Susceptibility to corn leaf aphid in maize is associated with changes in defense-inducing maize benzoxazinoid levels, with high HDMBOA-Glc levels and low DIMBOA-Glc levels leading to toxicity but a reduced plant aphid defense response of callose deposition. Variation in HDMBOA-Glc production is due to a transposon insertion that inactivates O-methyltransferases in lines with low HDMBOA-Glc levels.
Abstract
Plants differ greatly in their susceptibility to insect herbivory, suggesting both local adaptation and resistance tradeoffs. We used maize (Zea mays) recombinant inbred lines to map a quantitative trait locus (QTL) for the maize leaf aphid (Rhopalosiphum maidis) susceptibility to maize Chromosome 1. Phytochemical analysis revealed that the same locus was also associated with high levels of 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc) and low levels of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc). In vitro enzyme assays with candidate genes from the region of the QTL identified three O-methyltransferases (Bx10a-c) that convert DIMBOA-Glc to HDMBOA-Glc. Variation in HDMBOA-Glc production was attributed to a natural CACTA family transposon insertion that inactivates Bx10c in maize lines with low HDMBOA-Glc accumulation. When tested with a population of 26 diverse maize inbred lines, R. maidis produced more progeny on those with high HDMBOA-Glc and low DIMBOA-Glc. Although HDMBOA-Glc was more toxic to R. maidis than DIMBOA-Glc in vitro, BX10c activity and the resulting decline of DIMBOA-Glc upon methylation to HDMBOA-Glc were associated with reduced callose deposition as an aphid defense response in vivo. Thus, a natural transposon insertion appears to mediate an ecologically relevant trade-off between the direct toxicity and defense-inducing properties of maize benzoxazinoids.
INTRODUCTION
Plants in natural and agricultural ecosystems are constantly attacked by a multitude of insect herbivores, including leaf and root chewers, stem borers, leaf miners, gall formers, and phloem feeders. Most of these herbivores have distinct geographical distributions, resulting in divergent herbivore communities across latitudinal (Züst et al., 2012), longitudinal (Kozlov, 2008), and altitudinal gradients (Pélissier et al., 2008). The pronounced geographical heterogeneity of herbivore communities coincides with considerable variation in herbivore resistance and expression of defensive traits in many plants (Johnson, 2011). In some natural ecosystems, the geographical distribution of herbivores has been shown to covary with specific defensive phenotypes (Prasad et al., 2012; Züst et al., 2012). However, similar associations have rarely been described for crop species, and the extent to which pest occurrence may have shaped the defensive makeup of crops remains unknown.
Independent of geographical patterns, the high genetic diversity of plants provides an opportunity to map quantitative trait loci (QTL) conferring insect resistance (Kroymann et al., 2003; Klingler et al., 2009; Qiu et al., 2010). QTL mapping exploits genetically heritable within-species differences that can statistically be associated with a specific part of the genome. QTL mapping is often limited by the relatively high cost of genotyping and the difficulty of repeating experiments with a segregating population. To overcome these limitations, large sets of recombinant inbred lines have been created for several plant species, including Arabidopsis thaliana (Keurentjes et al., 2011) and maize (Zea mays) (Flint-Garcia et al., 2005; Yu et al., 2008; McMullen et al., 2009a). Once such recombinant inbred lines have been genotyped, they represent a permanent resource that can be used to genetically map loci that influence any phenotypic trait that varies in the population.
As a productive food crop that is attacked by more than 90 insect species (Steffey et al., 1999), maize is an attractive model for the identification of resistance factors. Geographical variation in the abundance of insect herbivores suggests that there may be associated variation in the level of pest resistance among different commercial maize lines. For instance, the western maize rootworm (Diabrotica virgifera virgifera) is a major pest in the midwestern US but has not been present in Europe until recently (Miller et al., 2005). The maize leaf aphid (Rhopalosiphum maidis) does not survive winters in North America but migrates north each summer from warmer areas, resulting in periodic outbreaks of this species (Steiner et al., 1985).
Recent genome sequencing of the maize inbred line B73 (Schnable et al., 2009) and the development of high-resolution physical and genetic maps (Wei et al., 2009; Zhou et al., 2009; Ganal et al., 2011) has opened up new research opportunities. A nested association mapping (NAM) population of ∼5000 recombinant inbred lines was generated by crossing a genetically diverse population of 25 maize inbred lines to B73 (Flint-Garcia et al., 2005; Yu et al., 2008; McMullen et al., 2009a). The NAM population has been used to map numerous maize traits, including resistance to northern leaf blight (Setosphaeria turcica; Poland et al., 2011) and southern leaf blight (Bipolaris maidis; Kump et al., 2011). In both cases, genome-wide association mapping revealed numerous QTL with relatively small additive effects. To date, the NAM population has not been employed to identify alleles involved in insect resistance.
Insect resistance factors in maize as well as other plants have been identified not only by genetic mapping approaches but also by phytochemical screening. Known maize antiherbivore defenses include protease inhibitors (Lawrence et al., 2012), the Maize insect resistance1 (Mir1) Cys protease (Pechan et al., 2000), ribosome-inactivating proteins (Walsh et al., 1991; Bass et al., 1995), and other protein-mediated defenses, as well as secondary metabolites such as chlorogenic acid (Cortés-Cruz et al., 2003), maysin (Rector et al., 2003), and benzoxazinoids (Frey et al., 1997). Benzoxazinoids have been demonstrated to confer resistance to aphids (Ahmad et al., 2011), chewing herbivores (Glauser et al., 2011), and a wide range of other pests, pathogens, and weeds (Niemeyer, 2009). They are found abundantly in many Poaceae, including maize, wheat (Triticum aestivum), and rye (Secale cereale) (Zuniga et al., 1983), as well as in several dicot species in the Plantaginaceae and Ranunculaceae (Schullehner et al., 2008). In maize seedlings, the predominant benzoxazinoid is 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA; Figure 1), which is stored as an inactive glucoside (DIMBOA-Glc). Constitutive benzoxazinoid levels tend to decline as the plants age (Cambier et al., 2000). In addition to DIMBOA-Glc, 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc), 2,4-dihydroxy-7,8-dimethoxy-1,4- benzoxazin-3-one glucoside (DIM2BOA-Glc), and 2-hydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (HMBOA-Glc) are commonly found in maize leaves.
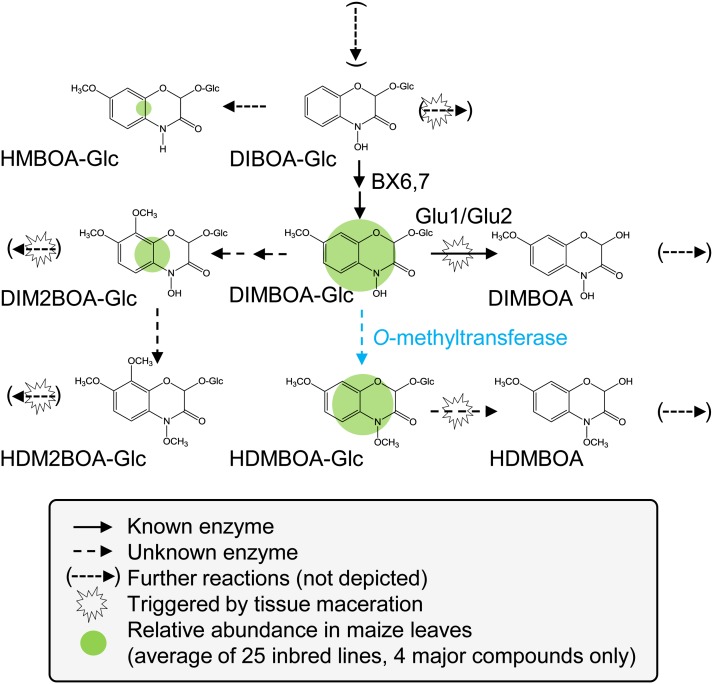
Figure 1.
Selected Steps of Benzoxazinoid Metabolism in Maize.
The major benzoxazinoids detected in the leaves of maize seedlings are DIMBOA-Glc, HDMBOA-Glc, DIM2BOA-Glc, and HMBOA-Glc. In contrast with DIMBOA-Glc, the biosynthetic steps leading to HDMBOA-Glc, DIM2BOA-Glc, and HMBOA-Glc are unknown. Upon tissue disruption by herbivores, the different glucosides are cleaved by β-glucosidases (Glu), leading to the release of active aglucones. The N-methoxyl group of HDMBOA decreases its stability and renders it more reactive than DIMBOA.
[See online article for color version of this figure.]
Benzoxazinoids are predominantly stored as glucosides in the cell vacuole. Tissue maceration by chewing herbivores results in the release of active aglucones by the action of endogenous β-glucosidases (Figure 1). The nonenzymatic breakdown of the major aglucone DIMBOA results in the formation of 6-methoxybenzoxalin-3-one, which is insect deterrent (Grambow et al., 1986). Insect feeding also induces enzymatic conversion of DIMBOA-Glc to HDMBOA-Glc (Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011). While HDMBOA-Glc is also activated by glucosidases during tissue maceration, further nonenzymatic breakdown is faster for HDMBOA than DIMBOA (Maresh et al., 2006). The conversion of DIMBOA-Glc to HDMBOA-Glc has been associated with increased resistance to both pathogens and herbivores (Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011). Thus, herbivory-induced HDMBOA-Glc accumulation creates a pool of reactive defense compounds that allows maize plants to respond more rapidly to subsequent herbivore attack. Although DIMBOA-Glc methyltransferase activity has been demonstrated enzymatically (Oikawa et al., 2002), it was unclear which genes encode this activity.
Unlike chewing herbivores, such as caterpillars, aphids feed by inserting their stylets into phloem sieve elements. Analysis of aphid honeydew and sap collected by stylectomy shows that DIMBOA-Glc, but not DIMBOA, is found in the phloem (Givovich et al., 1992, 1994; Caillaud and Niemeyer, 1996). Additionally, aphid feeding does not increase overall DIMBOA accumulation (Cambier et al., 2001), suggesting that glucosidase-mediated activation of DIMBOA-Glc does not occur in response to insects that feed exclusively from the phloem sieve elements. However, a recent study shows that DIMBOA is secreted into the apoplast upon aphid infestation (Ahmad et al., 2011).
In artificial diet experiments, DIMBOA-Glc and DIMBOA reduced feeding by five monocot-feeding aphid species (Givovich and Niemeyer, 1995). In the case of the rose-grain aphid (Metopolophium dirhodum) feeding from artificial diet, the HDMBOA-Glc LD50 (concentration to reduce aphid survival by 50%) was fivefold lower than the DIMBOA-Glc LD50 (Cambier et al., 2001), suggesting that, as in the case of chewing herbivores, conversion of DIMBOA-Glc to HDMBOA-Glc could provide a defensive benefit to the plants. Benzoxazinoids may also function to elicit the production of other plant defensive metabolites or proteins. The maize benzoxazinoneless1 (bx1) indole glycerol phosphate lyase1 (igl1) double mutant, which is blocked in the first step of benzoxazinoid biosynthesis, has reduced callose accumulation as a defense response (Ahmad et al., 2011). Infiltration of DIMBOA, but not HDMBOA-glucoside, into the maize apoplastic space induced callose formation, suggesting that differences in the structure or reactivity of benzoxazinoids leads to differential regulation of maize callose accumulation.
As chewing herbivores rather than aphids have been the focus of most prior research on natural variation in maize herbivore resistance (McMullen et al., 2009a; Meihls et al., 2012), we initiated experiments using R. maidis and recombinant inbred lines of the NAM population to identify resistance mechanisms. Concerted mapping of loci associated with aphid resistance, benzoxazinoid production, and callose deposition was followed by the molecular and biochemical characterization of candidate genes. Our results show that dominant aphid susceptibility in several NAM parental lines is associated with a DIMBOA-Glc methyltransferase that is inactivated by a transposon insertion in other inbred lines.
RESULTS
Mapping of R. maidis Resistance and HDMBOA-Glc QTL to the Same Interval on Maize Chromosome 1
To explore natural variation in maize aphid resistance, we measured R. maidis reproduction on 2-week-old seedlings of the NAM population parental lines (Figure 2). Reproduction was low on the inbred lines B97, M37W, Mo17, and Oh7B, intermediate on B73, and high on CML52, CML69, CML277, CML322, NC350, and NC358. Aphid progeny production varied more than 100-fold, suggesting that it should be possible to map aphid resistance as a quantitative trait using the recombinant inbred lines of the NAM population. Analysis of aphid reproduction on 124 recombinant inbred lines derived from B73 × CML322 identified a single significant QTL on maize Chromosome 1 (bin 1.04; see Supplemental Figure 1A online). On average, the CML322 allele increased aphid progeny production by approximately three nymphs per adult aphid per week (see Supplemental Figure 1B online), and aphid susceptibility was dominant in F1 progeny of crosses between B73 and CML322 (see Supplemental Figure 1C online). Measurement of R. maidis reproduction on recombinant inbred lines derived from B73 crossed to CML52, CML69, and CML277 also identified a significant aphid resistance QTL in maize bin 1.04 (see Supplemental Figure 2 online). Going on the assumption that aphid resistance is influenced by the same locus in all four of these inbred lines, we combined the data sets in an association mapping approach. This narrowed the genomic interval containing the aphid resistance QTL to ∼4 Mb containing 31 annotated genes (see Supplemental Table 1 online).
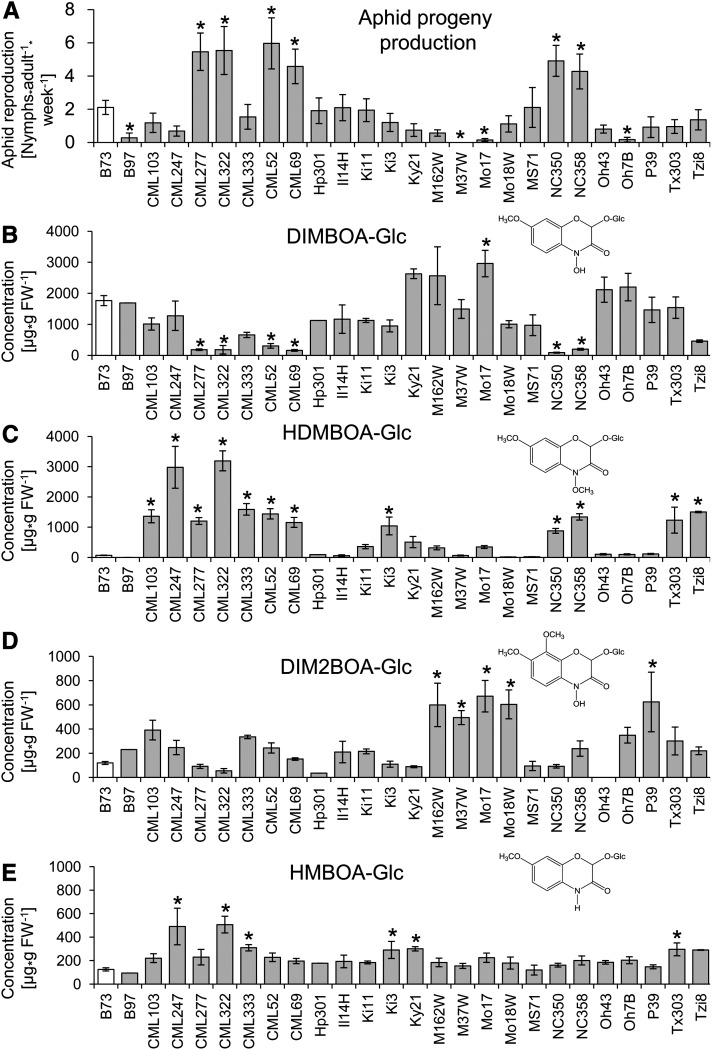
Figure 2.
Aphid Reproduction on and Benzoxazinoid Content in Parental Lines of the Maize NAM Population.
(A) The number of R. maidis progeny produced per adult aphid over 7 d is shown (mean ± se; n = 8; n = 18 for B73). Asterisks indicate significant differences relative to B73 (*P < 0.05, Dunnett’s test).
(B) to (E) DIMBOA-Glc, HDMBOA-Glc, DIM2BOA-Glc, and HMBOA-Glc abundance (mean ± se; n = 3 to 7, with the exception of single measurements for B97 and Hp301, for which no statistical calculations were done). Asterisks indicate significant differences relative to B73 (*P < 0.05, Dunnett’s test). FW, fresh weight.
Since benzoxazinoids are known to have a profound effect on maize resistance to insect herbivores (Cambier et al., 2001; Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011), we measured benzoxazinoid concentration in 2-week-old seedlings of the NAM parental lines as a possible explanatory factor for the observed aphid performance differences (Figure 2A). All of the detected benzoxazinoids showed considerable variation in the NAM parental lines (Figure 2). DIMBOA-Glc and HDMBOA-Glc concentrations were negatively correlated (P < 0.01, r = −0.51, n = 27; see Supplemental Figure 3A online), DIMBOA-Glc and DIM2BOA-Glc were positively correlated (P < 0.05, r = 0.39, n = 27; see Supplemental Figure 3B online), and no significant correlation was observed between DIMBOA-Glc and HMBOA-Glc concentrations (P > 0.05, r = 0.18, n = 27; see Supplemental Figure 3C online). Aphid reproduction was negatively correlated with DIMBOA-Glc (P < 0.01, r = −0.76, n = 27; see Supplemental Figure 4A online) and positively with HDMBOA-Glc (P < 0.05, r = 0.45, n = 27; see Supplemental Figure 4B online) across the NAM parental lines. In particular, the six NAM parental lines that were most suitable for aphid reproduction (CML52, CML69, CML277, CML322, NC350, and NC358; Figure 2A) had the lowest DIMBOA-Glc content (Figure 2B) and relatively high HDMBOA-Glc levels (Figure 2C).
To test whether a QTL for HDMBOA-Glc production colocalizes with the identified aphid resistance QTL, we quantified benzoxazinoids in B73 × CML322 recombinant inbred lines that had previously been used for aphid resistance mapping. The identified HDMBOA-Glc QTL coincided with the aphid resistance QTL (Figure 3). The hypothesis that CML322 has constitutively higher DIMBOA-Glc methyltransferase activity than B73 is supported by the observations that (1) the CML322 allele in bin 1.04 increased HDMBOA-Glc content by ∼1200 μg/g fresh weight relative to the B73 allele (see Supplemental Figure 5A online); (2) high HDMBOA-Glc content is dominant in F1 progeny from a B73 × CML322 cross (see Supplemental Figure 5B online); (3) a QTL for DIMBOA-Glc content is located on Chromosome 1 in the B73 × CML322 recombinant inbred population (see Supplemental Figure 5C online); and (4) DIMBOA-Glc and HDMBOA-Glc concentrations are negatively correlated in these lines (see Supplemental Figure 5D online).
Figure 3.
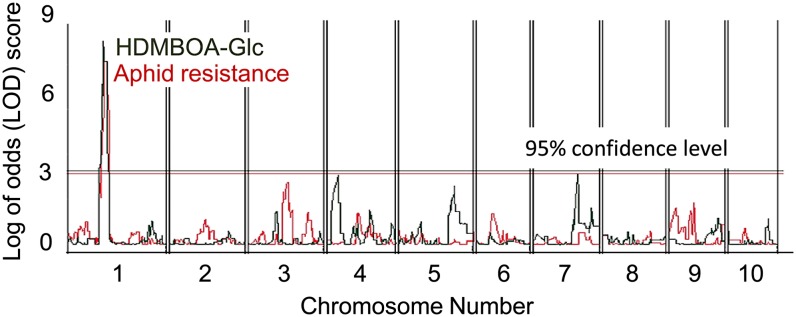
QTL for Aphid Resistance and HDMBOA-Glc Content Colocalize on Maize Chromosome 1 in the B73 × CML322 Recombinant Inbred Line Population.
[See online article for color version of this figure.]
Three Putative Methyltransferase Genes with Homology to Benzoxazinoneless7 Are Located in the Aphid Resistance/HDMBOA-Glc QTL
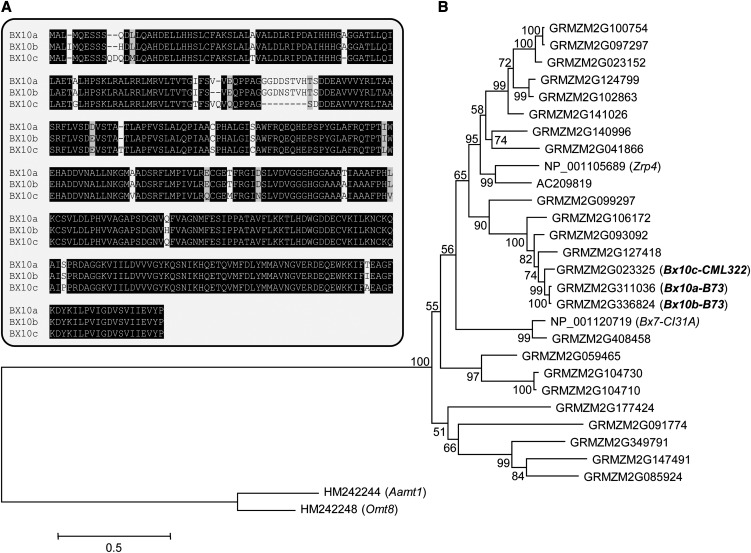
Given the colocalization of the aphid resistance and HDMBOA-Glc QTL (Figure 3), we hypothesized that variation in a DIMBOA-Glc methyltransferase would constitute the underlying genetic basis of these phenotypes. Three adjacent genes in the QTL interval (GRMZM2G311036, GRMZM2G336824, and GRMZM2G023325) cluster closely together in a phylogenetic tree of predicted maize O-methyltransferase sequences (Figure 4). These three genes showed >40% amino acid sequence identity to Benzoxazinoneless7 (BX7) (GenBank ID NP_001120719; TRIBOA-Glc O-methyltransferase), a known enzyme in the DIMBOA-Glc biosynthesis pathway (Jonczyk et al., 2008). Thus, the enzymes encoded by GRMZM2G311036, GRMZM2G336824, and GRMZM2G023325 were likely candidates for catalyzing the conversion of DIMBOA-Glc to HDMBOA-Glc. Following the convention used for the other nine known maize benzoxazinoid biosynthesis genes, we named the identified genes Benzoxazinoneless10a (Bx10a; GRMZM2G311036), Bx10b (GRMZM2G336824), and Bx10c (GRMZM2G023325).
Figure 4.
Sequence Alignment and Rooted Phylogenetic Tree with the Three Maize O-Methyltransferases Contained within the HDMBOA-Glc QTL.
(A) Protein sequence alignment (ClustalW) of the three O-methyltransferase homologs GRMZM2G311036, GRMZM2G336824, and GRMZM2G023325 (Bx10a-c).
(B) Rooted phylogenetic tree of putative maize Omt genes similar to Bx10a-c. The tree was inferred using the maximum likelihood method and n = 1000 replicates for bootstrapping. Bootstrap values are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The maize genes Aamt1 and Omt8 were used as an outgroup. Sequences used for the alignment are shown in Supplemental Data Set 1 online.
A Transposon Insertion in Bx10c Correlates with Low Levels of HDMBOA-Glc and Lower Aphid Reproduction
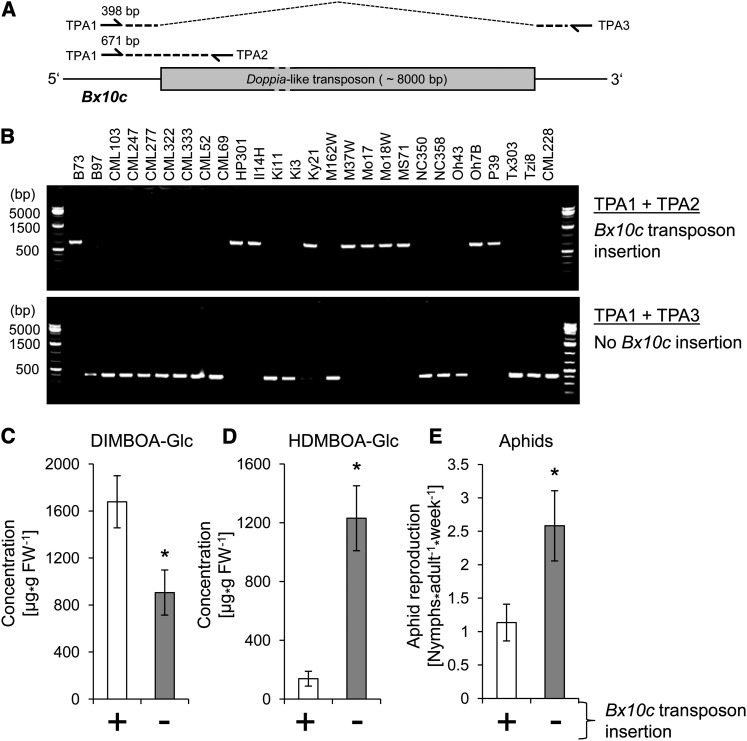
The Bx10c gene in inbred line B73 was found to contain an inserted CACTA family transposon (base pairs 66,501,488 to 66,509,673 on Chromosome 1 in the maize genome assembly v2), with ∼73% DNA sequence identity to the previously described Doppia4 transposon (GenBank ID AF187822.1; Bercury et al., 2001). The >8000-bp insertion in the first exon of Bx10c is likely to be a knockout mutation. Although Bx10a and/or Bx10b transcripts could be detected using quantitative RT-PCR (qRT-PCR) analysis, Bx10c was not expressed in B73 (Figure 5). Due to the 98% DNA sequence identity of Bx10a and Bx10b, attempts to design primers that would reliably amplify one but not the other were unsuccessful. Analysis of DNA sequence data from the maize HapMap2 project (Chia et al., 2012) showed that the Bx10c gene in CML322 does not contain the Doppia4-like transposon insertion. This observation was confirmed by PCR amplification and DNA sequencing of the complete gene (GenBank ID = KC754964). Consistent with this finding, Bx10c transcription was detectable by qRT-PCR in CML322 seedlings (Figure 5). To investigate the Bx10c transposon insertion presence or absence in all NAM parental lines, a PCR approach with gene-specific and transposon insertion-specific primers was conducted as shown in Figure 6A. Whereas 10 of the maize lines contained an insertion in Bx10c, the other 17 did not (Figure 6B). On average, NAM parental lines with the transposon insertion had significantly higher DIMBOA-Glc content (Figure 6C), lower HDMBOA-Glc content (Figure 6D), and lower aphid reproduction (Figure 6E) than lines without the insertion. However, due to reduced genetic recombination resulting from the proximity of Bx10c to the pericentromeric region of Chromosome 1 (Gore et al., 2009), we cannot rule out possible effects of other nearby genes on the observed phenotypes.
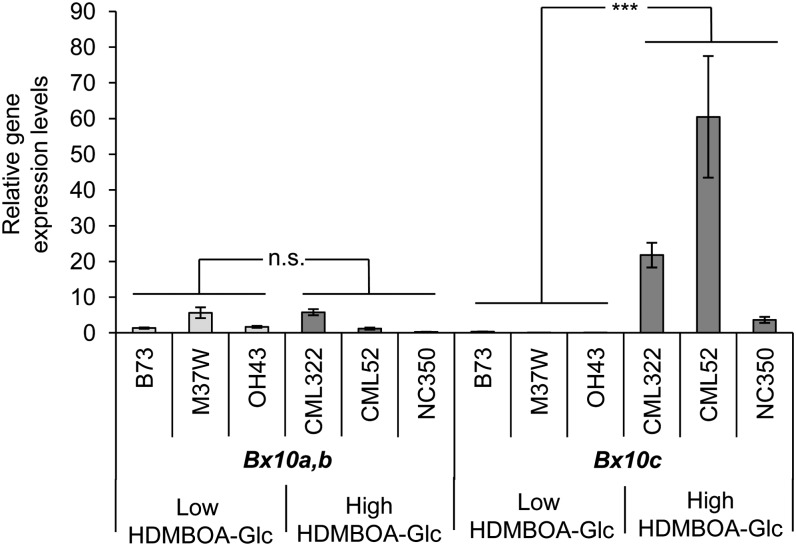
Figure 5.
Bx10c Is Expressed in the High HDMBOA-Glc Lines CML322, CML52, and NC350 but Not in the Low HDMBOA-Glc Lines B73, M37W, and Oh43.
Gene expression of uninfested plants was measured using qRT-PCR relative to actin (means ± se; n = 6). Asterisks indicate significant differences in expression levels between high and low HDMBOA-Glc genotypes (t test, ***P < 0.001; n.s., not significant).
Figure 6.
A Doppia-Like Transposon in Bx10c Is Associated with Reduced HDMBOA-Glc Production and Increased Aphid Resistance in the NAM Parental Lines.
(A) Strategy for PCR analysis to amplify fragments that are specific for either the insertion or the deletion allele.
(B) Detection of the transposon knockout and functional alleles of Bx10c by PCR using primers TPA1, TPA2, and TPA3 (described in Supplemental Table 2 online).
(C) to (E) Comparison of DIMBOA-Glc content, HDMBOA-Glc content, and aphid reproduction in NAM parental lines, with and without the transposon insertion (mean ± se; n = 10 or 17). Asterisks indicate significant differences (*P < 0.05; two-tailed Student’s t test). FW, fresh weight.
The Bx10a, Bx10b, and Bx10c Genes Encode DIMBOA-Glc O-Methyltransferases and Are Differentially Expressed in the NAM Lines
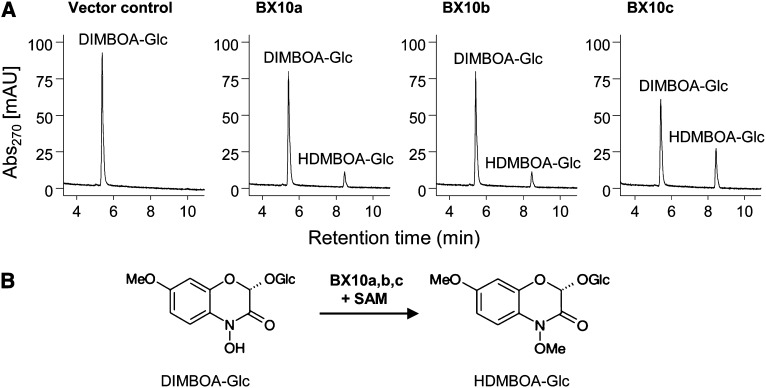
Gene cloning and in vitro enzyme assays showed that the B73 alleles of Bx10a and Bx10b encode an enzyme with DIMBOA-Glc O-methyltransferase activity (Figure 7). Enzyme activity depended on the presence of S-adenosyl-l-Met as a methyl donor in the reaction mixture. As Bx10c is interrupted by a transposon insertion in inbred line B73, we cloned this gene from CML322, which does not have an insertion (Figure 6B). Enzyme assays showed that BX10c from CML322 also has DIMBOA-Glc O-methyltransferase activity (Figure 7).
Figure 7.
BX10a-c Are Functional DIMBOA-Glc O-Methyltransferases That Produce HDMBOA-Glc.
(A) Equal amounts of purified recombinant BX10a, BX10b, and BX10c as well as a control without enzyme were incubated with the substrate DIMBOA-Glc and the cosubstrate S-adenosyl-l-Met. Products were extracted with methanol and analyzed by HPLC-UV. mAU: milli absorbance unit.
(B) Schematic representation of the reaction catalyzed by BX10 in the presence of the cosubstrate S-adenosyl-l-Met (SAM).
To test whether differences in Bx10 gene expression can explain the different HDMBOA-Glc levels in the NAM lines, we compared transcript abundance of Bx10a/b and Bx10c in three high HDMBOA-Glc lines (CML322, CML52, and NC350) and three low HDMBOA-Glc lines (B73, M37W, and Oh43). Whereas Bx10a/b transcripts could be detected in nearly all lines at low levels, Bx10c showed a strong transcript accumulation exclusively in the high HDMBOA-Glc lines (Figure 5). These results indicate that the constitutive conversion of DIMBOA-Glc to HDMBOA-Glc is mainly regulated by Bx10c gene expression.
Maize Lines Containing Less DIMBOA-Glc, Form Less Callose and Are More Susceptible to Aphids
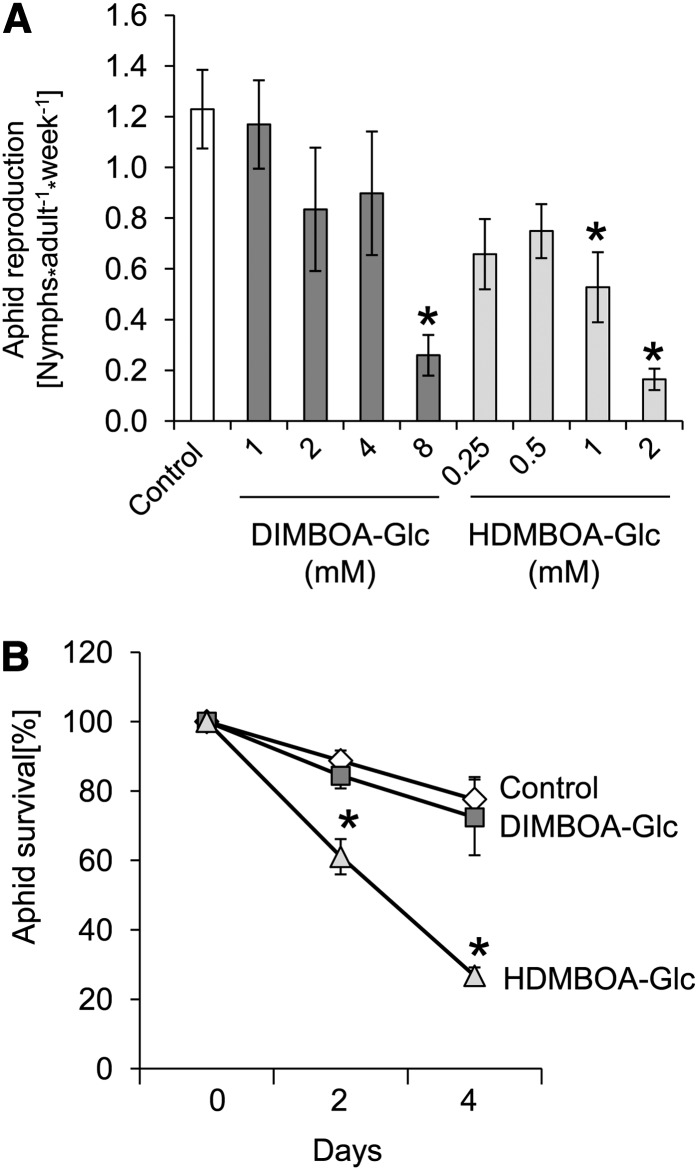
To obtain a more detailed understanding of the contrasting roles of DIMBOA-Glc and HDMBOA-Glc in aphid resistance, we conducted in vitro feeding assays with purified compounds. DIMBOA-Glc and HDMBOA-Glc were added to aphid artificial diet at concentrations similar to those observed in whole maize leaves (Figures 2B and 2C). Contrary to the positive correlation between DIMBOA-Glc and aphid resistance in planta (Figure 5), but similar to what has been observed with other herbivores and pathogens (Cambier et al., 2001; Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011), artificial diet assays showed that HDMBOA-Glc is more toxic for R. maidis than DIMBOA-Glc; whereas 2 mM HDMBOA-Glc significantly reduced aphid progeny production (Figure 8A) and survival (Figure 8B), 2 mM DIMBOA-Glc did not. Benzoxazinoid assays conducted with the remaining aphid diet showed intact benzoxazinoids and no breakdown products, suggesting that DIMBOA-Glc and HDMBOA-Glc remained intact over the course of the experiment.
Figure 8.
HDMBOA-Glc Reduces Aphid Performance in Vitro More Strongly Than DIMBOA-Glc.
(A) Aphid progeny production on diet with benzoxazinoids. Progeny per adult aphid were counted after 2 d on diet containing the indicated concentrations of DIMBOA-Glc and HDMBOA-Glc (mean ± se; n = 6; n = 12 for controls). Asterisks indicate significant differences (*P < 0.05, Dunnett’s test relative to control samples).
(B) Adult aphid survival on diet with benzoxazinoids. Diet contained no benzoxazinoids (control; diamonds), 2 mM DIMBOA-Glc (squares), or 2 mM HDMBOA-Glc (triangles) (mean ± se; n = 6; n = 12 for controls). Asterisks indicate significant differences (*P < 0.05, Dunnett’s test relative to control samples).
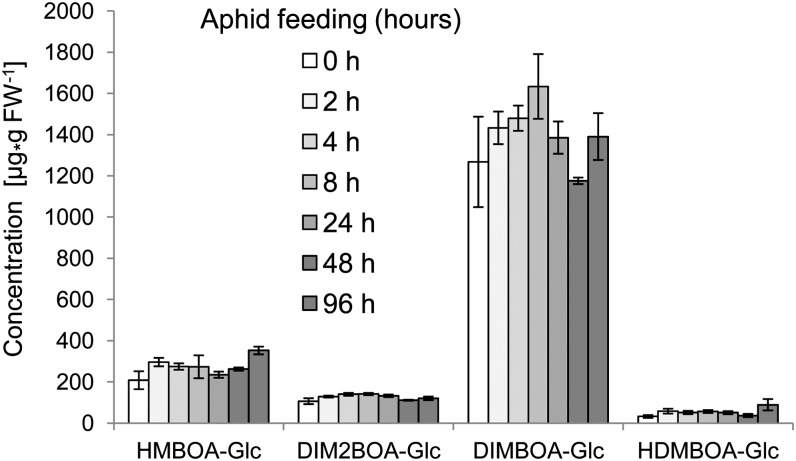
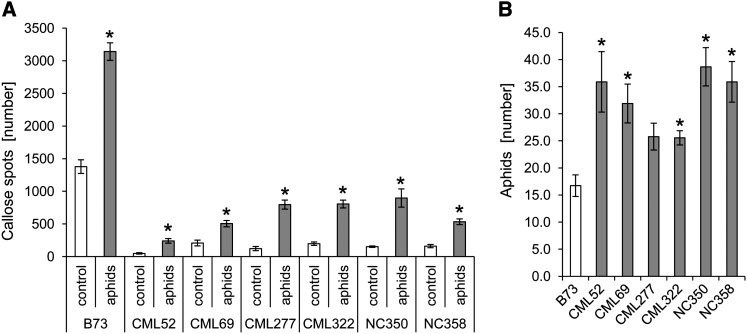
Although experiments with fungal pathogens and caterpillar feeding found induced conversion of DIMBOA-Glc to HDMBOA-Glc (Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011), this was not the case with R. maidis feeding (Figure 9), thus ruling out differential induction as a possible explanation for the above discrepancy. As an alternative hypothesis, we tested whether other defenses are induced by DIMBOA-Glc but not HDMBOA-Glc. Ahmad et al. (2011) demonstrated that infiltration of DIMBOA, but not HDMBOA-Glc, into maize leaves induced accumulation of callose, a common plant defense against aphid feeding (Dreyer and Campbell, 1987; Walling, 2000; Botha and Matsiliza, 2004). To determine whether similar effects can result from natural variation in maize benzoxazinoid content, we measured constitutive and aphid-induced callose accumulation in B73, CML52, CML69, CML277, CML322, NC350, and NC358. Aphid feeding increased callose formation on all seven tested maize lines (Figure 10A). However, both control and aphid-treated samples from the six maize inbred lines with low DIMBOA-Glc content had much lower callose levels than the corresponding B73 reference samples (P < 0.05, Tukey’s HSD test; Figure 10A). As in the case of aphids that were able to roam freely on whole maize seedlings (Figure 2A), aphids that were caged on individual leaves in this experiment produced more progeny on inbred lines with low DIMBOA-Glc content than on B73 (Figure 10B). Taken together, this suggests that DIMBOA-Glc increases aphid resistance by promoting callose deposition and that the increased production of HDMBOA-Glc by methylation of DIMBOA-Glc leads to aphid susceptibility via a reduction in callose defenses.
Figure 9.
Aphids Do Not Induce Benzoxazinoid Biosynthesis in Maize.
Accumulation of benzoxazinoids in B73 after 0, 2, 4, 8, 24, 48, and 96 h of aphid feeding (mean ± se; n = 5). No significant differences were detected (*P > 0.05; Dunnett’s test relative to 0-h control time point). FW, fresh weight.
Figure 10.
Aphid-Induced Callose Formation Is Reduced in Maize Inbred Lines with Low DIMBOA-Glc and High HDMBOA-Glc Concentrations.
(A) Number of callose spots in maize leaves with and without aphid feeding (mean ± se; n = 4 [controls], n = 18 [B73 + aphids], and n = 9 [other inbred lines + aphids]). Asterisks indicate significant differences relative to control for each genotype (P < 0.05; two-tailed Student’s t tests).
(B) Number of aphids present 3 d after 10 aphids were caged on a leaf blade (mean ± se; n = 18 [B73], and n = 9 [other inbred lines]). Asterisks indicate significant differences (*P < 0.06; two-tailed t test compared with B73 control, with Bonferroni correction for multiple comparisons).
When aphids were given a choice of host plants, there was no significant difference in their settling on B73 relative to NC350 or NC358 (see Supplemental Figure 6 online). In the case of pairwise comparisons with CML277 and CML52, aphids actually preferred the less suitable host, B73. Thus, it appears that different plant factors mediate R. maidis host plant choice and reproductive success on maize.
DISCUSSION
By mapping a QTL for natural variation in maize resistance to R. maidis, we identified a DIMBOA-Glc methyltransferase maize gene family representing important catalysts in the formation of plant defenses. Although several studies have demonstrated that herbivory and pathogen infection induce DIMBOA-Glc to HDMBOA-Glc conversion in maize (Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011; Huffaker et al., 2011), it was unclear which genes encode this enzyme activity. In addition to Bx10a, Bx10b, and Bx10c, other genes that are in the same cluster of the phylogenetic tree (Figure 4; GRMZM2G099297, GRMZM2G106172, GRMZM2G093092, and GRMZM2G127418) may also encode DIMBOA-Glc methyltransferases. Given the currently available DNA sequence data, it is not possible to determine whether the size of this gene family varies among the inbred lines of the NAM population.
The presence of multiple genes encoding the same DIMBOA-Glc methyltransferase activity may allow variable defense activation in different plant tissues or in response to different pests and pathogens. Whereas DIMBOA-Glc is produced constitutively in most maize lines studied so far, the conversion to HDMBOA-Glc can be both constitutive (as shown here) or induced by insect feeding and pathogen attack (Oikawa et al., 2004; Dafoe et al., 2011; Glauser et al., 2011; Huffaker et al., 2011). In the maize hybrid Delprim, HMDBOA-Glc is strongly inducible in the leaves, but produced constitutively in the roots (Marti et al., 2013). Across the maize inbred lines of the NAM population, the Bx10c transposon insertion strongly influences the relative constitutive abundance of DIMBOA-Glc and HDMBOA-Glc (Figures 6C and 6D). However, the lack of Bx10c transcripts in Oh43 (Figure 5), an inbred line which does not contain a Bx10c transposon insertion, suggests another regulatory mechanism for Bx10c downregulation in this line. Additionally, it is quite likely that the transcriptional regulation of other members of the DIMBOA-Glc methyltransferase gene family affects the constitutive or induced accumulation of HDMBOA-Glc. Such biochemical redundancy in key regulated steps of defense pathways may occur commonly in plants. For instance, although most reactions in Arabidopsis indole glucosinolate biosynthesis are encoded by single genes, induced indole glucosinolate methoxylation is encoded by multiple genes with the same enzymatic functions (Pfalz et al., 2009, 2011).
In addition to having toxic effects on herbivores and pathogens, DIMBOA has a further function in the induction of callose as a plant defense response. This was previously demonstrated by reduced callose accumulation in a bx1 igl1 maize double mutant, as well as by callose formation in response to infiltration of DIMBOA into the apoplastic space (Ahmad et al., 2011). Our results show that natural variation in this pathway has a significant effect on maize aphid resistance. Even though HDMBOA-Glc is more deleterious than DIMBOA-Glc to R. maidis in artificial diet assays (Figure 8), aphids grow better on maize with low DIMBOA-Glc and elevated HDMBOA-Glc (see Supplemental Figure 4 online). This can be explained by the observation that callose, which provides defense against aphids (Dreyer and Campbell, 1987; Walling, 2000; Botha and Matsiliza, 2004), is more abundant in B73 than in maize lines with low DIMBOA-Glc content (Figure 10A). Further research will be needed to identify the mechanisms by which DIMBOA and/or one of its breakdown products induces callose formation. It is quite possible that DIMBOA acts early in a pathway that induces not only callose formation, but also other plant defenses that provide protection against aphids.
Callose induction by benzoxazinoids in maize is similar to previously reported callose induction by indole glucosinolate breakdown in Arabidopsis (Clay et al., 2009). Specificity of these pathways is demonstrated by the fact that chemically similar transformations lead to opposite effects on callose formation. Whereas DIMBOA induces callose formation in maize, and O-methylation to HDMBOA abolishes this effect (Ahmad et al., 2011), hydrolysis of indol-3-ylmethylglucosnolate (or 1-methoxyindol-3-ylmethylglucosinolate) in Arabidopsis does not trigger callose formation but hydrolysis of the O-methylated product 4-methoxyindole-3-ylmethylglucosinolate creates an inducer of callose formation (Clay et al., 2009). For both plant species, activation of existing defense products, benzoxazinoids (maize) or glucosinolates (Arabidopsis) is an indicator of enemy attack and thus provides a reliable signal for inducing other defenses. It is likely that future research with other plant systems will provide additional examples of defensive metabolites that have a secondary function as signals in activating other defenses.
Although aphids produce more progeny on plants with low DIMBOA-Glc content (Figure 2), this was not reflected as a preference for these plants in aphid choice assays (see Supplemental Figure 6 online). In two cases, the aphids actually settled preferentially on inbred lines that are less suitable host plants. This suggests that different plant factors affect host plant choice and reproduction in the case of R. maidis. It is not known what plant cues affect host plant choice in this species. However, similar situations, where choices made by adult insects do not identify the best host plants for developing progeny have been reported in other plant–insect interactions (Mayhew, 2001).
The parental lines of the NAM population, which were chosen based on their high level of genetic diversity (Flint-Garcia et al., 2005; McMullen et al., 2009b; Chia et al., 2012), vary considerably in their accumulation of defense-related benzoxazinoids (Figure 2). The negative correlation between DIMBOA-Glc and HDMBOA-Glc content (see Supplemental Figure 3A online) suggests that there are divergent defense strategies in maize. B73 and other maize lines with constitutively high DIMBOA-Glc and low HDMBOA-Glc levels likely rely on inducible responses to increase HDMBOA-Glc accumulation when there is an herbivore or pathogen attack. Conversely, maize seedlings with high levels of HDMBOA-Glc are already provisioned to be more resistant to attack by chewing herbivores. However, such lines might also be more prone to false alarms and the production of phytotoxic benzoxazinoid breakdown products in the absence of attack. Thus, depending on the particular environment in which a plant is located, either constitutive or inducible HDMBOA-Glc production may be the better strategy.
Previous studies have mapped QTL for resistance to lepidopteran herbivores, the southwestern maize borer (Diatraea grandiosella) and the maize earworm (Helicoverpa zea), to bin 1.04 of the maize genome (Byrne et al., 1998; Groh et al., 1998; Brooks et al., 2005). This variation in chewing herbivore resistance may also be mediated by regulation of DIMBOA-Glc methyltransferase. A previously reported bin 1.04 QTL for DIMBOA-Glc abundance (Butrón et al., 2010) likely also reflects variation in DIMBOA-Glc methyltransferase activity. However, as HDMBOA-Glc was not measured in this assay, it is not possible to determine whether there was a concomitant increase in this metabolite. Since B73 × CML322 recombinant inbred lines, which we used in our study (Figure 3A), also were used by Butrón et al. (2010), it is quite likely that the same QTL was mapped.
It is perhaps significant that the six NAM parental lines with the highest constitutive HDMBOA-Glc/DIMBOA-Glc ratio (CML52, CML69, CML277, CML322, CML350, and CML358; Figure 2) represent tropical germplasm. Since pressure from insect herbivores and pathogens tends to be higher in tropical habitats (Rasmann and Agrawal, 2011), maize breeding in such regions may have selected for plants with constitutively elevated HDMBOA-Glc accumulation. The Egyptian cotton leafworm (Spodoptera littoralis) and the fall armyworm (Spodoptera frugiperda) are able to reglycosylate DIMBOA to DIMBOA-Glc (Glauser et al., 2011), which may render this compound ineffective as a defense. However, HDMBOA is highly unstable and immediately degrades into active catabolites, which may make it impossible for caterpillars to detoxify it effectively. The ability of maize plants to produce high amounts of HDMBOA-Glc may therefore be an important prerequisite for stable yields under heavy pressure by chewing arthropods. Aphids, on the other hand, tend to be a lesser problem than chewing herbivores at the maize seedling stage. Thus, the concomitant selection for aphid susceptibility due to a reduction in callose accumulation and perhaps other aphid-specific defenses may not be a significant problem for tropical maize farmers. It remains to be determined whether defenses elicited by DIMBOA are beneficial in temperate agroecosystems, which are characterized by a distinct pathogen community (Ullstrup and Renfro, 1976) and a few dominating herbivore pests, many of which are resistant to benzoxazinoids (Dafoe et al., 2011; Glauser et al., 2011; Robert et al., 2012).
The distinct induction patterns, phytochemical properties, and potential defense elicitation roles of benzoxazinoids suggest that they may have divergent functions in plant defenses against different herbivores and pathogens. Alternatively, benzoxazinoids may act synergistically at different biological levels to protect the plant against its enemies (Glauser et al., 2011). Deciphering the individual contribution of the different benzoxazinoid derivatives to insect resistance has remained difficult because of their unknown biochemical and genetic origins and the lack of genetic resources to study them in vivo. Data presented here will permit identification of additional genes involved in maize benzoxazinoid metabolism. For instance, investigation of natural variation in DIM2BOA-Glc accumulation (Figure 2D) may lead to the identification of the as yet unknown enzymes for the biosynthesis of this benzoxazinoid. GRMZM2G408458 for instance, which is similar to Bx7 (Figure 4), may also be part of the core DIMBOA-Glc biosynthesis pathway.
A single large-effect benzoxazinoid QTL causes most of the aphid resistance variation in our experiments (Figure 6E). However, it is likely that more extensive analysis of all recombinant inbred lines in the NAM population would identify additional aphid resistance mechanisms. For instance, other QTL may function in maize inbred lines that are more resistant to R. maidis than B73 (Figure 2A). Similarly, genetic mapping of resistance to chewing herbivores has identified QTL that are not linked to known benzoxazinoid-related genes (McMullen et al., 2009a; Meihls et al., 2012). Our results demonstrate that newly available maize genetic and genomic resources make it straightforward to proceed from the observation of such natural variation in insect resistance to elucidating the genetic and biochemical basis of this variation. The identification of these and other herbivore resistance QTL will open up new opportunities for improving maize through classical breeding and transgenic approaches.
METHODS
Plants and Growth Conditions
To grow maize (Zea mays) plants for experiments, single seeds were buried ∼1.5 cm deep in a 7.6 × 7.6-cm plastic pot (∼200 cm3) filled with moistened maize mix (produced by mixing 0.16 m3 Metro-Mix 360 [Scotts], 0.45 kg finely ground lime, 0.45 kg Peters Unimix [Scotts], 68 kg Turface MVP [Profile Products], 23 kg coarse quartz sand, and 0.018 m3 pasteurized field soil). Plants were grown in Conviron growth chambers under 16-h-light/8-h-dark photoperiod and 180 μmol photons m−2 s−1 light at constant 23°C and 60% humidity and were watered from below as needed.
Aphids and Growth Conditions
A maize leaf aphid (Rhopalosiphum maidis) colony was started with insects obtained from S. Gray (USDA Plant Soil and Nutrition Laboratory, Ithaca, NY), from a colony that was originally collected in New York State and had been maintained on barley (Hordeum vulgare). Aphids were reared on 4- to 8-week-old maize plants (variety B73) under 16-h-light/8-h-dark photoperiod at constant 23°C. Plants were watered from below as needed. Adult aphids were used for all experiments.
Whole-Plant Aphid Bioassays
NAM population parental lines, F1 progeny, and B73 × CML322, B73 × CML277, B73 × CML69, and B73 × CML52 recombinant inbred lines were screened for resistance to R. maidis infestation. Plants were used for aphid bioassays at the age of 2 weeks (V2-V3 stage). Ten adult aphids were confined on 2-week-old seedling plants using microperforated polypropylene bags (15.25 cm × 61 cm; PJP Marketplace). Seven days after infestation, the surviving adults and progeny were counted.
Aphid choice assays were performed under conditions similar to the whole-plant aphid bioassays. Seeds of the two inbred lines used for comparisons were planted in opposite corners of individual pots. Ten aphids were released in the center of the pots between the plants. The pots were covered with perforated bags in a manner that allowed contact between the leaves of the two plants. Pots were placed in the growth chamber in randomized orientation. After 24 h, the surviving aphids were counted, and the plant from which they were feeding was recorded.
Artificial Diet Assays
A previously described benzoxazinoid artificial diet assay (Cambier et al., 2001) was modified to measure the effects of benzoxazinoids on R. maidis reproduction and survival. DIMBOA-Glc and HDMBOA-Glc were purified from maize as described previously (Glauser et al., 2011). All other chemicals were purchased from Sigma-Aldrich.
Aphid artificial diet was adapted from Prosser and Douglas (1992). Five stock solutions were prepared as follows: Solution A, 4.1 mM FeCl3·6H2O, 1 mM CuCl2·4H2O, 2 mM MnCl2·6H2O, and 10.5 mM ZnSO4; Solution B, 0.1 mM biotin, 4.2 mM pantothenate, 0.9 mM folic acid, 16 mM nicotinic acid, 2.4 mM pyridoxine, 1.5 mM thiamine, 72 mM choline, and 56 mM myo-inositol; Solution C, 11 mM Ala, 19 mM Arg, 29 mM Asn, 29 mM Asp, 5 mM Cys, 33 mM Gln, 17 mM Glu, 2.4 mM Gly, 17 mM His, 17 mM Ile, 17 mM Leu, 17 mM Lys, 5.7 mM Met, 5.7 mM Phe, 11 mM Pro, 11 mM Ser, 17 mM Thr, 5.7 mM Trp, 1.2 mM Tyr, and 17 mM Val; Solution D, 19 mM ascorbic acid, 1.7 mM citric acid, 8.1 mM MgSO4·7H2O, and 1660 mM Suc; and Solution E, 660 mM K2HPO4. Diet for aphid bioassays was prepared by mixing 1 mL Solution A, 5 mL Solution B, 50 mL Solution C, 30 mL Solution D, 10 mL Solution E, and 4 mL water. DIMBOA-Glc was added to the diet at 1, 2, 4, and 8 mM concentrations. HDMBOA-Glc was added to the diet at 0.25, 0.5, 1, and 2 mM concentrations.
Fifteen to 20 adult R. maidis were added to 30-mL plastic cups, a sheet of Parafilm (Pechiney Packaging Company) was stretched across the 3-cm diameter opening, 80 μL of artificial diet, with and without benzoxazinoids, was pipetted onto the Parafilm, and another sheet of Parafilm was stretched across the top to keep the diet in place. The number of nymphs produced by the adult aphids was counted after 2 d on control diet, diet containing DIMBOA-Glc (1, 2, 4, or 8 mM), or diet containing HDMBOA-Glc (0.25, 0.5, 1, or 2 mM). The number of surviving adult aphids was counted after 2 and 4 d on control diet and diet with 2 mM DIMBOA-Glc or 2 mM HDMBOA-Glc. The continued presence of intact benzoxazinoids and absence of breakdown products in the aphid diet was confirmed at the end of the experiment.
Tissue Collection for Benzoxazinoid Assays
For measurement of leaf benzoxazinoid content, NAM parental lines, B73 × CML322 recombinant inbred lines, and B73 × CML322 F1 progeny were grown in the same manner as for aphid bioassays. When the plants were 2 weeks old, the distal half of the second leaf was harvested and snap-frozen in liquid nitrogen for benzoxazinoid assays.
For measuring benzoxazinoid changes induced by aphid feeding, cages were placed 1 cm away from the leaf tip on the dorsal side of the third leaf of 2-week-old maize seedlings. Ten aphids were added to each cage and, at the end of the feeding period, the ∼1-cm leaf segment contained within the cage was harvested for benzoxazinoid assays. To avoid the influence of diurnal cycles on benzoxazinoid abundance, aphids were added to the cages in a staggered manner (0, 2, 4, 8, 24, 48, and 96 h after the start of the experiment), and all tissue samples were harvested at noon, 96 h after the start of the experiment. The harvested tissue samples were snap-frozen in liquid nitrogen and stored at −80°C for later analysis of benzoxazinoids.
Extraction and Analysis of Benzoxazinoids
Benzoxazinoid concentrations were determined according to a previously described protocol (Glauser et al., 2011) with some modifications. Extraction solvents were HPLC-grade methanol (VWR), Milli-Q water (Millipore), and analytical-grade formic acid (Sigma-Aldrich). Fresh plant leaves were frozen in liquid nitrogen, stored at −80°C, and ground to a powder with a mortar and pestle under liquid nitrogen. Twenty milligrams of frozen powder were weighed in a 1.5-mL microcentrifuge tube, and 1 mL of extraction solvent (methanol/water/formic acid, 50:49.5:0.5, v/v) was added. The tubes were vortexed for ∼10 s, and five to 10 glass beads (2 mm diameter) were added to each tube. The samples were then extracted in a tissue lyser (Retsch MM300) at 30 Hz for 3 min and centrifuged at 14,000g for 3 min, and the supernatant was transferred to an HPLC vial and stored at −80°C before analysis.
Benzoxazinoid analysis was performed on an Acquity UPLC system (Waters) equipped with an eλ photodiode array detector and coupled to a Synapt G2 quadrupole time-of-flight mass spectrometer (Waters) through an electrospray interface. An Acquity BEH C18 column from Waters (2.1 × 50 mm, 1.7-μm particle size) was used. The following gradient program was employed at a flow rate of 400 μL/min: solvent A = water + formic acid 0.05%, solvent B = acetonitrile + formic acid 0.05%; 2 to 27.2% B in 3.5 min, 27.2 to 100% B in 1.0 min, holding at 100% B for 1.0 min, and reequilibration at 2% B for 1.0 min. The temperatures of the column and autosampler were maintained at 40 and 15°C, respectively. The injection volume was 2.5 μL. UV spectra were acquired over the range 210 to 400 nm at a frequency of 20 Hz and a resolution of 1.2 nm. The extracted trace at 264 nm was used for quantification of benzoxazinoids. The quadrupole time-of-flight mass spectrometer was operated in positive ion mode over a range of 85 to 600 D using a scan time of 0.4 s. Source parameters were as follows: capillary and cone voltages 2800 and 25 V, respectively, source temperature 120°C, desolvation gas flow and temperature 800 L/h and 330°C, respectively, cone gas flow 20 L/h. Accurate mass measurements were provided by infusing a solution of the synthetic peptide Leu enkephalin through the Lockspray ESI probe. The mobile phase was directed to waste before the mass spectrometry from 0.0 to 1.0 min and 4.0 to 6.5 min. The following extracted ion chromatograms were used with a mass window of ±0.01 D for quantification: mass-to-charge ratio (m/z) 194.045 for DIMBOA-Glc (retention time [RT] 2.14 min) and DIMBOA (RT 2.38 min), m/z 166.050 for HDMBOA-Glc (RT 2.71 min) and HDMBOA (RT 3.22 min), m/z 224.056 for DIM2BOA-Glc (RT 2.18 min), and m/z 178.051 for HMBOA-Glc (RT 2.07 min). Absolute concentrations of benzoxazinoids were determined using external calibration curves obtained from purified DIMBOA, DIMBOA-Glc, and HDMBOA-Glc standards (Glauser et al., 2011). The concentrations of the calibration points were 0.2, 1, 2, 5, 20, and 50 μg/mL for the three benzoxazinoids. Whenever benzoxazinoid concentrations in plant samples were equal to or <5 μg/mL, mass spectrometry traces were used for quantification. For concentrations above 5 μg/mL, the UV trace was employed since the signal exceeded the linear domain of the mass spectrometry. We verified that similar values were obtained with both detection modes for a signal corresponding to a concentration of 5 μg/mL.
Callose Assays
Maize inbred lines B73, CML52, CML69, CML277, CML322, NC350, and NC358 were grown in a growth chamber for 10 d. Callose formation was induced by caging 10 R. maidis 1 cm from the leaf tip on the dorsal side of the third leaf for 3 d. Control leaves received cages without aphids for 3 d. After 3 d, the number of aphid offspring in each cage was counted, and the portion of the leaf contained in the cage was stained according to a previously described protocol (Luna et al., 2011). Each leaf segment was decolorized for 48 h in 98% ethanol and stained for 2 h with 0.01% aniline-blue in 0.07 M, pH 9.0, phosphate buffer. Callose spots were counted by microscopy on both sides of the 14-mm leaf segment that was contained within the aphid cage.
Data Analysis
QTL analysis using individual sets of recombinant inbred lines was done by composite interval mapping using the Windows QTL Cartographer software version 2.5 (Wang et al., 2012). The experimental LOD threshold was determined by permutation tests with 500 repetitions at the significance level of 0.05. All analyses were performed using the default settings in the WinQTL program. The program settings were as follows: the CIM program module = Model 6: Standard Model, walking speed = 2 centimorgans, control marker numbers = 5, window size = 10 centimorgans, regression method = backward regression method. Maize genetic marker data were downloaded from www.panzea.org and were used for association mapping using multiple sets of recombinant lines from the NAM population. Statistical tests were conducted using JMP (www.jmp.com) and SAS (www.sas.com).
Preparation of Genomic DNA, RNA, and cDNA
Leaf material was harvested from 14-d-old seedlings, flash-frozen in liquid nitrogen, and stored at −80°C until sample preparation. After grinding of the frozen leaf material to a fine powder in a mortar filled with liquid nitrogen, DNA and RNA were extracted using the DNeasy plant mini kit (Qiagen) and RNeasy plant mini kit (Qiagen), respectively, according to the manufacturer’s instructions. Nucleic acid concentration, purity, and quality were assessed using a spectrophotometer (NanoDrop 2000c; Thermo Scientific) and an Agilent 2100 Bioanalyzer (Agilent Technologies). Prior to cDNA synthesis, 0.75 µg RNA was DNase treated using 1 μL DNase (Fermentas). Single-stranded cDNA was prepared from the DNase-treated RNA using SuperScript III reverse transcriptase and oligo(dT20) primers (Invitrogen).
Cloning and Expression of O-Methyltransferase Genes
The complete open reading frames of Bx10a-B73, Bx10b-B73, and Bx10c-CML322 were amplified from cDNA with the primer pairs listed in Supplemental Table 1 online. PCR products were cloned as blunt fragments into the sequencing vector pCR-Blunt II-TOPO (Invitrogen) and both strands were fully sequenced. For heterologous expression with an N-terminal His-tag, the genes were inserted as BsaI fragments into the expression vector pASK-IBA37 plus (IBA). The constructs were introduced into the Escherichia coli strain TOP10 (Invitrogen). Liquid cultures of the bacteria harboring the expression constructs were grown at 37°C to an OD600 of 0.6 to 0.8. Anhydrotetracycline (IBA) was added to a final concentration of 200 µg L−1, and the cultures were incubated for 20 h at 18°C. The cells were sedimented for 5 min at 5000g and 4°C. For breaking up the cells, the pellet was resuspended in ice-cold 4 mL 50 mM Tris-HCl, pH 8.0, containing 0.5 M NaCl, 20 mM imidazole, 50 mM mercaptoethanol, and 10% glycerol and subsequently exposed to ultrasonication (4 × 20 s; Bandelin UW2070). The debris was separated by centrifugation for 20 min at 16,100g and 4°C. N-terminal His-tagged proteins were purified using nickel-nitrilotriacetic acid spin columns (Qiagen) according to the manufacturer’s instructions. The purified proteins were eluted with 50 mM Tris-HCl, pH 8.0, containing 0.5 M NaCl, 250 mM imidazole, and 10% glycerol. The salt was removed by gel filtration using Illustra NAP-10 columns (GE Healthcare), and the proteins were redissolved in 50 mM MOPS, pH 7.0, containing 50% glycerol.
O-Methyltransferase Assays
The in vitro O-methyltransferase activity of BX10a-B73, BX10b-B73, and BX10c-CML322 was proven using enzyme assays containing purified recombinant protein, the substrate DIMBOA-Glc, and the cosubstrate S-adenosyl-l-Met. The assays were performed with 4 mM DTT, 0.4 mM S-adenosyl-l-Met, 0.2 mM DIMBOA-Glc, and 0.5 µg desalted enzyme buffered by 50 µM MOPS, pH 7.0, in a volume of 50 µL. The assays were performed in glass vials and incubated for 20 min at 25°C at 400 rpm using a ThermoMixer comfort 5355 (Eppendorf). The reaction was stopped by adding 1 volume of methanol, and the mixture was centrifuged for 10 min at 4200g and 4°C. HDMBOA-Glc formation was determined by reversed phase liquid chromatography using an Agilent 1200 HPLC system (Agilent Technologies). For separation, a Kinetex 2.6 μm C18 100 × 4.6-mm column was used (Phenomenex). Formic acid (0.05%) and acetonitrile were used as eluents A and B, respectively. Elution was initiated isocratically for 0.5 min at 10% eluent B and continued with linear gradients to 40 and 100% eluent B in 30 min and 1 min, respectively. The column was reequilibrated for 4 min. The flow rate was set to 1.1 mL/min. Five microliters of the sample was injected, and the absorbance was measured at the maximum of 270 nm. Metabolite identities were confirmed using authentic standard compounds.
Sequence Analysis and Phylogenetic Tree Reconstruction
A nucleotide sequence alignment of maize methyltransferase genes similar to Bx10a, Bx10b, and Bx10c was computed using the MUSCLE (codon) algorithm (see Supplemental Data Set 1 online; gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.2; clustering method, UPGMB) implemented in MEGA5 (Tamura et al., 2011). Based on the MUSCLE alignment, a phylogenetic tree was reconstructed with MEGA5 using a maximum likelihood algorithm (general time-reversible model, gamma distributed rates among sites). Codon positions included were 1st+2nd+3rd+Noncoding. All positions with <90% site coverage were eliminated. Ambiguous bases were allowed at any position. A bootstrap resampling analysis with 1000 replicates was performed to evaluate the tree topology. Two recently described maize methyltransferase genes, Anthranilic acid methyltransferase1 (Aamt1) and O-Methyltransferase8 (Omt8), which both belong to a different class of O-methyltransferases, were included as an outgroup.
Transposon Analysis
To test for the presence of a Doppia-like transposon in the first exon of Bx10c alleles, PCR was performed with genomic DNA as template and the gene specific primer TPA1 located on exon 1 and the transposon specific primer TPA2 (Supplemental Table 2 online). Conversely, the absence of the transposon was analyzed using two gene specific primers (TPA1 and TPA3) binding on gene regions upstream and downstream, respectively, of the transposon insertion site. The resulting PCR products (671 bp and 398 bp, respectively) were cloned as blunt fragments into the sequencing vector pCR®-Blunt II-TOPO® and both strands were fully sequenced.
qRT-PCR Analysis
For the amplification of Bx10 gene fragments with a length of ∼150 bp, primer pairs specific for Bx10a/b and Bx10c were designed having a Tm ≥ 58°C, a GC content between 35 and 55%, and a primer length in the range of 20 to 25 nucleotides (see Supplemental Table 2 online for primer information). Primer specificity was confirmed by agarose gel electrophoresis, melting curve analysis, and sequence verification of cloned PCR amplicons. Primer pair efficiency was determined using the standard curve method with fivefold serial dilution of cDNA and found to be between 90 and 105%. The actin gene Zm-Actin1 (accession number MZEACT1G) was used as a reference gene. The cDNA was prepared as described above, and 1 μL cDNA was used in all 20-μL reactions. Samples were run in triplicates using Brilliant III SYBR Green QPCR Master Mix (Stratagene) with ROX as the reference dye. The following PCR conditions were applied for all reactions: initial incubation at 95°C for 3 min followed by 40 cycles of amplification (95°C for 20 s and 60°C for 20 s). Plate reads were taken during the annealing and the extension step of each cycle. Data for the melting curves were gathered at the end of cycling from 55 to 95°C.
All samples were run on the same PCR machine (MxPro Mx3000P; Stratagene, Agilent Technologies) in an optical 96-well plate. Five biological replicates were analyzed as triplicates in the quantitative PCR for each of the two inbred lines. Data for the relative quantity to calibrator average (dRn) were exported from the MXPro Software.
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under the following accession numbers: Aamt1, HM242244; Omt8, HM242248; Zrp4, NP_001105689; Bx7-CI31A, NP_001120719; Bx10a-B73, KC754962; Bx10b-B73, KC754963; Bx10c-CML322, KC754964; GRMZM2G100754, AFW64970; GRMZM2G097297, AFW64969; GRMZM2G023152, AFW60893; GRMZM2G124799, AFW78713; GRMZM2G102863, AFW78712; GRMZM2G141026, AFW78711; GRMZM2G140996, AFW78710; GRMZM2G041866, AFW65591; GRMZM2G099297, AFW64033; GRMZM2G106172, AFW87991; GRMZM2G093092, AFW87990; GRMZM2G127418, DAA39165; GRMZM2G408458, AFW60754; GRMZM2G059465, DAA48819; GRMZM2G104730, AFW55836; GRMZM2G104710, AFW55834; GRMZM2G177424, DAA60533; GRMZM2G091774, AFW56785; GRMZM2G349791, AFW60116; GRMZM2G147491, DAA38843; and GRMZM2G085924, DAA38899.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Aphid Susceptibility Is Caused by a Dominant Allele on CML322 Chromosome 1.
Supplemental Figure 2. Maize Chromosome LOD Plots Showing Location of Aphid Resistance QTL.
Supplemental Figure 3. Correlation of DIMBOA-Glc Content with Other Benzoxazinoids in Parental Lines of the NAM Population.
Supplemental Figure 4. Correlation of Aphid Resistance and Benzoxazinoid Content of Maize Leaves.
Supplemental Figure 5. High HDMBOA-Glc Accumulation Is Caused by a Dominant Allele on CML322 Chromosome 1.
Supplemental Figure 6. Aphid Choice Experiments, Comparing B73 to More Susceptible Inbred Lines.
Supplemental Table 1. Predicted B73 Genes in the Area of the Bin 1.04 R. maidis Resistance QTL.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Nucleotide Sequence Alignment of Putative Methyltransferase Genes Similar to Bx10a, Bx10b, and Bx10c.
Acknowledgments
This research was funded by US National Science Foundation Award IOS-1139329, Defense Advanced Research Projects Agency Award W31P4Q-10-1-0011, and a Friedrich Wilhelm Bessel Research Award from the Alexander von Humboldt Foundation to G.J.; USDA Award 2011-67012-30675 to L.N.M.; and the USDA–Agricultural Research Service and US National Science Foundation Award DBI-0820619 to E.S.B. Research activities of J.G., V.H., T.G.K., G.G., and M.E. were supported by a Sinergia Grant of the Swiss National Science Foundation (SNF 136184). The work of M.E. is supported by a Marie Curie Intra European Fellowship (Grant 273107). We thank Gabriella Gómez for assistance with callose assays, Neil Villard for benzoxazinoid measurements, and I-Chun Chen for aphid bioassays. Part of this work was carried out using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation.
AUTHOR CONTRIBUTIONS
L.N.M., V.H., J.G., M.E., T.G.K., and G.J. designed the research. L.N.M., V.H., G.G., H.B., H.K., and M.M.H. performed research. L.N.M., V.H., J.G., H.K., A.E.L., E.S.B., M.E., T.G.K., and G.J. analyzed data. M.E., T.G.K., and G.J. wrote the article. L.N.M. and V.H. contributed equally to the study and share first authorship. M.E., T.G.K. and G.J. contributed equally to the study and share senior authorship.
Glossary
- QTL
quantitative trait loci
- NAM
nested association mapping
- DIMBOA
2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one
- HDMBOA-Glc
2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside
- DIM2BOA-Glc
2,4-dihydroxy-7,8-dimethoxy-1,4- benzoxazin-3-one glucoside
- HMBOA-Glc
2-hydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside
- qRT-PCR
quantitative RT-PCR
- m/z
mass-to-charge ratio
- RT
retention time
References
- Ahmad S., Veyrat N., Gordon-Weeks R., Zhang Y.H., Martin J., Smart L., Glauser G., Erb M., Flors V., Frey M., Ton J. (2011). Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol. 157: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass H.W., OBrian G.R., Boston R.S. (1995). Cloning and sequencing of a second ribosome-inactivating protein gene from maize (Zea mays L.). Plant Physiol. 107: 661–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercury S.D., Panavas T., Irenze K., Walker E.L. (2001). Molecular analysis of the Doppia transposable element of maize. Plant Mol. Biol. 47: 341–351 [DOI] [PubMed] [Google Scholar]
- Botha C.E.J., Matsiliza B. (2004). Reduction in transport in wheat (Triticum aestivum) is caused by sustained phloem feeding by the Russian wheat aphid (Diuraphis noxia). S. Afr. J. Bot. 70: 249–254 [Google Scholar]
- Brooks T.D., Willcox M.C., Williams W.P., Buckley P.M. (2005). Quantitative trait loci conferring resistance to fall armyworm and southwestern corn borer leaf feeding damage. Crop Sci. 45: 2430–2434 [Google Scholar]
- Butrón A., Chen Y.C., Rottinghaus G.E., McMullen M.D. (2010). Genetic variation at bx1 controls DIMBOA content in maize. Theor. Appl. Genet. 120: 721–734 [DOI] [PubMed] [Google Scholar]
- Byrne P.F., McMullen M.D., Wiseman B.R., Snook M.E., Musket T.A., Theuri J.M., Widstrom N.W., Coe E.H. (1998). Maize silk maysin concentration and corn earworm antibiosis: QTLs and genetic mechanisms. Crop Sci. 38: 461–471 [Google Scholar]
- Caillaud C.M., Niemeyer H.M. (1996). Possible involvement of the phloem sealing system in the acceptance of a plant as host by an aphid. Experientia 52: 927–931 [Google Scholar]
- Cambier V., Hance T., de Hoffmann E. (2000). Variation of DIMBOA and related compounds content in relation to the age and plant organ in maize. Phytochemistry 53: 223–229 [DOI] [PubMed] [Google Scholar]
- Cambier V., Hance T., De Hoffmann E. (2001). Effects of 1,4-benzoxazin-3-one derivatives from maize on survival and fecundity of Metopolophium dirhodum (Walker) on artificial diet. J. Chem. Ecol. 27: 359–370 [DOI] [PubMed] [Google Scholar]
- Chia J.M., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44: 803–807 [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés-Cruz M., Snook M., McMullen M.D. (2003). The genetic basis of C-glycosyl flavone B-ring modification in maize (Zea mays L.) silks. Genome 46: 182–194 [DOI] [PubMed] [Google Scholar]
- Dafoe N.J., Huffaker A., Vaughan M.M., Duehl A.J., Teal P.E., Schmelz E.A. (2011). Rapidly induced chemical defenses in maize stems and their effects on short-term growth of Ostrinia nubilalis. J. Chem. Ecol. 37: 984–991 [DOI] [PubMed] [Google Scholar]
- Dreyer D.L., Campbell B.C. (1987). Chemical basis of host-plant resistance to aphids. Plant Cell Environ. 10: 353–361 [Google Scholar]
- Flint-Garcia S.A., Thuillet A.C., Yu J., Pressoir G., Romero S.M., Mitchell S.E., Doebley J., Kresovich S., Goodman M.M., Buckler E.S. (2005). Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 44: 1054–1064 [DOI] [PubMed] [Google Scholar]
- Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R.B., Briggs S.P., Simcox K., Gierl A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Ganal M.W., et al. (2011). A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givovich A., Niemeyer H.M. (1995). Comparison of the effect of hydroxamic acids from wheat on 5 species of cereal aphids. Entomol. Exp. Appl. 74: 115–119 [Google Scholar]
- Givovich A., Morse S., Cerda H., Niemeyer H.M., Wratten S.D., Edwards P.J. (1992). Hydroxamic acid glucosides in honeydew of aphids feeding on wheat. J. Chem. Ecol. 18: 841–846 [DOI] [PubMed] [Google Scholar]
- Givovich A., Sandstrom J., Niemeyer H.M., Pettersson J. (1994). Presence of a hydroxamic acid glucoside in wheat phloem sap, and its consequences for performance of Rhopalosiphum padi (L) (Homoptera, Aphididae). J. Chem. Ecol. 20: 1923–1930 [DOI] [PubMed] [Google Scholar]
- Glauser G., Marti G., Villard N., Doyen G.A., Wolfender J.L., Turlings T.C., Erb M. (2011). Induction and detoxification of maize 1,4-benzoxazin-3-ones by insect herbivores. Plant J. 68: 901–911 [DOI] [PubMed] [Google Scholar]
- Gore M.A., Chia J.M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer J.A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S. (2009). A first-generation haplotype map of maize. Science 326: 1115–1117 [DOI] [PubMed] [Google Scholar]
- Grambow H.J., Luckge J., Klausener A., Muller E. (1986). Occurrence of 2-(2-hydroxy-4,7-dimethoxy-2h-1,4-benzoxazin-3-One)-beta-D-glucopyranoside in Triticum aestivum leaves and its conversion into 6-methoxy-benzoxazolinone. Zeitschrift für Naturforschung 41: 684–690 [Google Scholar]
- Groh S., Khairallah M.M., González-de-León D., Willcox M., Jiang C., Hoisington D.A., Melchinger A.E. (1998). Comparison of QTLs mapped in RILs and their test-cross progenies of tropical maize for insect resistance and agronomic traits. Plant Breed. 117: 193–202 [Google Scholar]
- Huffaker A., Dafoe N.J., Schmelz E.A. (2011). ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155: 1325–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.T.J. (2011). Evolutionary ecology of plant defences against herbivores. Funct. Ecol. 25: 305–311 [Google Scholar]
- Jonczyk R., Schmidt H., Osterrieder A., Fiesselmann A., Schullehner K., Haslbeck M., Sicker D., Hofmann D., Yalpani N., Simmons C., Frey M., Gierl A. (2008). Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: Characterization of Bx6 and Bx7. Plant Physiol. 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J.J.B., Willems G., van Eeuwijk F., Nordborg M., Koornneef M. (2011). A comparison of population types used for QTL mapping in Arabidopsis thaliana. Plant Gen. Res. 9: 185–188 [Google Scholar]
- Klingler J.P., Nair R.M., Edwards O.R., Singh K.B. (2009). A single gene, AIN, in Medicago truncatula mediates a hypersensitive response to both bluegreen aphid and pea aphid, but confers resistance only to bluegreen aphid. J. Exp. Bot. 60: 4115–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov M. (2008). Losses of birch foliage due to insect herbivory along geographical gradients in Europe: A climate-driven pattern? Clim. Change 87: 107–117 [Google Scholar]
- Kroymann J., Donnerhacke S., Schnabelrauch D., Mitchell-Olds T. (2003). Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc. Natl. Acad. Sci. USA 100 (suppl. 2): 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump K.L., Bradbury P.J., Wisser R.J., Buckler E.S., Belcher A.R., Oropeza-Rosas M.A., Zwonitzer J.C., Kresovich S., McMullen M.D., Ware D., Balint-Kurti P.J., Holland J.B. (2011). Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168 [DOI] [PubMed] [Google Scholar]
- Lawrence S.D., Novak N.G., Kayal W.E., Ju C.J.T., Cooke J.E.K. (2012). Root herbivory: Molecular analysis of the maize transcriptome upon infestation by Southern corn rootworm, Diabrotica undecimpunctata howardi. Physiol. Plant. 144: 303–319 [DOI] [PubMed] [Google Scholar]
- Luna E., Pastor V., Robert J., Flors V., Mauch-Mani B., Ton J. (2011). Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Maresh J., Zhang J., Lynn D.G. (2006). The innate immunity of maize and the dynamic chemical strategies regulating two-component signal transduction in Agrobacterium tumefaciens. ACS Chem. Biol. 1: 165–175 [DOI] [PubMed] [Google Scholar]
- Marti G., Erb M., Boccard J., Glauser G., Doyen G.R., Villard N., Robert C.A., Turlings T.C., Rudaz S., Wolfender J.L. (2013). Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ. 36: 621–639 [DOI] [PubMed] [Google Scholar]
- Mayhew P.J. (2001). Herbivore host choice and optimal bad motherhood. Trends Ecol. Evol. (Amst.) 16: 165–167 [DOI] [PubMed] [Google Scholar]
- McMullen, M., Frey, M., and Degenhardt, J. (2009a). Genetics and biochemistry of insect resistance in maize. In Handbook of Maize: Its Biology, J.L. Bennetzen and S. Hake, eds (New York: Springer), pp. 271–289 [Google Scholar]
- McMullen M.D., et al. (2009b). Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Meihls L.N., Kaur H., Jander G. (December 6, 2012). Natural variation in maize defense against insect herbivores. Cold Spring Harb. Symp. Quant. Biol.(online) /10.1101/sqb.2012.77.014662. [DOI] [PubMed] [Google Scholar]
- Miller N., Estoup A., Toepfer S., Bourguet D., Lapchin L., Derridj S., Kim K.S., Reynaud P., Furlan L., Guillemaud T. (2005). Multiple transatlantic introductions of the western corn rootworm. Science 310: 992. [DOI] [PubMed] [Google Scholar]
- Niemeyer H.M. (2009). Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: Key defense chemicals of cereals. J. Agric. Food Chem. 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Oikawa A., Ishihara A., Iwamura H. (2002). Induction of HDMBOA-Glc accumulation and DIMBOA-Glc 4-O-methyltransferase by jasmonic acid in poaceous plants. Phytochemistry 61: 331–337 [DOI] [PubMed] [Google Scholar]
- Oikawa A., Ishihara A., Tanaka C., Mori N., Tsuda M., Iwamura H. (2004). Accumulation of HDMBOA-Glc is induced by biotic stresses prior to the release of MBOA in maize leaves. Phytochemistry 65: 2995–3001 [DOI] [PubMed] [Google Scholar]
- Pechan T., Ye L.J., Chang Y.M., Mitra A., Lin L., Davis F.M., Williams W.P., Luthe D.S. (2000). A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell 12: 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier H.C., Peters W.S., Collier R., van Bel A.J., Knoblauch M. (2008). GFP tagging of sieve element occlusion (SEO) proteins results in green fluorescent forisomes. Plant Cell Physiol. 49: 1699–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Mikkelsen M.D., Bednarek P., Olsen C.E., Halkier B.A., Kroymann J. (2011). Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23: 716–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M., Vogel H., Kroymann J. (2009). The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21: 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.A., Bradbury P.J., Buckler E.S., Nelson R.J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K.V., et al. (2012). A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337: 1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser W.A., Douglas A.E. (1992). A test of the hypotheses that nitrogen is upgraded and recycled in an aphid Acyrthosiphon pisum symbiosis. J. Insect Physiol. 38: 93–99 [Google Scholar]
- Qiu Y.F., Guo J.P., Jing S.L., Zhu L.L., He G.C. (2010). High-resolution mapping of the brown planthopper resistance gene Bph6 in rice and characterizing its resistance in the 9311 and Nipponbare near isogenic backgrounds. Theor. Appl. Genet. 121: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Rasmann S., Agrawal A.A. (2011). Latitudinal patterns in plant defense: Evolution of cardenolides, their toxicity and induction following herbivory. Ecol. Lett. 14: 476–483 [DOI] [PubMed] [Google Scholar]
- Rector B.G., Liang G.M., Guo Y. (2003). Effect of maysin on wild-type, deltamethrin-resistant, and Bt-resistant Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 96: 909–913 [DOI] [PubMed] [Google Scholar]
- Robert C.A., et al. (2012). A specialist root herbivore exploits defensive metabolites to locate nutritious tissues. Ecol. Lett. 15: 55–64 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schullehner K., Dick R., Vitzthum F., Schwab W., Brandt W., Frey M., Gierl A. (2008). Benzoxazinoid biosynthesis in dicot plants. Phytochemistry 69: 2668–2677 [DOI] [PubMed] [Google Scholar]
- Steffey, K., Rice, M.E., All, J., Andrew, D.A., Gray, M.E., and Van Duyn, J.W. (1999). Handbook of Corn Insects. (Lanham, MD: Entomological Society of America). [Google Scholar]
- Steiner W.W.M., Voegtlin D.J., Irwin M.E. (1985). Genetic differentiation and its bearing on migration in North-American populations of the corn leaf aphid, Rhopalosiphum maidis (Fitch) (Homoptera, Aphididae). Ann. Entomol. Soc. Am. 78: 518–525 [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullstrup A.J., Renfro B.L. (1976). A comparison of maize diseases in temperate and in tropical environments. Int. J. Pest Manage. 22: 491–498 [Google Scholar]
- Walling L.L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19: 195–216 [DOI] [PubMed] [Google Scholar]
- Walsh T.A., Morgan A.E., Hey T.D. (1991). Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kilodalton internal peptide segment. J. Biol. Chem. 266: 23422–23427 [PubMed] [Google Scholar]
- Wang, S.M., Basten, C.J., and Zeng, Z.B. (2012). Windows QTL Cartographer 2.5. (Raleigh, NC: North Carolina State University).
- Wei F., et al. (2009). The physical and genetic framework of the maize B73 genome. PLoS Genet. 5: e1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Holland J.B., McMullen M.D., Buckler E.S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., et al. (2009). A single molecule scaffold for the maize genome. PLoS Genet. 5: e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga G.E., Argandona V.H., Niemeyer H.M., Corcuera L.J. (1983). Hydroxamic acid content in wild and cultivated Gramineae. Phytochemistry 22: 2665–2668 [Google Scholar]
- Züst T., Heichinger C., Grossniklaus U., Harrington R., Kliebenstein D.J., Turnbull L.A. (2012). Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]