Abstract
Purpose
Genetic variation may influence chemotherapy response and overall survival in cancer patients.
Experimental design
We conducted a genome-wide scan in 535 advanced-stage non–small cell lung cancer (NSCLC) patients from two independent cohorts (307 from Nanjing and 228 from Beijing). A replication was carried out on an independent cohort of 340 patients from Southeastern China followed by a second validation on 409 patients from the Massachusetts General Hospital (Boston, MA).
Results
Consistent associations with NSCLC survival were identified for five single-nucleotide polymorphisms (SNP) in Chinese populations with P values ranging from 3.63 × 10−5 to 4.19 × 10−7 in the additive genetic model. The minor allele of three SNPs (rs7629386 at 3p22.1, rs969088 at 5p14.1, and rs3850370 at 14q24.3) were associated with worse NSCLC survival while 2 (rs41997 at 7q31.31 and rs12000445 at 9p21.3) were associated with better NSCLC survival. In addition, rs7629386 at 3p22.1 (CTNNB1) and rs3850370 at 14q24.3 (SNW1-ALKBH1-NRXN3) were further replicated in the Caucasian population.
Conclusion
In this three-stage genome-wide association studies, we identified five SNPs as markers for survival of advanced-stage NSCLC patients treated with first-line platinum-based chemotherapy in Chinese Han populations. Two of these SNPs, rs7629386 and rs3850370, could also be markers for survival among Caucasian patients.
Introduction
Lung cancer, predominantly non–small cell lung cancer (NSCLC), is the leading cause of cancer-related deaths worldwide; 5-year survival rates are only 15.1% in China (1). The current standard treatment of patients with early stage NSCLC is surgical resection, with or without adjuvant chemotherapy (2, 3). However, most patients are diagnosed at advanced stage, where platinum-based chemotherapy is the mainstay of treatment. Response to therapy varies considerably between patients (4).
The traditional tumor–node–metastasis staging system is not useful for predicting response to therapy, and molecular biomarkers can add value in this setting (5, 6). It has been suggested that the discovery and application of molecular biomarkers to incorporate with traditional clinical prognostic factors could guide individualized treatment and improve the outcomes of cancer patients (7). Many studies have been conducted to explore single-nucleotide polymorphisms (SNP) in candidate genes and overall survival in NSCLC, including studies from our groups (8–12). Genome-wide association studies (GWAS), which make no assumptions about the genomic location of the causal variants, are expected to complement the candidate gene approach.
Here, we conducted a GWAS scan by genotyping 906,703 SNPs for association with survival in 535 Han Chinese with advanced NSCLC. Fast track replication was conducted in an independent NSCLC cohort from Southeastern China followed by further validation in NSCLC patients from the Massachusetts General Hospital (Boston, MA).
Materials and Methods
Study populations
The discovery phase of the study included 2 groups of advanced NSCLC patients from Nanjing and Beijing described previously in our GWAS study on lung cancer susceptibility (13). To construct a relatively homogenous population with same treatment, our current study was restricted to stage III or IV NSCLC patients treated with first-line platinum-based chemotherapy without surgery. Specifically, 307 patients recruited at the Affiliated Cancer Hospital and the First Affiliated Hospital of Nanjing Medical University (Nanjing Study), and 228 patients from the Cancer Hospital, Chinese Academy of Medical Sciences (Beijing Study), were included in the discovery set. The first replication included 340 NSCLC patients recruited from Nanjing Thoracic Hospital and Shanghai Chest Hospital (Southeastern China; 14), and the second replication included 409 NSCLC patients from the Massachusetts General Hospital (Boston, MA; Harvard cohort; 15). Subjects in the discovery phase and the first replication phase were all unrelated Han Chinese, and those in the second replication set were Caucasian. All patients had histopathologically or cytologically confirmed NSCLC, which was reviewed by at least 2 local pathologists. Informed consent was obtained from each subject at the time of recruitment, and this study was approved by the Institutional Review Boards of each participating institution.
Quality control in GWAS
Before the survival analysis, we conducted systematic quality control on the raw genotyping data to filter both unqualified samples and SNPs, as described previously (13). SNPs were excluded if: (i) SNPs were not mapped on autosomal chromosomes; (ii) SNPs had a call rate <95%; or (iii) SNPs had minor allele frequency (MAF) <0.05. Samples with low call rates (<95%), ambiguous gender, familial relationships (PI_HAT > 0.25), outliers in the principal component analysis and extreme heterozygote rate (>6 SD from nearest neighbor) were removed. Finally, 303 cases were used for the Nanjing study, and 225 cases for the Beijing study with 576,351 overlapped SNPs.
SNPs selection and genotyping
Genome-wide association analysis was conducted using the additive model by multivariate Cox regression analysis with adjustment for age, gender, smoking status, histology, and stage in each of the discovery studies. Meta-analysis on Nanjing and Beijing discovery samples were then carried out based on fixed or random models. For fast track replication, we selected 33 top SNPs that had (i) P < 1 ×10−4 for all GWAS samples and (ii) a consistent association at P < 0.05 in both the Nanjing study and the Beijing study (Supplementary Table S1). Twelve SNPs were used in the first validation, whereas 21 SNPs were excluded because of high-linkage disequilibrium (LD) with the 12 selected SNPs. Five SNPs that were found to be associated with overall survival at P values lower than 0.05 (in any genetic model) and had the same association direction as the GWAS scan were further genotyped in the second validation set. Genotyping for the validation populations was done using TaqMan allelic discrimination assay on an ABI 7900 system (Applied Biosystems), except one (rs3850370) by SNP chip (Illumina 610k Quad, Call rate: 100%), and one (rs41997) by imputation analysis using the LD information from the 1000 Genomes Project database [by MACH, the estimated squared correlation (r2) between imputed and true underlying genotypes is 1.00, indicating an excellent imputation quality] for the Harvard cohort. The information on primers and probes are available upon request. A total of 10% random samples were reciprocally tested on the TaqMan assay, and the reproducibility was 100%.
Statistical analysis
Overall survival time was calculated from the time of diagnosis until death or the latest follow-up. We used PLINK 1.07 for general statistical analysis (16). The survival package in R (PLINK plug-in) was used to conduct the analyses of lung cancer-related death. HR and their 95% confidence intervals (CI) were calculated by multivariate Cox regression analyses adjusted for age, gender, histology, stage, and smoking status. The Cochrane Q statistics test was used for the assessment of heterogeneity. The fixed and random effects meta-analyses were used to calculate the pooled HR. If the result of the Q test was not significant, the fixed effects model was chosen. Otherwise, the pooled HR was estimated using the random effects model. The MACH 1.0 software (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) was used to impute untyped SNPs by the LD information from the HapMap phase II database (CHB+JPT was used as reference set, released July 17, 2006; 17). Region plot was generated using an online tool, LocusZoom 1.1 (http://csg.sph.umich.edu/locuszoom/). Analyses were also conducted using SAS version 9.1.3 (SAS Institute) or Stata version 9.2 (StataCorp LP).
Results
The characteristics of the NSCLC patients in the 2 discovery cohorts and the 2 replication cohorts are shown in Table 1. The median survival times (MST) were 14.2, 23.0, 23.8, and 11.9 months, and 206 (68.0%), 101 (44.9%), 120 (35.3%), as well as 353 (86.3%) patients died from lung cancer in these cohorts, respectively. Only the Harvard cohort had NSCLC patients other than squamous cell carcinoma and adenocarcinoma. The Harvard cohort also had more stage IV patients, female and ever smokers (Table 1).
Table 1.
Characteristics of study subjects
| Characteristics | Nanjing study (N =303) | Beijing study (N =225) | Replication I (N =340) | Replication II (N =409) |
|---|---|---|---|---|
| Death | 206 (68.0) | 101 (44.9) | 120 (35.3) | 353 (86.3) |
| MST (mo) | 14.2 | 23.0 | 23.8 | 11.9 |
| MFT (mo) | 18.8 | 21.9 | 14.6 | 37.4 |
| Age (y) | 61.16 ±10.50 | 57.69 ±10.36 | 60.02 ±10.70 | 63.79 ±10.84 |
| Gender | ||||
| Male | 211 (69.64) | 174 (77.33) | 226 (66.47) | 195 (47.68) |
| Female | 92 (30.36) | 51 (22.67) | 114 (33.53) | 214 (52.32) |
| Smoking status | ||||
| Never | 120 (39.60) | 76 (33.78) | 154 (45.29) | 42 (10.27) |
| Ever | 183 (60.40) | 149 (66.22) | 186 (54.71) | 367 (89.73) |
| Histologic type | ||||
| Squamous cell | 100 (33.00) | 111 (49.33) | 98 (28.82) | 63 (15.40) |
| Adenocarcinoma | 203 (67.00) | 114 (50.67) | 242 (71.18) | 220 (53.79) |
| Othersa | 0 | 0 | 0 | 126 (30.81) |
| Stage | ||||
| III | 127 (41.91) | 147 (65.33) | 183 (53.82) | 146 (35.70) |
| IV | 176 (58.09) | 78 (34.67) | 157 (46.18) | 263 (64.30) |
Abbreviations: MST, median survival time; MFT, median follow-up time for censored patients.
Other carcinomas include large cell, mixed cell, or undifferentiated carcinomas.
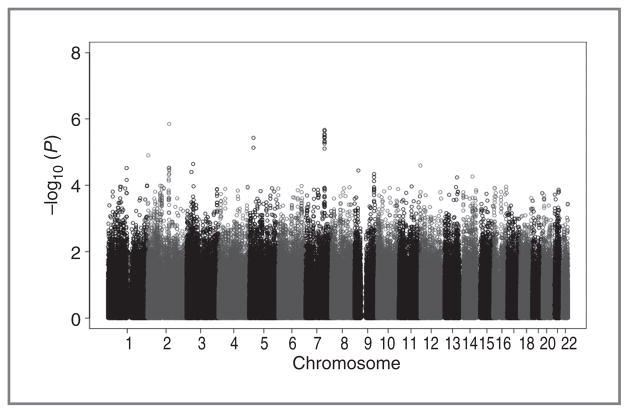
P values for the 2 discovery cohorts by meta-analysis are presented in the scatter plot with multiple suggestive associations (P < 10−4; Fig. 1 and Supplementary Table S1). We selected 12 SNPs (21 SNPs excluded) to conduct the validation using an independent NSCLC cohort from Southeastern China. For the 12 SNPs, 5 SNPs at 3p22.1 (rs7629386), 5p14.1 (rs969088), 7q31.31 (rs41997), 9p21.3 (rs12000445), and 14q24.3 (rs3850370) were found to be associated with overall survival in the same direction as the GWAS scan with P values lower than 0.05 (rs41997 and rs12000445 in additive genetic model, while rs7629386, rs969088, and rs3850370 in homozygote comparison; Table 2). For all Chinese samples, the 5 SNPs were associated with NSCLC survival with Pmeta (additive) values ranging from 3.63 ×10−5 to 4.19 ×10−7, with 3 (the minor allele of rs7629386, rs969088, and rs3850370) being associated with worse survival whereas 2 (the minor allele of rs41997 and rs12000445) were associated with improved survival (Table 2).
Figure 1.
Genome-wide association results for NSCLC overall survival in Han Chinese individuals. Scatter plot of P values in −log10 scale from meta-analysis results of the additive model on 576,351 SNPs from 2 discovery cohorts.
Table 2.
Result for 12 selected SNPs in Replication I samples and meta-analysis for the 3 Chinese cohorts
| Variable | Replication I
|
All Chinese
|
||||||
|---|---|---|---|---|---|---|---|---|
| Patients | Deaths | MST (mo) | Pa | HR (95% CI)a | Pb | HR (95% CI)b | Q-Pc | |
| rs12067801 (1p13.1) | ||||||||
| GG | 309 | 110 | 23.8 | |||||
| AG | 30 | 10 | 19.0 | 8.74 × 10−1 | 1.06 (0.54–2.08) | 5.40 × 10−4 | 1.71 (1.26–2.33) | 0.21 |
| AA | 1 | 0 | NA | 9.88 × 10−1 | NA | 9.60 × 10−3 | NA | NA |
| ADD | 8.79 × 10−1 | 1.05 (0.54–2.07) | 1.18 × 10−4 | 1.72 (1.30–2.26) | 0.29 | |||
| rs17019595 (2p25.2) | ||||||||
| TT | 262 | 94 | 21.2 | |||||
| CT | 74 | 23 | 32.0 | 2.99 × 10−1 | 0.78 (0.49–1.24) | 2.63 × 10−1 | 1.29 (0.83–2.02) | 0.03 |
| CC | 4 | 3 | 24.5 | 6.78 × 10−1 | 1.29 (0.38–4.38) | 1.81 × 10−3 | 3.51 (1.6–7.72) | 0.11 |
| ADD | 5.50 × 10−1 | 0.88 (0.59–1.32) | 1.36 × 10−1 | 1.39 (0.9–2.15) | 0.02 | |||
| rs7589111 (2q21.1) | ||||||||
| AA | 184 | 64 | 23.5 | |||||
| GA | 133 | 49 | 23.8 | 7.41 × 10−1 | 0.94 (0.64–1.37) | 9.10 × 10−2 | 1.19 (0.97–1.46) | 0.34 |
| GG | 23 | 7 | 34.2 | 1.37 × 10−1 | 1.85 (0.82–4.18) | 5.63 × 10−6 | 2.47 (1.67–3.65) | 0.62 |
| ADD | 6.61 × 10−1 | 1.07 (0.78–1.47) | 1.84 × 10−4 | 1.37 (1.16–1.61) | 0.21 | |||
| rs494905 (2q21.3) | ||||||||
| CC | 259 | 100 | 23.5 | |||||
| TC | 74 | 20 | 23.8 | 5.41 × 10−1 | 0.86 (0.53–1.40) | 1.34 × 10−1 | 1.45 (0.89–2.34) | 0.02 |
| TT | 7 | 0 | NA | 9.79 × 10−1 | NA | 2.63 × 10−1 | NA | 0.74 |
| ADD | 1.89 × 10−1 | 0.74 (0.47–1.16) | 2.80 × 10−1 | 1.33 (0.79–2.23) | <0.01 | |||
| rs7629386 (3p22.1) | ||||||||
| CC | 301 | 106 | 24.1 | |||||
| TC | 32 | 9 | 20.8 | 7.48 × 10−1 | 0.89 (0.45–1.78) | 1.04 × 10−3 | 1.56 (1.20–2.04) | 0.18 |
| TT | 3 | 3 | 12.2 | 2.30 × 10−2 | 3.92 (1.21–12.70) | 2.95 × 10−4 | 4.71 (2.04–10.89) | 0.26 |
| ADD | 3.49 × 10−1 | 1.27 (0.77–2.10) | 3.63 × 10−5 | 1.65 (1.30–2.09) | 0.47 | |||
| rs969088 (5p14.1) | ||||||||
| GG | 187 | 68 | 24.1 | |||||
| CG | 127 | 41 | 25.5 | 4.34 × 10−1 | 1.17 (0.79–1.74) | 8.68 × 10−4 | 1.42 (1.16–1.74) | 0.41 |
| CC | 26 | 11 | 17.4 | 2.80 × 10−2 | 2.10 (1.08–4.08) | 1.49 × 10−5 | 2.14 (1.52–3.01) | 0.80 |
| ADD | 6.60 × 10−2 | 1.31 (0.98–1.76) | 1.75 × 10−6 | 1.43 (1.24–1.66) | 0.55 | |||
| rs41997 (7q31.31) | ||||||||
| AA | 175 | 62 | 20.8 | |||||
| GA | 147 | 52 | 29.9 | 1.57 × 10−1 | 0.76 (0.52–1.11) | 2.67 × 10−5 | 0.64 (0.52–0.79) | 0.55 |
| GG | 17 | 6 | 21.2 | 3.32 × 10−2 | 0.36 (0.14–0.92) | 1.23 × 10−4 | 0.44 (0.29–0.67) | 0.73 |
| ADD | 4.69 × 10−2 | 0.73 (0.53–1.00) | 4.19 × 10−7 | 0.66 (0.56–0.78) | 0.61 | |||
| rs12000445 (9p21.3) | ||||||||
| AA | 188 | 74 | 20.8 | |||||
| GA | 133 | 39 | 33.0 | 1.39 × 10−1 | 0.74 (0.50–1.10) | 1.09 × 10−4 | 0.66 (0.54–0.82) | 0.81 |
| GG | 17 | 5 | 35.4 | 1.12 × 10−1 | 0.47 (0.18–1.19) | 2.37 × 10−3 | 0.48 (0.29–0.77) | 1.00 |
| ADD | 4.20 × 10−2 | 0.72 (0.52–0.99) | 7.12 × 10−6 | 0.67 (0.57–0.80) | 0.88 | |||
| rs2109664 (9q33.1) | ||||||||
| GG | 102 | 35 | 24.5 | |||||
| AG | 159 | 49 | 29.9 | 7.96 × 10−1 | 1.06 (0.67–1.68) | 1.31 × 10−2 | 1.35 (1.07–1.71) | 0.44 |
| AA | 76 | 33 | 18.0 | 1.30 × 10−1 | 1.46 (0.90–2.37) | 3.18 × 10−5 | 1.81 (1.37–2.39) | 0.57 |
| ADD | 1.53 × 10−1 | 1.20 (0.94–1.53) | 3.47 × 10−5 | 1.33 (1.16–1.53) | 0.59 | |||
| rs4980976 (12p13.33) | ||||||||
| CC | 262 | 99 | 22.8 | |||||
| TC | 73 | 17 | 25.5 | 1.70 × 10−1 | 0.70 (0.41–1.17) | 4.07 × 10−1 | 1.26 (0.73–2.19) | 0.01 |
| TT | 5 | 4 | 22.0 | 5.27 × 10−1 | 1.41 (0.49–4.03) | 8.54 × 10−3 | 2.52 (1.27–5.00) | 0.23 |
| ADD | 4.59 × 10−1 | 0.86 (0.57–1.29) | 1.68 × 10−1 | 1.38 (0.87–2.17) | 0.02 | |||
| rs9516673 (13q32.1) | ||||||||
| TT | 178 | 65 | 25.5 | |||||
| CT | 121 | 41 | 23.5 | 3.32 × 10−1 | 0.82 (0.55–1.22) | 1.19 × 10−1 | 1.18 (0.96–1.46) | 0.07 |
| CC | 35 | 11 | 20.2 | 5.76 × 10−1 | 0.83 (0.44–1.59) | 1.97 × 10−1 | 1.54 (0.80–2.97) | 0.03 |
| ADD | 4.34 × 10−1 | 0.89 (0.67–1.19) | 1.87 × 10−1 | 1.22 (0.91–1.65) | 0.02 | |||
| rs3850370 (14q24.3) | ||||||||
| AA | 152 | 55 | 24.5 | 0.71 | ||||
| CA | 155 | 51 | 21.2 | 4.32 × 10−1 | 1.17 (0.79–1.72) | 9.88 × 10−3 | 1.32 (1.07–1.62) | 0.96 |
| CC | 25 | 9 | 15.3 | 4.38 × 10−2 | 2.20 (1.02–4.73) | 1.79 × 10−5 | 2.13 (1.51–3.01) | 0.63 |
| ADD | 1.54 × 10−1 | 1.25 (0.92–1.70) | 2.92 × 10−5 | 1.38 (1.19–1.60) | 0.07 | |||
Abbreviations: MST, median survival time; ADD, mutational homozygote versus heterozygote versus wild homozygote; NA, not available.
Adjusted for age, gender, histology, stage, and smoking status.
Results from the meta-analysis.
P value for Cochrane Q statistics test.
We further validated these 5 SNPs in Harvard cohort samples. Four SNPs (rs7629386, rs969088, rs12000445, and rs3850370) had same direction of association (Table 3, Supplementary Fig. S1), but only rs7629386 at 3p22.1 (homozygote comparison) and rs3850370 at 14q24.3 (additive model) had significant association with NSCLC survival (Table 3, Supplementary Figs. S3 and S4). For rs969088, the MAF in Caucasians is too low to show a successful replication. The results of stratification analyses on the 5 SNPs (for Chinese samples) are shown in Supplementary Fig. S2. We only observed slightly heterogeneity in the HRs for rs41997 among histologic subtypes (P =0.050).
Table 3.
Result for 5 selected SNPs in Replication II samples and meta-analysis for all participants
| Variable | Replication II
|
All participants
|
||||||
|---|---|---|---|---|---|---|---|---|
| Patients | Deaths | MST (mo) | Pa | HR (95% CI)a | Pb | HR (95% CI)b | Q-Pc | |
| rs7629386 (3p22.1) | ||||||||
| CC | 170 | 140 | 10.5 | |||||
| TC | 160 | 137 | 12.4 | 9.77 × 10−1 | 1.00 (0.78–1.27) | 1.47 × 10−1 | 1.31 (0.91–1.87) | 0.03 |
| TT | 34 | 33 | 11.5 | 4.40 × 10−2 | 1.50 (1.01–2.23) | 1.68 × 10−2 | 3.23 (1.24–8.45) | 0.04 |
| ADD | 2.02 × 10−1 | 1.12 (0.94–1.34) | 1.24 × 10−2 | 1.43 (1.08–1.88) | 0.05 | |||
| rs969088 (5p14.1) | ||||||||
| GG | 337 | 287 | 11.5 | |||||
| CG | 18 | 15 | 10.2 | 9.30 × 10−1 | 0.98 (0.58–1.65) | 2.14 × 10−3 | 1.35 (1.12–1.64) | 0.33 |
| CC | 1 | 1 | 5.1 | 1.29 × 10−1 | 4.75 (0.64–35.49) | 5.90 × 10−6 | 2.19 (1.56–3.07) | 0.79 |
| ADD | 7.53 × 10−1 | 1.08 (0.66–1.76) | 3.16 × 10−6 | 1.40 (1.22–1.61) | 0.50 | |||
| rs41997d (7q31.31) | ||||||||
| AA | 169 | 143 | 12.3 | |||||
| GA | 195 | 172 | 11.9 | 3.32 × 10−1 | 1.12 (0.89–1.40) | 1.19 × 10−1 | 0.76 (0.54–1.07) | <0.01 |
| GG | 45 | 38 | 11.1 | 3.60 × 10−1 | 1.19 (0.82–1.71) | 1.21 × 10−1 | 0.60 (0.31–1.15) | 0.01 |
| ADD | 2.32 × 10−1 | 1.10 (0.94–1.30) | 1.19 × 10−1 | 0.77 (0.56–1.07) | <0.01 | |||
| rs12000445 (9p21.3) | ||||||||
| AA | 291 | 248 | 11.5 | |||||
| GA | 57 | 48 | 10.5 | 7.14 × 10−1 | 0.94 (0.69–1.29) | 6.04 × 10−4 | 0.74 (0.62–0.88) | 0.29 |
| GG | 5 | 4 | 13.4 | 9.78 × 10−1 | 1.01 (0.36–2.83) | 5.96 × 10−3 | 0.54 (0.35–0.84) | 0.63 |
| ADD | 7.50 × 10−1 | 0.96 (0.73–1.26) | 6.89 × 10−5 | 0.74 (0.64–0.86) | 0.19 | |||
| rs3850370 (14q24.3) | ||||||||
| AA | 247 | 203 | 13.4 | |||||
| CA | 142 | 130 | 10.0 | 2.30 × 10−2 | 1.30 (1.04–1.63) | 5.61 × 10−4 | 1.31 (1.12–1.53) | 0.88 |
| CC | 20 | 20 | 10.7 | 2.60 × 10−1 | 1.32 (0.81–2.16) | 3.18 × 10−5 | 1.82 (1.37–2.41) | 0.48 |
| ADD | 2.80 × 10−2 | 1.22 (1.02–1.45) | 4.22 × 10−6 | 1.31 (1.17–1.47) | 0.63 | |||
Abbreviations: MST, median survival time; ADD, mutational homozygote versus heterozygote versus wild homozygote.
Adjusted for age, gender, histology, stage and smoking status.
Results from the meta-analysis.
P value for Cochrane Q statistics test.
By imputation.
Discussion
In this study, we conducted a 3-stage (4 cohorts) analytical approach to better understand the genetic factors modulating survival in advanced NSCLC patients treated with first-line platinum-based chemotherapy. During the preparation of the manuscript, a GWAS on a Caucasian population was published using a discovery scan of 327 advanced NSCLC patients from the Texas MD Anderson Cancer Center (18). They replicated the association between rs1878022 and NSCLC survival in a Spanish cohort (420 patients, HR =1.23, P =0.05) but not in the Mayo Clinic cohort (315 patients, HR =1.16, P =0.15), whereas rs10937823 was replicated in the Mayo Clinic cohort (HR = 1.45, P =0.04) but not in the Spanish cohort (HR =0.96, P =0.84) (18). However, rs1878022 and rs10937823 were both nonsignificantly associated with NSCLC death risk in our GWAS scan samples (rs1878022: HR =1.10, P =0.204; rs10937823: HR =0.89, P =0.336; Supplementary Fig. S5A and S5B).
The SNP rs3850370 at 14q24.3 can modify survival in NSCLC across Chinese and Caucasian populations. rs3850370 is located among 3 genes: Ski-interacting protein (SKIIP, known also as SNW1, upstream 307kb), alkylation repair homolog 1 (ALKBH1, downstream 360kb), and Neurexin 3 (NRXN3, upstream 1210kb; Supplementary Fig. S6E). SNW1 is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing (19). Using a library of endoribonuclease-prepared short interfering RNAs (esiRNA), Kittler and colleagues identified that SNW1 is among the 37 genes required for cell division (20). Recently, it was also shown that SNW1 is required for TGF-β1 induced epithelial–mesenchymal transition and invasiveness in transformed cells (21). To date, 8 human homologues of the Escherichia coli DNA repair enzyme ALKB (ALKBH1-8) have been identified, which are involved in repair of alkylation damage in DNA and RNA (22). Among them, ALKBH3 was reported to be significantly overexpressed in lung adenocarcinoma cells and contributes to NSCLC cell survival (23), although combined treatment modalities with cisplatin- and lentivirus-mediated ALKBH2 downregulation were significantly more potent in inhibiting lung cell growth and inducing apoptosis than monochemotherapy (24). However, little is known for the role of ALKBH1 in NSCLC survival and/or therapy response.
In this study, rs7629386 (3p22.1) was also replicated across Chinese and Caucasian populations for homozygote comparison. rs7629386 is relatively rare in Chinese (MAF =0.07) but common in Caucasians (MAF =0.31); therefore, the combined P value for all participants was only 1.24 × 10−2 in the additive model. Many researchers have reported that deletion of 3p22.1 was early and frequent events in lung tumorigenesis (25, 26). rs7629386 is about 274 kb upstream of the β-catenin gene (coded by CTNNB1), a key component of the Wnt signaling pathway (Supplementary Fig. S6A). Somatic mutations and elevated levels of β-catenin have been observed in most common forms of human malignancies (27, 28).
rs969088, rs12000445, and rs41997 were not replicated in the Harvard cohort. The reasons for the discrepancy may be as follows: First, marker SNPs might be different between ethnic populations (because of different allele frequency and LD structure). Second, the demographic and clinic characteristics of Chinese NSCLC patients were different from those in the Harvard cohort (gender, stage, smoking rate, and histologic types) and these factors are survival related. Finally, the relatively small sample size in each participant studies may yield insufficient power. The real causative SNPs need to be identified by fine mapping and comparative analyses on both Chinese and Caucasian populations.
The SNP rs969088 at 5p14.1 is located about 490 kb downstream of Cadherin 9 (CDH9; Supplementary Fig. S6B). CDH9 belongs to the cadherin family that are involved in calcium-dependent cell–cell junctions in the nervous system (29). It was reported that hypermethylation of its family members, CDH1 and CDH13, was associated with better survival of patients with NSCLC (30, 31). rs12000445 is located at 9p21.3, a region frequently deleted in lung cancer (32), about 273 kb downstream of Hu-antigen B (HuB), one of the tumor antigens (Supplementary Fig. S6D). rs41997 is located at a site with suggested tumor suppressor genes (TSG) and frequent loss of heterozygosity in a variety of tumors of epithelial origin (33, 34). Three well known TSGs were close to rs41997 (about 700 to 1100 kb): Cystic fibrosis transmembrane conductance regulator (CFTR), wingless-type mouse mammary tumor virus integration site family, member 2 (WNT2), and suppressor of tumorigenicity 7 (ST7; Supplementary Fig. S6C). Promoter hypermethylation of CFTR was reported as an important prognostic factor in younger patients with NSCLC (35). The Wnt/β-catenin signal pathway has relationships with both lung cancer development and prognosis (36), while ST7 is a highly conserved TSG with reported mutations in breast and colon carcinomas (37).
To further evaluate possible influence of the 5 SNPs on gene expression, we searched publicly available cis–expression quantitative trait loci (eQTL) information from in lymphoblastoid cell lines (38–40). We only found that rs7799229 (in highly LD with rs41997, R2 =0.90) is closely associated with the expression of N(alpha)-acetyltransfer-ase 38 (NAA38; P =2.7 × 10−7; 39), a member of the like-Sm protein family, which transiently binds U6 small nuclear RNAs and is involved in the general maturation of RNA in the nucleus (41). We did not obtain information on the genotype–phenotype correlations of the 5 SNPs and gene expression in lung cancer tissues or cell lines (42, 43).
Our study has a number of strengths. First, this is the largest GWAS study to date to investigate overall survival in advanced NSCLC patients. Second, we used a 3-stage study design with 4 independent cohorts. SNPs that are associated with overall survival across all study populations have a high probability of being true-positive findings, thus reducing the need for strict multiple comparison correction. Third, we identified 5 candidate SNPs as marker SNPs for Chinese, suggesting important biologic pathways, and largely improved current notions on NSCLC survival. Several limitations of our study also need to be addressed. Considering the heterogeneity in genetic background and treatment to NSCLC between the Han Chinese and Caucasian populations, the validation power might be limited in replication II. Another limitation of the study is that the effect of SNP rs7629386 might be unstable because of low frequency of homozygote when a HR of 3.92 was observed in Han population but the HR was much lower at 1.50 in Caucasian population. Finally, eQTL analysis did not reveal significant findings. Therefore, further validations incorporating different ethnic background populations, together with the resequencing of the marked region and functional evaluations are warranted.
Supplementary Material
Translational Relevance.
Chemotherapy with platinum-based regimens is the standard of care for advanced-stage non–small cell lung cancer (NSCLC). However, reliable genetic variations for the prediction of response and overall survival to platinum-based therapy are still not well established. In this three-stage genome-wide association study, we identified five single-nucleotide polymorphisms as markers for survival of advanced-stage NSCLC patients treated with first-line platinum-based chemotherapy in Chinese Han populations. Two of these SNPs, rs7629386 and rs3850370, could also be markers for survival among Caucasian patients.
Acknowledgments
Grant Support
This work is funded by the National Natural Science Foundation of China (81270044, 30972541, 30901233, 30730080, and 30425001), the China National High-Tech Research and Development Program Grant (2009AA022705), the National Key Basic Research Program Grant (2011CB503805), NIH (CA092824-07, U19-CA148127-01), Jiangsu Key Discipline of Medicine (XK200718), and Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: T. Wu, D. Lu, D.C. Christiani, D. Lin, Z. Hu, H. Shen
Development of methodology: R.S. Heist, J. Dai, D.C. Christiani, Z. Hu, H. Shen
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Hu, X. Zhao, R.S. Heist, L. Su, B. Han, S. Cao, M. Chu, J. Dong, Y. Shu, L. Xu, Y. Chen, Y. Wang, F. Lu, H. Chen, W. Tan, H. Ma, J. Chen, G. Jin, T. Wu, D. Lu, D.C. Christiani, Z. Hu, H. Shen
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Hu, C. Wu, R.S. Heist, Y. Zhao, B. Han, S. Cao, M. Chu, J. Dai, J. Dong, Y. Shu, L. Xu, Y. Chen, Y. Wang, F. Lu, D. Yu, H. Chen, H. Ma, J. Chen, G. Jin, D.C. Christiani, Z. Hu, H. Shen
Writing, review, and/or revision of the manuscript: L. Hu, R.S. Heist, D.C. Christiani, Z. Hu, H. Shen
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L. Hu, Y. Jiang, D.C. Christiani, Z. Hu, H. Shen
Study supervision: D. Lin, Z. Hu, H. Shen
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–60. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, Ichinose Y, Ohta M, Hata E, Tsubota N, Tada H, et al. A randomized trial of adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. N Engl J Med. 2004;350:1713–21. doi: 10.1056/NEJMoa032792. [DOI] [PubMed] [Google Scholar]
- 4.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 6.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Shu Y, Chen Y, Chen J, Dong J, Liu Y, et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am J Respir Crit Care Med. 2011;183:641–8. doi: 10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- 10.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, et al. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol. 2004;22:2594–601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 11.Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–7. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heist RS, Zhai R, Liu G, Zhou W, Lin X, Su L, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:856–62. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12. 2 in Han Chinese. Nat Genet. 2011;43:792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 14.Wu W, Zhang W, Qiao R, Chen D, Wang H, Wang Y, et al. Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clin Cancer Res. 2009;15:3889–95. doi: 10.1158/1078-0432.CCR-08-2715. [DOI] [PubMed] [Google Scholar]
- 15.Heist RS, Zhou W, Wang Z, Liu G, Neuberg D, Su L, et al. Circulating 25-hydroxyvitamin D, VDR polymorphisms, and survival in advanced non-small-cell lung cancer. J Clin Oncol. 2008;26:5596–602. doi: 10.1200/JCO.2008.18.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Ye Y, Rosell R, Amos CI, Stewart DJ, Hildebrandt MA, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst. 2011;103:817–25. doi: 10.1093/jnci/djr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, et al. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem. 2003;278:35325–36. doi: 10.1074/jbc.M305191200. [DOI] [PubMed] [Google Scholar]
- 20.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–40. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 21.Villar V, Kocic J, Bugarski D, Jovcic G, Santibanez JF. SKIP is required for TGF-beta1-induced epithelial mesenchymal transition and migration in transformed keratinocytes. FEBS Lett. 2010;584:4586–92. doi: 10.1016/j.febslet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, et al. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer. 2011;104:700–6. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu SS, Xu W, Liu S, Chen B, Wang XL, Wang Y, et al. Down-regulation of ALKBH2 increases cisplatin sensitivity in H1299 lung cancer cells. Acta Pharmacol Sin. 2011;32:393–8. doi: 10.1038/aps.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yendamuri S, Vaporciyan AA, Zaidi T, Feng L, Fernandez R, Bekele NB, et al. 3p22.1 and 10q22. 3 deletions detected by fluorescence in situ hybridization (FISH): a potential new tool for early detection of non-small cell lung Cancer (NSCLC) J Thorac Oncol. 2008;3:979–84. doi: 10.1097/JTO.0b013e3181834f3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz RL, He W, Khanna A, Fernandez RL, Zaidi TM, Krebs M, et al. Genetically abnormal circulating cells in lung cancer patients: an antigen-independent fluorescence in situ hybridization-based case-control study. Clin Cancer Res. 2010;16:3976–87. doi: 10.1158/1078-0432.CCR-09-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JS, Crooks H, Dracheva T, Nishanian TG, Singh B, Jen J, et al. Oncogenic beta-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–8. [PubMed] [Google Scholar]
- 28.Chen MW, Vacherot F, De La Taille A, Gil-Diez-De-Medina S, Shen R, Friedman RA, et al. The emergence of protocadherin-PC expression during the acquisition of apoptosis-resistance by prostate cancer cells. Oncogene. 2002;21:7861–71. doi: 10.1038/sj.onc.1205991. [DOI] [PubMed] [Google Scholar]
- 29.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–61. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 30.Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12:7329–38. doi: 10.1158/1078-0432.CCR-06-0894. [DOI] [PubMed] [Google Scholar]
- 31.Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110:2785–92. doi: 10.1002/cncr.23113. [DOI] [PubMed] [Google Scholar]
- 32.Merlo A, Gabrielson E, Askin F, Sidransky D. Frequent loss of chromosome 9 in human primary non-small cell lung cancer. Cancer Res. 1994;54:640–2. [PubMed] [Google Scholar]
- 33.Bièche I, Champème MH, Matifas F, Hacène K, Callahan R, Lidereau R. Loss of heterozygosity on chromosome 7q and aggressive primary breast cancer. Lancet. 1992;339:139–43. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 34.Zenklusen JC, Bieche I, Lidereau R, Conti CJ. (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci U S A. 1994;91:12155–8. doi: 10.1073/pnas.91.25.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011;16:1203–9. doi: 10.1111/j.1440-1843.2011.01994.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu X, Sun PL, Li JZ, Jheon S, Lee CT, Chung JH. Aberrant Wnt1/beta-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. J Thorac Oncol. 2011;6:716–24. doi: 10.1097/JTO.0b013e31820c5189. [DOI] [PubMed] [Google Scholar]
- 37.Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–8. doi: 10.1038/86891. [DOI] [PubMed] [Google Scholar]
- 38.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 39.Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–22. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 40.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spiller MP, Boon KL, Reijns MA, Beggs JD. The Lsm2–8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–9. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullaughey K, Chavarria CI, Coop G, Gilad Y. Expression quantitative trait loci detected in cell lines are often present in primary tissues. Hum Mol Genet. 2009;18:4296–303. doi: 10.1093/hmg/ddp382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321–30. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.