This work examines the function and conservation of chloroplast ClpS in plants and identifies candidate Clp protease substrates in Arabidopsis plastids that are recognized by ClpS. The identification of direct candidate substrates for the Clp protease system in chloroplasts in this study represents a significant advancement because no substrates were identified previously.
Abstract
Whereas the plastid caseinolytic peptidase (Clp) P protease system is essential for plant development, substrates and substrate selection mechanisms are unknown. Bacterial ClpS is involved in N-degron substrate selection and delivery to the ClpAP protease. Through phylogenetic analysis, we show that all angiosperms contain ClpS1 and some species also contain ClpS1-like protein(s). In silico analysis suggests that ClpS1 is the functional homolog of bacterial ClpS. We show that Arabidopsis thaliana ClpS1 interacts with plastid ClpC1,2 chaperones. The Arabidopsis ClpS1 null mutant (clps1) lacks a visible phenotype, and no genetic interactions with ClpC/D chaperone or ClpPR core mutants were observed. However, clps1, but not clpc1-1, has increased sensitivity to the translational elongation inhibitor chloramphenicol suggesting a link between translational capacity and ClpS1. Moreover, ClpS1 was upregulated in clpc1-1, and quantitative proteomics of clps1, clpc1, and clps1 clpc1 showed specific molecular phenotypes attributed to loss of ClpC1 or ClpS1. In particular, clps1 showed alteration of the tetrapyrrole pathway. Affinity purification identified eight candidate ClpS1 substrates, including plastid DNA repair proteins and Glu tRNA reductase, which is a control point for tetrapyrrole synthesis. ClpS1 interaction with five substrates strictly depended on two conserved ClpS1 residues involved in N-degron recognition. ClpS1 function, substrates, and substrate recognition mechanisms are discussed.
INTRODUCTION
Proteolysis is crucial for regulation of various cellular processes as well as protein homeostasis and fitness of the cell. The regulated degradation of cellular proteins is performed by processive macromolecular enzymes, including the ATP-dependent 26S proteasome and caseinolytic peptidase (Clp) proteases (reviewed in Striebel et al., 2009; Sauer and Baker, 2011). These degradation machineries consist of large multisubunit proteolytic complexes whose active sites are sequestered within an internal chamber and the AAA+ (for ATPase associated with various cellular activities) chaperone complexes that recognize, unfold, and translocate substrates into the proteolytic cavity for selective degradation.
Clp proteases are found in almost all bacteria, mitochondria, and plastids (Yu and Houry, 2007). The bacterial Clp machine is composed of a peptidase core that forms two heptameric rings of proteolytic subunits (ClpP) stacked back to back in association with a ring-shaped AAA+ hexamer (ClpA, ClpX in Escherichia coli; ClpC, ClpE in Bacillus subtilis). ATP binding by these chaperones is sufficient for hexamer formation, whereas ATP hydrolysis is necessary for unfolding and translocation of substrates into the proteolytic chamber (Wickner et al., 1994; Weber-Ban et al., 1999; Reid et al., 2001). Importantly, AAA+ proteins use adaptor proteins to enhance or expand substrate recognition ability (reviewed in Kirstein et al., 2009). For example, stringent starvation proteinB (SspB) enhances the ClpX-mediated degradation of C-terminal small stable RNA A (SsrA)-tagged substrates, ClpC uses MecA (for medium-independent expression of competence A) for developmentally programmed protein degradation, and ClpA uses ClpS.
ClpS was discovered a decade ago as an adaptor protein specific for ClpA in E. coli (Dougan et al., 2002a; reviewed in Dougan et al., 2012). ClpS has been implicated as a key factor in the N-end rule pathway in which the regulation of the half-life of a protein is related to the identity of its N-terminal residue (Varshavsky, 1996, 2011). ClpS binds directly to N-terminal destabilizing residues (N-degron) to deliver substrates to ClpAP for degradation (Erbse et al., 2006; Schmidt et al., 2009; Schuenemann et al., 2009). ClpS has a folded C-terminal core domain for binding to the N-degron as well as interaction with ClpA through its N-terminal domain (N-domain) and an unstructured N-terminal extension for delivery of N-end rule substrates (Guo et al., 2002; Zeth et al., 2002; Erbse et al., 2006; Wang et al., 2008b; Schuenemann et al., 2009). The N-terminal portion of ClpS is also indispensable for inhibition of binding of SsrA-tagged proteins. The SsrA-tag (11 amino acids encoded by a small RNA that acts as both tRNA and mRNA) is attached covalently to the C- terminus of nascent peptide chains stalled on ribosomes, often due to truncated mRNA (Baker and Sauer, 2012). It has been suggested that binding of a single ClpS protein to the hexameric ClpA chaperone leads to the conformational changes that enable N-end rule substrate translocation into the pore and prevent SsrA-tagged protein binding (Baker and Sauer, 2012). Finally, ClpS can also enhance ClpAP-mediated removal of aggregates, probably in a N-degron independent manner (Dougan et al., 2002a). Despite the efforts from multiple labs over several years, only two natural substrates for E. coli ClpS have been discovered so far; these are putrescine aminotransferase and DNA protection during starvation protein (Ninnis et al., 2009; Schmidt et al., 2009). Therefore, the physiological significance of ClpS in bacteria remains to be understood (Dougan et al., 2010).
ClpS is also found in actinobacteria and cyanobacteria, which lack ClpA (Dougan et al., 2002a; Lupas and Koretke, 2003). The photosynthetic bacterium Synechococcus sp PCC 7942 possesses three catalytic ClpP proteins (ClpP1 to ClpP3) and one noncatalytic ClpR protein and uses two chaperone components ClpX and ClpC, as well as two ClpS paralogs (ClpS1 and ClpS2). Both ClpS1 and ClpS2 bind to ClpC but not to ClpX (Stanne et al., 2007). ClpS1 and ClpS2 were found in the soluble phase, whereas ClpS2 clearly also associated with membranes. Using gel filtration of the soluble cellular fraction, native ClpS1 eluted in a mass range up to ∼150 kD, whereas native ClpS2 also eluted at a higher mass range (>500 kD) (Stanne et al., 2007).
The chloroplast Clp protease system has evolved from the above-mentioned bacterial prototype and cyanobacterial ancestral machineries (Olinares et al., 2011a). In Arabidopsis thaliana chloroplasts, the Clp protease core complex consists of five ClpP subunits (ClpP1 and ClpP3 to ClpP 6) and four ClpR subunits (ClpR1 to ClpR4) in a known stoichiometry (Olinares et al., 2011b) as well as the AAA+ chaperones ClpC1/2 and ClpD. ClpC1/2 and ClpD can potentially be modulated by ClpS1. Multiple lines of evidence indicate that the ClpP/R core components have distinct functional contributions and influence embryogenesis, plastid biogenesis, and plant development (Shikanai et al., 2001; Rudella et al., 2006; Sjogren et al., 2006; Koussevitzky et al., 2007; Kim et al., 2009, 2013; Zybailov et al., 2009b; Olinares et al., 2011a). Chloroplast ClpC1/2 proteins have 88% sequence identity to each other and share 71 to 73% identities with cyanobacterial ClpC, while ClpD has only ∼45% sequence identity to ClpC proteins in chloroplasts or cyanobacteria. The ATPase activity of Arabidopsis ClpD was much lower than that of ClpC2 (Rosano et al., 2011), and the ClpD mRNA expression pattern is different from that of ClpC1/2 (reviewed in Olinares et al., 2011a). Thus, ClpD appears to have diversified substantially from the ClpC chaperones. Genetic and biochemical studies show that ClpC1/2 proteins coordinately contribute to plant fitness, chloroplast biogenesis, and function and that ClpC1 is also involved in chloroplast protein import (Constan et al., 2004; Sjogren et al., 2004; Kovacheva et al., 2005; Rosano et al., 2011; Bruch et al., 2012). These findings illustrate the physiological importance of the regulation of plastid proteomes via the Clp system. However, the mechanisms of substrate recognition and delivery to the Clp protease in plastids are unknown.
Studies on the N-end rule pathway have focused on bacteria and the eukaryotic cytosol, but hardly on mitochondria and chloroplasts (Dougan et al., 2010). A different type of the N-end rule pathway independent of ClpS appears to exist in mitochondria (Vogtle et al., 2009). Recent analysis of stability determinants of chloroplast proteins in tobacco (Nicotiana tabacum) suggests the presence of a prokaryotic N-end rule-like pathway in the chloroplast (Apel et al., 2010), but no N-end rule has been established for plastids, nor has a possible involvement of ClpS in this process been studied. We successfully detected a homolog of bacterial ClpS (AT1G68660) in chloroplast stroma of Arabidopsis (Olinares et al., 2010). In this study we (1) analyze the phylogeny and sequence features of the ClpS protein, (2) determine ClpS protein expression pattern, (3) show ClpS interaction with the N-domain of ClpC1 and with ClpC2, (4) determine the physiological significance of ClpS in the chloroplast through quantitative comparative proteomics of a ClpS null mutant (clps1) and comparison to a ClpC chaperone null mutant (clpc1-1), and (5) identify a set of candidate ClpS substrates based on affinity purifications. We also investigate the genetic interactions between ClpS and the ClpC/D chaperones. Finally, we show that clps1, but not clpc1-1, has increased sensitivity to the translational elongation inhibitor chloramphenicol (CAP), which suggests a link between translational capacity and ClpS. We discuss the identified candidate substrates and substrate recognition mechanisms in the chloroplast.
RESULTS
Evolutionary Lineage and Protein Structure of ClpS in Plants
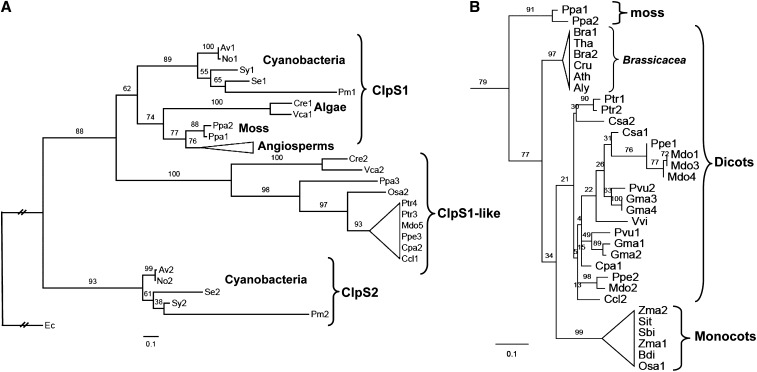
Previous phylogenetic analyses of the ClpS protein showed the evolutionary linkages among bacteria, some cyanobacteria, and a few plant species (Dougan et al., 2002b; Lupas and Koretke, 2003). To better define the evolutionary lineage of plant ClpS proteins and determine conservation of ClpS residues in angiosperms, we performed an in-depth phylogenetic analysis using a sequence alignment of 10 sequences from five cyanobacterial species, four sequences from two algal species, two moss sequences, 39 sequences from 19 vascular plant species, and E. coli ClpS as the outgroup (see Supplemental Figure 1 and Supplemental Data Set 1 online). ClpS sequences fall into distinct phylogenetic branches following the general outline of the tree of life (Figure 1A). As shown in the cladogram, the prototype ClpS first splits into two cyanobacterial derivatives, named ClpS1 and ClpS2 (Figure 1A). The cyanobacterial ClpS1 further evolved through the green lineage from eukaryotic unicellular algae to plants including moss and angiosperms, while the ClpS2 proteins are limited to cyanobacteria. Interestingly, an alternative type of ClpS1, which we assigned ClpS1-like protein, is found in eukaryotic organisms, including both algae and moss, and some of the angiosperms (e.g., rice [Oryza sativa] but not maize [Zea mays] and Arabidopsis) (Figure 1A).
Figure 1.
Phylogenetic Analysis of ClpS.
(A) Schematic Randomized Axelerated Maximum Likelihood (RAxML) phylogeny of 57 ClpS proteins from E. coli, five cyanobacterial species, twp algal species, moss, and 19 angiosperms. RAxML bootstrap support values are shown at the nodes of the tree, in which the E. coli ClpS is designated as the outgroup and ClpS relatives are classified into six clades: ClpS1 proteins from cyanobacteria, algae, moss, and angiosperms, ClpS2 protein from cyanobacteria, and ClpS1-like protein. ClpS1 and ClpS1-like proteins (39 in total) in angiosperms are collapsed and indicated as a triangle. Details for this angiosperm clade are shown in (B). The complete sequence alignment is shown in Supplemental Figure 1 online (excluding the cTPs) and is available as Supplemental Data Set 1 online. The species included are Physcomitrella patens (Ppa), Chlamydomonas reinhardtii (Cre), Volvox carteri (Vca), Anabaena variabilis ATCC 29413 (Av), Nostoc sp PCC 7120 (No), Synechocystis sp PCC 6803 (Sy), Synechococcus elongatus PCC 7942 (Se), Prochlorococcus marinus subsp marinus str. CCMP1375 (Pm), Escherichia coli str. K12 substr. W3110 (Ec). The 19 angiosperm species included are the 14 dicotyledons Populus trichocarpa (Ptr), Phaseolus vulgaris (Pvu), Glycine max (Gma), Cucumis sativus (Csa), Prunus persica (Ppe), Malus domestica (Mdo), Arabidopsis thaliana (Ath), Arabidopsis lyrata (Aly), Capsella rubella (Cru), Brassica rapa (Bra), Thellungiella halophila (Tha), Carica papaya (Cpa), Citrus clementina (Ccl), Vitis vinifera (Vvi), and the five monocotyledons Sorghum bicolor (Sbi), Zea mays (Zma), Setaria italica (Sit), Oryza sativa (Osa), and Brachypodium distachyon (Bdi). Distance is indicated as substitutions per site.
(B) Phylogeny of the ClpS1 clade for the angiosperms and moss. ClpS1 proteins in Brassicaceae are separated in a single clade distinct from the others, including those in monocots. Distance is indicated as substitution rate per site.
Figure 1B shows the phylogenetic analysis across the 33 mature ClpS1 sequences in moss, monocots, and dicots. A clear diversification in ClpS1 between the monocots and dicots is observed. Furthermore, ClpS1 in the five Brassicaceae species is distinct from those of other dicot species. Interestingly, the number of ClpS1 orthologs within each species varies between one (e.g., Arabidopsis and rice) and four (Glycine max); this could perhaps reflect specialization of ClpS paralogs toward specific substrates.
Predictions for Interactions between ClpS, Substrates, and Clp Chaperones
The various studies on E. coli ClpS have defined the regions and specific residues that are important for interaction with the ClpA chaperone and substrates (see Introduction). We evaluated to what extent these critical regions and residues are conserved across the ClpS family and possible implications for ClpS functions in plastids. Previous analysis of the E. coli ClpS protein distinguished between the short and unstructured N-terminal extension (M1-P25) and the ClpS core region (see Supplemental Figure 1 online).
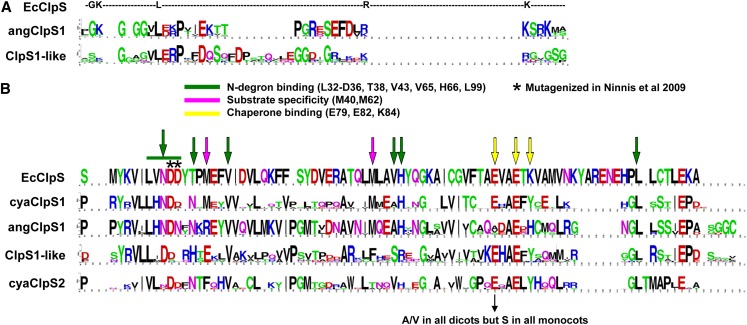
The N-terminal extension is important for inhibition of binding of SsrA-tagged proteins and for delivery/transfer of N-degron substrates to the Clp chaperone (Baker and Sauer, 2012). The N-terminal extension is of similar length in cyanobacterial ClpS1,2, but much longer in the eukaryotic ClpS1 and ClpS1-like proteins, suggesting that substrate delivery from ClpS to the Clp chaperones is different between eukaryotes and bacteria. Indeed, unlike nonphotosynthetic bacteria, photosynthetic eukaryotes are not (yet) known to possess SsrA tagging systems. The N-terminal extensions of the eukaryotic ClpS1 and ClpS1-like proteins each have clear consensus sequences that partially overlap (Figure 2A; see Supplemental Figure 1 online), such as several Gly residues and a conserved Pro. This conservation will allow testing the functional role of this N-terminal extension in substrate selection and delivery through mutagenesis.
Figure 2.
Sequence Conservation of ClpS Proteins in the Angiosperms and Comparison to Bacterial ClpS Proteins.
(A) Sequence logo showing sequence conservation of the N-terminal extensions of ClpS1 (angClpS1; 31 proteins) and ClpS1-like proteins (11 proteins) in the angiosperms and comparison to E. coli ClpS (EcClpS).
(B) Sequence logo indicating sequence conservation of the ClpS core region for ClpS1 in the cyanobacteria (cyaClpS1), ClpS1 in the angiosperms (angClpS1), ClpS1-like in the angiosperms (ClpS1-like), and ClpS2 in cyanobacteria (cyaClpS2). E. coli ClpS residues are marked that are known to be involved N-degron binding (in green), substrate specificity (in purple), or chaperone binding (in yellow). Asterisks indicate E. coli residues Asp-35 and Asp-36, which were mutated by Ninnis et al. (2009), resulting in loss of substrate interaction.
The ClpS core region showed conservation across the ClpS protein family, but with significant diversification between the various ClpS types (see Supplemental Figure 1 online). To better understand this diversification, we determined the ClpS core consensus sequences for ClpS1 and ClpS2 in the cyanobacteria as well as ClpS1 and ClpS1-like in the angiosperms (Figure 2B).
The residues Asn-34, Asp-35, and His-66 constitute the substrate pocket and are critical for binding to the α-amino group of the first N-terminal destabilizing residue of substrates in E. coli; they are 100% conserved in angiosperm ClpS1, but not in ClpS1-like or ClpS2. Of the four additional residues (Thr-38, Val-43, Val-65, and Leu-99; marked in green) involved in N-degron binding, two (Val-43 and Leu-99) are conserved in ClpS1 and ClpS1-like. Furthermore, two Met residues (Met-40 and Met-62) contribute to substrate specificity in E. coli. Met-62 is 100% conserved in ClpS1, but not in ClpS1-like or ClpS2 proteins. By contrast, E. coli residue Met-40 is replaced by Arg in all eukaryotic ClpS1 proteins and by Glu in all ClpS1-like and Phe in all ClpS2 proteins. These observations suggest that the ClpS substrate recognition mechanism for substrates with N-degrons is partially retained throughout evolution for ClpS1 and diverged more strongly in ClpS1-like and ClpS2. We note that the presence of conserved N-degrons has not been established for plastids, but the considerable conservation of residues involved in N-degron binding in E. coli provides a strong incentive to search for a plastid N-end rule (see Discussion).
The ClpS E. coli chaperone binding region has three critical residues (Glu-79, Glu-82, and Lys-84) (Figure 2B). Mutagenesis of both Glu-79 and Lys-84 together resulted in a loss of interaction with ClpA, whereas Glu-82 did not seem to be important for chaperone interaction (Zeth et al., 2002). Glu-82 is conserved across all ClpS proteins, whereas Glu-79 is not conserved in angiosperm ClpS1, but conserved across cyanoClpS1,2 and ClpS1-like proteins. Lys-84 is not conserved in photosynthetic ClpS proteins, but either replaced by Tyr or His. (We note that Glu-79 is replaced by Ala or Val in the dicotyledons and by Ser in the monocotyledons.) The absence of strong conservation in the chaperone binding region of ClpS prompted us to consider conservation of ClpS binding residues in the ClpA/C/D chaperones and structural information. In E. coli, the N-domain of ClpA is sufficient for ClpS interaction (Zeth et al., 2002). Based on high-resolution x-ray structures and functional analysis, the E. coli ClpS–ClpA interaction was suggested to require two hydrogen bonds formed between the ClpS residue Glu-79 and the ClpA residues Glu-28 and Thr-81, in addition to two salt bridges between the ClpS residues Glu-82 and Lys-84 and the ClpA residues Arg-86 and Glu-23 (Zeth et al., 2002) (see Supplemental Figure 2 online). One of the salt bridges is likely maintained between Arabidopsis ClpS1 and its ClpC chaperones because corresponding residues to Glu-82 (in E. coli ClpS) and Arg-86 (E. coli ClpA) are 100% conserved (Glu-137 in ClpS1 and Arg-179/Arg-199 in ClpC1/2) (see Supplemental Figure 2 online). In fact, these ClpS1 and ClpC residues are conserved in all 19 angiosperm species examined (Figures 2B and 3). However, the Arabidopsis ClpS1 residue at position 134 corresponding to E. coli Glu-79, conferring the hydrogen bond with the chaperone, appears to be eliminated by Ala substitution (Figure 2B), whereas the corresponding Thr residue (Thr-81 in E. coli ClpA) in ClpC1/2 (Thr-179 or Thr-194) is perfectly conserved (Figure 3). The salt bridge present in E. coli ClpS at position Lys-84 in Arabidopsis ClpS1 can thus no longer be formed within the ClpS1–ClpC interaction; instead, there appears to be a hydrogen bond at the ClpS1-ClpC1/2 interface, as the ClpS1 residue is changed to His (His-139) (see Supplemental Figure 2 online). The ClpA (charged) residue (Glu-23) is substituted with an uncharged polar residue (Asn-119 or Asn-139) in Arabidopsis (and other angiosperms) ClpC1/2 but is not conserved in angiosperm ClpD (Figure 3). The lack of conservation of the chaperone binding region makes it seem less likely that ClpS1 (or ClpS1-like) interacts with ClpD; this is consistent with the very different mRNA expression patterns between CLPS1 and CLPD in Arabidopsis, peaking respectively in young and senescing leaves. In conclusion, in silico analysis suggests that ClpS1 can interact with the chaperone modules ClpC1/C2 in a similar manner as observed for ClpS and the ClpA chaperone in eubacteria; however, experimental testing of such interaction is needed.
Figure 3.
In Silico Analysis of the Clp Chaperone Interactions with ClpS.
Sequence logo indicating sequence conservation of repeat 1 and repeat 2 of ClpC in cyanobacteria (cyaClpC), angiosperms (angClpC), and of ClpD in angiosperm (angClpD) and comparison to E. coli ClpA (ecoClpA). Residues known to be important for E. coli ClpA with ClpS are indicated.
So far, our in silico analysis suggests that ClpS1 is a functional homolog of ClpS in E. coli. In the remainder of this article, we will examine the functional role of ClpS1 in Arabidopsis chloroplasts, determine genetic and physical interactions with ClpC1,2, and identify candidate substrates through affinity purification.
ClpS1 Protein Accumulation Is Strictly Regulated in a Spatiotemporal Manner
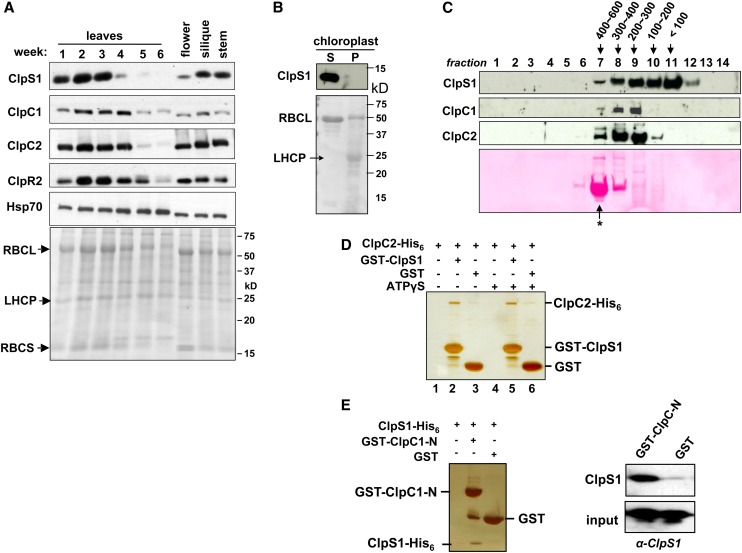
To understand when and where ClpS1 plays a role in Arabidopsis, we determined the endogenous levels of ClpS1 in stems, flowers, and siliques, as well as in leaves at several points during leaf development for Arabidopsis grown on soil under continuous light (Figure 4A). ClpS1 protein accumulated exclusively in photosynthetic green tissues with high levels in young, developing leaf tissues and was nearly absent in senescing rosettes (5 to 6 weeks old). Comparable accumulation patterns were observed for its chaperone partners, ClpC1 and ClpC2, although developmental downregulation of ClpC1/2 in leaves was delayed as compared with ClpS1 (Figure 4A). By contrast, chloroplast chaperone cpHSP70 (not associated with the Clp protease) was expressed stably throughout all the green tissues examined. The proteolytic core subunit ClpR2 also showed highest accumulation in the younger rosettes (Figure 4A). Consistent with these data, the transcript levels of CLPS1 and CLPC genes show similar tissue-specific and developmental expression profiles (Olinares et al., 2011a). ClpS1 was found only in the stromal fraction and not the chloroplast membranes (Figure 4B). These observations are consistent with a role for ClpS1 mediating regulated proteolysis in cooperation with ClpC1,2, in particular during plastid biogenesis, but not during senescence.
Figure 4.
Spatiotemporal Accumulation of ClpS1 and Interaction between ClpS1 and ClpC Proteins.
(A) Arabidopsis plants were grown on soil under continuous light. Leaves from the two outer rows of the rosette were harvested after 1 to 6 weeks. Flowers, siliques, and stems were collected after 6 weeks. Total proteins were extracted and analyzed by immunoblotting using anti-ClpS1, ClpC1/C2/R2, and cpHsp70 antibodies. Each lane contains 20 μg proteins, and the Ponceau-stained blot is shown as the loading control. Loss of RBCL during senescence (4 to 6 weeks) can be observed from the stained blot.
(B) ClpS1 is exclusively located in the stroma and is absent in the chloroplast membranes. To determine the intraplastid location of ClpS1, chloroplasts were isolated from soil-grown wild-type. The stromal (S) and membrane fractions (P) were separated by centrifugation and analyzed by SDS-PAGE and immunoblotting. The filter was also stained with Ponceau S (bottom panel).
(C) In vivo sizes of native ClpS1 and ClpC1/2 in the chloroplast stroma. Chloroplasts were isolated from soil-grown wild-type plants at leaf stage 1.07-1.08. The stromal proteins were prepared in the presence of ATPγS and separated using a Superose (gel filtration) column. The eluates were collected and pooled into 14 fractions. Proteins in each fraction were TCA precipitated, and equal volumes were analyzed by immunoblotting with antibodies against ClpS1, ClpC1, and ClpC2. The blot was also stained by Ponceau S, showing RBCL eluting as part of the 550-kD holocomplex (marked with an asterisk).
(D) Direct interaction of ClpS1 with ClpC2. ClpC2-His6 protein was incubated with or without ATPγS and subsequently combined with GST-ClpS1 or ClpS. Proteins were bound to the GST affinity resin and then eluted with reduced glutathione. Eluates were analyzed by SDS-PAGE and silver staining.
(E) Recombinant his6-tagged ClpS1 was incubated with recombinant GST or the N-domain of ClpC1 fused to GST. Proteins were bound to the GST affinity resin and then eluted with laemmli buffer. Eluates were analyzed by SDS-PAGE and silver staining, and ClpS1 was also detected by immunoblot with anti-ClpS1 antiserum.
[See online article for color version of this figure.]
ClpS1 Interacts with ClpC Chaperones
To test in vivo interaction between ClpS1 and ClpC1/2, we performed size-exclusion chromatography analysis of the chloroplast stromal proteome isolated from young wild-type Arabidopsis seedlings (Figure 4C). ClpC1/2 started to elute at ∼600 kD (fraction 7; marked by the 550-kD ribulose-1,5-bis-phosphate carboxylase/oxygenase [Rubisco] holocomplex), likely corresponding to ClpC hexamers, reaching a peak in fraction 9 at ∼200 kD (corresponding to dimers) and then rapidly declining to undetectable levels. We note that ClpC1/2 were previously detected as 200-kD complexes using native gels of chloroplast stroma (Peltier et al., 2006), whereas recombinant ClpC2 and ClpD each formed 180- to 220-kD homodimers and, upon incubation with 5 mM ATP, also ∼500- to 700-kD homohexamers (Rosano et al., 2011). Stromal ClpS1 eluted in fractions from ∼600 kD (fraction 7) to small monomeric proteins (fractions 11 and 12). These elution profiles are compatible with an in vivo interaction between ClpS1 and ClpC1 and/or ClpC2 and show that a significant amount of ClpS1 accumulated in complexes.
To investigate direct interaction between ClpS1 and ClpC chaperones and the influence of ATP, we generated recombinant ClpS1 N-terminally fused with glutathione S-transferase (GST-ClpS) and recombinant ClpC2-(HIS)6 fusion protein. GST-ClpS was bound to a GST affinity column and used as bait. Recombinant GST was used as negative control for unspecific ClpC2 binding (Figure 4D). Proteins were eluted using laemmli buffer. These in vitro pull-down experiments showed that ClpS1 does specifically bind to ClpC2 in an ATPγS-independent manner (Figure 4D). An SDS-PAGE gel showing the input in these assays is shown in Supplemental Figure 3A online. In case of ClpC1, we used recombinant N-terminal domain of ClpC1 (residues 39 to 252) to test interaction with ClpS1 because the complete ClpC1 protein was unstable in the E. coli, as reported previously (Rosano et al., 2011). Importantly, this N-domain contains the predicted sites for interaction with ClpS1 (Figure 3). Using recombinant ClpS1-His6, we tested its interaction with GST-ClpC1-N-domain using GST as a negative control (Figure 4E and inputs in Supplemental Figure 3B online). Silver staining and immunoblots of the eluates clearly show a specific interaction between the N-terminal domain of ClpC1 and ClpS1 (Figure 4E), thus providing further support for a conserved ClpS1 function in substrate delivery.
Interactions between ClpS1 and ClpT1,ClpT2
ClpT1 and ClpT2 are two small proteins peripherally associated with the ClpPR core and have strong similarity to the N-domain of ClpC1,2. Based on this similarity, we previously hypothesized that ClpS1 could interact with ClpT1,2 (Peltier et al., 2004; Olinares et al., 2011a). To test this hypothesis, we generated recombinant ClpT1 and ClpT2 and probed for interactions with ClpS1 using the GST-ClpS1 affinity column (see Supplemental Figure 4 online). However, no interactions were observed. Furthermore, when we previously analyzed the affinity-purified ClpPRT complex using in vivo StrepII-tagged ClpR4 or StrepII-tagged ClpP3, we did identify each ClpPR subunit as well as ClpT1 and ClpT2, but we never observed ClpS1 (Olinares et al., 2011b). Finally, we also did not observe ClpT1 or ClpT2 in the ClpS-GST experiments using stroma from wild-type chloroplasts or stroma from mutants lacking ClpS1 or both ClpS1 and ClpC1 (see below). In conclusion, the hypothesized interaction between ClpS1 and ClpT1,2 is (so far) not supported by experimental evidence.
Loss of ClpS1 and Genetic Interaction of ClpS1 with the Chloroplast Clp System
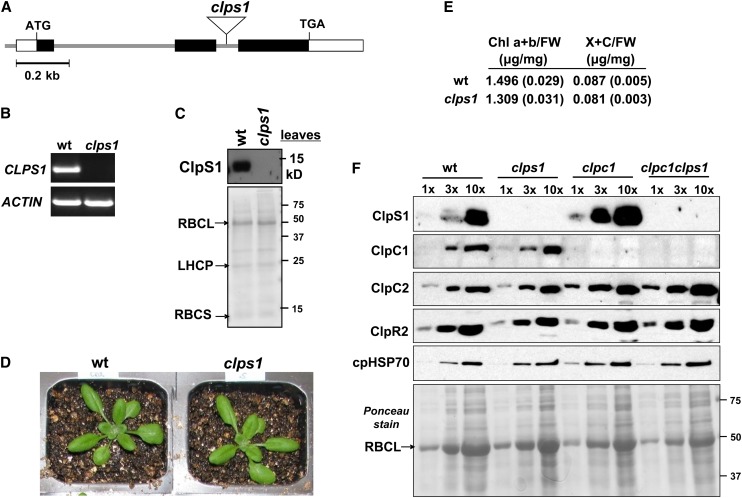
To explore the physiological significance of ClpS in Arabidopsis chloroplasts, we obtained a CLPS1 null mutant (clps1) with a confirmed T-DNA insertion in the second intron (Figure 5A). The loss of the CLPS1 mRNA accumulation was determined by RT-PCR (Figure 5B), and immunoblotting using affinity-purified anti-ClpS1 antibodies showed a complete loss of ClpS1 protein (Figure 5C). Growth and development of clps1 when grown on soil were not visibly different from the wild type (Figure 5D). However, true leaves of soil-grown clps1 seedlings (stage 1.07) under short-day conditions contained 13% less chlorophyll a+b on a fresh weight basis than did those of the wild type (Figure 5E). This indicates a weak chloroplast phenotype.
Figure 5.
Analysis of a ClpS1 Null Mutant in Arabidopsis.
(A) Gene model structure and position of T-DNA insertion in the CLPS1 null mutant used in this study. Exons (black boxes for coding sequence), 5′ and 3′ untranslated regions (open boxes), and T-DNA insertion (triangle) are indicated.
(B) CLPS1 transcript accumulation in the wild type and clps1. Transcripts were extracted from wild-type and clps1 seedlings and amplified by RT-PCR with gene-specific primers and analyzed on an agarose gel. ACTIN mRNA was used as an internal control.
(C) To determine if ClpS1 was absent in clps1, total leaf protein extracts from the wild type and clps1 were analyzed by SDS-PAGE and immunoblotting. The filter was also stained with Ponceau S (bottom panel).
(D) Comparison of wild-type (wt) and clps1 phenotypes. Plants were grown for 20 d on soil under 16/8-h light/dark cycle at ∼120 µmol photons m−2 s−1. No visible differences were observed.
(E) Pigment accumulation in soil-grown wild-type and clps1 seedlings at developmental stage 1.07. Plants were grown under a 10-h-light/14-h-dark period at ∼100 μmol photons m−1 s−2. Chlorophyll a+b (Chl a+b) and total xanthophyll and other carotenoid (X+C) contents were determined on a fresh weight basis; n = 6. se is indicated.
(F) Upregulation of ClpS1 and ClpC2 proteins in the clpc1-1 mutant background. Stromal proteins were isolated from wild-type, clps1, clpc1-1, and clps1 clpc1-1 seedlings and analyzed by immunoblotting with anti-ClpS1, ClpC1, ClpC2, ClpR2, and cpHSP70 antibodies. A titration of proteins was loaded for each genotype, as indicated. The filter was stained with Ponceau S (bottom panel).
[See online article for color version of this figure.]
We then exposed clps1 and wild-type plants to various light regimes (short/long daylength or continuous light at a range of growth light intensities between 40 and 250 μmol photons m−2 s−1), continuous dark for several days, heat (90 min or 3 h at 38°C; 38°C for 90 min, followed by 45°C for 30 min), and salinity (50, 100, 150, and 200 mM NaCl), but no significant effects were observed for the visible phenotype of clps1 compared with the wild type (data not shown). Moreover, overexpression of ClpS1 (confirmed at the ClpS1 protein level; up to 3.5-fold ClpS1 overaccumulation) in the wild-type background did not result in a visible growth phenotype (data not shown).
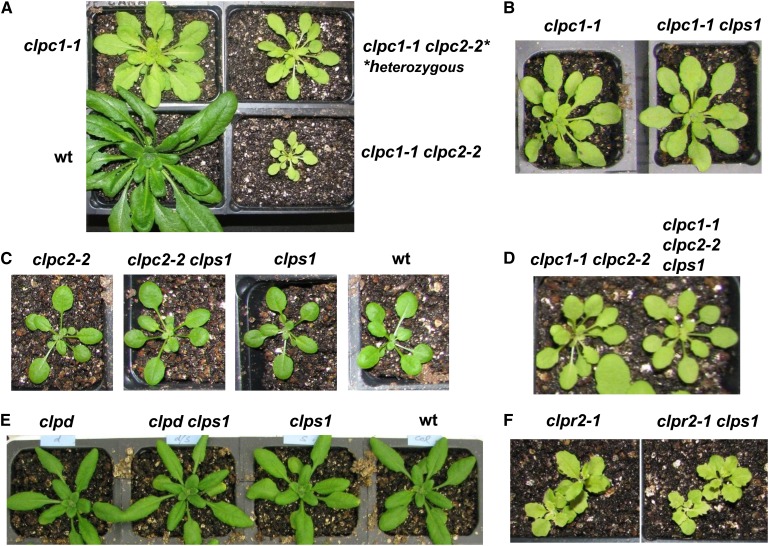
We then tested for genetic interactions between ClpS1, the three Clp chaperones (C1, C2, and D) and the ClpPR core complex (Figure 6). T-DNA insertion mutants for CLPC1, CLPC2, CLPD were obtained from publicly available T-DNA collections and genotyped; the CLPC alleles were described in previous studies (see Table 1 for details and references). With regard to the ClpPR complex, null mutations are embryo lethal for ClpP4 and ClpP5 and seedling lethal on soil for ClpR2, ClpR4, and ClpP3 but cause only a mild phenotype for ClpR1 (summarized in Olinares et al., 2011a). Therefore, we selected the virescent clpr2-1 knockdown mutant, which is viable on soil and has been previously characterized in detail (Rudella et al., 2006; Zybailov et al., 2009b). Double mutants of clps1 with these clpr2-1, clpc1-1, clpc2-1, and clpd mutants were then generated by crossing and screening the resulting F2 and F3 progenies for homozygous mutants. We also generated double mutants of clpc1-1 clpc2-2, using this leaky allele for CLPC2 because the double null allele for CLPC1 and CLPC2 is embryo or seedling lethal (Kovacheva et al., 2007). There was a clear dosage effect of CLPC2 on clpc1-1 (Figure 6A). The homozygous double mutant clpc1-1 clpc2-2 was crossed with clps1. These five ClpS1 double or triple homozygous mutant lines were then evaluated for growth and developmental phenotypes, along with the respective single and double parental lines and the wild type (Figures 6B to 6F). Despite these extensive efforts, we did not observe obvious visible effects of the clps1 null background in these other clp single or double mutants. We conclude that ClpS1 is not required for normal plant growth and development.
Figure 6.
Genetic Interactions of clps1 with other clp Mutants.
(A) Dosage effect of clpc2 on clpc1. clpc1-1 plants were crossed with clpc2-2, and progenies with homozygous clpc1-1 and either homozygous or heterozygous clpc2-2 are shown. Plants were grown for 45 d under 10/14-h light/dark cycle at 120 µmol photons m−2 s−1. We note that the reciprocal dosage effect was not observed (data not shown). wt, the wild type.
(B) Effect of clps1 on clpc1-1. Homozygous single and double mutant plants were grown for 37 d under 10/14-h light/dark cycle at 250 µmol photons m−2 s−1. No visible differences were observed.
(C) Effect of clps1 on clpc2-2. Homozygous single and double mutant plants and the wild type were grown on soil for 23 d under 10/14-h light/dark cycle at 120 µmol photons m−2 s−1. No visible differences were observed.
(D) Effect of clps1 on clpc1 clpc2. clpc1-1 clpc2-2 homozygous plants were crossed with clps1. Growth and development are shown for the resulting homozygous double and triple mutants grown for 45 d under 10/14-h light/dark cycle at 120 µmol photons m−2 s−1. No visible differences were observed.
(E) Effect of clps1 on clpd. Homozygous single and double mutants as well as wild-type plants were grown for 23 d on soil under 16/8-h light/dark cycle at 120 µmol photons m−2 s−1. No visible differences were observed.
(F) Effect of clps1 on clpr2-1. Homozygous single and double mutants were grown for 25 d on soil under 16/8-h light/dark cycle at 120 µmol photons m−2 s−1. No visible differences were observed.
Table 1. Clp Mutant Lines Used in This Study.
| Gene | Locus | Mutant Name | Germplasma | Source | Position T-DNA | Phenotype |
|---|---|---|---|---|---|---|
| CLPR2 | At1g12410 | clpr2-1b | SALK_046378 | SALK | 5′ UTR (-7) | Pale green, yellow |
| CLPC1 | At5g50920 | clpc1-1c | SALK_014058 | SALK | 4/9 Exon | Pale green, yellow |
| CLPC2 | At3g48870 | clpc2-2d | SAIL_622_B05 | Syngenta | 9/9 Exon | Wild-type like |
| CLPD | At5g51070 | clpd | SAIL_77_G05 | Syngenta | 1/12 Exon | Wild-type like |
| CLPS | At1g68660 | clps | SAIL_326B_G12 | Syngenta | 2/2 Intron | Wild-type like |
UTR, untranslated region.
Germplasm starting with “SALK” are from the Salk Institute collections, while SAIL mutants originate from Syngenta. All lines are ecotype Colubmia-0, except clpd, which was Columbia-3.
This mutant was originally isolated and described by Rudella et al. (2006).
Previously named clpc1-1 in Sjögren et al. (2004), and hsp93-V-2 in Kovacheva et al (2005)
Previously named hsp93-III-1 in Kovacheva et al (2007)
To further evaluate functional interactions between ClpC1, ClpC2, and ClpS1, we compared the levels of ClpS1, ClpC1, and ClpC2 in clps1, clpc1, and clps1 clpc1 (Figure 5F). We also analyzed abundance of the stromal chaperone cpHSP70 and ClpR2 as a marker for the ClpPR core. The abundance of neither cpHSP70 nor ClpR2 was significantly altered in the mutants, but ClpS1 abundance increased approximately threefold in clpc1, supporting functional interaction between ClpS1 and ClpC1. ClpC2 increased approximately threefold in clpc1 and clps1 clpc1 (Figure 5F), consistent with the genetics and supporting the observed redundancy between the two ClpC chaperones. It has been observed in E. coli that ClpA (the ClpC homolog) is destabilized in ClpS-depleted cells (Dougan et al., 2002a), but our results indicate that in Arabidopsis chloroplasts ClpS1 accumulation levels do not influence stability of ClpC.
The Molecular Phenotype of the clps1 Null Mutant and Comparison to the clpc1 Null Mutant
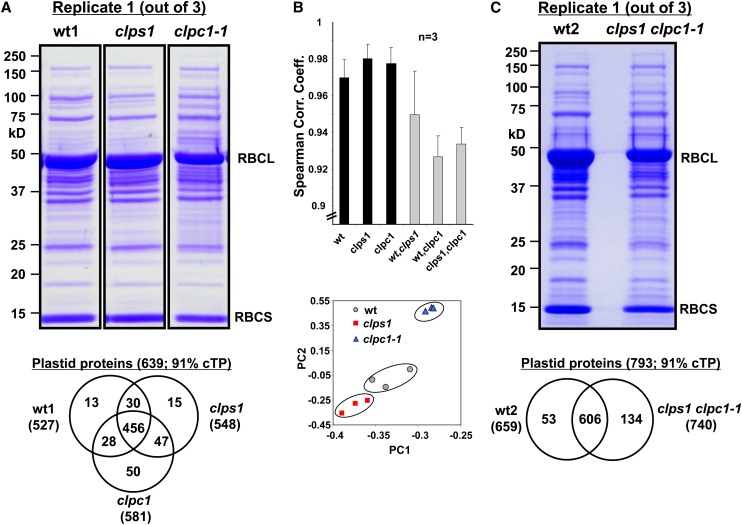
There was no significant visible growth phenotype in clps1, nor did we observe visible synergistic or suppression effects when clps1 was crossed with other clp mutants. However, there may well be a molecular phenotype at the protein level in clps1 that would indicate its functional role. Indeed, chlorophyll content was slightly reduced in clps1 compared with the wild type (Figure 5E). Therefore, we performed a comparative quantitative proteome analysis between the stromal chloroplast fractions of the wild type and clps1 employing label-free mass spectrometry (MS)–based quantification using an LTQ-Orbitrap and spectral counting, using an optimized workflow as previously described (Friso et al., 2011). For comparison, we also included the clpc1 single mutant (with virescent phenotype). Plants were grown on soil and harvested at leaf stages 1.08 to 1.09 during which ClpS1 is expressed with the highest levels in wild-type leaves. Chloroplasts were isolated and stromal proteomes extracted, separated by SDS-PAGE, and stained by Coomassie blue (Figure 7A). No obvious differences in protein patterns were observed between clps1 and the wild type, even when using larger gels (data not shown). However, accumulation levels of both large and small Rubisco subunits were clearly lower in clpc1 (Figure 7A). Protein abundances were quantified according to the number of matched adjusted tandem mass spectrometry (MS/MS) spectra (adjSPC). In total, 790 proteins were identified with two or more adjSPC with a total of 124,897 adjSPC (see Supplemental Data Set 2 online). Based on our most recent reference plastid proteome analysis (Huang et al., 2013; see The Plant Proteome Database [PPDB], http://ppdb.tc.cornell.edu), 633 proteins were located in the chloroplast and matched to 97% of all adjSPC (see Supplemental Data Set 2 online). This showed that the chloroplasts were highly purified. The Venn diagram of the identified plastid proteins shows excellent overlap between the three genotypes (Figure 7A). The biological replicates were very similar within the same genotype (correlation coefficient >0.97) but lower between genotypes (0.93 to 0.95), with the lowest (0.93) when comparing the wild type and clpc1 (Figure 7B). Furthermore, principle component analysis (PCA) also showed that the variation between genotypes was larger than between replicates within each genotype and that clps1 was more similar to the wild type than clpc1-1 (Figure 7B). Together this shows that (1) the quantitative proteome experiments are of high quality with little noise and (2) that clps1 has a measurable proteome phenotype but that it is different from the clpc1-1 proteome phenotype.
Figure 7.
Comparative Proteomics of the Wild Type, clps1, clpc1-1, and clps1 clpc1-1.
(A) Representative Coomassie blue–stained SDS-PAGE gel image of stromal proteins from the wild type (wt), clps1, and clpc1-1. RBCL and RBCS accumulation levels are visibly reduced in clpc1-1. Each gel lane was cut in 10 slices and subjected to in-gel trypsin digestion and MS. The Venn diagram compares plastid proteins identified in wild-type, clps1, and clpc1-1 leaves.
(B) Spearman correlation and PCA of the quantified proteomes of the wild type, clps1, and clpc1-1 are shown. The symbols in the PCA plot represent each of the biological replicates for each of the three genotypes. Error bars show the sd.
(C) Coomassie blue–stained SDS-PAGE gel image of chloroplast stroma from wild-type and clps1 clpc1-1 plants. RBCL and RBCS accumulation levels are visibly reduced in clps1 clpc1-1. The Venn diagram summarizes the identified proteins in the wild type and double mutant. Spearman correlation and PCA analyses are shown in Supplemental Figure 5 online.
[See online article for color version of this figure.]
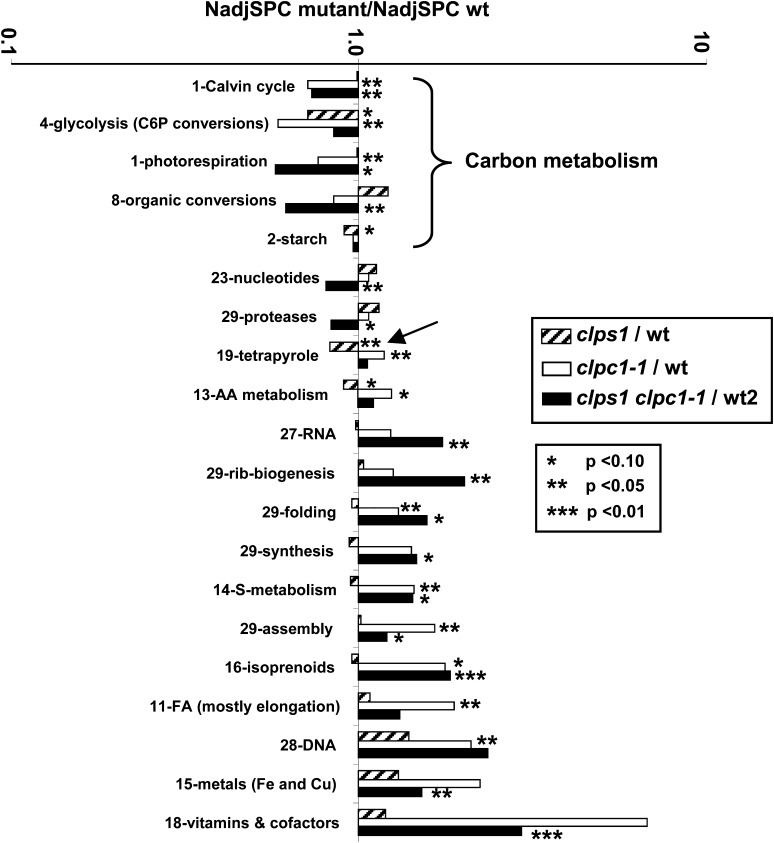
To evaluate general effects of loss of ClpS1 or ClpC1 on the plastid, we compared the chloroplast protein mass investments (based on the NadjSPC of assigned chloroplast proteins) across 34 functions between the three genotypes (see Supplemental Table 1 online). These functions were assigned using the MapMan bin system (Thimm et al., 2004) that we further curated (see PPDB). Figure 8 shows the 20 functions that were significantly affected (at P < 0.1, P < 0.05, or P < 0.01; Student’s t test) in one or more of the mutants. Clearly, clps1 showed very little proteome phenotype, with tetrapyrrole metabolism (18% down) being affected at P < 0.05 and three functions (starch metabolism, glycolysis [proteins involved in hexose phosphate conversions], and amino acid metabolism) significantly reduced at P < 0.1. By contrast, clpc1-1 showed significant effects for multiple functions (nine functions at P < 0.01), consistent with the visible growth phenotype. In particular, primary carbon metabolism in the plastid was downregulated at the level of the Calvin cycle (by 30%), glycolysis (by 41%), and photorespiration (by 24%). Functions that were upregulated included protein chaperones (by 30%), protein assembly factors (by 65%), DNA interacting proteins (by 110%), tetrapyrrole metabolism (by 18%), fatty acid metabolism (by 88%), and sulfur metabolism (by 44%).
Figure 8.
Protein Mass Investment in Specific Plastid Functions in the Wild Type, clps1, clpc1-1, and clps1 clpc1-1.
Functions that were significantly over- or underrepresented in the mutants are marked with asterisks. Three levels of significance are distinguished (P < 0.1, P < 0.05, or P < 0.01) and were determined using a Student’s t test (matched-paired samples). The arrow highlights the reduced investments in tetrapyrrole metabolism in clps1 compared with the wild type. C6P, hexose phosphates; AA, amino acid; FA, fatty acids; wt, the wild type.
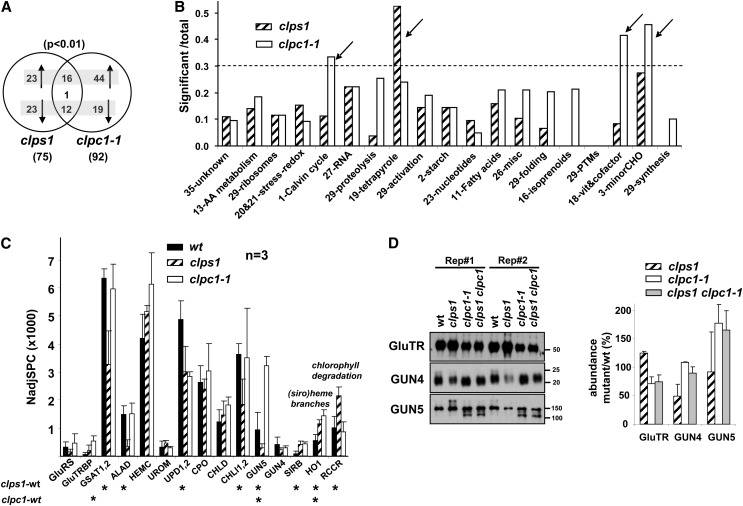
To further clarify these molecular phenotypes, we then identified significant (P < 0.01; GLEE algorithm) differential accumulation of individual proteins between the three genotypes. The differentially expressed proteins reflect indirect effects from the loss of ClpS1 or ClpC1, but upregulated proteins are also potential substrates for ClpS1- or ClpC1-dependent proteolysis. Thirty-nine stromal proteins were upregulated and 36 were downregulated in the clps1 mutant compared with the wild type. In clpc1-1, 61 proteins were upregulated and 31 were downregulated (Figure 9; see Supplemental Data Sets 3 and 4 online). Sixteen proteins were upregulated in both genotypes and 12 were downregulated in both genotypes (Figure 9A). Only one protein, Mg-protoporphyrin IX chelatase H (CHLH/GUN5), showed opposite behavior between the genotypes. In the case of clps1, but not clpc1, proteins in the tetrapyrrole pathway were strongly overrepresented among affected proteins (Figure 9B; see Supplemental Table 2 online). In case of clpc1, Calvin cycle proteins (nine out 27), metabolism of vitamins (five out of 12), and minor carbohydrate metabolism (5 out of 11) were overrepresented among affected proteins (Figure 9B), consistent with the findings in Figure 8 using total abundance per function. This clpc1 molecular phenotype points to reduced photosynthetic capacity and problems in protein homeostasis, consistent with the yellow phenotype, reduced growth, and the role of ClpC1 in protein import. The more modest clps1 molecular phenotype points to destabilization of the tetrapyrrole pathway (Figures 8 and 9B), in addition to more scattered effects (see Discussion).
Figure 9.
Proteins Significantly Up- or Downregulated in the Mutant Alleles.
(A) Venn diagram showing differentially (P < 0.01; determined by GLEE algorithm) expressed proteins in clps1 and clpc1-1. Upregulated and downregulated proteins are indicated with arrows. Only one protein, GUN5, showed opposite behavior between the two mutants.
(B) Overrepresentation analysis of chloroplast functions in the stromal proteomes, based on the number of significantly (P < 0.01; determined by GLEE algorithm) differentially accumulating proteins normalized to the number of proteins identified in each function. Functions marked with an arrow appear overrepresented. Only functions with 10 or more identified proteins are shown. Functions are ranked from high (left) to low (right) number of identified proteins (e.g., 35, unknown has the most proteins, 73; 29, synthesis has the least proteins, 10).
(C) Effect of loss of ClpS1 or ClpC1 on the tetrapyrrole pathway based on stromal proteome analysis. GluRS, Glu-tRNA synthase; PGR7, GluTR binding protein ; GSA1,2, Glu-1-semialdehyde 2,1-aminomutase 1,2; ALAD, porphobilinogen synthase-1 (δ-aminolevulinic acid dehydratase-1); HEMC, hydroxymethylbilane synthase; UROM, uroporphyrinogen-III synthase; UPD1,2, uroporphyrinogen decarboxylase 1,2; CPO, coproporphyrinogen III oxidase; CHLD, Mg-protoporphyrin IX chelatase D; CHLI1,2, Mg-protoporphyrin IX chelatase 1,2; GUN5, Mg-protoporphyrin IX chelatase H; SIRB, sirohydrochlorin ferrochelase; HO1, heme oxygenase 1; RCCR, red chlorophyll catabolite reductase. wt, the wild type. Error bars show the sd.
(D) Immunoblot analysis of proteins in the tetrapyrrole pathway using total leaf extracts from the wild type, clps1, and clpc1-1. Equal amounts of proteins were loaded. Error bars show the sd.
Figure 9C shows the quantitative response of the tetrapyrrole pathway, including 14 proteins involved in tetrapyrrole biosynthesis and one enzyme involved in chlorophyll degradation. Out of those 14 proteins, five proteins (GSAT, ALAD, UPD1, CHLI, and GUN5) were significantly (P < 0.01) downregulated and three (SIRB, HO1, and red chlorophyll catabolite reductase) were significantly (P < 0.01) upregulated in clps1. These upregulated proteins are specifically involved in siroheme (SIRB) and heme (HO1) biosynthesis, whereas the downregulated proteins are involved in the central tetrapyrrole pathway (GSAT, ALAD, and UPD1) or specifically chlorophyll biosynthesis (CHLI and GUN5). Red chlorophyll catabolite reductase involved in removal of toxic chlorophyll catabolite (Pruzinska et al., 2007) was upregulated in clps1 but not in clpc1-1. GluTRBP/PGR7 was 11-fold upregulated (P < 0.01) in the clpc1-1 mutant but was not significantly affected in clps1. GluTR-interacting protein (GluTRBP), also named PGR7 (Jung et al., 2010), helps to funnel ALA into the heme biosynthetic pathway, at the expense of chlorophyll (Czarnecki et al., 2011). GUN5 showed opposite behavior between the genotypes, with upregulation (3×) in clpc1 but downregulation (0.5×) in clps1 (Figure 9B). GUN5 was also strongly (>10×) upregulated in young clpr2-1 mutants (Zybailov et al., 2009b).
Immunoblot analysis using total leaf extracts of the three genotypes (Figure 9D) showed that ∼20% of GUN5 (a 144-kD protein) accumulated as an ∼110-kD breakdown protein in clpc1 and the double mutant but not in the wild type or clps1. When including the signal from this lower mass product, total GUN5 signal was ∼1.7× stronger in clpc1 and the double mutant. In case of the MS analysis, GUN5 was even more (3.4×) increased in clpc1. We have carefully evaluated the identified GUN5 peptides, and we can conclude only that GUN5 is clearly much higher in clpc1, but we did not see strong evidence for cleavage products by MS. GUN4 levels were reduced (0.4×) in clps1 and not much changed in clpc1 or the double mutant (Figure 9D), with similar tendencies (but no statistical significance) observed by MS. GluTR is the key control point for tetrapyrrole biosynthesis, whereas GUN4 improves Mg chelatase activity (Czarnecki and Grimm, 2012). Immunoblotting using total leaf extracts showed that GluTR is upregulated in clps1 but downregulated in clpc1-1. GluTR could not be quantified by MS.
The Molecular Phenotype of the clps1 clpc1 Double Null Mutant
To determine a possible molecular phenotype in clps1 clpc1, we performed quantitative comparative stromal proteome experiments as for the single mutants (Figure 7C). In total, 1077 proteins were identified with two or more adjSPC, giving rise to a total of 127,429 adjSPC (see Supplemental Data Set 5 online), and 793 proteins were located in the chloroplast and accounted for 95% of all adjSPC (see Supplemental Data Set 5 online). The Venn diagram shows the identified plastid proteins in the two genotypes, with 75% overlap (Figure 7C). The biological replicates were very similar, with Spearman correlation coefficients between replicates of the same genotype 0.96 (wt2) and 0.93 (clps1 clpc1), and PCA showed a clear double mutant proteome phenotype compared with the wild type (see Supplemental Figure 5 online).
We first evaluated the molecular phenotype of clps1 clpc1 based on chloroplast protein mass investments in 34 functions, including statistical analysis (Figure 8; see Supplemental Table 2 online) and compared that with the single mutant analysis, as discussed above. Consistent with the visible phenotype, the double mutant showed a generally similar molecular phenotype to that of the clpc1 single mutant, with significant (P < 0.01 or P < 0.01; t test) effects on eight functions. In particular, the double mutant showed downregulation of the Calvin cycle (by 30%), organic conversions (38%), and nucleotide metabolism (20%) and upregulation RNA metabolism (74%), isoprenoid metabolism (83%), vitamin biosynthesis (195%), ribosome biogenesis (100%), and metal homeostasis (52%) (Figure 8).
Significance analysis (P < 0.01; GLEE) of differential accumulation of individual proteins in clps1 clpc1 compared with wild-type plants showed that 65 stromal proteins were upregulated and 19 were downregulated (see Supplemental Data Set 6 online). Comparison of the differentially expressed proteins in clps1 clpc1 to those of the single mutants shows that 22 were upregulated and four were downregulated in both clpc1 and clps1 clpc1 (see Supplemental Data Sets 3 and 6 online). This confirms that the phenotype of the double mutant strongly resembles clpc1, but with an interesting apparent molecular interaction effect on RNA metabolism and ribosome biogenesis (Figure 8), reflected by upregulation of proteins in both categories. In particular, RH3 DEAD box RNA helicase, RNA splice factor P67 (also named SVR7), PPR proteins AT5G46580 and pTAC2, rpoC2 RNA polymerase β (PEP-β), and several editing and splice factors were upregulated (respectively 8×, 3×, >50×, 14×, and 10×) in the double mutant. Interestingly, SVR7, pTAC2, and AT5G46580 are PPR proteins that also have a small MutS-related domain, often associated with endonuclease activity (on single strands) that might have a role in mismatch repair or genetic recombination (see Discussion). We note that RH3 was also strongly upregulated in the ClpPR core mutants clpr2-1, clpr4-1, and clpp3-1 (Kim et al., 2009; Zybailov et al., 2009b; Kim et al., 2013), that RH3 is involved in RNA splicing and possibly ribosome biogenesis (Asakura et al., 2012), and that RNA splice factor SVR7 is a suppressor of a thylakoid FtsH protease mutant (Liu et al., 2010) (see Discussion).
ClpS1-Regulated Protein Degradation Is Coupled with Translational Capacity in Plastids
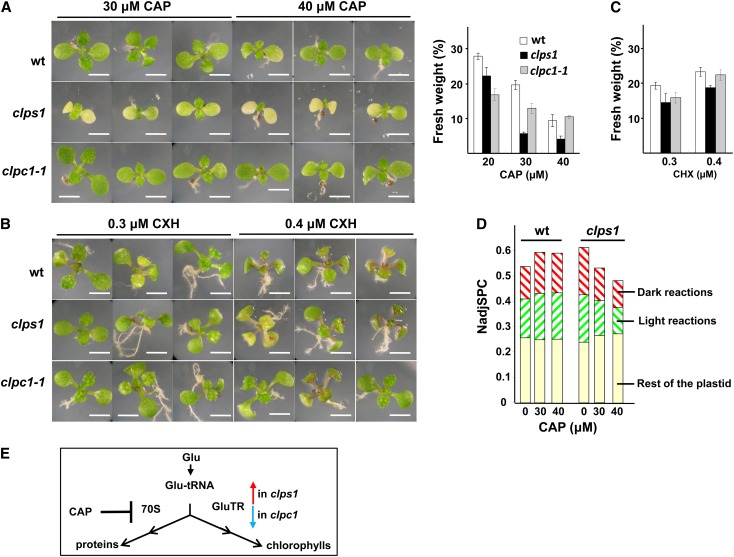
ClpS1 showed the highest accumulation in developing rosettes where plastid protein synthesis is high. Furthermore, the proteomics phenotype of clps1 clpc1 showed a phenotype in RNA metabolism and ribosome biogenesis factors (Figure 8), perhaps due to a synergistic clps1 and clpc1 interaction effect. Finally, one of the key functions of the Clp protease in E. coli (in particular ClpXP) is the removal of (SsrA-tagged) proteins resulting from stalled translation (Baker and Sauer, 2012). E. coli mutants that lack functional SsrA tagging showed increased sensitivity to several protein synthesis inhibitors, such as CAP, lincomycin (LIN), and spectinomycin (SPN) (de la Cruz and Vioque, 2001). Although no SsrA tags have been found in plants, we postulated that perhaps the plastid Clp system is important to remove stalled nascent protein chains and that clps1 may show increased sensitivity to a translational defect in the chloroplast. To test the hypothesis, we grew wild-type, clps1, and clpc1 plants on agar plates (with 2% Suc) containing different concentrations of the prokaryotic-type translational inhibitors LIN (50, 75, and 100 μM), SPN (1, 2, 4, 6, and 8 μg/mL), or CAP (5, 10, 20, 30, and 40 μM) (Figure 10A; data not shown). These three inhibitors differ in their molecular mechanism (Harms et al., 2003): (1) LIN interacts with the A- and P-site on the 50S subunit, hampering positioning of both tRNA molecules and directly inhibiting peptide bond formation; (2) SPN binds to the head region of the 30S subunit inhibiting EF-G catalyzed translocation of the peptidyl-tRNA from A- to P-site; and (3) CAP occupies the position of the amino acid attached to the A-site tRNA, preventing peptide bond formation and resulting in stalled polysomes. Because of the capacity of CAP to stabilize/trap polysomes, it is frequently used in purification and analysis of plastid polysomes. As control, we also grew these plants on plates containing the cytosolic (80S) translation inhibitor cycloheximide (CHX) (0.3 to 0.4 μM) (Figure 10B). As expected, all three genotypes showed decreased plant growth and bleaching for the prokaryotic type inhibitors with increasing effects at increasing concentrations. However, only in the case of CAP did we observe visible and reproducible differences between clps1 and the other genotypes (the wild type and clpc1). Fresh rosette weight of wild-type seedlings was approximately fivefold reduced at 20 µM CAP and further decreased at high CAP concentrations (see Supplemental Figure 7 online). Chlorophyll and carotenoid content based on fresh weight in the wild type increased by ∼25% at 20 µM CAP and then gradually decreased (chlorophyll) or remained unchanged (carotenoids) with increasing CAP concentrations (see Supplemental Figure 6 online). Importantly, growth on CAP resulted in reduced growth, development, and fresh weight (Figures 10A and 10C), as well as increased virescence and measurable loss of chlorophyll (per fresh weight) in clps1 compared with the wild type and clpc1 (see Supplemental Figure 6 online). No such differences between the wild type, clps1, and clpc1 were observed on CHX (Figure 10C). These results demonstrated a conditional phenotype in clps1 upon application of CAP, suggesting a functional linkage between protein synthesis and ClpS1-mediated proteolysis in the chloroplast, perhaps with ClpS1 playing a role in removing nascent chains from stalled ribosomes or through GluTR degradation (see also further below). It is not clear why LIN or SPN did not show similar genotype-specific effects.
Figure 10.
Effects of Translation Inhibitors on clps1, clpc1-1, and the Wild Type.
(A) and (B) Seedling phenotypes after treatment with CAP or CHX. Representative visible phenotypes of wild-type (wt), clps1, and clpc1-1 plants grown for 20 d under short days on half-strength Murashige and Skoog medium containing 2% Suc and CAP or CHX. Bars = 5 mm.
(C) Effect of antibiotic treatments on fresh weight of wild-type and clps1 seedlings. Plants were grown for 21 d on agar plates containing 20, 30, or 40 μM CAP (left-hand panel) or 0.3 and 0.4 μM CHX (right-hand panel). Error bars show the sd.
(D) Effect of CAP treatment on investments in the plastid proteome. Total cellular proteins were extracted from wild-type and clps1 seedlings after growth on 0, 30, or 40 μM CAP. The proteomes were loaded on a SDS-PAGE gels and proteins identified and quantified by MS/MS analysis after in-gel tryptic digestion. Total NadjSPC were calculated for all identified plastid proteins, for proteins involved in the Calvin cycle and proteins that are part of the thylakoid photosynthetic electron transport chain.
(E) Effect of CAP and the clps1 and clpc1 null mutants on the branch point for Glu-tRNA utilization. 70S, plastid 70S ribosomes.
To investigate this conditional phenotype in more detail, we performed a comparative proteome analysis of total leaf protein extracts of the wild type and clps1 after growth on 0, 30, or 40 μM CAP, using label-free MS-based quantification, similar to the previous experiments described above. In total, 2250 proteins were identified with two or more adjSPC, with a total of 101,634 matched adjSPC, and 767 proteins were located in the chloroplast and matched 56,857 adjSPC (56%) (see Supplemental Data Set 7 online). Increasing concentrations of CAP clearly decreased the relative plastid protein mass (by 31% at 40 μM CAP) in clps1 but not at all in the wild type, mostly by decreased investments in the light (43%) and dark (44%) reactions of photosynthesis (Figure 10D). The loss of plastid proteome is consistent with the visual and chlorophyll phenotypes, but the proteomics data did not provide an explanation for the higher sensitivity of clps1 for CAP.
Identification of Candidate ClpS1 Substrates by in Vitro Affinity Purification
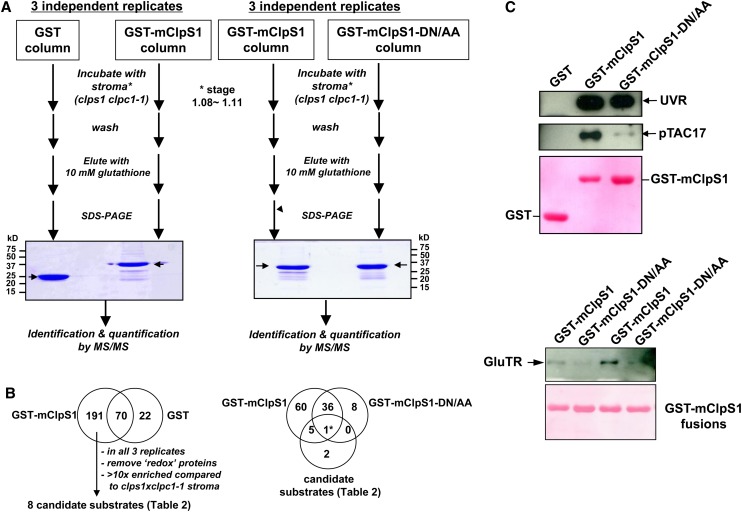
To identify direct candidate substrates for ClpS, we developed affinity-based experiments using recombinant GST-ClpS1 fusion protein with recombinant GST as control and the stromal proteome of clps1 clpc1 plants as source for potential ClpS1 substrates and/or ClpS1 interactors (Figure 11). We reasoned that substrates for ClpS1 should be overrepresented in stroma of clps1 clpc1 mutants, but not in the wild type. We incubated both GST and GST-ClpS1 affinity columns with isolated stromal proteome from chloroplasts isolated from young clps1 clpc1 seedlings and eluted potential interactors with reduced glutathione. The protein eluates were run on SDS-PAGE gels, stained with Coomassie blue, and the complete lane processed for protein identification and quantification by nano-liquid chromatography–MS/MS on the LTQ-Orbitrap. Figure 11A shows the SDS-PAGE image of one of three (pairwise) biological replicates. Using a similar bioinformatics workflow as for the previous proteome analysis, we identified 283 proteins (231 in plastids), with 22 proteins unique to GST controls and 191 proteins unique to the ClpS1-GST fusion affinity chromatography, and an overlap of 70 proteins (see Supplemental Data Set 8 online). The number of matched adjSPC was on average 4.5 times higher using ClpS1-GST compared with the negative control using GST, suggesting that we isolated a number of (putative) ClpS1 interactors. As expected, proteins identified with both ClpS1-GST and GST were dominated by proteins with known affinity for glutathione (e.g., glutathione transferase and glutaredoxin) and some of the very abundant stromal proteins (e.g., RBCL and Rubisco activase) and chaperones (e.g., CPN60). We further considered only the proteins uniquely identified in the ClpS1-GST affinity steps, removed additional redox proteins (various thioredoxins, and glutathione reductase and peroxidase), and required identification in each of the three replicates. This resulted in 51 remaining stroma-localized proteins. To select the most likely candidate substrates, affinity-purified proteins were required to have a 10-fold higher relative abundance (calculated based on NadjSPC) in the affinity eluates (see Supplemental Data Set 8 online) than in the stromal proteome of clps1 clpc1 or be absent in the unfractionated stroma (see Supplemental Data Set 5 online). Using this enrichment criterion, eight out of these 51 proteins remained as candidate substrates (Table 2, Figure 11B). These eight proteins were also enriched when compared with the wild type and single mutant stromal proteomes (Table 2). We consider those eight proteins as the best candidates for ClpS1 interactors and/or ClpS1 substrates; comments on their function are provided in the Discussion.
Figure 11.
Identification of Candidate ClpS1 Substrates by Affinity Chromatography.
(A) The workflow for the ClpS1 affinity purification and subsequent MS-based protein identification of ClpS1 substrate candidates. Representative gel images of pull-down eluates using wild-type and mutant (DN/AA) versions of GST-ClpS1 fusions, or the negative control GST, as bait proteins. The arrow indicates eluted GST or GST-ClpS1 (bait). Some of the visible bands represent breakdown products of recombinant GST or GST-ClpS1.
(B) The workflow used to choose candidate ClpS1 substrates. The Venn diagram on the left shows those proteins identified using either GST or GST-ClpS1 as bait. From the 191 proteins only identified in the GST-ClpS1 affinity experiments, we considered only those candidate substrates that were observed in all three independent replicates that were also at least 10-fold enriched compared with the stroma of clps1 clpc1-1. Moreover, proteins with glutathione binding domains or thioredoxins were removed. The resulting eight candidate substrates were then compared with the proteins identified in a separate set of GST-ClpS1 and GST-ClpS1-DN/AA affinity assays as indicated in the Venn diagram on the right. Details of the candidate substrate proteins can be found in Table 2.
(C) Confirmation of ClpS1 interactions by immunoblotting. Eluates were transferred to blots and probed with antisera raised against pTAC17, the UVR protein, or GluTR. Ponceau stains of the blots are shown and visualize the recombinant bait.
[See online article for color version of this figure.]
Table 2. Candidate Plastid Substrates for ClpS Interactors and/or ClpS Substrates Based on Affinity Enrichment Using Chloroplast Stroma from clps clpc1 Double Mutant Leaves.
| Locusa | Annotation | Function | ClpS-GST (Sum adjSPC)b | ClpS-GST/wt1c | ClpS-GST/clpsc | ClpS-GST/clpc1c | ClpS-GST/wt2c | ClpS-GST/clps clpc1c | ClpS-GST and ClpS-DNAA-GSTd |
|---|---|---|---|---|---|---|---|---|---|
| AT2G03390.1 | uvrB/uvrC motif-containing protein (UVR) | 28-DNA | 105 | 131 | 14775 | 197 | nd | nd | 0.38* |
| AT1G80480.1 | PRLI-interacting factor L (pTAC17) | 28-DNA | 46 | 22 | 24 | 9 | 47 | 16 | Only in wild-type ClpS (3x) |
| AT1G22410.1 AT4G33510.1 AT4G39980.1 | 3-Deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthetase (DHS) | 13-Aromatic AA metabolism | 45 | 6364 | 6364 | 12 | 2121 | 11 | Only in wild-type ClpS (2x) |
| AT1G48850.1 | Chorismate synthase (CS) | 13-Aromatic AA metabolism | 32 | 63 | 51 | 27 | 17 | 16 | Only in wild-type ClpS (2x) |
| AT3G03890.1 | Pyridox_oxidase domain protein (PYROX) | 18-Vitamin b6 metabolism | 25 | 13 | 9 | 4 | 19 | 11 | Only in wild-type ClpS (1x) |
| AT1G58290.1 | Glutamyl-tRNA reductase (GluTR) | 19-Tetrapyrole metabolism | 18 | nd | nd | nd | nd | nd | Only in wild-type ClpS (2x) |
| AT5G52960.1 | Unknown protein | 35-Unknown | 14 | 14 | 38 | 89 | nd | nd | In neither |
| AT5G47870.1 | RAD52-2 | 28-DNA | 7 | nd | nd | nd | nd | nd | In neither |
Asterisk indicates ratio mutant ClpS/wild-type ClpS calculated from NadjSPC. nd, not determined because the protein was not detected in the stroma.
Proteins present in all of the three replicates of ClpS-GST affinity purification against stroma from clpsxclps, but never in the three replicates with GST as bait.
Sum of the adjSPC across the three replicates of ClpS-GST affinity purification.
Fold enrichment in the average of the three replicates of ClpS-GST affinity purification compared with the abundance in stroma from wt1, wt2, clps, clpc1, or clps clpc1 (from Supplemental Tables 1 and 5 online). NadjSPC was used to calculate the enrichment.
Comparison of candidate proteins using wild-type ClpS-GST versus mutant Clps-GST (average of three replicates).
As described earlier, critical residues in ClpS1 for substrate interaction and N-degron binding have been identified in E. coli. Using this information, in vitro pull-down experiments in E. coli using ClpS1 mutated in these critical residues (D35A/D36A) allowed identification of two N-end rule substrates from ClpAS-depleted cell lysates (Ninnis et al., 2009). We therefore created a mutant form of Arabidopsis ClpS1 harboring a double mutation in the corresponding residues D89A/N90A (Figure 2B) and repeated the affinity experiment using clps1 clpc1 stroma with three biological replicates (Figure 11; see Supplemental Data Set 8 online). In total, 110 proteins were identified (after removing glutathione interactors and thioredoxins), with 65 unique to the wild-type ClpS1 bait, 37 shared between both, and only eight unique to the ClpS1 D89A/N90A mutant construct (Figure 11). Out of the eight candidate substrates listed in Table 2, we identified five only with the wild-type ClpS1 bait, one (UVR domain protein) was identified with both baits, and the two least abundant proteins in Table 2 were not identified. This suggests that five of the candidate substrates require the conserved ClpS1 substrate binding site for interaction, whereas the interaction between the UVR protein and ClpS1 is governed by a different region.
To verify the MS/MS-based analysis, we performed immunoblot analysis of the GST-ClpS1 affinity eluates using antisera against pTAC17, the UVR protein, and GluTR (Figure 11C). This confirmed that each of these is specifically enriched by the GST-ClpS1 affinity experiments, but not by GST alone. Furthermore, in agreement with the MS/MS-based quantification, pTAC17, and GluTR, but not the UVR protein, showed a near absolute requirement for the presence of the conserved DN residues in ClpS1.
Features of the N-Terminal Region of Candidate Substrates Recognized by ClpS1
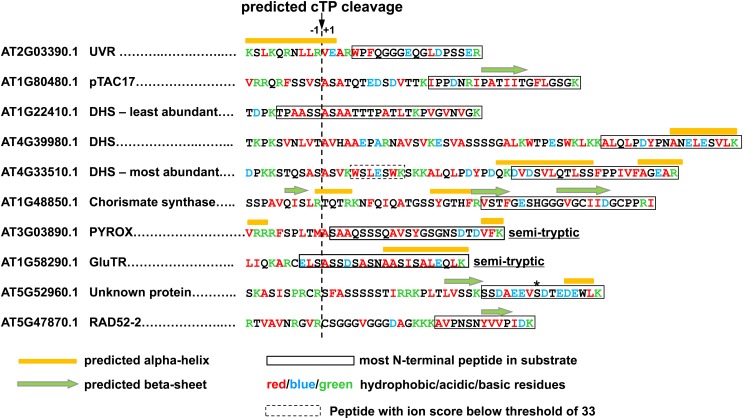
In E. coli, efficient degradation of a ClpS substrate is dependent on the N-degron, the presence of a short, unstructured region after the N-degron, and a short hydrophobic region in this unstructured linker region (Erbse et al., 2006; Ninnis et al., 2009). We therefore explored the N-terminal region of the eight Arabidopsis candidate substrates. Because each of the substrates have predicted N-terminal chloroplast transit peptides that are removed after import into the chloroplast, we used the predicted cTP processing site (by TargetP) and mapped the peptides identified in the GST-ClpS affinity purifications onto the sequences (Figure 12). In an effort to identify N-terminal peptides with posttranslational modifications, such as deamidation or acetylation, or even an additional N-terminal residue postribosomally attached by an amino acid transferase (as in E. coli; see Introduction), we also did so-called error-tolerant searches using MASCOT. However, no additional N-terminal peptides were detected. We then predicted the secondary structure (using Jpred, http://www.compbio.dundee.ac.uk/www-jpred/) and estimated the end points of the unstructured regions (immediately before α-helices or β-sheets). Finally, we color-coded hydrophobic, acidic, and basic residues (Figure 12). This collective information best describes the current information for the N-terminal sequence of the candidate substrates.
Figure 12.
Primary Sequences, Predicted Secondary Structure, and Experimental Information about the N-Terminal Regions of the Candidate ClpS1 Substrates.
Because each of the substrates have predicted N-terminal chloroplast transit peptides that are removed after import into the chloroplast, we used the predicted cTP processing site (by TargetP) and mapped the peptides identified by MS in the eluates from GST-ClpS affinity purifications onto the sequences. The asterisk indicates a phosphorylation site.
In case of the pyroxide oxidase domain (PYROX) and GluTR proteins, we likely identified the true N terminus by MS/MS as a semitryptic peptide (identified five times for GluTR and one time [with high ion score of 61] for PYROX), suggesting that these start with SAAQSS and ELSASS, respectively. For the other candidate substrates, the most N-terminal detected peptides were full tryptic, and the true N termini are likely upstream. Most of the candidate substrates had no predicted secondary structure around the N-terminal region and acidic residues (E/D) were underrepresented. These findings are consistent with patterns observed for ClpS substrates in E. coli. At this point, however, it would be premature to predict rules or motifs for chloroplast N-degrons recognized by ClpS1, given the uncertainty of the location of the N termini. Our efforts are now focused on the challenging task of determining these N-terminal residues, but also testing other recognition sites in ClpS1 candidate substrates.
DISCUSSION
Substrates and Substrate Recognition Hold the Key to Understanding Protease Function
The selection mechanism for proteolysis in plants is best understood for the 26S proteasome; it involves the covalent attachment of a small protein molecule, ubiquitin, to an internal Lys on the substrate resulting in the ATP-dependent degradation of the ubiquitylated protein (Finley, 2009; Vierstra, 2009). However, chloroplasts do not have a proteasome, and mechanisms or tagging systems for protease substrate selection within the chloroplast are not understood. Nevertheless, it is known that chloroplasts (and plastids in general) contain several dozen proteases in each of the plastid compartments (thylakoid lumen, integral thylakoid, stroma, envelope, and plastoglobules); most of these proteases evolved from bacteria. In the case of the Clp protease system (reviewed in Olinares et al., 2011a), much can be learned from the many mechanistic and structural studies regarding the Clp system in E. coli and B. subtilus (reviewed in Kirstein et al., 2009; Kress et al., 2009; Sauer and Baker, 2011; Baker and Sauer, 2012; Dougan et al., 2012). Surprisingly, even for these well-studied bacterial systems, only two physiological substrates for ClpS have been firmly identified, indicating the challenge of finding substrate and substrate selection mechanisms.
The central objective of this study was to determine the function and conservation of chloroplast ClpS in plants and identify candidate Clp protease substrates in Arabidopsis plastids that are recognized by ClpS. Bacterial ClpS is a protein specialized in selecting substrates with N-degrons, but other regions in the substrate also contribute, and ClpS may also be involved in degradation of substrates independent of the N-end rule. Generation of such N-degrons typically involves active modification of the N terminus, through deformylation (by a deformylase) and removal of one or more amino acids (by endopeptidases), followed by the (nonribosomal) addition of an amino acid through various types of amino transferases (Varshavsky, 2011; Dougan et al., 2012). Because of this complexity, it has been difficult to predict N-degrons in bacteria; consequently, identification of possible plastid N-degrons in plants will need to come from experimentation. An N-end rule and N-degrons have not been established for plastids, but it has been demonstrated that plastid protein N termini contribute to protein half-life (Apel et al., 2010).
ClpS1 Interacts with ClpC1 and ClpC2 and Is a Conserved Substrate Selector for the Chloroplast Clp Protease
Our phylogenetic analysis clearly suggested that most CLPS genes in vascular plants are derived from cyanobacterial CLPS1 and not cyanobacterial CLPS2; therefore, we assigned them as ClpS1. Moreover, several algae, moss, and vascular plants contain a second ClpS-derived gene, which we assigned ClpS1-like. Arabidopsis has only one CLPS gene, and it is part of the ClpS1 clade. Detailed comparison of the well-studied E. coli ClpS and Arabidopsis ClpS1 identified conservation of conserved elements, suggesting that Arabidopsis ClpS1 is a functional homolog of E. coli ClpS. However, ClpS1 and ClpS1-like both have extended N termini (downstream of the cTP) indicative of their diversification. Importantly, in vitro reconstitution using recombinant ClpS1 with (the N-terminal part of) ClpC1 and ClpC2 showed that ClpS1 interacted with both ClpC chaperones. This was further supported by gel filtration analysis of the oligomeric state of the chloroplast stroma proteome. Together, this suggests that Arabidopsis ClpS1 (and ClpS1-like proteins in other species) likely has a conserved function in substrate selection for the plastid Clp protease system.
The Role of ClpS1 as Part of the Plastid Clp System
Given the presence and strong conservation of ClpS genes across all tested plant and algal species, it was surprising that the Arabidopsis clps1 null mutant did not show a visible growth or developmental phenotype. However, in the presence of the plastid translation elongation inhibitor CAP, the clps1 null mutants showed a conditional phenotype compared with the wild type as well as clpc1 null mutants. Proteome analysis of total leaf extracts of these CAP-treated plants showed a loss of plastid proteins compared with the wild type, consistent with the loss of chlorophyll and reduced biomass, suggesting that ClpS1 plays a role in plastid biogenesis in particular when chloroplast protein synthesis capacity is a limiting factor. Perhaps the loss of ClpS1, and the consequent effects on the tetrapyrrole pathway, has effects on the distribution of Glu-tRNA or Glu-tRNA synthase between protein synthesis and tetrapyrrole synthesis (Figure 10E). We generated double mutants between ClpS1 with mutants in the ClpPR core (clpr2-1) and each of the ClpC/D chaperones, as well as a triple mutant between ClpS1, ClpC1, and ClpC2 with a leaky allele for ClpC2 (to avoid lethality). No contribution of the ClpS1 background to the visible phenotypes was found. Similarly, loss of ClpS in E. coli also does not result in a growth phenotype. The lack of visible phenotype is in line with a role of ClpS1 as a Clp protease adaptor that enhances the delivery of a subset of substrates to the Clp chaperones, while suppressing delivery of other substrate classes. Consistent with a functional relation between ClpS and ClpC1,2, ClpS1 levels increased threefold in the clpc1 and clpc2 null mutants.
Comparative quantitative analysis of the stromal proteome, as well as immunoblotting of total leaf extracts, of the wild type, clps1, clpc1, and clps1 clpc1 did show a molecular phenotype for clps1, which was different and less dramatic than for clpc1 and the double mutant. In particular, the tetrapyrrole pathway was affected in clps1, though only subtly reducing the chlorophyll content. For instance, the level of GluTR was increased (by 25% ± 3%) in clps1, whereas GUN4 was decreased in clps1 (by 51% ± 21%), but unchanged in clpc1. The molecular phenotype of clpc1 and the double mutant was much stronger due to the role of ClpC1 in protein import (Constan et al., 2004; Kovacheva et al., 2007), in addition to a likely role in proteolysis, in agreement with the virescent phenotypes, with significant decrease of the Calvin cycle, increased abundance of various proteins involved in protein synthesis and folding, and altered accumulation of proteins involved in various aspects of metabolism.
Candidate ClpS1 Substrates Are Involved with Central Plastid Metabolic Pathways and Plastid DNA Quality Control
The ClpS1 affinity experiments identified eight candidate substrates (11 when counting the three isoforms of DHAP synthase [DHS] separately). Three of these (RAD52-2, UVR protein, and pTAC17) have functions related to plastid DNA organization/quality/repair. RAD52-2 (AT5G47870) was recently shown to be localized in plastids and functions in DNA repair (Samach et al., 2011), and we identified RAD52-2 in Arabidopsis nucleoids (Huang et al., 2013). UVR has both UVR and YccV domains (AT2G03390) and has not been studied in plants. However, the YccV protein in bacteria functions as an initiator for DNA replication (Leonard and Grimwade, 2011; Scholefield et al., 2011), and UVR domains are found in the bacterial Uvr-ABC machinery involved in nucleotide excision repair (Truglio et al., 2006). pTAC17 (AT1G80480) is a protein with CobW domains and derives its name from identification in plastid transcriptionally active chromosome (pTAC) preparations. The UVR protein was identified with very high MS scores in the affinity purification, but it is a low abundance protein in chloroplasts. pTAC17 was abundant in stroma, but its abundance did not appear to be changed in clps1. Research into a possible role of the Clp system in plastid DNA organization and quality control is warranted.
The identification of GluTR as ClpS1 interactor matches well to the increased abundance in the clps1 mutant observed through immunoblotting. Similar to RAD52-2 and UVR protein, GluTR was not sufficiently abundant to be quantified/observed in the stroma proteome using MS; this makes its identification with high MS scores in the affinity purifications even more significant. GluTR is a key regulatory enzyme in tetrapyrrole biosynthesis (Schmied et al., 2011; Czarnecki and Grimm, 2012) in complex with one or more proteins (Czarnecki et al., 2011). Interestingly, GluTR in the bacterium Salmonella typhimurium was shown to be a substrate for ClpAP and/or LON proteases (Wang et al., 1999a, 1999b). GluTR in bacteria is more stable and more abundant in heme-limited cells but unstable and less abundant in normal growing cells. The turnover of GluTR in bacteria was dependent on the N-terminal 18 amino acids of GluTR. We therefore compared this N-terminal region of bacterial GluTR with Arabidopsis GluTR as part of an extensive phylogenetic analysis of GluTR in (cyano)bacteria, algae, and angiosperms, but whereas there is significant sequence conservation for this 18-residue domain (data not shown), the most N-terminal peptide identified in ClpS1 affinity purifications for Arabidopsis GluTR was upstream of this 18-residue domain.
Candidate substrates, the DHAP synthases and chorismate synthase, are involved in the shikimate pathway, which is a major plastid-localized pathway providing precursors for the aromatic amino acids and all phenylpropanoids, flavonoids, and lignin. It was striking that all three DHSs (AT1G22410, AT4G33510, and AT4G39980) were identified as ClpS1 interactors, and each of the paralogs was identified with unique peptides. The DAHP synthase family carries out the first step in the plastid-localized shikimate pathway (converting PEP and erythrose 4-phosphate into DHAP), while the candidate substrate chorismate synthase (AT1G48850) carries out the last step of the pathway, generating chorismic acid, a major intermediate branch point metabolite (Tzin and Galili, 2010). Consequently, the pathway must be carefully regulated in order to meet varying demands. Both DHS and chorismate synthase are under negative feedback control (Tzin and Galili, 2010); ClpS-regulated proteolysis could provide a control mechanism to reduce the activity of the shikimate pathway, in particular after leaf development is complete, and avoid unnecessary consumption of costly reduced carbohydrates.
Finally, the two other candidate substrates are AT5G52960 and PYROX protein (AT3G03890) possibly involved in vitamin B6 metabolism. At5g52960 has no predicted functional domains, but it has a short Ser N-terminal repeat followed by Arg residues. As frequently observed for Ser/Arg repeat proteins, this protein has been shown to be phosphorylated (Figure 12) in several studies (Sugiyama et al., 2008; Kline et al., 2010; Reiland et al., 2011) (see http://phosphat.mpimp-golm.mpg.de), which may relate to protein stability or function.
In summary, most of the candidate substrates are involved either in DNA organization/repair or in key secondary metabolic pathways, consistent with the accumulation of ClpS1 in young developing tissue. The notion that the Clp protease system is by far the most abundant soluble protease in the chloroplast (Peltier et al., 2004, 2006) is consistent with a broad range of substrates.
Additional Candidate Substrates
The selection of the eight candidate substrates was in part determined by a minimal required enrichment (10-fold) compared with the stroma of clps1 clpc1-1. When reducing this minimal requirement to fivefold enrichment, while maintaining the other requirements (in all three replicates, but never in the negative GST control), there were five additional candidate substrates. These were 5′-adenylylsulfate reductase-2 (APR2) involved in sulfate metabolism, porphobilinogen synthase-1 (ALAD-1) involved in tetrapyrrole biosynthesis, isopropylmalate isomerase (SSU1) involved in amino acid metabolism, as well as protein Tyr phosphatase (DSP4 or SEX4) and the small subunit of ADP-Glc pyrophosphorylase (APS1), both involved in starch metabolism. APS1 and ALAD1 were also identified three times in the second set of experiments using GST-ClpS but never using the ClpS-DN/AA mutant construct (Figure 11B). In particular, the latter two proteins (APS1 and ALAD1) could also be considered candidate substrates, thereby adding another enzyme of the tetrapyrrole biosynthetic pathway to the list of putative ClpS1 targets and expanding the substrate list to starch metabolism.
Features of the N-Terminal Region of Candidate Substrates Recognized by ClpS1
In E. coli, efficient degradation of a substrate bearing an N-degron is dependent on the presence of a short unstructured region between the folded domain and the N-degron (Erbse et al., 2006). Moreover, the composition of the linker affects substrate turnover (Erbse et al., 2006; Wang et al., 2008a; Ninnis et al., 2009).
The only established example of a degron for plastid proteins is for the enzyme chlorophyllide a oxygenase (CAO), which converts chlorophyll a into chlorophyll b (Sakuraba et al., 2009). The activity of CAO is regulated at the level of protein stability via a negative feedback mechanism through chlorophyll b. ClpC1 and an amino acid sequence (97)QDLLTIMILH(106) in the N-terminal domain (designated the A domain) of CAO are both essential for this regulatory mechanism (Nakagawara et al., 2007). Moreover, this sequence induced the destabilization of green fluorescent protein, suggesting that this sequence serves as a degradation signal that is recognized by proteases functioning in the chloroplast (Sakuraba et al., 2009). However, this sequence is not located at the very N terminus of the mature CAO protein, indicating that it is not an N-degron and perhaps not recognized by ClpS1 but rather by ClpC1 directly or via a hypothetical adaptor protein. Using tobacco plants with transgenic chloroplasts that express (plastid-encoded) reporter proteins whose N and C termini were systematically modified, major stability determinants were found to be located in the N terminus (Apel et al., 2010). Testing of all 20 amino acids in the position after the initiator Met (for these plastid encoded proteins) revealed strong differences in protein stability and indicated an important role of the penultimate N-terminal amino acid residue, in part through its influence on excision efficiency of the N-terminal Met, as well as the downstream sequence (Apel et al., 2010). Indeed, the most destabilizing residues (Cys and His) were absent at the penultimate position of plastid-encoded proteins. These observations are consistent with the general N-end rule (Varshavsky, 2011) but insufficient to define a plastid N-degron.
All candidate ClpS substrates that we identified in our experiments were nuclear encoded and not plastid encoded. The N termini of these nuclear-encoded proteins are generated by the action of the stromal processing peptidases, following import into the chloroplast (Richter and Lamppa, 1998; Trosch and Jarvis, 2011). Subsequent removal of one or more residues can occur through the action of various chloroplast amino peptidases, as suggested by systematic stromal proteome analysis (Zybailov et al., 2008). Additionally, many processed chloroplast proteins are N-terminally acetylated (Zybailov et al., 2008; Bischof et al., 2011; Finkemeier et al., 2011; Wu et al., 2011), but little is understood about regulation of these subsequent maturation steps. Moreover, there are no clear plastid candidates for L/F amino-transferases that could transfer a residue to the mature N terminus, thereby generating the primary N-degron residue. Accurate prediction of the N termini (and their possible modifications) of mature nuclear-encoded chloroplast proteins is therefore not possible, thus defining possible N-degrons must come from experimentation.
Among the eight candidate substrates that we identified, we likely identified the true N termini for the PYROX and GluTR proteins, and they are SAAQSSSQAV and ELSASSDSASN, respectively. These N termini have in common that they lack aromatic (Tyr, Phe, and Trp) and positively charged residues (Arg, Lys, and His). Furthermore, they are enriched in Ala and Ser (both hydrophobic) as well as negative charged residues (Gln, Asp, and Glu). The N-terminal residues are Ser or Glu, and the penultimate residues are Ala or Leu (both apolar hydrophobic). There is little obvious similarity to N-degrons in E. coli nor to the stability determinants observed in transgenic tobacco plastids (Apel et al., 2010). Efforts are now underway to identify the true N termini of these ClpS1 interactors and test their function, as well as more downstream regions, as substrate recognition sites.
Our affinity experiments showed that most plastid candidate substrates specifically require the two conserved residues in the N-degron motif of ClpS1, whereas one of them, the UVR protein, interacts with ClpS1 independently of these two tested residues. The observed conserved motifs in angiosperm ClpS1 (Figure 2) combined with the identification of several strong endogenous candidate CpS1 substrates will allow determination of the rules for ClpS1 substrate selection in Arabidopsis and other vascular plants.
A New Opening in Chloroplast Protease Research
The identification of direct candidate substrates for the Clp protease system in chloroplasts in this study represents a significant advancement because no substrates were identified previously. So far, suggested substrates have come from indirect, comparative proteome analysis of ClpPR core mutants and the wild type (Stanne et al., 2009; Zybailov et al., 2009b; Kim et al., 2013). However, it appears that many upregulated proteins in these mutants result from indirect, pleiotropic effects, such as a response to reduced ATP synthesis through photosynthesis (e.g., strong upregulation of the chloroplast envelope ATP/ADP transporter NTT2; Kim et al., 2009) or upregulation of protein chaperones (e.g., strong upregulation of stromal ClpB3 in clpr2-1, clpr4-1, and clpp3-1; Zybailov et al., 2008; Kim et al., 2009, 2013) or the chloroplast RNA splice factor DEAD box helicase RH3 (Asakura et al., 2012). Identification of direct substrates for the Clp system has been challenging even for the simpler and far better studied Clp system in E. coli. Similarly, solid identification of protease substrates for other chloroplast proteases is limited to the D1 protein of PSII, which appears to involve both thylakoid FtsH and lumenal DegP (Kato et al., 2012, and references therein). Now that we have identified direct candidate protease substrates recognized by ClpS1, identification of possible substrates tags or recognition sites should be feasible.
METHODS
Plant Growth, Mutant Isolation, and RT-PCR Analysis
Plant growth, genotyping, and RNA extraction were performed as described previously (Rudella et al., 2006), and additional growth conditions are detailed in the figure legends. The locations of T-DNA insertions were confirmed by DNA sequencing. For RT-PCR, total RNA was isolated with an RNeasy plant mini kit (Qiagen). The first strand was synthesized from equal amounts of total RNA with Superscript III reverse transcriptase (Invitrogen). We tested 15, 20, 25, and 30 cycles for the primer pairs. Fifteen cycles were insufficient to visualize all transcripts, while 20 and 25 cycles best allowed us to visualize the transcripts, and we observed good linearity for 20 and 25 cycles. Primers for genomic PCR or RT-PCR analysis are listed in Supplemental Table 3 online. Details and references for the T-DNA insertion lines described in Table 1.
Phylogenetic Analysis
For phylogenetic analysis of ClpS-related sequences, 57 ClpS proteins from Escherichia coli, five cyanobacterial species, two algal species, moss, and 19 angiosperms were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The aligned sequences were exported in Clustal format and viewed and edited (removal of gaps and/or variable extensions) in Jalview (www.jalview.org/). The alignment is available as Supplemental Data Set 1 online. Sequences were then converted in PHYLIP format, and phylogenetic trees were generated (1000 iterations) using the CIPRES Web portal (http://www.phylo.org/) using the tool RAxML HPC blackbox with the general time reversal model selected as the protein substitution matrix. RAxML bootstrap support values are shown at the nodes of the tree, in which the E. coli ClpS was designated as the outgroup, and ClpS relatives are classified into six clades. The resulting phylogenetic trees were annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Sequence logos were generated using http://weblogo.berkeley.edu/.
Growth on Antibiotics and Heat or Salt Stress Treatment of Arabidopsis Mutants
For antibiotic treatments, clps1, clpc1-1, and the wild type were grown with or without antibiotic (CAP and CHX) in half-strength Murashige and Skoog plates (0.5× Murashige and Skoog salts [Sigma-Aldrich], 1× Gamborg’s B5 vitamin [Sigma-Aldrich], 0.6% [w/v] Phytoblend [Caission Laboratories], and 2% [w/v] Suc, pH 5.7) under 10-h-light (40 µE m−2 s−1, 23°C)/14-h-dark (21°C) cycles for 14 or 20 to 21 days. For salt stress treatments, Arabidopsis seedlings were grown on half-strength Murashige and Skoog plates with 2% Suc and 50, 100, 150, or 200 mM NaCl. For heat shock treatments, 20-d-old Arabidopsis thaliana seedlings grown on half-strength Murashige and Skoog plates were exposed to the following temperature stress treatments: (1) 38°C for 90 min, (2) 38°C for 3 h, or (3) 38°C for 90 min and then 45°C for 30 min. Following these treatments, plants were returned to normal growth conditions and monitored for 5 d.
In Vivo ClpS1, ClpC1, and ClpC2 Interaction by Gel Filtration
To test in vivo interaction between ClpS1, ClpC1, and ClpC2, chloroplasts were isolated from wild-type plants at leaf stage 1.07-1.08. Stromal proteins were prepared in the presence of ATPγS (5 mM) and separated using a Superose gel filtration column as described (Olinares et al., 2010). The eluates were collected and pooled into 14 fractions. Proteins in each fraction were then trichloroethanoic acid precipitated, and equal volumes were analyzed by immunoblotting and Ponceau staining.
GST-ClpS1 and GST-ClpS1 DN/AA Cloning and Protein Expression
For testing the direct interaction of ClpS1 with ClpC1/C2/T1/T2, recombinant ClpS1-(His)6, ClpC2-(His)6, and ClpT1,T2-(His)6 proteins as well as GST, GST-ClpS1, and GST-N-domain of ClpC1 (residues 39 to 252) were generated in E. coli. Primer sequences used are listed in Supplemental Table 3 online. ClpS1 cDNA was amplified by GST-ClpS forward and reverse primers and cloned into pGEX5X-1(GE Health Sciences) using NotI and EcoRI. To generate the site-specific mutant for Arabidopsis ClpS1 D89A/N90A (DN/AA), two pieces of cDNA was amplified using primers GST-ClpS forward/ClpS_DNAA reverse, and ClpS_DNAA forward/GST-ClpS reverse, and connected into one cDNA using GST-ClpS forward and reverse primers. After cloning using the pCR8/GW/TOPO TA cloning kit (Invitrogen), mutations were confirmed by DNA sequencing and introduced into pGEX-5X-1 with NotI-EcoRI sites. For generation of recombinant GST, we used the pGEX-5X-1 vector. GST-ClpS1 and GST-ClpS1 DN/AA were expressed E. coli strain BL21 and Rosetta (DE3) cells, respectively, by adding 1 or 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h. Cells were broken by sonication in buffer containing PBS, pH 8.0, and 1 mM protease inhibitor phenylmethylsulfonyl fluoride. Both proteins were recovered in the soluble fraction and were further purified with a GST trap FF column (GE healthcare). After washing the GST trap column with PBS and GST elution buffer (50 mM Tris-HCl, pH 8.0, and 150 mM NaCl), proteins were eluted by 10 mM reduced GSH in the GST elution buffer and dialyzed at 4°C overnight in IPTG buffer. ClpT1 and ClpT2 were cloned out of an Arabidopsis cDNA library. The gene products were cloned into the expression vector pET-28b (Novagen). ClpT1/ClpT2 forward and reverse primers were used for cloning procedure. The plasmid pET28-ClpT1 and pET28-ClpT2 were transformed into BL21 (DE3) E. coli cells. Proteins were induced with IPTG (1 mM final concentration) at 37°C for 3.5 h and purified by the nickel-nitrilotriacetic acid (Ni-NTA) His bind resin (Novagen) with a step gradient (40 to 250 mM imidazole with 30 mM increments, 50 mM NaH2PO4, pH 8.0, and 300 mM NaCl). For ClpS1-(His)6, the gene product was amplified with ClpS-His6 primers and introduced into pET28b. The resulting vector was transformed into BL21 (DE3), and protein expression was induced at 37°C for 3.5 h with 1 mM IPTG. After purification using Ni-NTA resin, the fusion protein was dialyzed at 4°C overnight with PBS. The N-domain of ClpC1 was amplified from Arabidopsis cDNA library using ClpC1Ndomain forward and reverse primers, and the PCR product was directly cloned into EcoRI-SalI sites of pGEX-5X-1. The resulting plasmid DNA was introduced into Rosetta DE3 strain, and the ClpC1 N-domain was expressed at 37°C for 3 h with 1 mM IPTG. The N-domain protein was purified with glutathione sepharose 4B resin (GE) and 10 mM reduced glutathione and dialyzed in TBS buffer. Mature ClpC2 was amplified with Arabidopsis cDNA library and ClpC2 forward and reverse primers, and the gene product was directly introduced in NdeI-NcoI sites of the pET21a vector (Novagen). ClpC2 expression in the Rosetta DE3 strain was induced with 1 mM IPTG at 37°C for 3 h. The protein was found predominantly in inclusion bodies, which were solubilized with 8 M urea. The urea-soluble lysate was subjected to affinity purification with Ni-NTA resin and 1 M imidazole, and the eluates were dialyzed against 25 mM Tris-HCl, pH 8, 100 mM NaCl, and 2 mM EDTA.
In Vitro Interactions between ClpS1 and Clp Subunits
For ClpS1 interaction with ClpT1/T2 proteins or N-domain of ClpC1, GST fusions or GST and (His)6-tagged proteins were incubated for 90 min at room temperature in a buffer containing 25 mM Tris-HCl, pH 7.5, 2 mM EDTA, 100 mM NaCl, 15% glycerol, and 0.5% (v/v) Triton X-100, and then the mixtures were combined with glutathione sepharose 4B resin equilibrated with the same buffer for 30 min at 4°C. The protein-bound resin was washed five times. Proteins were directly eluted with 2× Laemmli buffer at 75°C for 5 min. The input and eluates were analyzed by SDS-PAGE and silver staining. ClpS1 interaction with ClpC2 was analyzed by the same procedures except that purified ClpC2 was incubated in the presence or absence of ATPγS (4 mM) and combined with GST-ClpS1 or GST proteins.
GST-ClpS1 and GST-ClpS1 DN/AA Pull-Down Assay Using Stroma from clps1 clpc1 Seedlings
Purified recombinant proteins were dialyzed against binding buffer [10 mM HEPES-KOH, pH 8.0, 5 mM Mg(OAc)2, 50 mM KOAc, and 10% glycerol] and then coupled (20 min) to a GSTrap FF column (GE Healthcare). Arabidopsis stromal proteins (2 to 4 mg proteins) from clps1 clpc1 seedlings were diluted with the binding buffer to 20 mL and applied to the GST-ClpS- or GST-ClpS1 DN/AA-coupled column and incubated for 2 h with peristaltic pump at 4°C. The columns were washed twice with 10 to 20 mL of the binding buffer. Proteins that specifically interact with GST-ClpS1 or GST-ClpS1 DN/AA were eluted with 6 mL of 10 mM reduced glutathione in the binding buffer and concentrated to 150 μL using Amicon ultra-4 centrifuge filters (10 NMWL; Millipore). Twenty-five microliters of eluates were denatured in 4× Laemmli buffer, heated at 70°C for 10 min, separated on 15% SDS-PAGE, and stained with Coommassie Brilliant Blue R 250.
Isolation of Proteins from Seedlings
Chloroplasts were isolated and separated into stromal and membrane fractions as described (Olinares et al., 2010). Total leaf proteins were extracted as described (Friso et al., 2011).
Pigment and Protein Assays
To quantify chlorophyll and carotenoid contents, seedlings were ground to a powder in liquid N2 and extracted with 80% acetone overnight in the dark at 4°C, followed by removal of cell debris by centrifugation for 10 min at ∼15,000g. The chlorophyll and carotenoid quantifications were according to Lichtenthaler (1987). Protein amounts were determined using the Bradford reagent (Bio-Rad) or the BCA protein assay kit (ThermoScientific).
SDS-PAGE for Proteome Analysis
Total leaf extracts or stromal protein extracts were solubilized in 1% SDS and 50 mM Tris-HCl, pH 8.25. Fifty micrograms total leaf or stromal proteins were each run on Bio-Rad Criterion Tris-HCl precast gels (10.5 to 14% acrylamide gradient). Gels were stained with Coomassie Brilliant Blue R 250. Each of the gel lanes was cut into 10 bands followed by reduction, alkylation, and in-gel digestion with trypsin as described (Shevchenko et al., 2006).
Antibody Production
The GST-ClpS fusion protein was purified over a 1-mL GST column. After binding of the fusion protein to the column and subsequent washing with PBS buffer, ∼1 mL Factor Xa (Prozyme) in Factor Xa buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM CaCl2) was loaded onto the column and allowed to incubate for 16 h at room temperature. The GST column was then assembled in tandem to an N-benzamidine column. Purified ClpS was eluted from the GST column (while GST remained bound) in Factor Xa buffer, and Factor Xa protease was purified over the N-benzamidine column to yield pure ClpS. This purified ClpS proteins was used for antisera production in rabbits using a commercial service (Cocalico Biologicals). For pTAC17 and UVR antibodies, the gene products were amplified with the specific primer sets and introduced into BamHI-XhoI (pTAC17) or EcoRI-XhoI (UVR) sites of the pET21a vector. Proteins were expressed in BL21 DE3 strain at 37°C for 3 h with 1 mM IPTG. pTAC17 protein was found in inclusion bodies and thus solubilized in 8 M urea, while UVR was soluble protein. After purification with the Ni-NTA affinity resin, both antigens were used for antisera production in rabbits (Alpha Diagnostics).
Antisera and Immunoblot Analysis
For immunoblots, proteins were blotted to nitrocellulose membranes and probed with antibodies using chemiluminescence for detection, following standard procedures. Antisera against ClpS1, pTAC17, and UVR proteins were generated in rabbits as described. Additional antisera were generous gifts from various colleagues as follows: GUN4, GUN5, CHLD, CHLI (R.M. Larkin), ClpC1 and ClpC2 (S. Rodermel), cpHSP70 (M. Nakai), GluTR (B. Grimm), and purchased from Agrisera.
MS
All MS data were collected using a LTQ-Orbitrap interfaced with a nano-liquid chromatography system and autosampler (Thermo) using data-dependent acquisition and dynamic exclusion (repeat count 2), with the Orbitrap portion operating at 100,000 resolution, essentially as described (Friso et al., 2011). Peak lists (in .mgf format) were generated from RAW files using DTA supercharge software, and all .mgf files were recalibrated as by Friso et al. (2011). Recalibrated files were searched with MASCOT v2.2 against TAIR10, including a small set of typical contaminants and the decoy (71,149 sequences; 29,099,754 residues). Three parallel searches (Mascot P value < 0.01; precursor ion window 700 to 3500 D) were performed: (1) full tryptic (6 ppm) with variable M-ox, Gln to pyro-Glu (N-termQ), N-term protein acetylation, and Fixed Cys-carbamido-methylation, two missed cleavages (in Mascot PR or PK does not count as missed cleavage); (2) semitryptic (3 ppm) with variable M-ox, N-term acetylation, Gln to pyro-Glu (N-termQ), and fixed Cys-carbamido-methylation, two missed cleavages; and (3) error-tolerant, full tryptic only (3 ppm) with variable M-ox and Fixed Cys-carbamido-methylation (no missed cleavage). Using a post-Mascot script, all search results were further filtered for minimum ion score of 33, but 35 for single peptide identifications. Proteins could be identified and quantified only with the full tryptic (6 ppm) search. Proteins identified by MS/MS spectra that were all shared with other proteins identified with unique peptides were discarded. The semitryptic and error-tolerant searches served to increase protein coverage; these additional search results were combined with the full tryptic search results using scripts as described (Zybailov et al., 2009a). The rationales for the selected modifications were as follows: variable M-ox is a common modification; fixed Cys-carbamido-methylation because all samples were alkylated; Gln to pyro-Glu (N-termQ) because it has been previously shown that this occurs very frequently (∼50%) (Zybailov et al., 2009a) and can be detected with great confidence due to the relatively large mass shift (−17.03 D); N-term protein acetylation in full tryptic because many proteins are N-terminally acetylated (note this occurs often after removal of N-terminal Met, but MASCOT allows for this removal of N-terminal Met, so these will be still full tryptic). N-term acetylation is semitryptic because many chloroplast proteins become N-terminally acetylated after removal cleavable transit peptide and it can be assigned with great confidence (Zybailov et al., 2008, 2009a).
PCA tests and Spearman rank correlation analyses were done using the statistical platform R. For statistical analysis of quantitative stromal protein comparison between the wild type, clps1, clpc1, and clps1 clpc1 based on the spectral counting, we used as input NadjSPC and the GLEE software, which was developed in MATLAB version 7 (MathWorks) (A. Poliakov, L. Ponnala, P.B.D. Olinares, Y. Asakura, and K.J. van Wijk, unpublished data). GLEE is essentially an improved version of the PLGEM software (Pavelka et al., 2008), benchmarked against QSpec (Choi et al., 2008), and tested by Kim et al. (2013). GLEE was run in a Windows platform with a cubic polynomial equation fitting and 1000 iterations for estimation of variation.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ClpS1, AT1G68660; ClpC1, AT5G50920; ClpC2, AT5G50920; ClpD, AT5G51070; ClpT1, AT4G25370; ClpT2, AT4G12060; Aly, XP_002888676; Ath, NP_564937; Av1, YP_321794; Av2, YP_323351; Bdi, XP_003574520; Bra1, Bra038361; Bra2, Bra004024; Ccl1, Ciclev10032846m; Ccl2, Ciclev10033097m; Ccl3, Ciclev10032846m; Cpa1, evm.model.supercontig_1.209; Cpa2, evm.TU.supercontig_953.2; Cre1, XP_001691804; Cre2, XP_001702813; Cru, EOA35838; Csa1, XP_004157424; Csa2, XP_004135108; Ec, BAA35600; Gma1, NP_001235414; Gma2, NP_001234970; Gma3, XP_003534179; Gma4, NP_001237811; Mdo1, MDP0000165546; Mdo2, MDP0000385620; Mdo3, MDP0000329721; Mdo4, MDP0000262682; Mdo5, MDP0000269988; No1, NP_488365; No2, NP_484448; Osa1, NP_001061868; Osa2, NP_001044714; Pm1, NP_876045; Pm2, NP_875946; Ppa1, CAK19331; Ppa2, XP_001759963; Ppa3, XP_001767468 ; Ppe1, EMJ10924; Ppe2, EMJ27174; Ppe3, EMJ24948; Ptr1, XP_002315955; Ptr2, XP_002311446; Ptr3, XP_002315814; Ptr4, XP_002311579; Pvu1, Phvul.003G028500; Pvu2, Phvul.004G160300; Sbi, XP_002445542; Se1, YP_399709; Se2, YP_171753; Sit, Si014569m.g; Sy1, NP_441749; Sy2, NP_440999; Tha, Thhalv10019264m; Vca1, XP_002949013; Vca2, XP_002946530 ; Vvi, XP_002274839; Zma1, ACN28518; and Zma2, ACN25974.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Sequence Alignment of ClpS across Prokaryotes, Algae, Moss, and Angiosperms.
Supplemental Figure 2. In silico analysis of the chaperone interactions between ClpS and ClpC/A chaperones in E. coli and Arabidopsis.
Supplemental Figure 3. Inputs for Pull-Down Assays Probing the Interactions between ClpS and ClpC1,C2.
Supplemental Figure 4. Pull-Down Assay of GST-ClpS with ClpT1/2.
Supplemental Figure 5. Spearman Correlation and Principle Component Analyses of the Quantified Proteomes of the Wild Type and clps1 clpc1-1.
Supplemental Figure 6. Fresh Weight and Pigment Analysis of Wild-Type and clps1 Seedlings Grown on Different CAP Concentrations.
Supplemental Table 1. Chloroplast Investments in 34 Functions in clps1, clpc1-1, clps1 clpc1, and the Wild-Type Samples Based on Curated MapMan Bin Distributions.
Supplemental Table 2. Functional Enrichment in Differential Protein Expression in the Single and Double Mutants.
Supplemental Table 3. The Oligonucleotide Sequences Used for Genotyping and RT-PCR of clp Mutants and Production of Recombinant Proteins.
Supplemental Data Set 1. ClpS Sequences Used for the Phylogeny.
Supplemental Data Set 2. Comparative Proteomics Data Sets of wt1, clps1, and clpc1-1 before Grouping.
Supplemental Data Set 3. Statistical Analysis of Comparative Proteomics Data Sets of wt1, clps1, and clpc1-1 after Grouping.
Supplemental Data Set 4. Differentially Expressed Plastid Proteins in Either the Single and Double Mutants Determined through Comparative Proteome Analysis of Isolated Stromal Proteomes.
Supplemental Data Set 5. Comparative Proteomics Data Sets of wt2 and clps1 clpc1.
Supplemental Data Set 6. Statistical Analysis of Comparative Proteomics Data Sets of wt1 clps1 clpc1-1 after Grouping.
Supplemental Data Set 7. Effect of CAP on the Proteomes of the Wild Type and clps1.
Supplemental Data Set 8. Identification of Candidate ClpS1 Substrates by Affinity Chromatography.
Acknowledgments
This work was supported by the National Science Foundation (MCB-1021963) to K.J.V.W. Part of this work was carried out by using the resources of the Computational Biology Service Unit of Cornell University, which is partially funded by the Microsoft Corporation. We thank colleagues Robert Larkin, Steve Rodermel, Masato Nakai, and Bernhard Grimm for sending us specific antisera. We thank Qi Sun from the Computational Biology Service Unit of Cornell University for all his support with the PPDB and proteomics pipeline. We thank other members of the van Wijk lab for help and discussions.
AUTHOR CONTRIBUTIONS
K.N., Y.A., G.F., J.K., S.O., and H.R. performed the experimental analysis. L.P. carried out the statistical and correlation analyses. G.F. carried out all MS analyses. K.J.V.W., Y.A., and K.N. designed the experiments. K.N. and K.J.V.W. wrote the article. K.J.V.W. obtained the funding for this project, recruited the coauthors into the project, and provided general oversight.
Glossary
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- GST
glutathione S-transferase
- MS/MS
tandem mass spectrometry
- adjSPC
adjusted MS/MS spectra
- PPDB
The Plant Proteome Database
- PCA
principle component analysis
- MS
mass spectrometry
- LIN
lincomycin
- CAP
chloramphenicol
- CHX
cycloheximide
- SPN
spectinomycin
- DHS
DHAP synthase
- CAO
chlorophyllide a oxygenase
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- Ni-NTA
nickel-nitrilotriacetic acid
References
- Apel W., Schulze W.X., Bock R. (2010). Identification of protein stability determinants in chloroplasts. Plant J. 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Galarneau E.R., Watkins K.P., Barkan A., van Wijk K.J. (2012). Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 159: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.A., Sauer R.T. (2012). ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof S., Baerenfaller K., Wildhaber T., Troesch R., Vidi P.A., Roschitzki B., Hirsch-Hoffmann M., Hennig L., Kessler F., Gruissem W., Baginsky S. (2011). Plastid proteome assembly without Toc159: Photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch E.M., Rosano G.L., Ceccarelli E.A. (2012). Chloroplastic Hsp100 chaperones ClpC2 and ClpD interact in vitro with a transit peptide only when it is located at the N-terminus of a protein. BMC Plant Biol. 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Fermin D., Nesvizhskii A.I. (2008). Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics 7: 2373–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D., Froehlich J.E., Rangarajan S., Keegstra K. (2004). A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki O., Grimm B. (2012). Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63: 1675–1687 [DOI] [PubMed] [Google Scholar]
- Czarnecki O., Hedtke B., Melzer M., Rothbart M., Richter A., Schröter Y., Pfannschmidt T., Grimm B. (2011). An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23: 4476–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Vioque A. (2001). Increased sensitivity to protein synthesis inhibitors in cells lacking tmRNA. RNA 7: 1708–1716 [PMC free article] [PubMed] [Google Scholar]
- Dougan D.A., Micevski D., Truscott K.N. (2012). The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+proteases. Biochim. Biophys. Acta 1823: 83–91 [DOI] [PubMed] [Google Scholar]
- Dougan D.A., Mogk A., Zeth K., Turgay K., Bukau B. (2002b). AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529: 6–10 [DOI] [PubMed] [Google Scholar]
- Dougan D.A., Reid B.G., Horwich A.L., Bukau B. (2002a). ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9: 673–683 [DOI] [PubMed] [Google Scholar]
- Dougan D.A., Truscott K.N., Zeth K. (2010). The bacterial N-end rule pathway: Expect the unexpected. Mol. Microbiol. 76: 545–558 [DOI] [PubMed] [Google Scholar]
- Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D.A., Bukau B. (2006). ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439: 753–756 [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A.J., Sweetlove L.J. (2011). Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. (2009). Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso, G., Olinares, P.D.B., and van Wijk, K.J. (2011). The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. In Chloroplast Research in Arabidopsis, R.P. Jarvis, ed (New York: Humana Press), pp. 265–282. [DOI] [PubMed] [Google Scholar]
- Guo F., Esser L., Singh S.K., Maurizi M.R., Xia D. (2002). Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J. Biol. Chem. 277: 46753–46762 [DOI] [PubMed] [Google Scholar]
- Harms J.M., Bartels H., Schlünzen F., Yonath A. (2003). Antibiotics acting on the translational machinery. J. Cell Sci. 116: 1391–1393 [DOI] [PubMed] [Google Scholar]
- Huang M., Friso G., Nishimura K., Qu X., Olinares P.D., Majeran W., Sun Q., van Wijk K.J. (2013). Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J. Proteome Res. 12: 491–504 [DOI] [PubMed] [Google Scholar]
- Jung H.S., Okegawa Y., Shih P.M., Kellogg E., Abdel-Ghany S.E., Pilon M., Sjölander K., Shikanai T., Niyogi K.K. (2010). Arabidopsis thaliana PGR7 encodes a conserved chloroplast protein that is necessary for efficient photosynthetic electron transport. PLoS ONE 5: e11688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Sun X., Zhang L., Sakamoto W. (2012). Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 159: 1428–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Olinares, P.B.D., Oh, S.H., Ghisaura, S., Poliakov, A., Ponnala, L., and van Wijk, K.J. (2013). Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis thaliana Plant Physiol. 162: 157–179. [DOI] [PMC free article] [PubMed]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J., Molière N., Dougan D.A., Turgay K. (2009). Adapting the machine: Adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7: 589–599 [DOI] [PubMed] [Google Scholar]
- Kline K.G., Barrett-Wilt G.A., Sussman M.R. (2010). In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjögren L.L., Zhao Y., Clarke A.K., Chory J. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bédard J., Patel R., Dudley P., Twell D., Ríos G., Koncz C., Jarvis P. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kress W., Maglica Z., Weber-Ban E. (2009). Clp chaperone-proteases: Structure and function. Res. Microbiol. 160: 618–628 [DOI] [PubMed] [Google Scholar]
- Leonard A.C., Grimwade J.E. (2011). Regulation of DnaA assembly and activity: Taking directions from the genome. Annu. Rev. Microbiol. 65: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Liu X., Yu F., Rodermel S. (2010). An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A.N., Koretke K.K. (2003). Bioinformatic analysis of ClpS, a protein module involved in prokaryotic and eukaryotic protein degradation. J. Struct. Biol. 141: 77–83 [DOI] [PubMed] [Google Scholar]
- Nakagawara E., Sakuraba Y., Yamasato A., Tanaka R., Tanaka A. (2007). Clp protease controls chlorophyll b synthesis by regulating the level of chlorophyllide a oxygenase. Plant J. 49: 800–809 [DOI] [PubMed] [Google Scholar]
- Ninnis R.L., Spall S.K., Talbo G.H., Truscott K.N., Dougan D.A. (2009). Modification of PATase by L/F-transferase generates a ClpS-dependent N-end rule substrate in Escherichia coli. EMBO J. 28: 1732–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares P.D., Kim J., Davis J.I., van Wijk K.J. (2011b). Subunit stoichiometry, evolution, and functional implications of an asymmetric plant plastid ClpP/R protease complex in Arabidopsis. Plant Cell 23: 2348–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares P.D., Kim J., van Wijk K.J. (2011a). The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807: 999–1011 [DOI] [PubMed] [Google Scholar]
- Olinares P.D., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Fournier M.L., Swanson S.K., Pelizzola M., Ricciardi-Castagnoli P., Florens L., Washburn M.P. (2008). Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol. Cell. Proteomics 7: 631–644 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ripoll D.R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Pruzinská A., Anders I., Aubry S., Schenk N., Tapernoux-Lüthi E., Müller T., Kräutler B., Hörtensteiner S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B.G., Fenton W.A., Horwich A.L., Weber-Ban E.U. (2001). ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl. Acad. Sci. USA 98: 3768–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S., Finazzi G., Endler A., Willig A., Baerenfaller K., Grossmann J., Gerrits B., Rutishauser D., Gruissem W., Rochaix J.D., Baginsky S. (2011). Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. (1998). A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95: 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano G.L., Bruch E.M., Ceccarelli E.A. (2011). Insights into the Clp/HSP100 chaperone system from chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 286: 29671–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A., Friso G., Alonso J.M., Ecker J.R., van Wijk K.J. (2006). Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Tanaka R., Yamasato A., Tanaka A. (2009). Determination of a chloroplast degron in the regulatory domain of chlorophyllide a oxygenase. J. Biol. Chem. 284: 36689–36699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Melamed-Bessudo C., Avivi-Ragolski N., Pietrokovski S., Levy A.A. (2011). Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23: 4266–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R.T., Baker T.A. (2011). AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80: 587–612 [DOI] [PubMed] [Google Scholar]
- Schmidt R., Zahn R., Bukau B., Mogk A. (2009). ClpS is the recognition component for Escherichia coli substrates of the N-end rule degradation pathway. Mol. Microbiol. 72: 506–517 [DOI] [PubMed] [Google Scholar]
- Schmied J., Hedtke B., Grimm B. (2011). Overexpression of HEMA1 encoding glutamyl-tRNA reductase. J. Plant Physiol. 168: 1372–1379 [DOI] [PubMed] [Google Scholar]
- Scholefield G., Veening J.W., Murray H. (2011). DnaA and ORC: More than DNA replication initiators. Trends Cell Biol. 21: 188–194 [DOI] [PubMed] [Google Scholar]
- Schuenemann V.J., Kralik S.M., Albrecht R., Spall S.K., Truscott K.N., Dougan D.A., Zeth K. (2009). Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS. EMBO Rep. 10: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Sjögren L.L., MacDonald T.M., Sutinen S., Clarke A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L., Stanne T.M., Zheng B., Sutinen S., Clarke A.K. (2006). Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne T.M., Pojidaeva E., Andersson F.I., Clarke A.K. (2007). Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J. Biol. Chem. 282: 14394–14402 [DOI] [PubMed] [Google Scholar]
- Stanne T.M., Sjögren L.L., Koussevitzky S., Clarke A.K. (2009). Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem. J. 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Striebel F., Kress W., Weber-Ban E. (2009). Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 19: 209–217 [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Nakagami H., Mochida K., Daudi A., Tomita M., Shirasu K., Ishihama Y. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Trösch R., Jarvis P. (2011). The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS ONE 6: e23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio J.J., Croteau D.L., Van Houten B., Kisker C. (2006). Prokaryotic nucleotide excision repair: The UvrABC system. Chem. Rev. 106: 233–252 [DOI] [PubMed] [Google Scholar]
- Tzin V., Galili G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3: 956–972 [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996). The N-end rule: Functions, mysteries, uses. Proc. Natl. Acad. Sci. USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (2011). The N-end rule pathway and regulation by proteolysis. Protein Sci. 20: 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Vögtle F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. (2009). Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439 [DOI] [PubMed] [Google Scholar]
- Wang K.H., Oakes E.S., Sauer R.T., Baker T.A. (2008a). Tuning the strength of a bacterial N-end rule degradation signal. J. Biol. Chem. 283: 24600–24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.H., Roman-Hernandez G., Grant R.A., Sauer R.T., Baker T.A. (2008b). The molecular basis of N-end rule recognition. Mol. Cell 32: 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Elliott M., Elliott T. (1999b). Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 181: 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wilson S., Elliott T. (1999a). A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J. Bacteriol. 181: 6033–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Ban E.U., Reid B.G., Miranker A.D., Horwich A.L. (1999). Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401: 90–93 [DOI] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M.R. (1994). A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91: 12218–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Oh M.H., Schwarz E.M., Larue C.T., Sivaguru M., Imai B.S., Yau P.M., Ort D.R., Huber S.C. (2011). Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A.Y., Houry W.A. (2007). ClpP: A distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581: 3749–3757 [DOI] [PubMed] [Google Scholar]
- Zeth K., Ravelli R.B., Paal K., Cusack S., Bukau B., Dougan D.A. (2002). Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat. Struct. Biol. 9: 906–911 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Friso G., Kim J., Rudella A., Rodríguez V.R., Asakura Y., Sun Q., van Wijk K.J. (2009b). Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 8: 1789–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov, B., Rutschow, H., Friso, G., Rudella, A., Emanuelsson, O., Sun, Q., and van Wijk, K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed]
- Zybailov B., Sun Q., van Wijk K.J. (2009a). Workflow for large scale detection and validation of peptide modifications by RPLC-LTQ-Orbitrap: Application to the Arabidopsis thaliana leaf proteome and an online modified peptide library. Anal. Chem. 81: 8015–8024 [DOI] [PubMed] [Google Scholar]