This study shows that AMSH1, an AMSH3-related deubiquitinating enzyme, interacts with the endocytosis machinery through the ESCRT-III subunit VPS2.1. The results demonstrate the importance of AMSH1 and VPS2.1 in autophagic degradation and in the physiological processes related to it.
Abstract
In eukaryotes, posttranslational modification by ubiquitin regulates the activity and stability of many proteins and thus influences a variety of developmental processes as well as environmental responses. Ubiquitination also plays a critical role in intracellular trafficking by serving as a signal for endocytosis. We have previously shown that the Arabidopsis thaliana ASSOCIATED MOLECULE WITH THE SH3 DOMAIN OF STAM3 (AMSH3) is a deubiquitinating enzyme (DUB) that interacts with ENDOSOMAL COMPLEX REQUIRED FOR TRANSPORT-III (ESCRT-III) and is essential for intracellular transport and vacuole biogenesis. However, physiological functions of AMSH3 in the context of its ESCRT-III interaction are not well understood due to the severe seedling lethal phenotype of its null mutant. In this article, we show that Arabidopsis AMSH1, an AMSH3-related DUB, interacts with the ESCRT-III subunit VACUOLAR PROTEIN SORTING2.1 (VPS2.1) and that impairment of both AMSH1 and VPS2.1 causes early senescence and hypersensitivity to artificial carbon starvation in the dark similar to previously reported autophagy mutants. Consistent with this, both mutants accumulate autophagosome markers and accumulate less autophagic bodies in the vacuole. Taken together, our results demonstrate that AMSH1 and the ESCRT-III-subunit VPS2.1 are important for autophagic degradation and autophagy-mediated physiological processes.
INTRODUCTION
Reversible posttranslational modification by the small modifier protein ubiquitin is a critical step for regulating protein activities and abundance in many plant signaling pathways and cellular processes (reviewed in Vierstra, 2009). Thus, ubiquitinating as well as deubiquitinating enzymes (DUBs) play key roles in diverse cellular functions. Whereas soluble proteins can be degraded by the 26S proteasome upon polyubiquitination, plasma membrane–bound proteins are degraded by vacuolar proteases following ubiquitin-dependent endocytosis (reviewed in Zelazny et al., 2011). In plants, the auxin efflux facilitator PIN-FORMED2 (PIN2), the flagellin receptor FLAGELLIN-SENSITIVE2, the water channel PLASMA MEMBRANE INTRINSIC PROTEIN2, the iron transporter IRON-REGULATED TRANSPORTER1, and the boron transporter REQUIRES HIGH BORON1 have been shown to be ubiquitinated prior to endocytosis (Abas et al., 2006; Göhre et al., 2008; Lee et al., 2009; Barberon et al., 2011; Kasai et al., 2011). Furthermore, translational fusion of monoubiquitin to PLASMA MEMBRANE PROTON ATPASE (PMA) was shown to be sufficient for triggering endocytosis and vacuolar transport via multivesicular bodies (MVBs) (Herberth et al., 2012). However, how exactly ubiquitin-dependent endocytosis is regulated at the molecular level in plants and contributes to different physiological processes remains to be elucidated.
We have recently shown that the Arabidopsis thaliana DUB, ASSOCIATED MOLECULE WITH THE SH3 DOMAIN OF STAM3 (AMSH3), interacts with the endocytosis machinery and is essential for plant development (Isono et al., 2010; Katsiarimpa et al., 2011). AMSH3 is closely related to the human DUBs AMSH and AMSH-LP, which belong to the class of eukaryotic DUBs and metalloproteases with an MPR1, PAD1 N-terminal+ (MPN+) domain (Tanaka et al., 1999; Maytal-Kivity et al., 2002; Komander et al., 2009). Two MPN+ domain proteins, namely, REGULATORY PARTICLE NON-ATPASE11 (RPN11) (Glickman et al., 1998; Verma et al., 2002) and COP9 SIGNALOSOME5 (CSN5) (Chamovitz et al., 1996; Cope et al., 2002), are subunits of stable multiprotein complexes. By contrast, Arabidopsis AMSH3 is not part of a stable higher molecular weight complex (Isono et al., 2010), and, in contrast with RPN11 and CSN5, AMSH proteins are active as monomers (McCullough et al., 2004). AMSH proteins have essential functions in the development of animals and plants, since AMSH-deficient mice die postnatum with loss of neurons in the hippocampus (Ishii et al., 2001), and Arabidopsis amsh3 null mutations are seedling lethal and cause a number of intracellular trafficking defects (Isono et al., 2010).
Autophagy (macroautophagy) is another vacuolar degradation pathway, by which cytosolic components or organelles are selectively or nonselectively transported to the vacuole/lysosome for degradation (reviewed in Klionsky and Ohsumi, 1999). Since the discovery of AUTOPHAGY-RELATED (ATG) genes from yeast in the 1990s, intensive genetic and molecular analyses have identified over 30 autophagy-related genes. Autophagy is also implicated in a diverse array of physiological and pathological effects in mammals (reviewed in Mizushima and Levine, 2010). ATG genes are highly conserved also in plants, and mutant analyses have shown them to have central functions in nutrient remobilization during starvation and senescence (Doelling et al., 2002; Yoshimoto et al., 2004, 2009; Sláviková et al., 2005; Thompson et al., 2005; Inoue et al., 2006; Phillips et al., 2008; Chung et al., 2009).
The individual steps of autophagosome formation have been revealed by ultrastructural and biochemical studies (reviewed in Klionsky and Ohsumi, 1999). First, isolation membranes or phagophores are formed, probably from the endoplasmic reticulum, which will then engulf parts of the cytosol in autophagosomes with characteristic double-membrane structures. Autophagosomes are then targeted to vacuoles or lysosomes in which their contents are degraded by resident proteases. Alternatively, depending on organism and cell type, autophagosomes may undergo fusion with late endosomes or MVBs to form amphisomes. Amphisomes can then fuse to vacuoles/lysosomes to become autolysosomes, in which the autophagosomal contents are degraded. In mammals and flies, several studies have reported that intact MVBs as well as a functional ENDOSOMAL COMPLEX REQUIRED FOR TRANSPORT-III (ESCRT-III), a core complex in MVB sorting, are necessary for proper autophagosomal degradation (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007; Han et al., 2012). However, a role for ESCRT-III in the plant autophagy pathway has not been demonstrated yet.
We have previously shown that Arabidopsis AMSH3 interacts with the ESCRT-III subunits VACUOLAR PROTEIN SORTING2.1 (VPS2.1) and VPS24.1 (Katsiarimpa et al., 2011). The embryo- and seedling-lethal phenotypes of vps2.1 and amsh3 mutations, respectively, have prevented us so far from analyzing the physiological functions of VPS2.1 and AMSH3. In this study, we characterize the Arabidopsis amsh1-1 mutant, which has reduced levels of the AMSH3-related gene, AMSH1.
Unlike the amsh3 null mutants, the amsh1-1 knockdown mutant does not have an apparent growth defect. However, amsh1-1 accumulates ubiquitinated proteins and shows chlorosis when transferred to the dark, a phenotype reminiscent of autophagy mutants. Furthermore, amsh1-1 accumulates ATG8 in the dark and accumulates less autophagic bodies in the vacuole. AMSH1, like AMSH3, directly interacts with the ESCRT-III subunit VPS2.1. Plants that overexpress a dominant-negative form of VPS2.1 are deficient in endocytosis, accumulate ATG8, and show hypersensitivity to dark treatment. Similar to previously identified autophagy-defective atg mutants from Arabidopsis, amsh1-1 shows also altered susceptibility toward pathogen infection. Together, the results presented here demonstrate the importance of AMSH1 and VPS2.1 in autophagic degradation and in the physiological processes related to it.
RESULTS
Arabidopsis AMSH Genes Are Evolutionary Conserved and Belong to Independent Clades within the AMSH Gene Family
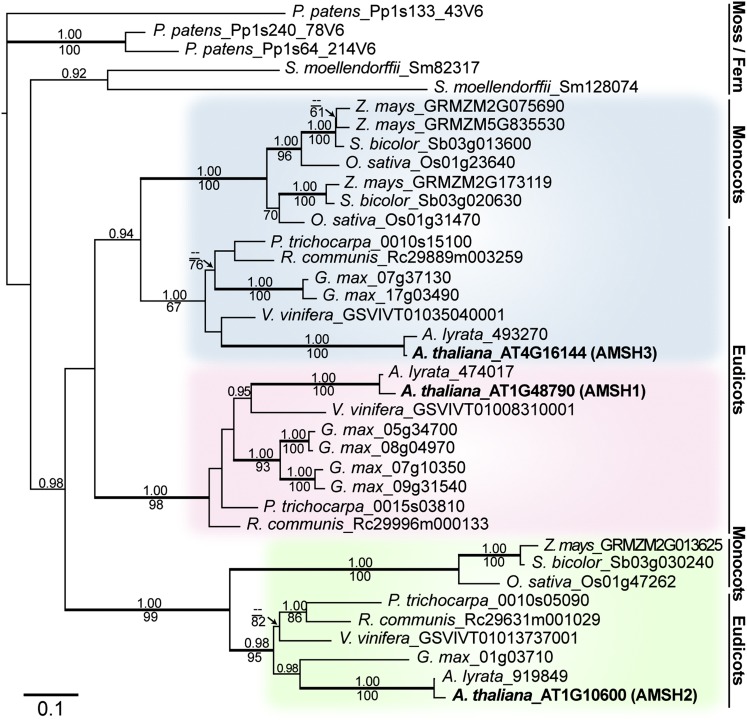
AMSH proteins are widely conserved in eukaryotes. In the Arabidopsis genome, three AMSH genes can be found according to sequence similarity in the catalytic domain (Maytal-Kivity et al., 2002; Isono et al., 2010). To understand the evolutionary origin of the three Arabidopsis AMSH genes, we identified sequences related to Arabidopsis AMSHs in the genomes of 10 fully sequenced plant species (Physcomitrella patens, Selaginella moellendorffii, maize [Zea mays], sorghum [Sorghum bicolor], rice [Oryza sativa], Populus trichocarpa, castor bean [Ricinus communis], soybean [Glycine max], grape [Vitis vinifera], and Arabidopsis lyrata). Phylogenic analysis based on nucleotide sequences of the resulting 37 genes showed that each of the three Arabidopsis AMSH genes is part of an independent clade with other eudicot homologs, suggesting an evolutionary conservation of the three genes in eudicot species (Figure 1; see Supplemental Data Set 1 online).
Figure 1.
AMSH1, AMSH2, and AMSH3 Belong to Independent Clades.
Phylogenetic analysis of AMSH genes from P. patens, S. moellendorffii, maize, sorghum, rice, P. trichocarpa, castor bean, soybean, grape, A. lyrata, and Arabidopsis using P. patens_Pp133_43V6 as an outgroup. A maximum likelihood tree (-ln L = 9462.34) based on the region surrounding the MPN+ domain (549 bp) is shown. The alignment used for generating the phylogenic tree is available as Supplemental Data Set 1 online. Values above branches indicate posterior probabilities (>0.9) calculated by Bayesian analysis, and those below branches indicate maximum parsimony bootstrap values (>60). Thick branches are highly supported (posterior probabilities P > 0.95 and bootstrap values >90). Bar = 0.1 amino acid substitutions per site.
[See online article for color version of this figure.]
The monophyly of AMSH2 with high support allowed us to infer that the hitherto uncharacterized AMSH2 genes have been conserved at least from ancestral angiosperms. By contrast, AMSH1 and AMSH3 may have originated from a eudicot-specific gene duplication event after the separation from monocots. Alternatively, AMSH1 and AMSH3 may have already been present before the separation of monocots and eudicots, and monocots may have lost their copy of the AMSH1 gene and gone through an independent gene duplication event in the course of evolution.
AMSH1 and AMSH3 Show Synergistic Interaction
We next wanted to establish whether the three Arabidopsis AMSH genes have redundant functions. We previously identified two amsh3 mutant null alleles (Isono et al., 2010) and now investigated additional T-DNA insertion lines of AMSH1 and AMSH2. For AMSH1, we identified one T-DNA insertion line, which we named amsh1-1, with reduced AMSH1 transcript level. amsh1-1 carried an insertion in the 5′-untranslated region of the gene (Figure 2A), and the transcript level of AMSH1 was reduced more than 10-fold compared with the wild type (see Supplemental Figure 1A online). AMSH1 protein levels were also strongly decreased in this mutant (Figure 2F). Since AMSH1 transcripts were still detectable, and since immunoblotting with the anti-AMSH1 antibody showed a weak band in amsh1-1, it is likely that amsh1-1 is not a complete loss-of-function mutant but rather a weak mutant allele with compromised AMSH1 function. By contrast, we found that the available T-DNA insertion line of AMSH2 did not have significantly reduced AMSH2 transcript levels (data not shown).
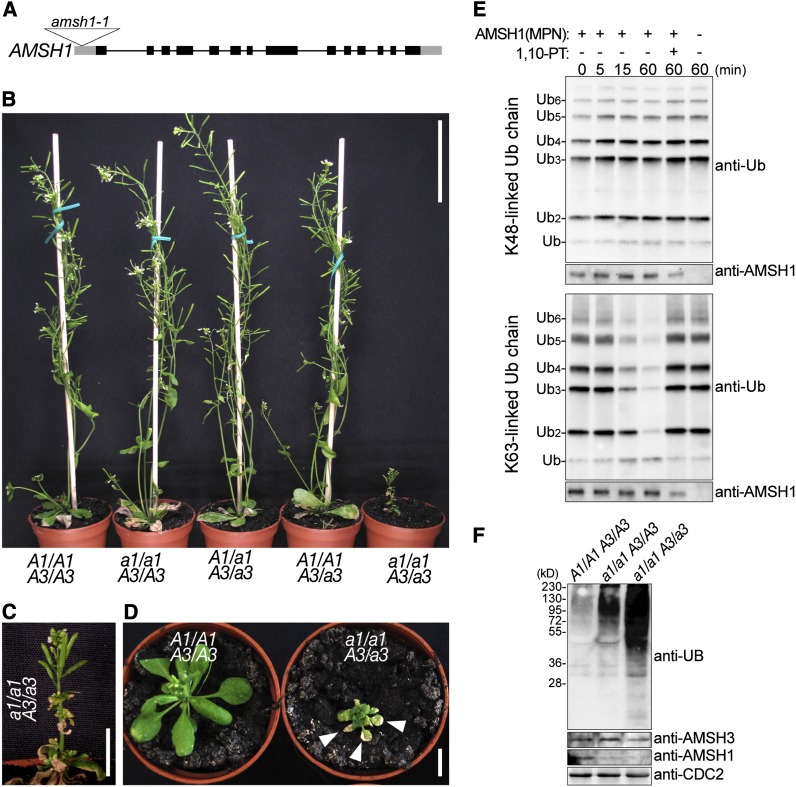
Figure 2.
amsh1/amsh1 AMSH3/amsh3 Is Dwarf and Accumulates Ubiquitinated Proteins.
(A) T-DNA insertion site of amsh1-1. Lines indicate introns and boxes indicate exons (black boxes, coding region; gray boxes, untranslated regions). The triangle indicates the site of T-DNA insertion.
(B) Phenotypes of progeny from an amsh1-1 amsh3-2 double heterozygous (A1/a1 A3/a3) plant. The amsh1-1 homozygous mutant in an amsh3-2 heterozygous background (a1/a1 A3/a3) shows dwarfism. Bar = 6 cm.
(C) Magnification of the a1/a1 A3/a3 plant shown in (B). Bar = 1 cm.
(D) Photographs of an a1/a1 A3/a3 plant in comparison with a wild-type (A1/A1 A3/A3) plant of the same age. Note that the a1/a1 A3/a3 mutant plant exhibits early senescence, indicated by arrowheads. Bar = 1 cm.
(E) DUB assay with K48- or K63-linked ubiquitin chains. The MPN+ domain of AMSH1 was incubated with or without the metalloprotease inhibitor 1,10-phenanthroline (1,10-PT). The reactions were terminated at the indicated time points, and hydrolysis of ubiquitin chains was detected by immunoblotting using an anti-UB antibody. The amount of AMSH1 (MPN+) in each reaction was verified by immunoblotting with an anti-AMSH1 antibody.
(F) Immunoblots with anti-UB, anti-AMSH3, and anti-AMSH1 antibodies on total protein extracts from the wild type (A1/A1 A3/A3), homozygous amsh1-1 (a1/a1 A3/A3), and a1/a1 A3/a3. CDC2 is used as a loading control.
When grown in continuous light, homozygous amsh1-1 plants did not show obvious developmental phenotypes (Figure 2B). However, when we introduced the amsh3-2 allele into amsh1-1, the resulting amsh1-1/amsh1-1 AMSH3/amsh3-2 mutant (a1/a1 A3/a3) showed severe growth defects and early senescence (Figures 2B to 2D), indicating a synergistic interaction between AMSH1 and AMSH3.
AMSH1 is an active enzyme and its catalytic MPN+ domain possesses DUB activity toward K63-linked, but not K48-linked, ubiquitin chains. The activity of the AMSH1 MPN+ domain was inhibited by the metalloprotease inhibitor 1,10-phenanthroline (Figure 2E), indicating that like other MPN+ domain proteins, AMSH1 activity is also dependent on metal ions coordinated in the MPN+ domain. Molecular analysis showed that the amsh1-1 homozygous mutants, in spite of their normal growth, accumulated ubiquitinated proteins at a higher level than the wild type, and amsh1-1/amsh1-1 AMSH3/amsh3-2 accumulated ubiquitin conjugates at an even higher level (Figure 2F). Interestingly, amsh1-1 did not show obvious decreases in its monoubiquitin levels (see Supplemental Figure 1B online), suggesting that the depletion of free ubiquitin molecules is probably not the cause of its phenotype, in contrast with the yeast DUB mutant doa4 (Swaminathan et al., 1999).
AMSH1 and AMSH3 Have Distinct Expression Patterns
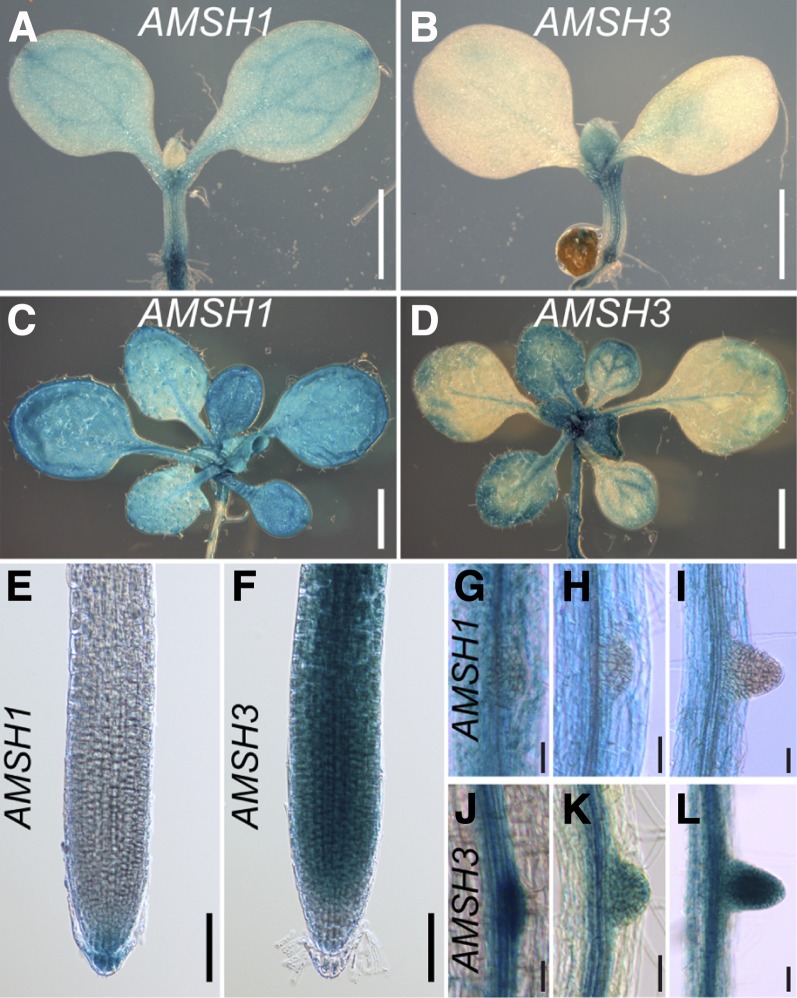
To test whether AMSH1 and AMSH3 are expressed in the same tissues, we generated promoter–β-glucuronidase (GUS) fusions for both genes and analyzed the expression patterns. During seedling development, the expression of both AMSH1 and AMSH3 could be observed in leaves and hypocotyls as well as in roots, though their expression patterns overlapped only partially (Figures 3A to 3F). Furthermore, AMSH3 was strongly expressed in emerging lateral roots, whereas AMSH1 expression seemed to be excluded from this region (Figures 3G to 3L). The largely differential expression pattern of AMSH1 and AMSH3 implies that the two genes might be under different spatio-temporal regulation and that AMSH1 and AMSH3 functions are not interdependent.
Figure 3.
AMSH1 and AMSH3 Are Differentially Expressed during Plant Development.
Histochemical assay showing expression patterns of AMSH1pro:GUS ([A], [C], [E], and [G] to [I]) and AMSH3pro:AMSH3-GUS ([B], [D], [F], and [J] to [L]).
(A) and (B) Seven-day-old seedlings. Bars = 0.5 mm.
(C) and (D) Fourteen-day-old seedlings. Bars = 1 mm.
(E) and (F) Root tips of 7-d-old seedlings. Bars = 0.1 mm.
(G) to (L) Emerging lateral roots of 14-d-old seedlings.
amsh1-1 Shows Hypersensitivity to Dark Treatment
The senescence phenotype of amsh1/amsh1 AMSH3/amsh3 caught our attention since we observed that amsh1-1 also showed early senescence when grown under short-day conditions with 8 h light (110 to 120 µmol m−2 s−1 light) and 16 h dark. This phenotype was not apparent in amsh1-1 plants that had been grown in 10 h light/14 h dark (see Supplemental Figure 1E online), indicating that daylength has a critical effect on the physiology of amsh1-1.
Early senescence is, among others, a hallmark of autophagy mutants (Doelling et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Yoshimoto et al., 2009). Since autophagy is also associated with intracellular trafficking and protein degradation (Rojo et al., 2001; Surpin et al., 2003; Zouhar et al., 2009), a function associated with AMSH3 (Isono et al., 2010; Katsiarimpa et al., 2011), we examined the autophagy pathway in amsh1-1 in more detail.
We first analyzed the response of amsh1-1 to artificial carbon starvation upon transfer to the dark. Wild-type seedlings normally survive the prolonged dark treatment, probably due to a functional autophagic nutrient recycling pathway, whereas mutants of autophagic components show chlorosis upon prolonged dark treatment (Doelling et al., 2002; Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Yoshimoto et al., 2009; Chung et al., 2010). To investigate whether amsh1-1 also has altered response to dark treatment, we transferred wild-type and amsh1-1 seedlings for 5 d to the dark. Indeed, while wild-type seedlings were still green after 5 d of dark treatment, amsh1-1 seedlings became yellowish and had reduced chlorophyll content (51.7% in comparison with the wild type) (Figures 4A and 4B). This result suggests that amsh1-1, like previously reported atg mutants, may have also defects in the autophagic pathway. The dark-induced chlorosis and the accumulation of ubiquitinated proteins in amsh1-1 could be complemented by a genomic fragment of AMSH1, indicating that the downregulation of AMSH1 in amsh1-1 is indeed the cause of these phenotypes (see Supplemental Figures 1C and 1D online).
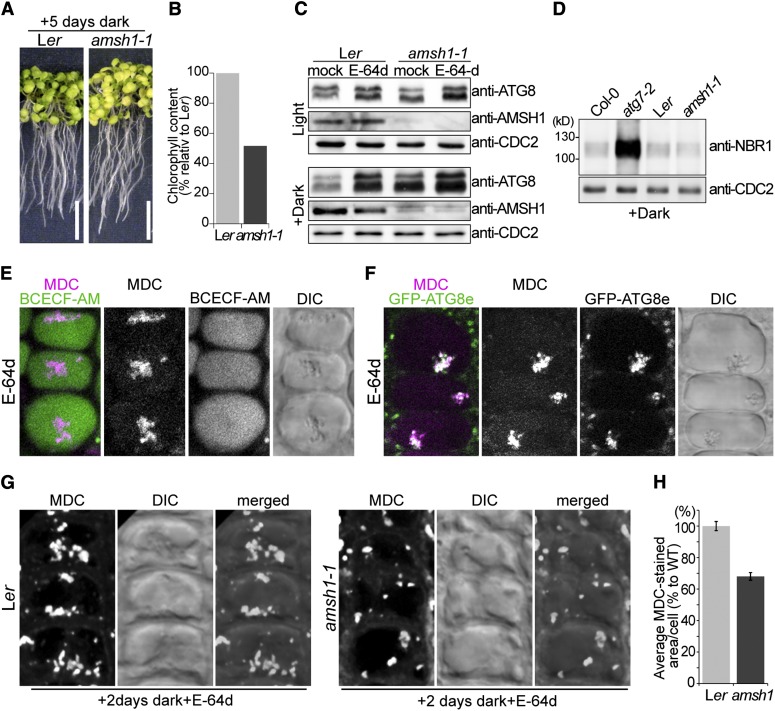
Figure 4.
amsh1-1 Is Deficient in Autophagic Degradation.
(A) Photographs of wild-type (Ler) and amsh1-1 seedlings after 5 d in the dark (left panel). Seedlings were grown 7 d on half-strength MS under long-day conditions before transfer to dark. Note that starvation-induced chlorosis is enhanced in amsh1-1 compared with the wild type. Bars = 1 cm.
(B) Total chlorophyll content of seedlings in (A). Value of the wild type was set to 100%.
(C) Immunoblot with an anti-ATG8 antibody. Total proteins were extracted from seedlings grown in light for 12 d (Light, top panel) or as in (A) (Dark, bottom panel) before treatment with either DMSO (mock) or E-64d.
(D) Total proteins were extracted from seedlings grown as in (A) and subjected to immunoblot with an anti-NBR1 antibody. atg7-2 is used as a positive control and CDC2 is used as loading control.
(E) Confocal images of MDC-stained wild-type root epidermal cells. BCECF-AM was used to visualize the vacuoles. Seedlings were grown as in (A) and treated with E-64d for 1 h before staining with MDC. Note that upon E-64d treatment, MDC positive autophagic bodies accumulate in the BCECF-AM–stained vacuole.
(F) Confocal images of MDC-stained GFP-ATG8e–expressing root epidermal cells. Seedlings were grown as in (A) and treated 6 h with E-64d before staining with MDC.
(G) Wild-type (Ler) and amsh1-1 seedlings were grown as in (A). Seedlings were subsequently treated with E-64d for 1 h and stained with MDC. Confocal images (maximal projection) of MDC-stained root epidermal cells of the wild type (Ler, left panel) and amsh1-1 (right panel) are shown.
(H) Quantification of MDC-staining positive area per cell in the wild type (Ler) and amsh1-1 (n = 831 for Ler and n = 749 for amsh1-1). Photographs taken in (G) were analyzed by the FluoView software, and the values of the wild type were set to 100%. Error bars indicate se.
amsh1-1 Is Defective in Autophagic Degradation
To further assess the autophagy pathway in amsh1-1, we monitored the abundance of ATG8, which is a structural component of autophagosomes (Yoshimoto et al., 2004). ATG8 can be used as an autophagy marker, since its accumulation implies defect in the autophagy pathway. ATG8 accumulates either upon inhibition of degradation by the application of protease inhibitors like E-64d (Inoue et al., 2006) or in mutants defective in autophagosome formation (Yoshimoto et al., 2004; Thompson et al., 2005; Phillips et al., 2008; Chung et al., 2010). Indeed, when wild-type and atg7-2 (Hofius et al., 2009) seedlings were incubated with E-64d, E64-d–induced accumulation of ATG8 was observed in the wild type, while atg7-2 accumulated ATG8 without E-64d (see Supplemental Figure 2C online).
The amount of ATG8 was comparable in light-grown wild type and amsh1-1 (Figure 4C, top panel). However, when the seedlings were grown in the dark for 5 d to induce autophagy, high amounts of ATG8 accumulated in amsh1-1 in comparison with the wild type (Figure 4C, bottom panel), suggesting that amsh1-1 is less efficient in autophagic degradation. The accumulation of ATG8 was enhanced in both genotypes upon E-64d treatment, indicating that even in amsh1-1, autophagic degradation was not completely inhibited. Transcript levels of all ATG8 isoforms in amsh1-1 were comparable to the wild type under this condition (see Supplemental Figure 2E online), inferring that the accumulation of ATG8 is not a consequence of transcriptional misregulation in amsh1-1.
We next wanted to examine whether selective autophagy is also impaired in amsh1-1. As opposed to bulk autophagy, during selective autophagy, specific cargo proteins are recognized by cargo adaptors and degraded via the autophagy pathway. NEIGHBOR of BRCA1 GENE1 (NBR1) is a cargo receptor and substrate of selective autophagy (Svenning et al., 2011). NBR1 accumulated in atg7-2 as previously reported, but not in amsh1-1 (Figure 4D; see Supplemental Figure 2D online), suggesting that NBR1-mediated selective autophagy is not defective in amsh1-1. The fact that amsh1-1 does not show bulk and selective autophagic degradation defects in light suggests that residual AMSH1 in amsh1-1 is sufficient for both processes in light. However, the low amount of AMSH1 probably becomes limiting in the dark, where bulk autophagy, but not NBR1-mediated selective autophagy, is highly activated.
To distinguish between the defects in autophagic protein degradation and autophagosome formation, we made use of the dye monodansylcadaverine (MDC), which stains autophagic bodies (Contento et al., 2005) upon treatment with the vacuolar protease inhibitor E-64d. E-64d inhibits the degradation of autophagic bodies in the vacuole and thus causes accumulation of MDC-stained autophagic bodies in the vacuoles stained with 2′,7′-bis-(2-carboxyethyl)-5-(and 6)carboxyfluorescein acetoxymethyl ester (BCECF-AM) (Figure 4E). The accumulation of MDC-positive aggregates was not detected in atg7-2 (see Supplemental Figure 2A online), indicating that although it accumulates ATG8, the formation of autophagosomes is not visible in this mutant. Furthermore, E-64d–induced MDC-positive vacuolar aggregates colocalized with the autophagosome marker GFP-ATG8e (Contento et al., 2005) (Figure 4F), but not with the late-endosome marker YFP-ARA7 (Geldner et al., 2009) (see Supplemental Figure 2B online), corroborating the specificity of MDC staining under our experimental conditions.
amsh1-1, in contrast with atg7-2, is not impaired in the formation of autophagosomes, since MDC-stained compartments were visible in amsh1-1. However, accumulation of E-64d–induced aggregates was less apparent in amsh1-1 compared with the wild type when maximal projection images of z-stacks obtained by confocal microscopy were analyzed (Figure 4G). Subsequent quantification of the thus imaged MDC-positive signal area in individual cells (n = 831 for the wild type, and n = 749 for amsh1-1) showed that amsh1-1 had reduced accumulation of autophagic bodies in the vacuole when compared with the wild type (68% of average MDC-stained area per cell compared with the wild type) (Figure 4H). This result indicates that autophagosomes are less efficiently targeted to the vacuole in amsh1-1.
AMSH1 Interacts with the ESCRT-III Subunit VPS2.1 through Its MIT Domain
We next investigated whether the inhibition of bulk autophagic degradation in amsh1-1 is a consequence of altered intracellular trafficking. For this purpose, we examined the interaction of AMSH1 with the endocytosis machinery. AMSH1, like AMSH3, contains an N-terminal microtubule interacting and trafficking (MIT) domain, which we had previously shown to be important for the interaction with the MIT interacting motif (MIM) present in ESCRT-III subunits (Katsiarimpa et al., 2011). We therefore tested in a directed yeast two-hybrid (Y2H) assay whether AMSH1 can also interact with the ESCRT-III core subunits VPS2.1, VPS20.1, VPS24.1, and SUCROSE NON-FERMENTING7.1 (SNF7.1) (Winter and Hauser, 2006; Richardson et al., 2011; Shahriari et al., 2011). Among the four tested subunits, AMSH1 interacted only with VPS2.1 (Figure 5A; see Supplemental Figure 3A online).
Figure 5.
AMSH1 Interacts with ESCRT-III Subunit VPS2.1 through the MIT Domain.
(A) Y2H analysis of GBD-AMSH1 with GAD-fused ESCRT-III subunits VPS2.1, VPS20.1, VPS24.1, and SNF7.1. Transformants were plated on medium lacking Leu, Trp, and His (-LWH) supplemented with 5 mM 3-Amino-1,2,4-triazole (3-AT) (left panel) to test for their auxotrophic growth or on medium lacking Leu and Trp (-LW) (right panel).
(B) Y2H analysis of GBD-AMSH1 (FL; full-length) and GBD-AMSH1(ΔMIT) with GAD-VPS2.1. Transformants were grown on –LWH+6 mM 3-AT (left panel) or -LW (right panel) medium to test for their auxotrophic growth.
(C) In vitro binding assay of the MIT domain of AMSH1 with GST or GST-fused MIM domain of VPS2.1. After GST pulldown, bead-bound proteins were analyzed by immunoblotting using anti-AMSH1 and anti-GST antibodies. Arrowheads indicate the positions of AMSH1(MIT), and asterisks indicate degradation products.
[See online article for color version of this figure.]
The MIT domain is necessary for the interaction of AMSH1 with VPS2.1 since deletion of this domain led to loss of interaction (Figure 5B; see Supplemental Figure 3B online). Moreover, AMSH1 and VPS2.1 interact directly through the 154–amino acid MIT and 17–amino acid MIM region, respectively, as shown by an in vitro binding assay (Figure 5C). Altogether, these data indicate that AMSH1, like AMSH3, may play a role in ESCRT-III-mediated intracellular trafficking pathway by interacting with VPS2.1.
Overexpression of VPS2.1-GFP Causes Inhibition of Endocytosis
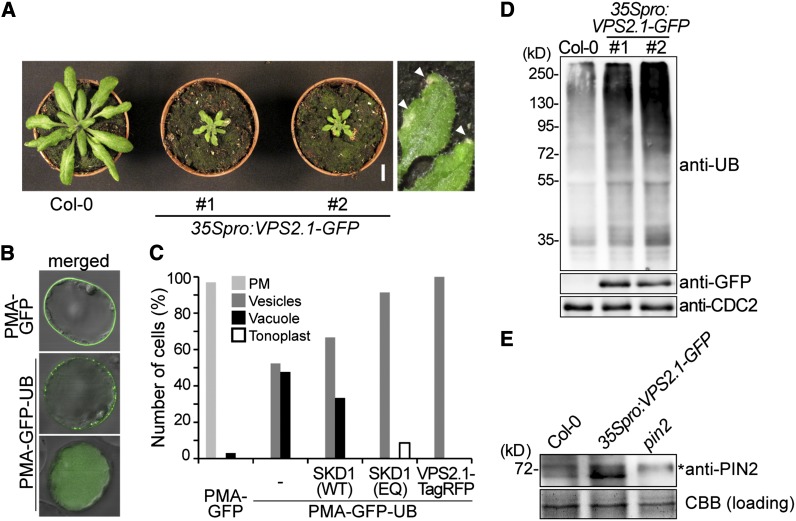
To investigate whether VPS2.1 is also involved in the same physiological pathway as AMSH1, we generated transgenic plants that express VPS2.1 as a C-terminal fusion with green fluorescent protein (35Spro:VPS2.1-GFP). C-terminal fusions of ESCRT-III subunits, including VPS2.1, with large proteins, such as GFP, have been shown to disturb ESCRT-III function in other organisms due to their aggregation in class-E compartments (Howard et al., 2001; Teis et al., 2008; Teis et al., 2010). Indeed, we found that 35Spro:VPS2.1-GFP plants occasionally showed abnormal growth already at the seedling stage (see Supplemental Figure 4A online) and that part of the overexpressed VPS2.1-GFP localized to large aggregates in the cytosol that may represent class-E compartments (see Supplemental Figure 4B online). After transfer to soil, adult 35Spro:VPS2.1-GFP plants, similarly to amsh1-1/amsh1-1 amsh3-2/AMSH3 plants, showed early leaf senescence and a severe dwarf phenotype (Figure 6A) and were mostly sterile (data not shown).
Figure 6.
VPS2.1-GFP–Overexpressing Seedlings Are Deficient in Endocytosis and Accumulate PIN2.
(A) VPS2.1-GFP–overexpressing plants are dwarf and exhibit an early senescence phenotype. Plants of two independent transgenic lines (#1 and #2) are shown together with a wild-type plant (Col-0) of the same age. Magnification of senescing leaves (arrowheads) is shown on the far right. Bar = 1cm.
(B) Analysis of the differential intracellular distribution of PMA-GFP and PMA-GFP-UB. PMA-GFP shows predominantly plasma membrane localization (top panel), while PMA-GFP-UB signals are found in vesicles and the vacuolar lumen (middle and bottom panels, respectively). Photographs are merged with differential interference contrast pictures of the corresponding cell.
(C) Localization of PMA-GFP and PMA-GFP-UB upon coexpression with SKD1 and VSP2.1 constructs. Cell culture–derived protoplasts expressing PMA-GFP alone (n = 33), PMA-GFP-UB (n = 128) alone or with 35Spro:HA-SKD1(WT) (n = 21), 35Spro:HA-SKD1(EQ) (n = 116), and 35Spro:VPS2.1-TagRFP (n = 40) were analyzed by confocal microscopy. Relative number of cells showing each localization was scored. Light-gray bars, plasma membrane; dark-gray bars, vesicles; solid bars, vacuole lumen; and open bars, tonoplast.
(D) VPS2.1-GFP–overexpressing plants accumulate ubiquitinated proteins. Total extract of wild-type (Col-0) and 35Spro:VPS2.1-GFP seedlings were subjected to immunoblotting using anti-UB, anti-GFP, and anti-CDC2 antibodies. CDC2 was used as a loading control.
(E) 35Spro:VPS2.1-GFP seedlings accumulate PIN2. Membrane protein extract from the roots of wild-type (Col-0), 35Spro:VPS2.1-GFP, and pin2 seedlings were subjected to immunoblotting using an anti-PIN2 antibody. A representative immunoblot is shown. Coomassie blue (CBB)–stained protein bands were used as a loading control. The asterisk indicates an unspecific band.
We then tested whether the expression of VPS2.1-GFP has an inhibitory effect on endocytosis of plasma membrane cargo. For this purpose, we examined the endocytosis of an artificial MVB cargo PMA-GFP-UB (Herberth et al., 2012) upon coexpression with 35Spro:VPS2.1-TagRFP, a construct expressing VPS2.1 with a C-terminal TagRFP in Arabidopsis cell culture–derived protoplasts. Fusion of monoubiquitin to PMA-GFP alters the intracellular distribution of this normally plasma membrane–localized protein, which then becomes visible in vesicles and in the vacuolar lumen (Figure 6B). SKD1/Vps4p is an AAA-ATPase required for the disassembly of ESCRT-III (Babst et al., 1997). As previously reported, coexpression of an inactive SKD1(EQ), but not that of wild-type SKD1(WT), results in the inhibition of vacuolar targeting of PMA-GFP-UB (Figure 6C). Similarly, we found PMA-GFP-UB signals to be excluded from the vacuolar lumen upon coexpression with 35Spro:VPS2.1-TagRFP, suggesting that the C-terminal fluorophore fusion of VPS2.1 has an inhibitory effect similar to SKD1(EQ) (Figure 6C).
We next analyzed the vacuolar degradation of the auxin efflux facilitator PIN2, a well-characterized cargo of the MVB pathway (Abas et al., 2006; Spitzer et al., 2009). While no apparent accumulation of PIN2 was observed in amsh1-1 (see Supplemental Figure 4C online), ubiquitinated proteins as well as PIN2 accumulated in 35Spro:VPS2.1-GFP seedlings at a higher level compared with wild-type seedlings (Figures 6D and 6E). Since the transcript levels of PIN2 were not consistently and strongly upregulated in 35Spro:VPS2.1-GFP seedlings (see Supplemental Figure 4D online), accumulation of PIN2 is probably a consequence of a posttranscriptional mechanism. Together, these results indicate that the overexpression of the dominant-negative VPS2.1-GFP has an inhibitory effect on the degradation of (ubiquitinated) MVB cargo, probably due to impaired ESCRT-III and MVB function.
VPS2.1-GFP–Overexpressing Plants Are Defective in Autophagic Degradation
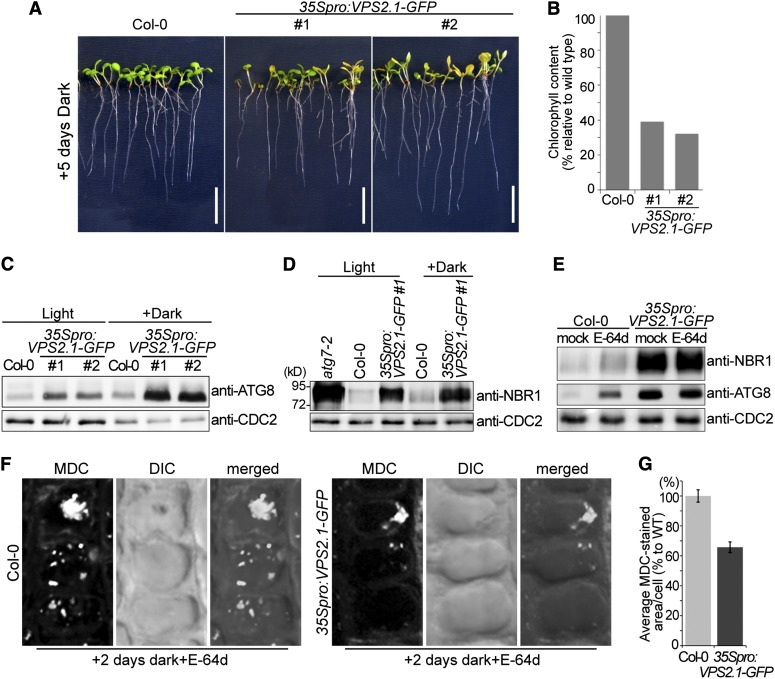
Since our studies had implicated the VPS2.1-interactor AMSH1 in autophagy, we examined the response of VPS2.1-GFP–overexpressing plants to dark treatment. After 5 d in the dark, 35Spro:VPS2.1-GFP seedlings, like amsh1-1 seedlings, showed strong chlorosis and reduction in chlorophyll content (39 and 32% compared with the wild type) (Figures 7A and 7B).
Figure 7.
VPS2.1-GFP–Overexpressing Seedlings Have Defects in Both Nonselective and Selective Autophagy.
(A) 35Spro:VPS2.1-GFP seedlings exhibit starvation-induced chlorosis after dark treatment. Long-day-grown wild-type (Col-0) and 35Spro:VPS2.1-GFP seedlings were transferred to the dark for 5 d. Note that the 35Spro:VPS2.1-GFP lines segregate for the transgene. Bars = 1 cm.
(B) Chlorophyll content of seedlings shown in (A).
(C) 35Spro:VPS2.1-GFP seedlings accumulate ATG8. Immunoblot using an anti-ATG8 antibody on total protein extract from wild-type (Col-0) and 35Spro:VPS2.1-GFP seedlings grown under long-day conditions for 12 d (Light) or transferred to dark after 7 d for 5 d (Dark). CDC2 was used as loading control.
(D) NBR1 accumulates in 35Spro:VPS2.1-GFP seedlings. Total protein extracts of atg7-2, the wild type (Col-0), and 35Spro:VPS2.1-GFP grown as in (C) were analyzed by immunoblotting using an anti-NBR1 antibody. CDC2 was used as loading control.
(E) E-64d does not enhance the accumulation of NBR1 and ATG8 in 35Spro:VPS2.1-GFP. Seedlings grown in the dark for 5 d were treated with E-64d for 6 h, and total extracts were submitted to immunoblotting with anti-NBR1 and anti-ATG8 antibodies. CDC2 was used as loading control. Note that E-64d enhances the accumulation of NBR1 and ATG8 only in the wild type (Col-0).
(F) Wild-type (Col-0) and 35Spro:VPS2.1-GFP seedlings were grown for 7 d in long days and subsequently 2 d in the dark. Seedlings were then treated with E-64d for 1 h and stained with MDC. Confocal images (maximal projection) of MDC-stained root epidermal cells of the wild type (Col-0, left panel) and 35Spro:VPS2.1-GFP (right panel) are shown. DIC, differential interference contrast.
(G) Quantification of MDC-stained area per cell in the wild type (WT; Col-0) and 35Spro:VPS2.1-GFP (n = 547 for Col-0, and n = 376 for 35Spro:VPS2.1-GFP). Photographs taken in (F) were analyzed by the FluoView software and the value of wild type was set to 100%. Error bars indicate se.
When we analyzed ATG8 protein abundance in 35Spro:VPS2.1-GFP seedlings, more ATG8 was detected even under normal growth conditions and a further increase was observed after transfer to the dark (Figure 7C). Expression of ATG8a-i was not increased in these lines, indicating that the accumulation of ATG8 is not a consequence of transcriptional misregulation of ATG8 genes (see Supplemental Figure 4F online).
Next, we examined the protein level of NBR1 in VPS2.1-GFP–overexpressing plants. In contrast with amsh1-1, in which NBR1 accumulation was not observed, NBR1 accumulated at high levels in 35Spro:VPS2.1-GFP plants (Figure 7D), while its transcript level remained unchanged (see Supplemental Figure 4E online). In contrast with ATG8, however, the amount of accumulated NBR1 remained unaltered after dark treatment, suggesting that NBR1-dependent selective autophagy is not dark induced. Treatment with E-64d did not enhance the accumulation of either NBR1 or ATG8 (Figure 7E), indicating that 35Spro:VPS2.1-GFP is severely impaired in autophagic degradation. Since, in contrast with amsh1-1, 35Spro:VPS2.1-GFP plants are incapable of dealing with basal levels of bulk and selective autophagic degradation in the light, we can conclude that overexpression of VPS2.1-GFP has a much stronger inhibitory effect on autophagic degradation than does the knockdown of AMSH1.
A report on Caenorhabditis elegans ESCRT-III RNAi mutants suggests that the accumulation of autophagosomes in ESCRT-III mutants is a consequence of the activation of autophagy due to a protective mechanism (Djeddi et al., 2012). In Arabidopsis, activation of autophagy, for example, after dark treatment, was shown to be coupled with transcriptional upregulation of ATG8 (Doelling et al., 2002; Thompson et al., 2005). Thus, the comparable transcript levels of ATG8 isoforms in amsh1-1 and 35Spro:VPS2.1-GFP with their corresponding wild types imply that autophagic flux is not increased in these mutants.
To examine the behavior of autophagosomes in dark-treated 35Spro:VPS2.1-GFP, we stained E-64d–treated seedlings with MDC. We reasoned that if VPS2.1 function is necessary for the autophagic degradation, cells should accumulate less MDC-stained autophagic bodies in the vacuoles of 35Spro:VPS2.1-GFP. MDC-stained vacuolar aggregates were observed in 35Spro:VPS2.1-GFP, indicating that autophagosomes formation is not inhibited (Figure 7F). Further quantification showed that the area of MDC-stained autophagic bodies per cell was smaller in 35Spro:VPS2.1-GFP cells (65.8% of average MDC-stained area per cell compared with the wild type; n = 547 for the wild type and n = 376 for 35Spro:VPS2.1-GFP) (Figure 7G), implying that VPS2.1-GFP overexpression and the partial loss of AMSH1 function lead to similar defects in autophagic degradation.
amsh1-1 Displays Altered Pathogen Susceptibility
Recent studies on atg mutants showed the involvement of autophagy in plant innate immunity and pathogen defense (Liu et al., 2005; Patel and Dinesh-Kumar, 2008; Hofius et al., 2009; Yoshimoto et al., 2009; Lai et al., 2011; Lenz et al., 2011; Wang et al., 2011). As amsh1-1 and 35Spro:VPS2.1-GFP showed impaired autophagic degradation, we hypothesized that these mutants may also exhibit altered pathogen response. Since VPS2.1-GFP–overexpressing plants were dwarf and had altered leaf size and morphology, we decided to conduct the pathogen assays only with amsh1-1. We used 5- to 6-week-old amsh1-1 plants grown under 10-h-light/14-h-dark conditions because under this condition amsh1-1 did not show early senescence (see Supplemental Figure 1E online).
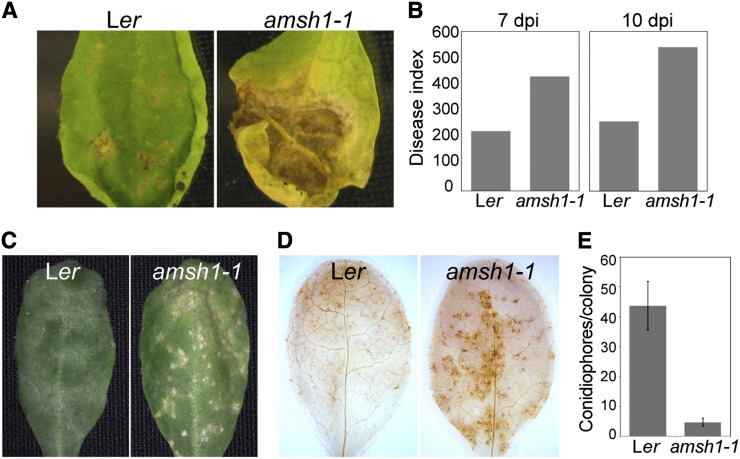
Wild-type and amsh1-1 plants were inoculated with the Arabidopsis pathogens Alternaria brassicicola or Erysiphe cruciferarum. A. brassicicola is a necrotrophic fungus that induces cell death upon infection. In comparison with the wild type, in which cell lesion was restricted to the inoculation spots, amsh1-1 showed increased susceptibility, visible by the complete wilting and spreading necrosis along the entire leaf 10 to 11 d after infection (Figures 8A and 8B). Thus, similarly to previously characterized atg mutants, amsh1-1 is also hypersensitive to A. brassicicola infection.
Figure 8.
amsh1-1 Displays Altered Susceptibility to Mildew Infection.
(A) Wild-type (Ler) and amsh1-1 plants were grown under short-day conditions (10 h light/14 h dark). Rosetta leaves of 5.5-week-old plants were drop-inoculated with 106 spores/mL A. brassicicola, and a representative leaf was photographed after 11 d of inoculation.
(B) Disease indices from experiments shown in (A) were calculated 7 d (7 dpi) and 10 d (10 dpi) after inoculation with A. brassicicola spores.
(C) Disease symptoms of wild-type (Ler) and amsh1-1 plants upon infection with E. cruciferarum spores. Plants were grown as in (A), and 6-week-old plants were inoculated with E. cruciferarum spores. A representative leaf was photographed 7 d after inoculation.
(D) amsh1-1 plants accumulate hydrogen peroxide. Leaves of wild-type (Ler) and amsh1-1 plants 2 d after infection with E. cruciferarum spores were stained with DAB for hydrogen peroxide accumulation, which is indicated by brown stain.
(E) Quantification of conidiophores per spore. Leaves were stained with trypan blue 6 d after inoculation with E. cruciferarum spores, and the number of conidiophores per colony was counted. Error bars indicate se.
E. cruciferarum is an obligate biotroph that requires living cells for growth. In the wild type, E. cruciferarum infection did not cause visible cell lesions on leaves 5 d after infection (Figure 8C, left panel). However, at the same stage, amsh1-1 showed spontaneous cell death on leaves (Figure 8C, right panel). This pathogen-induced cell death in amsh1-1 was accompanied by an increased production of reactive oxygen species (Figure 8D). We also counted fewer conidiophores per spores on amsh1-1 leaves when compared with the wild type (Figure 8E; see Supplemental Figures 5A to 5C online), indicating that amsh1-1 is more resistant against E. cruciferarum as an indirect consequence of programmed cell death, which is triggered in the mutant.
Transcriptional Upregulation of Defense-Related Genes May Be Responsible for the Early Senescence of amsh1-1 and 35Spro:VPS2.1-GFP
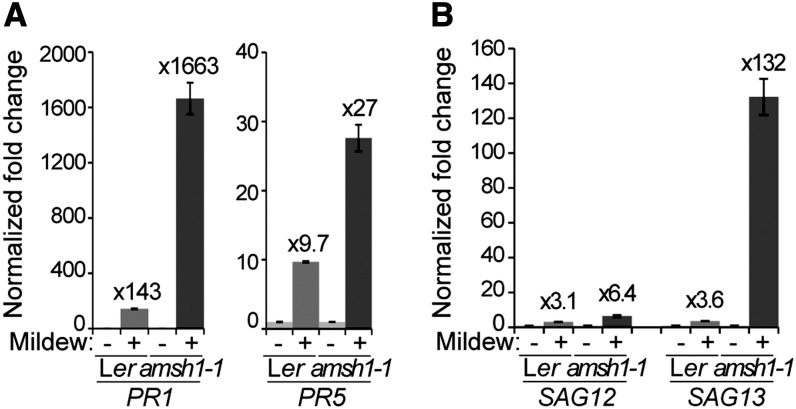
To further investigate the molecular mechanism underlying the amsh1-1 pathogen response phenotype, we examined the expression of defense-related genes in wild-type and amsh1-1 plants after infection with E. cruciferarum. Upon pathogen infection, basal immune responses, which are dependent on salicylic acid (SA), are upregulated and the expression of typical SA marker genes, such as PATHOGENE-RELATED GENE1 (PR1) and PR5 (Ward et al., 1991) as well as SENESCENCE ASSOCIATED GENE12 (SAG12) and SAG13 (Morris et al., 2000; Brodersen et al., 2002), is induced (reviewed in Wiermer et al., 2005).
Two days after infection with E. cruciferarum spores, a strong induction of PR1, PR5, SAG12, and SAG13 was detected both in the wild type and in amsh1-1 (Figures 9A and 9B). However, the induction of PR1, PR5, and SAG13 was much stronger in amsh1-1 (Figures 9A and 9B), suggesting that the SA signaling pathway is hyperactivated in amsh1-1. High PR gene transcript accumulation in association with programmed cell death may be the cause for the enhanced resistance phenotype toward E. cruciferarum while conferring enhanced susceptibility to a necrotrophic pathogen such as A. brassicicola.
Figure 9.
Transcript Levels of Pathogen- and Senescence-Related Genes Are Highly Upregulated in amsh1-1 after Mildew Infection.
(A) and (B) Relative gene expression of pathogen-related genes PR1 and PR5 (A) and senescence-related genes SAG12 and SAG13 (B) in wild-type (Ler) and amsh1-1 plants uninfected or infected with E. cruciferarum spores. Total RNA was extracted from plants 2 d after infection. Expression levels were normalized to the reference gene ACT8 and the expression levels in mock-treated Ler and amsh1-1 plants were set to 1 in each experiment. Error bars indicate se.
We then tested the possibility that the early senescence phenotype of amsh1-1 under short-day conditions (8 h light/16 h dark) is also a consequence of the upregulation of SA-induced genes. Indeed, a strong upregulation of PR1, PR5, and SAG13 was observed in 8-week-old amsh1-1, specifically when grown under 8 h light/16 h dark, but not under 10 h light/14 h dark (see Supplemental Figures 6A and 6B online). This suggests a correlation of the photoperiod-dependent early senescence phenotype (see Supplemental Figure 1D online) with the hyperactivated SA signaling in amsh1-1. Similar transcriptional upregulation of PR genes and SAG13 was also found in the 35Spro:VPS2.1-GFP seedlings (see Supplemental Figure 6C online).
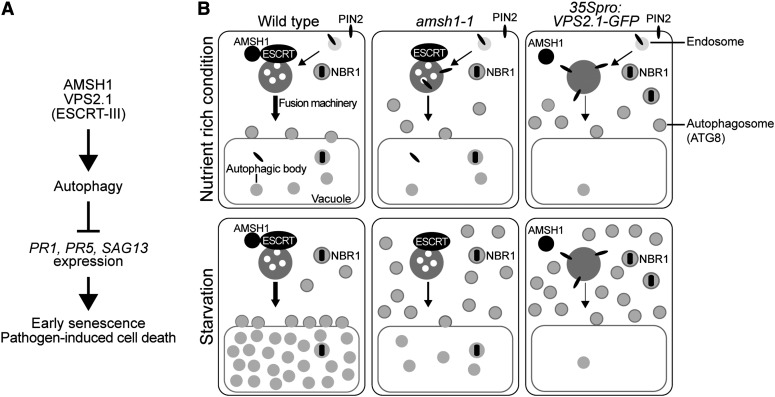
Altogether, these results suggest that AMSH1 and VPS2.1 are necessary for proper autophagy in plants and, thus, for physiological processes depending on an intact autophagy pathway (Figures 10A and 10B).
Figure 10.
AMSH1 and ESCRT-III Are Important for Autophagy and Autophagy-Mediated Physiological Responses in Plants.
(A) AMSH1 and VPS2.1 functions are important for autophagic degradation. As previously reported, functional autophagy represses PR and SAG gene expression, induction of which causes early senescence and pathogen-induced cell death.
(B) amsh1-1 and 35Spro:VPS2.1-GFP are impaired in the degradation of autophagosomes to different extents. In wild-type cells, plasma membrane–localized PIN2 is endocytosed via the MVB pathway. The selective autophagy cargo receptor and substrate NBR1 is delivered to the vacuole via autophagosomes. Autophagosomes fuse with the vacuole in order to degrade its contents. Factors required for the recognition and fusion of autophagosomes with the vacuolar membrane as well as proteases responsible for the degradation of autophagic bodies may be transported via an MVB-dependent pathway. Upon carbon deprivation in the dark, autophagic recycling is highly upregulated. The weak amsh1-1 knockdown mutant is still capable of endocytic and autophagic degradation under optimal growth condition. However, when bulk autophagy is highly activated upon dark-induced carbon starvation, amsh1-1 accumulates ATG8 and shows less autophagic bodies in the vacuole, indicative for impaired autophagic degradation. When ESCRT-III function is disturbed by overexpressing VPS2.1-GFP, both endocytosis and autophagic recycling is strongly inhibited even under optimal growth conditions, leading to the accumulation of ATG8, NBR1, and PIN2. Accumulation of ATG8 increases when bulk autophagy is activated in the dark. Taken together, intact AMSH1 and ESCRT-III (VPS2.1) are essential for proper autophagic degradation.
DISCUSSION
In this study, we showed that AMSH1, an AMSH3-related DUB, interacts with the endocytosis machinery through the ESCRT-III subunit VPS2.1. AMSH1 and AMSH3 possess an N-terminal MIT domain and a C-terminal MPN domain (Isono et al., 2010) but share only 47% overall amino acid identity, suggesting that the two proteins might have similar biochemical properties but also specific physiological functions. The fact that AMSH1 and AMSH3 are not products of a recent duplication event in Arabidopsis, that they show in part mutually exclusive expression patterns, and that in contrast with AMSH3, AMSH1 does not interact with VPS24.1 in Y2H interaction studies might further support this hypothesis.
Mutant analyses showed that AMSH1 and VPS2.1 are part of the autophagic degradation pathway and are important for plant survival in the dark, regulation of senescence, and pathogen defense. Phenotypes of amsh1-1 and 35Spro:VPS2.1-GFP include early senescence and chlorosis in the dark. At the molecular level, they both accumulate ubiquitinated proteins, and though to different degrees, both accumulate ATG8 and show defects in autophagosome delivery to the vacuole, indicating the involvement of AMSH1 and VPS2.1 in autophagic degradation. However, in contrast with 35Spro:VPS2.1-GFP, amsh1-1 does not show accumulation of PIN2. This might be due to the nature of the amsh1-1 allele, which allows a residual amount of AMSH1 to still exist. It is a future challenge to investigate whether AMSH1 directly targets ubiquitinated MVB cargos or rather components of the trafficking machinery. Taken together, our results imply that an intact MVB pathway with its associated DUB AMSH1 is also essential for autophagy in plants and, hence, for autophagy-mediated physiological processes.
Although there is a strong indication that SA-dependent basal immunity and cell death are indirectly deregulated in amsh1-1, we cannot exclude the possibility that AMSH1 and also VPS2.1 may directly affect plant defense. Receptor-mediated endocytosis and exocytosis are known to play important roles in plant immune response, and many specific and general players involved in this pathway have been identified. For example, PENETRATION1 (PEN1)/SYNTAXIN OF PLANTS121 (SYP121) is a SNARE protein that contributes to the delivery of antimicrobial compounds to the infection site (Collins et al., 2003). PEN1 and multiple other components of the intracellular trafficking pathway, including VAMP72 proteins, GNOM, an ESCRT-I subunit VPS28, and MVBs, have been shown to relocate to pathogen infection sites (Assaad et al., 2004; An et al., 2006; Kwon et al., 2008; Lu et al., 2012; Nielsen et al., 2012).
In contrast with syp122, syp42/syp43, which is defective in trans-Golgi dependent intracellular trafficking, shows increased susceptibility to the obligate biotroph Golovinomyces orontii (Zhang et al., 2007; Uemura et al., 2012). Similarly, ESCRT-I mutants are also more susceptible to infection with the obligate biotrophic oomycete Hyaloperonospora arabidopsidis (Lu et al., 2012), in contrast with amsh1-1, which is more resistant to the obligate biotroph E. cruciferarum. These apparently conflicting results might reflect the complexity of regulatory mechanisms with a different outcome depending on the mutant-pathogen combination.
Dysfunction of AMSH1 and VPS2.1 may inhibit trafficking of autophagosomes to the vacuole. The process of heterotypic membrane fusion by which autophagosome membranes are combined with other membranes is not yet well understood. One group of proteins important for proper and efficient fusion between membranes is the SNAREs, which reside on membranes and form heteromeric complexes. A recent article reported the identification of human Syntaxin 7 as an autophagosomal SNARE, essential for autophagosome fusion to endosomes and lysosomes (Itakura et al., 2012). Similar fusion mechanisms involving SNAREs might also take place in plants. A family of Rab GTPases, Ypt7p/Rab7/RAB7, was also shown to affect fusion of autophagosomes with endosomes and vacuoles in yeast (Balderhaar et al., 2010), human cell culture (Gutierrez et al., 2004; Jäger et al., 2004), and plants (Kwon et al., 2010). In amsh1-1 and VPS2.1-GFP–overexpressing plants, proteins required for docking and/or fusion between autophagosomes and the target membrane might be mistargeted or reduced, leading to fusion defects. Alternatively, the ESCRT machinery and AMSH1 may be directly involved in the heterotypic fusion event. Future experiments are needed to identify and reveal the identity of the factors involved in this process and elucidate the molecular mechanism of their regulation.
METHODS
Biological Material
All experiments were performed with Arabidopsis thaliana (Columbia-0 [Col-0] or Landsberg erecta [Ler] background). T-DNA insertion lines of AMSH1, designated amsh1-1 (CSHL_ET8678; Ler ecotype) and AMSH2 (CSHL_ET4018; Ler ecotype) were obtained from the Martienssen Lab (Cold Spring Harbor Laboratory). The T-DNA insertion site of amsh1-1 was identified using the specific T-DNA primer DS3.1 in combination with the gene-specific AMSH1 reverse primer and the wild type with the combination of AMSH1 forward and AMSH1 reverse primers. T-DNA insertion mutants atg7-2 (Hofius et al., 2009) and pin2 (Willige et al., 2011) in the Col-0 background were described previously.
Plant transformations were performed using the floral dip method (Clough and Bent, 1998). Seedlings were grown in continuous light, long-day (16 h light and 8 h dark), or short-day (8 h light and 16 h dark or 10 h light and 14 h dark) conditions, as indicated for each experiment, at 110 to 120 µmol m−2 s−1 light intensity. Standard Murashige and Skoog (MS) growth medium (Duchefa Biochemie) supplemented with 1% Suc or half-strength MS (2.15 g/L MS and 2.3 mM MES, pH 5.7) was used to grow seedlings, and adult plants were grown in soil.
For exposure to artificial starvation or carbon deprivation, 7-d-old seedlings grown on half-strength MS or 3-week-old plants grown on soil under long-day (16 h daylight/8 h dark) conditions were transferred to the dark for 2 or 5 d as indicated for each experiment.
Cloning Procedures
All primers used for cloning and subcloning are listed in Supplemental Table 1 online. Detailed cloning procedures are described in Supplemental Methods 1 online.
Molecular Phylogeny
The nucleotide sequences from various species were identified in Phytozome (http://www.phytozome.net) according to their similarity to Arabidopsis AMSH genes, aligned with ClustalX (Thompson et al., 1997), and then improved manually. Phylogenetic analyses were performed by maximum likelihood with PAUP using the sequence from Physcomitrella patens (Pp1s133_43V6) as an outgroup. All characters were equally weighted, and gaps were treated as missing data. The nucleotide substitution model was set as GTR + I + G by MrModeltest 2.2 (Nylander, 2004), and a heuristic search was implemented with 100 random addition sequence replicates involving TBR branch swapping.
To estimate clade credibility, bootstrap values by maximum parsimony method, and posterior probabilities by Bayesian analysis were calculated. Bootstrap values were calculated from 1000 pseudo-replicates, each with 100 random additions. For posterior probabilities, the Bayesian search was conducted by MCMC with two independent sets of four chains, each run for 10 million generations, sampling every 100 generations by MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). The nucleotide substitution model was set as GTR + I + G. The program Tracer (Tracer v1.4; available from http://beast.bio.ed.ac.uk/Tracer) was used to check the runs had reached stationarity and effective sample size of all the parameters was high (>100). The first 2.5 million generations before sufficient stationary generations were discarded as burn-in periods and the rest of trees were used to calculate posterior probabilities.
GUS Assay
Excised mature embryos from seeds as well as 7- and 14-d-old seedlings were treated with heptan (Roth) for 15 min. After removing heptan, the embryos and seedlings were incubated in GUS substrate solution (50 mM sodium phosphate, pH 7, 10 mM EDTA, 0.5 mM K4[Fe(CN)6], 0.5 mM K3[Fe(CN)6], 0.5 mM X-Gluc, and 0.02% Silvett) at 37°C. Tissues were cleared with an ethanol:acetic acid solution (6:1) at 37°C for 1 h and subsequently with a series of decreasing percentage of ethanol. Photographs were taken using a MZ16 (Leica) or BX61 (Olympus) microscope equipped with a charge-coupled device camera.
Chlorophyll Content Measurement
Twenty seedlings were weighed and immediately incubated in 1 mL N,N-dimethylformamide at 4°C under agitation in dark. After 48 h, the absorbance of the supernatant was measured at 664 and 647 nm. Subsequently, total chlorophyll content was determined according to total chlorophyll = [(OD664*7.04) + (OD647*20.27)]/fresh weight (Porra et al., 1998).
Quantitative RT-PCR
All primers used for quantitative RT-PCR are listed in Supplemental Table 2 online. Total RNA was extracted with a NucleoSpin RNA plant kit (Machery-Nagel), and 1 µg of total RNA was reverse transcribed with an oligo(dT) primer and M-MulV reverse transcriptase (Fermentas) following the manufacturer’s instructions. Quantitative real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a CFX96 real-time system cycler (Bio-Rad). A 45-cycle two-step amplification protocol (10 s at 95°C, 25 s at 60°C) was used for all measurements.
Protein Extraction, Immunoblotting, and Antibodies
Yeast total proteins were extracted as described previously (Kushnirov, 2000). SDS-PAGE and immunoblotting were performed according to standard methods.
Total protein extracts were prepared in extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, and protease inhibitor cocktail [Roche]). For PIN2 immunoblot analysis, roots of 10-d-old seedlings were homogenized in extraction buffer. Extracts were centrifuged for 20 min at 9000g, and the supernatant was further centrifuged for 1 h at 100,000g in a Sorvall MTX 500 benchtop centrifuge (ThermoScientific), and the P100 fraction was subjected to immunoblotting.
An anti-AMSH1 antibody was raised against the full-length protein 6xHis-AMSH1 expressed and purified from Escherichia coli Rosetta(DE3) strain (Novagen). Six hundred micrograms of purified protein was used to raise antibodies in rabbits (Eurogentec). The serum was used at a 1:1000 dilution. The specificity of the antibody was verified using total extracts from amsh1-1.
Additional antibodies used were as follows: anti-AMSH3 (Isono et al., 2010), anti-ATG8 (Thompson et al., 2005), anti-GFP (Invitrogen), anti-CDC2 (Santa Cruz), anti-GAL4BD (Santa Cruz), anti-PIN2 (Agrisera), anti-NBR1 (Svenning et al., 2011), anti-UB(P4D1) (Santa Cruz), and horseradish peroxidase–conjugated anti-HA (Sigma-Aldrich).
In Vitro DUB Assay, Glutathione S-Transferase Pulldown, and Y2H Analysis
Glutathione S-transferase (GST), GST-VPS2.1(MIM), GST-AMSH1(MPN), and GST-AMSH1(MIT) were expressed in E. coli Rosetta(DE3) cells (Merck) and purified with Glutathione Sepharose 4B (GE Healthcare). After purification, the GST moiety of GST-AMSH1(MPN) and GST-AMSH1(MIT) was removed by digestion with PreScission Protease (GE Healthcare). DUB assays and GST pull-down assays were performed as described previously (Isono et al., 2010; Katsiarimpa et al., 2011). Y2H analysis was performed as described previously (Katsiarimpa et al., 2011).
MDC and BCECF-AM Staining and E-64d Treatment
Arabidopsis seedlings were stained with 50 µM MDC (Sigma-Aldrich) in PBS for 10 min at room temperature to visualize autophagosomes. Subsequently, the seedlings were washed twice with PBS to remove excess stain. To visualize the vacuole, seedlings were incubated with 5 µM BCECF-AM (Molecular Probes) for 1 h. To inhibit vacuolar proteases, seedlings were incubated with 100 µM E-64d (Santa Cruz) for 1, 6, or 12 h as indicated for each experiment.
Microscopy
GFP-fused proteins, BCECF staining and MDC staining were analyzed with an FV-1000/IX81 confocal laser scanning microscope (Olympus) with a UPlanSApo ×60/1.20 (Olympus) objective using the 488- and 405-nm laser line, respectively. For maximal projection images of MDC-stained root cells, sequential Z-stack images were collected with 0.48-µm plane distance. Images were subsequently processed using FluoView (Olympus) and Photoshop CS6 (Adobe).
Pathogen Assays
For Alternaria brassicicola infection, 5.5-week-old plants grown under short-day conditions (10 h light/14 h dark) were drop inoculated with 106 spores/mL of A. brassicicola. Disease indices were calculated 7 and 10 d after inoculation according to Epple et al. (1997).
For the Erysiphe cruciferarum assay, 6-week-old plants grown under short-day conditions (10 h light/14 h dark) were inoculated with a density of 3 to 5 spores per mm2. Susceptibility of plants to mildew was scored by visual examination after 5 and 7 d of inoculation. To quantify fungal growth, the number of hyphae and of conidiophores per spore was counted under the microscope 5 and 7 d after inoculation, respectively, after staining the leaves with trypan blue (Pelikan).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: AMSH1 (AT1G48790), AMSH2 (AT1G10600), AMSH3 (AT4G16144), CDC2 (AT3G48750), SKD1 (AT2G27600), VPS2.1 (AT2G06530), VPS20.1 (AT5G63880), VPS24.1 (AT5G22950), SNF7.1 (AT4G29160), ATG8a (AT4G21980), ATG8b (AT4G04620), ATG8c (AT1G62040), ATG8d (AT2G05630), ATG8e (AT2G45170), ATG8f (AT4G16520), ATG8g (AT3G60640), ATG8h (AT3G06420), ATG8i (AT3G15580), PR1 (AT2G14610), PR5 (AT1G75040), SAG12 (AT5G45890), SAG13 (AT2G29350), ACT8 (AT1G49240), PIN2 (AT5G57090), and NBR1 (AT4G24690). Sequence accession numbers (Phytozome) for AMSH homologs are as follows: P. patens (Pp1s133_43V6, Pp1s240_78V6, and Pp1s64_214V6), Selaginella moellendorffii (Sm82317 and Sm128074), Zea mays (GRMZM2g075690, GRMZM5g835530, and GRMZM2g173119), Sorghum bicolor (Sb13g013600 and Sb3g020630), Oryza sativa (Os1g23640 and Os1g31470), Populus trichocarpa (0010s15100, 0015s03810, and 0010s05090), Ricinus communis (Rc29889m003259, Rc29996m000133, and Rc29631m001029), Glycine max (07g37130, 17g03490, 05g34700, 08g04970, 07g10350, 09g31540, and 01g03710), Vitis vinifera (GSVIVT01035040001, GSVIVT010083100001, and GSVIVT01013737001), and Arabidopsis lyrata (493270, 474017, and 919849).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. amsh1-1 Shows Early Senescence under 8-h Daylight Short-Day Conditions.
Supplemental Figure 2. Autophagosomes Accumulate in Wild-Type Plants after Dark Treatment.
Supplemental Figure 3. Expression of Y2H Constructs.
Supplemental Figure 4. Phenotypes of VPS2.1-GFP–Overexpressing Plants.
Supplemental Figure 5. E. cruciferarum Growth Is Restricted in amsh1-1 Plants.
Supplemental Figure 6. PR1, PR5, and SAG13 Are Upregulated in 35Spro:VPS2.1-GFP Plants under 8-h Daylight Conditions in amsh1-1 Plants.
Supplemental Table 1. List of Primers Used for Cloning.
Supplemental Table 2. List of Primers Used for qRT-PCR.
Supplemental Data Set 1. Nucleotide Sequence Alignment of Arabidopsis AMSH1, AMSH2, and AMSH3 with Their Counterparts from Other Plant Species.
Supplemental Methods 1. Cloning Procedure.
Acknowledgments
We thank Richard Vierstra (University of Wisconsin–Madison) for the anti-ATG8 antibody, Swen Schellmann (University of Cologne) for the PMA-GFP and PMA-GFP-UB constructs, Diane Bassham (Iowa State University) for the GFP-ATG8e line, Niko Geldner (University of Lausanne) for the YFP-ARA7 (Wave2y) line, Tsuyoshi Nakagawa (Shimane University) for the pGWB vectors, and the Martienssen lab (Cold Spring Harbor Laboratory) and Nottingham Arabidopsis Stock Centre for providing seeds. We also thank Natsumaro Kutsuna (University of Tokyo), Melina Zourelidou and Björn Willige (Technische Universität München) for technical advice and Marie-Kristin Nagel (Technische Universität München) for critical reading of the article. This work was supported by the following grants from the Deutsche Forschungsgemeinschaft: SCHW 751/7-1 to C.S., BR 3875/1-1 to F.B., SFB924 (B08) to R.H., and IS 221/2-2 (SPP1365/2) to E.I.
AUTHOR CONTRIBUTIONS
A.K., K.K., C.S., F.B., R.H., and E.I. designed the experiments. A.K., K.K., F.A., C.W., M.O., C.T., F.B., and E.I. performed the experiments. A.K., K.K., C.T., F.B., R.H., and E.I. analyzed the experiments. E.I. wrote the article.
Glossary
- DUB
deubiquitinating enzyme
- MVB
multivesicular body
- GUS
β-glucuronidase
- MDC
monodansylcadaverine
- BCECF-AM
2′,7′-bis-(2-carboxyethyl)-5-(and 6)carboxyfluorescein acetoxymethyl ester
- MIT
microtubule interacting and trafficking
- MIM
MIT interacting motif
- Y2H
yeast two-hybrid
- SA
salicylic acid
- Col-0
Columbia-0
- Ler
Landsberg erecta
- MS
Murashige and Skoog
- GST
glutathione S-transferase
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 Erratum. Nat. Cell Biol. 8: 424. [DOI] [PubMed] [Google Scholar]
- An Q., Ehlers K., Kogel K.H., van Bel A.J., Hückelhoven R. (2006). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172: 563–576 [DOI] [PubMed] [Google Scholar]
- Assaad F.F., Qiu J.L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., Emr S.D. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H.J., Arlt H., Ostrowicz C., Bröcker C., Sündermann F., Brandt R., Babst M., Ungermann C. (2010). The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J. Cell Sci. 123: 4085–4094 [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Odum N., Jørgensen L.B., Brown R.E., Mundy J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz D.A., Wei N., Osterlund M.T., von Arnim A.G., Staub J.M., Matsui M., Deng X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Chung T., Suttangkakul A., Vierstra R.D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149: 220–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Cope G.A., Suh G.S., Aravind L., Schwarz S.E., Zipursky S.L., Koonin E.V., Deshaies R.J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Djeddi A., Michelet X., Culetto E., Alberti A., Barois N., Legouis R. (2012). Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. J. Cell Sci. 125: 685–694 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Epple P., Apel K., Bohlmann H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007). Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179: 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H., Rubin D.M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V.A., Finley D. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Munafó D.B., Berón W., Colombo M.I. (2004). Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 117: 2687–2697 [DOI] [PubMed] [Google Scholar]
- Han J.H., Ryu H.H., Jun M.H., Jang D.J., Lee J.A. (2012). The functional analysis of the CHMP2B missense mutation associated with neurodegenerative diseases in the endo-lysosomal pathway. Biochem. Biophys. Res. Commun. 421: 544–549 [DOI] [PubMed] [Google Scholar]
- Herberth S., Shahriari M., Bruderek M., Hessner F., Müller B., Hülskamp M., Schellmann S. (2012). Artificial ubiquitylation is sufficient for sorting of a plasma membrane ATPase to the vacuolar lumen of Arabidopsis cells. Planta 236: 63–77 [DOI] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Howard T.L., Stauffer D.R., Degnin C.R., Hollenberg S.M. (2001). CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114: 2395–2404 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Ishii N., Owada Y., Yamada M., Miura S., Murata K., Asao H., Kondo H., Sugamura K. (2001). Loss of neurons in the hippocampus and cerebral cortex of AMSH-deficient mice. Mol. Cell. Biol. 21: 8626–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E., Katsiarimpa A., Müller I.K., Anzenberger F., Stierhof Y.D., Geldner N., Chory J., Schwechheimer C. (2010). The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151: 1256–1269 [DOI] [PubMed] [Google Scholar]
- Jäger S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E.L. (2004). Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117: 4837–4848 [DOI] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A., Anzenberger F., Schlager N., Neubert S., Hauser M.T., Schwechheimer C., Isono E. (2011). The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15: 1–32 [DOI] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbé S. (2009). Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Jung J.H., Yoshimoto K., Shirasu K., Park O.K. (2010). The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 64: 151–164 [DOI] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968 [DOI] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. (2007). ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17: 1561–1567 [DOI] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Lu Y.J., Schornack S., Spallek T., Geldner N., Chory J., Schellmann S., Schumacher K., Kamoun S., Robatzek S. (2012). Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 14: 682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K., Glickman M.H. (2002). MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., Urbé S. (2004). AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166: 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B. (2010). Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K., MacKerness S.A., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Nielsen M.E., Feechan A., Böhlenius H., Ueda T., Thordal-Christensen H. (2012). Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 109: 11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander, J.A. (2004). MrModeltest 2.0. (Uppsala, Sweden: Uppsala University). [Google Scholar]
- Patel S., Dinesh-Kumar S.P. (2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Urzinger M., Winkler J., Bubenzer C., Scheer H. (1998). Biosynthesis of the 3-acetyl and 13(1)-oxo groups of bacteriochlorophyll a in the facultative aerobic bacterium, Rhodovulum sulfidophilum—The presence of both oxygenase and hydratase pathways for isocyclic ring formation. Eur. J. Biochem. 257: 185–191 [DOI] [PubMed] [Google Scholar]
- Richardson L.G., Howard A.S., Khuu N., Gidda S.K., McCartney A., Morphy B.J., Mullen R.T. (2011). Protein-protein interaction network and subcellular localization of the Arabidopsis thaliana ESCRT machinery. Front. Plant Sci. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E., Gillmor C.S., Kovaleva V., Somerville C.R., Raikhel N.V. (2001). VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev. Cell 1: 303–310 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., Stenmark H. (2007). ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17: 1817–1825 [DOI] [PubMed] [Google Scholar]
- Shahriari M., Richter K., Keshavaiah C., Sabovljevic A., Huelskamp M., Schellmann S. (2011). The Arabidopsis ESCRT protein-protein interaction network. Plant Mol. Biol. 76: 85–96 [DOI] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M., Zheng H., Morita M.T., Saito C., Avila E., Blakeslee J.J., Bandyopadhyay A., Kovaleva V., Carter D., Murphy A., Tasaka M., Raikhel N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Amerik A.Y., Hochstrasser M. (1999). The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Kaneko K., Asao H., Kasai H., Endo Y., Fujita T., Takeshita T., Sugamura K. (1999). Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J. Biol. Chem. 274: 19129–19135 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S.D. (2008). Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15: 578–589 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Judson B.L., Emr S.D. (2010). ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 29: 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Kim H., Saito C., Ebine K., Ueda T., Schulze-Lefert P., Nakano A. (2012). Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., III, Koonin E.V., Deshaies R.J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahl-Goy P., Metraux J.P., Ryals J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. (2005). Plant immunity: The EDS1 regulatory node. Curr. Opin. Plant Biol. 8: 383–389 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Isono E., Richter R., Zourelidou M., Schwechheimer C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter V., Hauser M.T. (2006). Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Barberon M., Curie C., Vert G. (2011). Ubiquitination of transporters at the forefront of plant nutrition. Plant Signal. Behav. 6: 1597–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M.A., Qiu J.L., Olesen K.L., Thordal-Christensen H. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49: 302–312 [DOI] [PubMed] [Google Scholar]
- Zouhar J., Rojo E., Bassham D.C. (2009). AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]