The maize pathogen C. graminicola rigorously reduces the β-1,3-glucan content of its cell walls during early phases of infection. This study shows that dynamic modification of the surface of infection structures represents a mode for masking pathogen attack and is required to avoid activation of broad-spectrum plant defense responses and establishment of compatibility.
Abstract
β-1,3-Glucan and chitin are the most prominent polysaccharides of the fungal cell wall. Covalently linked, these polymers form a scaffold that determines the form and properties of vegetative and pathogenic hyphae. While the role of chitin in plant infection is well understood, the role of β-1,3-glucan is unknown. We functionally characterized the β-1,3-glucan synthase gene GLS1 of the maize (Zea mays) pathogen Colletotrichum graminicola, employing RNA interference (RNAi), GLS1 overexpression, live-cell imaging, and aniline blue fluorochrome staining. This hemibiotroph sequentially differentiates a melanized appressorium on the cuticle and biotrophic and necrotrophic hyphae in its host. Massive β-1,3-glucan contents were detected in cell walls of appressoria and necrotrophic hyphae. Unexpectedly, GLS1 expression and β-1,3-glucan contents were drastically reduced during biotrophic development. In appressoria of RNAi strains, downregulation of β-1,3-glucan synthesis increased cell wall elasticity, and the appressoria exploded. While the shape of biotrophic hyphae was unaffected in RNAi strains, necrotrophic hyphae showed severe distortions. Constitutive expression of GLS1 led to exposure of β-1,3-glucan on biotrophic hyphae, massive induction of broad-spectrum defense responses, and significantly reduced disease symptom severity. Thus, while β-1,3-glucan synthesis is required for cell wall rigidity in appressoria and fast-growing necrotrophic hyphae, its rigorous downregulation during biotrophic development represents a strategy for evading β-glucan–triggered immunity.
INTRODUCTION
The fungal cell wall is a rigid but dynamic structure that determines the shape and function of vegetative and pathogenic hyphae. Polysaccharides account for more than 90% of the fungal cell wall, with β-1,3-glucan covalently cross-linked to chitin forming the primary scaffold, to which other β-linked polysaccharides and proteins are attached (Ruiz-Herrera, 1991; Bowman and Free, 2006; Latgé, 2007). In most fungal species, β-1,3-linked glucan is the dominating polymer, comprising between 65 and 90% of the cell wall glucan fraction (Bernard and Latgé, 2001; Bowman and Free, 2006). Studies with the human pathogenic fungus Aspergillus fumigatus have shown that β-1,3-glucan is synthesized by a large protein complex, consisting of β-1,3-glucan synthase (GLS) as the catalytic domain, the small regulatory GTPase Rho1, a membrane H+-ATPase, and an ABC-type glucan transporter (Beauvais et al., 2001). Attempts to delete GLS genes in filamentous fungi have failed, suggesting that β-1,3-glucan is an essential cell wall component (Latgé, 2007). Indeed, in A. fumigatus, gene silencing experiments employing RNA interference (RNAi) showed that the GLS gene FKS1 (for FK506 Sensitivity) is indispensable for vegetative growth and viability (Mouyna et al., 2004), and similar results have been obtained from RNAi studies with Fusarium solani (Ha et al., 2006).
In plant pathogenic fungi, delicate regulation of cell wall polymer formation is required, as changing structural requirements during infection-related morphogenesis must be met. For example, cell wall rigidity is of prime importance in ex planta–differentiated elaborate infection cells called appressoria, as enormous turgor pressure of up to 8.0 MPa (80 bar) must be contained (Howard et al., 1991; Bastmeyer et al., 2002; Wilson and Talbot, 2009). However, not only in appressoria, but also in infection hyphae formed in planta, cell wall polymers are essential structural compounds, as indicated by severe hyphal distortions and virulence defects of chitin synthase mutants of several pathogens (Madrid et al., 2003; Soulié et al., 2006; Weber et al., 2006; Werner et al., 2007; Kim et al., 2009; Larson et al., 2011; Kong et al., 2012).
In addition to their role in structural rigidity, fungal cell wall polymers are thought to play essential roles in interorganismic communication and establishment of compatibility (Nürnberger et al., 2004). Thus, in planta–differentiated infection hyphae of different fungi show modifications of their surfaces, which likely help masking pathogen-associated molecular patterns (PAMPs). Such cell wall modifications may be of particular importance in biotrophic pathogens and in hemibiotrophic fungi during biotrophic development (Freytag and Mendgen, 1991; O’Connell et al., 1996; El Gueddari et al., 2002; Fujikawa et al., 2009). These fungi, when invading the plant cell, do not breach but invaginate the host plasma membrane, leading to sealing of the in planta–formed biotrophic hyphae and the establishment of an intimate interaction (Kankanala et al., 2007). The narrow interfacial matrix layer separating the fungal hypha from the plant plasma membrane, however, may represent a detrimental environment for the pathogen, with apoplastic plant β-1,3-glucanases and chitinases challenging the integrity of hyphal walls and giving rise to elicitor-active chitin and β-1,3-glucan fragments (Deller et al., 2011). Indeed, PAMPs such as β-1,4-N-acetyl glucosamine oligomers are recognized by corresponding LysM motif containing plasma membrane–localized pattern recognition receptors, such as the chitin elicitor binding protein and the CHITIN ELICITOR RECEPTOR KINASE1, which cooperatively mediate chitin elicitor signaling and immunity in rice (Oryza sativa; Shimizu et al., 2010). In tomato (Solanum lycopersicum), subnanomolar concentrations of N-acetylchitooligosaccharides are sufficient to induce defense responses (Felix et al., 1993).
Plant pathogenic fungi have developed different mechanisms to compromise chitin detection and initiation of defense responses. For example, conversion of surface-exposed chitin to its nonacetylated derivative chitosan, specifically initiated at host invasion, has been shown to occur in different rust fungi and in the hemibiotroph Colletotrichum graminicola (El Gueddari et al., 2002). Compared with chitin, chitosan is a poor chitinase substrate, and chitosan fragments are significantly less elicitor-active than chitin fragments of corresponding degrees of polymerization (Barber et al., 1989; Vander et al., 1998). Therefore, deacetylation of surface-localized chitin may interfere with pathogen recognition by the host.
Furthermore, a series of elegant studies has recently shown that the hemibiotrophic rice blast fungus Magnaporthe oryzae and the biotrophic tomato pathogen Cladosporium fulvum circumvent chitin-triggered host immunity by secretion of LysM domain–containing effector proteins, such as Slp1, Avr4, or Ecp6, sequestering polymeric chitin and chitin oligomers (van Esse et al., 2007; de Jonge et al., 2010; Mentlak et al., 2012). Sequestration of chitin likely represents a common strategy of host immune suppression by fungal pathogens, as LysM effectors are widely conserved in the fungal kingdom (de Jonge and Thomma, 2009), including Colletotrichum species (Kleemann et al., 2012).
Not only chitin but also linear or branched β-1,3-glucan fragments are known as highly potent elicitors of defense responses in plants (Cosio et al., 1996; Klarzynski et al., 2000; Shetty et al., 2009). However, enzymatic modifications or sequestration of β-1,3-glucan polymers or fragments thereof, as well as mechanisms leading to evasion of β-1,3-glucan–triggered immune responses, are unknown.
Using RNAi and overexpression of the GLS gene GLS1, live-cell imaging, aniline blue fluorochrome staining, genome-wide RNA sequencing, and quantitative RT-PCR (qRT-PCR), we analyzed the role of β-1,3-glucan in infection structures of C. graminicola and investigated the mechanism used by this fungus to circumvent β-1,3-glucan–triggered immunity of its host, maize (Zea mays). To establish disease, C. graminicola sequentially differentiates highly specific infection structures. On the cuticle of its host, conidia germinate to form dome-shaped melanized appressoria, which generate enormous turgor pressure (Bechinger et al., 1999; Deising et al., 2000). After penetration into the epidermal host cell, this hemibiotroph differentiates biotrophic infection structures (i.e., voluminous infection vesicles and primary hyphae). At this stage of infection, macroscopically visible disease symptoms do not occur. However, after perception of as yet unknown signal(s), the fungus switches to form highly destructive fast-growing necrotrophic hyphae, which rapidly colonize and kill the host tissue (Horbach et al., 2011).
In this study, we demonstrate that β-1,3-glucan is indispensable in cell walls of functional appressoria and necrotrophic hyphae and that significant downregulation of β-1,3-glucan synthesis in biotrophic infection structures is essential for evading β-1,3-glucan–triggered immunity in maize. This study functionally characterizes a GLS gene of a plant pathogenic fungus and reveals a mechanism for establishing compatibility with the host plant.
RESULTS
GLS1 of C. graminicola Is a Single-Copy Gene Encoding a Membrane-Integral GLS
BLASTX searches performed with GLS proteins of the yeast Saccharomyces cerevisiae (FKS1), and the filamentous Ascomycota M. oryzae, Aspergillus nidulans, Paracoccidioides brasiliensis, Neurospora crassa, and Fusarium solani (FKS1) suggested that, like in most other filamentous fungi (Latgé, 2007), only a single GLS gene exists in the annotated genome of C. graminicola (http://www.broadinstitute.org/annotation/genome/colletotrichum_group/FeatureSearch.html). BLASTX searches performed with more distantly related GLS proteins (i.e., GLSs of Ustilago maydis, Pichia pastoris, S. cerevisiae [FKS3], and F. solani [FKS2]) did not identify further homologs in the proteome of C. graminicola. We designated the single GLS gene of C. graminicola GLS1. The phylogenetic tree calculated on the basis of the derived amino acid sequences shows that the proteins of filamentous ascomycetes form a single clade, distinct from the GLSs of yeasts and basidiomcetes (Figure 1A). On the amino acid level, GLS1 of C. graminicola shares 77.7, 75.5, and 78.9% sequence identity with the GLSs of N. crassa, M. oryzae, and F. solani (FKS1), respectively, but only 59.3, 59.9, and 45.0% identity with FKS1, FKS2, and FKS3 of S. cerevisiae, respectively. However, plants are also capable of synthesizing a β-1,3-linked glucan polymer called callose. Callose synthase-like proteins of plants (e.g., GLS5 and GLS7 of Arabidopsis thaliana and CSLF3 of maize) share only 32, 30, and 17% identity with GLS1 of C. graminicola, respectively.
Figure 1.
GLS1 of C. graminicola Encodes a Functional GLS.
(A) Phylogenetic tree indicates close relatedness of GLS1 of C. graminicola with other GLSs of filamentous fungi: A. capsulatus, Ajellomyces capsulatus; A. dermattidis, Ajellomyces dermatitidis; A. flavus, Aspergillus flavus; A. oryzae, Aspergillus oryzae; A. fumigatus, Aspergillus fumigatus; A. nidulans, Aspergillus nidulans; C. albicans, Candida albicans; C. glabrata, Candida glabrata; C. paraplosis, Candida parapsilosis; C. gattii, Cryptococcus gattii; C. immitis, Coccidioides immitis; C. posadasii, Coccidioides posadasii; C. graminicola, Colletotrichum graminicola; C. cinerea, Coprinopsis cinerea; C. neoformans, Cryptococcus neoformans; E. dermatitis, Exophiala dermatitidis; F. solani (FKS1), Fusarium solani (ABC59463); F. solani (FKS2), Fusarium solani (XP003040299.1); L. bicolor, Laccaria bicolor; M. oryzae, Magnaporthe oryzae; M. acridum, Metarhizium acridum; N. crassa, Neurospora crassa; P. brasiliensis, Paracoccidioides brasiliensis; P. marneffei, Penicillium marneffei; P. pastoris, Pichia pastoris; P. graminis, Puccinia graminis f. sp tritici; S. cerevisiae (FKS1), Saccharomyces cerevisiae; S. cerevisiae (FKS2), Saccharomyces cerevisiae; S. cerevisiae (FKS3), Saccharomyces cerevisiae; S. pombe (Bgs1), Schizosaccharomyces pombe; S. pombe, Schizosaccharomyces pombe (Bgs2); S. pombae (Bgs3), Schizosaccharomyces pombe (NP594766.1); S. pombae (Bgs4), Schizosaccharomyces pombe; S. sclerotiorum, Sclerotinia sclerotiorum; T. stipitatus, Talaromyces stipitatus; U. maydis, Ustilago maydis; and Z. rouxii, Zygosaccharomyces rouxii. Red, filamentous Ascomycota; blue, unicellular Ascomycota (yeasts); green, Basidiomycota.
(B) Complementation of S. cerevisiae Δfks1 by GLS1 cDNA of C. graminicola suggests that GLS1 encodes an active GLS. WT, S. cerevisiae reference strain Y00000; Δfks1, FKS1-deficient S. cerevisiae strain Y05251; TpAG300, Δfks1 transformant harboring the empty binary vector pAG300; TCgGLS11 - 4, independent Δfks1 transformants expressing the GLS1 cDNA of C. graminicola; YPD, YPD medium; YPDS, YPD medium supplemented with 1 M sorbitol. Number of yeast cells inoculated were (left to right) 5 × 104, 5 × 103, 5 × 102, and 5 × 10.
The size of the predicted fungal GLS proteins ranges from 1729 (Bgs1 of Schizosaccharomyces pombe) to 1955 amino acids (GLS1 of N. crassa and FKS1 of Cryptococcus neoformans), with GLS1 of C. graminicola containing 1940 amino acids. The vast majority of GLSs, including GLS1 of C. graminicola, have 16 predicted transmembrane domains flanking the glucan synthase domain (see Supplemental Figure 1 online). GLS genes of filamentous ascomycetes contain two introns at conserved sites at their 5′- and 3′-ends, respectively. Gene sizes range from 5184 (Bgs1 of S. pombe) to 5865 bp (GLS1 of N. crassa and FKS1 of C. neoformans). GLS1 of C. graminicola consists of 5820 bp (see Supplemental Figure 1 online).
The function of GLS1 of C. graminicola was confirmed by complementation of yeast strain Y05251, carrying a deletion of the GLS gene FKS1. Due to the presence of three GLS genes, deletion of FKS1 is not lethal in S. cerevisiae but results in severe growth defects on osmotically nonstabilized medium (Figure 1B, YPD). Growth defects of the Δfks1 strain were fully rescued by osmotically stabilizing the YPD medium with 1 M sorbitol (Figure 1B, YPDS). Approximately one million yeast transformants carrying the C. graminicola GLS1 cDNA were generated, four of which (TCgGLS1-4) were randomly chosen and used in growth assays. All yeast transformants expressing the C. graminicola GLS1 cDNA showed growth rates comparable to those of the reference strain Y00000, irrespective of osmotic support. The Δfks1 strain transformed with the empty expression vector pAG300 (TpAG300) was indistinguishable from the Δfks1 strain and showed poor growth on nonstabilized YPD plates (Figure 1B, compare Δfks1 and TpAG300).
Collectively, the yeast complementation experiments show that GLS1 of C. graminicola represents a functional GLS gene. Programs such as TMHMM Server v. 2.0 predict that GLS1 of C. graminicola is a plasma membrane–integral protein. To study the localization of GLS1, four independent C. graminicola replacement strains with single GLS1:eGFP (for enhanced green fluorescent protein) integrations were randomly chosen, expressing the GLS1:eGFP fusion under the control of the native GLS1 promoter (see Supplemental Figures 2A and 2B online). The GLS1:eGFP fusion was fully functional in the replacement strain as (1) osmotic stabilization was not required for vegetative growth, (2) growth and (3) sporulation rates did not differ from those of the wild-type strain CgM2 (see Supplemental Figures 2C to 2E online), and (4) cell wall defects did not occur (see below). The GLS1:eGFP replacement strains revealed strong eGFP fluorescence associated with the apical plasma membrane (see Supplemental Figure 3A online, left panel, arrowhead), slightly extending into subapical regions (see Supplemental Figure 3A online, left panel, short arrow). Fluorescence of GLS1:eGFP colocalized with that of the red-fluorescing membrane marker dye FM-464 only at the apical and subapical plasma membrane (see Supplemental Figure 3A online, central and right panels, arrows), indicating a tip-oriented gradient of GLS1. To further show that the GLS1:eGFP fusion protein is a plasma membrane–integral protein, protoplasts were generated, using fungal cell wall lysing enzymes from Trichoderma harzianum (see Supplemental Figure 3B online, DIC). Clearly, these protoplasts showed eGFP fluorescence in their plasma membranes (see Supplemental Figure 3B online, GLS1:eGFP, arrows).
RNAi-Mediated Reduction of GLS1 Expression Causes Severe Cell Wall Defects and Hypermelanization of Vegetative Hyphae
Like in experiments performed with other filamentous fungi (Latgé, 2007), attempts to delete the GLS gene of C. graminicola failed (see Supplemental Figure 4A online). PCR screens performed with more than 100 single-spore isolates of transformants always led to amplification of a 1-kb fragment of the GLS1 gene (see Supplemental Figure 4B online), suggesting ectopic integration of the deletion cassette. As average rates of homologous integration in this fungus rarely are below 30%, these results suggest that deletion of the GLS1 gene of C. graminicola is lethal.
To circumvent the lethal phenotype of a GLS1 deletion, an RNAi-based knockdown strategy was adopted (Kück and Hoff, 2010) (Figure 2). The RNAi vector consisted of the trpC promoter, a 475-bp sense and antisense fragment of the second exon of the GLS1 gene of C. graminicola, separated by 135 bp of the second intron of the Cut2 gene from M. oryzae, followed by the trpC terminator of A. nidulans. The nourseothricin resistance gene from Streptomyces noursei (Malonek et al., 2004) was used as the selection marker (Figure 2A).
Figure 2.
RNAi-Mediated Reduction of GLS1 Transcript Abundance Causes Reduction of β-1,3-Glucan Contents of Cell Walls, Hyphal Distortion, and Retarded Growth.
(A) RNAi construct transformed into C. graminicola. Bar indicates probe used for DNA gel blots. PtrpC, trpC promoter; TtrpC, trpC terminator, Nat-1, Nourseothricin acetyl transferase gene. Not to scale.
(B) DNA gel blot of XhoI-digested genomic DNA of C. graminicola wild-type (WT) and RNAi strains. A digoxygenin-labeled fragment of the Nat-1 gene served as the probe (bar in Figure 3A).
(C) GLS1 transcript abundance measured by qRT-PCR. Means of four replicate are shown. Bars represent ±sd.
(D) and (E) Morphology of colonies of the wild type (D) and a class I RNAi strain (E) on OMA supplemented with 0.15 M KCl.
(F) Radial growth of wild-type, class I, class II, and class III RNAi strains on OMA supplemented with 0.15 M KCl. Means of four replicates are shown. Bars represent ±sd.
(G) Vegetative hypha of the wild-type strain.
(H) Vegetative hypha of a class I RNAi strain showing hyphal swellings (arrowheads). Inset shows pigmentation of swellings.
(J) Severely distorted vegetative hyphae of a class II RNAi strain. Even on OMA supplemented with 0.15 M KCl the mycelium consists of swellings (asterisks), some of which are strongly melanized (arrow). Bars in (G) to (J) = 5 µm.
(K) eGFP fluorescence and quantification of fluorescence intensities in the wild type and a class I RNAi strain carrying a GLS1:eGFP replacement construct.
(L) β-1,3-Glucan distribution and quantification of fluorescence in a wild type and a class I RNAi strain carrying a GLS1:eGFP replacement construct. Hyphae were stained by aniline blue fluorochrome. Bars in micrographs in (K) and (L) = 10 µm. Three times 100 cells were measured bars on columns represent ±sd.
XhoI-digested genomic DNA of the wild-type strain and of nine RNAi strains was analyzed by DNA gel blot hybridization. Different numbers of RNAi constructs had integrated into the genomes of different transformants (Figure 2B), and the copy number of the RNAi construct correlated with the reduction of GLS1 transcript abundance, as indicated by qRT-PCR (cf. Figures 2B and 2C). According to different degrees of reduction of GLS1 transcript abundance, the nine RNAi strains were grouped into three classes. Compared with the wild-type strain, class I (strains 1 to 3) showed transcript abundances between 55 and 67%, class II (strains 4 to 6) between 33 and 37%, and class III (strains 7 to 9) below 25% (Figure 2C). All transformants exhibited severely reduced growth rates (Figures 2D to 2F).
Compared with the wild-type strain (Figure 2D), all RNAi strains showed a strongly altered colony phenotype and formed compact hypermelanized colonies (Figure 2E). While the wild-type strain developed normal filaments (Figure 2G), vegetative hyphae of class I RNAi strains exhibited severe hyphal swellings (Figure 2H, arrowheads), which often were strongly pigmented (Figure 2H, inset, arrowheads). Class II RNAi strains showed even more pronounced cell wall distortions (Figure 2J, asterisks), and many of the swellings were strongly melanized (Figure 2J, arrow).
To demonstrate that reduction of transcript abundance led to reduced GLS1 protein contents, the GLS1 gene of the wild-type strain and of the class I RNAi strain 1 was replaced by the GLS1:eGFP construct (see Supplemental Figures 2 and 5 online). Two representative transformants and the GLS1:eGFP replacement strain lacking the RNAi construct were comparatively analyzed by quantitative fluorescence microscopy (Figures 2K and 2L). Vegetative hyphae of the two class I RNAi strains showed a reduction in eGFP fluorescence of 66.4% ± 18.9% and 70.4% ± 29.4% (Figure 2K) and a reduction of aniline blue fluorescence by 56.7% ± 23.4% and 60.1% ± 13.2% (Figure 2L), clearly indicating downregulation of GLS1:eGFP expression and, accordingly, synthesis of β-1,3-glucan by RNAi.
These data show that RNAi enables the characterization of essential genes in the maize anthracnose fungus C. graminicola.
GLS1 Expression and Cell Wall β-1,3-Glucan Contents Are Drastically Downregulated in Biotrophic Hyphae
GLS genes are thought to be constitutively expressed during vegetative hyphal growth (Mouyna et al., 2004; Ha et al., 2006). During pathogenesis, infection structure–specific regulation of GLS gene may be required to support the function of the specialized pathogenic cells or hyphae. We wanted to analyze the expression of GLS genes and synthesis of β-1,3-glucan during the fungal infection process. Unfortunately, as infection structure differentiation of C. graminicola on maize leaves does not occur in a synchronized fashion, neither qRT-PCR–based quantification of infection structure–specific GLS1 transcript abundance nor chemical quantification of β-1,3-glucan is feasible. We therefore used the GLS1:eGFP replacement strains of C. graminicola (see Supplemental Figure 2 online) to quantify GLS1 expression and β-1,3-glucan contents in infection structures of C. graminicola by measuring eGFP signals (Figures 3A and 3C) and β-1,3-glucan–specific fluorescence after aniline blue fluorochrome staining (Figures 3B and 3D). Virulence of the GLS1:eGFP replacement strains did not differ from that of the wild-type strain (see Supplemental Figure 2F online). Strong eGFP fluorescence was observed in nongerminated conidia, appressoria, and necrotrophic secondary hyphae (Figure 3A, 0 h after inoculation [HAI], co; 12 HAI, ap; 72 HAI, sh; Figure 3C). Melanization of appressoria only slightly reduced eGFP fluorescence (Figure 3C). Unexpectedly, biotrophic primary hyphae showed only background eGFP fluorescence (Figures 3A, 24 HAI, ph; Figure 3C). Note that the fluorescence signal marked with an arrowhead was emitted by the appressorium on the cuticle, not by biotrophic hyphae (Figure 3A, 24 HAI). Accordingly, nongerminated conidia, nonmelanized immature appressoria, and necrotrophic secondary hyphae showed strong aniline blue fluorescence (Figure 3B, 0 HAI, co; 12 HAI, ap; 72 HAI, sh; Figure 3D). Mature appressoria emitted low levels of fluorescence (Figure 3D), likely due to the fluorescence-scavenging activity of melanin (Henson et al., 1999). Importantly, biotrophic infection vesicles and primary hyphae exhibited strongly reduced fluorescence intensities (Figure 3B, 24 HAI, ph; Figure 3D). To exclude the possibility that the lack of aniline blue labeling in biotrophic hyphae was due to apposition of α-1,3-glucan, as suggested for M. oryzae (Fujikawa et al., 2009, 2012), we stained cross sections of biotrophic and necrotrophic hyphae with aniline blue. This strategy was applied, rather than treatment of in planta–differentiated hyphae with different cell wall degrading enzymes, as the nature of the putative apposition, if any, is unknown in C. graminicola. Clearly, cross-sectioned biotrophic infection vesicles and primary hyphae did not stain with aniline blue (Figure 3E, iv and ph). As expected (Figure 3C), mature appressoria only showed background labeling (Figure 3D, ap). Cross-sectioned necrotrophic secondary hyphae exhibited intensive aniline blue labeling (Figure 3E, sh). Furthermore, specimens were treated with alkaline (0.1 n NaOH, 20 min at 60 or 121°C), which dissolves polymeric α-1,3-glucan (Sietsma et al., 1985) (see Supplemental Figure 6 online). Alkaline treatment did not restore aniline blue labeling, further indicating that differential expression of GLS1, and not masking of β-1,3-glucan by α-1,3-glucan apposition, caused the lack of aniline blue fluorescence in biotrophic structures.
Figure 3.
Infection Structure–Specific Expression of GLS1:eGFP and Synthesis of β-1,3-Glucan in C. graminicola.
(A) GLS1:eGFP is strongly expressed in conidia (co; 0 HAI), appressorium (ap; 12 HAI), and necrotrophic secondary hyphae (sh; 72 HAI) but not in biotrophic primary hyphae (ph; 24 HAI).
(B) Staining of infection structures with β-1,3-glucan–specific aniline blue fluorochrome shows β-1,3-glucan in conidia (co; 0 HAI), appressorium (ap; 12 HAI), and necrotrophic secondary hyphae (sh; 72 HAI), but not in biotrophic primary hyphae (ph; 24 HAI). Bars (A) and (B) = 10 µm.
(C) Quantification of eGFP fluorescence in infection structures. co, conidia; gt, germ tubes; nma, nonmelanized appressoria; ma, melanized appressoria; ph, biotrophic primary hyphae; sh, necrotrophic secondary hyphae. Three times 100 infection structures were measured.
(D) Quantification of β-1,3-glucan in infection structures using aniline blue fluorochrome. co, condia; gt, germ tubes; nma, nonmelanized appressoria; ma, melanized appressoria; ph, biotrophic primary hyphae; sh, necrotrophic secondary hyphae. Three times 100 infection structures were measured. Bars in (C) and (D) represent ±sd.
(E) and (F) Aniline blue fluorochrome labeling of biotrophic (E) and necrotrophic (F) hyphae. ap, appressorium; iv, infection vesicle; ph, primary hypha; sh, secondary hypha. Bars = 20 µm.
In conclusion, quantitative fluorescence microscopy revealed that GLS1 expression and β-1,3-glucan contents of cell walls are prominent in conidia, appressoria, and necrotrophic hyphae but are lacking in biotrophic infection structures of C. graminicola.
GLS1 of C. graminicola Is Required for Asexual Sporulation, Adhesion, and Differentiation of Functional Appressoria
Intense fluorescence of conidia in transformants expressing a GLS1:eGFP fusion and after aniline blue fluorochrome staining (Figure 3) suggested that GLS1 expression and β-1,3-glucan synthesis are required for formation of asexual spores. Indeed, reduction of GLS1 transcript levels by RNAi strongly affected asexual sporulation rates and the conidial shape (see Supplemental Figure 7 online).
Osmotically stabilized conidia of the wild-type strain and the class I RNAi strains (Figures 4A and 4B, asterisk) germinated and differentiated appressoria on artificial substrata (Figures 4A and 4B, arrow). Intriguingly, while appressoria of the wild-type strain subsequently remained unaltered, ∼15% of all appressoria formed by class I RNAi strains exploded (Figure 4B, arrows; Figure 4F) and released cell components such as lipid droplets (Figure 4B, arrowheads). Interestingly, several appressoria of class I RNAi strains that did not explode (Figure 4C, white arrow; Figure 4F), but they developed hyphae reminiscent of biotrophic primary hyphae (Figure 4C, black arrow). Hyphae developing from appressoria of class I RNAi strains on the plant surface had a diameter of 7.8 ± 3.2 µm, which is in good agreement with and statistically not different (P > 0.05) from diameters of 8.1 ± 3.2 µm measured for biotrophic primary hyphae formed in planta (Politis and Wheeler, 1973; Horbach et al., 2009). As tight adhesion of appressoria is required for directed growth of the penetration peg into the plant cell wall, failure in penetration and development of primary hyphae on the cuticle is likely due to compromised adhesion. Indeed, compared with the wild-type strain, adhesion of infection structures of the class I RNAi strains tested was 36.7-, 34.5-, and 26.9-fold reduced on onion epidermal cells, polyester, and glass, respectively (Figure 4G).
Figure 4.
Appressoria of RNAi Strains of C. graminicola Lack Cell Wall Rigidity, Have Melanization Defects, and Are Nonadhesive.
(A) On polyester, conidia (asterisk) the wild-type (WT) strain germinate and form melanized appressoria (arrow).
(B) Conidia (asterisk) of class I RNAi strains germinate and form appressoria, many of which explode (arrow) and release lipid bodies (arrowhead).
(C) Appressorium (white arrow) of a class I RNAi strain with a voluminous hypha (black arrow) reminiscent of a primary hypha. Inset shows an irregularly melanized appressorium surrounded by a ring of melanin (arrowheads). A hypha with large diameter reminiscent of a primary hypha is marked by a black arrow.
(D) Conidia (asterisk) of class II RNAi strains are able to form germ tubes (arrow) but fail to differentiate appressoria.
(E) Germling of a class II strain surrounded by a melanin precipitate. Bars in (A) to (E) = 10 µm.
(F) Quantification of infection structure differentiation on polyester. In each of the three independent experiments performed, 100 infection structures were counted.
(G) Adhesion of infection structures formed by wild-type and three independent class I RNAi strains on onion epidermis or artificial surfaces 24 HAI. In each of the three independent experiments performed, 100 infection structures were counted. Bars in (F) and (G) represent sd.
While appressoria of the wild-type strain were strongly melanized (Figure 4A, arrow), melanization was clearly reduced in the appressoria of class I RNAi strains (Figures 4B and 4C, inset). Interestingly, melanin often formed a ring on the underlying polyester and onion epidermis, surrounding the appressorium (Figure 4C, inset, arrowheads), suggesting that an intact β-1,3-glucan network is required for incorporation of melanin into the cell wall. Melanization appeared to be severely deregulated in class II RNAi strains. Conidia of these strains were able to germinate (Figure 4D, arrow) but failed to form appressoria (Figure 4F). While germlings of wild-type conidia never melanized, those of class II RNAi strains produced massive amounts of pigment that was secreted into the medium and precipitated on the substratum surrounding the hypha (Figure 4E, class II RNAi strains).
When class I RNAi strains formed appressoria in sterile distilled water, ∼75% of these cells ruptured (Figures 4B and 4F). We measured the appressorial diameters of those appressoria that did not explode(Figures 5A and 5B). In distilled water, class I RNAi strains showed significantly increasing appressorial diameters. Interestingly, addition of the osmolyte polyethylene glycol 6000 (PEG 6000) (400 mg/mL) caused shrinking of the appressoria of the class I RNAi strains to the wild-type level, indicating that appressorial cell walls of the class I RNAi strains were highly elastic. The appressorial diameter of the wild-type strain was unaffected by addition of the osmolyte (Figures 5A and 5B). Increased appressorial cell wall elasticity was expected to affect the appressorial turgor pressure. Indeed, wild-type appressoria showed equilibrium cell collapse called cytorrhizis at a PEG 6000 concentration of more than 400 mg/mL, whereas ∼190 mg/mL was sufficient in the class I RNAi strains (Figure 5C), indicating significantly reduced appressorial turgor pressure of class I RNAi strains, compared with the wild type.
Figure 5.
Downregulation of Appressorial β-1,3-Glucan Contents Increases Cell Wall Elasticity and Reduces Turgor Pressure.
(A) Appressoria of the wild-type (WT) strain show comparable sizes in water and in the osmolyte PEG 6000 (400 mg/mL), as indicated by the double arrow. Appressoria of class I RNAi strains had larger diameters in water than in PEG 6000. Bar = 10 µm.
(B) Quantification of appressorial diameters of the wild type and three class I RNAi strains in water and after addition of PEG 6000 (400 mg/mL). Three times 100 appressoria were measured.
(C) Incipient cytorrhizis indicates that the appressorial turgor pressure of class I RNAi strains is considerably lower than that of the wild-type strain. Three times 100 appressoria were counted. Bars in (B) and (C) represent ±sd.
These data show that synthesis of β-1,3-glucan is indispensable for appressorial adhesion, cell wall rigidity, and melanization and for generation of turgor pressure.
GLS1 Is Indispensable for Appressorial Penetration, Development of Necrotrophic Hyphae, and Anthracnose Disease Symptoms in Maize
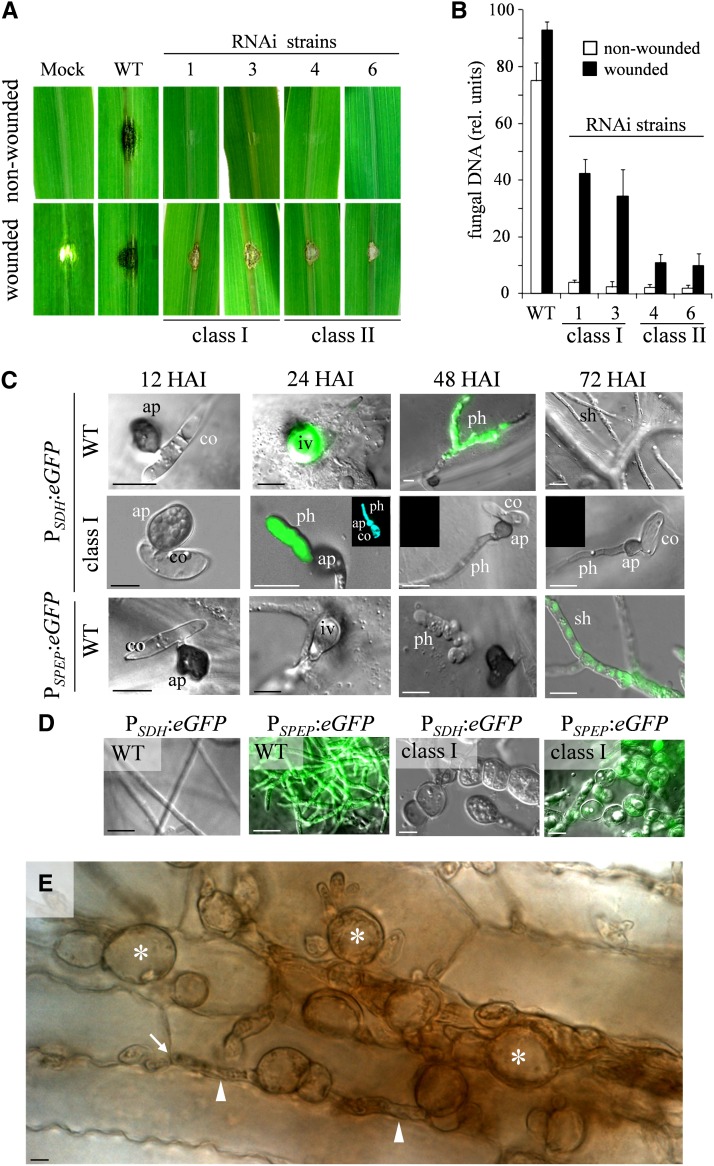
In order to investigate the role of GLS1 of C. graminicola in plant infection, conidia of the wild-type strain and of class I and class II RNAi strains were inoculated onto intact and wounded segments of the youngest fully expanded leaves of 2- to 3-week-old maize plants, and virulence was evaluated 7 d after inoculation. The wild-type strain caused disease symptoms on both intact and wounded leaves. By contrast, the RNAi strains were unable to invade intact leaves. On wounded leaves, however, the RNAi strains caused minor necroses at the margins of the wounds (Figure 6A). Fungal development, assessed by quantitative PCR (qPCR) using internal transcribed spacer two primers (Behr et al., 2010), fully confirmed macroscopically observed disease development (Figure 6B). Thus, fungal virulence correlated with GLS1 transcript abundance (cf. Figures 2C and 6B).
Figure 6.
RNAi Strains of C. graminicola Form Severely Distorted Necrotrophic Hyphae and Are Nonpathogenic on Maize.
(A) Disease symptoms on wounded and nonwounded maize leaves after inoculation with the wild type (WT) and independent class I (1 and 3) and class II RNAi strains (4 and 6). Mock-inoculated leaves were treated with 0.01% (v/v) Tween 20.
(B) Quantification of fungal development on intact or wounded maize leaves after inoculation with the strains used in (A). Three biological repeats, with two technical repeats each, were analyzed; bars represent sd.
(C) Development of infection structures of the wild type and a class I RNAi strain expressing the eGFP gene under the control of the biotrophy-specific SDH promoter (PSDH) and of the wild-type strain expressing the eGFP gene under the control of the necrotrophy-specific SPEP promoter (PSPEP) on intact maize leaves. ap, appressorium; co, conidium; ph, primary hypha; sh, secondary hypha. Insets in class I, 24 to 72 HAI show viability staining with fluorescein diacetate.
(D) Development of infection structures of the wild type and a class I RNAi strain on wounded maize leaves. eGFP gene expression is under control of the biotrophy-specific SDH (PSDH) or the necrotrophy-specific SPEP promoter (PSPEP).
(E) Necrotrophic hyphae of a class I RNAi strain in a maize leaf after wound inoculation. Hyphae show severe swellings (asterisks), connected by narrow hyphae (arrowheads). Arrow indicates penetration point in an anticlinal plant cell wall. Bar in (C) to (E) = 10 µm.
In order to understand the role of GLS in the infection process of the hemibiotroph C. graminicola, differential interference contrast microscopy, in combination with eGFP-based fluorescence microscopy, was employed. Discrimination of biotrophic and necrotrophic infection structures in planta is usually based on morphology, but as morphology of infection hyphae may be severely compromised in RNAi strains exhibiting cell wall defects, additional morphology-independent criteria were needed.
Therefore, to construct biotrophy reporter strains, we took advantage of biotrophy-specific genes identified in a cDNA library prepared from intracellular biotrophic hyphae of the Arabidopsis pathogen Colletotrichum higginsianum (Takahara et al., 2009). Based on the expression level in C. higginsianum, we selected nine homologous genes of C. graminicola, fused ∼1.5 kb of their 5′-upstream regions, likely harboring their promoters, to the eGFP gene, transformed these constructs into the C. graminicola wild-type strain, and analyzed the transformants for biotrophy-specific eGFP expression. Based on fluorescence intensities and biotrophy specificity of eGFP expression, we selected the promoters of a saccharopine dehydrogenase gene (SDH) and of a gene encoding a NmrA-like family protein. A necrotrophy-specific gene encoding a secreted peptidase of the class of subtilases (SPEP) had previously been identified in a YSST library of C. graminicola (Krijger et al., 2008), and the promoter of this gene was used for necrotrophy-specific eGFP expression (see Supplemental Figure 8 online).
The promoter-eGFP fusions chosen were transformed into the wild type and all class I RNAi strains, and morphology of infection structures and eGFP fluorescence were compared after inoculation onto nonwounded and wounded maize leaves (Figures 6C and 6D). On intact leaves, the wild-type strain had formed appressoria on the cuticle after 12 HAI, penetrated the epidermal cell wall by 24 HAI, and brightly fluorescing biotrophic infection vesicles had formed in the epidermal host cell, due to eGFP expression under control of the biotrophy-specific saccharopine dehydrogenase promoter (PSDH) (Figure 6C, PSDH:eGFP, WT, 24 HAI). Also, primary hyphae fluoresced intensively (Figure 6C, PSDH:eGFP, WT, 48 HAI). In necrotrophic secondary hyphae, no fluorescence was detected, and fluorescence in primary hyphae had ceased when necrotrophic hyphae had formed (Figure 6C, PSDH:eGFP, WT, 72 HAI). In comparison, under control of the necrotrophy-specific promoter of the gene encoding a secreted peptidase (PSPEP), eGFP fluorescence was only observed in fast growing necrotrophic hyphae (Figure 6C, PSPEP:eGFP, WT, 72 HAI). Thus, using these promoters allowed us to discriminate between biotrophic and necrotrophic hyphae on a morphology-independent basis.
Class I RNAi strains harboring the PSDH:eGFP construct were allowed to form appressoria on intact leaves (Figure 6C, PSDH:eGFP, class I, 12 HAI). As observed on artificial surfaces (Figure 4C), several appressoria of these strains developed voluminous hyphae on the cuticle of the host plant, reminiscent of biotrophic primary hyphae (Figure 6C, PSDH:eGFP, class I, 24 HAI). These hyphae fluoresced brightly, indicating that they indeed represent biotrophic hyphae. Viability staining with fluorescein diacetate showed that on the cuticle biotrophic hyphae were viable at 24 HAI but not at later time points after inoculation (Figure 6C, PSDH:eGFP, class I, 24 to 72 HAI, insets). These observations were confirmed by experiments using eGFP fusions with the second biotrophy-specific promoter, controlling the expression of an NmrA-like gene (see Supplemental Figure 9 online).
When the wild-type strain was inoculated onto wounded leaves, biotrophic hyphae were not observed, and fast-growing thin necrotrophic hyphae formed immediately after wound inoculation (Figure 6D, WT). In agreement with hyphal morphology of the wild-type strain, fluorescence was observed in strains expressing eGFP under the control of the necrotrophy-specific promoter (Figure 6D, PSPEP:eGFP, WT), but not under the control of the biotrophy-specific promoter (Figure 6D, PSDH:eGFP, WT). When class I RNAi strains were inoculated onto wounded leaves, hyphae with large swellings formed (Figure 6E, asterisks). These hyphae, in contrast with necrotrophic hyphae of the wild-type strain (Figure 6C, WT, 72 HAI), were strongly melanized and grew slowly, but were able to penetrate the anticlinal maize cell walls (Figure 6E, arrow). The hyphal swellings observed in host cells were connected by thin hyphae (Figure 6E, arrowheads), suggesting that these hyphae are necrotrophic hyphae with severe cell wall distortions. Indeed, the distorted hyphae showed eGFP fluorescence under the control of the necrotrophy-specific promoter (Figure 6D, PSPEP:eGFP, class I) but not of the biotrophy-specific SDH promoter (Figure 6D, PSDH:eGFP, class I). Formation of necrotrophic hyphae by class I RNAi strains is in agreement with the necroses observed after inoculation of these strains onto wounded leaves (see Figure 6A, wounded, class I RNAi strains). Importantly, formation of necrotrophic hyphae is significantly affected if GLS1 transcript abundance is reduced by only ∼40% (Figure 2C).
Forced Expression of GLS1 during Biotrophic Development Affects Hyphal Shape, Exposes β-1,3-Glucan, and Triggers Defense Responses in Maize
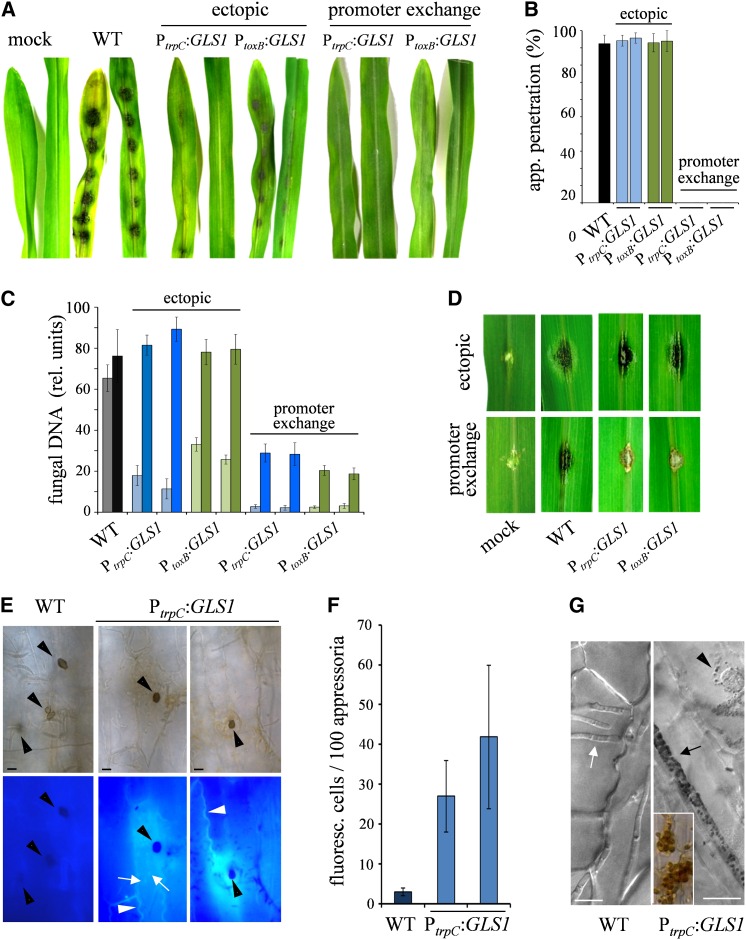
The profiles of GLS1 expression and synthesis of β-1,3-glucan (Figure 3) suggest that downregulation of GLS1 causes dilatation of biotrophic hyphae and that exposition of β-1,3-glucan must be avoided during biotrophic growth of C. graminicola in order to evade β-1,3-glucan–triggered immunity. To test these hypotheses, two different approaches were taken. First, the GLS1 promoter (PGLS1) was replaced by either of the two constitutively active promoters, including the trpC promoter (PtrpC) of Aspergillus nidulans (Pöggeler et al., 2003) or the toxB promoter (PtoxB) of the wheat (Triticum aestivum) pathogen Pyrenophora tritici-repentis (Andrie et al., 2005). Second, the GLS1 gene fused to PtrpC or PtoxB was ectopically integrated into the genome of C. graminicola as an additional copy of GLS1 (see Supplemental Figure 10A online). Infection assays testing for the exposition of β-1,3-glucan were performed with strains harboring an additional ectopic copy of GLS1 controlled by PtrpC (Figure 7A). As both promoter exchange strains showed lower vegetative growth rates than the wild-type strain (see Supplemental Figure 10D online), likely due to the fact that the promoter activity of PGLS1 is stronger than that of PtrpC and PtoxB, the infection process was studied in detail with the strains harboring an ectopic GLS1 copy controlled by PtrpC. The wild-type and PtrpC:GLS1 strains used for infection assays also harbored the PSDH:eGFP biotrophy reporter construct, allowing us to visualize biotrophic development and to demonstrate that β-1,3-glucan is indeed synthesized in biotrophic infection structures. Interestingly, the diameters of biotrophic infection structures formed by two independent transformants carrying the ectopically integrated PtrpC:GLS1 construct were significantly thinner (PtrpC:GLS1-1, 5.1 ± 1.9; PtrpC:GLS1-2, 4.2 ± 2.4 µm) than those of the wild-type strain (8.4 ± 3.3 µm) (Figure 7A, WT, PtrpC:GLS1, and PSDH:eGFP, arrowheads). Necrotrophic hyphae of the wild-type strain had a diameter of 3.8 ± 2.3 µm, which is statistically not different (P < 0.05) from the diameter of the biotrophic hyphae of the PtrpC:GLS1 strains. aniline blue fluorochrome staining clearly showed that β-1,3-glucan was exposed on the surface of biotrophic hyphae of the PtrpC:GLS1 strains but not on those of the wild-type strain (Figure 7A, biotrophy, WT and PtrpC:GLS1, aniline blue, arrowheads; Figure 7B, WT and PtrpC:GLS1, light-blue bars). Necrotrophic hyphae of both wild-type and PtrpC:GLS1 strains contained substantial amounts of β-1,3-glucan, but PtrpC:GLS1 strains showed stronger aniline blue fluorescence than the wild-type strain (Figure 7A, necrotrophy, WT and PtrpC:GLS1, aniline blue; Figure 7B, WT and PtrpC:GLS1, dark-blue bars). The presence of β-1,3-glucan in biotrophic infection structures was further confirmed by aniline blue staining of cross sections (Figure 7C, biotrophy, iv). Secondary hyphae, as expected, showed significant aniline blue labeling (Figure 7C, necrotrophy, sh).
Figure 7.
Forced Expression of GLS1 in Biotrophic Hyphae of C. graminicola Affects Hyphal Morphology.
(A) Comparison of biotrophic infection vesicles (top panel, arrowhead) formed by the wild-type (WT) strain and strains harboring an ectopically integrated GLS1 copy controlled by PtrpC indicated that forced expression of GLS1 led to reduction of hyphal diameters. Both wild-type and PtrpC:GLS1 strains carried a PSDH:eGFP construct confirming biotrophic lifestyle of fluorescing structures. While aniline blue fluorescence indicated that β-1,3-glucan was present in biotrophic hyphae of the PtrpC:GLS1 strain (biotrophy; aniline blue; PtrpC:GLS1; arrowhead), no fluorescence was visible in the wild-type strain (biotrophy; aniline blue; WT; arrowhead). Necrotrophic hyphae of both the wild type and PtrpC:GLS1 strain showed aniline blue fluorescence (necrotrophy; aniline blue; WT and PtrpC:GLS1; arrowhead). DIC, differential interference contrast micrographs. Bars = 20 µm.
(B) Quantification of aniline blue fluorescence in biotrophic (light blue) and necrotrophic hyphae (dark blue) of the wild type and two independent PtrpC:GLS1 strains. Three times 100 measurements were performed with each strain; bars are ±sd. RFU, relative fluorescence units.
(C) Staining of cross sections of biotrophic and necrotrophic hyphae of the PtrpC:GLS1 strain with aniline blue fluorochrome. ap, appressorium; iv, infection vesicle; sh, secondary hyphae. Bars = 20 µm.
Infection assays performed with the promoter exchange strains and with the strains harboring an additional ectopic copy of GLS1 showed that these strains were severely impaired in virulence (Figure 8A). Strains harboring an ectopic copy of PtrpC:GLS1 or PtoxB:GLS1 exhibited significantly increased GLS1 transcript abundance, whereas in promoter exchange strains, transcript concentration was only ∼50% of that of the wild-type strain (see Supplemental Figure 10C online). Accordingly, appressorial penetration rates of independent strains harboring an ectopic copy of PtrpC:GLS1 or PtoxB:GLS1 did not differ from those of the wild-type strain. By contrast, appressoria of all promoter exchange strains tested were unable to breach maize epidermal cell walls (Figure 8B). Deterioration of virulence on intact leaves was confirmed by quantifying fungal DNA (Figure 8C, gray, light-blue, and light-green bars). The fact that strains harboring an ectopic copy of PtrpC:GLS1 or PtoxB:GLS1 did not have a penetration defect but still exhibited reduced virulence suggested that defense responses were induced by β-1,3-glucan oligomers generated in the interfacial matrix. This assumption implies that, if strains of C. graminicola expressing GLS1 during biotrophic development were inoculated onto wounded leaves, where only necrotrophic hyphae are differentiated, virulence should not be impaired. Indeed, symptom severity caused by independent strains harboring an ectopic copy of PtrpC:GLS1 or PtoxB:GLS1 on wounded leaves was comparable to that caused by the wild type (Figure 8D, ectopic), and this was confirmed by quantifying fungal DNA in infected leaves (Figures 8C, ectopic, black, dark-blue, and dark-green bars). Both promoter exchange strains showed reduced virulence on wounded leaves, likely due to reduced growth rates (see Supplemental Figure 10D online). These strains were not further considered in infection assays.
Figure 8.
Forced Expression of GLS1 in Biotrophic Hyphae of C. graminicola Induces Defense Responses in Maize and Causes Reduced Fungal Virulence.
(A) Disease symptoms on nonwounded maize leaves after inoculation with the wild-type (WT) strain, two independent strains harboring a single ectopically integrated extra copy of GLS1 controlled by the trpC or toxB promoter of A. nidulans or P. tritici-repentis, respectively (ectopic, PtrpC:GLS; PtoxB:GLS), and strains with the GLS1 promoter exchanged by the trpC or toxB promoter (promoter exchange, PtrpC:GLS; PtoxB:GLS). Mock-inoculated leaves were treated with 0.01% (v/v) Tween 20.
(B) Appressorial penetration rates of the wild-type strain, two independent strains harboring a single ectopically integrated extra copy of GLS1 controlled by the trpC or toxB promoter, respectively (ectopic, PtrpC:GLS; PtoxB:GLS), and strains with the GLS1 promoter exchanged by the trpC or toxB promoter (promoter exchange, PtrpC:GLS; PtoxB:GLS).
(C) Quantification of fungal development on nonwounded (gray, WT; light blue, PtrpC:GLS; light green, PtoxB:GLS) and wounded maize leaves (black, WT; dark blue, PtrpC:GLS; dark green, PtoxB:GLS) by qPCR. Three independent measurements were performed for each strain in (B) and (C); bars are ±sd.
(D) Disease symptoms on wounded maize leaves after inoculation with the wild-type strain or representative promoter exchange or ectopic PtrpC:GLS or PtoxB:GLS strains. Mock inoculated leaves were treated with 0.01% (v/v) Tween 20.
(E) Both wild-type and PtrpC:GLS1 strains differentiated melanized appressoria (black arrowheads) and invaded intact maize leaves, but only the PtrpC:GLS1 strains caused whole-cell (white arrows) or cell wall fluorescence (white arrowheads) in maize under UV light, indicative of defense responses. Bars = 10 µm.
(F) Quantification of fluorescing maize cells decorated with single appressoria of the wild type or PtrpC:GLS1 strains. Three times 100 cells were measured. Bars represent ±sd.
(G) Maize cells infected by hyphae of PtrpC:GLS1 strains formed vesicles (arrowhead) that turned dark brown (insert) and densely decorated the invading hyphae (arrow). The wild-type strain formed hyphae (white arrow) rarely associated with vesicles. Bars = 10 µm.
Importantly, host cells attacked by appressoria of the C. graminicola wild-type strain only rarely induced defense responses, indicating effective evasion of recognition. By contrast, microscopy revealed that PtrpC:GLS1 strains triggered whole-cell and/or cell wall fluorescence in 30 to 40% of the host cells decorated by a single appressorium, as compared with only ∼2% of those attacked by a single wild-type appressorium (Figures 8E and 8F). The assumption that exposition of β-1,3-glucan on the surface of biotrophic hyphae contributed to induction of cell wall fluorescence was further supported by leaf infiltration assays. Both the long chain β-1,3-glucan laminarin and β-1,3-glucan fragments of a degree of polymerization (DP) of 15 to 25 Glc units, but not glucans of a DP ≤ 7 or water, caused significant cell wall fluorescence (see Supplemental Figure 11A online).
Furthermore, PtrpC:GLS1 strains, but not the wild-type strain, massively elicited formation of infection-related brown vesicles in the plant (Vargas et al., 2012), possibly containing phytoalexins (Figure 8G, cf. WT and PtrpC:GLS1). Vesicles formed in the cytoplasm were initially colorless (Figure 8G, PtrpC:GLS1, arrowhead), but turned dark brown (Figure 8G, PtrpC:GLS1, insert) and eventually decorated the hypha completely (Figure 8G, PtrpC:GLS1, arrow).
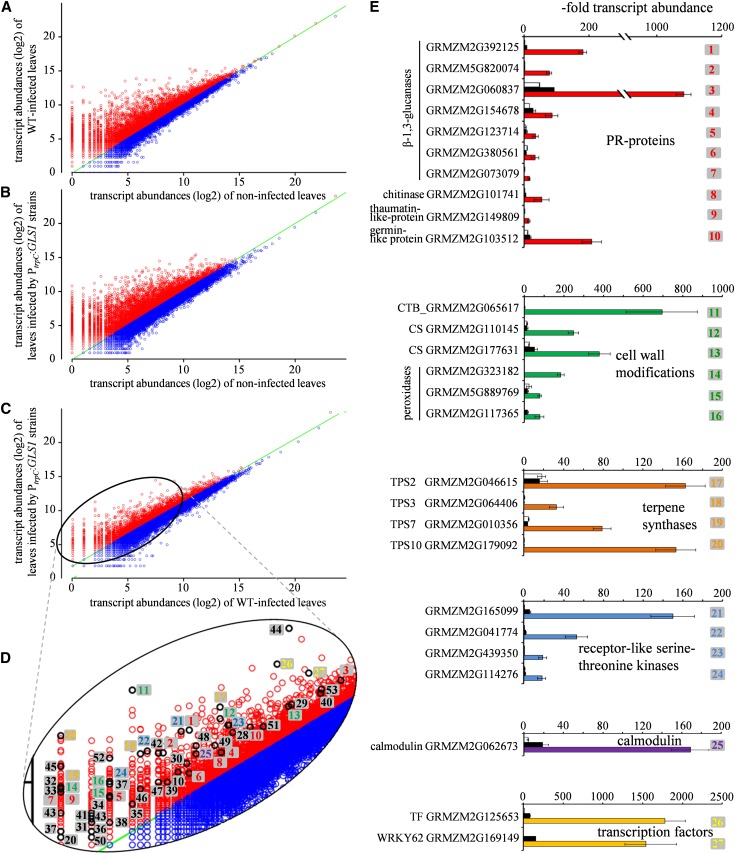
In order to characterize defense responses not only cytologically, but also on the transcript level, genome-wide sequencing of mRNA isolated from noninfected control leaves and from leaves inoculated either with the wild type or with PtrpC:GLS1 strains was performed using Illumina next-generation sequencing technology. Scatterplots depicting the transcript abundance of wild-type-infected and PtrpC:GLS1-infected leaves versus transcript levels in noninoculated control leaves (Figures 9A and 9B) revealed massive upregulation of genes in both interactions. In order to identify transcripts specifically induced in leaves infected by β-1,3-glucan–exposing PtrpC:GLS1 strains, likely enriched in β-1,3-glucan–responsive genes, transcript patterns of leaves inoculated with the PtrpC:GLS1 strains were compared with those of the wild type (Figure 9C). In PtrpC:GLS1-inoculated leaves, a total of 2179 genes were more than 2.5-fold increased, with many genes known as genes typically upregulated in PAMP-triggered defense responses (Figure 9D; see Supplemental Data Set 1 online). These genes include PR proteins, such as 12 β-1,3-glucanases, one chitinase, three thaumatin- and one germin-like proteins, 21 enzymes involved in cell wall reinforcement, and four terpene synthases possibly involved in phytoalexin synthesis. Furthermore, increased transcript abundance of more than 50 genes encoding Ser-Thr receptor-like kinases, nine genes encoding calmodulin, as well as 15 genes encoding zinc-finger and eight encoding WRKY transcription factors have been identified. Other upregulated genes encode proteins involved in protein degradation (i.e., proteases, ubiquitin ligases, as well as enzymes involved in synthesis of auxin or cytokinin phytohormones) (see Supplemental Data Set 1 online). In comparison, 2164 genes were more than 2.5-fold downregulated in maize leaves infected by PtrpC:GLS1 strains compared with wild-type-infected leaves. Several of the encoded proteins are known susceptibility factors. Forty-six downregulated genes code for proteins containing iron or manganese or are involved in uptake of these ions, suggesting major rearrangement of the redox status in maize leaves after β-glucan perception. Seven manganese binding proteins belong to the cupin protein superfamily, representing members of different germin subfamilies. Furthermore, a protein Tyr phosphatase 1 and an alcohol dehydrogenase 1 were 10.5- and 12-fold downregulated, respectively (see Supplemental Data Set 2 online).
Figure 9.
Analysis of the Transcriptional Response of Z. mays to Infection by C. graminicola Wild-Type and PtrpC:GLS1 Strains by Illumina mRNA Sequencing and qRT-PCR.
(A) and (B) Comparison of transcript abundances of wild-type-infected (A) and PtrpC:GLS1-infected (B) leaves with that of noninfected controls. Red, transcript abundance of upregulated genes; blue, transcript abundance of downregulated genes. Changes are given on a log2 scale.
(C) Comparison of transcript abundances of PtrpC:GLS1-infected with that of wild-type-infected leaves. Red, transcript abundance of upregulated genes; blue, transcript abundance of downregulated genes. WT, the wild type.
(D) Magnification of the region encircled in (C). Transcripts of genes marked by black circles were quantified by qRT-PCR (E). Colors of numbers correspond to gene categories ([E]; see Supplemental Data Set 1 online). Green lines in (A) to (D) indicate identical transcript abundance levels under both conditions.
(E) Quantification of transcripts putatively involved in defense responses. White, noninfected control leaves; black, wild-type-infected leaves; colors, leaves infected by PtrpC:GLS1 strains. Colors correspond to gene categories shown in Supplemental Data Set 1 online, and numbers correspond to those in (D). Error bars are standard deviations. Three independent repeats of infection assays were performed.
To confirm data obtained by Illumina mRNA sequencing for those genes putatively involved in PAMP-triggered immunity, qRT-PCRs were performed with transcripts indicated in Figure 9D. Strikingly, nine β-1,3-glucanases were dramatically upregulated in leaves infected by β-1,3-glucan–exposing PtrpC:GLS1 strains, one by more than 1000-fold, compared with transcript levels of histone H2B and β-tubulin (Figure 9E, PR-proteins). Infiltration of leaves with the long chain β-1,3-glucan laminarin and with β-1,3-glucan fragments of a DP of 15 to 25 also increased transcript abundance of four of these β-1,3-glucanases massively (see Supplemental Figure 11B online).
Likewise, other genes encoding PR proteins, such as a chitinase and a thaumatin- and a germin-like protein, were strongly upregulated in leaves infected by PtrpC:GLS1 strains (Figure 9E, PR-proteins).
As cell wall reinforcement, indicated by whole-cell and cell wall fluorescence, was prominent after inoculation with PtrpC:GLS1 strains (Figures 8D and 8E), we also quantified transcript abundance of a gene encoding a protein belonging to the callose synthase complex, as well as of two genes encoding cellulose synthases and of 10 genes encoding cell wall–localized peroxidases (Figure 9B; see Supplemental Figure 12 online, cell wall modifications). All of these genes showed significantly increased transcript abundance in leaves infected by the strains expressing GLS1 during biotrophy. The transcriptional responses of genes involved in cell wall modification to infection by PtrpC:GLS1 strains are supported by increased cell wall fluorescence after infiltration of maize leaves with laminarin and β-1,3-glucan fragments of a DP of 15 to 25 (see Supplemental Figure 11A online).
Sesqui- and diterpene phytoalexins have recently been identified in maize (Huffaker et al., 2011; Schmelz et al., 2011), and brown vesicles decorating the hyphae of PtrpC:GLS1 strains suggest that phytoalexin synthesis may also contribute to the defense response against these strains. Supporting this assumption, four genes encoding terpene synthases (i.e., TPS2, TPS3, TPS7, and TPS10) showed massive increases in transcript abundance specifically after inoculation with strains exposing β-1,3-glucan during biotrophy, with TPS2 and TPS10 upregulated by more than 150-fold (Figure 9E, terpene synthases).
Furthermore, strong upregulation after infection by PtrpC:GLS1 strains of several genes involved in signal perception and genes encoding transcription factors, including zinc finger and WRKY transcription factors, was confirmed by qRT-PCR (Figure 9E; see Supplemental Figure 12 online). Transcript abundances of all genes tested by qRT-PCR were significantly increased (P < 0.05) in leaves infected with PtrpC:GLS1 strains compared with wild-type-infected and noninfected control leaves.
In conclusion, downregulation of GLS1 expression and surface exposure of β-1,3-glucan in C. graminicola leads to a reduction of cell wall rigidity and the formation of voluminous biotrophic hyphae but is required to evade broad spectrum β-glucan–triggered immune responses in maize.
DISCUSSION
Apical growth is a hallmark of fungi and is required for pathogenicity (Wessels, 1993; Stoldt et al., 1997). During hyphal growth, a delicate balance between apical plasticity and subapical rigidity is required and is mediated by tight regulation of enzymes involved in cell wall biogenesis (Sietsma et al., 1985; Walker et al., 2008). In fungal infection structures, tight regulation of the composition of the β-1,3-glucan-chitin core may be of particular importance, as different pathogenic hyphae have specific functions in plant infection. Remarkably, although β-1,3-glucan is the most prominent polymer in fungal cell walls, the role of GLSs and of β-1,3-glucan in pathogenesis is unknown.
The data shown here indicate that β-1,3-glucan is required for appressorial infection and rapid, destructive growth of necrotrophic hyphae and that infection structure–specific GLS1 expression is a key factor in the establishment of a compatible interaction between C. graminicola and its host plant, maize.
GLS1 Is Required for Appressorial Penetration
In plant pathogenic fungi, appressorial host infection is turgor driven, and rigid appressorial cell walls are therefore required for containing turgor pressure, targeted force exertion, and plant invasion (Deising et al., 2000; Bastmeyer et al., 2002; Wilson and Talbot, 2009). Conceivably, mutants exhibiting defects in their β-1,3-glucan-chitin scaffold are affected in appressorium function and virulence. For example, mutants of C. graminicola defective in the class V myosin motor domain-CHS develop appressorial initials, but cell walls lyse when appressoria mature, and ΔchsV strains are unable to cause disease on intact maize leaves (Werner et al., 2007). Also, in M. oryzae and U. maydis, myosin motor domain-CHSs are essential for appressorial plant invasion (Weber et al., 2006; Kong et al., 2012).
As melanized appressoria of C. graminicola generate a turgor pressure of ∼5.5 MPa (55 bar) (Bechinger et al., 1999), it is not surprising that these cells have rigid walls, that GLS1 is strongly expressed, and that β-1,3-glucan is a prominent cell wall polymer (Figure 4). In contrast with ΔchsV strains of this fungus, however, appressoria of class I RNAi strains with reduced GLS1 transcript abundance did not show cell wall autolysis but swelled in osmotically nonstabilized media (Figures 6A and 6B) and exploded (Figure 5B). Intriguingly, upon addition of an osmolyte to swollen appressoria, their diameter shrunk by approximately half of their size (i.e., to the size of wild-type appressoria), clearly depicting the enormous elasticity of appressorial cell walls of class I RNAi strains and the role of GLS1 in cell wall rigidity.
Appressoria of class I RNAi strains were unable to adequately integrate melanin into their cell walls, and melanin formed a ring surrounding these appressorium (Figure 5C, inset). In the human pathogens A. fumigatus and Wangiella dermatitidis, deletion of genes encoding α-1,3-glucan synthase and a CHS caused hypermelanization (Liu et al., 2004; Maubon et al., 2006). In W. dermatitidis, melanization of cell walls increases cell wall rigidity, as indicated by stab inoculation experiments testing for invasive growth competence (Brush and Money, 1999). Appressoria of RNAi strains of C. graminicola, in contrast with vegetative hyphae, did not hypermelanize, and leakage of melanin from the appressorial cell wall reported here suggests that an intact β-1,3-glucan polymer network is essential for incorporation and linking of melanin to the cell wall. However, insufficient melanization of the appressorial wall of RNAi strains is unlikely to cause lack of rigidity, as melanin-deficient mutants of C. graminicola (Horbach et al., 2009), C. lagenarium (Kubo et al., 1991), and M. oryzae (Chida and Sisler, 1987) did not show increased elasticity.
Not only generation of appressorial turgor pressure, but also tight adhesion of infection cells to the plant surface is required for plant invasion (Nicholson and Epstein, 1991; Howard and Valent, 1996). Appressoria of class I RNAi strains were unable to adhere to all substrata tested (Figure 5C). In six different Colletotrichum species, glycoproteins contributing to adhesion have been detected in the appressorial mucilage by a monoclonal antibody (Hutchison et al., 2002). Interestingly, some fungal cell wall proteins are covalently linked to a hydroxyl group of a β-1,3-glucan chain (De Groot et al., 2005; Ecker et al., 2006), and glycoproteins mediating appressorial adhesion may likewise be linked to and require an intact β-1,3-glucan core.
Our experiments strongly suggest that β-1,3-glucan is required for appressorial adhesion, penetration competence, and pathogenicity on intact maize leaves (Figures 5C, 5F, and 7A to 7C).
Differential Roles of GLS1 in Biotrophic and Necrotrophic Infection Structures
After penetration of the plant cell wall, biotrophic and hemibiotrophic fungi invaginate the plant plasma membrane, leading to the establishment of a biotrophy-specific interfacial matrix (O'Connell et al., 1996; Kankanala et al., 2007). Fungal cell walls exposed to this matrix exhibit drastically modified surface carbohydrate polymer compositions (Freytag and Mendgen, 1991; Mendgen and Deising, 1993; O’Connell et al., 1996; El Gueddari et al., 2002; Fujikawa et al., 2009; Treitschke et al., 2010), and these modifications may be required for both cell wall integrity and interaction with the host plant.
Synthesis of β-1,3-glucan is thought to be constitutively required in fungal hyphae (Mouyna et al., 2004; Ha et al., 2006). Surprisingly, voluminous biotrophic infection structures of C. graminicola exhibited dramatically reduced synthesis of β-1,3-glucan, as indicated by extremely low levels of GLS1:eGFP expression and staining of infection vesicles and primary hyphae with the β-1,3-glucan–specific dye aniline blue fluorochrome. Overexpression of GLS1 in infection vesicles and primary hyphae significantly reduced the diameter of these biotrophic hyphae, which was similar to that of necrotrophic hyphae (Figure 8B, top panel). As thin hyphae have a higher surface-to-volume ratio, favoring efficient nutrient uptake (Bhadauria et al., 2011), we assume that the large volume of biotrophic infection structures does not reflect their function in nutrient uptake, but rather indicates that cell walls are poor in or devoid of β-1,3-glucan and therefore dilate. Indeed, members of the oomycete family Saprolegniaceae (i.e., Saprolegnia ferex, Achlya bisexualis, and Achlya ambisexualis) are unable to adjust their hyphal turgor pressure to increasing extracellular osmotic potentials and secrete endoglucanases to reduce the strength of their cell walls. Endoglucanase-mediated reduction of cell wall strength caused a drastic increase of the hyphal diameter and strongly affected the hyphal shape (Money and Hill, 1997).
In sharp contrast to biotrophic hyphae, β-1,3-glucan represents a major cell wall polymer in necrotrophic secondary hyphae of C. graminicola (Figure 4), and, conceivably, necrotrophic hyphae of class I RNAi strains exhibit severe cell wall defects (Figures 7D and 7E). As expected, RNAi strains with defects in β-1,3-glucan synthesis exhibit severely reduced virulence (Figures 7A, 7B, and 7E).
Taken together, these data indicate that expression of GLS1 and synthesis of β-1,3-glucan are of significant importance in necrotrophic but not in biotrophic hyphae of C. graminicola.
Forced Expression of β-1,3-Glucan in Biotrophic Hyphae of C. graminicola Triggers Immunity in Maize
Downregulation of GLS1 expression and of synthesis of β-1,3-glucan during formation of biotrophic hyphae of C. graminicola suggests that evasion of PAMP recognition may be fundamental for escaping PAMP-triggered immunity. Fragments of structural cell wall polymers, including linear and/or branched β-1,3-glucans, function as PAMPs in plants and mammals and trigger innate immune responses in both host backgrounds (Nürnberger et al., 2004). In humans, the membrane-bound nonclassical C-type lectin receptor Dectin-1 recognizes either linear β-1,3-glucan or β-1,3–1,4-glucan polymers or linear β-1,3-glucan polymers with short linear β-1,6-glucose side chains (Latgé, 2010, and references therein). In plants, β-glucan recognition receptors have as yet not been functionally characterized. However, structurally different β-glucans are differentially recognized (Klarzynski et al., 2000; Yamaguchi et al., 2000). A putative high-affinity β-glucan receptor of soybean (Glycine max) exhibiting an endo-cleaving β-1,3-glucanase domain has been characterized biochemically in detail (Fliegmann et al., 2004). This protein has β-1,3-glucanase, but no β-1,6-glucanase activity, and specific disintegration of β-1,3-glucan chains may thus amplify elicitor-active β-glucan elicitor molecules with a structure specifically recognized by a putative β-glucan receptor complex. Intriguingly, genes encoding proteins related to the β-glucan receptor from soybean exist in presumably all plant species (Fliegmann et al., 2004).
The majority of pattern recognition receptors are plasma membrane located (Postel and Kemmerling, 2009), and exposure of PAMPs may therefore be particularly critical during biotrophic development (i.e., when pathogenic hyphae develop in the biotrophy-specific interfacial matrix in close vicinity of the host membrane). Indeed, infiltration of maize leaves with laminarin as well as β-1,3-glucan oligomers (DP 15 to 25) elicit defense responses directed against this structural cell wall polymer (i.e., a drastic increase of transcript abundances of four β-1,3-glucanase genes) (see Supplemental Figure 11B online). As enzymatic modification and/or high-affinity binding of β-1,3-glucan has not been reported, masking or downregulation of β-1,3-glucan synthesis during critical stages of the infection process, or a combination thereof, may be employed to evade recognition of this PAMP.
In the human pathogen A. fumigatus, coating by the surface hydrophobin RodA and melanin apposition renders conidia immunologically inert, although several immunogenic molecules are localized beneath the surface (Aimanianda et al., 2009; Chai et al., 2010). Based on enzymatic digestion of infection hyphae of the rice blast fungus M. oryzae, in combination with immunohistochemistry, Fujikawa et al. (2009) suggested that apposition of α-1,3-glucan contributes to masking of PAMPs, such as chitin and β-1,3-glucan. However, in their study, the authors did not discriminate between biotrophic and necrotrophic hyphae, so conclusions on masking of biotrophic hyphae during initial stages of the infection process cannot be drawn. Staining of cross sections of biotrophic hyphae of the maize pathogen C. graminicola (Figure 3D) clearly showed that β-1,3-glucan is not a major component of the cells of biotrophic hyphae. Interestingly, expression of a bacterial α-1,3-glucanase in rice plants mediated strong resistance not only against M. oryzae but also against Cochlioborus miyabeanus and the basidiomycete Rhizoctonia solani (Fujikawa et al., 2012), strongly arguing that hyphal masking by α-1,3-glucan is an important mechanism of establishment of compatibility, which may be particularly relevant at later stages of pathogenesis.
The data reported here provide evidence that biotrophy-associated downregulation of GLS1 expression and synthesis of β-1,3-glucan are required for evading β-glucan–triggered immunity. In biotrophic hyphae of C. graminicola, β-1,3-glucan is not detectable, and in wild-type-inoculated maize leaves, <5% of epidermal cells attacked by a single appressorium exhibit defense responses, as indicated by whole-cell or cell wall fluorescence (Figures 8D and 8E). By contrast, strains overexpressing GLS1 during biotrophic growth expose significant amounts of β-1,3-glucan (Figures 8B and 8C) and evoke dramatic defense responses (i.e., whole-cell or cell wall fluorescence) in 30 to 40% of the epidermal cells attacked (Figures 8D and 8E). Furthermore, in maize cells attacked by hyphae exposing β-1,3-glucan, but not in those infected by wild-type hyphae, formation of large numbers of brown vesicles possibly containing antifungal compounds such as the recently identified sesqui- and diterpenoid phytoalexins (Huffaker et al., 2011; Schmelz et al., 2011) occurs. Recent studies have shown that transcript concentrations of the terpene synthases genes TPS6 and TPS11 as well as concentrations of the terpene phytoalexins zealexins and kauralexins were strongly upregulated after infection of maize by various fungal pathogens such as Rhizopus microsporus, Cochliobolus heterostrophus, Colletotrichum sublineolum, Aspergillus flavus, and Fusarium graminearum (Huffaker et al., 2011; Schmelz et al., 2011). Also, in the compatible interaction between maize and the biotrophic smut fungus U. maydis, TPS6 and TPS11 appear to contribute to plant defense, as indicated by downregulation of terpene synthase by virus-induced gene silencing (van der Linde et al., 2011). Remarkably, C. graminicola induced neither upregulation of terpene synthase transcripts nor of terpene phytoalexins (Huffaker et al., 2011; Schmelz et al., 2011). Comparative Illumina mRNA sequencing of maize leaves infected by C. graminicola wild-type and PtrpC:GLS1 strains and qRT-PCR experiments revealed that four different terpene synthase genes were dramatically upregulated in PtrpC:GLS1-infected, but not in wild-type-infected, leaves (Figure 9). Accordingly, in PtrpC:GLS1 strains, transcript abundance of PR genes was dramatically increased, most notably those of β-1,3-glucanases, as were those of genes involved in cell wall modifications (Figure 9), all of which can be regarded as classical PAMP responses. This also holds true for various components of signal perception and signal transduction (e.g., more than 50 Ser-Thr receptor-like kinases, almost 10 calmodulins, and several calmodulin-dependent protein kinases) (Figure 9; see Supplemental Data Set 1 online) (Zimmermann et al., 2006; Afzal et al., 2008; Almagro et al., 2009). In addition to genes actively involved in defense responses, downregulation of susceptibility genes can contribute to resistance. Transient RNAi-mediated silencing experiments suggested that the germin genes GER3 or GER5 (Zimmermann et al., 2006) and alcohol dehydrogenase 1 (Pathuri et al., 2011) mediate compatibility between barley (Hordeum vulgare) and the biotrophic barley pathogen Blumeria graminis f. sp hordei and its host. Likewise, several genes strongly downregulated in maize leaves in response to infection by PtrpC:GLS1 strains may represent susceptibility factors, and downregulation of these factors may contribute to β-glucan–triggered immunity (see Supplemental Data Set 2 online).
Taken together, our data strongly suggest that biotrophy-specific downregulation of GLS is required for the establishment of a compatible interaction between C. graminicola and maize.
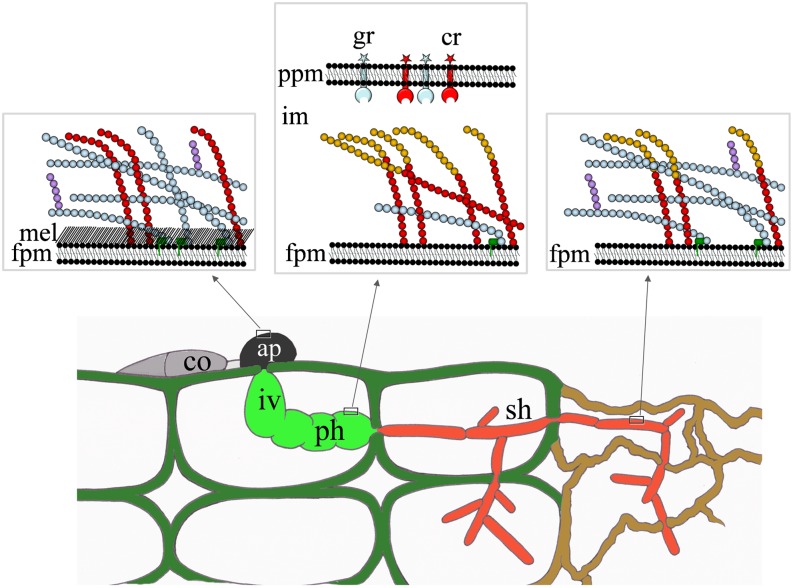
Live-cell imaging and aniline blue fluorochrome staining indicated that cell wall composition of the maize pathogen C. graminicola is highly dynamic during infection structure development and is required for host infection and compatibility (Figure 10). Massive β-1,3-glucan and chitin synthesis, providing the cell wall rigidity required for containing turgor pressure, occurs in appressoria (Figures 2 and 10), and polymeric β-1,3-glucan is likely to carry β-1,6-glucan side chains. Infection vesicles and primary hyphae encased by the plant plasma membrane do not contain significant amounts of β-glucan and develop into voluminous hyphae, probably due to insufficient cell wall rigidity. Furthermore, these hyphae secrete chitin deacetylases into the interfacial matrix layer, yielding chitosan, which is both a poor substrate for plant chitinases and a poor elicitor (El Gueddari et al., 2002, and references therein). Thus, at this delicate stage of pathogenesis, neither elicitor-active β-1,3-glucan nor chitin is exposed on the surface of biotrophic hyphae, and defense responses are circumvented. When necrotrophic development is initiated, GLS1 is massively expressed and β-1,3-glucan is a prominent cell wall polymer in secondary hyphae (Figure 10).
Figure 10.
Model of Cell Wall Modifications in Infection Structures of C. graminicola.
In appressoria (ap), cell walls consist of significant amounts of chitin (red circles), β-1,3-glucan (light-blue circles), and β-1,6-glucan (violet), forming a rigid scaffold, with melanin (mel) forming a layer in the vicinity of the fungal plasma membrane (fpm). Biotrophic infection vesicles (iv) and primary hyphae (ph; green) are located within the interfacial matrix and surrounded by the plant plasma membrane (ppm) containing β-glucan (gr) and chitin receptors (cr). The walls of these voluminous hyphae show no or very low amounts of β-1,3-glucan, and surface-localized chitin is deacetylated to yield chitosan (brown circles). Cell walls of thin secondary hyphae (sh; orange) exhibit chitin (red circles), chitosan (brown circles), β-1,3- (light-blue circles), and β-1,6-glucan (violet). These hyphae are highly destructive and cause necrosis of host cells.
The data presented here show that modification of cell wall components during the early infection process is crucial for the establishment of compatibility between C. graminicola and maize. This finding is surprising, as fungal plant pathogens, including Colletotrichum species, harbor large numbers of effectors in their genomes. However, defining Colletotrichum-specific effectors as predicted extracellular proteins without any homology to proteins outside this genus, genome mining of C. higginsianum and C. graminicola revealed that 365 effector-encoding genes exist in C. higginsianum, but only 177 exist in C. graminicola (i.e., <50%; O’Connell et al., 2012). Thus, one may speculate that fungi equipped with a small effector repertoire have developed other mechanisms to establish compatibility (e.g., dynamic infection-related cell wall modifications) to avoid PAMP exposition.
METHODS
Fungal Strains, Culture Conditions, Infection Structure Differentiation, and Infection Assays
The wild-type strain M2 of Colletotrichum graminicola (teleomorph Glomerella graminicola) and RNAi strains generated in this study were cultivated on oatmeal agar (OMA; Werner et al., 2007), complete medium (Leach et al., 1982), potato dextrose (Difco Laboratories), synthetic minimal medium [10 g Glc, 1 g Ca(NO3)2, 0.2 g KH2PO3, 0.25 g MgSO4, and 0.054 g NaCl per liter], or synthetic complete medium (without amino acids; Becton Dickinson), with amino acids added according to Treco and Lundblad (1993). To grow RNAi strains, the media were supplemented with 0.15 M KCl, 1 M sorbitol, or 0.5 M Suc.
In liquid media, strains were grown in an incubation shaker (Unitron; Infors) at 110 rpm and 23°C. On solidified media, containing 1.5% (w/v) agar-agar (Difco Laboratories), strains were grown at 23°C under continuous fluorescent light (Climas Control CIR; UniEquip; Werner et al., 2007).
The Saccharomyces cerevisiae reference strain Y00000 (parental S288C) (Mat a, his3D1, leu2D0, met15D0, ura3D0) and the Δfks1 mutant Y05251 (BY4741; Mat a; his3D1; leu2D0; met15D0; ura3D0; YLR342w::kanMX4) (Euroscarf) were grown at 30°C and 150 rpm in liquid YPD (Difco) or yeast synthetic complete medium (Difco) lacking uracil. Solidified media contained 1.5% (w/v) agar (Difco). Y05251 cells were grown in the presence of 1 M sorbitol.
Infection structure differentiation of the wild-type and RNAi strains was induced on polyester, glass, or onion epidermis as described (Horbach et al., 2009), with 0.15 M KCl added as an osmolyte.
Twelve-day-old whole maize (Zea mays cv Nathan) plants, 7-cm-long leaf segments of the 2nd and 3rd leaf, depending on the experiment, or epidermal cell layers from onion (Allium cepa cv Grano) bulbs were used to assess virulence of the wild-type and RNAi strains. Maize leaf segments and onion epidermal layers were inoculated with 10-μL droplets containing 104 conidia in 0.01% (v/v) Tween 20. Wound inoculation was performed as described (Horbach et al., 2009). Leaf segments were incubated in sealed Petri dishes (diameter of 9 cm) in a BOD 400 incubator (Uni Equip) in darkness at 25°C. For each treatment and time point analyzed, 12 leave segments were evaluated. Mock inoculation was performed with 0.01% (v/v) Tween 20. Symptoms were photographed 6 d after inoculation. For qPCR, inoculation was done with 10 detached 2nd or 3rd leaves per repetition, treatment, and time point, in which each leaf received a single 10-μL droplet containing104 conidia. For qRT-PCR and Illumina RNA sequencing, whole maize plants were inoculated with one drop (10 µL) of a spore suspension containing 106 spores/mL. For each treatment, the 2nd and 3rd leaf of nine plants were used per repetition. Mock inoculation was performed using sterile 0.01% (v/v) Tween 20. Plants were incubated in a Percival AR-75L growth chamber (CLF Laborgeräte) (12 h light; 200 mE; 70% relative humidity; 25°C).
All experiments were performed in triplicate and with four repetitions.
Molecular Phylogenetic Analyses
Multiple sequence alignments were done using ClustalW (http://embnet.vital-it.ch/software/ClustalW.html; Larkin et al., 2007). Phylogenetic dendograms were constructed using MEGA 5 (www.megasoftware.net; Tamura et al., 2011), with the minimum evolution algorithms using 1000 bootstrap replications. Sequences of all fungal GLS genes were obtained from the database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Complementation of the S. cerevisiae Δfks1 Mutant with the C. graminicola GLS1 cDNA
In order to synthesize cDNA of GLS1, total RNA was extracted (Chirgwin et al., 1979) from vegetative mycelium of C. graminicola. mRNA was isolated using the Nucleotrap mRNA purification kit, and cDNA synthesis was performed using the Creator SMART cDNA library construction kit (BD Biosciences Clontech). The primers CgGLS1NotI-Fw and CgGLS1NotI-Rv were used to amplify the GLS1 cDNA, which was cloned into the NotI site of the yeast cDNA expression vector pAG300 (www.addgene.org; Horbach et al., 2009). These primers, and others mentioned here, are listed in Supplemental Data Set 1 online. Correct orientation of the DNA was confirmed by sequencing using primer pAG300-Fw. The empty vector (pAG300) and the vector containing the GLS1 cDNA were transformed into S. cerevisiae strain Y05251 using the lithium acetate procedure (Becker and Lundblad, 2001) to yield the complemented yeast transformants TCgGLS11-4. As a control, strain Y05251 was also transformed with empty pAG300, yielding TpAG300. Yeast cells were grown on yeast synthetic complete medium agar lacking uracil.
Targeted Deletion, Promoter Exchange, and Overexpression of GLS1, Construction of RNAi Strains, and Generation of C. graminicola GLS1:eGFP Replacement Strains
For targeted deletion of the 5940-bp C. graminicola GLS1 gene, the Nourseothricin acetyltransferase gene Nat-1 from Streptomyces noursei was PCR amplified from pNR1 (Malonek et al., 2004) using primers Nourse1pNR1-Fw and Nourse1pNR1-Rv. The 1009-bp 5′ and the 1003-bp and 3′ flanking regions of the GLS1 gene were amplified from genomic DNA using primers CgPGLS1-fw and CgP1GLS15'-flank-rv, and CgTGLS13'-flank-fw and CgTGLS1-rv, respectively. The products were fused by joint PCR (Yu et al., 2004), and nested primers CgPGLS1nest-fw and CgTGLS1nest-rv were used to amplify the 4210-bp knockout construct, which was transformed into conidial protoplasts as described (Werner et al., 2007). Tests for homologous integration of the knockout construct was done with primers CgPGLS1test-fw and CgTGLS1test-rv.
The RNAi cassette from plasmid pRedi (Janus et al., 2007) was used to generate an RNAi construct targeting GLS1 transcripts. The 475-bp GLS1 sense and antisense fragments were amplified from genomic DNA of C. graminicola, using the primers RNAi(GLS1)-fw and RNAi(GLS1)-Rv, and RNAi(GLS1)i-fw and RNAi(GLS1)i-Rv, respectively. The sense and antisense fragments were used to replace the XhoI-SnaBI and BglII-ApaI fragments of pRedi and were thus separated by 135 bp of the intron of the Magnaporthe oryzae Cut2 gene (National Center for Biotechnology Information: XM_365241.1), existing in pRedi, as a linker (Janus et al., 2007). The resulting 6.24-kb RNAi construct was excised from pRedi by DraI digestion, purified by gel elution, and transformed into conidial protoplasts of C. graminicola, and single spore isolates were generated (Werner et al., 2007).
To study cell specificity of expression of GLS1 and localization of the protein, a GLS1:eGFP replacement construct, consisting of the 1-kb 3′-end of the coding region of the GLS1 gene fused in frame to the eGFP gene, followed by the HygR resistance cassette and the 1-kb 5′- noncoding sequence of GLS1 containing the terminator, was transformed into the C. graminicola wild-type strain (see Supplemental Figure 2 online). The 5′-coding region of GLS1 was amplified with the primers CgGLS1GFP-Fw and CgGLS1GFP5′-flank-Rv, using genomic DNA as the template. The eGFP gene and the HygR cassette were amplified using primers EGFP-Fw and HygR-Rv, with plasmid pSH1.6EGFP, kindly provided by Amir Sharon, Tel Aviv University, Israel, as template. Using genomic DNA as template, the 3′-flank of GLS1 was amplified with primers CgGLS1GFP3′-flank-Fw and CgTGLS1GFP-Rv. The GLS1:eGFP construct was fused by double-joint PCR (Yu et al., 2004), and the complete 6.2-kb fragment was amplified with nested primers CgGLS1:GFP.nest-fw and CgGLS1:GFP.nest-rv. Phusion High-Fidelity DNA Polymerase (New England Biolabs) was used in all PCR reactions. The GLS1:eGFP construct was transformed into conidial protoplasts (Werner et al., 2007), and single spore isolates were tested for site-specific integration and replacement of the wild-type GLS1 gene by DNA gel blot hybridization.
For overexpression of GLS1, the trpC promoter of Aspergillus nidulans was amplified from pSM1 (Pöggeler et al., 2003) using primers PtrpC-Sac1-Fw and PtrpC-Sac1-Rv. The toxB promoter of Pyrenophora tritici-repentis was amplified from pCM29 (Andrie et al., 2005) using primers PtoxB-Sac1-Fw and PtoxB-Sac1-Rv. The PCRs products were digested by SacI, purified, and ligated into SacI-digested pNR1. The complete GLS1 gene was amplified with the primers CgGLS1NotI-Fw and CgTGLS1NotI-Rv. The 7050-bp PCR product was NotI-digested, purified, and ligated into pNR1. The resulting 10,032-bp (PtrpC:GLS1:NatR) and 10,058-bp (PtoxB:GLS1:NatR) constructs were amplified using primes NatOvExp.nest-Fw and NourspNR1-Rv and transformed into conidial protoplasts (Werner et al., 2007). Single spore isolates were tested for numbers of the GLS1 overexpression constructs integrates by DNA gel blot hybridization.
For targeted promoter exchange, PtrpC, PtoxB, and the Nourseothricin resistance cassette were amplified as described above. The 1009-bp 5′ and the 1003-bp and 3′ flanking regions of the GLS1 promoter and the 5′-part of GLS1 gene coding sequence were amplified from genomic DNA using primers P-TrpCGLS5′flank-Fw, P-TrpCGLS5′flank-Rv, CgGLS3′flank-Fw, and P-TrpCGLS3′flank-Rv, respectively. The products were fused by double-joint PCR (Yu et al., 2004). Nested primers P-TrpCGLS1nest-Fw and P-TrpCGLS1nest-Rv were used to amplify the 4210-bp promoter replacement construct, which was transformed into conidial protoplasts (Werner et al., 2007). Homokariotic strains were tested for site-specific integration by DNA gel blot hybridization.
DNA Extraction and Genomic DNA Gel Blot Analysis
Extraction of genomic DNA and DNA gel blot analyses were performed as described (Werner et al., 2007). To analyze transformants for the number of integrations of the RNAi construct, 10 mg of DNA digested with the appropriate restriction endonuclease was used. The 511-bp alkali-labile DIG-dUTP–labeled probe (Roche Diagnostics) specific for the nourseothricin acetyl transferase gene (Nat-1) was amplified from plasmid pNRl using primers NatR probe-Fw and NatR probe-Rv. To analyze transformants for the correct integration of a single copy of the GLS1:eGFP cassette into genomic DNA of wild-type and RNAi strains, DNA gel blots were hybridized with a 500-bp DIG-dUTP–labeled hygromycin phosphotransferase (HygR)–specific probe, which was amplified from the GLS1:eGFP construct as template using primers HygR probe-fw and HygR probe-rv.
qRT-PCR and PCR
qRT-PCR was performed in a total volume of 20 µL, with 100 ng of total RNA pretreated with RQ1 RNase-free DNase (Promega), using the Power SYBR Green RNA-to-CT 1-step kit (Applied Biosystems) according to the instructions of the manufacturer. Primers used are shown in Supplemental Data Set 3 online. Melting curve and agarose gel analyses confirmed amplification of a single product. Transcript abundance was calculated and normalized as described (Gutjahr et al., 2008), using α-actin and histone H3 for C. graminicola and histone H2B and β-tubulin for maize. Cycle threshold values of three independent replicates were used to calculate mean values and standard deviations.
To quantify fungal development, qPCR was performed with 100 ng of template DNA, 100 nM of each primer (CgITS2-q1 and CgITS2-q2), and 10 μL of iQ SYBR Green Supermix (Bio-Rad Laboratories) in a total volume of 20 µL. DNA used for qPCR was isolated using a peqGOLD Plant DNA Mini Kit (Peqlab). All values are standardized to the average threshold cycle value obtained with DNA extracted from nonwounded leaves inoculated with the C. graminicola wild-type strain at 0 HAI. Cycle threshold values of three independent replicates were used to calculate mean values and standard deviations.
Expression of eGFP under Control of Biotrophy- and Necrotrophy-Specific Promoters
To identify biotrophy-specific promoters of C. graminicola, 1.2 to 1.3 kb of the 5′ nontranslated flanks of homologs of biotrophy-specific genes of Colletotrichum higginsianum encoding a NmrA-like family protein, a saccharopine dehydrogenase, a pentafunctional AROM protein, an argininosuccinate lyase, an argininosuccinate synthase, and the nonannotated unigene 3 (Takahara et al., 2009) were amplified using primers containing EcoRV restriction sites in the forward primer and Eco47III sites in the reverse primers for unidirectional cloning into plasmid pSH1.6EGFP, 5′ of eGFP. The promoter:eGFP fusions, together with the HygR gene controlled by the GPDA promoter of A. nidulans, were PCR amplified from pSH1.6EGFP using primers PSH1-Fw and Hyg.treminator-rev, transformed into C. graminicola, and microscopically screened for specific eGFP expression in infection vesicles and primary hyphae. Of the putative biotrophy-specific promoters tested, the 1.2 kb 5′-nontranslated region of the gene encoding a saccharopine dehydrogenase (SDH) and the 1.3 kb-5′-nontranslated region of a gene encoding an NmrA-like family protein mediated biotrophy-specific expression of eGFP. To identify necrotrophy-specific promoters of C. graminicola, putative promoters of genes that had previously shown necrotrophy-specific expression in qRT-PCR experiments (Krijger et al., 2008) (i.e., the 5′ noncoding regions of the genes encoding a secreted peptidase and an extracellular Ser-rich protein) were tested, and the 1.2-kb 5′ nontranslated region of the secreted peptidase gene mediated expression specifically in secondary hyphae. eGFP fusions with the promoters of the genes encoding the NmrA-like family protein, the saccharopine dehydrogenase, and the secreted peptidase were also transformed into the C. graminicola class I RNAi strains. Primers used to produce promoter:eGFP fusions for these genes were CgPrNmrA-Fw and CgPrNmrA-Rv (NmrA-like family protein), CgPrSacDh-Fw and CgPrSacDh-Rv (saccharopine dehydrogenase), and CgPrSecPep-Fw and CgPrSecPep-Rv (secreted peptidase).
Microscopy and Imaging
Bright-field, differential interference contrast microscopy, and fluorescence microscopy were performed using a Nikon Eclipse 600 or a Nikon Eclipse 90i confocal laser scanning microscope. For fluorescence microscopy, a Plan Apo 60/1.4 oil lens and the following settings were used: excitation wavelength, 488 nm; laser light transmittance, 25% (ND4 in; ND8 out); and pinhole diameter, 30 mm.
To analyze β-1,3-glucan contents of cell walls, infected maize leaves were harvested at 0, 12, 24, 48, and 72 HAI and stained with aniline blue fluorochrome (Biosupplies Australia) as described (Vogel and Somerville, 2000). To exclude the possibility that differential aniline blue fluorochrome staining was due to masking of β-1,3-glucan by apposition of α-glucan, specimens were incubated at 60°C or autoclaved in 0.1 n NaOH for 20 min and subsequently stained with aniline blue fluorochrome.
Quantitative fluorescence levels of GLS1:eGFP-expressing transformants of C. graminicola were evaluated at 0, 12, 24, and 72 HAI using a Zeiss Observer Z1 inverted microscope equipped with a Plan Apochromat ×63/1.40 oil immersion objective and an AxioCam MRm camera. Epi-illumination analyses employed filter set 49 for aniline blue fluorochrome and filter set 38HE for eGFP. Image acquisition and analysis were performed using Zeiss AxioVision 4.8.2 (June, 2010) software with the Physiology module (all from Carl Zeiss).
Cross sections of biotrophic and necrotrophic infection hyphae (20 µm) were made using Plaque GP-low-melting agarose-embedded specimens cut with a Carl Zeiss microtome (Hyrax V50).
Illumina mRNA Sequencing and Bioinformatics
Illumina NG mRNA sequencing was performed with samples from noninfected maize leaves and from leaves infected by the wild-type and the PtrpC:GLS1 strains. Identical amounts of mRNA from three independent samples of each treatment were mixed, and Illumina sequencing was performed by ServiceXS using the paired-end sequencing protocol. Data sets of 100-bp reads yielding a minimum of 4.5 Gb of Illumina-filtered sequence data per sample were obtained. For further analyses, reads ≥70 bp with a phred score ≥ Q20 were selected. Bases with a phred score below this level were removed, and reads containing theses bases were split. The filtering was performed by Fast qFilter version 2.05 (ServiceXS). The obtained reads were aligned to the maize B73 reference genome (ftp.maizegdb.org) using BWA version 0.5.9 (Li and Durbin, 2009) and TopHat version 1.4.0 (Trapnell et al., 2009) and corresponded to a minimum of 36,020 unique transcripts. The counts obtained were normalized according to Mortazavi et al. (2008). Downstream processing was performed by Philip de Groot, R-Consultancy, Apeldoorn, The Netherlands, using the core packages of R software version 2.15.3. To confirm β-glucan–triggered defense responses, a total of 50 candidate genes were selected for qRT-PCR. In RNaseq analyses, these genes showed a minimum of 3.5-fold increased transcript abundance in PtrpC:GLS1, compared with wild-type-infected leaves, and were selected as members of different functional categories.
Other Methods
Appressorial turgor pressure was measured as incipient cytorrhizis using PEG 6000 as described (Howard et al., 1991).
Fluorescein diacetate staining was performed as described (Söderström, 1977).
To measure conidiation, Petri dishes (diameter of 9 cm) with 14-d-old fungal cultures grown on OMA were washed on a rotary shaker with 10 mL 0.01% (v/v) Tween 20 for 10 min to yield homogenous conidial suspensions. Conidia were counted in a Thoma chamber.
Statistics
Calculations (t test, analysis of variance) were performed with the software XLSTAT version 2009.4.02 (Addinsoft).
Accession Numbers
Sequence data for genes of C. graminicola used in this article can be found in the GenBank/EMBL database under the following accession numbers: GLS1, EFQ30502; saccharopine dehydrogenase (SDH), EFQ35895; NmrA-like family protein (NmrA), GLRG_07923; secreted peptidase (SPEP), EFQ25677; and developmentally regulated MAPK-interacting protein, EFQ32584. RNAseq data are available under GEO accession number GSE48208. Accession numbers of glucan synthases of other fungal species are as follows: Ajellomyces capsulatus, XP001539822.1; Ajellomyces dermatitidis, XP002629106.1; Aspergillus flavus, XP002383406.1; Aspergillus oryzae XP001816673.1; A. fumigatus, XP751118.1; A. nidulans, XP661333.1; Candida albicans, XP721429.1; Candida glabrata, XP446406.1; Candida paraplosis, ABX80511.1; Candida gattii, XP003197612.1; Candida paraplosis imitis, XP001247982.1; Coccidioides posadasii, XP003065511.1; Coprinopsis cinerea, XP001837755.2; Cryptococcus neformans, XP771791.1; Exophiala dermatitidis, ABL63820.1; Fusarium solani (FKS1), ABC59463; F. solani (FKS2), XP003040299.1; Laccaria bicolor, XP001878782.1; M. oryzae, XP368379.2; Metarhizium acridum, EFY92417.1; Neurospora crassa, XP957980.1; Paracoccidioides brasiliensis, XP002792935.1; Penicillium marneffei, XP002147495.1; Pichia pastoris, XP002491155.1; P. graminis f. sp tritici, XP003307175.1; S. cerevisiae (FKS1), NP013446.1; S. cerevisiae (FKS2), NP011546.1; S. cerevisiae (FKS3), NP014036.1; Schizosaccharomyces pombe (Bgs1), NP595971.1; S. pombe (Bgs2), NP594032.1; S. pombe (Bgs3), NP594766.1; S. pombe (Bgs4), CAA20125.1; Sclerotinia sclerotiorum, XP001586992.1; Talaromyces stipitatus, XP002481640.1; Ustilago maydis, XP757786.1; Zygosaccharomyces rouxii, XP002494904.1. Plasmid pSH1.6EGFP is available under Addgene deposit number 69980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene (Left) and Protein Structure (Right) of GLSs of Different Asco- and Basidiomycota.
Supplemental Figure 2. Generation of GLS1:eGFP Replacement Strains and Comparison of the Wild-Type and the Replacement Strains.
Supplemental Figure 3. GLS1 of C. graminicola Is Plasma Membrane Localized.
Supplemental Figure 4. Strategy for Targeted Deletion of GLS1 and Screen for Transformants with Homologous Integration of the Deletion Cassette.
Supplemental Figure 5. Replacement of GLS1 by an GLS1:eGFP Fusion in Three RNAi Strains and Comparison of the Wild-Type, the RNAi, and the Replacement Strains.
Supplemental Figure 6. β-1,3-Glucan Is Not Masked by Alkali-Soluble Cell Wall Appositions.
Supplemental Figure 7. Formation of Asexual Spores by the C. graminicola Wild-Type and Independent Class I and II RNAi Strains.
Supplemental Figure 8. Construction of Biotrophy- and Necrotrophy-Specific Promoter:eGFP Fusions and Confirmation of the Integration of a Single Copy of the Different Constructs into the Genome.
Supplemental Figure 9. Expression of the Biotrophy-Specific PNmrA:eGFP Reporter Construct in the Wild Type and a Class I RNAi Strain of C. graminicola.
Supplemental Figure 10. Forced Expression of GLS1 by Promoter Exchange and Ectopic Introduction of an Additional Copy of GLS1 Controlled by Constitutive Promoters and Comparison of the Wild-Type Strain and Transformants.
Supplemental Figure 11. β-1,3-Glucan–Induced Defense Responses in Maize Leaves.
Supplemental Figure 12. Analysis of the Transcriptional Response of Z. mays to Infection by C. graminicola Wild-Type and PtrpC:GLS1 Strains by qRT-PCR.
Supplemental Data Set 1. Transcript Abundance of Genes More Than 2.5-Fold Upregulated in Maize Leaves Infected by PtrpC:GLS1 Strains Compared with Wild Type–Infected Leaves.
Supplemental Data Set 2. Transcript Abundance of Genes More Than 2.5-Fold Downregulated in Maize Leaves Infected by PtrpC:GLS1 Strains Compared with Wild Type–Infected Leaves.
Supplemental Data Set 3. PCR Primers Used in This Study.
Acknowledgments
We thank the Deutscher Akademischer Austauschdienst (Grant A/08/71733 to E.O.-G.) for financial support. The skillful technical assistance of Andrea Beutel, Doris Jany, and Elke Vollmer (Martin-Luther-University Halle-Wittenberg, Germany) is highly appreciated. We thank Bettina Tudzynski (Westfälische Wilhelms-Universität Münster, Germany) for providing plasmid pNR1, Stefanie Pöggeler and Ulrich Kück (Ruhr-Universität Bochum, Germany) for providing plasmid pSM1, Amir Sharon (Tel Aviv University, Israel) for providing pSH1.6EGFP, Axel Mithöfer (Max-Planck-Institute for Chemical Ecology, Jena, Germany) for providing β-1,3-glucan oligomers, and Darja Deising for artwork (Figure 10). We also thank Edgar Peiter and Kathrin Thor (Martin-Luther-University Halle-Wittenberg) for help with microscopy and stimulating discussions as well as Gero Steinberg (University of Exeter, UK) and Ralf Horbach and Ivo Schliebner (Martin-Luther-University Halle-Wittenberg) for critically reading the article.
AUTHOR CONTRIBUTIONS
E.O.-G. and H.B.D. designed the experiments, E.O.-G. performed research. E.O.-G. and H.B.D. analyzed data and wrote the article.
Glossary
- GLS
β-1,3-glucan synthase
- RNAi
RNA interference
- PAMP
pathogen-associated molecular pattern
- qRT-PCR
quantitative RT-PCR
- HAI
hours after inoculation
- qPCR
quantitative PCR
- DP
degree of polymerization
- OMA
oatmeal agar
- PEG 6000
polyethylene glycol 6000
References
- Afzal A.J., Wood A.J., Lightfoot D.A. (2008). Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 21: 507–517 [DOI] [PubMed] [Google Scholar]
- Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S.R., Clavaud C., Paris S., Brakhage A.A., Kaveri S.V., Romani L., Latgé J.-P. (2009). Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460: 1117–1121 [DOI] [PubMed] [Google Scholar]
- Almagro L., Gómez Ros L.V., Belchi-Navarro S., Bru R., Ros Barceló A., Pedreño M.A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60: 377–390 [DOI] [PubMed] [Google Scholar]
- Andrie R.M., Martinez J.P., Ciuffetti L.M. (2005). Development of ToxA and ToxB promoter-driven fluorescent protein expression vectors for use in filamentous ascomycetes. Mycologia 97: 1152–1161 [DOI] [PubMed] [Google Scholar]
- Barber M.S., Bertram R.E., Ride J.P. (1989). Chitin oligosaccharides elicit lignification in wounded wheat leaves. Physiol. Mol. Plant Pathol. 34: 3–12 [Google Scholar]
- Bastmeyer M., Deising H.B., Bechinger C. (2002). Force exertion in fungal infection. Annu. Rev. Biophys. Biomol. Struct. 31: 321–341 [DOI] [PubMed] [Google Scholar]
- Beauvais A., Bruneau J.M., Mol P.C., Buitrago M.J., Legrand R., Latgé J.P. (2001). Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 183: 2273–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger C., Giebel K.-F., Schnell M., Leiderer P., Deising H.B., Bastmeyer M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285: 1896–1899 [DOI] [PubMed] [Google Scholar]
- Becker, D.M., and Lundblad, V. (2001). Introduction of DNA into yeast cells. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.17.11–13.17.10. [DOI] [PubMed] [Google Scholar]
- Behr M., Humbeck K., Hause G., Deising H.B., Wirsel S.G.R. (2010). The hemibiotroph Colletotrichum graminicola locally induces photosynthetically active green islands but globally accelerates senescence on aging maize leaves. Mol. Plant Microbe Interact. 23: 879–892 [DOI] [PubMed] [Google Scholar]
- Bernard M., Latgé J.-P. (2001). Aspergillus fumigatus cell wall: Composition and biosynthesis. Med. Mycol. 39 (Suppl 1): 9–17 [PubMed] [Google Scholar]
- Bhadauria V., Banniza S., Vandenberg A., Selvaraj G., Wei Y. (2011). Cataloging proteins putatively secreted during the biotrophy-necrotrophy transition of the anthracnose pathogen Colletotrichum truncatum. Plant Signal. Behav. 6: 1457–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.M., Free S.J. (2006). The structure and synthesis of the fungal cell wall. Bioessays 28: 799–808 [DOI] [PubMed] [Google Scholar]
- Brush L., Money N.P. (1999). Invasive hyphal growth in Wangiella dermatitidis is induced by stab inoculation and shows dependence upon melanin biosynthesis. Fungal Genet. Biol. 28: 190–200 [DOI] [PubMed] [Google Scholar]
- Chai L.Y., Netea M.G., Sugui J., Vonk A.G., van de Sande W.W., Warris A., Kwon-Chung K.J., Kullberg B.J. (2010). Aspergillus fumigatus conidial melanin modulates host cytokine response. Immunobiology 215: 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida T., Sisler H.D. (1987). Restoration of appressorial penetration ability by melanin precursors in Pyricularia oryzae treated with anti-penetrants and in melanin-deficient mutants. J. Pest Sci. 12: 49–55 [Google Scholar]
- Chirgwin J.M., Przybyla A.E., MacDonald R.J., Rutter W.J. (1979). Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18: 5294–5299 [DOI] [PubMed] [Google Scholar]
- Cosio E.G., Feger M., Miller C.J., Antelo L., Ebel J. (1996). High affinity binding of fungal β-glucan elicitors to cell membranes of species of the plant family Fabaceae. Planta 200: 92–99 [Google Scholar]
- De Groot P.W.J., Ram A.F., Klis F.M. (2005). Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42: 657–675 [DOI] [PubMed] [Google Scholar]
- de Jonge R., Thomma B.P.H.J. (2009). Fungal LysM effectors: Extinguishers of host immunity? Trends Microbiol. 17: 151–157 [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H.P., Kombrink A., Shinya T., Desaki Y., Bours R., van der Krol S., Shibuya N., Joosten M.H.A.J., Thomma B.P.H.J. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953–955 [DOI] [PubMed] [Google Scholar]
- Deising H.B., Werner S., Wernitz M. (2000). The role of fungal appressoria in plant infection. Microbes Infect. 2: 1631–1641 [DOI] [PubMed] [Google Scholar]
- Deller S., Hammond-Kosack K.E., Rudd J.J. (2011). The complex interactions between host immunity and non-biotrophic fungal pathogens of wheat leaves. J. Plant Physiol. 168: 63–71 [DOI] [PubMed] [Google Scholar]
- Ecker M., Deutzmann R., Lehle L., Mrsa V., Tanner W. (2006). Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 281: 11523–11529 [DOI] [PubMed] [Google Scholar]
- El Gueddari N.E., Rauchhaus U., Moerschbacher B.M., Deising H.B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156: 103–112 [Google Scholar]
- Felix G., Regenass M., Boller T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Fliegmann J., Mithöfer A., Wanner G., Ebel J. (2004). An ancient enzyme domain hidden in the putative β-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 279: 1132–1140 [DOI] [PubMed] [Google Scholar]
- Freytag S., Mendgen K. (1991). Surface carbohydrates and cell wall structure of in vitro -induced uredospore infection structures of Uromyces viciae-fabae before and after treatment with enzymes and alkali. Protoplasma 161: 94–103 [Google Scholar]
- Fujikawa T., Kuga Y., Yano S., Yoshimi A., Tachiki T., Abe K., Nishimura M. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73: 553–570 [DOI] [PubMed] [Google Scholar]
- Fujikawa T., Sakaguchi A., Nishizawa Y., Kouzai Y., Minami E., Yano S., Koga H., Meshi T., Nishimura M. (2012). Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8: e1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y.S., Covert S.F., Momany M. (2006). FsFKS1, the 1,3-beta-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryot. Cell 5: 1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J.M., Butler M.J., Day A.W. (1999). The dark side of the mycelium: Melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37: 447–471 [DOI] [PubMed] [Google Scholar]
- Horbach R., Navarro-Quesada A.R., Knogge W., Deising H.B. (2011). When and how to kill a plant cell: Infection strategies of plant pathogenic fungi. J. Plant Physiol. 168: 51–62 [DOI] [PubMed] [Google Scholar]
- Horbach R., Graf A., Weihmann F., Antelo L., Mathea S., Liermann J.C., Opatz T., Thines E., Aguirre J., Deising H.B. (2009). Sfp-type 4′-phosphopantetheinyl transferase is indispensable for fungal pathogenicity. Plant Cell 21: 3379–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R.J., Ferrari M.A., Roach D.H., Money N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88: 11281–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard R.J., Valent B. (1996). Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50: 491–512 [DOI] [PubMed] [Google Scholar]
- Huffaker A., Kaplan F., Vaughan M.M., Dafoe N.J., Ni X., Rocca J.R., Alborn H.T., Teal P.E., Schmelz E.A. (2011). Novel acidic sesquiterpenoids constitute a dominant class of pathogen-induced phytoalexins in maize. Plant Physiol. 156: 2082–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison K.A., Green J.R., Wharton P.S., O'Connell R.J. (2002). Identification and localisation of glycoproteins in the extracellular matrices around germ-tubes and appressoria of Colletotrichum species. Mycol. Res. 106: 729–736 [Google Scholar]
- Janus D., Hoff B., Hofmann E., Kück U. (2007). An efficient fungal RNA-silencing system using the DsRed reporter gene. Appl. Environ. Microbiol. 73: 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-E., Lee H.-J., Lee J., Kim K.W., Yun S.-H., Shim W.-B., Lee Y.-W. (2009). Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 55: 449–459 [DOI] [PubMed] [Google Scholar]
- Klarzynski O., Plesse B., Joubert J.-M., Yvin J.-C., Kopp M., Kloareg B., Fritig B. (2000). Linear β-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann J., Rincon-Rivera L.J., Takahara H., Neumann U., Ver Loren van Themaat E., van der Does H.C., Hacquard S., Stüber K., Will I., Schmalenbach W., Schmelzer E., O’Connell R.J. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8: e1002643 Erratum. PLos Pathog. 8:/10.1371/annotation/0f398a0c-dfda-4277-b172-4ff9cb31aec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L.-A., Yang, J., Li, G.-T., Qi, L.-L., Zhang, Y.-J., Wang, C.-F., Zhao, W.-S., Xu, J.-R., and Peng, Y.-L. (2012). Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae PLoS Pathog. 8: e1002526. [DOI] [PMC free article] [PubMed]
- Krijger J.-J., Horbach R., Behr M., Schweizer P., Deising H.B., Wirsel S.G.R. (2008). The yeast signal sequence trap identifies secreted proteins of the hemibiotrophic corn pathogen Colletotrichum graminicola. Mol. Plant Microbe Interact. 21: 1325–1336 [DOI] [PubMed] [Google Scholar]
- Kubo Y., Nakamura H., Kobayashi K., Okuno T., Furusawa I. (1991). Cloning of a melanin biosynthetic gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant Microbe Interact. 4: 440–445 [DOI] [PubMed] [Google Scholar]
- Kück U., Hoff B. (2010). New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 86: 51–62 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Larson T.M., Kendra D.F., Busman M., Brown D.W. (2011). Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr. Genet. 57: 177–189 [DOI] [PubMed] [Google Scholar]
- Latgé J.P. (2007). The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 66: 279–290 [DOI] [PubMed] [Google Scholar]
- Latgé J.-P. (2010). Tasting the fungal cell wall. Cell. Microbiol. 12: 863–872 [DOI] [PubMed] [Google Scholar]
- Leach J., Lang B.R., Yoder O.C. (1982). Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J. Gen. Microbiol. 128: 1719–1729 [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kauffman S., Becker J.M., Szaniszlo P.J. (2004). Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell 3: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid M.P., Di Pietro A., Roncero M.I. (2003). Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47: 257–266 [DOI] [PubMed] [Google Scholar]
- Malonek S., Rojas M.C., Hedden P., Gaskin P., Hopkins P., Tudzynski B. (2004). The NADPH-cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem. 279: 25075–25084 [DOI] [PubMed] [Google Scholar]
- Maubon D., Park S., Tanguy M., Huerre M., Schmitt C., Prévost M.C., Perlin D.S., Latgé J.-P., Beauvais A. (2006). AGS3, an alpha(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 43: 366–375 [DOI] [PubMed] [Google Scholar]
- Mendgen K., Deising H. (1993). Infection structures of fungal plant pathogens - A cytological and physiological evaluation. New Phytol. 124: 193–213 [DOI] [PubMed] [Google Scholar]
- Mentlak T.A., Kombrink A., Shinya T., Ryder L.S., Otomo I., Saitoh H., Terauchi R., Nishizawa Y., Shibuya N., Thomma B.P.H.J., Talbot N.J. (2012). Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24: 322–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Money N.P., Hill T.W. (1997). Correlation between endoglucanase secretion and cell wall strength in oomycete hyphae: Implications for growth and morphogenesis. Mycologia 89: 777–785 [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mouyna I., Henry C., Doering T.L., Latgé J.P. (2004). Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 237: 317–324 [DOI] [PubMed] [Google Scholar]
- Nicholson, R.L., and Epstein, L. (1991). Adhesion of fungi to the plant surface. In The Fungal Spore and Disease Initiation in Plants and Animals, G.T. Cole and H.C. Hoch, eds (New York, London: Plenum Press), pp. 3–23. [Google Scholar]
- Nürnberger T., Brunner F., Kemmerling B., Piater L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198: 249–266 [DOI] [PubMed] [Google Scholar]
- O’Connell R.J., Pain N.A., Hutchison K.A., Jones G.L., Green J.R. (1996). Ultrastructure and composition of the cell surfaces of infection structures formed by the fungal plant pathogen Colletotrichum lindemuthianum. J. Microsc. 181: 204–212 [Google Scholar]
- O’Connell R.J., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathuri I.P., Reitberger I.E., Hückelhoven R., Proels R.K. (2011). Alcohol dehydrogenase 1 of barley modulates susceptibility to the parasitic fungus Blumeria graminis f.sp. hordei. J. Exp. Bot. 62: 3449–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S., Masloff S., Hoff B., Mayrhofer S., Kück U. (2003). Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43: 54–61 [DOI] [PubMed] [Google Scholar]
- Politis D.J., Wheeler H. (1973). Ultrastructural study of the penetration of maize leaves by Colletotrichum graminicola. Physiol. Plant Pathol. 3: 465–471 [Google Scholar]
- Postel S., Kemmerling B. (2009). Plant systems for recognition of pathogen-associated molecular patterns. Semin. Cell Dev. Biol. 20: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J. (1991). Biosynthesis of beta-glucans in fungi. Antonie van Leeuwenhoek 60: 72–81 [DOI] [PubMed] [Google Scholar]
- Schmelz E.A., Kaplan F., Huffaker A., Dafoe N.J., Vaughan M.M., Ni X., Rocca J.R., Alborn H.T., Teal P.E. (2011). Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 108: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty N.P., Jensen J.D., Knudsen A., Finnie C., Geshi N., Blennow A., Collinge D.B., Jørgensen H.J.L. (2009). Effects of β-1,3-glucan from Septoria tritici on structural defence responses in wheat. J. Exp. Bot. 60: 4287–4300 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sietsma J.H., Sonnenberg A.M.S., Wessels J.G.H. (1985). Localization by autoradiography of synthesis of (1∅3)-β and (1∅6)-β linkages in a wall glucan during hyphal growth of Schizophyllum commune. J. Gen. Microbiol. 131: 1331–1337 [Google Scholar]
- Söderström B.E. (1977). Vital staining of fungi in pure cultures and in soil with fluorescein diacetate. Soil Biol. Biochem. 9: 59–63 [Google Scholar]
- Soulié M.C., Perino C., Piffeteau A., Choquer M., Malfatti P., Cimerman A., Kunz C., Boccara M., Vidal-Cros A. (2006). Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell. Microbiol. 8: 1310–1321 [DOI] [PubMed] [Google Scholar]
- Stoldt V.R., Sonneborn A., Leuker C.E., Ernst J.F. (1997). Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16: 1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara H., Dolf A., Endl E., O’Connell R. (2009). Flow cytometric purification of Colletotrichum higginsianum biotrophic hyphae from Arabidopsis leaves for stage-specific transcriptome analysis. Plant J. 59: 672–683 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treco, D.A., and Lundblad, V. (1993). Basic Techniques of Yeast Genetics. (New York: John Wiley & Sons). [Google Scholar]
- Treitschke S., Doehlemann G., Schuster M., Steinberg G. (2010). The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22: 2476–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse H.P., Bolton M.D., Stergiopoulos I., de Wit P.J.G.M., Thomma B.P.H.J. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20: 1092–1101 [DOI] [PubMed] [Google Scholar]
- Vander P., V rum K.M., Domard A., Eddine El, Gueddari N, Moerschbacher B.M. (1998). Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118: 1353–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K., Kastner C., Kumlehn J., Kahmann R., Doehlemann G. (2011). Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol. 189: 471–483 [DOI] [PubMed] [Google Scholar]
- Vargas W.A., Martín J.M., Rech G.E., Rivera L.P., Benito E.P., Díaz-Mínguez J.M., Thon M.R., Sukno S.A. (2012). Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 158: 1342–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Somerville S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97: 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Munro C.A., de Bruijn I., Lenardon M.D., McKinnon A., Gow N.A.R. (2008). Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4: e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I., Assmann D., Thines E., Steinberg G. (2006). Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Sugui J.A., Steinberg G., Deising H.B. (2007). A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Microbe Interact. 20: 1555–1567 [DOI] [PubMed] [Google Scholar]
- Wessels J.G.H. (1993). Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 123: 397–413 [DOI] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Yamada A., Hong N., Ogawa T., Ishii T., Shibuya N. (2000). Differences in the recognition of glucan elicitor signals between rice and soybean: beta-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. Plant Cell 12: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Hamari Z., Han K.H., Seo J.A., Reyes-Domínguez Y., Scazzocchio C. (2004). Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981 [DOI] [PubMed] [Google Scholar]
- Zimmermann G., Bäumlein H., Mock H.P., Himmelbach A., Schweizer P. (2006). The multigene family encoding germin-like proteins of barley. Regulation and function in basal host resistance. Plant Physiol. 142: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]