This work explores the transient morphogenetic activity in developing leaves that underlies some of the variability in leaf shape and size. It finds that AP1/FUL genes, previously characterized mainly by their role in reproductive development, are negative targets of TCPs during tomato leaf development. The work also shows a role for TCPs in flowering induction.
Abstract
Flexible maturation rates underlie part of the diversity of leaf shape, and tomato (Solanum lycopersicum) leaves are compound due to prolonged organogenic activity of the leaf margin. The CINCINNATA -TEOSINTE BRANCHED1, CYCLOIDEA, PCF (CIN-TCP) transcription factor LANCEOLATE (LA) restricts this organogenic activity and promotes maturation. Here, we show that tomato APETALA1/FRUITFULL (AP1/FUL) MADS box genes are involved in tomato leaf development and are repressed by LA. AP1/FUL expression is correlated negatively with LA activity and positively with the organogenic activity of the leaf margin. LA binds to the promoters of the AP1/FUL genes MBP20 and TM4. Overexpression of MBP20 suppressed the simple-leaf phenotype resulting from upregulation of LA activity or from downregulation of class I knotted like homeobox (KNOXI) activity. Overexpression of a dominant-negative form of MBP20 led to leaf simplification and partly suppressed the increased leaf complexity of plants with reduced LA activity or increased KNOXI activity. Tomato plants overexpressing miR319, a negative regulator of several CIN-TCP genes including LA, flower with fewer leaves via an SFT-dependent pathway, suggesting that miR319-sensitive CIN-TCPs delay flowering in tomato. These results identify a role for AP1/FUL genes in vegetative development and show that leaf and plant maturation are regulated via partially independent mechanisms.
INTRODUCTION
Plant leaves are flat lateral organs that are produced repeatedly by the shoot apical meristem (SAM). While most leaf growth is determinate, young leaves feature transient indeterminate growth, during which they maintain organogenic activity in specific regions at their margins, termed marginal blastozones (MBs). The temporal and spatial extent of this organogenic activity underlies some of the morphological diversity of leaf form. An extended organogenic window enables the formation of elaborated leaf forms, such as compound leaves, which are composed of multiple leaflets, each resembling a simple leaf (Hagemann and Gleissberg, 1996; Dengler and Tsukaya, 2001; Kaplan, 2001; Efroni et al., 2010; Floyd and Bowman, 2010).
Studies on leaf development have identified several groups of transcription factors and hormonal cues that are involved in defining the organogenic window and its flexibility (Burko and Ori, 2013; Fambrini and Pugliesi, 2013). Class I knotted like homeobox (KNOXI) proteins play important roles in the maintenance of the organogenic activities of both the SAM and the MB (Hake et al., 2004; Hay and Tsiantis, 2009, 2010; Blein et al., 2010). Downregulation of KNOXI activity in Cardamine hirsuta and tomato (Solanum lycopersicum) resulted in accelerated leaf maturation and decreased leaf complexity (Hay and Tsiantis, 2006; Shani et al., 2009). Conversely, their overexpression in tomato mutants, such as Mouse-ear and Curl, or in transgenic lines in several species led to enhanced organogenic activity and delayed maturation (Hareven et al., 1996; Chen et al., 1997; Parnis et al., 1997; Janssen et al., 1998; Bharathan et al., 2002; Tsiantis et al., 2002; Müller et al., 2006; Kimura et al., 2008; Barth et al., 2009; Shani et al., 2009). The hormone cytokinin (CK) has been shown to promote the extended organogenic activity of tomato and lettuce (Lactuca sativa) leaves downstream of KNOXI proteins (Frugis et al., 2001; Shani et al., 2010).
CINCINNATA -TEOSINTE BRANCHED1, CYCLOIDEA, PCF (CIN-TCP) transcription factors, a subset of class II TCPs (Martín-Trillo and Cubas, 2010), restrain the organogenic activity of the MB by promoting leaf maturation. Their downregulation in Antirrhinum majus and Arabidopsis thaliana resulted in leaf rumpling due to delayed maturation of the leaf margin and in delayed senescence (Nath et al., 2003; Palatnik et al., 2003; Koyama et al., 2007; Efroni et al., 2008; Schommer et al., 2008), while their overexpression led to precocious maturation, smaller leaf size, and senescence (Palatnik et al., 2003; Efroni et al., 2008; Schommer et al., 2008; Sarvepalli and Nath, 2011). In tomato, the CIN-TCP protein LANCEOLATE (LA) promotes leaf maturation together with additional related CIN-TCPs (LA-like proteins). LA-like genes are regulated by miR319, and as a result, LA and miR319 show opposite expression dynamics during leaf development: miR319 expression is high in emerging leaf primordia and is downregulated at the P5 stage, when LA expression is upregulated (Shleizer-Burko et al., 2011). Gain-of-function La mutants show accelerated leaf maturation and differentiation, while downregulation of LA-like genes by overexpression of miR319 leads to indeterminate leaf growth, especially at the margin (Mathan and Jenkins, 1962; Stettler, 1964; Dengler, 1984; Ori et al., 2007; Shleizer-Burko et al., 2011). LA activity was recently shown to be partly mediated by gibberellin (GA; Yanai et al., 2011). Several potential CIN-TCP targets have been identified in Arabidopsis. These include jasmonate biosynthesis genes, miR167, genes involved in auxin signal transduction, miR164 and ASYMMETRIC LEAVES1 (Kosugi and Ohashi, 2002; Schommer et al., 2008; Koyama et al., 2010; Danisman et al., 2012; Rubio-Somoza and Weigel, 2013). Recently, CIN-TCPs were shown to modify CK response in Arabidopsis leaves via an interaction with the chromatin remodeling ATPase BRAHMA (BRM) and activation of the CK response inhibitor ARABIDOPSIS RESPONSE REGULATOR 16 (ARR16) (Efroni et al., 2013).
MADS box transcription factors are involved in many developmental processes in plants (Rounsley et al., 1995; Ng and Yanofsky, 2001; Becker and Theissen, 2003; Hileman et al., 2006; Dornelas et al., 2011; Smaczniak et al., 2012a). APETALA1/FRUITFULL (AP1/FUL) MADS box genes (also called SQUA) play conserved roles in the specification of floral meristem identity (Huijser et al., 1992; Bowman et al., 1993; Theissen et al., 1996, 2000; Ferrándiz et al., 2000; Litt and Irish, 2003; Parenicová et al., 2003; Smaczniak et al., 2012b) and are involved in the induction of flowering in many species (Immink et al., 1999; Yan et al., 2003; Ellul et al., 2004; Samach and Lotan, 2007; Ruokolainen et al., 2010; Kobayashi et al., 2012; Park et al., 2012). Their expression is induced by the flowering promoting factor FLOWERING LOCUS T (FT) upon induction to flowering (Mandel et al., 1992; Teper-Bamnolker and Samach, 2005; Berbel et al., 2012). In the core eudicots, this family is represented by three distinct clades, euAPETALA1 (euAP1), euFRUITFULL (euFUL), and AGAMOUS-like 79 (AGL79), thought to have evolved from a common eudicot through several duplication events (Litt and Irish, 2003; Shan et al., 2007). These clades and specific family members have acquired additional specific functions. For example, AP1 is involved in specification of organ identity (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993; Benlloch et al., 2006), and FUL functions in carpel development and promotes determinate growth. Loss-of-function mutations in the Arabidopsis FUL gene and suppression of FUL-like genes in poppy (Papaver somniferum) affect cauline leaf shape (Gu et al., 1998; Ferrándiz et al., 2000; Teper-Bamnolker and Samach, 2005; Pabón-Mora et al., 2012). The pea (Pisum sativum) AGL79-like gene VEGETATIVE1 (VEG1) was found to be involved in specifying secondary inflorescence meristem identity (Berbel et al., 2012). Recently, FUL-like genes were shown to play a role in compound-leaf morphogenesis in Aquilegia coerulea, a basal eudicot that contains only FUL-like genes. Suppression of A. coerulea FUL-like genes resulted in reduced leaf complexity (Pabón-Mora et al., 2013).
The tomato genome contains five AP1/FUL MADS box genes, MACROCALYX (MC)/AP1, MBP7 (FUL), MBP20 (AGL79), MBP10 (AGL79L), and TM4 (FULL) (Busi et al., 2003; Butler, 1952; Pnueli et al., 1991; Vrebalov et al., 2002; Litt and Irish, 2003; Hileman et al., 2006; Leseberg et al., 2008; Park et al., 2012). MC was shown to be involved in sepal development and in fruit abscission and ripening (Vrebalov et al., 2002; Nakano et al., 2012). TM4 and MBP7 are involved in fruit ripening, likely via interaction with Ripening-Inhibitor (Bemer et al., 2012). The role of MBP20 and MBP10 in tomato development has not been described.
To explore the nature of the transient organogenic window in tomato leaf development, we compared gene expression between genotypes with different levels of LA-like activity. The expression of the AP1/FUL MADS box genes positively correlated with the extent of the organogenic window. Genetic analysis showed that MBP20 is involved in leaf development downstream of LA. LA binds to regulatory sequences upstream of MBP20 and TM4. These findings reveal a role for MBP20 and TM4, and possibly additional AP1/FUL genes, in the regulation of compound leaf development.
RESULTS
Expression of Tomato AP1/FUL MADS Box Genes Is Correlated with the Organogenic Activity of the Leaf Margin
Developing tomato leaves retain a window of organogenic activity that enables the elaboration of leaf shape (Burko and Ori, 2013). To identify genes that mediate this activity, we used microarray analysis to compare gene expression among tomato genotypes that vary in the extent of the organogenic window due to differences in the activity of the LA gene. These included the gain-of-function allele La-2, mutated in the miR319-recognition site, wild-type tomato (S. lycopersicum cv M82, sp), and transgenic plants overexpressing the Arabidopsis miR319 in leaves (Ori et al., 2007; Shleizer-Burko et al., 2011) under the control of the FIL promoter (Lifschitz et al., 2006; Shani et al., 2009). To minimize expression differences that are secondary to the developmental alterations, we collected shoot apices containing the SAM and the two youngest leaf primordia, the phenotype of which is very similar among these genotypes (Figure 1A).
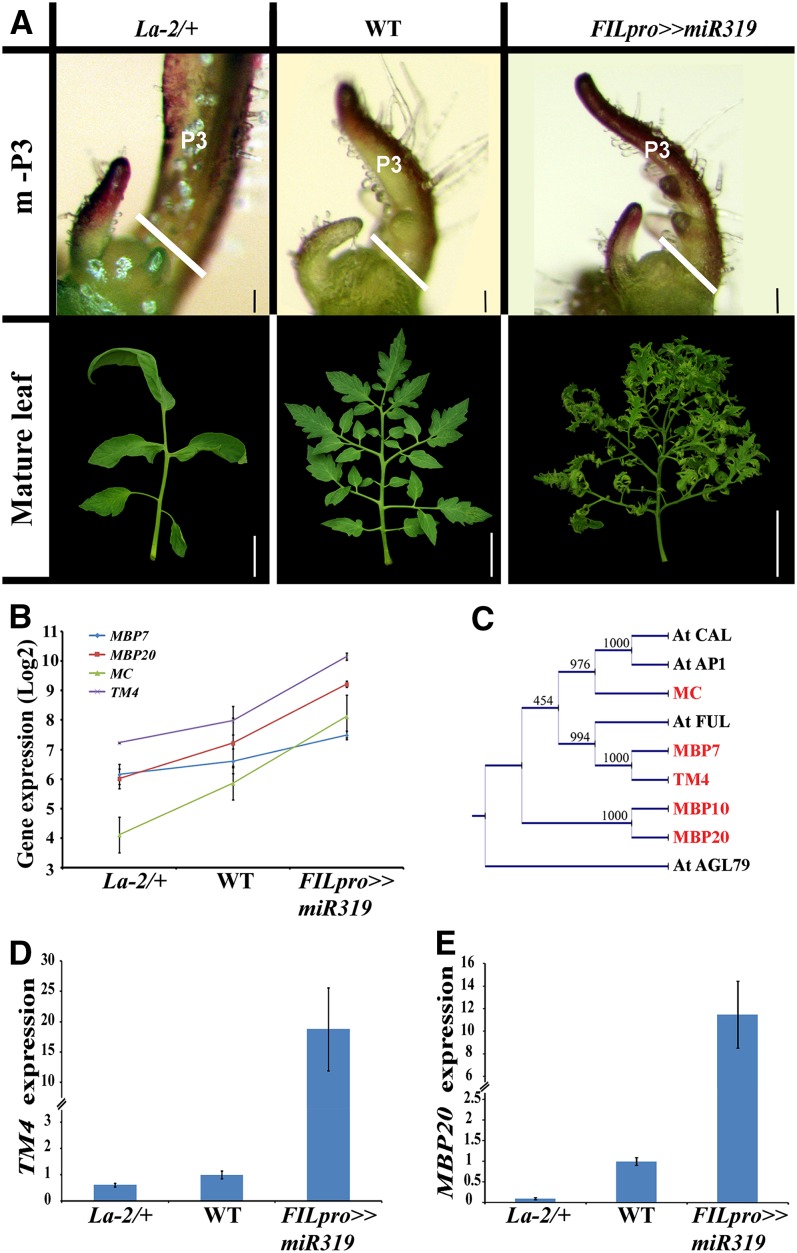
Figure 1.
Expression of AP1/FUL Genes Is Positively Correlated with the Organogenic Activity of the Leaf Margin.
(A) Phenotypes of young leaf primordia and mature leaves of the genotypes used for the expression profiling. m-P3, SAM and three youngest leaf primordia. Genotypes are indicated at the top of each column. For microarray analysis, tissue was collected from the SAM and two youngest leaf primordia, as indicated by the white line in the top row. Bars = 0.1 mm (top row) and 5 cm (bottom row). WT, the wild type.
(B) Microarray expression data for four AP1/FUL genes, shown as an average of three biological repeats (±se).
(C) Phylogenetic analysis of tomato and Arabidopsis AP1/FUL genes.
(D) and (E) Expression of TM4 (D) and MBP20 (E) in the fifth leaf at the P5 stage of the indicated genotypes. Expression was assayed by qRT-PCR relative to the reference gene EXP and is shown as an average of three biological repeats (±se). FILpro>>miR319 plants expresses miR319 under the control of the FIL promoter.
[See online article for color version of this figure.]
The expression of four closely related MADS box genes from the AP1/FUL subfamily was positively correlated with the extent of the organogenic activity of the leaf margin and negatively correlated with the level of LA activity (Figure 1B). These included MBP7 (FUL2), MBP20 (AGL79), TM4 (FUL1), and MC. These results were verified by quantitative RT-PCR (qRT-PCR) in apices containing the SAM and the four youngest leaf primordia (see Supplemental Figure 1A online). qRT-PCR also showed similar behavior of the fifth member of this gene family, MBP10 (AGL79L), which was not present on the microarray, indicating that the expression of all members of this gene family correlates with the organogenic window (Figure 1C; see Supplemental Figures 1A and 2A online). Since in other species the expression of most AP1/FUL genes was reported mainly in reproductive organs, we examined the expression of MBP20 and TM4 in dissected P5 primordia. Their expression was decreased in La-2 and increased in FILpro>>miR319 primordia (Figures 1D and 1E). The leaf-specific effect of LA on the expression of these genes was further examined using plants expressing a mutated form of LA with reduced sensitivity to miR319 (LAm) (Ori et al., 2007) or miR319 under the control of the BLS promoter, expressed specifically in leaves starting at the P4 stage (Shalit et al., 2009; Shani et al., 2009). Leaf primordia at the P4 stage were collected before flowering. At this stage, any observed expression changes are expected to be immediate effects of the changes in LA expression. The expression of MBP20 and TM4 was upregulated in BLSpro>>miR319 and that of MBP20 was also downregulated in BLSpro>>LAm (see Supplemental Figure 1B online). In conclusion, LA-like proteins negatively regulate the expression of AP1/FUL genes.
miR319 Affects Leaf and Plant Maturation in an Opposite Manner and via Separate Pathways
Often a genetic alteration that affects the rate of leaf maturation similarly affects plant maturation, as manifested by flowering time, for example (Teper-Bamnolker and Samach, 2005; Lifschitz et al., 2006; Shalit et al., 2009). Maturation is accelerated in La-2 leaves, while leaves overexpressing miR319 show delayed maturation and indeterminate growth (Ori et al., 2007; Shleizer-Burko et al., 2011). Interestingly, the effect of these genotypes on plant maturation was the opposite: FILpro>>miR319 plants and BLSpro>>miR319 plants flower with fewer leaves than the wild type, and the number of leaves produced by La-2 mutants until flowering is slightly increased (Figure 2A). It should be noted that the effect of these genotypes on flowering is complex, as leaf production is faster in La-2 and slower in FILpro>>miR319 (Shleizer-Burko et al., 2011), resulting in La-2 plants flowering after a shorter time and FILpro>>miR319 after a longer time from planting relative to wild type.
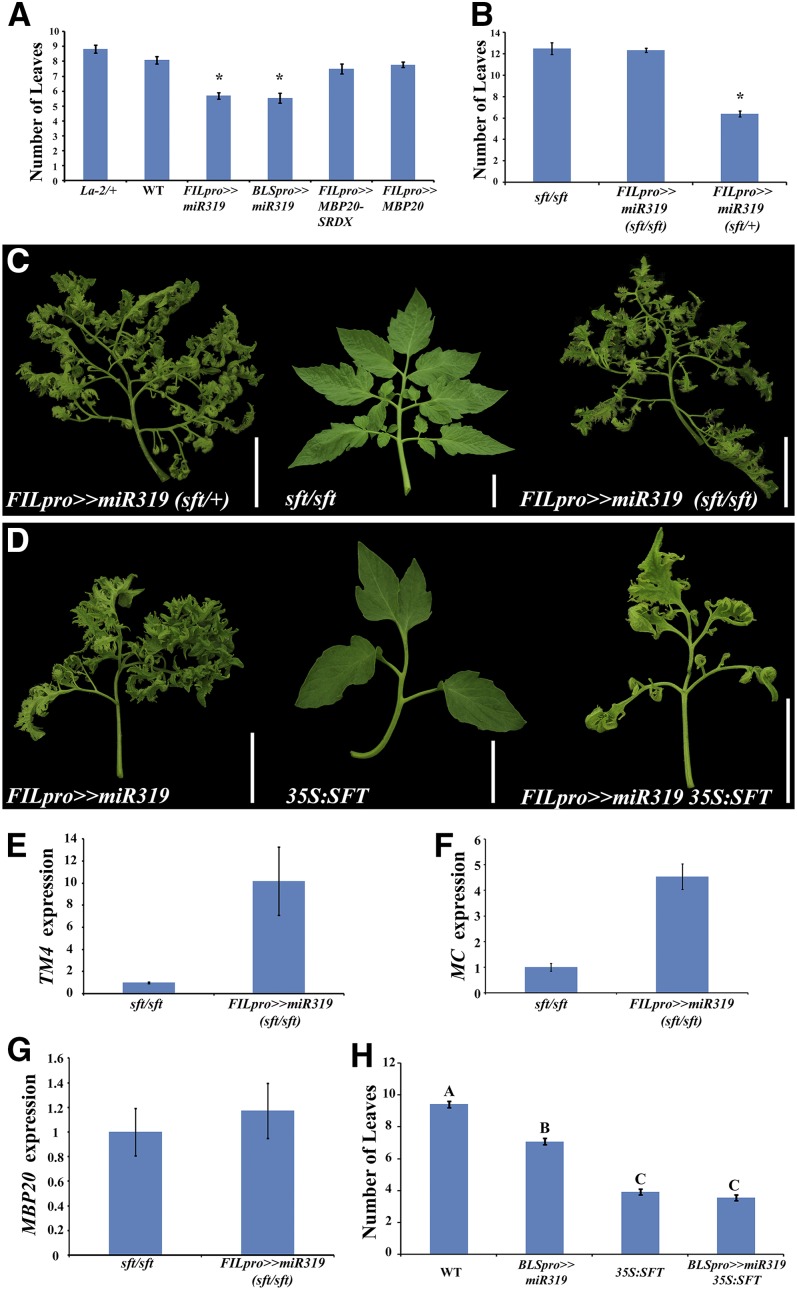
Figure 2.
miR319 Affects Plant and Leaf Maturation via Separate Pathways.
(A) and (B) Number of leaves until the first visible flower. Shown are averages ± se (for FILpro>>MBP20-SRDX, n = 5; for all other genotypes, n = 10). Asterisks indicate statistically significant differences from the wild type (A) or sft (B) at P < 0.05.
(C) and (D) Mature leaves of the indicated genotypes. 35S:SFT plants express SFT under the control of the 35S promoter by direct fusion; FILpro>>miR319 plants express miR319 under the control of the FIL promoter using the transactivation system (Moore et al., 1998). Bars = 5 cm.
(E) to (G) Expression of TM4 (E), MC (F), and MBP20 (G) in apices containing the SAM and six youngest leaf primordia, in which the third leaf of the plant was at the P6 stage. Relative expression was assayed using qRT-PCR relative to the reference gene EXP and is shown as an average of four biological repeats ±se.
(H) Number of leaves until the first visible flower. Shown are averages ±se (n > 7). Letters indicate statistically significant differences at P < 0.05.
[See online article for color version of this figure.]
SINGLE FLOWER TRUSS (SFT) is the tomato ortholog of the flowering promoting factor FT. sft mutants flower late and have more intercalary leaflets than the wild type, whereas 35S:SFT shows early flowering and its leaves are simplified due to accelerated maturation (Molinero-Rosales et al., 2004; Lifschitz and Eshed, 2006; Lifschitz et al., 2006; Shalit et al., 2009). To examine whether the effect of FILpro>>miR319 on flowering timing and AP1/FUL expression depends on SFT activity and whether it affects plant and leaf determination via common or separate pathways, we introduced the FILpro>>miR319 transgene into the sft and 35S:SFT backgrounds. sft FILpro>>miR319 plants flowered after producing the same number of leaves as single sft mutants (Figure 2B), but their leaves resembled those of FILpro>>miR319 (Figure 2C). 35S:SFT FILpro>>miR319 leaves showed indeterminate growth similar to FILpro>>miR319 leaves and an intermediate phenotype with respect to leaf complexity (Figure 2D). Because of the very low fertility of FILpro>>miR319 plants, we monitored flowering time in BLSpro>>miR319 35S:SFT plants. BLSpro>>miR319 35S:SFT plants flowered after three to four leaves, similar to 35S:SFT plants (Figure 2H; see Supplemental Figures 3A to 3C online). Therefore, sft and 35S:SFT were epistatic to FILpro>>miR319 with respect to plant maturation, and FILpro>>miR319 was epistatic to 35S:SFT and sft with respect to leaf determination. This indicates that the effect of miR319 on flowering time is mediated by SFT and that miR319 and its target genes affect leaf development and flowering time via separate pathways.
The effect of miR319 overexpression on the expression of TM4 and MC was milder in the sft background than in the wild type (Figures 2E and 2F), and the expression of MBP20, MBP7, and MBP10 was similar between sft and sft FILpro>>miR319 (Figure 2G; see Supplemental Figures 3D and 3E online). This suggests that the expression of these genes is positively regulated by SFT and negatively by LA-like proteins and that the effect of LA on their expression is at least partially dependent on functional SFT.
MBP20 Is Involved in Tomato Leaf Patterning
To further understand the role of AP1/FUL genes in leaf development, we examined the dynamics of their expression in the fifth leaf produced by the plant at successive developmental stages using qRT-PCR. Early stages were sampled with the SAM and younger primordia. In contrast with AP1/FUL genes from other species, all family members were expressed in tomato leaves (Figure 3A; see Supplemental Figure 2B online). MBP20 mRNA was transiently and substantially upregulated in the P3 and P4 stage primordia. Its expression was then sharply downregulated at the P5 stage (Figure 3A, bars). Interestingly, LA expression is sharply upregulated at this stage (Figure 3A, dashed line; Shleizer-Burko et al., 2011). The closest homolog of MBP20, MBP10, showed similar but less profound expression dynamics (see Supplemental Figure 2B online). We chose to focus on MBP20 for further investigation of its role downstream of LA in leaf patterning due to the negative correlation between its expression and LA expression and because we identified a putative LA binding site in its promoter (see below). In agreement with the mRNA expression, a 3209-bp MBP20 promoter drove expression at early stages of leaf development, starting from the P1 stage and in the leaflet primordia, but not in the SAM (Figures 3B, 3D, and 3E). The expression was downregulated in the P2 and P3 primordia of La-2 plants (Figures 3C, 3F, and 3G). Thus, expression of MBP20 is negatively correlated with that of LA.
Figure 3.
Expression and Function of MBP20 in Tomato Leaf Development.
(A) Expression dynamics of MBP20 (columns, left axis) and LA (red dashed line, right axis) along the development of the fifth leaf of wild-type tomato plants, assayed by qRT-PCR relative to the reference gene EXP. Shown are averages ± se (n = three to six biological repeats). m-P2, m-P3, and m-P4 represent SAM and two, three, or four youngest leaf primordia, respectively. The LA expression data is illustrated according to Shleizer-Burko et al. (2011).
(B) and (C) Fluorescence of the mRFP protein expressed under the control of the MBP20 promoter (red) at stages P1 to P3 in the wild type (B) and La-2/+ (C) viewed with a stereomicroscope using a Nuance camera and software (CRi).
(D) to (G) Histochemical staining of β-glucuronidase (GUS) activity (blue). β-Glucuronidase was expressed under the control of the MBP20 promoter. Shown are SAM and five youngest leaf primordia ([D] and [F]) and magnifications of the SAM and three youngest primordia ([E] and [G]) of the wild type ([D] and [E]) and La-2/+ ([F] and [G]).
(H) Fifth leaves of the indicated genotypes. WT, the wild type.
(I) MBP20-SRDX and MBP20 expression in apices containing the SAM and five youngest leaf primordia of FILpro>>MBP20 and FILpro>>MBP20-SRDX transgenic plants compared with the wild type. Expression was assayed by qRT-PCR relative to the reference gene EXP and is shown as an average of four biological repeats (±se).
(J) Number of primary, secondary, and intercalary leaflets on the fifth leaves of the indicated genotypes, shown as an average of seven to nine leaves from different plants (±se).
Bars = 250 μm (B), (C), (E), and (G), 1 mm (D) and (F), and 5 cm in (H).
To understand the role of MBP20 in leaf development, we overexpressed in leaves wild-type MBP20 or a fusion of MBP20 with the SRDX repression motif (Hiratsu et al., 2003), using the trans-activation system (Moore et al., 1998) and the FIL promoter (Shani et al., 2009). We reasoned that as LA activity similarly affects the entire tomato AP1/FUL clade, genes in this clade may act redundantly during leaf development. The MBP20-SRDX fusion was expected to downregulate their redundant targets, affecting the downstream developmental pathways. Overexpression was verified using qRT-PCR. Overexpression of MBP20 in leaves had a mild effect on leaf shape in four out of eight independent lines (Figures 3H to 3J; see Supplemental Figures 4A, 4C, and 4D online). Leaves overexpressing MBP20-SRDX were simplified, with fewer primary leaflets, a lack of higher order leaflets, and smooth leaflet margins (Figures 3H to 3J; see Supplemental Figures 4A, 4B, and 4D online), in agreement with the observation that downregulation of FUL-like genes in A. coerulea results in simplified leaves (Pabón-Mora et al., 2013). A similar phenotype was observed in crosses of eight independent transgenic lines to the FIL promoter, with three lines showing a strong phenotype, two lines showing an intermediate phenotype, and three lines showing a weak phenotype. One of the strong lines was used in further analyses. To verify the relevance of this phenotype to MBP20 function, we coexpressed MBP20 and MBP20-SRDX in leaves. While MBP20 overexpression had a very mild effect on leaf shape, it substantially suppressed the FILpro>>MBP20-SRDX phenotype (see Supplemental Figure 4D online), implying that the FILpro>>MBP20-SRDX phenotype results at least in part from impaired MBP20 function. FILpro>>MBP20 and FILpro>>MBP20-SRDX plants flowered after producing a similar number of leaves as wild-type plants (Figure 2A). It should be noted that the MBP20-SRDX transgene could possibly affect other genes in addition to the natural targets of the AP1/FUL family, as they may share binding sites with additional MADS box transcription factors (Riechmann et al., 1996; Melzer et al., 2006) and may be engaged in unnatural interactions. 35Spro>>MBP20-SRDX plants showed severe developmental alterations, including enlarged sepals (see Supplemental Figure 4E online), suggesting that the activity of MC is compromised by this transgene, as predicted. Together, these results suggest that MBP20, and possibly additional AP1/FUL genes, are involved in compound-leaf development.
LA Binds to Sequences in the MBP20 and TM4 Regulatory Regions
xamination of the MBP20 and TM4 promoters revealed several putative TCP binding sites (Kosugi and Ohashi, 2002; Schommer et al., 2008; Aggarwal et al., 2010; Viola et al., 2012), which were designated sites I-IV and sites I-II, respectively (Figures 4A and 4B). We tested whether LA binds to these sequences using electrophoretic mobility shift assay (EMSA) and a recombinant, bacterially expressed LA protein, fused to maltose binding protein (MBP-LA). MBP-LA bound to a 60-bp-long probe that contained MBP20 site IV but not to a probe that contained a mutation in the putative binding site (Figure 4A). A nonlabeled probe competed with the binding, indicating that the binding of LA to site IV is specific. Furthermore, inclusion of anti-MBP antibodies in the reaction resulted in a supershift (Figure 4A). No binding to sites I-III could be detected (see Supplemental Figure 5A online). Recombinant MBP-LA also bound to site II in the TM4 promoter, and competition and supershift assays confirmed the specificity of this binding (Figure 4B). We verified the binding of LA to site IV in the MBP20 promoter by a yeast one-hybrid assay (Li and Herskowitz, 1993; Pruneda-Paz et al., 2009), in which a fusion of LA to the GAL4 activation domain (prey) showed binding to a wild-type 500-bp fragment (bait) surrounding site IV but not to a fragment with two point mutations in site IV (see Supplemental Figure 5B online).
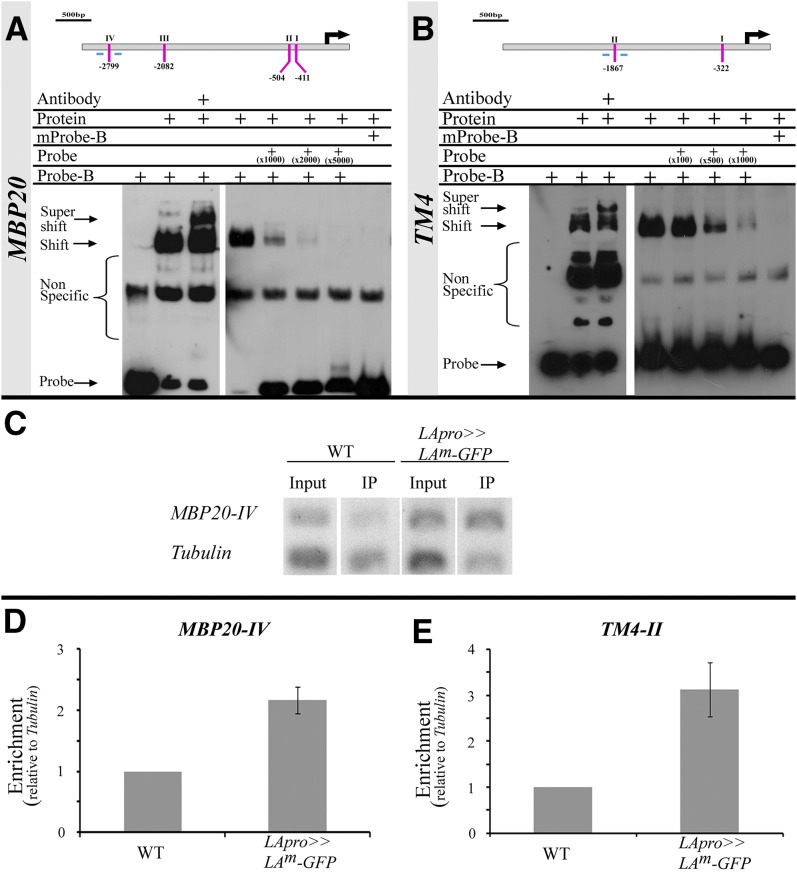
Figure 4.
LA Binds to the MBP20 and TM4 Promoters.
(A) and (B) Top: Schematic diagrams of the MBP20 (A) and TM4 (B) promoters and potential core TCP binding sites (GGNCC, indicated with lines and roman numerals). Black arrows indicate the translation start sites. Bottom: EMSA performed with biotin-labeled probes (Probe-B) and recombinant LA protein fused to MBP (Protein). The components included in each reaction are indicated above each lane. Probe, unlabeled probe (folds of the amount of labeled probe indicated); mProbe and mProbe-B, unlabeled and labeled mutated probe (…GGNaCt…), respectively; Antibody, antibodies against MBP.
(C) to (E) PCR and quantitative PCR analyses of a ChIP assay, performed with wild-type plants (WT) or plants expressing a LAm-GFP fusion under the control of the LA promoter (LApro>>LAm-GFP) and anti-GFP antibodies.
(C) PCRs were performed with specific primers for MBP20-IV (lines below the gene diagrams in [A]) and Tubulin. Input, nonimmunoprecipitated samples; IP, samples after ChIP.
(D) and (E) Quantitative PCR reactions were performed with specific primers for MBP20-IV (D) or TM4-II (E) (lines below the gene diagrams in [A] and [B], respectively) or Tubulin. Shown are averages (±se) of fold enrichment, compared with the wild type (n = two technical and three biological repeats in [D] and three biological repeats in [E]).
[See online article for color version of this figure.]
We further verified in vivo binding of LA to the identified sites in the promoters of MBP20 and TM4 using chromatin immunoprecipitation (ChIP) assays. We used transgenic plants expressing a fusion of LAm and green fluorescent protein (GFP) under the control of the LA promoter (LApro>>LAm-GFP) and anti-GFP antibodies. ChIP samples were tested using PCR and quantitative PCR with primer pairs spanning the MBP20-IV and TM4-II binding sites and the TUBULIN gene as a negative control. An enrichment of the MBP20-IV and TM4-II sites was detected in the LApro>> LAm-GFP immunoprecipitation sample compared with the input sample and the wild-type samples (Figures 4C to 4E). In conclusion, LA binds specifically to the MBP20 and the TM4 promoters.
MBP20 Genetically Mediates LA Activity
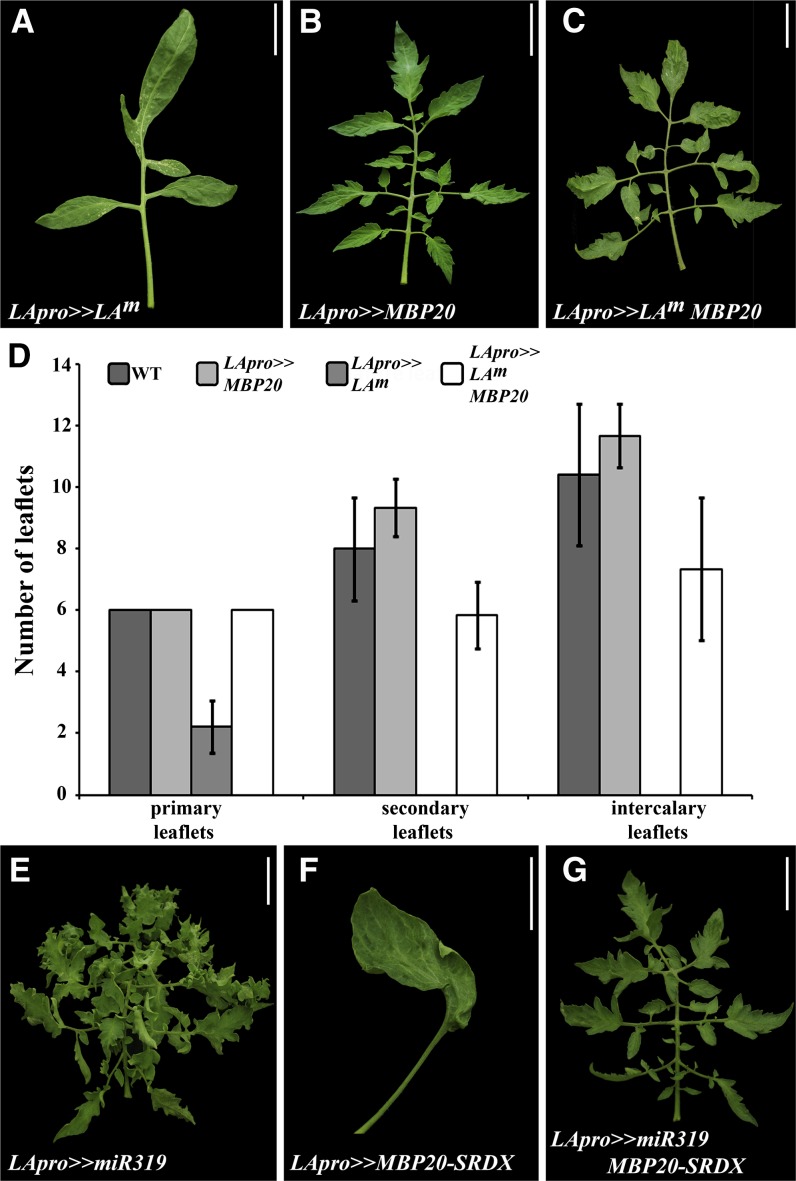
To test whether MBP20 mediates LA-like activity, we introduced LApro>>MBP20 and LApro>>MBP20-SRDX into genotypes with altered LA activity by crosses. Expression of LAm under the control of the LA promoter (LApro>>LAm) led to simple, reduced leaves with fused primary leaflets and no secondary leaflets (Figures 5A and 5D; see Supplemental Figure 6A online). Coexpression of MBP20 via the same promoter did not affect the phenotype of the first few leaves, but substantially suppressed the LApro>>LAm phenotype of later leaves, reinstating the formation of separated primary, secondary, and intercalary leaflets (Figures 5A to 5D; see Supplemental Figure 6A online). Analysis of LA expression confirmed that this suppression did not result from silencing of the LA transgene (see Supplemental Figure 6B online). Curiously, LApro>>MBP20 did not suppress the phenotype of the La-2 gain-of-function allele, which could result from inaccurate expression of the LA promoter. Coexpression of miR319 and MBP20-SRDX under the control of the LA promoter resulted in an intermediate phenotype between the LApro>>miR319 and LApro>>MBP20-SRDX phenotype (Figures 5E to 5G). Together, these results suggest that MBP20 partly mediates the effects of LA and miR319 on leaf shape but that additional factors are also involved, likely including the closely related gene MBP10 and additional tomato AP1/FUL genes.
Figure 5.
MBP20 Genetically Mediates LA Activity.
(A) to (C) and (E) to (G) Mature leaves of the indicated genotypes. All the transgenes were expressed using the transactivation system. All leaves shown are from plants heterozygous for all transgenes, which include the promoter and the expressed gene. Bars = 5 cm.
(D) Number of primary, secondary, and intercalary leaflets on mature leaves of the indicated genotypes, shown as an average of four to five leaves from different plants (±se). WT, the wild type.
[See online article for color version of this figure.]
MBP20 Modulates the Effects of KNOXI and CK in Tomato Leaf Development
To further understand the context of MBP20 activity in leaf development, we tested its genetic interaction with the tomato KNOXI gene Tkn2 and the hormone CK, both of which promote the organogenic activity of the tomato leaf margin. Leaves overexpressing Tkn2-SRDX are simple (Figure 6D; Shani et al., 2009). Notably, coexpression of MBP20 suppressed this phenotype, restoring the formation and separation of primary and secondary leaflets (Figures 6B, 6D, and 6G), and coexpression of MBP20-SRDX enhanced the Tkn2-SRDX phenotype (see Supplemental Figure 6C online). Whereas overexpression of Tkn2 resulted in filamentous leaves, leaf expansion was restored by coexpression of MBP20-SRDX (Figures 6C, 6E, and 6H). MBP20-SRDX also partly suppressed the increased leaf complexity phenotype of the Me/+ mutant (Figures 6C, 6F, and 6I). FILpro>>CKX3 leaves, overexpressing the CK deactivation gene CYTOKININ OXIDASE3 (CKX3) (Werner and Schmülling, 2009; Shani et al., 2010), are simplified with only primary leaflets and smooth margins. Coexpression of MBP20 partly suppressed this phenotype, whereas coexpression of MBP20-SRDX enhanced it (Figures 6A to 6C and 6J to 6L). Cumulatively, these results suggest that MBP20, possibly along with additional family members, play a role downstream of LA and KNOXI genes in maintaining the organogenic activity of the leaf margin.
Figure 6.
MBP20 and MBP20-SRDX Modify the Leaf Phenotypes Caused by Altered KNOXI and CK Activity.
Mature leaves of the indicated genotypes. All transgenes were expressed using the transactivation system. WT, the wild type. Bars = 5 cm.
[See online article for color version of this figure.]
DISCUSSION
This study shows that some of the tomato AP1/FUL genes promote indeterminate growth in tomato leaves and are negatively regulated by LA-like proteins.
Role of AP1/FUL in Tomato Leaf Development
Our results identify a role for AP1/FUL transcription factors during vegetative development, in modulating the extent of the indeterminate growth of developing tomato leaves. Overexpression of several MADS box genes affected leaf shape and fate in Arabidopsis (Goodrich et al., 1997; Honma and Goto, 2001). AP1/FUL genes in core eudicots have been divided into three distinct lineages, euFUL, euAP1, and AGL79, that evolved following two duplication events (Litt and Irish, 2003; Shan et al., 2007). Whereas AP1-like genes are expressed and function mainly during reproductive development, FUL is also expressed in leaves. ful mutants have wider cauline leaves (Gu et al., 1998; Teper-Bamnolker and Samach, 2005), and ful soc double mutants show variable phenotypes related to indeterminate growth, including reversion of inflorescence meristems to vegetative growth (Melzer et al., 2008). The role of the AGL79-like clade is less characterized, but recently the pea AGL79-like gene VEG1 was shown to be involved in secondary inflorescence meristem identity (Berbel et al., 2012). Basal eudicots contain only one clade of FUL-like genes, most closely related to the FUL lineage of core eudicots. These FUL-like genes are expressed more broadly, similar to FUL-like genes of core eudicots. Interestingly, all three classes of AP1/FUL genes from tomato are expressed in leaves (Hileman et al., 2006; Shalit et al., 2009; this study), suggesting that leaf expression of AP1-like and AGL79-like genes has either been maintained or reacquired in tomato and possibly other species with compound leaves. Recently, FUL-like genes have been shown to be expressed in leaves and involved in compound-leaf morphogenesis in A. coerulea but not in other basal eudicot species (Pabón-Mora et al., 2013). Pabón-Mora et al. suggested that this unique role of FUL-like gene may have been acquired specifically in the lineage leading to A. coerulea. The role shown here for the ALG79-like gene MBP20 and likely additional AP1/FUL genes in tomato compound-leaf development could be explained by two alternative scenarios: (1) The role in compound-leaf development has evolved independently at least twice. (2) This role evolved early but has been lost or is redundant with other genes in most of the species, which may be supported by the leaf expression of FUL-like genes in basal eudicots and the FUL clade in core eudicots. Analysis of the role of these genes in additional species with compound leaves may help distinguish between these possibilities.
MADS box transcription factors have been implicated in the regulation of growth and differentiation in Arabidopsis floral organs, being expressed both during the initiation of floral organs and at later developmental stages (Dornelas et al., 2011; Johnson and Lenhard, 2011). The effect of MC on sepal size in tomato is in agreement with this proposed role. Floral organs are leaf derivatives, and many common factors are involved in the growth and development of these organs (Johnson and Lenhard, 2011), including class II TCPs and MADS box genes (Wellmer et al., 2006; Nag et al., 2009). Therefore, AP1/FUL genes may have evolved diverse functions in the regulation of leaf and flower organ growth, which differ among species and organs.
Interestingly, to date, AP1/FUL genes have been found to promote determinate growth and to be upregulated as the plant matured. These results suggest that during tomato leaf development they act to promote indeterminate growth and are upregulated transiently during early leaf development. This suggests that this gene family has adopted distinct roles in specific developmental contexts. Tomato AP1/FUL proteins show both conserved and unique protein–protein interaction profiles compared with Arabidopsis (Leseberg et al., 2008). These unique interactions may be involved in the species- and process-specific roles of these proteins.
AP1/FUL Genes Mediate LA-Like Activity during Leaf Development
Our results identify MBP20 and TM4 as targets of LA-like proteins. Whereas MBP20 was dramatically upregulated in FILpro>>miR319 plants relative to the wild type, leaf overexpression of MBP20 in an otherwise wild-type background only slightly affected leaf shape. This suggests that its activity is not a limiting factor in the wild-type leaf and that the indeterminate growth phenotype of FILpro>>miR319 results from altered expression of additional genes. These may include other AP1/FUL genes that were also upregulated in FILpro>>miR319, as well as additional pathways. Recently, LA was shown to act in part by positive regulation of GA homeostasis (Yanai et al., 2011). In Arabidopsis, several genes that are positively regulated by miR319-sensitive CIN-TCPs have been identified (Schommer et al., 2008; Koyama et al., 2010; Danisman et al., 2012; Rubio-Somoza and Weigel, 2013). Interestingly, the class I TCP TCP15 binds to the regulatory sequences of a partially overlapping set of genes (Uberti-Manassero et al., 2012). This suggests that class I and class II TCPs may have common targets, which is also supported by the finding that they bind similar DNA sequences (Kosugi and Ohashi, 2002; Danisman et al., 2012; Viola et al., 2012). It has been previously suggested that class I and class II TCPs affect growth in an antagonistic manner (Li et al., 2005; Hervé et al., 2009; Koyama et al., 2010; Martín-Trillo and Cubas, 2010). Class I TCPs were recently shown to promote CK responses in both Arabidopsis and tomato (Steiner et al., 2012a, 2012b), and Arabidopsis CIN-TCP were shown to dampen CK response by activating ARR16 via an interaction with the chromatin-modifying complex BRM (Efroni et al., 2013). Moreover, BRM was recently shown to affect GA biosynthesis and response (Archacki et al., 2013). As CK promotes and GA restricts the window of indeterminate growth in the tomato leaf margin, TCPs possibly regulate the extent of this window by modulating the balance between CK and GA, and class I and class II TCPs may affect this balance antagonistically. Therefore, LA-like proteins likely promote leaf maturation via the regulation of an array of genes, such that the alteration of the activity of only one of these targets is not expected to mimic the FILpro>>miR319 phenotype.
TCP genes are involved in floral organ development, and class II TCPs and FUL show opposite expression dynamics during flower development (Wellmer et al., 2006; Nag et al., 2009; Rubio-Somoza and Weigel, 2013). Therefore, the interaction between TCPs and MADS box genes may be conserved in leaves and floral organs. TCPs were recently identified as potential SEP3 and AP1 targets in Arabidopsis (Kaufmann et al., 2009) Thus, TCPs and MADS box transcription factors may be involved in a regulatory feedback loop, as has been suggested for additional AP1 regulators (Kaufmann et al., 2009, 2010).
The finding that coexpression of MBP20-SRDX modifies both the Tkn2 and miR319 overexpression phenotypes suggests that MBP20, or the pathway it regulates, may be an antagonistic target of LA-like and KNOXI proteins. As KNOXI genes have been shown to affect the balance between GA and CK (Hay et al., 2002, 2004; Jasinski et al., 2005; Yanai et al., 2005; Shani et al., 2010), this balance is a possible additional common target of KNOXI and LA-like proteins. Alternatively, the suppression of the Tkn2 overexpression phenotype by MBP20-SRDX could result from a requirement for both increased KNOXI activity and decreased LA-like activity to promote organogenesis.
MBP20 and TM4 are shown here to be repressed by LA. Other genes have been reported to be activated by CIN-TCPs (Schommer et al., 2008; Koyama et al., 2010; Martín-Trillo and Cubas, 2010; Danisman et al., 2012). Thus, it appears that CIN-TCPs can act as either activators or repressors, depending on the species, the specific target, and the developmental contexts.
Independent Programs Regulate Leaf and Plant Determination
Downregulation of miR319-sensitive CIN-TCPs results in indeterminate leaf growth (Ori et al., 2007; Shleizer-Burko et al., 2011) and in reduced numbers of leaves produced before flowering (Figure 2). These results indicate that in tomato, organ and plant determination are regulated via independent pathways that can be separated. Interestingly, in Arabidopsis, TCP proteins promote determination of both the leaf and the plant. Downregulation of Arabidopsis CIN-TCP activity caused late flowering (Palatnik et al., 2003; Schommer et al., 2008; Koyama et al., 2010), and expression of an activated form of TCP4 from its endogenous promoter induced early flowering (Sarvepalli and Nath, 2011). Therefore, while the effect of TCPs on leaf maturation is conserved between these species, their effect on flowering varies between tomato and Arabidopsis.
Unlike TCPs, the ratio between the activities of SFT and SP similarly affects all aspects of determinate growth in tomato (Lifschitz et al., 2006; Shalit et al., 2009; Efroni et al., 2010). High SFT/SP ratio promotes flowering and accelerates leaf maturation, leading to simpler leaves. In this research, we show that overexpression of miR319 is epistatic to both reduced and increased SFT activity with respect to leaf shape, but SFT is epistatic to miR319 overexpression with respect to flowering time. This further supports the notion that LA-like proteins influence leaf and plant maturation via separate pathways. The effect on leaf maturation is either downstream of or partly independent of SFT, while the effect on flowering requires intact SFT.
METHODS
Plant Materials and Genetics
Tomato (Solanum lycopersicum cv M82, sp) plants were grown as described previously (Shleizer-Burko et al., 2011). Except for the 35S:SFT plants (Lifschitz et al., 2006), all the described transgenic plants were produced using the LhG4 transactivation system (Moore et al., 1998; Shani et al., 2009). The following tomato driver and responder lines have been previously described: FILpro:LhG4, BLSpro:LhG4, OP:NLS-mRFP (Shalit et al., 2009; Shani et al., 2009), OP:GUS (Lifschitz et al., 2006; Efroni et al., 2008), OP:miR319, OP:LAm (Ori et al., 2007), LApro:LhG4 (Shleizer-Burko et al., 2011), OP:Tkn2, OP:Tkn2-SRDX (Shani et al., 2009), and OP:CKX (Shani et al., 2010). 35Spro:LhG4 is a gift from Yuval Eshed (Weizmann Institute). The tomato lines OP:MBP20, OP:MBP20-SRDX, OP:LAm-GFP, and MBP20pro:LhG4 were generated during this research as described below.
Plasmids and cDNA Clones
To generate OP:MBP20-SRDX, assembly PCR was used to introduce the 36-nucleotide-long SRDX motif CTCGATCTGGATCTAGAACTCCGTTTGGGTTTCGCT + TAA stop codon (Hiratsu et al., 2003) into the C terminus of the MBP20 protein. MBP20-SRDX, LAm-GFP, and MBP20 were cloned downstream to an OP array (Moore et al., 1998) and subsequently cloned into the pART27 binary vector (Gleave, 1992). The MBP20 promoter (3209 bp) was amplified from the BAC clone CO2Le0092m23 (Tomato Functional Genomics Database) and cloned into pART27, upstream to the LhG4 array.
For the yeast one-hybrid assay, the MBP20 promoter fragment IV (504 bp), DNA Bait, was cloned into the pLacZi plasmid and introduced into the yeast strain YM-4271. LA, protein prey, was cloned into the pDEST22 plasmid to produce translational fusions with the GAL4 activation domain (GAL4-AD) and introduced into the yeast strain YU-187 (Pruneda-Paz et al., 2009). MBP20, MBP20-IV, MBP20m-IV, and LA were cloned using the gateway homologous recombination system (Hartley et al., 2000) starting with the pENTR/D-TOPO cloning kit (Invitrogen). MBP20m-IV was generated by a two-step PCR using the following primer combinations: 1, pMBP20-IV-f + pMBP20m-IV-r and pMBP20-IV-r + pMBP20m-IV-f; 2, pMBP20-IV-f + pMBP20-IV-r. Primers used for cloning are listed in Supplemental Table 1 online.
Analysis of Transgenic Lines
Phenotypic analyses were performed with progeny of crosses between a driver (promoter:LhG4) and a responder (OP:GENE) line. For the initial characterization of the transgenic lines, at least seven independent driver or responder lines were crossed to OP:NLS-mRFP or FILpro:LhG4, and a representative line was selected for further analysis. Leaflet number was counted on fully expanded leaves from four to 10 different plants.
Tomato and Yeast Transformation
Tomato cotyledon transformation was performed according to McCormick (1991). Yeast was transformed by the lithium-acetate method (Daniel Gietz et al., 2002). When pLacZi plasmids were used, they were linearized at the ApaI site prior to transformation.
β-Galactosidase Activity Assay
β-Galactosidase activity was determined as described (Pruneda-Paz et al., 2009). Briefly, transformed yeast cells were grown for 24 h at 30°C in 400 μL SD medium lacking Trp and uracil. Then, 100 μL of the culture was combined with 400 μL YPD and grown for 6.5 h at 30°C. A total of 150 μL of this culture was used to determine the OD600, and 300 μL was centrifuged and washed with Z buffer (10 mM KCl and 1 mM MgSO4 in phosphate buffer, pH 7.0), resuspended in 30 μL of the same buffer, and lysed by four freeze/thaw cycles. Enzymatic reaction was initiated by addition of 170 μL Z buffer + BME (0.27 mL/100 mL), with 120 μg 2-nitrophenyl-β-d-galactopyranoside (Sigma-Aldrich), followed by a 19 h incubation at 30°C. The enzymatic reaction was stopped by addition of 80 μL 1 M Na2CO3, followed by centrifugation. A total of 150 μL of the supernatant was used to determine the OD420 and to calculate the β-galactosidase activities.
Phylogenetic Analysis
Amino acid sequences were aligned using a progressive alignment algorithm (see Supplemental Data Set 1 online). A maximum likelihood tree was constructed using a distance-based method and a neighbor-joining algorithm with 1000 bootstrap replicates, using the CLC Main Workbench 5.6.1 program (www.clcbio.com).
Tissue Collection, RNA Analysis, and Statistical Analysis
P1 designates the youngest leaf primordium; it becomes P2 when a new primordium initiates, etc. For leaf primordia at the P1 to P3 stages, the leaf at the respective developmental stage was collected with younger leaf primordia and the SAM. At least three biological repeats, each consisting of at least five plants, were used for RNA expression analysis. RNA was extracted using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions, except that samples were incubated for 30 min at room temperature after addition of the lysis buffer. cDNA synthesis was performed using the Verso cDNA kit (Thermo Scientific) with 1 μg RNA. Quantitative PCR and qRT-PCR analysis was performed using a Corbett Rotor-Gene 6000 real-time PCR machine, with TaqMan probes (PrimerDesign) and Premix Ex Taq (TaKaRa) for MBP20 and LA or SYBR Premix Ex Taq II (TaKaRa) for all other genes. Levels of mRNA were calculated relative to the EXPRESSED (EXP) gene as an internal control as follows: In each biological repeat, the levels of the analyzed gene (GENE) and EXP were separately calculated relative to a standard curve obtained by a dilution series of a reference sample. GENE/EXP ratio was calculated for each sample, and the average expression values of all repeats are presented as “relative expression.” Primers are listed in Supplemental Table 1 online.
Microarray Analysis
Microarray expression analysis was performed with total RNA from 12-d-old apices that contained the SAM and the two youngest leaf primordia. Labeled RNA was hybridized to an Affymetrix GeneChip Tomato Genome Array (900738). Three biological repeats were analyzed for the wild-type and La-2/+ genotypes and two for the FILpro>>miR319 genotype. Affymetrix Microarray Suite version 5.0 was used to simultaneously normalize data from all groups (Hubbell et al., 2002). One-way analysis of variance was applied to compare the three genotypes (P < 0.05). Each pair of genotypes was compared based on t tests and false discovery rates to identify differentially expressed genes.
Imaging, Microscopy, and GUS Staining
Fluorescence imaging was performed using a SMZ1500 fluorescence stereomicroscope (Nikon) equipped with a Nuance camera (CRi) as described previously (Shleizer-Burko et al., 2011). Scanning electron microscopy was performed using a JEOL 5410 LV microscope as described previously (Brand et al., 2007). β-Glucuronidase staining was performed as described previously (Ori et al., 2000).
Expression and Purification of the MBP-Tagged LA Protein
The LA coding region was amplified from cDNA (primers listed in Supplemental Table 1 online) and cloned into the pMal plasmid (New England Biolabs) at the XhoI and EcoRI sites to generate a fusion with MBP (MBP-LA) and transformed into the Escherichia coli strain Rosetta (EMD-Novagen). Sixteen milliliters of overnight-grown culture starter was introduced into 1600 mL of fresh Luria-Bertani and grown at 37°C until ∼0.6 OD600. Recombinant protein expression was induced with 0.4 mM isopropyl β-d-1 thiogalactoside. Cells were harvested by centrifugation and incubated for 30 min on ice with 160 mL lysis buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 0.2 mg/mL Lysozim), and then disrupted by three freeze/thaw cycles in liquid nitrogen and 37°C, respectively, and by sonication. The lysate was centrifuged, and the protein purified from the supernatant using amylose/agarose beads (NEB E8021S). Elution of bound protein was performed with 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 10 mM maltose.
EMSA
DNA probe was generated by end labeling of a 60-base single-stranded oligonucleotide using the DNA 3′ End Biotinylation Kit (Pierce 89818) and hybridization to complementary synthetic oligonucleotides (see Supplemental Table 1 online). EMSAs were performed using the LightShift chemiluminescent EMSA kit (Pierce 20148). Briefly, 10 μL of purified recombinant MBP-LA fusion protein was incubated at room temperature in 1× binding buffer, 50 ng/μL poly(dI/dC), 2.5% glycerol, 0.05% Nonidet P‑40, 50 fmol biotin-labeled probe, and 3.75 μg BSA for 30 to 40 min. For the Supershift, after 30 to 40 min of incubation, 3 μg of antibody against MBP was added and the reaction incubated for an additional 20 to 30 min. The samples were resolved on 6% nondenaturing polyacrylamide gels, electrotransferred onto 0.45 μm Biodyne B nylon membrane (Pierce 7701), and cross-linked to the membrane. The migration of the biotin-labeled probe was detected on x-ray film (5-h exposure) using streptavidin–horseradish peroxidase conjugates and chemiluminescent substrate according to the manufacturer’s protocol.
ChIP
ChIP was performed on dissected shoots (containing the SAM and young leaves) of 21-d-old plants grown on nitch medium as described (Ricardi et al., 2010). Briefly, 1.5 g plant tissue was cross-linked by 30 min of vacuum infiltration in 1% formaldehyde. Cross-linking was stopped with Gly and vacuum infiltration for an additional 5 min. Nuclei were isolated and chromatin was fragmented by sonication, calibrated to reach an average fragment size of 0.4 kb. ChIP reactions were performed using anti-GFP antibody (Abcam ab290) prebound to protein A-agarose beads (Santa Cruz Biotechnology sc-2001) and salmon sperm (Sigma-Aldrich B1626), followed by DNA elution, cross-linking reversal with Proteinase K (Roche 03115879), and DNA recovery. Enrichment for LA-bound sequences was assayed by PCR or quantitative PCR on the immunoprecipitated DNA. Quantitative PCR enrichment was calculated by normalizing to TUBULIN and to the total input of each sample. Two technical repeats and three biological replicates per genotype were analyzed for the enrichment of MBP20-IV and three biological replicates per genotype were analyzed for the enrichment of TM4-II.
Accession Numbers
Sequence data for genes used in this study can be found in the Arabidopsis Genome Initiative, Sol Genomics Network, or the National Center for Biotechnology Information under the following accession numbers: MBP20/AGL79 (Solyc02g089210, BT013126.1), TM4/TDR4/FULL/FUL1 (Solyc06g069430, AY098732.1), MBP7/FUL/FUL2 (Solyc03g114830, AY306156.1), MBP10/AGL79L (Solyc02g065730), MC/AP1 (Solyc05g012020, AF448521.1), LA (Solyc07g062680, EF091571), Tkn2 (Solyc02g081120, U76407.1), SFT (Solyc03g063100, AY186735.1), Tubulin (Solyc04g077020), EXP (Solyc07g025390), FUL (AT5G60910), AP1 (AT1G69120), CAL (AT1G26310), and AGL79 (AT3G30260).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of AP1/FUL Genes Is Negatively Correlated with LA Expression.
Supplemental Figure 2. Sequence Comparison and Dynamic Expression of AP1/FUL Genes.
Supplemental Figure 3. miR319 Affects Leaf Maturation and Flowering Time via Separate Pathways.
Supplemental Figure 4. Overexpression of MBP20-SRDX Leads to Reduced Formation of Secondary and Intercalary Leaflets.
Supplemental Figure 5. LA Binds the MBP20 and TM4 Promoters.
Supplemental Figure 6. Genetic Interaction of MBP20 with LA and Tkn2-SRDX.
Supplemental Table 1. Primers Used in This Work.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 1C.
Acknowledgments
We thank Eilon Shani (University of California, San Diego) and Moran Farhi (Hebrew University) for help with the yeast one-hybrid assay, Yuval Eshed (Weizmann Institute) for plant material, Tsafi Danieli (Protein Expression Facility Wolfson Centre, Hebrew University) for advice and vectors, and Gideon Rechavi (Chaim Sheba Medical Center) for help with the microarray. We also thank Yuval Eshed, Alon Samach (Hebrew University), David Weiss (Hebrew University), and members of our lab for helpful discussions and Smadar Harpaz-Saad (Hebrew University) and Eilon Shani (University of California, San Diego) for critical reading of the article. This work was supported by grants from the Israel Science Foundation (60/10), the U.S.–Israel Binational Agricultural Research and Development Fund (IS 04140-08C and IS-4531-12C), and German-Israel Project Cooperation Foundation (OR309/1-1; FE552/12-1).
AUTHOR CONTRIBUTIONS
Y.B., S.S.-B., O.Y., L.-E.W., and N.O. designed the research. Y.B., S.S.-B., I.S., I.D.Z., and O.Y. performed research and analyzed the data. O.Y., J.J.-H., and I.K. performed microarray and did the statistical analysis. Y.B., S.S.-B., and N.O. wrote the article.
Glossary
- SAM
shoot apical meristem
- MB
marginal blastozone
- CK
cytokinin
- qRT-PCR
quantitative RT-PCR
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- GFP
green fluorescent protein
- GA
gibberellin
References
- Aggarwal P., Das Gupta M., Joseph A.P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R., et al. (2013). BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS ONE 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S., Geier T., Eimert K., Watillon B., Sangwan R.S., Gleissberg S. (2009). KNOX overexpression in transgenic Kohleria (Gesneriaceae) prolongs the activity of proximal leaf blastozones and drastically alters segment fate. Planta 230: 1081–1091 [DOI] [PubMed] [Google Scholar]
- Becker A., Theissen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Bemer M., Karlova R., Ballester A.R., Tikunov Y.M., Bovy A.G., Wolters-Arts M., Rossetto Pde.B., Angenent G.C., de Maagd R.A. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R., d’Erfurth I., Ferrandiz C., Cosson V., Beltrán J.P., Cañas L.A., Kondorosi A., Madueño F., Ratet P. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel, A., Ferrándiz, C., Hecht, V., Dalmais, M., Lund, O.S., Sussmilch, F.C., Taylor, S.A., Bendahmane, A., Ellis, T.H.N., Beltrán, J.P., Weller, J.L., and Madueño, F. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3: 797. [DOI] [PubMed]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860 [DOI] [PubMed] [Google Scholar]
- Blein T., Hasson A., Laufs P. (2010). Leaf development: What it needs to be complex. Curr. Opin. Plant Biol. 13: 75–82 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Alvarez J., Weigel D., Meyerowitz E.M., Smyth D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Brand A., Shirding N., Shleizer S., Ori N. (2007). Meristem maintenance and compound-leaf patterning utilize common genetic mechanisms in tomato. Planta 226: 941–951 [DOI] [PubMed] [Google Scholar]
- Burko, Y., and Ori, N. (2013). The tomato leaf as a model system for organogenesis. In The Plant Organogenesis, I. de Smet, ed (New York, USA: Humana Press), pp. 1–19. [DOI] [PubMed] [Google Scholar]
- Busi M.V., Bustamante C., D’Angelo C., Hidalgo-Cuevas M., Boggio S.B., Valle E.M., Zabaleta E. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52: 801–815 [DOI] [PubMed] [Google Scholar]
- Butler L. (1952). The linkage map of the tomato. J. Hered. 43: 25–36 [Google Scholar]
- Chen J.J., Janssen B.J., Williams A., Sinha N. (1997). A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell 9: 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S., van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., van Dijk A.D.J., Muino J.M., Cutri L., Dornelas M.C., Angenent G.C., Immink R.G.H. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler N.G. (1984). Comparison of leaf development in Normal (+/+), Entire (E/E), and Lanceolate (La/+) plants of tomato, Lycopersicon esculentum Ailsa Craig. Bot. Gaz. 145: 66–77 [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: Current issues. Int. J. Plant Sci. 162: 459–464 [Google Scholar]
- Dornelas M.C., Patreze C.M., Angenent G.C., Immink R.G.H. (2011). MADS: The missing link between identity and growth? Trends Plant Sci. 16: 89–97 [DOI] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: A critical review. Plant Cell 22: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.-K., Kim H.J., Wu M.-F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul P., Angosto T., Garcia-Sogo B., Garcia-Hurtado N., Martin-Trillo M., Salinas M., Moreno V., Lozano R., Martinez-Zapater J. (2004). Expression of Arabidopsis APETALA1 in tomato reduces its vegetative cycle without affecting plant production. Mol. Breed. 13: 155–163 [Google Scholar]
- Fambrini M., Pugliesi C. (2013). Usual and unusual development of the dicot leaf: Involvement of transcription factors and hormones. Plant Cell Rep. 32: 899–922 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C., Gu Q., Martienssen R., Yanofsky M.F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Floyd S.K., Bowman J.L. (2010). Gene expression patterns in seed plant shoot meristems and leaves: Homoplasy or homology? J. Plant Res. 123: 43–55 [DOI] [PubMed] [Google Scholar]
- Frugis G., Giannino D., Mele G., Nicolodi C., Chiappetta A., Bitonti M.B., Innocenti A.M., Dewitte W., Van Onckelen H., Mariotti D. (2001). Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/singlestranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Gu Q., Ferrándiz C., Yanofsky M.F., Martienssen R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hagemann W., Gleissberg S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199: 121–152 [Google Scholar]
- Hake S., Smith H.M.S., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hartley J.L., Temple G.F., Brasch M.A. (2000). DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Craft J., Tsiantis M. (2004). Plant hormones and homeoboxes: Bridging the gap? Bioessays 26: 395–404 [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2009). A KNOX family TALE. Curr. Opin. Plant Biol. 12: 593–598 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: Versatile regulators of plant development and diversity. Development 137: 3153–3165 [DOI] [PubMed] [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M.C., Ramboer A., Lacout F., Tremousaygue D. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L.C., Sundstrom J.F., Litt A., Chen M.Q., Shumba T., Irish V.F. (2006). Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23: 2245–2258 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hubbell E., Liu W.M., Mei R. (2002). Robust estimators for expression analysis. Bioinformatics 18: 1585–1592 [DOI] [PubMed] [Google Scholar]
- Huijser P., Klein J., Lönnig W.E., Meijer H., Saedler H., Sommer H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink R.G., Hannapel D.J., Ferrario S., Busscher M., Franken J., Lookeren Campagne M.M., Angenent G.C. (1999). A petunia MADS box gene involved in the transition from vegetative to reproductive development. Development 126: 5117–5126 [DOI] [PubMed] [Google Scholar]
- Irish V.F., Sussex I.M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B.J., Lund L., Sinha N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Johnson K., Lenhard M. (2011). Genetic control of plant organ growth. New Phytol. 191: 319–333 [DOI] [PubMed] [Google Scholar]
- Kaplan D.R. (2001). Fundamental concepts of leaf morphology and morphogenesis: A contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162: 465–474 [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Yasuno N., Sato Y., Yoda M., Yamazaki R., Kimizu M., Yoshida H., Nagamura Y., Kyozuka J. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg C.H., Eissler C.L., Wang X., Johns M.A., Duvall M.R., Mao L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59: 2253–2265 [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.J., Herskowitz I. (1993). Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262: 1870–1874 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57: 3405–3414 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A., Irish V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M.A., Gustafson-Brown C., Savidge B., Yanofsky M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Mathan D.S., Jenkins J.A. (1962). A morphogenetic study of Lanceolate, a leaf shape mutant in the tomato. Am. J. Bot. 49: 504–514 [Google Scholar]
- McCormick, S. (1991). Transformation of tomato with Agrobacterium tumifaciens. In Plant Tissue Culture Manual, K. Lindsey, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–9.
- Melzer, R., Kaufmann, K., and TheissŸen, G. (2006). Missing links: DNA-binding and target gene specificity of floral homeotic proteins. In Advances in Botanical Research, D.E. Soltis, J.H. LeebensMack, P.S. Soltis, and J.A. Callow, eds (Massachusetts,USA: Academic Press), pp. 209–236.
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N., Latorre A., Jamilena M., Lozano R. (2004). SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218: 427–434 [DOI] [PubMed] [Google Scholar]
- Moore I., Gälweiler L., Grosskopf D., Schell J., Palme K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K.J., He X., Fischer R., Prüfer D. (2006). Constitutive knox1 gene expression in dandelion (Taraxacum officinale, Web.) changes leaf morphology from simple to compound. Planta 224: 1023–1027 [DOI] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Kimbara J., Fujisawa M., Kitagawa M., Ihashi N., Maeda H., Kasumi T., Ito Y. (2012). MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Ng M., Yanofsky M.F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2: 186–195 [DOI] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Ori N., Eshed Y., Chuck G., Bowman J.L., Hake S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532 [DOI] [PubMed] [Google Scholar]
- Pabón-Mora N., Ambrose B.A., Litt A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158: 1685–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Mora N., Sharma B., Holappa L.D., Kramer E.M., Litt A. (2013). The Aquilegia FRUITFULL-like genes play key roles in leaf morphogenesis and inflorescence development. Plant J. 74: 197–212 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Schatz M.C., Lippman Z.B. (2012). Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnis A., Cohen O., Gutfinger T., Hareven D., Zamir D., Lifschitz E. (1997). The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9: 2143–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L., Abu-Abeid M., Zamir D., Nacken W., Schwarz-Sommer Z., Lifschitz E. (1991). The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1: 255–266 [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardi M.M., González R.M., Iusem N.D. (2010). Protocol: Fine-tuning of a chromatin immunoprecipitation (ChIP) protocol in tomato. Plant Methods 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S.D., Ditta G.S., Yanofsky M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2013). Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruokolainen S., Ng Y.P., Broholm S.K., Albert V.A., Elomaa P., Teeri T.H. (2010). Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol. 10: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Lotan H. (2007). The transition to flowering in tomato. Plant Biotechnol. 24: 71–82 [Google Scholar]
- Sarvepalli K., Nath U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H., Zhang N., Liu C., Xu G., Zhang J., Chen Z., Kong H. (2007). Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol. Phylogenet. Evol. 44: 26–41 [DOI] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., Sharon E., Ori N. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G.H., Angenent G.C., Kaufmann K. (2012a). Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012b). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.-S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012b). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Yanai O., Efroni I., Ori N., Eshed Y., Weiss D. (2012a). Class I TCPs modulate cytokinin-induced branching and meristematic activity in tomato. Plant Signal. Behav. 7: 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler R.F. (1964). Dosage effects of the Lanceolate gene in tomato. Am. J. Bot. 51: 253–264 [Google Scholar]
- Teper-Bamnolker P., Samach A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Becker A., Di Rosa A., Kanno A., Kim J.T., Münster T., Winter K.U., Saedler H. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42: 115–149 [PubMed] [Google Scholar]
- Theissen G., Kim J.T., Saedler H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43: 484–516 [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Hay, A., Ori, N., Kaur, H., Henderson, I., Holtan, H., McCormick, S., and Hake, S. (2002). Developmental signals regulating leaf form. In Developmental Genetics and Plant Evolution, Q.C.B. Cronk, R.M. Batemen, and J.A. Hawkins, eds (London, UK: Taylor & Francis), pp. 418–430. [Google Scholar]
- Uberti-Manassero N.G., Lucero L.E., Viola I.L., Vegetti A.C., Gonzalez D.H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Viola I.L., Reinheimer R., Ripoll R., Manassero N.G., Gonzalez D.H. (2012). Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 287: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wellmer F., Alves-Ferreira M., Dubois A., Riechmann J.L., Meyerowitz E.M. (2006). Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12: 527–538 [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68: 571–582 [DOI] [PubMed] [Google Scholar]