | TPS21 | TPS41 | TPS20a | TPS45b | |
| GPP | − | +/− | − | − | − | − | − |
| NPP | − | + | + | − | − | + | +/− |
| E,E-FPP | − | +/− | − | − | − | − | - |
| Z,Z-FPP | − | +/− | − | +/− | − | − | +c |
| GGPP | − | − | − | - | − | − | - |
| NNPP | − | − | − | +c | − | − | +/− |
| CLPPd | − | NDe | ND | ND | ND | ND | ND |
In vitro assays were carried out as described in Methods. The “+” indicates preferred substrate, “−” indicates no significant activity with the substrate, and “+/−” indicates some activity but kcat/Km is >10-fold lower than with the preferred substrate, except in the case of Sh-TPS45 (SBS) and Sl-TPS21 (see footnote c).
From accession LA2100.
From accession LA1393.
Since NNPP is not commercially available, coupled assays with Sl-CPT2, IPP, and DMAPP were performed to achieve a concentration of NNPP equivalent to the concentrations used for the other substrates. Preferred substrate was defined as the substrate that the enzyme converted to product at a rate >2.5-fold greater than the other substrates during the 2-h assay period.
Since CLPP is not commercially available, shown here are the results of a coupled assay with Sl-TPS18 and Cistus creticus CLS (Falara et al., 2010) and GGPP.
ND, not determined.
In tobacco, CPS2 (encoding a protein most similar to the Solanum TPS41 proteins) encodes CLS, the enzyme that supplies the substrate for Nt-ABS (Sallaud et al., 2012; Figure 1C). The TPS41 genes encode proteins with a complete open reading frame in S. lycopersicum, S. pimpinellifolium, and S. tuberosum, but not in S. pennellii and S. habrochaites. However, the putative TPS41 proteins in S. lycopersicum, S. pimpinellifolium, and S. tuberosum have a nonconserved substitution in the DXDD motif shown to be essential for CPS-like enzymes (these three TPS41 have EVDT instead) and are therefore unlikely to be functional. We tested this hypothesis by expressing Sl-TPS41 in Escherichia coli and assaying the purified enzyme for activity with geranylgeranyl diphosphate (GGPP), and no activity was observed. In addition, no significant activity was observed with GPP, NPP, E,E-FPP, Z,Z-FPP, and nerylneryl diphosphate (NNPP; Table 1). Since NNPP is not commercially available, we performed coupled assays with Sl-CPT2, which was recently shown to be an NNPP synthase that uses isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (Akhtar et al., 2013; see Methods).
TPS18 genes in S. lycopersicum, S. pimpinellifolium, S. pennellii, and S. habrochaites all have an intact open reading frame, although the gene may be missing in S. tuberosum. The Sl-TPS18 enzyme was produced by expression in E. coli, purified, and tested with GPP, NPP, E,E-FPP, Z,Z-FPP, GGPP, NNPP, and CLPP (the latter two substrates via a coupled assay; see Methods), but no activity was observed (Table 1).
TPS20 from S. lycopersicum (=PHS1), S. pennellii, and S. habrochaites was previously shown to catalyze the formation of phellandrenes and several other monoterpenes from NPP (Schilmiller et al., 2009; Gonzales-Vigil et al., 2012). Sl-TPS20, with a Km value of 9.1 µM and a kcat value of 4.1 s−1 for NPP, did not have significant activity with any other substrate tested (Table 1). Although we previously reported the enzymatic activities of proteins encoded by several alleles at the S. habrochaites TPS20 locus, all of which function as monoterpene synthases, they were not originally tested with the C20 substrates GGPP and NNPP (Gonzales-Vigil et al., 2012). Since Sh-TPS20 from LA1778 is not expressed, we expressed a cDNA for Sh-TPS20 from accession LA2100 and tested the protein, produced in E. coli, for activity with multiple substrates. It had activity only with NPP, catalyzing the formation of β-phellandrene as the dominant product.
The Sl-TPS19 enzyme catalyzed the formation of β-myrcene and β-ocimene isomers from either NPP or GPP (but at different ratios) and β- and α-farnescene from Z,Z-FPP and E,E-FPP (see Supplemental Figure 6 online). However, analysis of the kinetic parameters of the purified enzyme produced in E. coli indicated a Km value for NPP of 29.6 µM and a kcat value of 1.38 s−1, Km values for GPP and E,E-FPP too high to be determined, and a Km value for Z,Z-FPP of 59.8 µM and a kcat value of 0.017 s−1 (Table 2). Sl-TPS19 was unable to use either GGPP or NNPP as a substrate. With a kcat/Km value for NPP being >150-fold higher than the corresponding value for Z,Z-FPP, we concluded that the specific substrate of Sl-TPS19 is NPP (Table 1).
Table 2. Kinetic Analysis of TPSs in Solanum.
| Enzyme | Substrate | Km (µM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| Sl-TPS19 | NPP | 29.6 ± 1.1 | 1.38 ± 0.08 | 46.6 |
| Sl-TPS19 | Z,Z-FPP | 59.8 ± 7.3 | 0.017 ± 0.001 | 0.3 |
| Sl-TPS21 | Z,Z-FPP | 25.0 ± 2.5 | 0.017 ± 0.001 | 0.7 |
| Sh-TPS45a | Z,Z-FPP | 3.3 ± 0.4 | 0.59 ± 0.18 | 181.0 |
From accession LA1393.
S. pennellii (LA0716) has a single gene in the Sl-TPS19-Sl-TPS20 clade, now renamed Spe-TPS19 (see above). We have previously shown that an Spe-TPS19 allele from accession LA0716 encodes an enzyme that catalyzes the formation of α-phellandrene as the main monoterpene product (Falara et al., 2011). Here, we tested a second allele, from accession LA2560, and found that it encodes a protein with a similar substrate and product specificity as Sl-TPS19, producing β-myrcene and β-ocimene from NPP (see Supplemental Figure 6 online). Analysis of the volatile terpene profile of leaf trichome dips from plants of this accession indicated that this accession indeed preferentially synthesizes β-myrcene and β-ocimene (see Supplemental Figure 6 online).
Because of the very high identity between S. pimpinellifolium and S. lycopersicum TPS19 and TPS20 proteins, the S. pimpinellifolium TPS19 and TPS20 proteins were not tested for activity. In potato, all the TPS-e/f genes contain mutations that prevent the formation of functional proteins.
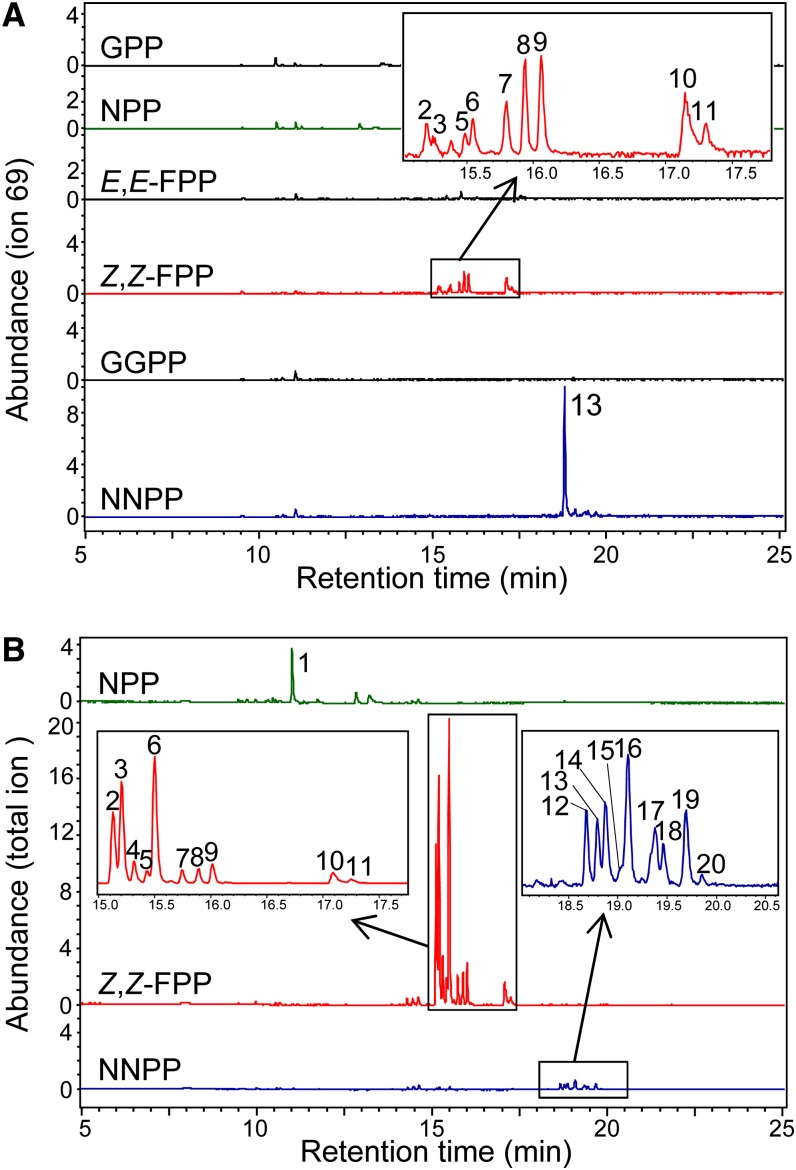
TPS21 appears to encode a functional protein only in S. lycopersicum and S. pimpinellifolium. In the coupled assay of Sl-TPS21 with Sl-CPT2 using DMAPP and IPP as substrates, Sl-TPS21 catalyzed the formation of an unidentified diterpene based on retention time (Figure 6A) and mass spectrometry (see Supplemental Figure 7 online). When incubated with Z,Z-FPP, it catalyzed the formation of several sesquiterpenes, including santalene and bergamotene (Figure 6A), with a Km value of 25.0 µM and a kcat value of 0.017 s−1 (Table 2). Sl-TPS21 exhibited little activity with any other substrate. Although kinetics parameters could not be determined for Sl-TPS21 with NNPP due to the lack of a commercially available substrate, a comparison of the products produced in the coupled reaction utilizing the substrates (IPP and DMAPP) and the Sl-CPT2 enzyme at concentrations that led to the production of NNPP concentration equal to the concentration of Z,Z-FPP used to assay Sl-TPS21 indicated that Sl-TPS21 appears to be 2.5-fold more active with NNPP than with Z,Z-FPP (Table 1, Figure 6A).
Figure 6.
Sl-TPS21 and Sh-TPS45-LA1393 (SBS) in Vitro Enzymatic Assays with Various Substrates.
(A) Gas chromatograms of the products obtained in reactions catalyzed by Sl-TPS21 with various substrates.
(B) Gas chromatograms of the products obtained in reactions catalyzed by Sh-TPS45-SBS from accession LA1393 with NPP, Z,Z-FPP, and NNPP (this protein has one amino acid difference with Sh-SBS from accession LA1777 and was shown to give the same products as Sh-SBS from accession LA1777 with NPP and Z,Z-FPP; Sallaud et al., 2009; Gonzales-Vigil et al., 2012). The enzymatic assay solutions with all substrates other than NNPP contained 1.1 or 0.9 µg purified recombinant protein (Sl-TPS21 or Sh-TPS45, respectively) and 1.3 nmol of commercially available substrate in 50 μL CPT/TPS assay buffer (50 mM HEPES, 100 mM KCl, 7.5 mM MgCl2, 5% [v/v] glycerol, and 5 mM DTT, pH 7.5) and were incubated at 30°C for 2 h. Since NNPP is not commercially available, to ascertain that NNPP concentration was equivalent to the concentration of the commercially available substrates in the parallel assays, Sl-CPT2 activity was first measured using 14C-IPP and DMAPP as substrates by scintillation counting. Assays were then set up using 2.5 µg of Sl-CPT2 protein in the CPT/TPS assay buffer but with 3.3 nmol IPP and 1.7 nmol DMAPP and the reaction incubated for 2 h, after which Sl-CPT2 was denatured at 65°C for 10 min and 1.1 µg Sl-TPS21 or 0.9 µg Sh-TPS45 proteins were added to the reaction mixture. Peak 1, limonene; 2, (−)-endo-α-bergamotene; 3, (+)-α-santalene; 4, (−)-exo-α-bergamotene; 5, (−)-epi-β-santalene; 6, (+)-endo-β-bergamotene; 7 to 11, unidentified sesquiterpenes; 12 to 20, unidentified diterpenes. Mass spectra of all compounds are shown in Supplemental Figure 7 online.
Sh-TPS45-Sh-SBS enzymes (from accessions LA1777 and LA1393) were previously shown not to work with the trans-prenyldiphosphates GPP, E,E-FPP, and GGPP and to use NPP and, preferentially, Z,Z-FPP (Sallaud et al., 2009; Gonzales-Vigil et al., 2012). Here, we observed that Sh-SBS (from accession LA1393) also has activity with NNPP, producing a number of diterpenes (Figure 6B; see Supplemental Figure 7 online). However, its preferred substrate is Z,Z-FPP (Figure 6B), with a Km value of 3.3 µM and a kcat value of 0.59 s−1 (Table 2).
As described above, Sl-CPT2, which is located within the cluster (Figure 2; see Supplemental Figure 1 online), was previously shown to encode a protein that catalyzed the formation of NNPP using DMAPP and IPP as substrates (Akhtar et al., 2013). Sl-CPT2 is also able to use NPP and IPP or Z,Z-FPP and IPP as substrates. Here, the in vitro enzymatic activities of Sh-CPT9 and St-CPT1 were also determined. Sh-CPT9 catalyzed the formation of a mixture of Z,Z-FPP and NNPP (at an ∼2:1 ratio), using NPP and IPP as substrates (Table 3; see Supplemental Figure 8 online). St-CPT1 catalyzed the formation of NNPP using DMAPP and IPP or NPP and IPP as substrates (Table 3; see Supplemental Figure 8 online). St-CPT1 was also able to use Z,Z-FPP as a substrate but not as efficiently (Table 3).
Table 3. Products of Solanum CPTs in Vitro Assays with Various Substrates.
| Substrate | S. lycopersicum | S. habrochaites | S. tuberosum | ||
|---|---|---|---|---|---|
| CPT1 | CPT2 | CPT1 | CPT9 | CPT1 | |
| IPP + DMAPP | NPPa | NNPPb | Z,Z-FPPc | – | Z,Z-FPP, NNPP |
| IPP + NPP | – | Z,Z-FPP, NNPPb | Z,Z-FPPc | Z,Z-FPP, NNPP | Z,Z-FPP, NNPP |
| IPP + Z,Z-FPP | – | NNPPb | – | NNPP | NNPP |
The most abundant reaction product is indicated in bold. “-” indicates no product.
Originally reported by Schilmiller et al. (2009).
Originally reported by Akhtar et al. (2013).
Originally reported by Sallaud et al. (2009).
Expression of TPS and CPT Genes in the Chromosome 8 Cluster of S. lycopersicum
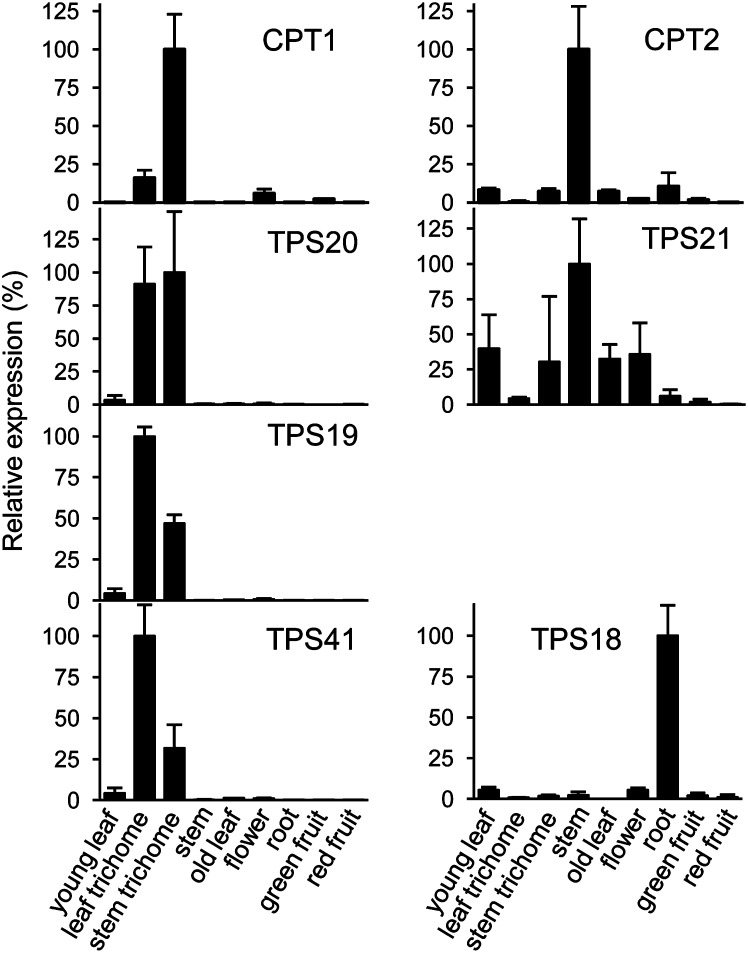
We examined relative transcript levels of Sl-CPT1 (NDPS1), Sl-CPT2, Sl-TPS18, Sl-TPS19, Sl-TPS20, Sl-TPS21, and Sl-TPS41 in different parts of the plant by quantitative RT-PCR (qRT-PCR) (Figure 7) (similar data for Sl-CPT1 and Sl-CPT2 were also reported in Akhtar et al., 2013). The data indicate that Sl-CPT1, Sl-TPS19, and Sl-TPS20 are almost exclusively expressed in stem and leaf trichomes, consistent with the presence in trichomes of monoterpenes derived from NPP (Schilmiller et al., 2009). Sl-TPS41 is also highly expressed in trichomes, although so far no activity has been demonstrated for the protein it encodes. On the other hand, Sl-CPT2, which encodes NNPP synthase, and Sl-TPS21, which uses NNPP to make an unidentified diterpene, are both expressed maximally in the stems. Sl-TPS18, for which no enzymatic activity has yet been identified, is maximally expressed in roots (Figure 7).
Figure 7.
Gene Expression Analysis by qRT-PCR.
TPS and CPT gene expression in different tissues of S. lycopersicum. PCR conditions and gene-specific primers are described in Methods and Supplemental Table 3 online. Error bars represent se. Values are from three biological replicates.
DISCUSSION
Phylogenetic and Biochemical Evidence Suggests That the Ancestral Solanaceae Gene Cluster Contained Diterpene Synthases
It has been hypothesized that functional gene clusters in plant specialized metabolism confer selective advantage due to the increased probability of inheriting all the genes for a complete pathway and expressing them together and, thus, the ability to synthesize a final product with defensive properties (Wegel et al., 2009; Field et al., 2011; Kliebenstein and Osbourn, 2012).
The phylogenetic analysis (Figure 3; see Supplemental Figure 3 online) indicates that the TPS-c and TPS-e/f genes clustered together at the tip of chromosome 8 in present day Solanum species are orthologs of the tobacco TPS genes CPS2 and ABS, respectively. While a genetic study previously concluded that Z-abienol synthesis is controlled by a single locus in tobacco (Vontimitta et al., 2010), suggesting that Nt-CPS2 and Nt-ABS are physically linked, the highly fragmented nature of the tobacco genomic sequences available to the public has precluded a determination of physical linkage between Nt-CPS2 and Nt-ABS. Furthermore, while Nt-CPS2 was physically mapped to this locus, the lack of polymorphism has so far prevented the mapping of the Nt-ABS gene (Sallaud et al., 2012).
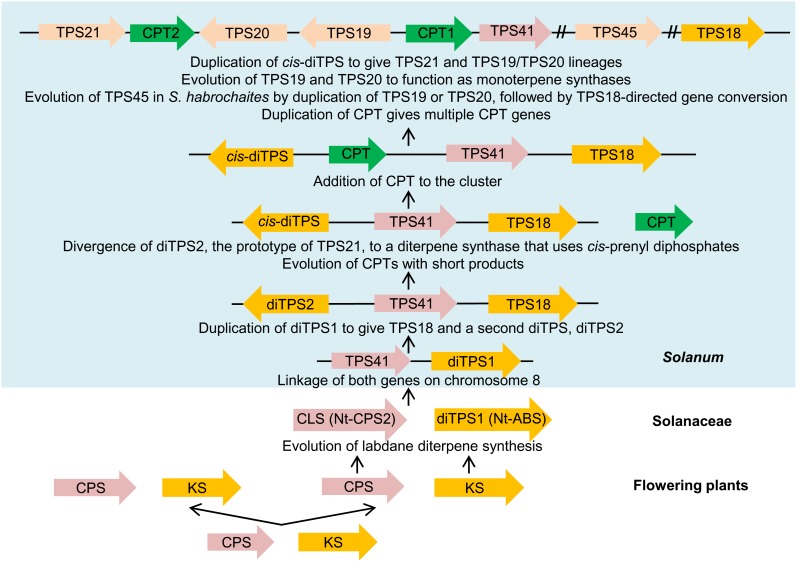
Thus, while it is presently unclear whether the clustering of these genes predates the split between the Solanum and Nicotiana lineages, it is likely that these genes were initially involved in diterpene biosynthesis using GGPP as the starting substrate. However, of the species analyzed here, only CPS2 and ABS in tobacco appear to retain this function, where the combined activity of the two encoded enzymes leads to the production of the diterpene Z-abienol (Sallaud et al., 2012). The inference from phylogenetic analysis that the original TPS-e/f gene in this cluster more closely resembled Nt-ABS (Figure 3) is strengthened by the observation that Nt-ABS is more similar to KS than any of the TPS-e/f genes in the Solanum cluster. Indeed, Nt-ABS does not have the deletions in exon 4 and exon 5 that are characteristic of the Solanum TPS-e/f genes (a deletion in exon 4 for TPS18 genes and a deletion in exon 5 for TPS21/19/20 genes; Figure 4). The early divergence of this part of the TPS-e/f clade for specialized diterpene biosynthesis (for a similar conclusion, see Sallaud et al., 2012) is in contrast with the situation where TPS-e/f genes for specialized diterpene synthesis evolved from KS more recently within the monocot lineage (Zhou et al., 2012).
If Nt-CPS2 and Nt-ABS are indeed linked in N. tabacum, then these genes already constituted a functional cluster, since the product of the enzyme encoded by the former gene is the substrate of the enzyme encoded by the latter. Each gene is derived from a diterpene synthase, CPS and KS, respectively, that is involved in primary metabolism (Figures 1C and 8). These enzymes also work in tandem (Duncan and West, 1981), but the genes encoding them are usually unlinked (see Falara et al., 2011 for S. lycopersicum and Mayer et al., 1999 and Theologis et al., 2000 for Arabidopsis).
Figure 8.
A Model for the Evolution of the Functional Gene Cluster.
ABS, Z-abienol synthase.
Evolution of the Functional Gene Cluster on Chromosome 8 in Solanum
The evidence obtained in this investigation reveals that this functional gene cluster has undergone further evolution in Solanum (Figure 8). The Solanum TPS41 genes, encoding proteins that are most similar to Nt-CPS2, are either completely or partially deleted or mutated, and thus may encode nonfunctional proteins (although the gene is still expressed; Figure 7). In addition, further duplications and divergence of the ancestral TPS-e/f gene in the cluster occurred in Solanum, leading to the TPS18 gene lineage (for which no enzymatic activity is currently assigned) and a second gene lineage. This second gene lineage further duplicated, leading to two additional TPS genes that encode proteins with different catalytic activities. In S. lycopersicum, TPS21 encodes an enzyme that acts most efficiently as a diterpene synthase, albeit one that uses NNPP, rather than GGPP, while TPS19 and TPS20 encode enzymes that use NPP to synthesize monoterpenes. It is worth noting that Sl-CPT2, which catalyzes the formation of NNPP, and Sl-TPS21, which uses this substrate, are close to each other in the S. lycopersicum cluster and also show similar expression patterns with maximal expression in stem (a search for the diterpene product of Sl-TPS21 in stem is currently underway). The expression patterns of Sl-CPT1, which catalyzes the formation of NPP, and Sl-TPS20 and Sl-TPS19, which use NPP to make monoterpenes, are also similar to each other with maximal expression in trichomes, and these three genes are also adjacent to one another. Thus, it appears that divergence of both biochemical and tissue-specific expression pattern is occurring with different components of this gene cluster in S. lycopersicum.
Finally, in S. habrochaites, TPS45, which encodes an enzyme that uses Z,Z-FPP to make sesquiterpenes, probably arose via an initial duplication event of Sh-TPS19 or Sh-TPS20 followed by a Sh-TPS18–directed gene conversion event of a short segment of the new gene (Figure 8). Our genomic analysis (see Supplemental Figure 5 online) shows that Sh-ZIS and Sh-SBS are both alleles of Sh-TPS45. As the Sh-ZIS allele is much more widespread in S. habrochaites populations than the Sh-SBS allele, which is limited to a central region of the geographic range of the species (Gonzales-Vigil et al., 2012), it is likely that the Sh-ZIS allele arose first.
The evolution of the TPS21/19/20 prototype gene that encoded an enzyme that used a cis-prenyl diphosphate as a substrate must have occurred concurrently with or subsequently to the evolution of a CPT that made short cis-prenyl diphosphates (≤C20), at least as some of its product. CPTs that make short-chain cis-prenyl diphosphates evolved recently, possibly within Solanaceae, from CPTs making longer cis-prenyl diphosphates (Akhtar et al., 2013). At some point, a gene encoding such substrates was integrated into this locus via a chromosomal rearrangement (Figure 8). Such a rearrangement must have occurred before the split between S. tuberosum and the other Solanum species investigated here, since an NNPP synthase (encoded by St-CPT1) is part of the locus in S. tuberosum (Figure 2). The CPT gene in the cluster duplicated and the copies diverged, so that each CPT protein evolved to catalyze the formation of distinct short cis-prenyl diphosphates. Interestingly, the evolution of a Z,Z-FPP synthase (zFPS) in S. habrochaites appears not to have involved a gene duplication, but rather the evolution of a new allele within the same gene, as Sl-NDPS1 (=Sl-CPT1) and zFPS (=Sh-CPT1) are orthologous loci (Figure 2).
It is possible that the gene duplications and divergence that gave rise to TPSs that use cis-prenyl diphosphates occurred before the Nicotiana and Solanum lineages split, and subsequently these genes were lost in tobacco. However, this scenario is less parsimonious (requiring both gain and loss events) and is also unlikely because TPS enzymes that use cis-prenyl diphosphates have not been identified in any other species in Solanaceae outside Solanum.
The switch to a cis-prenyl diphosphate substrate may have made TPS41 superfluous, a hypothesis supported by the accumulation of mutations in its catalytic domains in S. lycopersicum, S. pimpinellifolium, and S. tuberosum together with complete or partial deletion in S. pennellii accessions and S. habrochaites (LA1778) (Figure 2; see Supplemental Figures 1 and 2 online).
Which cis-Prenyl Diphosphate Substrate Was First Used by the TPS Genes in the Cluster?
As TPS21 genes and TPS19/20 genes are on sister clades, it is not obvious whether the most recent common ancestral gene encoded a protein that used NNPP or NPP as the substrate (or both). Since this entire TPS branch first arose from diterpene synthases that use trans-prenyl diphosphates, the most parsimonious scenario is a change from trans- to cis-prenyl diphosphate–utilizing diterpene synthases, thus making TPS21 most likely the original cis-prenyl diphosphate–utilizing TPS in the cluster. Furthermore, the single CPT gene found in the potato cluster encodes NNPP synthase rather than NPP synthase.
While the direction of change in substrate specificity between NNPP and NPP cannot be determined with certainty, it is interesting to note that the change in the length of the cis-prenyl diphosphate substrates among the TPS genes in the cluster parallels similar evolution among TPSs that use trans-prenyl diphosphates in that it appears to depend on the general flexibility of the enzymes in accepting substrates of different lengths. For example, TPSs can often utilize multiple substrates (GPP, farnesyl diphosphate [FPP], or GGPP) in vitro even though in vivo they may use mainly, or exclusively, a single substrate (reviewed in Chen et al., 2011).
Thus far, the in vivo switch of TPSs from linear trans-prenyl diphosphates to linear cis-prenyl diphosphate substrates has been documented only for TPS-e/f enzymes in Solanum (this work; Sallaud et al., 2009; Schilmiller et al., 2009). However, in vitro testing has shown that several tomato TPSs from other clades that use trans-prenyl diphosphates in vivo are capable of using cis-prenyl diphosphate substrates in vitro (Falara et al., 2011), and some sesquiterpene synthases from tobacco can use cis-trans FPP in addition to all-trans FPP as substrates (O’Maille et al., 2006). More recently, a CPT gene that catalyzes the head-to-middle condensation of two DMAPP molecules to produce lavendulyl diphosphate has been described in lavender (Lavendula x. intermedia) (Demissie et al., 2013). Lavendulyl diphosphate is predicted to be the precursor of the irregular monoterpene lavandulol. While the TPS that uses lavendulyl diphosphate remains unknown, these data are suggestive of convergent evolution of short-chain CPT activity.
In conclusion, our combined genomic, phylogenetic, and biochemical analyses of a functional gene cluster for terpene biosynthesis in several Solanum species indicate dynamic processes of gene accretion and divergent biochemical evolution.
METHODS
Plant Material and Growth Conditions
The seeds of Solanum lycopersicum, Solanum pennellii, Solanum pimpinellifolium, and Solanum habrochaites were obtained from the Tomato Genetic Resource Center (http://tgrc/ucdavis.edu). Throughout the text, when not specifically indicated, the S. lycopersicum plants used are of cultivar MP1. Plants were grown as previously described (Falara et al., 2011).
Isolation of Tomato Genomic DNA Fragments to Fill in Sequence Gaps
To further improve the genomic DNA sequence of the S. lycopersicum chromosome 8 TPS/CPT cluster presented by Falara et al. (2011), the BAC clone of the cluster (LE_HBa,0137M19) was obtained from the Arizona Genomics Institute (University of Arizona, Tucson, AZ). The BAC clone of 119 kb (SH_Ba, 0202J04) was also identified for S. habrochaites accession LA1778 to include much of the chromosome 8 cluster. Whole sequences were obtained by PCR using BACs as template or by subcloning into pBluescript vector with several restriction enzymes. Parts of the Sh-TPS41 and Sh-P450-2 sequences were obtained by DNA walking using the Genome Walker Universal Kit (Clontech). For S. pimpinellifolium, the recently published draft genome sequence of accession LA1589 (Tomato Genome Consortium, 2012) was utilized. Likewise, the S. pennellii genomic assembly was aided largely by the draft genome sequence and a scaffold assembly of accession LA0716 (http://www.usadellab.org/cms/index.php?page=projects-some). These resources, along with the nearly complete genomic DNA sequence of S. lycopersicum, formed our initial sequence assemblies. BLASTN searches with sequences from the S. lycopersicum chromosome 8 cluster identified homologous contigs or scaffolds of high nucleotide sequence identity. Those sequences were assembled and used as a framework for primer design, with the remaining gaps closed through PCR and subsequent sequencing. Homologous sequence alignments also aided primer design, especially for the highly similar TPS19 and TPS20 genes. PCR amplification was performed with KOD DNA polymerase (Novagen) or Taq DNA polymerase (New England BioLabs). Fragments obtained from PCR were gel purified and the DNA extracted with the MinElute gel extraction kit (Qiagen) or Wizard SV PCR and gel purification kit (Promega) and then verified by sequencing. Purified PCR fragments from regions of high similarity, such as TPS19 and TPS20, were cloned into the pCR 4Blunt-TOPO vector (Invitrogen) for sequencing.
Phylogenetic Analysis
Maximum likelihood trees in Figures 3 and 5 were constructed using MEGA version 5 (Tamura et al., 2011). Sequence alignments were first constructed in MEGA using Muscle (Edgar, 2004). Maximum likelihood trees were then inferred using a General Time Reversible model with gamma distribution (five categories) and invariable sites (GTR+G+I) to help model evolutionary rate differences among sites. Bootstrap values were performed with 1000 replications (values shown next to branches). Nucleotide sequences in Figures 3 and 5 were inferred using a General Time Reversible model with gamma distribution (+G = 3.2635 and +G = 5.3324) and invariable sites ([+I] = 6.1511% sites and [+I] = 11.5688% sites). Evolutionary analysis based on Bayesian inference in Supplemental Figure 3 online was conducted with the Mr. Bayes (Huelsenbeck and Ronquist, 2001) plug-in to Geneious 6.0.2 (http://www.geneious.com/). Initial alignments were again completed with Muscle (Edgar, 2004). A General Time Reversible model was utilized with four gamma rate variation categories (GR+G). The chain length was 1,100,000 (four heated chains with a length of 0.2), and trees were sampled every 200. Branch lengths were unconstrained. Sequence alignments for all analyses used in the construction of phylogenetic trees are provided as Supplemental Data Sets 1 to 3 online. Sequence alignments in Figure 4 and Supplemental Data Set 4 online were constructed using ClustalW (Thompson et al., 1994).
Isolation of Full-Length TPS and CPT cDNAs
The full-length optimized cDNA of Sl-TPS19 and St-CPT1 was synthesized and ligated into pUC57 by GenScript USA. For recombinant protein expression, truncated versions of Sl-TPS19 and St-CPT1 (without the transit peptide coding region) were amplified and subcloned into the bacterial expression vector pEXP5-NT/TOPO and pET28a(+), respectively. For recombinant protein expression of Sl-TPS18, Sl-TPS21, and Sl-TPS41, the entire open reading frames of Sl-TPS18, Sl-TPS21, and Sl-TPS41 were amplified from cDNA prepared from M82 leaves and flower tissues and subsequently ligated into the pEXP5-NT/TOPO vector. For recombinant protein expression of Sh-CPT9, full-length Sh-CPT9 was amplified using cDNA prepared from LA1777 leaves and cloned into pGEM-T Easy vector and then subcloned into pEXP5-NT/TOPO vector without the predicted transit peptide. Spe-TPS19-LA2560 was amplified by RT-PCR from cDNA isolated from trichomes of S. pennellii and cloned into the pCR 4-TOPO vector (Life Technologies). Subsequently, a synthetic codon optimized version of Spe-TPS19-LA2560, lacking the chloroplast transit peptide, was synthesized by GenScript. The synthetic gene contained BamHI and SalI linkers at the 5′ and 3′ ends, respectively, and these were utilized to subclone the insert into the pHIS8 vector, as previously described (Gonzales-Vigil et al., 2012). All primers for cloning cDNAs are shown in Supplemental Table 2 online.
The other TPS and CPT genes were described by Schilmiller et al. (2009) (Sl-TPS20), Akhtar et al. (2013) (Sl-CPT1 and Sl-CPT2), Sallaud et al. (2009) (Sh-CPT1, zFPS), and Gonzales-Vigil et al. (2012) (Sh-TPS20-LA2100 and Sh-TPS45-LA1393 [Sh-SBS]).
Recombinant Protein Expression in Escherichia coli and TPS and CPT Enzyme Assays
E. coli BL21 (DE3) or BL21-CodonPlus (DE3) cells (Stratagene) containing the plasmid pEXP5-NT/TOPO expressing Sl-TPS18, Sl-TPS19, Sl-TPS20, Sl-TPS21, Sl-TPS41, Sl-CPT1, Sl-CPT2, and Sh-CPT9, pHIS8 expressing Sh-TPS20-LA2100, Spe-TPS19-LA2560, Sh-TPS45-LA1393, and Sh-CPT1, or pET28a(+) expressing St-CPT1 individually were grown in Luria-Bertani medium containing 100 ng/μL ampicillin or 50 ng/μL kanamycin until the OD600 reached 0.6 to 1.0 and then induced with 0.4 to 1.0 mM isopropyl 1-thio-β-d-galactopyranoside at 16 or 18°C for 16 h. Cell pellets were resuspended in binding buffer for Ni2+ column containing 100 mM sodium phosphate and 300 mM NaCl, pH 8.0. After sonication, the E. coli crude protein extracts in the binding buffer were passed through Ni2+ columns and recombinant His6-tagged proteins were purified via Ni2+ affinity chromatography, according to the manufacturer’s instructions (Qiagen). Partially purified recombinant proteins were desalted on PD-10 columns equilibrated with assay buffer A containing 50 mΜ HEPES, 7.5 mM MgCl2, 100 mM KCl, 5 mM DTT, and 10% glycerol (v/v), pH 7.0 (for TPS assay) or in assay buffer B containing 50 mΜ HEPES, 7.5 mM MgCl2, 100 mM KCl, 5 mM DTT, and 10% glycerol (v/v), pH 8.0 (for CPT assay) or changed to these buffers using Amicon Ultra centrifugal filter (Millipore). Protein concentration was estimated by measuring the OD595 or by running the samples in parallel with a known amount of BSA as the standard. Enzymatic assays were performed in assay buffer, containing 0.2 to 5 μg of protein and 26 to 200 μM of substrates in a volume of 50 to 250 μL. All commercially available prenyl-diphosphate substrates, including DMAPP, IPP, GPP, NPP, E,E-FPP, Z,Z-FPP, and GGPP, were used (Echelon Biosciences). Assay mixtures were incubated at 30°C for 30 min to 2 h. For CPT assays, the enzyme was denatured at 65°C for 10 min after the CPT assay, and the reaction products were converted from diphosphates to the related alcohol by incubation with rAPid alkaline phosphatase (Roche) at 37°C for 30 min. The resultant mixture was directly exposed to polydimethylsiloxane solid-phase microextraction (SPME) fiber (Supelco) at 42°C for 15 min. For the assay with NNPP or CLPP as substrate, the assay mixture containing Sl-CPT2 or Cistus creticus CLS (Cc-CLS; Falara et al., 2010) was incubated together with TPS protein using DMAPP and IPP as substrates in the assay buffer C containing 50 mΜ HEPES, 7.5 mM MgCl2, 100 mM KCl, 5 mM DTT, and 10% glycerol (v/v), pH 7.5 (for CPT/TPS coupled assay). Regarding the analyses of GGPP, NNPP, and CLPP, assay mixtures were extracted with 100 μL ethyl acetate or hexane containing 100 ng/μL tetradecane. After concentration of the extraction mixture under the N2 gas to <10 µL, 3 μL was injected and analyzed by gas chromatography–mass spectrometry. For the Km and kcat values of Sl-TPS19, Sl-TPS21, and Sh-TPS45 accession LA1393, reaction mixtures containing 0.2 to 0.8 µg of purified protein were incubated with various substrate concentration (10 to 300 µM GPP, NPP, or Z,Z-FPP) for 10 to 15 min at 30°C. After denaturation of the enzyme at 80°C for 10 min, the products were analyzed using SMPE fiber. Calibration curves of mono- and sesquiterpene standards (2-carene and E,E-farnesene) were generated using the SPME fiber to determine the range of linearity. Data of initial rates versus substrate concentration were analyzed by nonlinear regression of the Michaelis-Menten equation and obtained Km and Vmax values. kcat values were calculated from the Vmax value and the concentration of the enzyme in the assay.
Gas Chromatography–Mass Spectrometry Analysis of TPS and CPT Enzyme Assay Products
Terpene standards, geraniol, nerol, E,E-farnesol, and geranylgeraniol were obtained from Sigma-Aldrich. Enzyme assays were performed in a 1-mL glass vial, and the volatile compounds produced from the assays were extracted by the SPME fiber. The fiber was then injected into CP-sil 5 CB column (25-m length, 0.25-µm film thickness, and 0.25-mm ID; Agilent Technologies) on a GC17-A coupled to a GCMS-QP5000 (Shimadzu). Injector temperature was 310°C, and interface temperature was 280°C. After a 5-min isothermal hold at 45°C, the column temperature was increased by 15°C/min to 240°C and then 10°C/min to 240°C. Splitless mode was used.
Gene Expression Analysis by qRT-PCR
Different tissues of S. lycopersicum plants (cultivar MP1) were collected in liquid nitrogen. Total RNA was isolated with E.Z.N.A. Plant RNA MiniKit (Omega Bio-tek), treated with DNA-free kit (Ambion) to remove genomic DNA contamination, and used for first-strand cDNA synthesis using the High Capacity cDNA reverse transcription kit and random primers (Applied Biosystems) according to the manufacturer’s protocol. The resulting cDNA was diluted 10-fold, and 1 μL was used as template for PCR amplification in a 20-μL reaction using Fast SYBR green PCR master mix (Applied Biosystems) and gene-specific primers (see Supplemental Table 3 online). Reactions were performed on a StepOnePlus real-time PCR system (Applied Biosystems) with the following cycles: 95°C for 20 s, followed by 40 cycles (95°C for 3 s and 60°C for 30 s). A final dissociation step was performed to assess the quality of amplified product. Relative expression levels of each target transcript in different tissues were calculated using the relative quantification method normalized to the expression levels of tomato elongation factor-1α (EF-1 α; GenBank: X14449). Alignment of the tomato TPS cDNA sequences allowed the design of gene-specific primers for their PCR amplification (see Supplemental Table 3 online). Three biological replicates were used for each point, and triplicates of each sample were done.
Accession Numbers
Genomic and cDNA sequence data from this article can be found in the GenBank/EMBL data libraries under accession number KC807995 for S. lycopersicum, KC807996 for S. pimpinellifolium-LA1589, KC807997 for S. pennellii-LA0716, KC807998 for S. habrochaites-LA1778, KC807999 for S. habrochaites-LA1696, KC808000 for S. habrochaites-LA2104, KC808001 for S. habrochaites-LA2167, and KC808002 for SpeTPS19-LA2560 and KF000066 for Nt-ABS. Other TPS gene accession numbers are as follows: Os-KS, Q0JA82; At-KS, Q9SAK2; Pg-KS, GU045756; Pg-CPS, GU045755; At-CPS, Q38802; Os-CPS1, Q6ET36; Nt-CPS2, HE588139; Nt-ABS, HE588140, Sl-TPS24 (KS), JN412086; Sl-TPS40 (CPS), JN412074; Sh-TPS20-LA2100, JN990689; and Spe-TPS19-LA2560 and KC808002. The CPT gene accession number for At-CPT1 is NP_565551.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nucleotide Sequences of the Genomic Regions in Solanum lycopersicum, S. pimpinellifolium, S. pennelli, S. habrochaites, and S. tuberosum Containing the Functional TPS/CPT Gene Cluster, and Annotated Nicotiana tabacum ABS Gene.
Supplemental Figure 2. Determination of TPS41 Presence or Absence in S. pennellii Accessions.
Supplemental Figure 3. Evolutionary Relationships Based on Bayesian Inference of TPS18, 19, 20, 21, and 45 in Solanum.
Supplemental Figure 4. GC-MS Analysis of Volatile Terpene Compounds in S. habrochaites LA1778 Leaves.
Supplemental Figure 5. Sequences of Genomic Segments Containing Sh-TPS45 (ZIS and SBS) and CPT9 from Several Solanum habrochaites Accessions.
Supplemental Figure 6. In Vitro Enzymatic Assay of Sl-TPS19 and Spe-TPS19-LA2560 and Monoterpene Analysis of S. pennellii LA2560 Leaf Dips.
Supplemental Figure 7. Mass Spectrometry Analysis of the Numbered Peaks in Figure 6.
Supplemental Figure 8. Analysis of Reaction Products Obtained from in Vitro Assay of Sh-CPT9 and St-CPT1 Using TLC.
Supplemental Table 1. Sequences of the Oligonucleotide Primers Used for Supplemental Figure 2 Online.
Supplemental Table 2. Sequences of the Oligonucleotide Primers Used for Gene Cloning.
Supplemental Table 3. Sequences of the Oligonucleotide Primers Used in qRT-PCR Analysis.
Supplemental Data Set 1. TPS Coding Nucleotide Sequences Used for Construction of the Phylogenetic Tree in Figure 3.
Supplemental Data Set 2. TPS Coding Nucleotide Sequences Used for Construction of the Phylogenetic Tree in Supplemental Figure 3 Online.
Supplemental Data Set 3. CPT Coding Nucleotide Sequences Used for Construction of the Phylogenetic Tree in Figure 5.
Supplemental Data Set 4. TPS Protein Sequences Used for the Alignment in Figure 4.
Acknowledgments
We thank Dina St. Clair (University of California, Davis) for permission to screen the S. habrochaites BAC library. This research was supported by National Science Foundation Award IOS-1025636 to E.P. and C.S.B. and a Strategic Partnership Award from the Michigan State University Foundation to C.S.B. The genomic sequencing of S. pennellii was supported by an exceptional grant of the Max-Planck-Society to A.R.F.
AUTHOR CONTRIBUTIONS
E.P. and C.S.B. designed the research. Y.M., T.T.H.N., K.W., V.F., E.G.-V., B.L., A.M.B., P.S., B.U., and A.T. performed analyses of sequences and biochemistry of TPSs and CPTs. D.K. and R.A.W. screened the BAC clones for S. lycopersicum and S. habrochaites. A.R.F. provided novel materials. E.P., C.S.B., Y.M., and T.T.H.N. wrote the article.
Glossary
- TPS
terpene synthase
- CPT
cis-prenyl transferase
- Z,Z-FPP
2Z,6Z-farnesyl diphosphate
- GPP
geranyl diphosphate
- CLS
copal-8-ol diphosphate synthase
- CLPP
copal-8-ol diphosphate
- KS
ent-kaurene synthase
- CPS
copalyl diphosphate synthase
- NNPP
nerylneryl diphosphate
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- qRT-PCR
quantitative RT-PCR
- SPME
solid-phase microextraction
- GGPP
geranylgeranyl diphosphate
- NPP
neryl diphosphate
- FPP
farnesyl diphosphate
References
- Akhtar T.A., Matsuba Y., Schauvinhold I., Yu G., Lees H.A., Klein S.E., Pichersky E. (2013). The tomato cis-prenyltransferase gene family. Plant J. 73: 640–652 [DOI] [PubMed] [Google Scholar]
- Aubourg S., Lecharny A., Bohlmann J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Cho R.J., Campbell M.J., Winzeler E.A., Steinmetz L., Conway A., Wodicka L., Wolfsberg T.G., Gabrielian A.E., Landsman D., Lockhart D.J., Davis R.W. (1998). A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Demissie Z.A., Erland L.A., Rheault M.R., Mahmoud S.S. (2013). The biosynthetic origin of irregular monoterpenes in Lavandula: Isolation and biochemical characterization of a novel cis-prenyl diphosphate synthase gene, lavandulyl diphosphate synthase. J. Biol. Chem. 288: 6333–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J.D., West C.A. (1981). Properties of kaurene synthetase from Marah macrocarpus endosperm: Evidence for the participation of separate but interacting enzymes. Plant Physiol. 68: 1128–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V., Akhtar T.A., Nguyen T.T., Spyropoulou E.A., Bleeker P.M., Schauvinhold I., Matsuba Y., Bonini M.E., Schilmiller A.L., Last R.L., Schuurink R.C., Pichersky E. (2011). The tomato terpene synthase gene family. Plant Physiol. 157: 770–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V., Pichersky E., Kanellis A.K. (2010). A copal-8-ol diphosphate synthase from the angiosperm Cistus creticus subsp. creticus is a putative key enzyme for the formation of pharmacologically active, oxygen-containing labdane-type diterpenes. Plant Physiol. 154: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Fiston-Lavier A.S., Kemen A., Geisler K., Quesneville H., Osbourn A.E. (2011). Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B., Osbourn A.E. (2008). Metabolic diversification—Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Field B., Osbourn A. (2012). Order in the playground: Formation of plant gene clusters in dynamic chromosomal regions. Mobile Genet. Elements 2: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Gonzales-Vigil E., Hufnagel D.E., Kim J., Last R.L., Barry C.S. (2012). Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J. 71: 921–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Hurst L.D., Pál C., Lercher M.J. (2004). The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Hurst L.D., Williams E.J., Pál C. (2002). Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet. 18: 604–606 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J., Osbourn A. (2012). Making new molecules - Evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 15: 415–423 [DOI] [PubMed] [Google Scholar]
- Koonin E.V. (2009). Evolution of genome architecture. Int. J. Biochem. Cell Biol. 41: 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S., Tang H. (2000). Regulation of adjacent yeast genes. Trends Genet. 16: 109–111 [DOI] [PubMed] [Google Scholar]
- Martin D.M., Aubourg S., Schouwey M.B., Daviet L., Schalk M., Toub O., Lund S.T., and Bohlmann J. (2010). Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10: 226. [DOI] [PMC free article] [PubMed]
- Mayer K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402: 769–777 [DOI] [PubMed] [Google Scholar]
- Nakazato T., Warren D.L., Moyle L.C. (2010). Ecological and geographic modes of species divergence in wild tomatoes. Am. J. Bot. 97: 680–693 [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag). [Google Scholar]
- Olmstead R.G., Bohs L., Migid H.A., Santiago-Valentin E., Garcia V.F., Collier S.M. (2008). A molecular phylogeny of the Solanaceae. Taxon 57: 1159–1181 [Google Scholar]
- O’Maille P.E., Chappell J., Noel J.P. (2006). Biosynthetic potential of sesquiterpene synthases: Alternative products of tobacco 5-epi-aristolochene synthase. Arch. Biochem. Biophys. 448: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. (2010). Secondary metabolic gene clusters: Evolutionary toolkits for chemical innovation. Trends Genet. 26: 449–457 [DOI] [PubMed] [Google Scholar]
- Osbourn A.E., Field B. (2009). Operons. Cell. Mol. Life Sci. 66: 3755–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.J. (2006). Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317 [DOI] [PubMed] [Google Scholar]
- Pichersky E. (1990). Nomad DNA—A model for movement and duplication of DNA sequences in plant genomes. Plant Mol. Biol. 15: 437–448 [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004). A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C., Giacalone C., Töpfer R., Goepfert S., Bakaher N., Rösti S., Tissier A. (2012). Characterization of two genes for the biosynthesis of the labdane diterpene Z-abienol in tobacco (Nicotiana tabacum) glandular trichomes. Plant J. 72: 1–17 [DOI] [PubMed] [Google Scholar]
- Sallaud C., Rontein D., Onillon S., Jabès F., Duffé P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissier A. (2009). A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21: 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Schauvinhold I., Larson M., Xu R., Charbonneau A.L., Schmidt A., Wilkerson C., Last R.L., Pichersky E. (2009). Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA 106: 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282: 34013–34018 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M.S., Olsen C.E., Sato S., Tabata S., Jørgensen K., Møller B.L., Rook F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68: 273–286 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A., et al. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408: 816–820 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vontimitta V., Danehower D.A., Steede T., Moon H.S., Lewis R.S. (2010). Analysis of a Nicotiana tabacum L. genomic region controlling two leaf surface chemistry traits. J. Agric. Food Chem. 58: 294–300 [DOI] [PubMed] [Google Scholar]
- Wegel E., Koumproglou R., Shaw P., Osbourn A. (2009). Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell 21: 3926–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P.R., Xu M., Jin Y., Coates R.M., Peters R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T., et al. (2012). A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Wong S., Wolfe K.H. (2005). Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 37: 777–782 [DOI] [PubMed] [Google Scholar]
- Zhou K., Xu M., Tiernan M., Xie Q., Toyomasu T., Sugawara C., Oku M., Usui M., Mitsuhashi W., Chono M., Chandler P.M., Peters R.J. (2012). Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Köllner T.G., Zhao N., Li G., Jiang Y., Zhu L., Ma J., Degenhardt J., Chen F. (2012). Dynamic evolution of herbivore-induced sesquiterpene biosynthesis in sorghum and related grass crops. Plant J. 69: 70–80 [DOI] [PubMed] [Google Scholar]