This article investigates the developmental roles of a pre-mRNA splicing factor by analyzing ROOT INITIATION DEFECTIVE1 (RID1), a DEAH-box RNA helicase. The results show that the requirement for RID1 is not constitutive, but changes dynamically throughout development, suggesting that robust levels of pre-mRNA splicing are critical for several specific aspects of plant development.
Abstract
Pre-mRNA splicing is a critical process in gene expression in eukaryotic cells. A multitude of proteins are known to be involved in pre-mRNA splicing in plants; however, the physiological roles of only some of these have been examined. Here, we investigated the developmental roles of a pre-mRNA splicing factor by analyzing root initiation defective1-1 (rid1-1), an Arabidopsis thaliana mutant previously shown to have severe defects in hypocotyl dedifferentiation and de novo meristem formation in tissue culture under high-temperature conditions. Phenotypic analysis in planta indicated that RID1 is differentially required during development and has roles in processes such as meristem maintenance, leaf morphogenesis, and root morphogenesis. RID1 was identified as encoding a DEAH-box RNA helicase implicated in pre-mRNA splicing. Transient expression analysis using intron-containing reporter genes showed that pre-mRNA splicing efficiency was affected by the rid1 mutation, which supported the presumed function of RID1 in pre-mRNA splicing. Our results collectively suggest that robust levels of pre-mRNA splicing are critical for several specific aspects of plant development.
INTRODUCTION
In higher eukaryotes, most primary transcripts of genes (i.e., pre-mRNAs) contain sequences, named introns, that intervene coding sequences. Pre-mRNAs become translatable mature forms only after the introns are removed by precisely regulated processing. This process, called pre-mRNA splicing, is executed by the spliceosome. The major type of spliceosome contains five RNA-protein complexes, named U1, U2, U4, U5, and U6 small ribonucleoprotein particles (snRNPs), each of which comprises a specific kind of uridine-rich small nuclear RNA (UsnRNA) and its tightly associated proteins as core factors and more than 50 non-snRNP proteins as additional factors (reviewed in Burge et al., 1999; Will and Lührmann, 2011). The spliceosome components are assembled on pre-mRNAs in a highly ordered sequence, precisely regulating the step-by-step reactions of pre-mRNA splicing. At the preliminary stage, the 5′ splice site of pre-mRNA is recognized by U1 snRNP through a base-pairing interaction with the 5′ end of U1 small nuclear RNA (snRNA). U2 snRNP subsequently binds to the branching point sequence of the U1 snRNP-associated pre-mRNA, and then the other three snRNPs join the assembly as U4/U6-U5 tri-snRNP to form a complete spliceosome that catalyzes trans-esterification reactions (Will and Lührmann, 2011). The activated spliceosome drives the two steps of trans-esterification to produce ligated exons and lariat-form introns. The ligated exons are immediately released as mRNA from the spliceosome and the intron lariats are released at the final stage of pre-mRNA splicing. The progression of these processes is accompanied by a cascade of dynamic changes of the spliceosomal supercomplex, such as compositional and conformational changes of snRNPs, association and dissociation of accessory proteins, and rearrangements of RNA–RNA and RNA–protein interactions (reviewed in Wahl et al., 2009; Will and Lührmann, 2011).
Of the non-snRNP spliceosomal proteins, a notably important group is the DExD/H-box protein family, the members of which contain ATP-dependent RNA helicase activity and participate in a wide range of RNA-related events (reviewed in Tanner and Linder, 2001; Will and Lührmann, 2011). In budding yeast, eight DExD/H-box proteins Pre-RNA Processing2 (Prp2), Prp5, Prp16, Prp22, Prp28, Prp43, Suppressor of BRR1 protein2, and Bad Response to Refrigeration2 were identified as key players in pre-mRNA splicing and shown to act at different events of RNA–RNA and RNA–protein rearrangements (reviewed in Staley and Guthrie, 1998). Studies with human cells have shown that proteins homologous to these yeast proteins are also present in the human spliceosome and that each of the homologous pairs mediates the same rearrangement events (Rappsilber et al., 2002; Will and Lührmann, 2011), suggesting that the DExD/H-box proteins have evolutionarily conserved roles in pre-mRNA splicing. Consistent with this view, a database search analysis detected genes encoding homologs of the yeast spliceosomal DExD/H-box proteins in plant genomes (Wang and Brendel, 2004; http://www.plantgdb.org/SRGD/index.php). However, information on plant spliceosomal DExD/H-box proteins is still quite limited. The only experimentally examined member of this family in plants is ENHANCED SILENCING PHENOTYPE3 (ESP3), a DEAH-box RNA helicase of Arabidopsis thaliana that is orthologous to Prp2. Missense mutations of this protein enhanced RNA silencing and caused weak growth defects and an early flowering phenotype (Herr et al., 2006), implying a connection between pre-mRNA splicing, RNA silencing, and developmental regulation, although the function of ESP3 in pre-mRNA has not been verified.
We have been investigating plant organogenesis using temperature-sensitive mutants of Arabidopsis. One such mutant, shoot redifferentiation defective2-1 (srd2-1), is characterized by the extreme temperature sensitivity of hypocotyl dedifferentiation and de novo meristem formation (Yasutani et al., 1994; Ozawa et al., 1998; Ohtani and Sugiyama, 2005). SRD2 encodes an activator of snRNA transcription, and several experiments using srd2-1, the point mutation mutant of this gene, indicated that the SRD2-mediated upregulation of snRNA transcription is essential for the acquisition of cell proliferation competence (Ohtani and Sugiyama, 2005) and for several particular developmental processes, including the establishment of apical meristems (Ohtani et al., 2008, 2010). Another mutant, root initiation defective1-1 (rid1-1), was identified as being temperature sensitive for adventitious root formation from hypocotyl explants (Konishi and Sugiyama, 2003). At the restrictive temperature, rid1-1 mutant plants in tissue culture exhibit a phenotype that is very similar to that of srd2-1 (Konishi and Sugiyama, 2003), which implies that the functions of SRD2 and RID1 are closely related at the molecular level. In this article, we describe the results of a detailed analysis of the rid1-1 mutant. Phenotypic characterization of rid1-1 with reference to srd2-1 showed that RID1 and SRD2 play similar roles in various stages of plant development and that the requirement for these factors is not constitutive, but changes dynamically throughout development. Chromosome mapping followed by sequence analysis revealed that RID1 encodes a DEAH-box RNA helicase similar to the yeast splicing factor Prp22. Experiments with a reporter gene (In-YFP) designed to monitor pre-mRNA splicing efficiency demonstrated that splicing was suppressed in the rid1 mutant. It was also shown that several alternative splicing events were affected by the rid1 mutation. Our data provide evidence for the developmental significance of the DEAH-box RNA helicase RID1, which regulates the efficiency of pre-mRNA splicing.
RESULTS
Temperature Sensitivity of rid1-1 in Tissue Culture
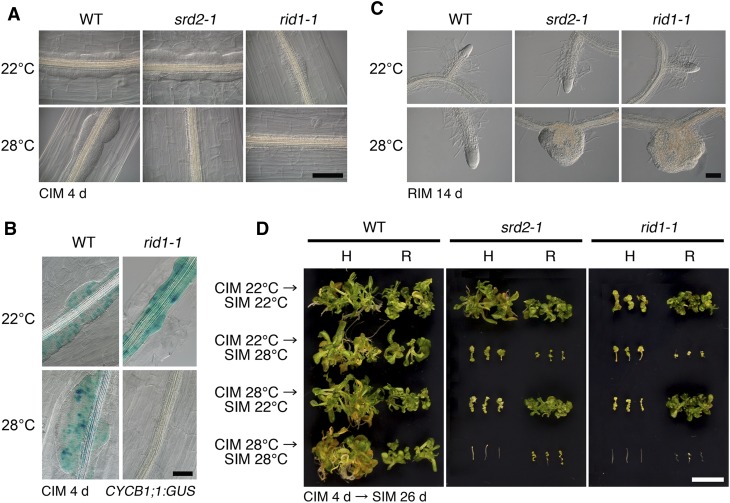
An initial characterization of rid1-1 in tissue culture showed that it is temperature sensitive for adventitious root formation, lateral root formation, and callus formation from hypocotyl explants (Konishi and Sugiyama, 2003; see Supplemental Figure 1 online). In this study, we compared the temperature sensitivity of rid1-1 with that of srd2-1. Figure 1A shows the temperature-dependent defect in callus initiation from rid1-1 and srd2-1 hypocotyl explants. After 4 d of culture on callus-inducing medium (CIM), callus formation was initiated in the stele at 22°C (permissive temperature). By contrast, these mutants did not exhibit any morphological sign of callus initiation when the explants were cultured at 28°C (restrictive temperature). Compared with the wild type and srd2-1, callus cell proliferation at 22°C was poorer in rid1-1. The CYCB1;1:GUS (for β-glucuronidase) reporter gene, an indicator of active cell division, demonstrated that cell division was not initiated in the rid1-1 hypocotyl at 28°C (Figure 1B), as was the case for srd2-1 (Ohtani and Sugiyama, 2005). To examine the effect of the rid1-1 and srd2-1 mutations on lateral root development, primary root segments of rid1-1 and srd2-1 were cultured on root-inducing medium to induce lateral root formation (Figure 1C). When cultured at 28°C, both the rid1-1 and srd2-1 explants produced knob-like lateral roots that appeared to lack structural organization. Malformation of the lateral roots was more profound in rid1-1 than srd2-1. Shoot regeneration phenotypes were also examined. In this experiment, shoot regeneration was induced from hypocotyl and root explants by a two-step culture procedure, which consists of preculture on CIM and subsequent culture on shoot-inducing medium (SIM), and the temperature sensitivity of the hypocotyls and roots was tested. In both the rid1-1 and srd2-1 mutants, shoot regeneration from hypocotyl explants was highly sensitive to the restrictive temperature, both on CIM and SIM, whereas shoot regeneration from root explants was inhibited only at the restrictive temperature during culture on SIM, but not on CIM (Figure 1D). This finding indicated that the rid1-1 mutation, like the srd2-1 mutation, severely affected both cell dedifferentiation and shoot organogenesis processes during shoot regeneration from hypocotyl explants, but affected only the shoot organogenesis process in the case of root explants. In summary, these results demonstrate that the phenotypes of rid1-1 and srd2-1 observed in tissue culture are highly similar but that rid1 displayed more severe defects than srd2-1 in some cases.
Figure 1.
Effects of the srd2-1 and rid1-1 Mutations on Callus Initiation, Lateral Root Formation, and Adventitious Shoot Formation.
(A) Hypocotyl explants of the wild type (WT), srd-1, and rid1-1 were cultured on CIM for 4 d at 22 or 28°C. Bar = 100 µm.
(B) Expression patterns of CYCB1;1:GUS in hypocotyl explants of the wild type and rid1-1 cultured on CIM for 4 d at 22 or 28°C. Bar = 50 µm.
(C) Root explants of the wild type, srd2-1, and rid1-1 were cultured on root-inducing medium (RIM) for 14 d at 22 or 28°C. Bar = 100 µm.
(D) Hypocotyl (H) and root (R) explants of the wild type, srd2-1, and rid1-1 were cultured on CIM at 22 or 28°C for 4 d. Then, explants were transferred onto SIM and cultured at 22 or 28°C for 26 d. Bar = 1 cm.
Effects of the rid1-1 Mutation on the Meristem Activity of Seedlings
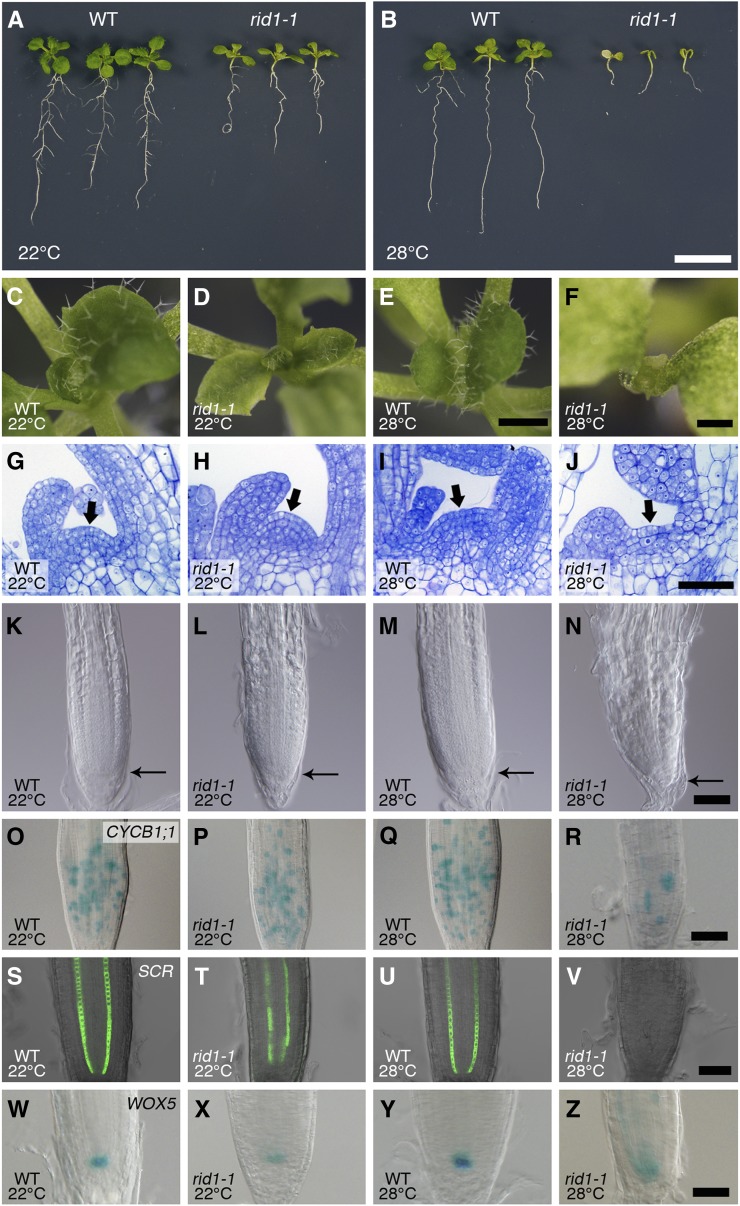
The effects of the rid1-1 mutation on meristem activity were assessed after seedlings were germinated and grown at 22 or 28°C for 12 d. The rid1-1 mutant germinated both at 22 and 28°C, but the subsequent growth was moderately and strongly inhibited at 22 and 28°C, respectively (Figures 2A to 2F). At 28°C, rid1-1 growth was arrested shortly after germination, and the rid1-1 seedlings died without forming visible true leaves (Figures 2B, 2E, and 2F). Histological observation of the shoot apex of rid1-1 seedlings grown at 28°C indicated that the shoot apical meristem (SAM) was initially activated, but not maintained, and was consumed in the production of abnormal leaf primordia (Figures 2G to 2J).
Figure 2.
Effects of the rid1-1 Mutation on the Apical Meristems of Seedlings.
(A) and (B) Twelve-day-old seedlings of the wild type (WT) and rid1-1 grown at 22°C (A) or 28°C (B). Bar = 1 cm.
(C) to (Z) Seedlings of the wild type ([C], [E], [G], [I], [K], [M], [O], [Q], [S], [U], [W], and [Y]) and the rid1-1 mutant ([D], [F], [H], [J], [L], [N], [P], [R], [T], [V], [X], and [Z]) were germinated and grown for 12 d at 22°C ([C], [D], [G], [H], [K], [L], [O], [P], [S], [T], [W], and [X]) or 28°C ([E], [F], [I], [J], [M], [N], [Q], [R], [U], [V], [Y], and [Z]).
(C) to (F) Top views of seedlings. Bars = 1 mm for (C) to (E) and 0.5 mm for (F).
(G) to (J) Longitudinal sections of the shoot apex. Resin-embedded tissues were sectioned and stained with toluidine blue O. The arrows indicate the SAM region. Bar = 50 µm.
(K) to (N) Differential interference contrast images of the root apical region. The distal end of the RAM is indicated by a thin arrow. Bar = 50 µm.
(O) to (R) Expression of CYCB1;1:GUS in the RAM. Bar = 50 µm.
(S) to (V) Expression of SCR:GFP in the RAM. Bar = 50 µm.
(W) to (Z) Expression of WOX5:GUS in the RAM. Bar = 50 µm.
The rid1-1 mutation also affected the structure of the root apical meristem (RAM) in a temperature-dependent manner (Figures 2K to 2N). In the primary roots of rid1-1 seedlings grown at 22°C, the size of the RAM and cell division activity, as gauged by CYCB1;1:GUS expression, were not different from those of the wild type (Figures 2K, 2L, 2O, and 2P), although the shape of the root tip was slightly altered (Figures 2K and 2L). When grown at 28°C, however, the RAM was much smaller and the rate of cell division much lower in the RAM of the mutant than of the wild type (Figures 2M, 2N, 2Q, and 2R). The tissue organization of the RAM was inspected using two marker genes, SCR:GFP (for green fluorescent protein) and WOX5:GUS, which are specifically expressed in endodermis cells and the quiescent center, respectively (Wysocka-Diller et al., 2000; Sarkar et al., 2007). In the RAM of rid1-1, the expression pattern of SCR:GFP was somewhat reduced at 22°C and completely absent at 28°C (Figures 2T and 2V; compare with the wild type in Figures 2S and 2U). The expression of WOX5:GUS was reduced at 22°C and diffuse at 28°C in rid1-1 (Figures 2X and 2Z; compare with the wild type in Figures 2W and 2Y). These abnormalities of marker gene expression were indicative of impaired cell specification in the RAM of rid1-1, which was further supported by near absence of columella cells (see Supplemental Figure 2 and Supplemental References 1 online). These results suggest that RID1 is required for the establishment and maintenance of SAM and RAM organization but that it is not involved in the initial activation of the SAM and RAM at the beginning of postgerminative growth.
Effects of the rid1 Mutation on Growth and Development in the Adult Phase
To examine the effects of the rid1-1 mutation on growth and development in the adult phase, the wild type and rid1-1 were cultured at 22°C for 2 weeks and then cultured at 22 or 28°C for an additional 2 weeks. The rid1-1 plants transferred to 28°C stopped growing, exhibited chlorosis, and eventually died (Figure 3A; compare with size of seedlings in Figure 2A). This phenotype is more severe than that of srd2-1 plants, which could survive to flowering at 28°C after 7 d of culture at 22°C (Ohtani et al., 2008). At a constant temperature of 22°C, the rid1-1 plants displayed relatively moderate defects in leaf development. Compared with the wild type, the number of leaves produced was decreased in rid1-1 (Figure 3B), and the leaves of rid1-1 were pale green in color and rounder in shape (Figure 3B). Microscopy observations of the leaf surface and cross sections of rid1-1 revealed morphological abnormalities in the guard cells of rid1-1 (Figure 3F) and faulty cell organization of palisade mesophyll and vascular tissue (Figures 3G and 3H). These phenotypes partially resemble those seen in srd2-1 plants exposed to a temperature of 28°C (Ohtani et al., 2008). Thus, the rid1-1 mutation had a more severe effect than srd2-1 in several aspects of growth and development in the adult phase.
Figure 3.
Effects of the rid1-1 Mutation on Vegetative Development.
(A) The wild type (WT) and rid1-1 were grown at 22°C for 2 weeks and then at 22 or 28°C for an additional 2 weeks. Bar = 1 cm.
(B) Morphological appearance of leaves from plants grown at 22°C for 4 weeks. The two cotyledons are indicated as Cot. Bar = 1 cm.
(C) to (H) Detailed observation of the second true leaves of the wild type ([C] to [E]) and rid1-1 ([F] to [H]) grown at 22°C.
(C) and (F) Differential interference contrast images of stomatal apparatuses. The red arrow indicates abnormal guard cells. Bar = 50 µm.
(D), (E), (G), and (H) Cross sections of the second true leaves of wild-type and rid1-1 plants grown at 22°C. Resin-embedded tissues were sectioned and stained with toluidine blue O. ep, epidermis; pm, palisade parenchyma; sm, spongy parenchyma. Vein structures are magnified in (E) and (H). Bars = 25 µm.
Map-Based Cloning of RID1
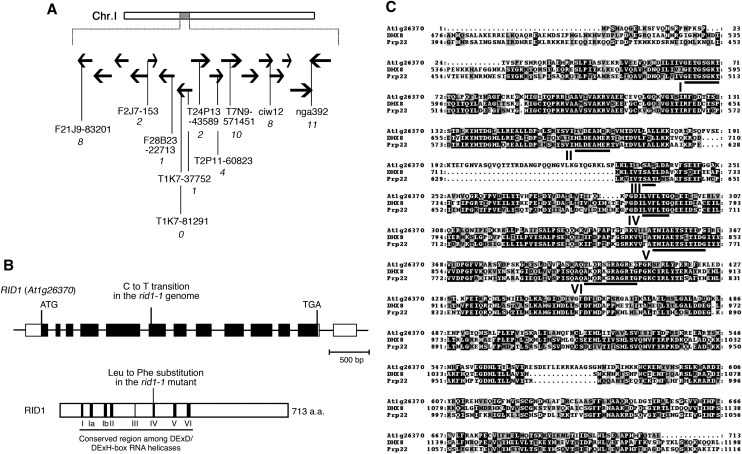
RID1 was previously mapped to the middle region of chromosome I (Konishi and Sugiyama, 2003). For positional cloning of RID1, we performed fine chromosome mapping of the rid1-1 mutation and precisely mapped rid1-1 to a 67-kb region at the 43-centimorgan position of chromosome I (Figure 4A). Sequence analysis of this region of the rid1-1 genome revealed a single-base (cytosine to thymine) substitution in the sixth exon of At1g26370 (Figure 4B). Complementation with a genomic fragment that encompassed the entire region of At1g26370 successfully rescued the temperature sensitivity of rid1-1 for adventitious root and callus formation. Therefore, we concluded that RID1 corresponds to At1g26370. RID1 encodes a protein that contains eight motifs characteristic of RNA helicases. The single-base substitution in the rid1-1 genome was deduced to change the conserved Leu-291 residue of the gene product, which lies in motif IV, to Phe (Figure 4B).
Figure 4.
Chromosome Mapping of the RID1 Locus and Alignment of Amino Acid Sequences.
(A) Chromosomal location of the rid1-1 mutation. Black horizontal arrows represent BAC clones around the RID1 locus on chromosome I. The number of recombination events between DNA polymorphism markers and the RID1 locus is given in italicized numerals beneath the marker names.
(B) Structure of RID1 (top) and its encoded protein (bottom). Black boxes represent exons, lines represent introns, and unfilled boxes represent 5′- and 3′-untranslated regions (top). The position of the single-base substitution in rid1-1 is indicated. The highly conserved DEAH-box RNA helicase region is underlined, and motifs I to VI are numbered (bottom). a.a., amino acids.
(C) Alignment of amino acid sequences of RID1, human DHX8, and yeast Prp22. Alignment was generated with the ClustalW program. Identical residues are highlighted on a black background, and similar residues are shaded in gray. The highly conserved DEAH-box RNA helicase domains are underlined and labeled motifs I to VI.
Eukaryotic RNA helicases constitute a large protein family and are classified into many subfamilies. RID1 belongs to the DEAH-box RNA helicase subfamily. A BLAST search using the RID1 sequence as query identified more than 20 genes encoding putative proteins of this subfamily in the Arabidopsis genome, including ESP3 (see Supplemental Figure 3 and Supplemental Data Set 1 online). RID1 showed a high level of sequence similarity with Prp22 of budding yeast and its human ortholog DHX8 (for DEAH box polypeptide8; also known as DDX8 or HRH1) (Figure 4C). Prp22 and DHX8 have been demonstrated to mediate the release of spliced mRNA from the spliceosome after completion of splicing (Company et al., 1991; Ohno and Shimura, 1996; Schwer and Gross, 1998). The sequence similarity between RID1 and Prp22 and DHX8 suggests that RID1 also has a role in pre-mRNA splicing.
Function of RID1 in Pre-mRNA Splicing
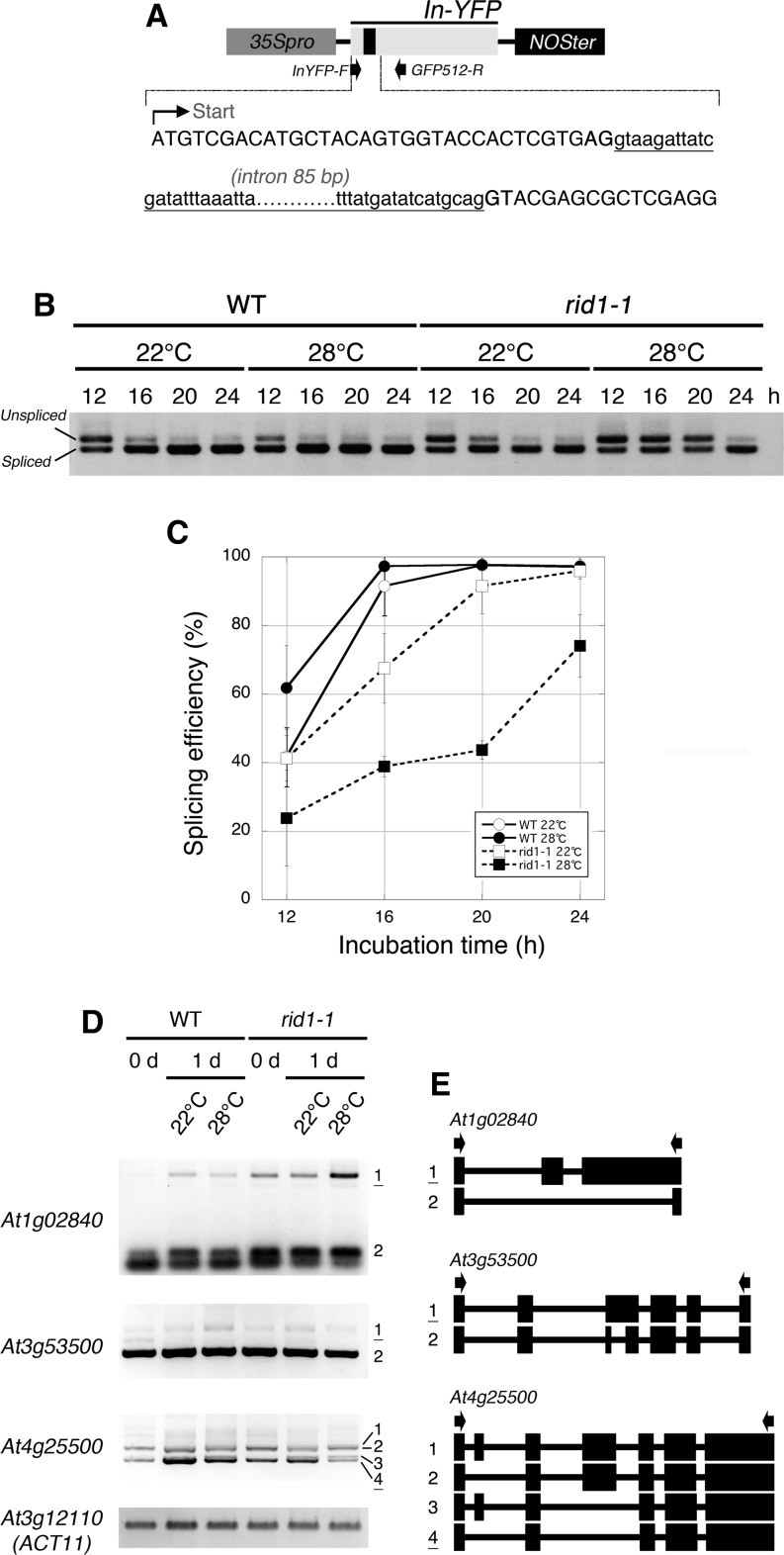
By analogy to Prp22 and DHX8, we speculated that RID1 is involved in pre-mRNA splicing. To test this possibility, we compared pre-mRNA splicing efficiencies between wild-type and rid1-1 mutant cells and evaluated the effect of the rid1-1 mutation on splicing efficiency. Pre-mRNA splicing efficiencies were measured by transient expression analysis using the In-YFP reporter gene, in which the yellow fluorescent protein (YFP) coding sequence is interrupted by a synthetic intron sequence (Figure 5A). The In-YFP reporter was introduced into mesophyll protoplasts of the wild type and rid1-1, and after incubation at 22 or 28°C for various periods of time, spliced and unspliced RNAs derived from the reporter gene were distinguished by RT-PCR. In the wild type, the spliced forms were dominant after incubation for 16 h or longer at either 22 or 28°C, and almost 100% of the RNAs were in the spliced form within 20 h of incubation (Figures 5B and 5C). In the case of rid1-1, the spliced forms accumulated more slowly during incubation. This effect of the rid1-1 mutation was much more apparent at 28°C than at 22°C (Figure 5B). At 28°C, the spliced form became the majority after only 24 h (Figure 5C). Thus, the rid1-1 mutation caused a marked reduction in the efficiency of pre-mRNA splicing, in a temperature-dependent manner. This finding demonstrated the involvement of RID1 in pre-mRNA splicing.
Figure 5.
Effects of the rid1-1 Mutation on Pre-mRNA Splicing Efficiency and Alternative Splicing Patterns.
(A) Structure of the synthetic reporter gene. The black rectangle in In-YFP indicates the inserted synthetic intron. The first part of the In-YFP sequence is shown; upper- and lowercase letters represent exon and intron sequences, respectively. Arrows below the In-YFP region indicate the positions of primers used for RT-PCR.
(B) Detection of spliced and unspliced forms of the In-YFP transcript by RT-PCR. The In-YFP reporter gene was introduced into mesophyll protoplasts of the wild type (WT) and rid1-1 and incubated at 22 or 28°C for the indicated periods of time. Total RNAs isolated from the protoplasts were subjected to RT-PCR analysis independently three times.
(C) Splicing efficiency of the In-YFP reporter gene. Data represent the mean and sd of three independent experiments.
(D) The effects of the rid1-1 mutation on alternative splicing patterns. Total RNAs were isolated from hypocotyl explants of the wild type and rid1-1 cultured on CIM at 22 or 28°C for the indicated periods of time and subjected to RT-PCR analysis independently three times. Splice variants are numbered, and underlined variants were either more or less abundant in rid1-1 at 28°C. At3g12100 (ACT11) was used as a control.
(E) Schematic diagrams of splice variants shown in (D). Exons are represented as filled rectangles and introns as thin lines. Arrows indicate the positions of primers used for RT-PCR.
The effects of the rid1 mutation on pre-mRNA splicing were also examined by monitoring changes in alternative pre-mRNA splicing patterns of endogenous transcripts during callus initiation of hypocotyl explants. We examined all of the alternative splicing events described by Palusa et al. (2007) and Simpson et al. (2008) (103 in total) and found that nine were substantially affected by the rid1-1 mutation (Figure 5D; see Supplemental Figure 4 online). Most of the effects of rid1 on alternative splicing patterns were more apparent at 28°C than at 22°C. Sequence analysis of the splice variants revealed that the rid1-1 mutation influenced multiple modes of alternative splicing events, such as exon skipping, alternative 3′ acceptor site selection, and intron retention (Figure 5E). These results likely reflect the impaired regulation of pre-mRNA splicing in the rid1-1 mutant.
In an attempt to further examine the molecular function of RID1, we also tried to establish whether RID1 is a functional analog of Prp22, by conducting a complementation analysis of two missense mutants of yeast, prp22T637A and prp22R805A, which showed severe cold-sensitive defects in cell growth (Schwer and Meszaros, 2000). When the wild-type PRP22 gene was introduced and expressed artificially in the prp22 mutants, their growth defects were completely suppressed. However, artificial inductions of RID1 or Prp22-RID1, which was designed to produce a chimeric protein consisting of the nonconserved N-terminal region of Prp22 and full-length RID1, did not recover the growth of the prp22 mutants (see Supplemental Figure 5 online). Thus, RID1 is not replaceable to Prp22, suggesting that R1D1 has a different function from Prp22 in pre-mRNA splicing at the molecular level.
Subcellular Localization of RID1
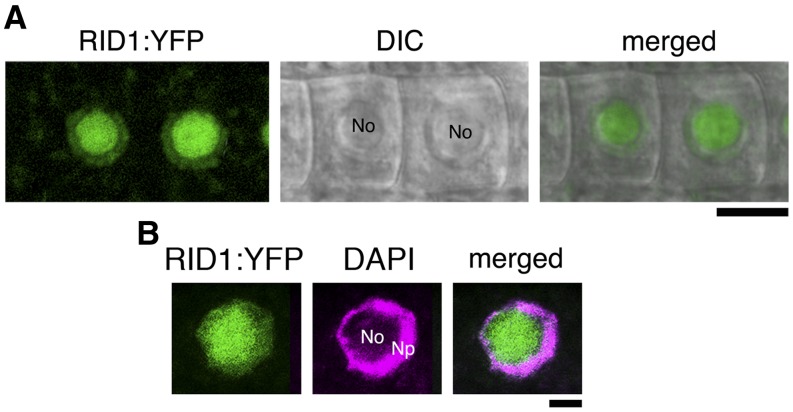
The subcellular localization of RID1 was examined using a fusion of YFP to the C terminus of RID1, RID1-YFP, which was expressed in rid1-1 under the control of the RID1 promoter. The chimeric RID1:RID1-YFP gene complemented the temperature sensitivity of adventitious root formation of rid1-1. In the root epidermal cells of transgenic plants expressing RID1-YFP, the YFP signals were exclusively detected in the nuclei (Figure 6A), particularly in the nucleolar region (Figure 6B). This observation indicated that RID1 is a nuclear protein with preferential localization to the nucleolus.
Figure 6.
Subcellular Localization of RID1:YFP.
Fluorescence signals of RID1:YFP in the RAM. The fluorescence images of RID1:YFP signal were merged with the differential interference contrast (DIC) image (A) and with the fluorescence image of DAPI staining (B). No, nucleoli region; Np, nucleoplasmic region. Bars = 10 µm in (A) and 5 µm in (B).
The nucleolus is the site of ribosome biogenesis, including transcription of rRNA genes, rRNA maturation, and ribosome assembly (reviewed in Shaw and Brown, 2012), and some DExD/H-box proteins are known to function in rRNA maturation (Tanner and Linder, 2001). To test the possibility that RID1 is also involved in rRNA maturation, we performed RNA gel blot analysis with a probe specific for the internal transcribed spacer region 1 of rRNA. The result showed no apparent differences between the wild type and rid1-1 in the accumulation of internal transcribed spacer regions of rRNA (see Supplemental Figure 6 online). Thus, the contribution of RID1 to rRNA maturation is expected to be negligibly small.
Expression Patterns of RID1
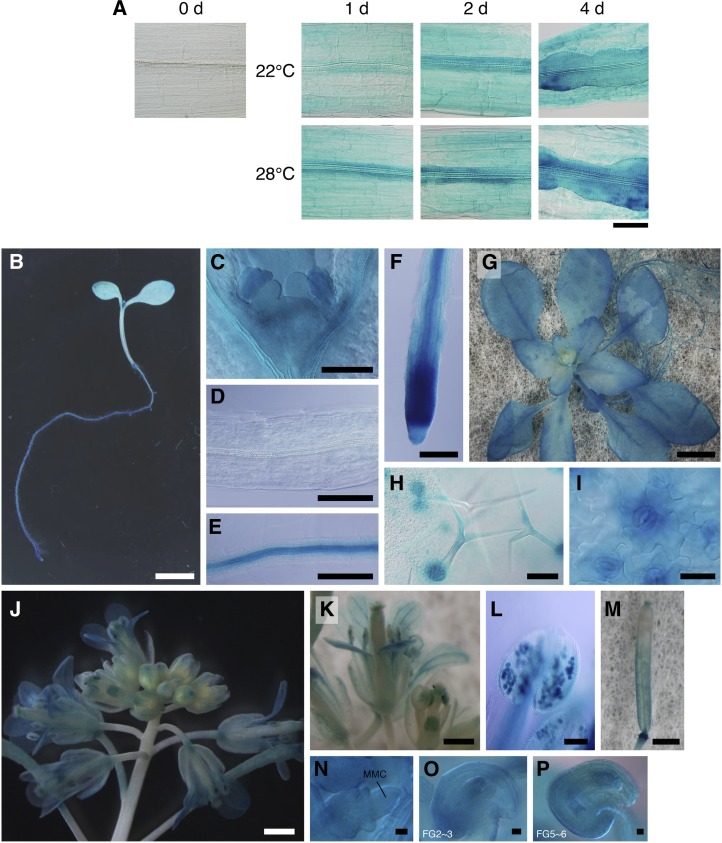
To monitor the expression patterns of RID1, we constructed the ProRID1:GUS reporter gene, consisting of the promoter region of RID1 fused to the structural gene for GUS. We first examined the expression patterns of RID1:GUS during callus initiation from hypocotyl explants. No GUS signal was detected in the hypocotyl tissue before culture on CIM; however, GUS activity increased in the stele rapidly upon CIM culture of hypocotyl explants, and cells in the stele started to proliferate (Figure 7A). These patterns of GUS activity indicated that RID1 undergoes dynamic changes in expression in hypocotyl tissues induced by CIM culture, which seem to be associated with the dedifferentiation process prior to the reinitiation of cell division.
Figure 7.
GUS Analysis of RID1 Promoter Activity.
(A) ProRID:GUS expression patterns during callus initiation. Hypocotyl explants of 12-d-old RID:GUS seedlings were cultured on CIM for the indicated times at 22 or 28°C and then subjected to histochemical detection of GUS activity. Bar = 100 µm.
(B) to (F) ProRID1:GUS expression patterns in 6-d-old seedlings. (C) to (F) show magnified images of the shoot apex, hypocotyl, root stele, and root tip tissues, respectively. Bars = 5 mm in (B) and 100 µm (C) to (F).
(G) to (I) RID1:GUS expression patterns in 24-d-old transgenic plants. Panels show the macroscopy image (G) and higher magnifications ([H] and [I]) of the leaf surface. Bars = 5 mm in (G), 100 µm in (H), and 25 µm in (I).
(J) to (P) ProRID1:GUS expression patterns in the reproductive phase. Panels show an inflorescence (J), opening flower (K), anther containing mature pollens (L), pistil after fertilization (M), megaspore mother cell–containing ovule (N), ovule at stage FG2/3 (O), and ovule at stage FG5/6 (P). MMC, megaspore mother cell. Bars = 1 mm in (J) and (M), 0.5 mm in (K), 100 µm in (L), and 10 µm in (N) to (P).
In seedlings harboring RID1:GUS, strong GUS activity was found at the shoot apex, leaf primordia, stipules, root tip, root stele, and lateral root primordia (Figures 7B to 7F). This expression pattern is almost the same as that seen for SRD2:GUS (Ohtani et al., 2008). It is of note that the root stele expressed RID1:GUS and SRD2:GUS strongly, while the hypocotyl stele did not. As discussed previously for SRD2, expression of these genes in the root stele may be related to the high competence of cell proliferation in this tissue (Ohtani and Sugiyama, 2005). During the adult vegetative phase, RID1:GUS plants showed GUS activity in developing and young organs (Figure 7G). A relatively high level of GUS activity was observed in the trichomes, trichome support cells, guard cells, and veins of young leaves as well as in the apical regions of the shoot and root (Figures 7G to 7I). These sites include cells and tissues that are highly vulnerable to the rid1-1 mutation (Figures 2 and 3).
At the reproductive stage, we found that RID1:GUS was expressed in various floral tissues and that the expression pattern was changed as the floral organs developed. In floral buds, just before anthesis, the GUS signals were substantially restricted to the pistils. In opened buds, the GUS activity in the pistil declined, but was elevated in mature pollen, filaments, and vascular tissues (Figures 7J to 7L). We also found that RID1:GUS was expressed in developing female gametophytes and ovules (Figures 7N to 7P). After pollination, RID1:GUS was again expressed in the pistils, in addition to the pedicels (Figures 7J and 7M). Thus, our expression analysis using the RID1:GUS reporter indicated that RID1 expression is not constitutive but is spatially and temporally regulated during development.
Knockout Analysis of RID1
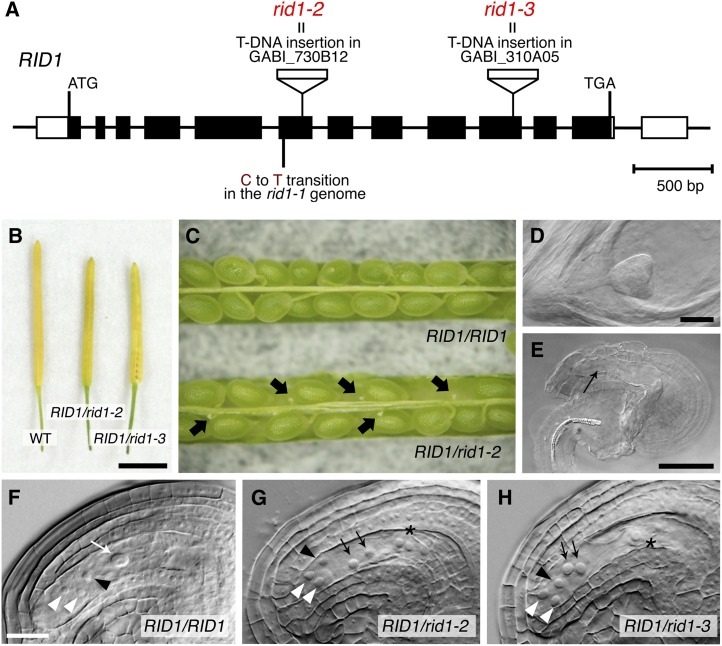
For phenotypic analysis of RID1 knockout plants, we used two T-DNA insertion lines, GABI_730B12 and GABI_310A05. T-DNA was inserted within the sixth exon of RID1 in GABI_730B12 and within the tenth exon in GABI_310A05 (Figure 8A). These mutations will hereafter be referred to as rid1-2 and rid1-3, respectively. Genotyping of plants derived from these lines revealed that there were no homozygous rid1-2 or rid1-3 offspring (Table 1). The rid1-2 or rid1-3 heterozygous plants showed almost normal growth and morphology throughout vegetative and reproductive development, except that the mature siliques were shorter than those of the wild type (Figure 8B).
Figure 8.
Effects of T-DNA Insertion Mutations of RID1 on Reproduction.
(A) T-DNA insertions in RID1. Black boxes represent exons, lines represent introns, and unfilled boxes represent 5′- and 3′-untranslated regions. The positions of the T-DNA insertions are indicated, as is the position of the point mutation in rid1-1.
(B) Mature siliques borne on wild-type (WT), RID1/rid1-2, and RID1/rid1-3 plants. Bar = 5 mm.
(C) Opened siliques of the RID1/RID1 and RID1/rid1-2 plants. Black arrows indicate aborted seeds.
(D) and (E) Normal (D) and aborted (E) seeds in the same young silique of the RID1/rid1-2 plant. The black arrow indicates the site at which embryogenesis normally occurs. Bars in (D) and (E) = 50 µm.
(F) to (H) Wild-type (F) and representative mutant ([G] and [H]) female gametophytes.
(F) Two synergid nuclei at the micropylar end (white arrowheads), an egg cell nucleus (black arrowhead), and the large nucleus of the central cell formed by the fusion of polar nuclei (white arrow) were detected. Bar = 50 µm.
(G) and (H) Synergid nuclei (white arrowheads) in mutant gametophytes were of similar size to the egg cell nucleus (black arrowheads). The polar nuclei were unfused (black arrows), and the antipodal cells were enlarged and protruded toward the center (asterisk). The antipodal cells and fused antipodal nuclei (asterisk) exhibited signs of disintegration in rid1-3 gametophytes (H).
Table 1. Segregation of the rid1-2 and rid1-3 Mutations.
| Plants | RID1/RID1 | RID1/rid1-2 | rid1-2/rid1-2 |
|---|---|---|---|
| Selfed progeny of RID1/rid1-2 | 109 | 51 | 0 |
| F1 progeny of RID1/rid1-2 (F) × RID1/RID1 (M) | 168 | 0 | 0 |
| F1 progeny of RID1/RID1 (F) × RID1/rid1-2 (M) | 100 | 56 | 0 |
| RID1/RID1 | RID1/rid1-3 | rid1-3/rid1-3 | |
| Selfed progeny of RID1/rid1-3 | 102 | 49 | 0 |
| F1 progeny of RID1/rid1-3 (F) × RID1/RID1 (M) | 81 | 0 | 0 |
| F1 progeny of RID1/RID1 (F) × RID1/rid1-3 (M) | 79 | 41 | 0 |
The “F” indicates the female parent, and the “M” indicates the male parent.
Reciprocal crossing between wild-type plants and rid1-2 or rid1-3 heterozygotes revealed that both mutations could be transmitted only through male gametophytes (Table 1). Therefore, we speculated that the rid1-2 and rid1-3 mutations have a serious effect on female gametophyte development. Microscopy observations demonstrated morphological abnormalities in the mutant female gametophytes. Specifically, the synergid nuclei and egg cell nucleus were similar in size, the polar nuclei were not fused, the antipodal cells were enlarged and protruded, and the antipodal nuclei were fused (Figures 8F to 8H). These findings imply that the rid1-2 and rid1-3 mutations affect the cellular specification of female gametophytes. It is notable that these phenotypes are quite similar to those of the splicing factor defective mutants, lachesis (lis) and clotho (clo) (Gross-Hardt et al., 2007; Moll et al., 2008). Self-pollination of RID1/rid1-2 and RID1/rid1-3 plants resulted in a 109:51 segregation of RID1/RID1 and RID1/rid1-2 and a 102:49 segregation of RID1/RID1 and RID1/rid1-3, respectively (Table 1). These data do not fit with the 1:1 segregation expected for simple female gametophytic lethality. We found that about half of the seeds were aborted in siliques borne on the RID1/rid1-2 or RID1/rid1-3 plants, in which no morphological signs of embryogenesis were visible (Figures 8D and 8E; see Supplemental Table 1 online). These findings suggested that the rid1-2 and rid-3 mutations from the male parent affect prefertilization processes, rather than postfertilization processes, probably by interfering with the formation and/or physiological activities of male gametophytes. Our results collectively show that RID1 has roles in gametophyte development that are important for successful reproduction.
DISCUSSION
Through the molecular genetic analysis of the Arabidopsis temperature-sensitive mutant rid1-1 we identified the underlying gene RID1 as At1g26370. This gene encodes a DEAH-box RNA helicase that is similar in sequence to human DHX8 and yeast Prp22. As these proteins are known to act at the final step of pre-mRNA splicing (Ohno and Shimura, 1996; Schwer and Gross, 1998), we speculated that RID1 has functions in pre-mRNA splicing. This hypothesis was strongly supported by the finding that the splicing efficiency of the transiently expressed reporter transcript was much lower in the rid1-1 mutant, particularly at the restrictive temperature, than in the wild type (Figures 5A to 5C). Furthermore, we found that alternative splicing patterns of endogenous transcripts were affected by the rid1-1 mutation (Figure 5D), which is also consistent with the involvement of RID1 in pre-mRNA splicing.
The similarity between RID1 and human DHX8 and yeast Prp22 first led us to presume that RID1 functions in releasing mRNA at the final step of pre-mRNA splicing, as do DHX8 and Prp22 (Ohno and Shimura, 1996; Schwer and Gross, 1998). However, our data showed that the rid1-1 mutation influenced not only intron removal but also the recognition of the splicing site (Figures 5D and 5E) and that RID1 did not possess the molecular activity to complement mutant Prp22 of yeast (see Supplemental Figure 5 online). These results imply that RID1 has a different role in pre-mRNA splicing than do DHX8 and Prp22, as suggested by the phylogenetic tree analysis that showed that RID1 is not located in the clade that contains Prp22 (see Supplemental Figure 3 online). Furthermore, the preferential localization of RID1-YFP in the nucleolus provided further clues as to the function of RID1 (Figure 6). Many studies on animals have shown that the nucleolus and its associated bodies, particularly Cajal bodies, have critical roles in snRNP biogenesis, which is required for spliceosome formation (reviewed in Patel and Bellini, 2008). Nucleolar proteome analysis of Arabidopsis revealed that a number of snRNP proteins are present in the nucleolus (Pendle et al., 2005). Several splicing factors were also found in the nucleolus (Lorković et al., 2004; Pendle et al., 2005; Tillemans et al., 2006), and the nucleolus and Cajal bodies are known to function in U1 snRNP maturation (Lorković and Barta, 2008) in Arabidopsis. These findings strongly suggest that, as in animals, the nucleolus is involved in snRNP biogenesis, and possibly in spliceosome formation, in plants (Shaw and Brown, 2012). A lot of rearrangements of RNA–RNA and RNA–protein are included in these processes, and RNA helicases should mediate at least a subset of them. Taking all of this into consideration, we speculate that RID1 might contribute to snRNP biogenesis and consequently spliceosome assembly. Future detailed molecular characterization of RID1 is needed to assess the validity of this hypothesis.
The rid1-1 mutant phenotypically resembled the srd2-1 mutant in terms of tissue culture responses (Figure 1; see Supplemental Figure 1 online; Ohtani and Sugiyama, 2005) and several aspects of plant development (Figures 2, 3, and 8; Ohtani et al., 2008, 2010). RID1 expression was regulated temporally and spatially in tissue culture and during development (Figure 7), and these expression patterns considerably overlapped those of SRD2 (Ohtani and Sugiyama, 2005; Ohtani et al., 2008, 2010). The similarities between rid1-1 and srd2-1 and between RID1 and SRD2 suggest that certain developmental processes depend on a molecular event that involves both SRD2 and RID1 as essential factors. Previously, SRD2 was shown to activate the transcription of snRNAs, including all spliceosomal UsnRNAs (Ohtani and Sugiyama, 2005); in this work, we have shown that RID1 is a DEAH-box RNA helicase involved in efficient pre-mRNA splicing. Therefore, the increase in pre-mRNA splicing, which is driven by RID1 and the SRD2-mediated production of UsnRNAs, is most likely the key factor in regulating the development processes that are highly sensitive to the rid1 and srd2 mutations. The importance of splicing factors during plant development has been reported by several research groups (Tsukaya et al., 2013). For instance, a knockout mutant of SmD3-b, the core component of snRNP, showed pleotropic phenotypes, including reduced root growth, defective leaf venation, and changed numbers of floral organs (Swaraz et al., 2011), and null mutants of DEFECTIVELY ORGANIZED TRIBUTARIES2/MERISTEM-DEFECTIVE, which encodes a member of the SART1 family implicated in spliceosome assembly, exhibited abnormalities in meristem organization and vein patterning (Petricka et al., 2008; Casson et al., 2009). Moreover, the specification of female gametophyte cells was disturbed in the splicing factor defective mutants lis, clo, and atropos (ato). LIS, GAMETOPHYTE FACTOR1 (GFA1)/CLO, and ATO encode Arabidopsis homologs of yeast splicing factor Prp4, Snu114p, and Prp9, respectively (Coury et al., 2007; Gross-Hardt et al., 2007; Moll et al., 2008). Arabidopsis plants with reduced expression of SAP130, a subunit of the U2 snRNP-associated complex, had defects in both female and male gametophyte formation (Aki et al., 2011). In the vaj mutant, a weak allele of GFA1/CLO, abnormalities in floral organ development were reported (Yagi et al., 2009). These studies, together with the phenotypes of rid1 and srd2, suggest that specific aspects of development, such as floral organogenesis, vein formation, and gametophyte development, are particularly susceptible to defects in spliceosome machinery.
Assuming that the capacity of pre-mRNA splicing regulates specific developmental processes, the next question is how it does so. One possibility is that the capacity of pre-mRNA splicing determines the overall level of gene expression and influences the cell's ability to undergo cell division and/or cell differentiation and to regulate specific processes at the smallest cost to the cell. The other possibility is that a certain set of genes involved in specific developmental processes might require a particularly high spliceosome activity and therefore be regulated selectively by the capacity of splicing. Our results, which showed that the rid1-1 mutation affected only a subset of alternative splicing events (Figures 5D and 5E), favor the latter hypothesis. A comprehensive search for genes that are selectively affected by rid1 and srd2 and contribute to the rid1 and srd2 phenotypes would elucidate the mechanisms underlying the pre-mRNA splicing-mediated regulation of plant development.
METHODS
Plant Materials and Growth Conditions
The rid1-1 mutant was derived from the Landsberg erecta (Ler) strain of Arabidopsis thaliana (Konishi and Sugiyama, 2003; identical to rid1 described in the previous article). The GABI-Kat 730B12 and 310A05 lines, which were generated from the Columbia (Col) strain by T-DNA insertion mutagenesis, were obtained from a population of GABI-Kat flanking sequence tags (http://www.gabi-kat.de). These lines were crossed twice with the Col wild type and then used for the phenotypic analysis. The CYCB1;1:GUS reporter line was generated by transforming the Ler strain with pCDG (Colón-Carmona et al., 1999). The SCR:GFP reporter lines in the Wassilewskija background were provided by Philip N. Benfey (Wysocka-Diller et al., 2000). The WOX5:GUS reporter line in the Ler background was provided by Thomas Laux (Sarkar et al., 2007). The rid1-1 mutation was introduced into the reporter lines by crossing, and the resultant progeny lines homozygous for both the reporter gene and the mutation were used for the subsequent analyses. The growth conditions for seedlings and plants were described by Ohtani et al. (2008).
Tissue Culture and Histological Observation
Tissue culture was performed as previously described (Ozawa et al., 1998). Observations of tissues, detection of GUS activity, and sectioning of tissues were performed according to the methods described by Ohtani et al. (2008).
Chromosome Mapping
The rid1-1 mutant was crossed with the Col strain, and the resultant F3 progeny were used for chromosome mapping of the RID1 locus. These plants were examined for adventitious root formation at 28°C and for DNA polymorphisms to genotype their parental F2 plants. DNA was extracted from the shoot apices of the plants to be tested by grinding them in 200 μL of extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% [w/v] SDS). DNA was recovered by ethanol precipitation and subjected to simple sequence length polymorphism (Bell and Ecker, 1994) and cleaved amplified polymorphic sequence analysis (Konieczny and Ausubel, 1993). The original cleaved amplified polymorphic sequence markers in Figure 4A are listed in Supplemental Table 2 online. The chromosomal location of RID1 was determined on the basis of linkage of the rid1-1 mutation to the Ler alleles of the polymorphic marker loci.
Plasmid Construction
The At1g26370 genomic fragment, which corresponded to the region from –2084 to +3336 (where +1 represents the first nucleotide of the start codon of At1g26370), was amplified by PCR from the genomic DNA of Ler and then cloned into the pCR8/GW/TOPO vector (Invitrogen). The cloned At1g26370 fragment was integrated into pHG and pHGY (Yamaguchi et al., 2008) using LR clonase (Invitrogen) to generate the binary vectors At1g26370/pHG and At1g26370/pHGY, respectively. pHG was prepared by removing the 35S promoter sequence from pH35GS (Kubo et al., 2005). The T-DNA construct of At1g26370/pHG was designed to express At1g26370 under the control of its own promoter and used for complementation analysis. The T-DNA construct of At1g26370/pHGY contained RID1:RID1-YFP. For the RID1:GUS reporter construct, the region from –2084 to +27 (with +1 being defined as the first nucleotide of the start codon) of At1g26370 was cloned into the pENTR/D-TOPO vector (Invitrogen) and then integrated into the binary vector pBGGUS (Kubo et al., 2005) by the LR reaction.
In-YFP, a reporter gene designed to monitor pre-mRNA splicing efficiency, was synthesized by a custom gene synthesis service (Texas Genomics Japan) and cloned into the pCR8/GW/TOPO vector. The intron sequence was followed by the syn7 intron, which was described previously by Goodall and Filipowicz (1989) (Figure 5A). The In-YFP sequence was integrated into p35SG (Yamaguchi et al., 2010) to generate the In-YFP/p35SG plasmid for transient expression under the 35S promoter.
Information about all primer sets used is provided in Supplemental Table 2 online.
Complementation Test
The At1g26370/pHG plasmid was transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) and then used for plant transformation. In the case of the complementation test against rid1-1, the At1g26370 genomic fragment was introduced into rid1-1 plants directly, and the temperature sensitivity of callus and adventitious root formation of T2 progeny hypocotyl explants was tested. For complementation tests with rid1-2 and rid1-3, the Col plants transformed with At1g26370/pHG were crossed with the GABI-Kat 730B12 and 310A05 lines, and the resultant F2 progenies were subjected to genotyping analysis.
Transient Expression Analysis
Protoplast preparation and transfection were performed by the tape-Arabidopsis sandwich method described by Wu et al. (2009). Protoplasts were prepared from the true leaves of 14-d-old wild-type and rid1-1 seedlings, transfected with the In-YFP/p35SG plasmid, and incubated at 22 or 28°C for 12, 16, 20, and 24 h. Total RNAs were isolated from protoplasts using the TRIzol Plus RNA purification kit (Invitrogen) and subjected to first-strand cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen). DNA fragments derived from the spliced and/or unspliced forms of the In-YFP transcripts were PCR amplified from the resultant first-strand cDNA samples. The PCR program consisted of 35 cycles of a 30-s denaturation phase (95°C), a 30-s annealing phase (56°C), and a 30-s extension phase (72°C). The PCR products were separated electrophoretically on agarose gels and stained with SYBR gold nucleic acid gel stain (Molecular Probes), and the gel images were analyzed using Image J software (http://rsbweb.nih.gov/ij/). The splicing efficiencies were calculated using the following formula: (signal intensity of spliced form)/(sum of signal intensities of spliced and unspliced forms) × 100 (%). The experiments were repeated independently three times.
RT-PCR Analysis
Total RNAs were extracted using a TRIzol Plus RNA purification kit (Invitrogen) from hypocotyl explants of the wild type and rid1-1, which were cultured on CIM (Ohtani et al., 2008) for 0 or 1 d at 22 or 28°C. The RNA samples were treated with DNaseI according to the manufacturer’s instructions (Invitrogen). DNase-treated RNA (3 µg) was used to synthesize first-strand cDNA with an oligo(dT) primer in a 20-μL reaction volume using SuperScript III reverse transcriptase (Invitrogen). The reverse transcription reaction was diluted to a final volume of 100 µL, of which 1-μL aliquots were used as templates for PCR, along with 1.5 µM of each gene-specific primer (see Supplemental Table 2 online) and Ex Taq polymerase (TaKaRa). The PCR conditions were the same as those by Palusa et al. (2007) (for At1g55310, At3g53500, At3g55460, At4g02430, and At4g25500: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 70°C for 1 min, and completed at 72°C with 10 min) or by Simpson et al. (2008) (for At1g02840, At2g40910, At3g55630, At5g15230, and At3g12110: 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 50°C for 30 s, and 70°C for 1 min, and completed at 72°C for 10 min). The amplified PCR products were resolved by electrophoresis on 2.5% (for At1g02840, At2g40910, At3g55630, At5g15230, and At3g12110) or 1% (for At1g55310, At3g53500, At3g55460, At4g02430, and At4g25500) agarose gels and the gel images were analyzed using the AE-9020 E-shot II (ATTO). The experiments were repeated independently three times. Information about all primer sets used is provided in Supplemental Table 2 online.
For cloning and sequencing of splice variants, individual bands corresponding to different sizes were gel purified with NucleoSpin gel and PCR Clean-up (Macherey-Nagel). Each PCR product was cloned into the pGEM-T Easy vector according to the manufacturer’s instructions (Promega).
Subcellular Localization Analysis of RID1:YFP
The Col plants were transformed with At1g26370/pHGY to generate transgenic plants carrying RID1:RID1-YFP. Root tips of 7-d-old seedlings of the T2 lines were analyzed. For 4′,6-diamidino-2-phenylindole (DAPI) staining, the seedlings were fixed with 4% (w/v) paraformaldehyde in PBS for 5 min at room temperature, rinsed with PBS three times, and incubated in DAPI solution (5 µg/mL in PBS) for 5 min. Observations were performed with an LSM 700 laser scanning confocal microscope (Zeiss).
Transformation of Plants
A simplified version of the floral dip method described on the website http://www.plantpath.wisc.edu/fac/afb/protocol.html was used for plant transformation (Clough and Bent, 1998).
Phylogenetic Analysis
A BLAST search of the TAIR10 protein database (http://www.Arabidopsis.org/index.jsp) was performed using the amino acid sequence of RID1 as query, and 20 Arabidopsis proteins containing the conserved RNA helicase domains were detected. The amino acid sequences corresponding to the conserved RNA helicase domains of these Arabidopsis proteins and yeast Prp proteins were aligned using ClustalW and then manually adjusted to make fine alignments (see Supplemental Data Set 1 online). An unrooted tree was established by the neighbor-joining method using PHYLIP software (http://evolution.gs.washington.edu/phylip.html). The bootstrap values were obtained from 1000 trials.
Complementation Analysis of Yeast prp22 Mutants
Yeast strain YBST1 (Mata ura3-52 trp1-63 his3-Δ200 leu2Δ1 ade2-101 lys2-801 prp22::LEU2) with p360-Prp22 (URA3 CEN) and TRP1 CEN plasmids carrying the PRP22-Ala mutant alleles prp22 T637A or prp22 R805A were kindly provided by Beate Schwer (Schwer and Meszaros, 2000). The point mutation mutants prp22 T637A and prp22 R805A were generated by plasmid shuffling methods according to Schwer and Meszaros (2000). The genomic fragment of yeast PRP22 was amplified by PCR. The RID1 cDNA fragment was amplified by PCR from first-strand DNA synthesized from total RNA of wild-type seedlings. To generate the PRP22-RID1 chimeric gene, the PRP22 fragment corresponding to the nonconserved N-terminal region of Prp22 (from nucleotide +1 to +1440, where +1 represents the first nucleotide of the start codon of Prp22) and the RID1 cDNA fragment corresponding to the region from +115 to the stop codon (where +1 represents the first nucleotide of the start codon of RID1 cDNA) were independently amplified by PCR and then integrated by another PCR amplification using PRP22 and RID1 fragments as templates. Amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen) and then integrated into pYES-DEST52 (Invitrogen) using LR clonase (Invitrogen). The resultant plasmids were transformed into the prp22 T637A and prp22 R805A mutants, and the temperature sensitivity of cell growth upon overexpression of PRP22, RID1, or PRP22-RID1 was determined according to the manufacturer’s protocol (Invitrogen). Information about all primer sets used is provided in Supplemental Table 2 online.
RNA Gel Blot Analysis
Total RNAs were extracted using a TRIzol Plus RNA purification kit (Invitrogen) from hypocotyl explants of the wild type and rid1-1 that had been cultured on CIM (Ohtani et al., 2008) for 0 or 1 d at 22 or 28°C. The RNA samples (2 µg) were separated by gel electrophoresis, stained with SYBR Gold nucleic acid gel stain (Invitrogen), blotted onto nylon membranes, and then subjected to hybridization with digoxigenin-labeled probe specific for internal transcribed spacer region 1 (ITS1). The images were analyzed using ImageQuant LAS 4010 (GE Healthcare). Information about all primer sets used is provided in Supplemental Table 2 and Supplementary References 1 online.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under the following accession numbers: RID1 (At1g26370; NM_102401), Prp22 (NM_001178904), and DHX8 (NM_004941).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Effects of the srd2-1 and rid1-1 Mutations on Callus and Root Formation from Hypocotyl and Root Explants.
Supplemental Figure 2. Effects of the rid1-1 Mutation on Root Cap Structure.
Supplemental Figure 3. Phylogenetic Tree of Arabidopsis DExH RNA Helicase Proteins.
Supplemental Figure 4. Effects of the rid1-1 Mutation on Alternative Splicing Patterns.
Supplemental Figure 5. Complementation Analysis of Yeast prp22 Mutants.
Supplemental Figure 6. Effects of the rid1-1 Mutation on Pre-rRNA Processing.
Supplemental Table 1. Number of Normal and Aborted Seeds in Siliques Borne on RID1/rid1-2 and RID1/rid1-3 Plants.
Supplemental Table 2. Oligonucleotides Used in This Study.
Supplemental Data Set 1. Amino Acid Sequences Used to Generate the Phylogenetic Tree of Arabidopsis DExH RNA Helicase Proteins.
Supplemental References 1. References for Supplemental Figure 2 and Supplemental Table 2.
Acknowledgments
We thank the Salk Institute and Peter Doerner (Edinburgh University) for providing pCDG, Philip N. Benfey (Duke University) and Hidehiro Fukaki (Kobe University) for providing the SCR reporter lines, Thomas Laux (University of Freiburg) and Minako Ueda (Nara Institute of Science and Technology) for providing the WOX5 reporter lines, and Beate Schwer (Cornell University) for providing the yeast strain YBST1 and the plasmids containing PRP22 genes. We also thank Ryoko Hiroyama and Nao Kume (Biomass Engineering Program, RIKEN) and Seiko Nomura and Sachiko Ooyama (Plant Science Center, RIKEN) for their technical assistance. This work was supported in part by the RIKEN Biomass Engineering Program and RIKEN Plant Science Center, by Grants-in-Aid from the Japan Society for the Promotion of Science (Grants 18870028 and 24770052 to M.O. and 18370016 and 22370015 to M.S.), and by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant 19060001 to M.S.).
AUTHOR CONTRIBUTIONS
M.O. and M.S. designed the project and wrote the article. M.O. performed the experiments and analyzed the data. T.D. suggested the experimental designs and contributed to critical discussions.
Glossary
- snRNP
small ribonucleoprotein particle
- UsnRNA
uridine-rich small nuclear RNA
- snRNA
small nuclear RNA
- CIM
callus-inducing medium
- SIM
shoot-inducing medium
- SAM
shoot apical meristem
- RAM
root apical meristem
- Ler
Landsberg erecta
- Col
Columbia
- DAPI
4′,6-diamidino-2-phenylindole
References
- Aki S., Nakai H., Aoyama T., Oka A., Tsuge T. (2011). AtSAP130/AtSF3b-3 function is required for reproduction in Arabidopsis thaliana. Plant Cell Physiol. 52: 1330–1339 [DOI] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. (1999). Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, 2nd ed, R.F. Geste-land, T.R. Cech, and J.F. Atkins, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 525–560. [Google Scholar]
- Casson S.A., Topping J.F., Lindsey K. (2009). MERISTEM-DEFECTIVE, an RS domain protein, is required for the correct meristem patterning and function in Arabidopsis. Plant J. 57: 857–869 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Company M., Arenas J., Abelson J. (1991). Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Coury D.A., Zhang C., Ko A., Skaggs M.I., Christensen C.A., Drews G.N., Feldmann K.A., Yadegiri R. (2007). Segregation distortion in Arabidopsis gametophytic factor 1 (gfa1) mutants is caused by a deficiency of an essential RNA splicing factor. Sex. Plant Reprod. 20: 87–97 [Google Scholar]
- Goodall G.J., Filipowicz W. (1989). The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R., Kägi C., Baumann N., Moore J.M., Baskar R., Gagliano W.B., Jürgens G., Grossniklaus U. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A.J., Molnàr A., Jones A., Baulcombe D.C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Konishi M., Sugiyama M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130: 5637–5647 [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J., Barta A. (2008). Role of Cajal bodies and nucleolus in the maturation of the U1 snRNP in Arabidopsis. PLoS ONE 3: e3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J., Hilscher J., Barta A. (2004). Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell 15: 3233–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll C., von Lyncker L., Zimmermann S., Kägi C., Baumann N., Twell D., Grossniklaus U., Gross-Hardt R. (2008). CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 56: 913–921 [DOI] [PubMed] [Google Scholar]
- Ohtani M., Demura T., Sugiyama M. (2008). Differential requirement for the function of SRD2, an snRNA transcription activator, in various stages of plant development. Plant Mol. Biol. 66: 303–314 [DOI] [PubMed] [Google Scholar]
- Ohtani M., Demura T., Sugiyama M. (2010). Particular significance of SRD2-dependent snRNA accumulation in polarized pattern generation during lateral root development of Arabidopsis. Plant Cell Physiol. 51: 2002–2012 [DOI] [PubMed] [Google Scholar]
- Ohtani M., Sugiyama M. (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis Plant J. 43: 479–490 [DOI] [PubMed] [Google Scholar]
- Ohno M., Shimura Y. (1996). A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 10: 997–1007 [DOI] [PubMed] [Google Scholar]
- Ozawa S., Yasutani I., Fukuda H., Komamine A., Sugiyama M. (1998). Organogenic responses in tissue culture of srd mutants of Arabidopsis thaliana. Development 125: 135–142 [DOI] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S.N. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Patel S.B., Bellini M. (2008). The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 36: 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle A.F., Clark G.P., Boon R., Lewandowska D., Lam Y.W., Andersen J., Mann M., Lamond A.I., Brown J.W.S., Shaw P.J. (2005). Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J.J., Clay N.K., Nelson T.M. (2008). Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56: 251–263 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A.I., Mann M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schwer B., Gross C.H. (1998). Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B., Meszaros T. (2000). RNA helicase dynamics in pre-mRNA splicing. EMBO J. 19: 6582–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P., Brown J. (2012). Nucleoli: Composition, function, and dynamics. Plant Physiol. 158: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C.G., Fuller J., Maronova M., Kalyna M., Davidson D., McNicol J., Barta A., Brown J.W.S. (2008). Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 53: 1035–1048 [DOI] [PubMed] [Google Scholar]
- Staley J.P., Guthrie C. (1998). Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Swaraz A.M., Park Y.-D., Hur Y. (2011). Knock-out mutations of Arabidopsis SmD3-b induce pleotropic phenotypes through altered transcript splicing. Plant Sci. 180: 661–671 [DOI] [PubMed] [Google Scholar]
- Tanner N.K., Linder P. (2001). DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell 8: 251–262 [DOI] [PubMed] [Google Scholar]
- Tillemans V., Leponce I., Rausin G., Dispa L., Motte P. (2006). Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18: 3218–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H., Byrne M.E., Horiguchi G., Sugiyama M., Van Lijsebettens M., Lenhard M. (2013). How do ‘housekeeping’ genes control organogenesis?—Unexpected new findings on the role of housekeeping genes in cell and organ differentiation. J. Plant Res. 126: 3–15 [DOI] [PubMed] [Google Scholar]
- Wang B.-B., Brendel V. (2004). The ASRG database: Identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., Lührmann R. (2009). The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Will C.L., Lührmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J.W., Helariutta Y., Fukaki H., Malamy J.E., Benfey P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Yagi N., Takeda S., Matsumoto N., Okada K. (2009). VAJ/GFA1/CLO is involved in the directional control of floral organ growth. Plant Cell Physiol. 50: 515–527 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T. (2008). Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutani I., Ozawa S., Nishida T., Sugiyama M., Komamine A. (1994). Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol. 105: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]