To understand the mechanisms of protein trafficking, it is essential to characterize the cellular machinery that performs the transport. Genetic analyses identified two genes required for vacuolar trafficking in Arabidopsis, MTV1 and AGD5. These genes appear to participate in the formation of clathrin-coated vesicles for the transport of vacuolar proteins from the trans-Golgi network.
Abstract
Many soluble proteins transit through the trans-Golgi network (TGN) and the prevacuolar compartment (PVC) en route to the vacuole, but our mechanistic understanding of this vectorial trafficking step in plants is limited. In particular, it is unknown whether clathrin-coated vesicles (CCVs) participate in this transport step. Through a screen for modified transport to the vacuole (mtv) mutants that secrete the vacuolar protein VAC2, we identified MTV1, which encodes an EPSIN N-TERMINAL HOMOLOGY protein, and MTV4, which encodes the ADP ribosylation factor GTPase-activating protein NEVERSHED/AGD5. MTV1 and NEV/AGD5 have overlapping expression patterns and interact genetically to transport vacuolar cargo and promote plant growth, but they have no apparent roles in protein secretion or endocytosis. MTV1 and NEV/AGD5 colocalize with clathrin at the TGN and are incorporated into CCVs. Importantly, mtv1 nev/agd5 double mutants show altered subcellular distribution of CCV cargo exported from the TGN. Moreover, MTV1 binds clathrin in vitro, and NEV/AGD5 associates in vivo with clathrin, directly linking these proteins to CCV formation. These results indicate that MTV1 and NEV/AGD5 are key effectors for CCV-mediated trafficking of vacuolar proteins from the TGN to the PVC in plants.
INTRODUCTION
Intracellular compartmentalization and multicellular development are two evolutionary innovations of pivotal importance for understanding the basic biology of many eukaryotic organisms, including all metazoans and land plants. An endomembrane system of near modern complexity may have been present in the last common eukaryotic ancestor (Dacks and Field, 2007). Indeed, most of the proteins responsible for trafficking are conserved throughout all eukaryotes, although they tend to be expanded in number in multicellular organisms (Dacks and Field, 2007; Sanderfoot, 2007). By contrast, multicellularity is not universal in eukaryotes and is thought to have evolved independently in plants and animals (Meyerowitz, 2002). However, the main model system for studying intracellular trafficking has been the unicellular yeast Saccharomyces cerevisiae, and so many of the unique features present in multicellular models remain relatively unexplored. In plants, the specific constrains imposed by multicellularity and their distinct developmental strategy have led to specific arrangements of the endomembrane system. A conspicuous peculiarity of the plant endomembrane system is the presence of large vacuoles, which in most plant cells occupy the majority of the cellular volume. The enlarged vacuoles of plants are necessary adaptations to autotrophy and immobility, as they provide an energy-efficient organ expansion mechanism for exploring the surroundings and a buffering organelle to maintain cellular homeostasis (Zouhar and Rojo, 2009). Plants may contain different types of vacuoles in a single cell, and they develop specialized vacuoles in certain cells, such as the protein storage vacuoles in seed tissues (Rojo and Denecke, 2008), which constitute the main source of proteins for human and livestock nutrition. Despite their importance, the molecular machinery responsible for the delivery of proteins and membrane to these distinct vacuoles is largely uncharacterized. Two different types of vesicles, clathrin-coated vesicles (CCVs) and dense vesicles, bud from the trans-Golgi network (TGN) and contain vacuolar cargo in plants (Otegui et al., 2006; Hinz et al., 2007; Kang et al., 2011), but their actual contribution to vacuolar trafficking remains unresolved. Moreover, a vesicle-independent compartmental maturation model for vectorial transport from the TGN to the prevacuolar compartment (PVC) has been recently proposed (Scheuring et al., 2011). To test the validity of these models and attain a mechanistic understanding of vacuolar transport in plants, it is essential to identify the trafficking machinery involved in the different steps of the pathway.
In S. cerevisiae, genetic analysis has been instrumental in revealing the role of many proteins in intracellular trafficking and in understanding the organization and function of the endomembrane system in this unicellular organism (Schekman and Novick, 2004). In comparison, our knowledge of the increased repertoire of trafficking components in multicellular eukaryotes is limited, in part because genetic dissection of intracellular trafficking is not as straightforward as in yeast. In plants, transient transport assays and, more recently, genetic analysis, have been used for studying vacuolar trafficking (Denecke et al., 2012). Through these combined efforts, several factors involved in vacuolar transport in plants have been characterized (Rojo and Denecke, 2008); in particular, the function of the vacuolar sorting receptors (VSRs) in this pathway is relatively well understood (De Marcos Lousa et al., 2012). However, the number of vacuolar trafficking components identified is still more than one order of magnitude lower than those known to function in this process in yeast (Bonangelino et al., 2002; Ricarte et al., 2011), and it is therefore evident that we still lack most information on the core functioning of the vacuolar pathway in plants. In particular, the machinery involved in anterograde trafficking from the TGN is uncharacterized, and it is unknown whether vesicle trafficking participates in this step.
Vesicular transport of proteins involves cargo recruitment to specific membrane domains followed by induction of vesicle budding and cargo loading into the incipient vesicles. VSRs are a plant-specific family of type I transmembrane proteins that recruit soluble vacuolar cargo through direct protein–protein interaction and have been shown genetically to be necessary for transport of the cargo to the vacuole (Shimada et al., 2003; Zouhar et al., 2010). VSRs cycle between the TGN and the PVC (Sanderfoot et al., 1998; Tse et al., 2004; Reichardt et al., 2007; Foresti et al., 2010; Saint-Jean et al., 2010), interact in vitro with µ-adaptins (Sanderfoot et al., 1998; Happel et al., 2004), and are found in CCV-enriched fractions (Hinz et al., 1999), indicating that they may mediate sorting and loading of soluble cargo into CCVs for their transport to the vacuole (De Marcos Lousa et al., 2012). However, since TGN factors coupling VSR cargo recruitment to CCV budding and cargo loading into the vesicles are unknown, this trafficking model remains speculative. It has been reported that Arabidopsis thaliana Epsin1 binds clathrin and VSR1 and has a role in trafficking of a chimeric vacuolar cargo (Song et al., 2006a); however, Epsin1 localization at the TGN or an in vivo role in VSR cycling and in trafficking of endogenous vacuolar proteins has not been documented. Epsin1 is one of 43 Arabidopsis EPSIN N-TERMINAL HOMOLOGY (ENTH) proteins that are characterized by a conserved phospholipid binding ENTH domain for insertion into membranes. ENTH proteins contain in many cases clathrin binding motifs (Legendre-Guillemin et al., 2004) that allow them to function as monomeric adaptors for clathrin coat recruitment to membranes (Horvath et al., 2007).
Another class of proteins that has been shown to bind clathrin in animal systems is the ADP ribosylation factor GTPase-activating protein (ARF GAP) family, whose members induce the hydrolysis of GTP bound to ARF and are essential factors to couple vesicle formation with cargo loading (Tanabe et al., 2005; Natsume et al., 2006; Spang et al., 2010; Bai et al., 2011). However, there are no prior reports of plant ARF GAPs binding to clathrin or having a role in vacuolar trafficking. Through a genetic screen, we identified the MODIFIED TRANSPORT TO THE VACUOLE1 (MTV1) and MTV4 Arabidopsis genes, which encode plant-specific members of the ENTH and ARF GAP protein families localized at the TGN and in CCVs. MTV1 and MTV4 bind clathrin and cooperatively participate in the transport of vacuolar cargo and VSRs, suggesting that they are key effectors coupling VSR-dependent cargo recruitment to cargo loading into CCVs for vectorial transport from the TGN to the PVC.
RESULTS
MTV1 and MTV4 Encode Plant-Specific Proteins with ENTH and ARF GAP Domains, Respectively
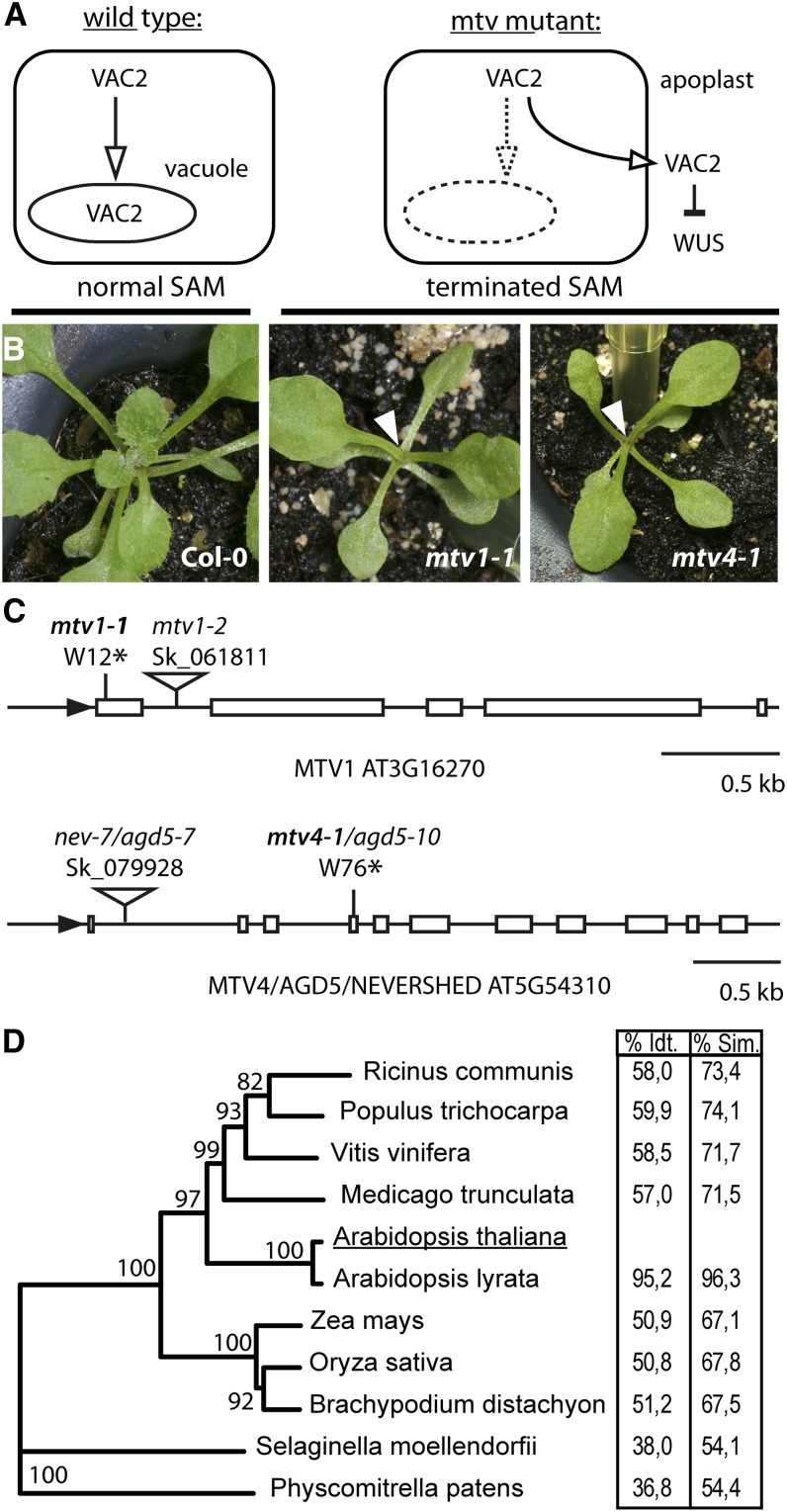
To identify components required for vacuolar trafficking of soluble cargo, we conducted a mutant screen, which has been previously described in more detail (Sanmartín et al., 2007). Briefly, a dodecapeptide derived from the CLAVATA3 (CLV3) protein is the extracellular ligand of the CLV receptor kinase complexes (Betsuyaku et al., 2011). Via this signaling pathway, CLV3 negatively regulates the activity of WUSCHEL, thus reducing the stem cell pool size in the shoot apical meristem. For the screen, CLV3 was fused to the vacuolar sorting signal of barley (Hordeum vulgare) lectin and expressed under the 35S promoter, creating a chimeric protein termed VAC2. VAC2 is recognized by the vacuolar sorting machinery and transported to the vacuole, which prevents its secretion into the extracellular space and effectively renders it inactive (Rojo et al., 2002). Mutations that disrupt vacuolar trafficking may result in VAC2 export into the extracellular space via the default secretion pathway. There, VAC2 activates the CLAVATA pathway and leads to meristem consumption (Rojo et al., 2002; Sanmartín et al. 2007). We used this highly sensitive assay to screen for mtv mutants that secrete VAC2 into the extracellular space, resulting in premature termination of the shoot apical meristem (Figure 1A).
Figure 1.
Isolation of mtv1 and mtv4 in a Screen for Vacuolar Trafficking Mutants.
(A) Schematic outline of the genetic screen.
(B) Normal shoot apical meristem in Col-0 and terminated meristem (arrowheads) in mtv1-1 and mtv4-1 mutants in the VAC2 background.
(C) Intron/exon structure and localization of point mutations and T-DNA insertions in MTV1 and NEV/AGD5/MTV4.
(D) Phylogenetic tree of MTV1 putative orthologs. Table shows percentage of amino acid identity (Idt.) and similarity (Sim.).
[See online article for color version of this figure.]
We isolated two mtv mutants from an ethyl methanesulfonate (EMS)–mutagenized VAC2 population, termed mtv1-1 and mtv4-1. Both mutants exhibit determinate growth of shoot apical meristems (Figure 1B) and premature termination of floral meristems, which in most cases did not produce carpels. These meristem phenotypes were dependent on the presence of the VAC2 transgene, indicating that they are due to mis-secretion of VAC2 and not to a direct activity of MTV1 and MTV4 on meristems. We identified the mutant loci by a map-based cloning strategy. The mutation in mtv1-1 was mapped to a region in chromosome 3, containing 24 genes (At3g16180 to At3g16410), and by sequencing of candidate genes we discovered a nonsense mutation in the coding sequence of the At3g16270 locus. The mtv1-1 mutation introduces a stop codon after only 11 amino acids of the protein sequence and is thus predicted to be a null allele (Figure 1C). At3g16270 had not been functionally characterized yet and is annotated in the TAIR10 database as an ENTH domain containing a protein of unknown function.
MTV4 was mapped to a region on chromosome 5 containing 59 genes (At5g54160 to At5g54630). Sequencing of candidate genes revealed a nonsense mutation in the coding sequence of the At5g54310 locus, converting Trp-76 to a premature stop. At5g54310 has been previously identified as NEVERSHED/ARF-GAP5 (NEV/AGD5) (Liljegren et al., 2009). mtv4-1 plants displayed floral organ abscission defects characteristic of the nev mutant phenotype and thus represent a novel allele of nev/agd5, which we designated as agd5-10 (Figure 1C).
To verify that the genes identified were the causative loci for the terminated meristem phenotype in the VAC2 background, we crossed the VAC2 reporter line with T-DNA insertion mutants mtv1-2 (Salk_061811) and mtv4-2/nev-7/agd5-7 (Salk_079928), both of which harbor insertions in the first intron of the respective genes (Figure 1C). In the F2 of these crosses, the meristem termination phenotype was recapitulated in homozygous mutant plants containing the VAC2 transgene, confirming that the mutations in MTV1/At3g16270 and NEV/AGD5/At5g54310 indeed cause secretion of the VAC2 marker (see Supplemental Figure 1A online). Immunoblot analysis showed that both the original EMS alleles and the T-DNA alleles are full loss-of-function mutants of MTV1 and NEV/AGD5 (see Supplemental Figure 1B online). For all subsequent analyses described in this article, we used the respective T-DNA alleles, unless otherwise stated.
MTV1 has not been characterized previously. Analysis of the primary amino acid sequence indicated that the predicted MTV1 protein contains only one identifiable conserved domain, an N-terminal (amino acids 1 to 134) ENTH/VHS domain (InterPro code IPR008942). Interestingly, an NMR spectrum of MTV1’s ENTH domain has been generated (López-Méndez et al., 2004), which structurally confirms its classification. Although this domain is predicted to be encoded in 43 other Arabidopsis proteins, WUBLAST analysis showed that full-length MTV1 did not have any significant homology to any other protein in the TAIR10 protein database. However, a search for orthologs in other plant species indicated that MTV1 is conserved throughout the angiosperms. We performed an analysis of protein identity/similarity to Arabidopsis MTV1 and found values between 95.2 and 50.9% identity in the analyzed members of the angiosperm clade (Figure 1D; see Supplemental Data Set 1 online). Notably, in Selaginella and Physcomitrella, we found orthologs with 36.8% identity and 54.1% similarity, suggesting that MTV1 may be conserved also outside of the spermatophytes. Furthermore, in most plant species, we could identify only one ortholog of MTV1, indicating that it is a well conserved and mostly single-copy gene. A phylogenetic analysis of orthologs of MTV1, Epsin1, Epsin2/2R, and Epsin3, clearly separates the genes into three distinct clades (see Supplemental Figure 2 and Supplemental Data Set 2 online). Taken together, MTV1 represents a novel and unique member of the group of ENTH domain containing proteins, which is potentially involved in vacuolar protein trafficking.
MTV4/NEV/AGD5 encodes a member of the ARF GAP protein family, which in Arabidopsis contains 15 members (Vernoud et al., 2003). Interestingly, in contrast with the other 14 Arabidopsis ARF GAPs, the homology of NEV/AGD5 with proteins outside the plant kingdom is restricted to the ARF GAP domain (amino acids 16 to 130), whereas in plant species, there are proteins sharing identity throughout the whole sequence with NEV/AGD5 (amino acids 1 to 483), suggesting that these latter form a clade of plant-specific ARF GAPs (Liljegren et al., 2009; this article).
MTV1 and NEV/AGD5 Exhibit Near-Identical Patterns of Expression and Interact Genetically
Previous functional analyses showed that NEV/AGD5 is required for correct abscission of floral organs, which is a specific developmental process restricted to a limited number of cells (Liljegren et al., 2009). However, our result demonstrating the secretion of VAC2 and the resulting shoot apical meristem termination in the nev/agd5 mutant points toward a more general role of NEV/AGD5 in trafficking events. Previous tissue-specific analyses of NEV/AGD5 expression revealed broad expression in flowers, leaves, stems, and roots (Liljegren et al., 2009).
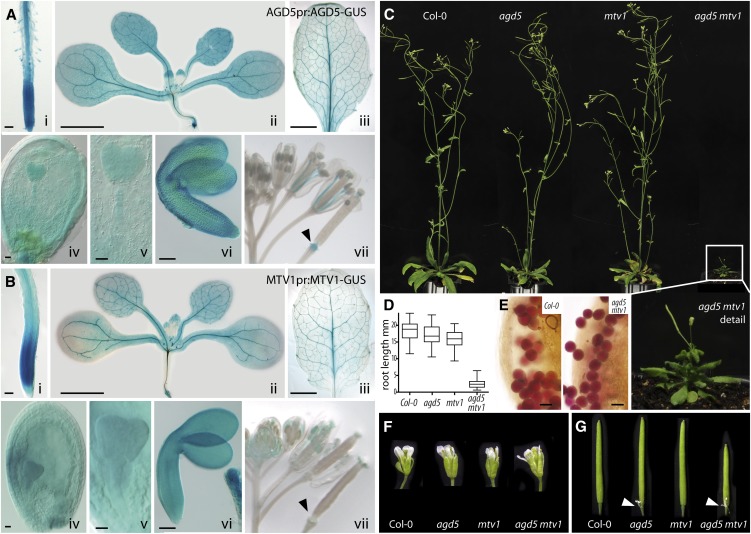
To further define the tissues and developmental stages in which NEV/AGD5 could play a role, we generated stable pAGD5:AGD5-GUS (for β-glucuronidase) transgenic lines using an additional upstream regulatory sequence and analyzed tissues of different developmental stages. We detected GUS signal in almost all tissues of the plant (Figure 2A). Strong signal was observed in the root meristem, root, shoot, and leaf vasculature with less expression in leaf ground tissue (Figure 2A, i to iii and vii). Importantly, we observed strong signal in developing and mature embryos (Figure 2A, iv-vi), which would be consistent with a role in the transport of storage proteins to protein storage vacuoles, which peaks during mid and late stages of embryogenesis (Santos Mendoza et al., 2008). Intriguingly, pMTV1:MTV1-GUS transgenic lines exhibit a nearly identical expression pattern (Figure 2B), although MTV1 expression was absent from the floral abscission zone (cf. Figures 2A, vii, and 2B, vii, arrowheads). Based on these expression data and the virtually identical meristem termination phenotype when in the VAC2 background, it appears possible that MTV1 and NEV/AGD5 act in a common trafficking pathway.
Figure 2.
Expression and Phenotypic Characterization of mtv1 and nev/agd5 Single and Double Mutants.
(A) Expression of NEV/AGD5 as revealed by the ProAGD5:AGD5-GUS transgene.
(B) Expression of MTV1 as revealed by the ProMTV1:MTV1-GUS transgene. Lowercase roman numerals indicate tissue: (i) primary seedling root, (ii) 11 DAG plant, (iii) expanded leaf of soil-grown plant, (iv) immature seed at heart embryo stage, (v) heart stage embryo, (vi) mature embryo, (vii) inflorescence with flowers of different developmental stages; arrowhead marks abscission zone.
(C) Phenotype of 4-week-old wild-type, nev/agd5, mtv1, and nev/agd5 mtv1 double mutant plants. A magnified image of the boxed nev/agd5 mtv1 plant is shown below.
(D) Root length of 5 DAG seedlings, as a box plot; whiskers indicate population range; n > 40.
(E) Pollen viability is not altered in the mtv1 nev/agd5 double mutant as indicated by Alexander stain.
(F) Flower size is not altered in the mtv1 nev/agd5 double mutant.
(G) Mature siliques. Arrowheads mark nonabscised floral organs in nev/agd5 single and mtv1 nev/agd5 double mutants.
Bars = 100 µm in (i) and (vi), 2 cm in (ii) and (iii), 25 µm in (iv), (v), and (E).
To test the hypothesis that MTV1 and NEV/AGD5 have associated functions, we analyzed the functional consequences of combining the mtv1 and nev/agd5 mutations. After crossing the single mutants, we noted in the segregating F2 population the occurrence of severely dwarfed plants (Figures 2C and 2D). These were subsequently identified as mtv1 nev/agd5 double homozygotes by genotyping. Double mtv1 nev/agd5 mutants have reduced growth of both aerial and underground organs (Figures 2C and 2D). However, despite the drastic reduction in vegetative plant size, they have a relatively normal development and show no obvious patterning defects. Interestingly, flowers were the only organs of normal size in the double mutant (Figure 2F), which may be due to higher expression levels of genes with redundant functions (i.e., other ENTH or AGD proteins) in those organs. Pollen viability was not affected, as demonstrated by vital Alexander stain (Figure 2E), but the siliques of the double mutant were consistently shorter than those of the wild type or single mutants and exhibited the same abscission defect as observed in the nev/agd5 single mutants (Figure 2G, arrowheads; Liljegren et al., 2009). As expected, the reduced plant stature, smaller number of siliques, and reduced silique size drastically reduce total seed yield in the double mutants. The synergistic genetic interaction observed suggests a functional relationship (Pérez-Pérez et al., 2009) and that MTV1 and NEV/AGD5 converge at a trafficking node that is essential for proper organ and plant growth. This agrees with prior reports showing similar synergistic impairment of plant growth when combining mutations in homologous, redundant trafficking genes (Surpin et al., 2003; Zhang et al., 2007; Uemura et al., 2012) or mutations in nonhomologous trafficking genes that encode components of a protein complex (Ebine et al., 2008).
MTV1 and NEV/AGD5 Function Specifically in the Transport of Vacuolar Cargo
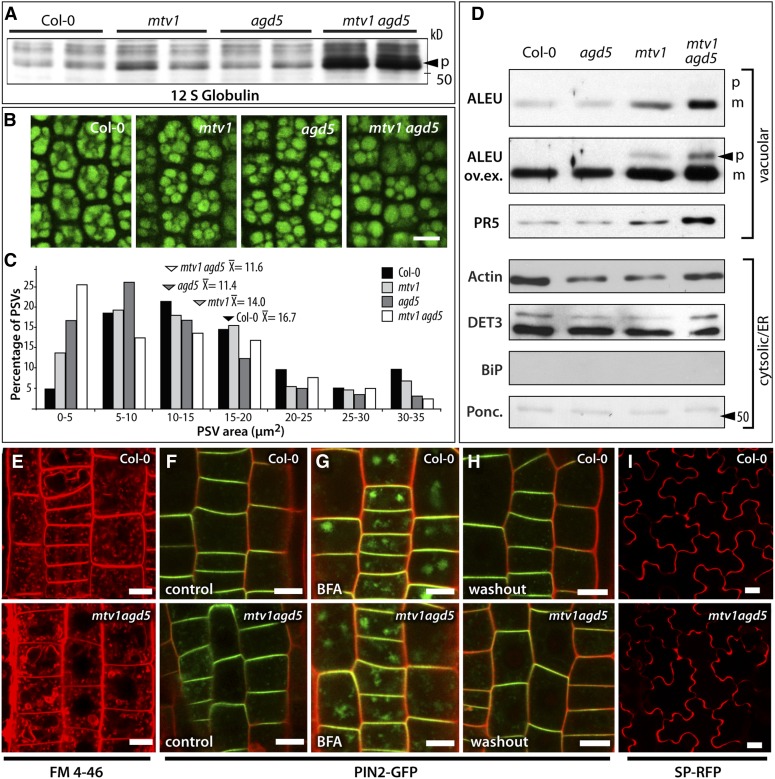
The finding that MTV1 and MTV4 encode ENTH and ARF GAP proteins suggests that VAC2 secretion in the respective mutants is due to a block in vesicular trafficking and implies that endogenous cargo using the same pathway should also be affected. 12S globulins and Aleurain are endogenous Arabidopsis vacuolar proteins that are secreted in other mutants affected in VAC2 trafficking, suggesting that they share a common trafficking pathway (Sanmartín et al., 2007; Zouhar et al., 2009, 2010). Importantly, the ubiquitous expression of MTV1 and NEV/AGD5 in seeds and in vegetative tissues is compatible with a role in 12S globulins and Aleurain trafficking. Processing of 12S globulins occurs between the PVC and the vacuole (Otegui et al., 2006), so the accumulation of precursor forms of larger size is indicative of defects in reaching these compartments (Shimada et al., 2003; Li et al., 2006). To test whether MTV1 and NEV/AGD5 are involved in 12S globulin transport, we compared the protein profile of wild-type and mutant seeds. In nev/agd5 and mtv1 single mutants, we detected a slight increase in the accumulation of ∼50-kD precursor forms of 12S globulins, which became pronounced in the double mtv1 nev/agd5 mutant (Figure 3A). Consistent with a reduced transport of these major storage proteins to the vacuole, microscopy analysis showed that both singles and the double mutant had smaller, more rounded PSVs with larger spaces in between (Figures 3B and 3C), indicative of defects in PSV filling during seed biogenesis. These results suggest that NEV/AGD5 and MTV1 cooperate in the trafficking of 12S globulins to the vacuole, likely at a stage prior to the transport of 12S globulin precursors from the TGN to the PVC.
Figure 3.
Mutations in mtv1 and nev/agd5 Specifically Affect Vacuolar Transport of Endogenous Proteins and Do Not Alter Endocytosis, Polar Targeting, Endosomal Recycling, or Secretion.
(A) Immunoblot analysis of seed protein extracts with 12S globulin antibody; arrowhead indicates the 52-kD precursor (p).
(B) Autofluorescence of protein storage vacuoles (PSVs) in hypocotyls of mature embryos from dry seeds.
(C) Size distribution of PSVs from each genotype is displayed in a bar graph (number of PSVs measured: n = 470 for Col-0, n = 566 for mtv1, n = 661 for agd5, and n = 660 for mtv1agd5). The mean PSV size (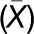 ) of the different genotypes is shown above the arrowheads. The PSVs were significantly smaller in mtv1 (
) of the different genotypes is shown above the arrowheads. The PSVs were significantly smaller in mtv1 (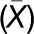 = 14.0 μm2, P = 4.4 × 10−5, unpaired t test), agd5 (
= 14.0 μm2, P = 4.4 × 10−5, unpaired t test), agd5 (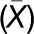 = 11.4 μm2, P = 1.4 × 10−16, unpaired t test), and mtv1agd5 (
= 11.4 μm2, P = 1.4 × 10−16, unpaired t test), and mtv1agd5 (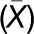 = 11.6 μm2, P = 1.6 × 10−15, unpaired t test) than in wild-type embryos (
= 11.6 μm2, P = 1.6 × 10−15, unpaired t test) than in wild-type embryos (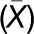 = 16.7 μm2).
= 16.7 μm2).
(D) Immunoblot analysis of extracellular fluid from fully expanded leaves of wild-type, single, and double mutants. ALEU, Aleurain proteinase; ALEU ov.ex., same as above but with increased exposure time to reveal an unprocessed precursor band (p, precursor form; m, mature form); PR5, thaumatin-like protein; DET3, subunit C of the V-ATPase; BiP, luminal binding protein BiP; Ponceau loading control.
(E) FM4-64 as marker for endocytotic uptake. The double mutant shows similar levels of intracellular (i.e., endocytosed) signal.
(F) to (H) PIN2-GFP signal (green) in propidium iodide (red) counterstained root epidermal cells.
(F) PIN2-GFP localizes to the apical polar domain of the wild type and the double mutant.
(G) PIN2-GFP is incorporated into BFA bodies after 1 h of BFA treatment in the wild type and double mutant.
(H) PIN2-GFP relocates to the apical plasma membrane 2 h after BFA was washed out in the wild type and the mutant.
(I) The secretory marker SP-RFP is secreted to the extracellular space of cotyledon epidermal cells in the wild type and the double mutant. Bars = 10 µm.
To determine whether NEV/AGD5 and MTV1 also affect transport of endogenous vacuolar cargo in vegetative tissues, we tested for mis-secretion of the vacuolar protease Aleurain (Ahmed et al., 2000). We analyzed the levels of Aleurain in extracellular fluid fractions from fully expanded leaves, contrasting them with the levels of actin, the V-ATPase subunit DET3, and the endoplasmic reticulum chaperone BiP as controls for contamination from intracellular proteins during extraction. In fractions from wild-type and nev/agd5 plants, we found trace amounts of Aleurain, actin, and DET3 and the absence of BiP (Figure 3D). Importantly, a moderate but specific increase in Aleurain levels in the extracellular fraction was observed in mtv1 plants, which was pronounced in the mtv1 nev/agd5 double mutants. Moreover, a precursor form of Aleurain was specifically found in the extracellular fluid fraction from mtv1 and mtv1 nev/agd5 plants, consistent with prior data showing incomplete processing of Aleurain when it is secreted into the apoplasm (Zouhar et al., 2010). To gain additional evidence for abnormal secretion of vacuolar cargo in the mutants, we used an antibody against PR5, which has been found in the vacuole proteome of Arabidopsis leaves (Carter et al., 2004). Similar to Aleurain, we found increased accumulation of PR5 in the apoplasm of mtv1 and mtv1 nev/agd5 leaves. To rule out that the higher levels of Aleurain and PR5 in the extracellular fractions were due to a potential upregulation of PR5 and Aleurain expression or increased protein stability, we also compared total protein extracts from the same leaf material. We did not find increased protein amounts in the mutants (see Supplemental Figure 3 online), indicating that PR5 and Aleurain accumulation in the extracellular fluid of the mutant leaves is indeed caused by the specific mistargeting of vacuolar cargo.
To explore whether the mtv1 and nev/agd5 mutations affect other trafficking pathways, we analyzed the fate of endocytic and secretory markers in the mtv1 nev/agd5 double mutant, where vacuolar phenotypes were most pronounced. We used the lipophilic styryl dye FM 4-64, which first labels the plasma membrane, is then internalized via endocytosis, passes through the endosomes, and ultimately reaches the tonoplast membrane (Bolte et al., 2004; Dettmer et al., 2006). We did not detect any changes in the dynamics of uptake and transit through the endosomes of FM4-64 in the roots of double mutants (Figure 3E; see Supplemental Figure 4 online). As an additional probe for endocytosis, we analyzed the internalization of the PIN1 and PIN2 auxin transporters in wild-type and double mutant roots. These transporters constitutively cycle between the plasma membrane and early endosomes, but treatment with brefeldin A (BFA), which inhibits the exocytotic step of recycling, leads to PIN accumulation in BFA bodies (Geldner et al., 2003; Langhans et al., 2011). We did not find any difference in BFA body accumulation of PIN1 and PIN2 between the wild type and double mutant (Figures 3F and 3G; see Supplemental Figures 5A and 5B online), suggesting that NEV/AGD5 and MTV1 do not play a role in these endocytic events. On the other hand, once BFA is removed, the endocytosed material is recycled from the BFA body to the plasma membrane in an exocytotic process similar or identical to secretion. We performed BFA washout experiments with PIN1-GFP (for green fluorescent protein) and PIN2-GFP and found no changes between the wild-type and double mutant plants (Figure 3H; see Supplemental Figure 5C online), ind ating that exocytotic processes were not affected. In these experiments, we also did not observe any defects in the polar targeting of PIN1-GFP or PIN2-GFP in the double mutant (Figures 3F and 3H), further corroborating that endo- and exocytosis, which govern PIN polarity (Geldner et al., 2003; Kleine-Vehn et al., 2008), are not dependent on the function of MTV1 or NEV/AGD5. As an additional test for secretion of a nonmembrane protein, we observed the secretory marker SP-RFP (for red fluorescent protein) (Hunter et al., 2007). We invariably detected it in the extracellular space of cotyledon epidermal cells of either the wild type or double mutants (Figure 3I). Taken together, our results suggest that MTV1 and NEV/AGD5 function specifically in vacuolar trafficking. Moreover, the synergistic phenotype of the double mutant indicates that MTV1 and NEV/AGD5 cooperate in a common trafficking step and the fact that secretion is not affected suggests that this is a post-Golgi step specific for the vacuolar pathway.
MTV1 and NEV/AGD5 Colocalize at the TGN and Redistribute to the Periphery of BFA Bodies
NEV/AGD5 has been shown to colocalize with bona fide TGN-associated proteins, such as VTI12 (by coimmunolocalization, Liljegren et al., 2009) and SYP61 (by transient coexpression assays in tobacco (Nicotiana tabacum) leaf cells; Stefano et al., 2010), suggesting that NEV/AGD5 may regulate ARF-dependent formation of specific vesicles at the TGN. By contrast, the localization of MTV1 is unknown. To study the subcellular distribution of MTV1, we stably transformed plants with a prMTV1:MTV1-GFP fusion construct and observed its subcellular localization by in vivo confocal imaging in root epidermal cells. This construct complements the mtv1 mutant, suggesting that it reproduces the activity of the endogenous gene. MTV1-GFP signal was found in punctate structures distributed broadly throughout the cytosol, which indicates that MTV1 resides at or is associated with an endomembrane compartment.
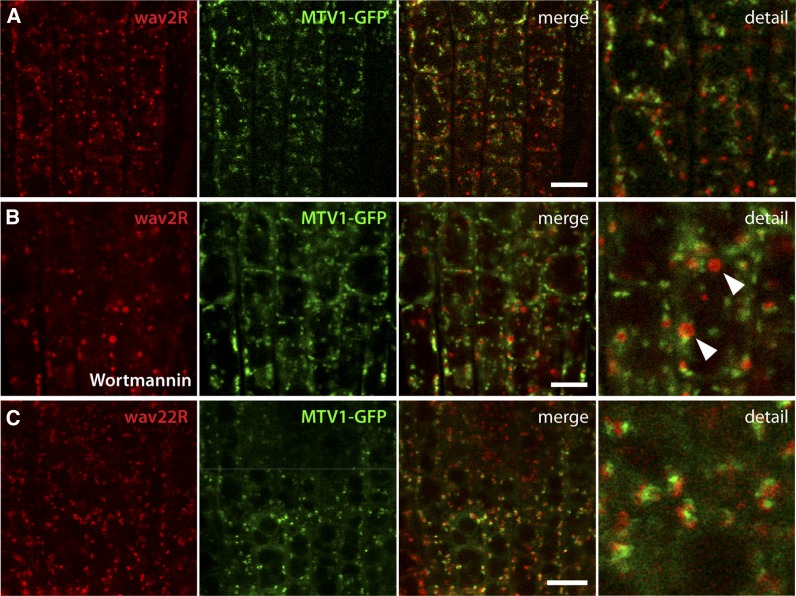
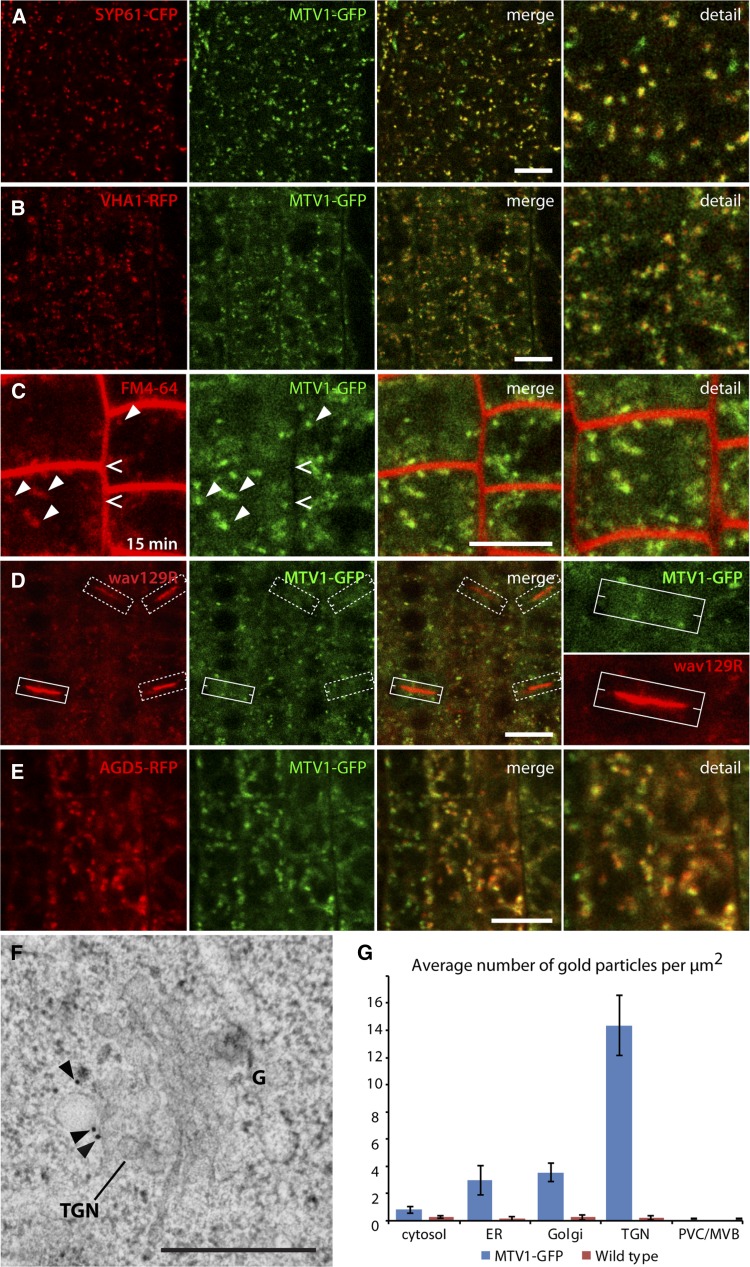
To define this compartment further, we crossed prMTV1:MTV1-GFP plants with a range of previously described fluorescent marker lines for various endomembrane compartment types. The LE/PVC marker wave2R (RabF2b/ARA7) (Geldner et al., 2009) did not show significant colocalization with MTV1 (Figure 4A). The phosphatidylinositol-3 kinase inhibitor wortmannin leads to swelling of the PVC into characteristic ring-like structures and is frequently used to identify this type of compartment (Tse et al., 2004; Wang et al., 2009). After wortmannin treatment, we observed wave2R-labeled ring-like structures, whereas MTV1-GFP fluorescence remained in a punctate pattern (Figure 4B, arrowheads), strongly suggesting that MTV1 is not associated with the PVC. Next we tested the Golgi marker wave22R (SYP32) (Geldner et al., 2009), and although there was no overlap with MTV1-GFP, the signals were frequently found in close proximity (Figure 4C; see Supplemental Figures 6A and 6B online). Time-lapse imaging showed that MTV1 comigrates with the Golgi marker (see Supplemental Movie 1 and Supplemental Figure 7A online), indicating that MTV1 likely resides on a Golgi-associated structure, such as the TGN (Viotti et al., 2010). Therefore, we crossed MTV1-GFP with the TGN markers SYP61-CFP (for cyan fluorescent protein; Robert et al., 2008) and VHA-a1-RFP (Dettmer et al., 2006) and found a very high degree of overlap and a nearly perfect comigration in time-lapse imaging (Figures 5A and 5B; see Supplemental Figures 6C, 6D, and 7B and Supplemental Movie 2 online), indicating that MTV1 localizes primarily to the TGN. The small fraction of MTV1 signal that comigrates but does not overlap with the signal from TGN markers may represent partial localization of the protein in trans-Golgi cisternae. Moreover, we observed strong colocalization of the MTV1-GFP–labeled structures with FM4-64 after short incubation times (Figure 5C, arrowheads), when the internalized dye has only reached the TGN (Dettmer et al., 2006; Gendre et al., 2011). By contrast, we could not detect any MTV1-GFP at the plasma membrane colocalizing with FM4-64 (Figure 5C, open arrows) or at the cell plate (Figure 5D) colocalizing with wave129R (RabA1g; Geldner et al., 2009). These results suggest that MTV1 resides at the TGN, as does NEV/AGD5, an observation that is consistent with their genetic and functional interaction. To test for their colocalization at the TGN directly, we generated an AGD5-RFP fusion and crossed it into plants carrying MTV1-GFP. We observed a strong degree of overlap, confirming that MTV1 and AGD5 reside on the same TGN compartment (Figure 5E; see Supplemental Figures 6E and 6F online). To further refine the localization of MTV1 at the ultrastructural level, we performed immunogold labeling on MTV1-GFP root cells using anti-GFP antibody and quantitatively analyzed gold particle localization. We observed the highest density of gold particles associated with the TGN (Figure 5F, arrowheads, and 5G; see Supplemental Figure 8 and Supplemental Table 1 online), confirming the results obtained by confocal light microscopy.
Figure 4.
MTV1 Is Separate from the PVC and the cis-Golgi.
(A) Live-cell imaging of root epidermal cells harboring the PVC marker wave2R (ARA7) and MTV1-GFP reveals no overlap.
(B) Incubation with 30 µmol wortmannin for 1 h caused formation of wave2R-labeled ring-like structures (arrowheads), and no colocalization with MTV1 was observed.
(C) The cis-Golgi marker wave22R (SYP32) did not colocalize with MTV1, but signals were typically found in close vicinity.
Bars = 20 µm.
Figure 5.
MTV1 and NEV/AGD5 Localize to the Early Endosome/TGN.
(A) and (B) The TGN markers SYP61-CFP ([A], signal in red) and VHA-a1-RFP (B) colocalize with MTV1-GFP.
(C) Pulse-chase labeling with FM4-64 reveals absence of MTV1-GFP at the plasma membrane (open arrowheads) and colocalization of MTV1-GFP with early endosome/TGN after 15 min (closed arrowheads).
(D) MTV1-GFP is absent from forming cell plates labeled with wave129R (RabA1g). White boxes outline four different forming cell plates, and the solid box on bottom left is magnified in the detail panel.
(E) AGD5-RFP and MTV1-GFP colocalize, confirming the previously described localization of AGD5 at the early endosome/TGN. Bars = 20 µm.
(F) Immunogold labeling of MTV1-GFP using anti-GFP antibody. Gold particles (arrowheads) are predominantly localized at the TGN. G, Golgi apparatus. Bar = 500 nm.
(G) Quantitative analysis of immunogold labeling experiment. Gold particle density at different subcellular compartments is given. Error bars represent se. Wild-type plants were used as negative controls to assess the specificity of the immunolabel.
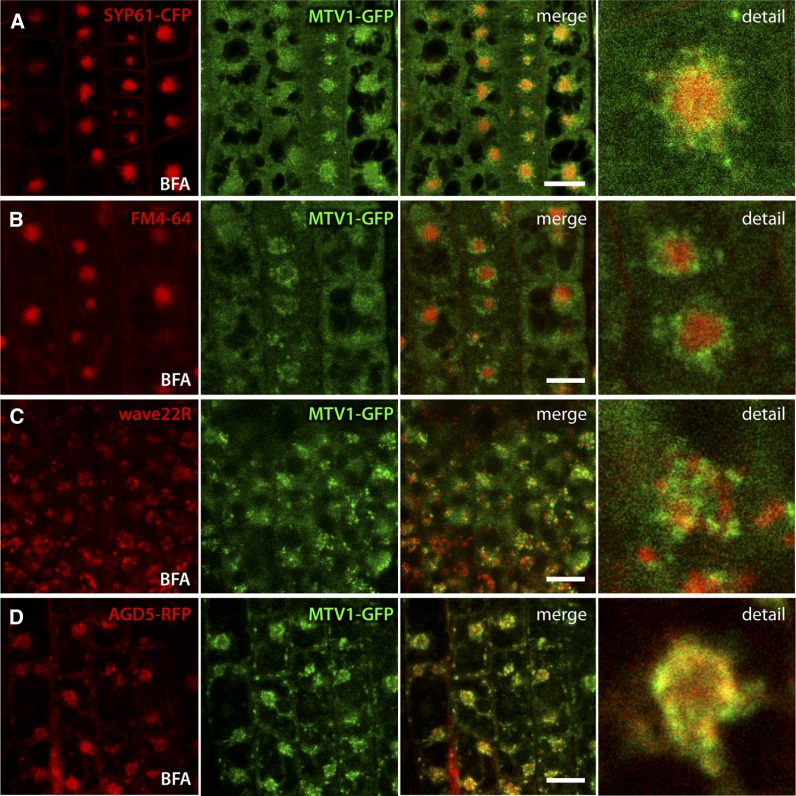
In Arabidopsis roots, treatment with the trafficking inhibitor BFA results in the redistribution of the TGN into aggregates called BFA bodies (Langhans et al., 2011). Surprisingly, we found that MTV1, while generally responsive to BFA, did not localize to the core of the BFA bodies, where TGN markers such as SYP61-CFP, VHA-a1-RFP, or endocytosed FM4-64 are found (Figures 6A and 6B; Dettmer et al., 2006; Lam et al., 2009). Instead, MTV1 was found in the outer periphery of the BFA body (Figures 6A and 6B). An apparently similar response to BFA was reported for cis-Golgi markers, such as wave22R (SYP32) (Geldner et al., 2009). However, we found that the peripheral SYP32-RFP– and MTV1-GFP–labeled structures after BFA treatment were practically mutually exclusive (Figure 6C), indicating that the peripheral BFA body localization of MTV1 does not correspond to the cis-Golgi. Importantly, NEV/AGD5 also exhibited this peculiar response to BFA treatment (Figure 6D). Taken together, our results demonstrate that MTV1 and NEV/AGD5 colocalize at the TGN, possibly in a subdomain characterized by its peripheral BFA body distribution.
Figure 6.
BFA Treatment Reveals That MTV1 and NEV/AGD5 Redistribute to the Periphery of BFA Bodies.
(A) to (D) Live-cell imaging of root epidermal cells from seedlings treated with 50 µM BFA for 1 h. Bars = 20 µm.
(A) SYP61-CFP (signal in red) labels the core of the BFA body, while MTV1-GFP labels the periphery.
(B) Five-minute pretreatment with FM4-64, followed by 1-h BFA incubation. FM4-64 labels the BFA body core and MTV1 the periphery.
(C) The cis-Golgi marker wave22R (SYP32) and MTV1-GFP do not overlap after BFA treatment.
(D) AGD5-RFP and MTV1-GFP both localize at the periphery of the BFA body.
MTV1 and NEV/AGD5 Function in Clathrin-Dependent Trafficking from the TGN
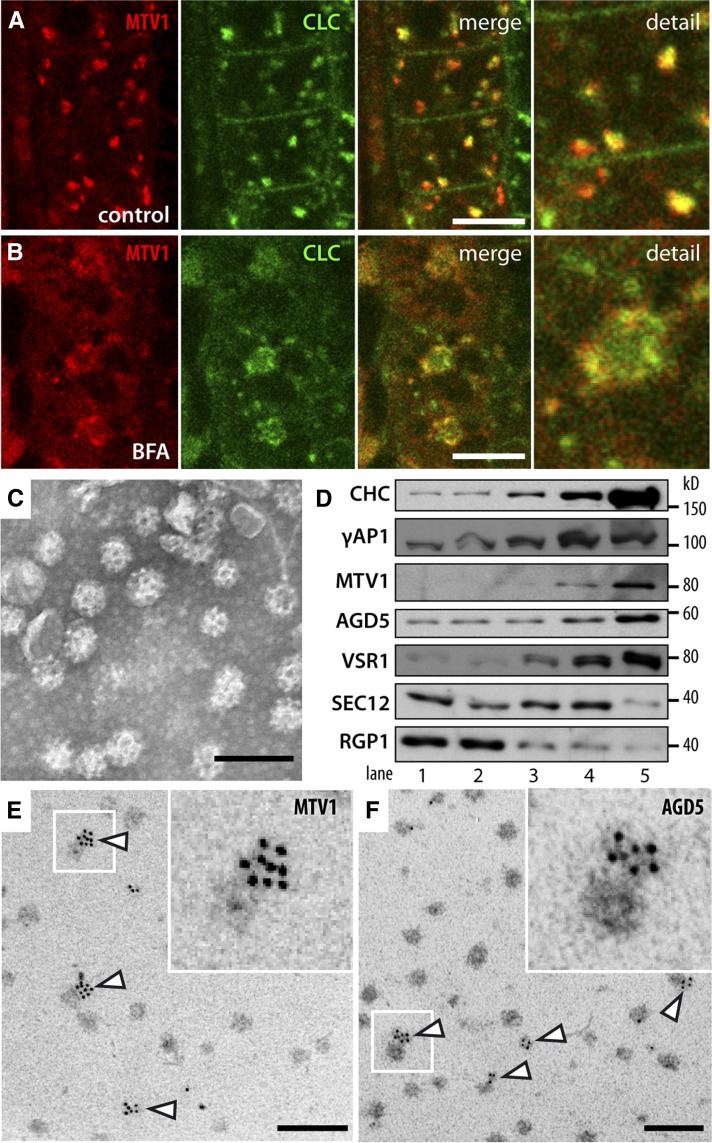
The presence of ARF GAP and ENTH domains in NEV/AGD5 and MTV1 suggests that these proteins may participate in vesicle formation. In particular, they could be involved in the biogenesis of CCVs that bud from the TGN in Arabidopsis and contain Aleurain (Otegui et al., 2006; Hinz et al., 2007). Interestingly, the TGN localization of MTV1 and NEV/AGD5 and the peculiar redistribution in response to BFA is very similar to that reported for clathrin (Ito et al., 2012). To directly assay for colocalization with clathrin, we analyzed plants expressing MTV1-mCherry and clathrin light chain-GFP (CLC-GFP). We observed a high degree of overlap between CLC-GFP and MTV1-RFP signals (Figure 7A), in line with the reported major localization of CLC to the TGN (Ito et al., 2012). Importantly, after treatment with BFA, CLC-GFP and MTV1-RFP were found together in the peripheral zone of the BFA bodies (Figure 7B). We conclude from these analyses that MTV1, NEV/AGD5, and CLC colocalize at the TGN, where MTV1 and NEV/AGD5 may participate in CCV formation. To determine whether NEV/AGD5 and MTV1 are associated with CCVs, we purified CCVs from Arabidopsis suspension culture cells. Electron microscopy analysis of the samples demonstrates that CCVs were efficiently isolated to high purity (Figure 7C). In protein gel blot analyses, we found very strong enrichment of clathrin heavy chain (CHC) in the CCV fraction and a corresponding depletion of markers from other cellular compartments, confirming a high level of purification (Figure 7D). Importantly, NEV/AGD5 and MTV1 were also highly enriched in the CCV fraction, in line with their suspected role in CCV formation. To further corroborate this result, we performed immunogold labeling on the isolated CCVs using antibodies against MTV1 and NEV/AGD5. In both cases, CCVs were clearly labeled by the antibodies (Figures 7E and 7F, arrowheads), which indicates that MTV1 and NEV/AGD5 are present on CCVs and thus probably participate in their formation at the TGN. Labeling was found only in part of the vesicles, which may represent the TGN-derived CCVs, while the unlabeled vesicles may represent the plasma membrane–derived CCVs.
Figure 7.
MTV1 and NEV/AGD5 Are Associated with CCVs.
(A) Live-cell imaging of root epidermal cells harboring MTV1-mCherry (in red) and CLC-GFP reveals partial overlap in the cell interior but not at the plasma membrane.
(B) After 1 h treatment with 50 µM BFA, MTV1-Cherry and CLC-GFP localize to the periphery of the BFA core.
(C) Purification of CCVs from Arabidopsis T87 cell cultures and electron microscopy image of purified fraction.
(D) Immunoblot analysis of enriched CCV fractions; 3 µg of protein was loaded in each lane. 1, Cell homogenate; 2, 30,000g supernatant; 3, Suc step gradient load; 4, linear D2O/Ficoll gradient load; 5, final enriched CCV fraction. Antibodies used: CHC, AP1γ-subunit (γ AP1), MTV1, NEV/AGD5, VSR1, SEC12 (endoplasmic reticulum), and RGP1 (Golgi/cytosolic).
(E) and (F) Immunogold labeling of embedded CCVs using anti-MTV1 (E) or anti-NEV/AGD5 (F) antibodies visualized with gold nanoparticle-conjugated secondary antibodies. Arrowheads point to CCVs with immunogold signal; inset is an enlargement of the region outlined by the white box.
Bars = 500 nm.
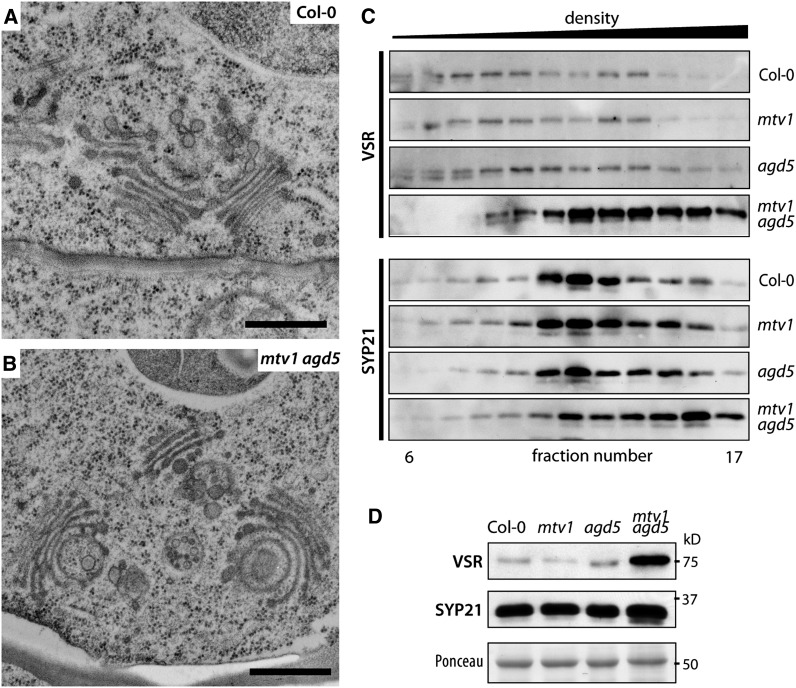
It follows that CCV formation may be disrupted in the mtv1 nev/agd5 mutants, and this could potentially lead to alterations in morphology or size of the TGN. To test this hypothesis, we analyzed the ultrastructure of root meristem cells of the mtv1 nev/agd5 double mutant by electron microscopy. We repeatedly observed morphological alterations at the trans-side of the Golgi and the TGN, with grossly curved cisternae that gave the TGN a bloated appearance (Figures 8A and 8B). This is very similar to what has been reported in the cells of nev/agd5 mutant flowers (Liljegren et al., 2009) or for multiple mutants in the TGN-localized SNAREs of the SYP4 group (Uemura et al., 2012). By contrast, we did not observe clear changes of the PVC, although we cannot exclude subtle morphological changes or alterations in size or number, which would only be detectable by a quantitative analysis of a large number of electron microscopy images. The changes in morphology support that MTV1 and NEV/AGD5 participate in CCV formation at the TGN. To further substantiate this hypothesis, we analyzed the subcellular distribution of VSRs, which are potentially incorporated into CCVs at the TGN (Hinz et al., 2007). We found that VSRs copurified with CLC, MTV1, and NEV/AGD5 in the isolated Arabidopsis CCV fractions (Figure 7D), consistent with prior evidence of VSR association with CCVs in pea (Pisum sativum) cotyledons (Kirsch et al., 1994; Hinz et al., 1999). We fractionated microsomes in Suc density gradients and compared the VSR profiles in samples from wild-type and mutant plants. In wild-type plants, VSRs were broadly distributed in the gradient (Figure 8C), consistent with a steady state localization primarily at the TGN (lighter fractions) and the PVC (denser fractions) (Rojo et al., 2003b). The distribution of VSRs was not significantly changed in mtv1 plants but was weakly altered in nev/agd5 plants and very strongly affected in mtv1 nev/agd5 plants, being displaced toward the bottom of the gradients. A shift to denser fractions was also observed for the PVC syntaxin SYP21 fractions in the mtv1 nev/agd5 mutant (Figure 8C), albeit to a lesser degree. The drastic change in VSR gradient distribution is consistent with defects in CCV-mediated transport of VSRs from the TGN to the PVC. Interestingly, the shift in VSRs distribution was accompanied by a strong increase in VSR abundance in mtv1 nev/agd5 plants (Figures 8C and 8D), a response that is common to other mutants with a disrupted VSR trafficking cycle (Yamazaki et al., 2008; Zouhar et al., 2009; Uemura et al., 2012). Taken together, the subcellular localization at the TGN and in CCVs, the defects in VSR and Aleurain distribution, and the observed morphological alterations of the TGN strongly suggest a role for MTV1 and NEV/AGD5 in CCV formation at the TGN.
Figure 8.
The mtv1 nev/agd5 Double Mutant Has Altered Post-Golgi Properties.
(A) and (B) Ultrastructure of Golgi apparatus and TGN in Col-0 (A) and mtv1 agd5 (B) root epidermal cells. Frequently, the trans-facing Golgi cisternae were bent into sickle shaped or fully circular structures. Bars = 200 nm.
(C) Suc density gradient of total (S1) protein extract of Col-0, mtv1, agd5, and mtv1 agd5, probing with VSR or SYP21 antiserum.
(D) Quantitative immunoblot comparing total protein extracts of Col-0, mtv1, nev/agd5, and mtv1 nev/agd5. Ponceau staining as loading control.
MTV1 and AGD5 Physically Interact with Clathrin
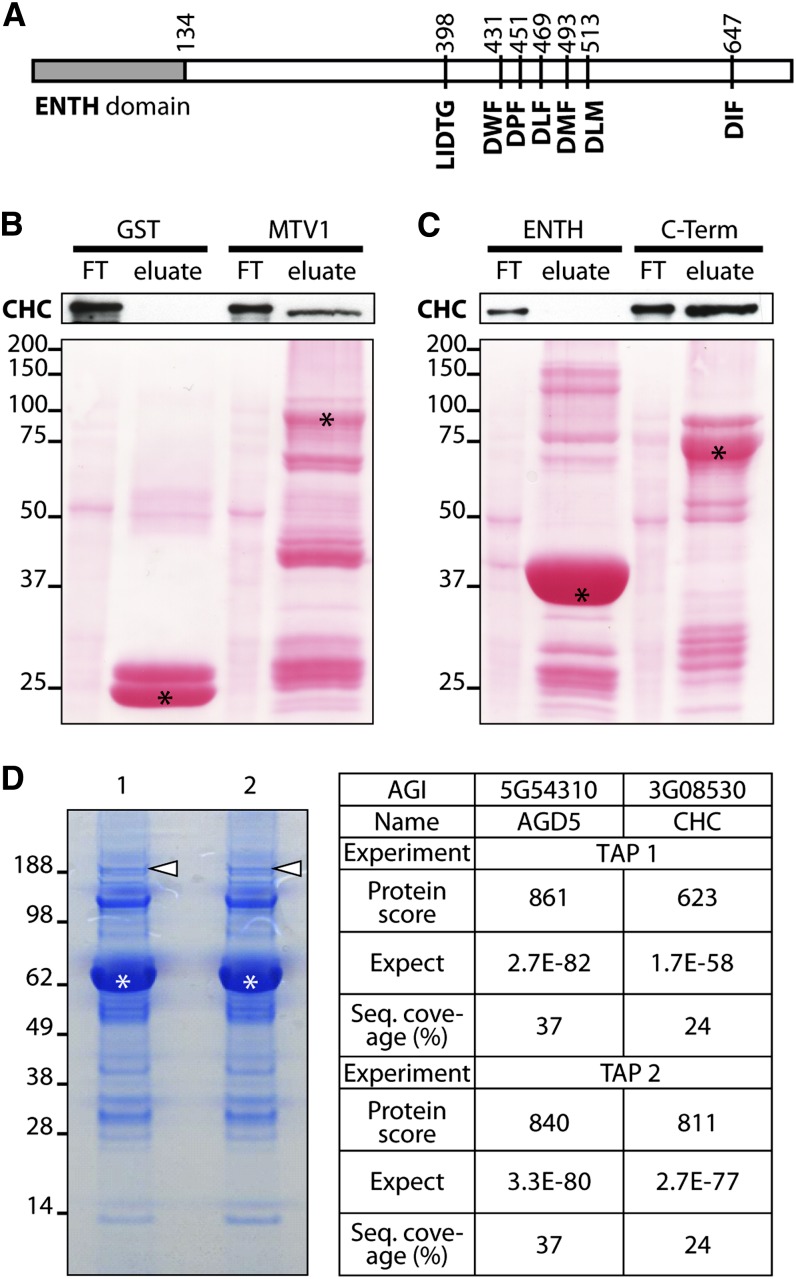
Several ENTH domain proteins, including EPSIN1, EPSINR2, and AtECA1 in Arabidopsis, contain specific motifs for direct binding to clathrin (Song et al., 2006a; Lee et al., 2007; Song et al., 2012). In addition, two ARF GAPs from mammals, SMAP1 and 2, have also been shown to interact physically with clathrin (Tanabe et al., 2005; Natsume et al., 2006). To elucidate whether NEV/AGD5 and MTV1 may interact with clathrin, we first searched for putative clathrin binding motifs in the primary amino acid sequence. Neither has a prototypical Lϕpϕp (L = Leu, P = polar; ϕ=bulky hydrophobic residue) clathrin binding box (Lafer, 2002), but MTV1 contains a slightly modified version (LIDTGD) that conforms to the clathrin binding motifs from the mammalian proteins GGA3, ACK1, and EPSIN1 (Dell’Angelica et al., 2000). Moreover, this motif is highly conserved among plant orthologs of MTV1 (see Supplemental Figure 9 online), suggesting that it is functionally relevant. In addition, MTV1 has five DLL or DxF-related motifs situated in the C-terminal half of the protein (Figure 9A; see Supplemental Figure 9 online), which were shown to be required for the clathrin binding activity of the human ENTH domain–containing protein Auxilin (Scheele et al., 2003) and are also found in other epsins.
Figure 9.
MTV1 and AGD5 Interact with CHC.
(A) Domain structure of MTV1 with the N-terminal ENTH domain, a putative clathrin binding box LIDTG, and 6 DxF (DLL) motifs implicated in clathrin binding.
(B) and (C) Pull-down experiments. GST-tagged bait proteins were bound to sepharose beads and incubated with Col-0 total protein extract. Flow-through was collected (FT), beads were washed extensively, and bound material (eluate) was analyzed by immunoblot using anti-CHC antibody. Asterisks in loading control mark the bait proteins. Clathrin binds GST-MTV1 full-length protein (B); clathrin binding occurs via the C-terminal part of the protein (C).
(D) NEV/AGD5 interacts with CHC in Tap-tag experiments. Left, gel separation of copurified proteins with AGD5-TAP from Arabidopsis cells in two independent experiments; asterisks mark bait protein and arrowheads the position of CHC. Right, table of mass spectrometry data from both experiments for peptides corresponding to AGD5 and CHC (see Supplemental Table 2 online).
[See online article for color version of this figure.]
To test whether MTV1 interacts with clathrin and to characterize in that case where the binding motifs reside, we performed pull-down experiments using three glutathione S-transferase (GST)–fused constructs of MTV1: the full-length protein, the N-terminal part (amino acids 1 to 134) containing the ENTH domain, and the rest of the protein, which contains the putative clathrin binding motifs. The pull-down assays showed clear binding of CHC to full-length MTV1 and to the C-terminal construct, but not to the N-terminal ENTH domain (Figures 9B and 9C). These results indicate that MTV1 directly binds clathrin, most likely through the clathrin binding motifs present in the C-terminal half of the protein, similar to what has been shown in canonical Epsins.
As clathrin binding motifs were not detected in the primary sequence of NEV/AGD5, we used tandem affinity purification to identify in vivo partners of NEV/AGD5. TAP-AGD5 was stably transformed in Arabidopsis cell cultures, and copurified proteins were identified by mass spectrometry. We performed two independent experiments and in both of them we could identify with very high confidence CHC copurifying with the bait (Figure 9D; see Supplemental Table 2 online). Interestingly, when the tag was fused to the C terminus of NEV/AGD5 (AGD5-TAP), clathrin interaction could not be identified in two independent experiments, which might indicate that the tag at this position masks important domains required for the interaction. Taken together, these experiments indicate that NEV/AGD5 and clathrin are closely associated in vivo, most likely through direct protein–protein interaction.
The interaction with clathrin fully supports the genetic and cell biological evidence, suggesting that MTV1 and NEV/AGD5 function in CCV formation at the TGN for transport of vacuolar cargo to the PVC.
DISCUSSION
The Molecular Mode of Action of MTV1 and NEV/AGD5
Our results show that the biological function of both MTV1 and NEV/AGD5 must lie in an anterograde, clathrin-dependent trafficking pathway from the TGN to the vacuole. The most likely molecular mode of action is a role in the productive formation of cargo-filled CCVs at the TGN. For MTV1, the literature about other ENTH domain–containing proteins, namely, epsins, supports this notion. Epsins have been shown to recognize and bind certain phosphoinositides in target membranes by their bulky ENTH domain. This domain partially enters into the lipid bilayer and dislodges the head groups, thereby introducing membrane curvature toward the cytosolic face or at least reducing the energy required for membrane bulging. Their long, disordered C-terminal part then serves as an adaptor for clathrin and adaptor complex subunits. Thus, epsins and at least part of other ENTH domain–containing proteins can be seen as adaptors involved in CCV formation. In the case of MTV1, our characterization strongly suggests such an adaptor role for CCV formation at the TGN.
For ARF GAPs, the current understanding of their molecular mode of action is not as undisputed. Most studies have centered on the prototypical mammalian ArfGAP1, which is involved in COPI vesicle formation. Knowledge about ARF GAPs involved in CCV formation is much more limited, although the basic mechanisms may well be the same (reviewed in Kon et al., 2011), and a recent study found an unexpected involvement of ArfGAP1 also in CCV formation (Bai et al., 2011). The initial hypothesis that ArfGAP1 triggers vesicle coat disassembly by the ARF-GTP to ARF-GDP conversion has been revisited over the last 10 years, and several, more refined models for ARF GAP function have been proposed (reviewed in Nie and Randazzo, 2006; Spang, et al., 2010).
All agree that GTP-bound ARFs recruit coat proteins and that ARF GAP–dependent GTP-GDP hydrolysis results in coat disassembly. The debate centers on the question of how the GTPase activity of ARFs is regulated, as early activity would result in premature coat dissociation and nonproductive vesicle formation. A recent model proposes a dual function for ARF GAP: (1) It acts in cargo recruitment at the membrane and facilitates interaction with GTP-bound ARF for coat assembly. (2) Once the coat is assembled and the vesicle has budded, ARF GAP activity commences, leading to GTP hydrolysis and coat disassembly (Spang et al., 2010). This implies that ARF GAPs act in cargo sorting and are part of the coat itself, which is in line with our observation that NEV/AGD5 is present on CCVs.
Arabidopsis NEV/AGD5 has been previously reported as a TGN-localized AGD that interacts with Arf1 (Liljegren et al., 2009; Stefano et al., 2010). Interestingly, these studies showed that NEV/AGD5 shares substantial sequence similarity in the ARF GAP domain to the mammalian Arf GAPs SMAP1 and SMAP2, both of which directly interact with CHC (Tanabe et al., 2005; Natsume et al., 2006). Our results suggest that NEV/AGD5 also interacts directly with CHC with an uncharacterized binding domain. The localization and interaction data of NEV/AGD5 and Arf1 as well as multiple, additional reports of Arf1 localization at the TGN (Tanaka et al., 2009; Stierhof and El Kasmi, 2010; reviewed in Robinson et al., 2011), add further weight to the hypothesis that Arf1 is involved in CCV formation at the TGN (Pimpl et al., 2003), in addition to its well established function in COPI vesicle formation at the Golgi (Lee et al., 2002; Takeuchi et al., 2002; Stefano et al., 2006). A recent report shows that Arf1 is recruited to the Golgi by the action of AGD8 and AGD9 (Min et al., 2013), so it may be possible that NEV/AGD5 recruits Arf1 to the TGN. Further experiments will be needed to clarify this question. Also, it needs to be determined whether NEV/AGD5 interacts with other ARFs in vivo, following up on the result that NEV/AGD5 in vitro can interact promiscuously with the plasma membrane–localized ARFB (Stefano et al., 2006).
Functional Redundancy for MTV1 and NEV/AGD5
Despite their presumed central roles in clathrin-dependent trafficking from the TGN to the vacuole, the single mtv1 and nev/agd5 mutants display relatively mild defects in vacuolar trafficking and no obvious overall growth or developmental aberrations. The most likely explanation for the absence of more severe phenotypes is functional redundancy with other members of the respective protein families. Based on sequence similarity and domain structure, NEV/AGD5 is a class II AGD and groups together with AGD6-AGD10 (Vernoud et al., 2003). Phenotypes that reveal their biological function might only manifest in multiple loss-of-function mutants, as is the case for AGD8 and AGD9 (Min et al., 2013), or in tissues where a single AGD is the only or the dominant AGD expressed, as for example RPA/AGD10 in root hairs (Song et al., 2006b). This is likely the case for NEV/AGD5 in the floral abscission zone, where the expression pattern and the trafficking defects of the single nev/agd5 mutant (Liljegren et al., 2009) suggest that NEV/AGD5 is the dominant AGD. In other plant tissues, functionally redundant AGDs may largely compensate for the loss of NEV/AGD5.
For MTV1, the situation is more complex, as the 43 ENTH domain–containing proteins in Arabidopsis show (with few exceptions) little sequence similarity among each other outside their ENTH domain, which makes predictions about their functional similarity difficult. So far, all other ENTH proteins analyzed (Epsin1, Epsin2R, ECA1, ECA2, and ECA4) interact with clathrin, even though they lack a canonical clathrin binding box. However, there is only limited data about their biological function, and the only known mutant with a vacuolar transport defect is epsin1 (Song et al., 2006a). This mutant has, like mtv1, no obvious overall phenotype (Song et al., 2006a), and so it is likely that also in this case other ENTH domain–containing proteins compensate for its lack. For Epsin2/2R, no direct functional data are available (Lee et al., 2007), whereas ECA1, 2, and 4 localize to the plasma membrane and cell plate (Song et al., 2012), making a direct involvement in vacuolar trafficking unlikely.
Contrary to the single mutants, the mtv1 nev/agd5 double mutant shows pronounced vacuolar trafficking defects and is severely affected in overall growth. Nevertheless, vacuolar trafficking is by no means completely abolished. For instance, mtv1 nev/agd5 seeds contain high levels of mature seed storage proteins. This can be explained in two ways, which are not mutually exclusive: (1) The abovementioned redundancy for both MTV1 and NEV/AGD5 with proteins of similar function, which may be more prevalent in certain tissues, such as in seeds, and (2) the presence of alternative machinery and pathways for TGN to PVC transport of vacuolar cargo. For example, in seeds, dense vesicles are abundant and may compensate for the reduced CCV-dependent transport of vacuolar cargo in mtv1 nev/agd5 mutants, which would also explain the relatively weak developmental phenotypes observed in these tissues.
METHODS
Plant Growth Conditions
Plants were grown on soil in the greenhouse or walk-in chambers at 22°C under a 16/8-h light/dark regime. For axenic culture, seeds were grown on vertical plates at 22°C in a 16/8-h light/dark cycle on 0.5× Murashige and Skoog (MS) medium with 1% Suc and 0.4% phytagel. For horizontal growth, phytagel was substituted for 0.8% agar.
Mutants and Transgenic Lines
mtv4-1 and mtv1-1 were isolated from an EMS-mutagenized population derived from the L1 transgenic line (Sanmartín et al., 2007). This population was previously described (Sohn et al., 2007). The mutant loci were identified by bulk segregant analysis and subsequent fine mapping using appropriate indel and cleaved-amplified polymorphic sequence markers. The mtv4-2 (Salk_079928; identical to nev-7 in Liljegren et al., 2009) and mtv1-2 (Salk_061811) SALK T-DNA lines were used for functional analyses, unless otherwise noted. The following subcellular marker lines have been described previously: wave2R (=mCherry-RabF2b/ARA7), wave22R (=mCherry-SYP32), wave129R (=mCherry-RabA1g) (Geldner et al., 2009); SYP61-CFP (Robert et al., 2008); Vha1-RFP (Dettmer et al., 2006); prPIN1:PIN1-GFP (Benková et al., 2003); prPIN2:PIN2-GFP (Abas et al., 2006); SP-RFP (Hunter et al., 2007); and CLC-GFP (Konopka et al., 2008). MTV1 and NEV/AGD5fusions to GFP, GUS, mRFP, mCherry, and TAP-Tag were realized using Gateway technology and the primers indicated in Supplemental Table 3 online for cloning into pDONR207 or pDONR221 entry vectors. An ∼500-bp promoter region upstream of ATG was used for pMTV1:MTV1-GUS and pMTV1:MTV1-GFP, and ∼1.5 kb was used for pAGD5:AGD5-GUS. Gateway subcloning into binary destination vectors pGWB04 and pGWB03 (Nakagawa et al., 2007) and pUbi N-RFP (Grefen et al., 2010) and Agrobacterium tumefaciens–mediated transformation was used to generate the stable transgenic plants or cell cultures indicated in the text. At least five independent transformants were analyzed for each stably transformed line.
Phylogenetic Analyses
Phylogenetic analyses were performed in Jalview (Waterhouse et al., 2009) using the Muscle alignment algorithm with default parameters. Neighbor joining was used to calculate the phylogenetic tree, and bootstrap values for 1000 iterations are indicated.
Protein Detection and Antibodies
The following previously described primary antibodies were used in immunoblots at 1:1000 concentration unless indicated otherwise: anti-NEV/AGD5 (Liljegren et al., 2009), 1:5000; anti-ALEURAIN (Ahmed et al., 2000); anti-CPY (Rojo et al., 2003a); anti-CHC (BD BioSciences 610499); anti-GST (Carl Roth 3998), 1:2000; anti-SYP21 (Da Silva Conceição et al., 1997); anti-VSR (Ahmed et al., 2000); anti-γAP-1 (kindly provided by I. Hwang); anti-SEC12 (Bar-Peled and Raikhel, 1997), anti-RGP1 (Delgado et al., 1998), anti-BiP (Santa Cruz Biotechnology sc-33757), antiactin (Sigma-Aldrich A04080), 1:2000; and anti-DET3 (kindly provided by K. Schumacher; Schumacher et al., 1999), 1:2000. Anti-MTV1 antiserum was raised in rabbit against an MBP-MTV1 full-length fusion protein. Secondary antibodies were horseradish peroxidase–conjugated anti-mouse (NA931V; GE Healthcare) and anti-rabbit (NA934V) at 1:10,000. Seed proteins were extracted and analyzed as described (Zouhar et al., 2010). Isolation and analysis of extracellular fluid were performed as described (Sanmartín et al., 2007). Density gradient fractionation was performed as described (Bassham and Raikhel, 1998). All immunoblot experiments were repeated at least three times.
CCV Isolation and Microscopy
T87W cells (60 to 90 mL packed cell volume) were lysed by nitrogen decompression, and CCVs were purified by differential and density gradient centrifugation, essentially as described (Depta and Robinson, 1986; Harley and Beevers, 1989). In brief, the cell homogenate (H) was subjected to sequential differential centrifugation at 100g, 1000g, and 30,000g. Microsomal membranes (SGL) from the 30,000g supernatant (S30) were concentrated by sedimentation (120,000g) and layered onto a Suc step gradient. The Suc step gradient was centrifuged (116,000g), and fractions corresponding to the 10 and 10/40% (w/v) Suc steps were harvested, from which membranes were concentrated by sedimentation (180,000g). The resulting pellet was resuspended (DFGL), layered over a linear D2O/Ficoll gradient, and centrifuged (80,000g). D2O/Ficoll gradient fractions containing enriched CCVs (equivalent to ∼14 to 16% [w/w] Suc) were harvested and concentrated by sedimentation (264,000g) to yield the final enriched CCV fraction, typically 100 to 300 µg. For immunoblot analysis, aliquots of subcellular fractions (H, S30, SG-load, DGF-load, LSG-load, and CCV) were solubilized in 1× Laemmli loading buffer. Protein concentration was quantified using Pierce 660 protein assay reagent containing ionic detergent compatibility reagent using BSA as a standard according to the manufacturer’s instructions (Thermo Fisher Scientific). For negative stain electron microscopy analysis, enriched CCV samples were adjusted to 2% (w/v) aqueous osmium tetroxide, dried as a thin layer onto the surface of a pioloform-coated 200 mesh nickel grid, and stained with Nano-W methylamine tungstate (catalog number 2018; Nanoprobes). Imaging was performed on a Philips CM120 transmission electron microscope (Philips/FEI) with a MegaView III side-mounted digital camera at the UW-Madison Medical School Electron Microscope Facility. Quantitative analysis of CCVs versus uncoated vesicles was performed on randomly selected electron microscopy images of negative-stained samples using the ImageJ (http://rsb.info.nih.gov/ij/) Cell Counter plug-in. Coated vesicles represented, on average, 76% (n = 11) of the total vesicles counted.
MTV1 Pulldown
The full-length coding sequence of MTV1, a region encompassing the ENTH domain (amino acids 1 to 134), or the C terminus (amino acids 135 to 699) was cloned into pDONR221 using primers indicated in Supplemental Table 3 online and subcloned into a Gateway modified version of pGEX-2T (GE Healthcare) to yield GST-MTV1, GST-ENTH, and GST-C-Term. Proteins were expressed in Escherichia coli BL21 cells, extracted in lysis buffer (1× PBS, 0.5% Triton X-100, 0.5 mM PMSF, and 1× complete inhibitor [Roche]), and bound to glutathione beads (Sigma-Aldrich G4502). Fresh extracts from liquid-grown Columbia-0 (Col-0) seedlings 8 d after germination (DAG) in the same lysis buffer were incubated with the beads, the flow-through was collected, and the beads were washed extensively. To elute bound proteins, beads were boiled in Laemmli loading buffer. Experiments were repeated four times.
AGD5 TAP-Tag
Transgenes encoding TAP tag fusions with AGD5 were cloned into pKNTAP and pKCTAP to yield TAPtag-AGD5 and AGD5-TAPtag under the control of the constitutive cauliflower tobacco mosaic virus 35S promoter (Van Leene et al., 2007). Constructs were transformed into Arabidopsis thaliana cell suspension cultures by Agrobacterium cocultivation (Van Leene et al., 2007). Tandem affinity purification of protein complexes was done using the GS tag (Van Leene et al., 2008), as described by Van Leene et al. (2011). Mass spectra were acquired using a 4800 Proteomics Analyzer (Applied Biosystems), and MS-based protein homology identification was based on The Arabidopsis Information Resource genomic database (Van Leene et al., 2010). Experimental background proteins were subtracted based on ∼40 TAP experiments on wild-type cultures and cultures expressing TAP-tagged mock proteins GUS, RFP, and GFP (Van Leene et al., 2010).
Ultrastructural Analysis and Immunogold Labeling
Ultrastructural analysis of high-pressure frozen, freeze-substituted (osmium tetroxide in acetone) and epoxy resin–embedded root tips was performed as previously described (Reichardt et al., 2007).
For immunolabeling, roots from 10-d-old wild-type and MTV1-GFP Arabidopsis seedlings were high-pressure frozen/freeze-substituted in a Baltec HPM010. Samples were then substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) plus 0.2% glutaraldehyde (Electron Microscopy Sciences) in acetone at −80°C for 72 h and warmed to −50°C for 24 h. After several acetone rinses, these samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences) for 72 h and polymerized at −50°C under UV light for 48 h. Sections were mounted on Formvar-coated nickel grids and blocked for 20 min with a 5% (w/v) solution of nonfat milk in Tris-buffered saline containing 0.1% Tween 20. The sections were incubated in the primary polyclonal antibodies against anti-GFP (Torrey Pines BioLabs) for 1 h, rinsed in Tris-buffered saline containing 0.5% Tween 20, and then transferred to the secondary antibody fragment (Fab’2-goat anti-rabbit 1:10; Electron Microscopy Sciences) conjugated to 15-nm gold particles for 1 h. Labeling quantification was performed using Fiji (http://fiji.sc/Fiji).
Chemical Treatments
At 4 to 5 DAG, vertically grown seedlings were incubated in 1 mL liquid 0.5× MS medium with 1% Suc in 24 microwell plates with gentle agitation. Drug concentrations used were 50 μM BFA for durations between 30 min and 2 h and 30 μM wortmannin for 1 h. FM4-64 was used at 2 μM, either as a prestain for 5 min on ice, followed by washing twice for 1 min in liquid medium on ice, or, for long-term treatments, remaining for 3 h in the incubation medium. All chemicals were dissolved in DMSO and controls carried equivalent volumes of DMSO.
Wide-Field and Confocal Microscopy
Alexander stain for pollen viability was performed as described (Peterson et al., 2010) and observed with a Leica DRE upright microscope with a ×20 0.75 apochromatic objective. Confocal imaging was performed on an inverted Leica SP5II, using a ×63 1.2 water immersion objective for subcellular features or a ×20 0.75 dry objective for overview images. The pinhole for colocalization experiments was adjusted to ensure confocality, photomultiplier tube settings were chosen to avoid amplifier saturation, and line averaging was employed to avoid between-frame registration errors due to mobile particles. Colocalization and drug treatment experiments were performed at least three times, in each observing at least 10 individual roots.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MTV1, AT3G16270; MTV4/NEV/AGD5, AT5G54310.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. T-DNA Insertion Mutants and the Original EMS Alleles of mtv1 and mtv4 Are Phenotypically Equivalent.
Supplemental Figure 2. Phylogenetic Analysis of MTV1 and the Previously Characterized ENTH Domain–Containing Proteins Epsin1, Epsin2/2R, and Epsin3.
Supplemental Figure 3. Seed Protein Profile and Analysis of Total Leaf Extracts.
Supplemental Figure 4. Endocytotic Uptake of FM4-64.
Supplemental Figure 5. PIN1-GFP Polar Localization, Formation of BFA Bodies, and Washout in Col-0 and mtv1 nev/agd5 Double Mutant.
Supplemental Figure 6. Colocalization of MTV1-GFP and Subcellular Markers in Cotyledon Pavement Cells.
Supplemental Figure 7. First, Middle, and Last Frames of Supplemental Movies 1 and 2 Online.
Supplemental Figure 8. Immunogold Labeling of MTV1-GFP.
Supplemental Figure 9. Multiple Sequence Alignment of MTV1 Orthologs.
Supplemental Table 1. Quantitative Analysis of Immunogold Labeling.
Supplemental Table 2. Protein Identification Data of the NEV/AGD5 TAP-Tag Experiment.
Supplemental Table 3. Primers Used for Cloning.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Figure 1D.
Supplemental Data Set 2. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 2 Online.
Supplemental Movie 1. Five-Minute Time-Lapse Video of MTV1-GFP (Green) and wave22R/SYP32-RFP (Red) in Root Epidermal and Cortical Cells.
Supplemental Movie 2. Five-Minute Time-Lapse Video of MTV1-GFP (Green) and SYP61-CFP (Red) in Root Epidermal and Cortical Cells.
Acknowledgments
We thank Dominique Eeckhout for expert assistance in the TAP-tag experiments. This work was in part funded by Human Frontier Science Program, EMBO, and Marie Curie Intra-European long-term fellowships to M.S., BIO2009-10784 and CSD2007-00057 to E.R., National Science Foundation IOS-1239311 to S.J.L., and National Science Foundation 1157824 to M.S.O.
AUTHOR CONTRIBUTIONS
M.S., E.R., S.Y.B., and M.S.O. designed the research. M.S., G.D.R., M.O.D., J.Z., S.Y.B., J.G.P., M.S.O., Y.-D.S., S.J.L., L.J., and G.D.J. performed the research. M.S. and E.R. wrote the article.
Glossary
- CCV
clathrin-coated vesicle
- TGN
trans-Golgi network
- PVC
prevacuolar compartment
- VSR
vacuolar sorting receptor
- EMS
ethyl methanesulfonate
- BFA
brefeldin A
- CHC
clathrin heavy chain
- MS
Murashige and Skoog
- Col-0
Columbia-0
- DAG
days after germination
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 Erratum. Nat. Cell Biol. 8: 424. [DOI] [PubMed] [Google Scholar]
- Ahmed S.U., Rojo E., Kovaleva V., Venkataraman S., Dombrowski J.E., Matsuoka K., Raikhel N.V. (2000). The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J. Cell Biol. 149: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M., et al. (2011). ARFGAP1 promotes AP-2-dependent endocytosis. Nat. Cell Biol. 13: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M., Raikhel N.V. (1997). Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 114: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C., Raikhel N.V. (1998). An Arabidopsis VPS45p homolog implicated in protein transport to the vacuole. Plant Physiol. 117: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Betsuyaku, S., Sawa, S. and Yamada, M. (2011). The function of the CLE peptides in plant development and plant-microbe interactions. In The Arabidopsis Book 9e0149, /10.1199/tab.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Bonangelino C.J., Chavez E.M., Bonifacino J.S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2486–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C., Pan S., Zouhar J., Avila E.L., Girke T., Raikhel N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks J.B., Field M.C. (2007). Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J. Cell Sci. 120: 2977–2985 [DOI] [PubMed] [Google Scholar]
- da Silva Conceição A., Marty-Mazars D., Bassham D.C., Sanderfoot A.A., Marty F., Raikhel N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9: 571–582 [PMC free article] [PubMed] [Google Scholar]
- Delgado I.J., Wang Z., de Rocher A., Keegstra K., Raikhel N.V. (1998). Cloning and characterization of AtRGP1. A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 116: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Puertollano R., Mullins C., Aguilar R.C., Vargas J.D., Hartnell L.M., Bonifacino J.S. (2000). GGAs: A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcos Lousa C., Gershlick D.C., Denecke J. (2012). Mechanisms and concepts paving the way towards a complete transport cycle of plant vacuolar sorting receptors. Plant Cell 24: 1714–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J., Aniento F., Frigerio L., Hawes C., Hwang I., Mathur J., Neuhaus J.-M., Robinson D.G. (2012). Secretory pathway research: The more experimental systems the better. Plant Cell 24: 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depta H., Robinson D.G. (1986). The isolation and enrichment of coated vesicles from suspension-cultured carrot cells. Protoplasma 130: 162–170 [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., Okatani Y., Uemura T., Goh T., Shoda K., Niihama M., Morita M.T., Spitzer C., Otegui M.S., Nakano A., Ueda T. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O., Gershlick D.C., Bottanelli F., Hummel E., Hawes C., Denecke J. (2010). A recycling-defective vacuolar sorting receptor reveals an intermediate compartment situated between prevacuoles and vacuoles in tobacco. Plant Cell 22: 3992–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D., Oh J., Boutté Y., Best J.G., Samuels L., Nilsson R., Uemura T., Marchant A., Bennett M.J., Grebe M., Bhalerao R.P. (2011). Conserved Arabidopsis ECHIDNA protein mediates trans-Golgi-network trafficking and cell elongation. Proc. Natl. Acad. Sci. USA 108: 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.-M., Paris N., Robinson D.G., Holstein S.E.H. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Harley S.M., Beevers L. (1989). Coated vesicles are involved in the transport of storage proteins during seed development in Pisum sativum L. Plant Physiol. 91: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz G., Colanesi S., Hillmer S., Rogers J.C., Robinson D.G. (2007). Localization of vacuolar transport receptors and cargo proteins in the Golgi apparatus of developing Arabidopsis embryos. Traffic 8: 1452–1464 [DOI] [PubMed] [Google Scholar]
- Hinz G., Hillmer S., Baumer M., Hohl I. (1999). Vacuolar storage proteins and the putative vacuolar sorting receptor BP-80 exit the Golgi apparatus of developing pea cotyledons in different transport vesicles. Plant Cell 11: 1509–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C.A.J., Vanden Broeck D., Boulet G.A.V., Bogers J., De Wolf M.J.S. (2007). Epsin: Inducing membrane curvature. Int. J. Biochem. Cell Biol. 39: 1765–1770 [DOI] [PubMed] [Google Scholar]
- Hunter P.R., Craddock C.P., Di Benedetto S., Roberts L.M., Frigerio L. (2007). Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 145: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., Nakano A. (2012). Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Kang B.H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kirsch T., Paris N., Butler J.M., Beevers L., Rogers J.C. (1994). Purification and initial characterization of a potential plant vacuolar targeting receptor. Proc. Natl. Acad. Sci. USA 91: 3403–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kon S., Funaki T., Satake M. (2011). Putative terminator and/or effector functions of Arf GAPs in the trafficking of clathrin-coated vesicles. Cell. Logist. 1: 86–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer E.M. (2002). Clathrin-protein interactions. Traffic 3: 513–520 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Cai Y., Tse Y.C., Wang J., Law A.H.Y., Pimpl P., Chan H.Y.E., Xia J., Jiang L. (2009). BFA-induced compartments from the Golgi apparatus and trans-Golgi network/early endosome are distinct in plant cells. Plant J. 60: 865–881 [DOI] [PubMed] [Google Scholar]
- Langhans M., Förster S., Helmchen G., Robinson D.G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62: 2949–2957 [DOI] [PubMed] [Google Scholar]
- Lee M.H., Min M.K., Lee Y.J., Jin J.B., Shin D.H., Kim D.H., Lee K.-H., Hwang I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.-J., Kim H., Kang H., Jang M., Lee D.W., Lee S., Hwang I. (2007). EpsinR2 interacts with clathrin, adaptor protein-3, AtVTI12, and phosphatidylinositol-3-phosphate. Implications for EpsinR2 function in protein trafficking in plant cells. Plant Physiol. 143: 1561–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V., Wasiak S., Hussain N.K., Angers A., McPherson P.S. (2004). ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 117: 9–18 [DOI] [PubMed] [Google Scholar]
- Li L., Shimada T., Takahashi H., Ueda H., Fukao Y., Kondo M., Nishimura M., Hara-Nishimura I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18: 3535–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren S.J., Leslie M.E., Darnielle L., Lewis M.W., Taylor S.M., Luo R., Geldner N., Chory J., Randazzo P.A., Yanofsky M.F., Ecker J.R. (2009). Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 136: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Méndez B., et al. (2004). NMR assignment of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana. J. Biomol. NMR 29: 205–206 [DOI] [PubMed] [Google Scholar]
- Meyerowitz E.M. (2002). Plants compared to animals: The broadest comparative study of development. Science 295: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Min M.K., Jang M., Lee M., Lee J., Song K., Lee Y., Choi K.Y., Robinson D.G., Hwang I. (2013). Recruitment of Arf1-GDP to Golgi by Glo3p-type ArfGAPs is crucial for Golgi maintenance and plant growth. Plant Physiol. 161: 676–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Natsume W., Tanabe K., Kon S., Yoshida N., Watanabe T., Torii T., Satake M. (2006). SMAP2, a novel ARF GTPase-activating protein, interacts with clathrin and clathrin assembly protein and functions on the AP-1-positive early endosome/trans-Golgi network. Mol. Biol. Cell 17: 2592–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z., Randazzo P.A. (2006). Arf GAPs and membrane traffic. J. Cell Sci. 119: 1203–1211 [DOI] [PubMed] [Google Scholar]
- Otegui M.S., Herder R., Schulze J., Jung R., Staehelin L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18: 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Micol J.L. (2009). Understanding synergy in genetic interactions. Trends Genet. 25: 368–376 [DOI] [PubMed] [Google Scholar]
- Peterson, R., Slovin, J.P. and Chen, C. (2010). A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1: 66–69 [Google Scholar]
- Pimpl P., Hanton S.L., Taylor J.P., Pinto-daSilva L.L., Denecke J. (2003). The GTPase ARF1p controls the sequence-specific vacuolar sorting route to the lytic vacuole. Plant Cell 15: 1242–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt I., Stierhof Y.-D., Mayer U., Richter S., Schwarz H., Schumacher K., Jürgens G. (2007). Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Ricarte F., Menjivar R., Chhun S., Soreta T., Oliveira L., Hsueh T., Serranilla M., Gharakhanian E. (2011). A genome-wide immunodetection screen in S. cerevisiae uncovers novel genes involved in lysosomal vacuole function and morphology. PLoS ONE 6: e23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Scheuring D., Naramoto S., Friml J. (2011). ARF1 localizes to the golgi and the trans-Golgi network. Plant Cell 23: 846–849, author reply 849–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Denecke J. (2008). What is moving in the secretory pathway of plants? Plant Physiol. 147: 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Sharma V.K., Kovaleva V., Raikhel N.V., Fletcher J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Zouhar J., Carter C., Kovaleva V., Raikhel N.V. (2003a). A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 7389–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Zouhar J., Kovaleva V., Hong S., Raikhel N.V. (2003b). The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol. Biol. Cell 14: 361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jean B., Seveno-Carpentier E., Alcon C., Neuhaus J.-M., Paris N. (2010). The cytosolic tail dipeptide Ile-Met of the pea receptor BP80 is required for recycling from the prevacuole and for endocytosis. Plant Cell 22: 2825–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A. (2007). Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 144: 6–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A.A., Ahmed S.U., Marty-Mazars D., Rapoport I., Kirchhausen T., Marty F., Raikhel N.V. (1998). A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 95: 9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín M., Ordóñez A., Sohn E.J., Robert S., Sánchez-Serrano J.J., Surpin M.A., Raikhel N.V., Rojo E. (2007). Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M., Dubreucq B., Baud S., Parcy F., Caboche M., Lepiniec L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Scheele U., Alves J., Frank R., Duwel M., Kalthoff C., Ungewickell E. (2003). Molecular and functional characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin. J. Biol. Chem. 278: 25357–25368 [DOI] [PubMed] [Google Scholar]
- Schekman R., Novick P. (2004). 23 genes, 23 years later. Cell 116 (2, suppl.) S13–S15, 1, S19 [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Vafeados D., McCarthy M., Sze H., Wilkins T., Chory J. (1999). The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100: 16095–16100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn E.J., Rojas-Pierce M., Pan S., Carter C., Serrano-Mislata A., Madueño F., Rojo E., Surpin M., Raikhel N.V. (2007). The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc. Natl. Acad. Sci. USA 104: 18801–18806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Lee M.H., Lee G.-J., Yoo C.M., Hwang I. (2006a). Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18: 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Jang M., Kim S.Y., Lee G., Lee G.-J., Kim D.H., Lee Y., Cho W., Hwang I. (2012). An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol. 159: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.-F., Yang C.-Y., Liu J., Yang W.-C. (2006b). RPA, a class II ARFGAP protein, activates ARF1 and U5 and plays a role in root hair development in Arabidopsis. Plant Physiol. 141: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Shiba Y., Randazzo P.A. (2010). Arf GAPs: Gatekeepers of vesicle generation. FEBS Lett. 584: 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G., Renna L., Chatre L., Hanton S.L., Moreau P., Hawes C., Brandizzi F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46: 95–110 [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Rossi M., Azzarello E., Pollastri S., Brandizzi F., Baluska F., Mancuso S. (2010). AGD5 is a GTPase-activating protein at the trans-Golgi network. Plant J. 64: 790–799 [DOI] [PubMed] [Google Scholar]
- Stierhof Y.-D., El Kasmi F. (2010). Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur. J. Cell Biol. 89: 285–297 [DOI] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Ueda T., Yahara N., Nakano A. (2002). Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31: 499–515 [DOI] [PubMed] [Google Scholar]
- Tanabe K., Torii T., Natsume W., Braesch-Andersen S., Watanabe T., Satake M. (2005). A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol. Biol. Cell 16: 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Kim H., Saito C., Ebine K., Ueda T., Schulze-Lefert P., Nakano A. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Eeckhout D., Persiau G., Van De Slijke E., Geerinck J., Van Isterdael G., Witters E., De Jaeger G. (2011). Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol. Biol. 754: 195–218 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Vernoud V., Horton A.C., Yang Z., Nielsen E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. (2009). Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Shimada T., Takahashi H., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2008). Arabidopsis VPS35, a retromer component, is required for vacuolar protein sorting and involved in plant growth and leaf senescence. Plant Cell Physiol. 49: 142–156 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M.-A., Qiu J.L., Olesen K.L., Thordal-Christensen H. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49: 302–312 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Muñoz A., Rojo E. (2010). Functional specialization within the vacuolar sorting receptor family: VSR1, VSR3 and VSR4 sort vacuolar storage cargo in seeds and vegetative tissues. Plant J. 64: 577–588 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E. (2009). Plant vacuoles: Where did they come from and where are they heading? Curr. Opin. Plant Biol. 12: 677–684 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]