Abstract
We previously reported molecular karyotype analysis of invasive breast tumour core needle biopsies by comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH) (Walker et al, Genes Chromosomes Cancer, 2008 May;47(5):405-17). That study identified frequently recurring gains and losses involving chromosome bands 8q22 and 8p21, respectively. Moreover, these data highlighted an association between 8q22 gain and typically aggressive grade 3 tumors. Here we validate and extend our previous investigations through FISH analysis of tumor touch imprints prepared from excised breast tumor specimens. Compared to post-surgical tumor excisions, core needle biopsies are known to be histologically less precise when predicting tumor grade. Therefore investigating these chromosomal aberrations in tumor samples that offer more reliable pathological assessment is likely to give a better overall indication of association. A series of 60 breast tumors were screened for genomic copy number changes at 8q22 and 8p21 by dual-color FISH. Results confirm previous findings that 8p loss (39%) and 8q gain (74%) occur frequently in invasive breast cancer. Both absolute quantification of 8q22 gain across the sample cohort, and a separate relative assessment by 8q22:8p21 copy number ratio, showed that the incidence of 8q22 gain significantly increased with grade (p = 0.004, absolute and p = 0.02, relative). In contrast, no association was found between 8p21 loss and tumor grade. These findings support the notion that 8q22 is a region of interest for invasive breast cancer pathogenesis, potentially harboring one or more genes that, when amplified, precipitate the molecular events that define high tumor grade.
Introduction
Despite recent advances in our understanding of the molecular basis of breast cancer, classical histological grading of breast cancer remains prominent in routine histopathological practice [1], [2], [3]. This is because pathological assessment of tumour grade offers a rapid and relatively inexpensive measure of tumor cell proliferation, differentiation and overall disease aggressiveness, assisting the clinical ascertainment of risk of recurrence and choice of adjuvant therapies through such algorithms as the Nottingham Prognostic Index [4]. Histological grade is established after microscopic evaluation of paraffin-embedded haematoxylin and eosin stained sections, and is typically represented by nuclear morphology, the number of mitoses and the degree of tubule formation. Patients with well differentiated (grade 1) tumors have significantly better survival than patients with poorly differentiated (grade 3) tumors [5]. Although routinely applied, issues of interobserver variability in the assessment of histological grade are a well recognised and ongoing challenge [6], [7], [8], [9], [10]. There is a need for advancement in the accuracy and reproducibility of routinely applied histopathological tools for better refined breast cancer diagnosis and prognosis.
Genome-wide profiling technologies have contributed much to current understanding of the association between breast cancer genotype and phenotype. Information learned from these technologies is progressively challenging the way in which conventional pathology protocols are applied, with an overall objective to stratify breast cancer patients at the time of diagnosis into more effective clinical risk groups for better targeted treatment interventions [1], [2], [3], [11], [12], [13]. However, the clinical translation of newly identified biologically relevant gene markers, including screening methods to allow their detection, in many cases requires further validative research.
Since development in 1992 by Kallioniemi and colleagues [14], metaphase comparative genomic hybridization (mCGH) and subsequent high-resolution array CGH (aCGH) adaptations [15], [16] have been widely applied to the interrogation of genomic copy number imbalances as they occur in breast cancer. Recurrent patterns of chromosomal loss and gain have been shown to associate with different histopathological subtypes, and their functional relevance assessed in turn by correlation with global gene expression signatures [12], [17], [18], [19], [20], [21], [22], [23], [24]. Amplification of discrete genomic regions in human breast carcinomas highlighted by these methodologies has resulted in the refined characterization of specific genes associated with breast tumourigenesis, including MYC at 8q24, CCND1 at 11q13 and ERBB2 at 17q12 [24], [25], [26], [27].
Our previously reported mCGH analysis of 42 diagnostic core needle biopsies from primary invasive carcinoma of the breast showed that copy number gain or amplification involving chromosome 8q occurred in 64% of all tumors, and selectively in 84% of grade 3 tumors [28]. These findings are in agreement with previous reports using mCGH that identified 8q gain in 41–65% of post-surgical breast tumor specimens [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], and in 68%–90% of grade 3 tumors [33], [34], [37], [40]. The patterns of 8q gain are not identical between different tumors, with some regions affected more frequently than others. Our study identified a recurring region of gain on chromosome 8 at band q22 (8q22), a finding of interest because it overlaps with common regions of gain reported independently by others, including 8q21–q23 [35], 8q21-qter [36], [37], [38], [39], [40], [41], [42], 8q22-qter [32] and 8q22–q23 [29], [31], [34], [41]. Higher resolution microarray studies using BAC and oligonucleotide platforms have in some cases allowed better definition of the precise genomic co-ordinates of regions of 8q gain, although the widely differing platforms, sampling characteristics and statistical algorithms for data analysis that have been applied have made consensus difficult [19], [20], [21], [22], [23], [43], [44], [45], [46], [47], [48], [49], [50]. Copy number gain at 8q22 has also been shown to correlate with increased expression levels of several genes across this region, including MTDH, LAPTM4B and YWHAZ, and associated with poor clinical outcome for breast cancer patients [51], [52].
In contrast to 8q, the short arm of chromosome 8 (8p) is reported to show copy number loss in breast tumors of high grade or in association with worse prognosis. However, the frequency of 8p loss is lower than for 8q gain overall (range 18–44%) [28], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], and in the high-grade tumor subsets (range 42–74%) [28], [33], [34], [37]. mCGH studies have identified various minimal regions of deletion, including 8p12-pter [31], 8p21-pter [42], 8p22-pter [34], [36], 8p21–p22 [28], [38] and 8p22–p23 [35], [40], [41]. Subsequent high-resolution profiling studies have confirmed that loss of 8p, from 8p12 to the telomere, sometimes with proximal amplification of 8p11–12, constitute the most frequent copy number alterations [22], [50], [53].
Our earlier described mCGH study of diagnostic core biopsies suggested significant association between 8q22 gain, 8p21 loss and grade 3 breast cancer. Subsequent FISH analysis of tumor touch imprints prepared from a subgroup of the same biopsies supported these findings [28]. For the present study, tumor touch imprints were prepared from a different larger cohort of 60 post-operative invasive breast tumor specimens, which offer the advantage of more precise scoring of histological grade from corresponding paraffin sections. Results confirm significant association of 8q22 gain with high-grade breast cancer, reinforcing need for future focussed interrogation of genomic features in this region.
Materials and Methods
Ethics Statement
All patients provided written consent to gift their tumor tissue to the Cancer Society Tissue Bank for future research. The use of banked samples for this research was approved by the Canterbury Ethics Committee.
Breast Tumor Samples
Tumor touch imprints were prepared from resected tumor samples of 60 breast cancer patients presenting for surgery at Christchurch Hospital. Pathological review was carried out as described previously [28]. This assessment includes macroscopic and microscopic measurement of tumor size, and grading according to a modified Bloom and Richardson protocol [5]. Immunohistochemistry was applied to determine hormone receptor status (ESR1, PGR) [H-score] [54], [55] and ERBB2 (HER-2) status (HercepTest, Dakocytomation, Glostrup). Any lymph nodes removed during surgery were examined macroscopically and microscopically for metastatic tumor infiltration.
Tumor Touch Slide Imprints
Touch slide imprints were prepared by briefly blotting surgically excised tissue on lint-free paper to remove excess moisture and then firmly pressing tissue samples against four separate pre-cleaned microscope slides. Cells were air dried and then fixed by soaking the slides in methanol:acetic acid (3∶1) for 20 minutes, air dried and stored at −20°C with desiccant.
Fluorescence in situ Hybridization
Bacterial artificial chromosome (BAC) DNA probes RP11-177H13 (8p21) and RP11-10G10 (8q22) contain sequences that map to human chr8:23,051,329-23,230,640 and chr8:101,209,852-101,362,596 (GRCh37/hg19 build), respectively, and were selected from a replicated 1 Mb Wellcome Trust Sanger Institute clone set [56] based on their central location within the 8p21 and 8q22 cytobands of interest [28]. RP11-177H13 spans coding domains of several genes, including TNFRSF10A, CHMP7, R3HCC1 and LOXL2. BAC RP11-10G10 contains SPAG1 and RNF19A. These two BAC clones were selected because of their central location within frequently lost or gained regions of interest at cytobands 8p21 and 8q22, respectively [28]. FISH methods, including probe preparation, slide preparation, hybridization and post-hybridization washes were performed as described [28]. Briefly, cells from tumor touch imprints were treated with pepsin, paraformaldehyde fixed, and genomic DNA was denatured by immersion for 4 min at 72–74°C in a solution of 70% formamide/2×SSC (pH 7.0) immediately prior to hybridization. BAC probes RP11-177H13 and RP11-10G10 were labeled by nick-translation using Spectrum Red and Spectrum Green (Vysis, Downers Grove, IL, USA), respectively. Labelled probes were hybridized at a concentration of 40–50 ng/µl in a 10 µl mix containing 50% formamide, 2×SSC, 10% dextran sulphate, 2 µg/µl herring sperm DNA and 1.5 µg/µl Cot-1 DNA. The probe mix was denatured at 75°C for 5 min, and allowed to preanneal at 37°C for 30 min, then immediately applied, under coverslip, to the denatured touch preparations and hybridized overnight at 37°C in a humidified chamber. Post-hybridization washes were carried out by washing two times, 5 min each, in 2×SSC (pH 7) at 45°C, followed by two 5 min washes in 50% formamide/2×SSC (pH 7) at 45°C, then two further washes, 5 min each, in 2×SSC at 45°C, and a final 15 min wash in 0.1×SSC at 60°C.
FISH Data Acquisition and Analysis
Following post-hybridization washes, slides were mounted in glycerol containing the antifade agent p-phenylene-diamine dihydrochloride (Sigma, 1 mg/ml) and 0.25 mg/ml DAPI (4′,6-diamidino-2-phenylindole) as counterstain for G-band visualization. Fluorescence images from interphase cells were digitally captured at selective bandwidths using a Leitz Aristoplan microscope equipped with a Photometrics KAF1400 CCD camera and QUIPS Smartcapture software (version 1.3; Vysis Inc, Downers Grove, IL). Areas with well-separated nuclei and overall good hybridization signal were selected for analysis by fluorescent microscopy. For each case, red (RP11-177H3, 8p21) and green (RP11-10G10, 8q22) fluorescent signals were counted at 100× magnification in at least 50 non-overlapping interphase nuclei. Two different approaches were applied to define chromosomal copy number changes. In the first analysis, absolute copy number gains and losses of 8p21 and 8q22 were scored as positive if more than 10% of analyzable nuclei harbored a copy number change. A signal score of 12 was assigned to nuclei displaying amplification of 8q22 when FISH signals were unable to be precisely enumerated. In the second approach, relative copy number for 8q22 was determined by calculating the mean copy number for RP11-10G10 specific signals and dividing by the respective value for RP11-177H13 at 8p21. To avoid the possibility that a greater green to red signal ratio was due solely to a deletion of chromosome 8p21, chromosome 8q22 gain was defined as relative 8q22/8p21 signal ratio greater than 2.0.
Statistical Analysis
For cross-tabulation of categorical variables two-sided mid-p Fisher's exact test was used or, where appropriate, the Extended Mantel-Haenzel chi square test for linear trend[57]. Survival analyses were carried out in SAS 9.2 (www.SAS.com) using the log-rank test, with and without stratification, or proportional hazards models.
Results
Patient Sample Characteristics
Tumor histopathology of the 60 breast tumor samples imprinted for FISH analysis in this study is summarized in Supporting Information Table S1 along with patient lymph node status. Fifty six of the 60 tumors were histopathologically classified as IDC and four were classified as invasive lobular carcinoma (ILC) (Supporting Information Table S1). Fifteen samples were classified as grade 1, 16 as grade 2, and 29 as grade 3. Tumor immunophenotype was determined for ESR1 (44 positive, 16 negative), PGR (35 positive, 25 negative), and ERBB2 (19 scored as negative or 1+, 11 scored as 2+, 11 scored as 3+ and 19 were not assayed). Lymph node status was confirmed in 56 samples, and 28 of these patients were found to have lymphatic spread of the disease (Supporting Information Table S1). Of the 15 grade 1 tumors, all were ESR1 positive and one of 10 tumors examined for ERBB2 expression showed 3+ staining. In comparison, 15 of the 29 grade 3 tumors (52%) were ESR1 negative and eight of the 24 tumors analysed for ERBB2 expression (33%) were characterized by 3+ staining.
Assessment of 8p21 and 8q22 Copy Number Status
BAC clones RP11-177H13 and RP11-10G10 hybridized cleanly to tumor nuclei in 54 of 60 breast tumor touch preparations. Data derived from the FISH analysis are detailed in Supporting Information Table S2 and summarized in Table 1. Of the 54 samples, the total cells analyzed ranged from 50 to 232 (Table 1). For the remaining six preparations (BT70, BT80, BT93, BT144, BT150 and BT172), analyzable cells were <50 (range 15–44) (Supporting Information Table S2), and low signal intensity was a complicating factor in two of these samples (BT93 and BT172). Data corresponding to the latter six tumors were therefore excluded from further analysis.
Table 1. Summary of signal patterns observed after FISH of the 8p21- and 8q22-band specific BAC probes, RP11-177H13 and RP11-10G10, respectively, to 54 breast tumor touch preparations.
| Case | Histological Type | Grade | Total cells analysed | Nuclei counts | 8q22/8p21 ratio | |||
| >2 copies of 8p21 | <2 copies of 8p21 | >2 copies of 8q22 | <2 copies of 8q22 | |||||
| BT9 | IDC | 1 | 103 | 0 | 81 | 5 | 2 | 1.68 |
| BT38 | IDC | 1 | 202 | 25 | 0 | 25 | 0 | 1.00 |
| BT39 | IDC | 1 | 206 | 11 | 1 | 5 | 8 | 0.97 |
| BT43 | IDC | 1 | 104 | 3 | 1 | 2 | 25 | 0.88 |
| BT45 | IDC | 1 | 100 | 0 | 82 | 0 | 15 | 1.57 |
| BT63 | IDC | 1 | 123 | 0 | 14 | 83 | 0 | 2.40 |
| BT66 | IDC | 1 | 95 | 53 | 0 | 53 | 0 | 1.00 |
| BT67 | IDC | 1 | 216 | 0 | 0 | 5 | 1 | 1.01 |
| BT69 | IDC | 1 | 203 | 3 | 0 | 3 | 0 | 1.00 |
| BT83 | IDC | 1 | 111 | 103 | 0 | 103 | 0 | 1.00 |
| BT207 | IDC | 1 | 63 | 4 | 5 | 62 | 0 | 3.88 |
| BT7* | IDC | 2 | 115 | 3 | 46 | 107 | 0 | 3.36 |
| BT19 | IDC | 2 | 102 | 98 | 0 | 99 | 0 | 1.03 |
| BT44 | IDC | 2 | 211 | 37 | 0 | 34 | 0 | 0.99 |
| BT52 | IDC | 2 | 161 | 31 | 0 | 36 | 0 | 1.01 |
| BT56* | IDC | 2 | 122 | 1 | 94 | 110 | 0 | 2.66 |
| BT68 | IDC | 2 | 154 | 0 | 137 | 8 | 1 | 1.85 |
| BT71 | IDC | 2 | 123 | 48 | 1 | 122 | 0 | 3.31 |
| BT109 | IDC | 2 | 116 | 94 | 0 | 95 | 0 | 1.06 |
| BT146 | IDC | 2 | 102 | 0 | 92 | 2 | 0 | 1.84 |
| BT173 | IDC | 2 | 152 | 2 | 0 | 122 | 0 | 3.20 |
| BT193 | IDC | 2 | 207 | 2 | 35 | 1 | 5 | 1.08 |
| BT208* | IDC | 2 | 111 | 0 | 91 | 107 | 0 | 2.89 |
| BT215 | IDC | 2 | 109 | 66 | 0 | 66 | 0 | 1.00 |
| BT2 | IDC | 3 | 108 | 59 | 3 | 74 | 1 | 3.35 |
| BT5 | IDC | 3 | 102 | 1 | 7 | 65 | 1 | 1.37 |
| BT30 | IDC | 3 | 142 | 50 | 48 | 118 | 0 | 3.08 |
| BT37* | IDC | 3 | 232 | 0 | 215 | 218 | 4 | 3.16 |
| BT40 | IDC | 3 | 111 | 3 | 0 | 109 | 0 | 3.05 |
| BT42* | IDC | 3 | 114 | 1 | 98 | 98 | 0 | 2.59 |
| BT46 | IDC | 3 | 50 | 2 | 1 | 21 | 3 | 1.24 |
| BT47* | IDC | 3 | 104 | 0 | 77 | 89 | 0 | 2.59 |
| BT57 | IDC | 3 | 111 | 129 | 2 | 87 | 0 | 1.59 |
| BT59 | IDC | 3 | 122 | 1 | 106 | 114 | 0 | 3.52 |
| BT61* | IDC | 3 | 128 | 1 | 107 | 115 | 0 | 2.55 |
| BT72 | IDC | 3 | 109 | 12 | 4 | 89 | 0 | 3.77 |
| BT76 | IDC | 3 | 104 | 4 | 0 | 84 | 0 | 4.76 |
| BT78 | IDC | 3 | 206 | 1 | 1 | 5 | 0 | 1.02 |
| BT91 | IDC | 3 | 113 | 3 | 0 | 86 | 0 | 1.62 |
| BT107 | IDC | 3 | 77 | 19 | 0 | 20 | 2 | 1.07 |
| BT108* | IDC | 3 | 102 | 2 | 66 | 50 | 0 | 2.04 |
| BT117 | IDC | 3 | 128 | 7 | 36 | 91 | 0 | 2.33 |
| BT119* | IDC | 3 | 161 | 0 | 64 | 75 | 0 | 1.62 |
| BT120 | IDC | 3 | 109 | 0 | 70 | 82 | 0 | 5.93 |
| BT139 | IDC | 3 | 89 | 2 | 0 | 2 | 0 | 1.00 |
| BT158 | IDC | 3 | 105 | 5 | 1 | 101 | 0 | 4.64 |
| BT182* | IDC | 3 | 130 | 1 | 58 | 109 | 1 | 2.33 |
| BT184 | IDC | 3 | 101 | 1 | 0 | 76 | 0 | 1.73 |
| BT188 | IDC | 3 | 112 | 0 | 93 | 97 | 0 | 3.95 |
| BT190 | IDC | 3 | 102 | 3 | 2 | 4 | 0 | 1.02 |
| BT191 | IDC | 3 | 101 | 0 | 0 | 98 | 0 | 1.98 |
| BT55 | ILC | 1 | 78 | 0 | 5 | 2 | 76 | 0.57 |
| BT124 | ILC | 2 | 151 | 1 | 0 | 1 | 0 | 1.00 |
| BT31 | ILC | 3 | 119 | 116 | 0 | 119 | 0 | 1.16 |
Abbreviations: IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma. Cases with likely i(8q) are marked with an asterisk.
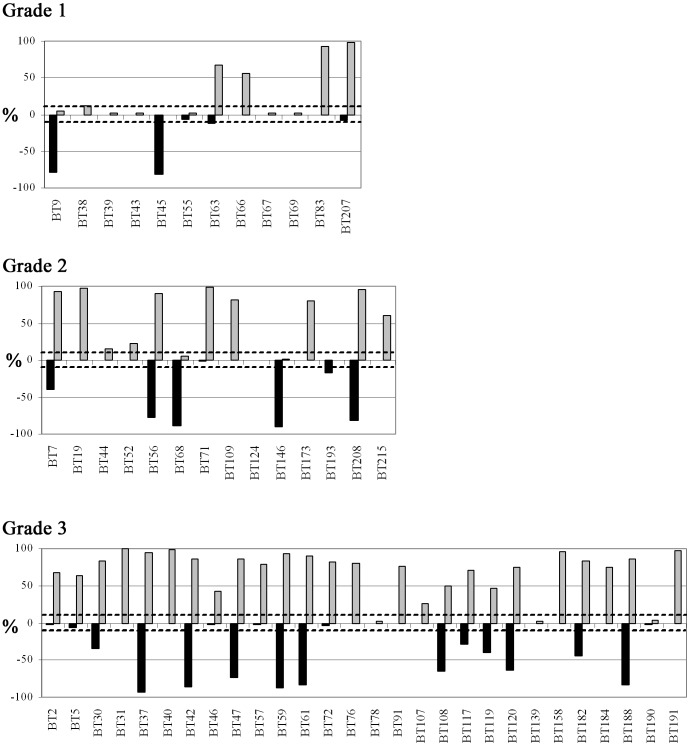
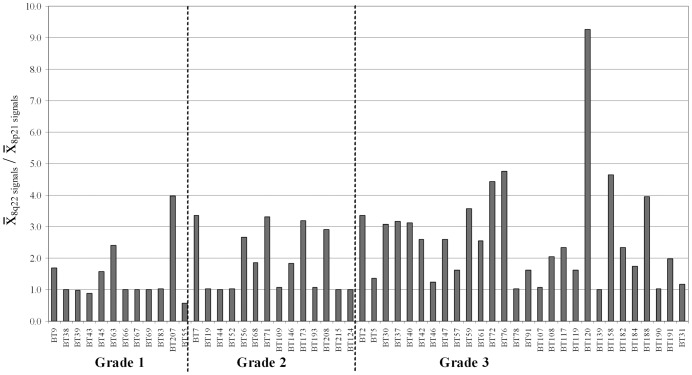
A representative example of signal patterns observed in interphase nuclei of the remaining 54 samples is shown in Supporting Information Figure S1. Absolute gain or loss of 8q22 (RP11-10G10) was detected in 74% (40/54) or 6% (3/54) of these tumors, respectively (Fig. 1). Thirty one percent (17/54) of tumors analyzed also harbored a high level gain of 8q22 (more than 5 copies in greater than 10% of nuclei scored) (Supporting Information Table S2). By comparison, assessment based on 8q22:8p21 copy number ratio revealed 8q22 gains in 43% (23/54) of tumors (Table 1 and Fig. 2).
Figure 1. Proportion of nuclei from 54 breast tumor touch preparations after FISH showing decreased copy number of BAC probe RP11-177H13 (8p21; black bars), and/or increased copy number of RP11-10G10 (8q22; gray bars).
Copy number changes are marked as loss or gain if at least 10% (marked by dashed lines) of nuclei show less than 2 signals from RP11-177H13 or more than 2 signals from RP11-10G10. Tumors are sorted into three graphs according to histological grade.
Figure 2. Relative copy number of 8q22 in 54 breast tumor touch preparations categorized by histological grade.
Relative copy number for 8q22 was determined by calculating the mean copy number for RP11-10G10 (8q22) specific signals and dividing by the respective value for RP11-177H13 at 8p21. Copy number gain is scored positive by a ratio greater than 2.0.
Absolute chromosomal gain or loss at 8p21 (RP11-177H13) was detected in 28% (15/54) or 39% (21/54) of the tumors, respectively (Fig. 1). All 15 tumors showing a gain at 8p21 also harbored a gain at 8q22, although we note variability in the number of nuclei scored for each of these chromosomal imbalances (Table 1). For 11 of these 15 tumor samples, 8q22:8p21 signal ratios <2.0 (range 1.0–1.6; mean 1.1) were consistent with whole chromosome 8 gain, and in nine of these cases (BT19, BT31, BT38, BT44, BT52, BT66, BT83, BT109, BT215), absolute copy number status including number and distribution of 8p21 and 8q22 signals across total cells analyzed supported this interpretation (Supporting Information Table S2, Table 1).
None of the tumors analyzed showed mixed populations of cells having both loss and gain of either 8p21 or 8q22. The formation of an isochromosome involving 8q with one remaining copy of chromosome 8, or alternatively unbalanced translocation(s) or isoderivatives resulting in 8p loss, was suggested in 10 tumor samples (BT7, BT37, BT42, BT47, BT56, BT61, BT108, BT119, BT182 and BT208) for which 31% to 79% of cell nuclei exhibited three copies of 8q22 but only one copy of 8p21 (Supporting Information Table S2). All specimens showing unbalanced 8p/8q signal patterns were scored as higher grade breast tumors (n = 3 grade 2; n = 7 grade 3). A tetraploid (4n) equivalent of the i(8q) signal pattern consistent with two copies of 8p21 and six copies of 8q22 was present in 10–50% of nuclei analyzed in four of these 10 samples (BT7, BT47, BT56 and BT182), and the latter pattern was also observed in samples BT40 and BT63 (Supporting Information Table S2).
For 52 of the 54 cases analyzed in detail, varying numbers of cells showed two signals only for each of the RP11-177H13 and RP11-10G10 probes applied, consistent with normal 8p21 and 8q22 copy number status. In 36 of these 52 cases, the proportion of nuclei that displayed this normal signal pattern ranged from 10%–99% (Supporting Information Table S2). For six cases, normal signal patterns were found in greater than 90% of cells analyzed, whereas in 12 cases these patterns were observed in less than 10% of cells analyzed (Supporting Information Table S2).
8p21 and 8q22 Copy Number Aberrations and Tumor Pathology
Copy number changes involving 8p21 and 8q22 were correlated with tumor pathology and the results are presented in Table 2. When tumors were grouped according to histological grade, the incidence of absolute and relative 8q22 gain significantly increased with grade (p = 0.004 and p = 0.02, respectively) (Table 2). In contrast, there was no association between histological grade and absolute 8p21 copy number change (Table 2). The proportion of tumors that harbored more than five copies of 8q22 was also higher in grade 3 tumors (11/28, 40%) when compared to grade 2 (4/14, 29%) and grade 1 (2/12, 17%) tumors (Supporting Information Table S2). Of the 10 cases with suggested i(8q)/ider(8q)/t(8q;?), 3/10 were classified as grade 2, and 7/10 were classified grade 3 (Supporting Information Table S2). Of six cases showing signal patterns consistent with duplication of one or other of these structural 8q imbalances, one was grade 1, two were grade 2 and three were grade 3 (Supporting Information Table S2, Table 1). In contrast, a higher proportion of grade 1 (3/12; 25%) or grade 2 tumors (5/14, 36%) than grade 3 tumors (1/28; 4%) showed 8q22:8p21 ratio and absolute copy number status consistent with gain of one or more copies of chromosome 8 (Supporting Information Table S2, Table 1). No significant association was seen between copy number changes at 8p21 or 8q22 and tumor type (IDC or ILC), ESR1 status, PGR status, ERBB2 status or nodal involvement (Table 2).
Table 2. Association of 8p21 loss and 8q22 gain with clinicopathologic and histologic features of 54 primary breast carcinomas.
| Pathology feature | 8p21 loss (number) | P | 8q22 gain (number) | P | Relative 8q22 gain (number) | P | |||||
| − | + | − | + | − | + | ||||||
| IDC | n = 51 | 30 | 21 | 12 | 39 | 28 | 23 | ||||
| ILC | n = 3 | 3 | 0 | 0.22a | 2 | 1 | 0.18a | 3 | 0 | 0.18a | |
| Grade | 1 | n = 12 | 9 | 3 | 7 | 5 | 10 | 2 | |||
| 2 | n = 14 | 8 | 6 | 4 | 10 | 9 | 5 | ||||
| 3 | n = 28 | 16 | 12 | 0.64b | 3 | 25 | 0.004b | 12 | 16 | 0.02b | |
| ESR1 | Negative | n = 16 | 11 | 5 | 3 | 13 | 8 | 8 | |||
| Positive | n = 38 | 22 | 16 | 0.48a | 11 | 27 | 0.47a | 23 | 15 | 0.49a | |
| PGR | Negative | n = 24 | 16 | 8 | 5 | 19 | 12 | 12 | |||
| Positive | n = 30 | 17 | 13 | 0.47a | 9 | 21 | 0.47a | 19 | 11 | 0.34a | |
| ERBB2 | 0/1+ | n = 19 | 13 | 6 | 4 | 15 | 11 | 8 | |||
| 2+ | n = 9 | 8 | 1 | 2 | 7 | 8 | 1 | ||||
| 3+ | n = 10 | 3 | 7 | 0.13b | 1 | 9 | 0.68b | 3 | 7 | 0.38b | |
| unknown | n = 16 | ||||||||||
| LN | Negative | n = 25 | 16 | 9 | 8 | 17 | 16 | 9 | |||
| Positive | n = 25 | 15 | 10 | 0.78a | 5 | 20 | 0.36a | 13 | 12 | 0.41a | |
Two-sided mid-p Fisher's exact test;
Extended Mantel-Haenszel chi square for linear trend.
Survival Analysis
Analysis of survival time until death from breast cancer was assessed for the 54 cases from this study and an additional nine cases from our previous study [28] where follow-up data were available. Forty-seven of these 63 cases showed 8q22 gain. All patients were potentially observed for at least five years (Supporting Information Table S3). The hazard ratio for death from breast cancer for those with 8q22 gain was 0.67 (compared with loss/no change) but this was not significant (p = 0.29). Gain was more common at high grades (39%, 71%, 91% across grades 1–3, respectively (p = 0.0003), but stratification by grade had little effect on the comparison of gain or no-gain (p = 0.20). Patients without 8q22 gain tended to be older than those with gain: 72.1 years (SD = 11.7) compared with 60.2 years (SD = 11.7), p = 0.01 (Supporting Information Table S3). The p-values for gain versus no-gain were slightly higher after accounting for age whether by stratification into those under or over 65 (p = 0.36) or by inclusion of age in a proportional hazards model (p = 0.46). With only 32 deaths, further exploration of these two possible confounders (grade and age) was not possible.
Discussion
Our previous interphase FISH analysis of core needle biopsy touch imprints had confirmed mCGH findings for the same sample set to show that 8q22 copy number changes occur frequently in breast cancer and are significantly associated with grade 3 tumors [28]. The present study extends this earlier work through application of FISH and an identical 8p21/8q22 probe set to tumor touch imprints prepared from a larger series of histologically more precise excised breast tumor biopsies. Results detailed here confirm that copy number changes affecting 8p and 8q occur frequently in invasive breast tumors. Moreover, this study adds new evidence to support an association between 8q22 copy number gain and grade 3 invasive carcinoma.
The 8q22 region was recently shown to harbor the metastasis gene, MTDH, which was overexpressed and amplified in poor-prognosis breast cancer [51]. Although characterization of MTDH revealed a function in both promoting metastasis and chemoresistance, Hu and colleagues did not explore a possible relationship between MTDH function and the development of grade 3 tumors. By comparison, Sotiriou et al explored the association of gene expression profiles with histological grade in a cohort of breast IDC tumors [58]. This study showed that 183 genes were expressed differently between 33 grade 1 and 31 grade 3 tumors. Interestingly, nine genes that mapped to 8q were significantly overexpressed in grade 3 tumors. Three of these nine genes (RAD54B, CCNE2, POLR2K) map as close as 113 kb (POLR2K) and up to almost 6 Mb (RAD54B) centromeric to the BAC probe RP11-10G10 used in this study. Previously, Pollack et al had shown that increased DNA copy number extending across an ∼11 Mb region within 8q21–q22, from NBN (NBS1) to YWHAZ (chr8:90,945,564-101,962,79), was associated with correspondingly altered mRNA levels for the genes found within the amplified region [19]. Although not represented on their array, both CCNE2 and RAD54B were noted genes of interest mapping within this region [19], as does MTDH. Our findings reported here using BAC clone RP11-10G10 are therefore consistent with these earlier studies. Our findings also align well with studies using higher density array platforms for genome-wide copy number assessment that report increased incidence of 8q22 gain in association with different biological characteristics that are commensurate with higher-grade malignancy, including ESR1-negative status, and a luminal B rather than luminal A profile [21], [43], [47]. In contrast to the findings of Hu et al [51], we were unable to demonstrate an association between shorter survival time and 8q22 gain with our cohort of 63 breast cancer patients. However, it is important to note that the patient cohort used by Hu et al was significantly younger (<50 years) than that used in our study, suggesting that the prognostic implication of 8q22 gain may only be relevant for early onset breast cancer.
CGH studies, both conventional and array, of pre-cancerous ductal carcinoma in situ (DCIS) have shown that 8q gains, including the 8q22 region, occur during early stages of breast tumorigenesis, but that this aberration is particularly associated with higher-grade DCIS [46], [59], [60], [61], [62]. Based on different frequencies of 16q loss between grade 1 and grade 3 breast tumors, it has been suggested that low grade and high grade tumors may evolve through different mechanistic pathways [34], [37], [63]. Similarly for 8q, it is possible that early over-representation of 8q22 may be a predetermining factor of grade 3 invasive breast cancer early in tumor development. However, the observation in our present study that absolute or relative gains involving the 8q22 region were also observed in 42% of grade 1 tumors supports the notion that a large proportion of invasive tumors share this aberration independently of histological grade. This result is consistent with other reports that have also highlighted shared chromosomal aberrations between low and high grade tumors, suggesting a model that predicts a significant number of breast tumors showing progression through the grades [46], [50], [64], [65]. Further studies are therefore required to refine this model and to understand the relationship between 8q22 gain, other recurring chromosomal imbalances and different morphological breast tumor subtypes.
In contrast to 8q22 gain, loss of 8p21 did not correlate significantly with tumor grade in our present FISH study. These results differ from our earlier report using mCGH, which described significantly increased frequency of 8p loss in high-grade IDC, and also from the reports of others which showed a similar trend when correlations with histological grade were assessed [33], [34], [66]. The study of Armes et al compared CGH and high-resolution gene expression profiles of 53 breast tumor samples, and showed that 8p loss was associated with grade 3 and ESR1-negative tumors. Microarray profiling identified 22 genes that mapped to 8p and for which gene expression levels were significantly lower in the grade 3 tumors. Three of these genes were selected for FISH analysis, and of these, PCM1 at 8p22 (∼5 Mb telomeric of BAC clone RP11-177H13) showed highest frequency of loss, affecting 45% of analyzable cases [66]. Using BAC arrays, Loo and coauthors have also reported increased incidence of 8p loss in ER-negative IDC compared with ER-positive IDC [43]. However, in a subsequent study, using Affymetrix GeneChip Mapping 10 K arrays but confined to ER-positive tumors, they reported that 8p loss occurs more frequently in IDC compared with ILC, and was prevalent in a higher grade ER-positive IDC subset [67]. Hwang and colleagues identified genomic regions within two BAC clones at 8p23.1 and 8p23.3 that separated recurrent from nonrecurrent IDC subgroups, but then found no difference in overall survival or systemic recurrence-free survival in the cohorts identified by these genomic segments [49]. In the same study, three of a total four regions showing loss in greater than 50% of the total 62 samples analyzed by aCGH mapped to 8p, including one region at 8p21.1–21.3 that overlaps BAC clone RP11-177H13 and spans CHMP2 and LOXL2 [49]. Thus, whereas 8p loss is clearly a significant and recurring feature of invasive breast cancer, there is much still to understand about its place in the hierarchy of genomic changes that accompany tumour initiation and progression.
In this study, 8q22 gain was determined using two methods: 1) scoring absolute signal frequency after FISH with RP11-10G10, and 2) calculating the mean FISH signal copy number ratio between RP11-10G10 and RP11-177H13 at 8p21. Although a correlation between 8q22 and grade 3 breast tumors was demonstrated using both methods, a higher frequency of absolute copy number gain was observed in grade 3 tumors (25/28) compared with relative 8q22 gain (16/28). One interpretation is that 8q22 copy number increase resulted from chromosome 8 polysomy in nine of the grade 3 tumors. However, gain of whole chromosome 8 was only apparent in one of these nine tumors as indicated by an 8q22:8p21 signal ratio less than 2.0 and a majority of abnormal cells showing identical signal copy numbers for the two FISH probes used. The discordancy shown in the remaining eight cases may be attributed to high numbers of contaminating normal cells and/or our conservative 8q22:8p21 ratio threshold for 8q22 gain.
FISH analysis of interphase nuclei from 10 tumors revealed sizeable cell populations showing simultaneous loss of chromosome 8p21 with duplication of 8q22. These signal patterns are consistent cytogenetically with presence of an isochromosome 8q, an isoderivative including 8q22, or unbalanced translocation(s) with breakpoints proximal to 8q22 incurring 8p loss. These and other structural aberrations of chromosome 8 have been reported at high frequency following detailed comparisons of mCGH profiles with multicolor spectral karyotyping (SKY) karyotypes in breast cancer cell lines [68], [69], [70]. Unbalanced 8p/8q signal patterns were more frequent in our grade 3 tumor subset, an observation consistent with the well recognized increasing genomic complexity of advanced malignancy. We note here that the various structural modalities by which 8q gain may arise would suggest that in some cases loss of 8p is mechanistically linked to these, and therefore a bystander to the biologically more impactful 8q gain. If the genes critical to tumor progression are located on 8q then the significance of genes deleted on 8p, at least in these cases, might be questioned.
In summary, using FISH analysis of tumor touch imprints prepared from excised breast tumor specimens, we have provided further evidence that a copy number gain at chromosome 8q22 is associated with typically aggressive grade 3 tumors. Because histological grade in breast cancer provides clinically important prognostic information, the association with copy number gain at 8q22 suggests the location of one or more genes or regulatory elements that may play a key role in determining grade and which may be candidate biological marker(s) for poor prognosis. Further research is necessary to identify such genes, which may lead to a better understanding of breast tumorigenesis as well as potential prognostic markers and novel targets for therapeutic intervention.
Supporting Information
Histopathological features of 60 invasive breast tumors imprinted for FISH analysis.
(XLS)
Supporting Information Table S1: FISH signal patterns observed using the 8p21- and 8q22-specific BAC probes RP11-177H13 (labelled red, R) and RP11-10G10 (labelled green, G), respectively,to assess copy number status in 60 breast tumour touch preparations. The number of nuclei scored with the various signal patterns is indicated. Cases with a total cell count <50 are highlighted in black.
(XLS)
Clinical follow-up data for 63 breast cancer patients.
(XLS)
Acknowledgments
We thank Campbell Sheen and Helen Morrin for expert laboratory assistance, and John Pearson for helpful advice. We are grateful to Dr Nigel Carter at The Wellcome Trust Sanger Institute for gifting the BAC clones. We are also grateful to the Cancer Society Tissue Bank for tumor samples and Val Davey (Christchurch Breast Cancer Patient Register) for access to clinical follow-up data. LW is a Sir Charles Hercus Health Research Fellow of the Health Research Council of New Zealand.
Funding Statement
The authors thank the New Zealand Breast Cancer Foundation, Lottery Health Research, and Cancer Society of New Zealand for funding that has enabled completion of this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rakha EA, Ellis IO (2011) Modern classification of breast cancer: should we stick with morphology or convert to molecular profile characteristics. Advances in anatomic pathology 18: 255–267. [DOI] [PubMed] [Google Scholar]
- 2. Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS (2011) Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res 13: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cummings MC, Chambers R, Simpson PT, Lakhani SR (2011) Molecular classification of breast cancer: is it time to pack up our microscopes? Pathology 43: 1–8. [DOI] [PubMed] [Google Scholar]
- 4. Lee AH, Ellis IO (2008) The Nottingham prognostic index for invasive carcinoma of the breast. Pathol Oncol Res 14: 113–115. [DOI] [PubMed] [Google Scholar]
- 5. Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19: 403–410. [DOI] [PubMed] [Google Scholar]
- 6. Komaki K, Sano N, Tangoku A (2006) Problems in histological grading of malignancy and its clinical significance in patients with operable breast cancer. Breast Cancer 13: 249–253. [DOI] [PubMed] [Google Scholar]
- 7. Paradiso A, Ellis IO, Zito FA, Marubini E, Pizzamiglio S, et al. (2009) Short- and long-term effects of a training session on pathologists' performance: the INQAT experience for histological grading in breast cancer. J Clin Pathol 62: 279–281. [DOI] [PubMed] [Google Scholar]
- 8. Robbins P, Pinder S, de Klerk N, Dawkins H, Harvey J, et al. (1995) Histological grading of breast carcinomas: a study of interobserver agreement. Hum Pathol 26: 873–879. [DOI] [PubMed] [Google Scholar]
- 9. Sloane JP, Amendoeira I, Apostolikas N, Bellocq JP, Bianchi S, et al. (1999) Consistency achieved by 23 European pathologists from 12 countries in diagnosing breast disease and reporting prognostic features of carcinomas. European Commission Working Group on Breast Screening Pathology. Virchows Arch 434: 3–10. [DOI] [PubMed] [Google Scholar]
- 10. Staradub VL, Messenger KA, Hao N, Wiley EL, Morrow M (2002) Changes in breast cancer therapy because of pathology second opinions. Ann Surg Oncol 9: 982–987. [DOI] [PubMed] [Google Scholar]
- 11. Geyer FC, Marchio C, Reis-Filho JS (2009) The role of molecular analysis in breast cancer. Pathology 41: 77–88. [DOI] [PubMed] [Google Scholar]
- 12. Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360: 790–800. [DOI] [PubMed] [Google Scholar]
- 13. Allred DC (2008) The utility of conventional and molecular pathology in managing breast cancer. Breast Cancer Res 10 Suppl 4S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, et al. (1992) Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818–821. [DOI] [PubMed] [Google Scholar]
- 15. Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, et al. (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer 20: 399–407. [PubMed] [Google Scholar]
- 16. Pinkel D, Segraves R, Sudar D, Clark S, Poole I, et al. (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 20: 207–211. [DOI] [PubMed] [Google Scholar]
- 17. van Beers EH, Nederlof PM (2006) Array-CGH and breast cancer. Breast Cancer Res 8: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reis-Filho JS, Simpson PT, Gale T, Lakhani SR (2005) The molecular genetics of breast cancer: the contribution of comparative genomic hybridization. Pathol Res Pract 201: 713–725. [DOI] [PubMed] [Google Scholar]
- 19. Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, et al. (2002) Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A 99: 12963–12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, et al. (2007) Integrated profiling of basal and luminal breast cancers. Cancer Res 67: 11565–11575. [DOI] [PubMed] [Google Scholar]
- 21. Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, et al. (2007) High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol 8: R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, et al. (2008) High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer 47: 530–542. [DOI] [PubMed] [Google Scholar]
- 23. Andre F, Job B, Dessen P, Tordai A, Michiels S, et al. (2009) Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451. [DOI] [PubMed] [Google Scholar]
- 24.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. [DOI] [PMC free article] [PubMed]
- 25. Ioannidis P, Mahaira L, Papadopoulou A, Teixeira MR, Heim S, et al. (2003) 8q24 Copy number gains and expression of the c-myc mRNA stabilizing protein CRD-BP in primary breast carcinomas. Int J Cancer 104: 54–59. [DOI] [PubMed] [Google Scholar]
- 26. Karlseder J, Zeillinger R, Schneeberger C, Czerwenka K, Speiser P, et al. (1994) Patterns of DNA amplification at band q13 of chromosome 11 in human breast cancer. Genes Chromosomes Cancer 9: 42–48. [DOI] [PubMed] [Google Scholar]
- 27. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, et al. (1989) Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712. [DOI] [PubMed] [Google Scholar]
- 28. Walker LC, Harris GC, Wells JE, Robinson BA, Morris CM (2008) Association of chromosome band 8q22 copy number gain with high grade invasive breast carcinomas by assessment of core needle biopsies. Genes Chromosomes Cancer 47: 405–417. [DOI] [PubMed] [Google Scholar]
- 29. Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, et al. (1994) Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A 91: 2156–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, et al. (1995) Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 147: 905–911. [PMC free article] [PubMed] [Google Scholar]
- 31. Hermsen MA, Baak JP, Meijer GA, Weiss JM, Walboomers JW, et al. (1998) Genetic analysis of 53 lymph node-negative breast carcinomas by CGH and relation to clinical, pathological, morphometric, and DNA cytometric prognostic factors. J Pathol 186: 356–362. [DOI] [PubMed] [Google Scholar]
- 32. Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, et al. (1998) Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 21: 177–184. [PubMed] [Google Scholar]
- 33. Buerger H, Otterbach F, Simon R, Schafer KL, Poremba C, et al. (1999) Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 189: 521–526. [DOI] [PubMed] [Google Scholar]
- 34. Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, et al. (1999) Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res 59: 1433–1436. [PubMed] [Google Scholar]
- 35. Loveday RL, Greenman J, Simcox DL, Speirs V, Drew PJ, et al. (2000) Genetic changes in breast cancer detected by comparative genomic hybridisation. Int J Cancer 86: 494–500. [DOI] [PubMed] [Google Scholar]
- 36. Richard F, Pacyna-Gengelbach M, Schluns K, Fleige B, Winzer KJ, et al. (2000) Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer 89: 305–310. [PubMed] [Google Scholar]
- 37. Buerger H, Mommers EC, Littmann R, Simon R, Diallo R, et al. (2001) Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol 194: 165–170. [DOI] [PubMed] [Google Scholar]
- 38. Hislop RG, Pratt N, Stocks SC, Steel CM, Sales M, et al. (2002) Karyotypic aberrations of chromosomes 16 and 17 are related to survival in patients with breast cancer. Br J Surg 89: 1581–1586. [DOI] [PubMed] [Google Scholar]
- 39. Rennstam K, Ahlstedt-Soini M, Baldetorp B, Bendahl PO, Borg A, et al. (2003) Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res 63: 8861–8868. [PubMed] [Google Scholar]
- 40. Weber-Mangal S, Sinn HP, Popp S, Klaes R, Emig R, et al. (2003) Breast cancer in young women (< or = 35 years): Genomic aberrations detected by comparative genomic hybridization. Int J Cancer 107: 583–592. [DOI] [PubMed] [Google Scholar]
- 41. Janssen EA, Baak JP, Guervos MA, van Diest PJ, Jiwa M, et al. (2003) In lymph node-negative invasive breast carcinomas, specific chromosomal aberrations are strongly associated with high mitotic activity and predict outcome more accurately than grade, tumour diameter, and oestrogen receptor. J Pathol 201: 555–561. [DOI] [PubMed] [Google Scholar]
- 42. Farabegoli F, Hermsen MA, Ceccarelli C, Santini D, Weiss MM, et al. (2004) Simultaneous chromosome 1q gain and 16q loss is associated with steroid receptor presence and low proliferation in breast carcinoma. Mod Pathol 17: 449–455. [DOI] [PubMed] [Google Scholar]
- 43. Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, et al. (2004) Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res 64: 8541–8549. [DOI] [PubMed] [Google Scholar]
- 44. Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, et al. (2005) High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res 7: R1186–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, et al. (2006) Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res 66: 4065–4078. [DOI] [PubMed] [Google Scholar]
- 46. Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, et al. (2006) Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, et al. (2006) Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 45: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 48. Han W, Jung EM, Cho J, Lee JW, Hwang KT, et al. (2008) DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer 47: 490–499. [DOI] [PubMed] [Google Scholar]
- 49. Hwang KT, Han W, Cho J, Lee JW, Ko E, et al. (2008) Genomic copy number alterations as predictive markers of systemic recurrence in breast cancer. Int J Cancer 123: 1807–1815. [DOI] [PubMed] [Google Scholar]
- 50. Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, et al. (2009) Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res 15: 2711–2722. [DOI] [PubMed] [Google Scholar]
- 51. Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, et al. (2009) MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, et al. (2010) Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med 16: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cooke SL, Pole JC, Chin SF, Ellis IO, Caldas C, et al. (2008) High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer 8: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barnes DM, Harris WH, Smith P, Millis RR, Rubens RD (1996) Immunohistochemical determination of oestrogen receptor: comparison of different methods of assessment of staining and correlation with clinical outcome of breast cancer patients. Br J Cancer 74: 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCarty KS Jr, Miller LS, Cox EB, Konrath J, McCarty KS Sr (1985) Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med 109: 716–721. [PubMed] [Google Scholar]
- 56. Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, et al. (2003) DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36: 361–374. [DOI] [PubMed] [Google Scholar]
- 57.Dean AG, Sullivan KM, Soe MM OpenEpi: Open Source Epidemiologic Statistics for Public Health, Version 2.3.1. Available: www.OpenEpi.com.
- 58. Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, et al. (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272. [DOI] [PubMed] [Google Scholar]
- 59. Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, et al. (1999) Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J Pathol 187: 396–402. [DOI] [PubMed] [Google Scholar]
- 60. Hwang ES, DeVries S, Chew KL, Moore DH 2nd, Kerlikowske K, et al. (2004) Patterns of chromosomal alterations in breast ductal carcinoma in situ. Clin Cancer Res 10: 5160–5167. [DOI] [PubMed] [Google Scholar]
- 61. Vos CB, ter Haar NT, Rosenberg C, Peterse JL, Cleton-Jansen AM, et al. (1999) Genetic alterations on chromosome 16 and 17 are important features of ductal carcinoma in situ of the breast and are associated with histologic type. Br J Cancer 81: 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vincent-Salomon A, Lucchesi C, Gruel N, Raynal V, Pierron G, et al. (2008) Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res 14: 1956–1965. [DOI] [PubMed] [Google Scholar]
- 63. Simpson PT, Reis-Filho JS, Gale T, Lakhani SR (2005) Molecular evolution of breast cancer. J Pathol 205: 248–254. [DOI] [PubMed] [Google Scholar]
- 64. Korsching E, Packeisen J, Helms MW, Kersting C, Voss R, et al. (2004) Deciphering a subgroup of breast carcinomas with putative progression of grade during carcinogenesis revealed by comparative genomic hybridisation (CGH) and immunohistochemistry. Br J Cancer 90: 1422–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roylance R, Gorman P, Papior T, Wan YL, Ives M, et al. (2006) A comprehensive study of chromosome 16q in invasive ductal and lobular breast carcinoma using array CGH. Oncogene 25: 6544–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Armes JE, Hammet F, de Silva M, Ciciulla J, Ramus SJ, et al. (2004) Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene 23: 5697–5702. [DOI] [PubMed] [Google Scholar]
- 67. Loo LW, Ton C, Wang YW, Grove DI, Bouzek H, et al. (2008) Differential patterns of allelic loss in estrogen receptor-positive infiltrating lobular and ductal breast cancer. Genes Chromosomes Cancer 47: 1049–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rummukainen J, Kytola S, Karhu R, Farnebo F, Larsson C, et al. (2001) Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping. Cancer Genet Cytogenet 126: 1–7. [DOI] [PubMed] [Google Scholar]
- 69. Kytola S, Rummukainen J, Nordgren A, Karhu R, Farnebo F, et al. (2000) Chromosomal alterations in 15 breast cancer cell lines by comparative genomic hybridization and spectral karyotyping. Genes Chromosomes Cancer 28: 308–317. [DOI] [PubMed] [Google Scholar]
- 70. Davidson JM, Gorringe KL, Chin SF, Orsetti B, Besret C, et al. (2000) Molecular cytogenetic analysis of breast cancer cell lines. Br J Cancer 83: 1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histopathological features of 60 invasive breast tumors imprinted for FISH analysis.
(XLS)
Supporting Information Table S1: FISH signal patterns observed using the 8p21- and 8q22-specific BAC probes RP11-177H13 (labelled red, R) and RP11-10G10 (labelled green, G), respectively,to assess copy number status in 60 breast tumour touch preparations. The number of nuclei scored with the various signal patterns is indicated. Cases with a total cell count <50 are highlighted in black.
(XLS)
Clinical follow-up data for 63 breast cancer patients.
(XLS)