Abstract
Despite its eradication over 30 years ago, smallpox (as well as other orthopoxviruses) remains a pathogen of interest both in terms of biodefense and for its use as a vector for vaccines and immunotherapies. Here we describe the application of mRNA-Seq transcriptome profiling to understanding immune responses in smallpox vaccine recipients. Contrary to other studies examining gene expression in virally infected cell lines, we utilized a mixed population of PBMCs in order to capture the essential intercellular interactions that occur in vivo and would otherwise be lost using single cell lines or isolated primary cell subsets. In this mixed cell population we were able to detect expression of all annotated vaccinia genes. On the host side, a number of genes encoding cytokines, chemokines, complement factors, and intracellular signaling molecules were downregulated upon viral infection, while genes encoding histone proteins and the interferon response were upregulated. We also identified a small number of genes that exhibited significantly different expression profiles in subjects with robust humoral immunity compared to those with weaker humoral responses. Our results provide evidence that differential gene regulation patterns may be at work in individuals with robust humoral immunity compared to those with weaker humoral immune responses.
Keywords: Next Generation Sequencing; mRNA-Seq; Vaccinia virus; Smallpox vaccine; High-Throughput Nucleotide Sequencing; Genome, Human; Gene Expression Profiling; Sequence Analysis, RNA; Transcriptome
Introduction
Vaccinia virus (VACV) is the immunologically cross-protective orthopox virus found in the smallpox vaccine used in the eradication of smallpox.1 Although smallpox has been eradicated, there continues to be significant public health interest in smallpox and other orthopox viruses for multiple reasons: biodefense against weaponized poxviruses; continuing outbreaks of zoonotic orthopox virus infections; the use of vaccinia virus as a vector for cancer immunotherapy; and vaccines against other infectious agents.2–9 Thus, there continues to be a need for an increased understanding of poxvirus biology, host response to infection, and the immunologic mechanisms behind immunity to poxviruses.
Next generation sequencing (NGS) is a powerful technology that holds tremendous promise in the areas of systems biology10 and vaccinomics11–15 for developing a deeper understanding of the host response to both vaccines and viral infections. Here we describe the use of NGS mRNA-Seq to analyze transcriptomic changes occurring in PBMCs from smallpox vaccine recipients after vaccinia virus stimulation, with a focus on early, innate responses to viral stimulation.
Materials and Methods
Subject Recruitment
Details regarding the cohort from which we selected subjects for use in this study have been previously published.16–19 Briefly, we selected 44 subjects from a cohort of 1,076 recipients of Dryvax®. All subjects had been vaccinated 1–48 months prior to enrollment, were generally healthy, had received no more than one dose of the smallpox vaccine, and were successfully immunized as evidenced by the characteristic vaccine “take.” Subjects were enrolled at the Mayo Clinic (Rochester, MN) and from the Naval Health Research Center (NHRC, San Diego, CA). IRB approval from both centers (Mayo and NHRC) was obtained prior to subject enrollment, and informed consent was obtained in writing from all subjects. We selected subjects from among those individuals with the highest (n=21) and those with the lowest (n=23) vaccinia-specific neutralizing antibody titers.
Viruses and cell lines
The NYCBOH strain of vaccinia virus was purchased from ATCC (Manassas, VA), while the vSC56 strain of vaccinia virus was graciously provided by B. Moss (NIAID, Bethesda, MD). All virus strains were grown and titered according to established protocols. Hela, Hela S3, and Vero cells were also obtained from ATCC.
VACV neutralization assay
VACV-specific, neutralizing antibody titers from each subject's serum sample were obtained using a high throughput neutralization assay developed at the FDA and further optimized in our lab as previously described.16, 20
Cell cultures, RNA extraction, mRNA-Seq
PBMCs were stimulated for eight hours with or without live VACV NYCBOH at an MOI (multiplicity of infection) of 5. Following the incubation, RNAprotect reagent (Qiagen, Valencia, CA) was added to each culture and total RNA was extracted by RNeasy Plus mini Kit (Qiagen). The quantity and quality of each RNA sample were determined by Nanodrop (Thermo Fisher Scientific, Wilmington, DE) and by an Agilent 2010 Bioanalyzer (Agilent, Palo Alto, CA). cDNA libraries were created using the mRNA-Seq 8 Sample Prep Kit (Illumina, San Diego, CA) according to the manufacturer's directions. Sample preparation was performed in the Advanced Genomics Technology Center, Gene Sequencing core facility at the Mayo Clinic. Poly-A RNA was isolated using two rounds of magnetic purification with olido-dT coated beads. The purified poly-A RNA was fragmented and reverse transcribed into double-stranded cDNA fragments, which were attached to Illumina adaptor sequences. Library validation and quantification was carried out using DNA 1000 Nano Chip kits on an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). cDNA libraries were loaded onto individual channels of each flow cell at a concentration of 5–7pM. Single end read sequencing was performed using the Genome Analyzer GAIIx (Illumina, San Diego, CA) with Illumina's Single Read Cluster Generation kit (v2) and 50 Cycle Illumina Sequncing Kit (v3). Flow cells were then analyzed with SCS v2.01 and v2.4. Image processing utilized the Illumina Pipeline Software v1.5 and FireCrest, bustard, ELAND, and CASAVA.21 Viral gene expression was determined by mapping sequencing reads to the vaccinia virus ACAM2000 (GenBank: AY313847.1) reference genome using Bowtie.
Statistical modeling and analyses
Randomized block principles were utilized to allocate specimens to flow cell and lane. Specifically, samples were randomly allocated to library preparation batch, flow cell, and lane, with the constraints that: 1) high and low responders were evenly balanced across flow cell and lane assignment; and 2) all samples for a given subject were assayed on the same flow cell.
Per-gene tests of statistical significance used generalized linear models22 assuming a Negative Binomial distribution.21 An offset of the 75th percentile23 was included as a normalization factor. Predictor variables were response status, stimulation status and the interaction of these two variables. The dispersion was allowed to vary across genes and was estimated via edgeR in an empirical Bayes-like manner sharing variance information across genes (moderated dispersion) with the prior.n parameter set to 3.24, 25 Due to the natural correlation between unstimulated and stimulated specimens within a subject, generalized estimating equations26 with an exchangeable correlation structure were utilized to properly estimate the variance. Genes with counts of at least 10 per response × stimulation combination or total counts of least 440 (i.e., an average of 5 counts/single lane) were analyzed; those with fewer counts were filtered out after calculating the normalization factor. False discovery rates (FDR) were computed as using standard methods.27, 28 Self-contained gene set tests29 were conducted using the Fisher's method30, 31 with restandardization.32 Gene sets were defined using immunologically based transcriptional modules.33 Both R34 and SAS35 computing packages were utilized.
Results
The high antibody group (n=21) had a median ID50 titer of 433.4 (IQR: 400.7 – 481.9), while the low antibody titer group (n=23) had a median ID50 titer of 35.5 (IQR: 29.5 – 40.2). Of note, 19 of the low group had antibody titers below the presumed protective threshold titer of 1:32.36 Each of these 44 subjects had two samples (uninfected, vaccinia-infected). We detected a similar number of reads between samples from high and low antibody (Ab) responders. The read counts between the stimulated and unstimulated samples were also comparable. We detected a mean of 11.3 million reads in the high titer, stimulated samples (10.9 million reads mapped to the human genome and 350,000 reads mapped to the vaccinia genome). In the high titer, unstimulated samples we detected a mean of 11.7 million reads (of which a mean of only 530 reads mapped to the viral genome).
Viral Stimulation and Host Gene Expression
Viral stimulation had a dramatic effect on host gene expression, with over 1,200 genes exhibiting significant upregulation or downregulation between unstimulated and stimulated samples from all subjects (p < 0.001 and false discovery rate [FDR] < 0.01). A small subset of these genes with p-values less than 0.001, an FDR less than 0.01, and fold changes in expression > 1.5 or < 0.75 are listed in Table 1 (The Supplemental Table contains the list of 1,000 genes, all with p<0.001, that were used in the pathway analysis). These genes encode for: a number of histone proteins, cytokines and growth factors (IL3, IL18, IFNG, BMP3); chemokines and receptors (CXCL6, XCR1); G protein-coupled receptors (GPBAR1, GPR84, ADORA3); genes involved in lipid metabolism (APO2, OLR1); heat shock proteins (HSPA4L, HSPA6); cellular receptors with immune function (CD14, PDCD1LG2, TNFRSF10D); as well as a number of proteins specifically expressed in antigen presenting cells (C5orf20, MPEG-1, TREML4).
Table 1.
Effect of vaccinia stimulation on cellular gene expression.
| Cellular Genes Downregulated upon Vaccinia Stimulation | ||||
|---|---|---|---|---|
| Gene Name | Gene Description | Fold Change | p-valuea | FDRb |
| CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 0.70 | < 1.00E-15 | < 1.00E-15 |
| ARNT2 | aryl-hydrocarbon receptor nuclear translocator 2 | 0.71 | < 1.00E-15 | < 1.00E-15 |
| PDPN | podoplanin | 0.72 | < 1.00E-15 | < 1.00E-15 |
| CD14 | CD 14 molecule | 0.73 | < 1.00E-15 | < 1.00E-15 |
| S100A8 | SI00 calcium binding protein A8 | 0.73 | < 1.00E-15 | < 1.00E-15 |
| TREML4 | triggering receptor expressed on myeloid cells-like 4 | 0.74 | < 1.00E-15 | < 1.00E-15 |
| S100A9 | S100 calcium binding protein A9 | 0.74 | < 1.00E-15 | < 1.00E-15 |
| PDCD1LG2 | programmed cell death 1 ligand 2 | 0.74 | < 1.00E-15 | < 1.00E-15 |
| THBS1 | thrombospondin 1 | 0.74 | < 1.00E-15 | < 1.00E-15 |
| SIRPB2 | signal-regulatory protein beta 2 | 0.75 | < 1.00E-15 | < 1.00E-15 |
| MPEG1 | macrophage expressed 1 | 0.75 | < 1.00E-15 | < 1.00E-15 |
| GPR84 | G protein-coupled receptor 84 | 0.72 | < 1.00E-15 | 8.86E-14 |
| C5orf20 | TRAF-interacting protein with forkhead-associated domain | 0.75 | 3.42E-14 | 2.66E-12 |
| IL18 | interleukin 18 (interferon-gamma-inducing factor) | 0.71 | 3.57E-14 | 2.77E-12 |
| OLR1 | oxidized low density lipoprotein (lectin-like) receptor 1 | 0.72 | 1.42E-11 | 7.73E-10 |
| HNMT | histamine N-methyltransferase | 0.75 | 2.89E-11 | 1.48E-09 |
| HAMP | hepcidin antimicrobial peptide | 0.67 | 3.73E-11 | 1.88E-09 |
| C15orf38 | chromosome 15 open reading frame 38 | 0.75 | 7.43E-09 | 2.68E-07 |
| MYEOV | myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas) | 0.72 | 1.76E-06 | 4.16E-05 |
| C20orf103 | chromosome 20 open reading frame 103 | 0.74 | 3.15E-06 | 6.96E-05 |
| SYT15 | synaptotagmin XV | 0.68 | 3.79E-06 | 8.22E-05 |
| TSP50 | Serine protease 50, involved in cellular proliferation | 0.55 | 7.23E-06 | 0.0001 |
| XCR1 | chemokine (C motif) receptor 1 | 0.75 | 1.50E-05 | 0.0003 |
| SEMA3A | semaphorin 3A,secreted | 0.73 | 4.04E-05 | 0.0007 |
| SIRPD | signal-regulatory protein delta | 0.55 | 5.34E-05 | 0.0009 |
| APOC2 | apolipoprotein C-II | 0.73 | 6.50E-05 | 0.0011 |
| BMP3 | bone morphogenetic protein 3 | 0.42 | 6.69E-05 | 0.0011 |
| ADORA3 | adenosine A3 receptor | 0.62 | 9.11E-05 | 0.0014 |
| KCNJ10 | potassium inwardly-rectifying channel, subfamily J, member 10 | 0.74 | 0.0001 | 0.0018 |
| NDP | Norrie disease (pseudoglioma) | 0.75 | 0.0002 | 0.0032 |
| C4BPB | complement component 4 binding protein, beta | 0.66 | 0.0002 | 0.0033 |
| GPBAR1 | G protein-coupled bile acid receptor 1 | 0.74 | 0.0003 | 0.0043 |
| PTGES | prostaglandin E synthase | 0.69 | 0.000364055 | 0.004745659 |
| CHST6 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 | 0.52 | 0.000621449 | 0.007433113 |
| MFAP5 | microfibrillar associated protein 5 | 0.74 | 0.000878039 | 0.009960752 |
| Cellular Genes Unregulated upon Vaccinia Stimulation | ||||
|---|---|---|---|---|
| Gene Name | Gene Description | Fold Change | p-value | FDR |
| TNFRSF10D | tumor necrosis factor receptor superfamily lOd, decoy with truncated death domain | 1.90 | < 2.00E-16 | < 2.00E-14 |
| HIST4H4 | histone cluster 1, H41; | 2.31 | < 2.00E-16 | < 2.00E-14 |
| HIST1H4H | histone cluster 1, H41; | 3.86 | < 2.00E-16 | < 2.00E-14 |
| HIST1H4C | histone cluster 1, H41; | 5.68 | < 2.00E-16 | < 2.00E-14 |
| HSPA4L | heat shock 70kDa protein 4-like | 5.74 | < 2.00E-16 | < 2.00E-14 |
| HIST1H4E | histone cluster 1, H41; | 6.91 | < 2.00E-16 | < 2.00E-14 |
| HIST1H1D | histone cluster 1, Hid | 9.37 | < 2.00E-16 | < 2.00E-14 |
| HIST1H1E | histone cluster 1, Hie | 40.45 | < 2.00E-16 | < 2.00E-14 |
| IL3 | interleukin 3 (colony-stimulating factor, multiple) | 3.35 | 2.22E-16 | 2.36E-14 |
| HIST1H2AM | histone cluster 1, H2ag | 3.77 | 1.34E-09 | 5.40E-08 |
| IFNB1 | interferon, beta 1, fibroblast | 4.49 | 1.56E-08 | 5.29E-07 |
| EPHB3 | EPH receptor B3 | 1.65 | 3.39E-08 | 1.09E-06 |
| HIST1H2AE | histone cluster 1, H2ae | 1.85 | 3.09E-07 | 8.55E-06 |
| HSPA6 | heat shock 70kDa protein 7 (HSP70B); heat shock 70kDa protein 6 (HSP70B') | 1.60 | 2.34E-06 | 5.34E-05 |
| HIST1H2BG | histone cluster 1, H2bi; | 2.46 | 2.85E-06 | 6.34E-05 |
| RNF152 | ring finger protein 152 | 1.82 | 4.33E-06 | 9.24E-05 |
| CH25H | cholesterol 25-hydroxylase | 2.66 | 6.58E-06 | 0.000137 |
| HIST2H2AC | histone cluster 2, H2ac | 1.90 | 3.11E-05 | 0.000545 |
| WFIKKN1 | WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 | 1.52 | 0.000187 | 0.002686 |
| LVRN | laeverin, aminopeptidase Q | 2.12 | 0.000247 | 0.003421 |
| HIST1H2BN | histone cluster 1, H2bn | 1.81 | 0.000419 | 0.005343 |
| IFNG | interferon, gamma | 2.57 | 0.00069 | 0.008117 |
p-value corrected for ____.
False discovery rate.
Interaction (High Responders vs Low Responders) Assessment of Gene Expression
Our study subjects were individuals from a large cohort who had the highest and lowest neutralizing antibody titers following smallpox vaccination, allowing us to compare high responder gene expression patterns following viral stimulation to the expression patterns in low responders. Given the large number of genes analyzed, we set a p-value cutoff of 5 × 10−5 and an FDR cutoff of 0.05. After applying these thresholds, three genes remained: KIR2DL3 (a killer cell immunoglobulin-like receptor); TPSD1 (a serine protease expressed in mast cells); and UNC13A (a phorbol ester receptor).
Pathway Analysis
Recognizing that both infection and immune responses are the result of a highly complex, ordered series of events, and that contributions of individual genes may be quite small, we conducted both Metacore- and Ingenuity-based pathway analyses of host gene expression, using the top 1,000 genes with lowest p-values (all genes were p<0.05. See Supplemental Table 1). Our analyses indicated that 15 Metacore pathways and five Ingenuity pathways were significantly enriched for differentially expressed genes upon viral stimulation (Table 3). These pathways involved innate pattern recognition receptors, FcR mediated phagocytosis, DC maturation, and oxidative stress responses. When comparing the high and low responders, we included the 593 genes with p< 0.05. The Ingenuity enrichment analysis indicated that five pathways (Table 4) were differentially activated by these two immune response groups.
Table 3.
Pathways enriched in differentially expressed genes upon viral stimulation.
| Ingenuity Pathway | p-value |
|---|---|
| TREM1 Signaling | 2.43 × 10−11 |
| Dendritic Cell Maturation | 6.17 × 10−9 |
| NRF2-mediated Oxidative Stress Response | 3.48 × 10−8 |
| Role of PRR in Recognition of Bacteria and Viruses | 3.67 × 10−8 |
| FcγR-mediated Phagocytosis in Macrophages and Monocytes | 1.04 × 10−7 |
| Metacore | p-value |
| Immune response - Alternative complement pathway | 1.03 × 10−7 |
| Immune response - Classical complement pathway | 2.08 × 10−7 |
| Immune response - Lectin-induced complement pathway | 1.79 × 10−6 |
| Immune response - FcγR-mediated phagocytosis in macrophages | 7.82 × 10−5 |
| Apoptosis and survival - Inhibition of ROS-induced apoptosis by 17b-estradiol | 2.01 × 10−4 |
| Cell adhesion - Chemokines and adhesion | 2.06 × 10−4 |
| Immune response - TREM1 signaling pathway | 2.98 × 10−4 |
| Inhibitory action of Lipoxins on Superoxide production in neutrophils | 4.83 × 10−4 |
| Immune response - Inhibitory action of lipoxins on superoxide production induced by IL-8 and Leukotriene B4 in neutrophils | 4.83 × 10−4 |
| Cell adhesion - ECM remodeling | 5.37 × 10−4 |
| Development - EPO-induced MAPK pathway | 7.92 × 10−4 |
| Immune response - CCR3 signaling in eosinophils | 9.21 × 10−4 |
| Chemotaxis - Lipoxin inhibitory action on fMLP-induced neutrophil chemotaxis | 1.07 × 10−3 |
| Immune response - HMGB1/RAGE signaling pathway | 1.16 × 10−3 |
Table 4.
Pathways enriched in differentially expressed genes comparing high/low responders.
| Ingenuity Pathway | p-value |
|---|---|
| Nitrogen Metabolism | 0.002 |
| Mitochondrial Dysfunction | 0.002 |
| Dopamine Receptor Signaling | 0.003 |
| EIF2 Signaling | 0.004 |
| Corticotropin Releasing Hormone Signaling | 0.008 |
| Biosynthesis of Steroids | 0.018 |
| Aldosterone Signaling in Epithelial Cells | 0.020 |
| Lysine Biosynthesis | 0.021 |
| Endothelin-1 Signaling | 0.028 |
| Starch and Sucrose Metabolism | 0.030 |
| Systemic Lupus Erythematosus Signaling | 0.030 |
| T Helper Cell Differentiation | 0.032 |
| Signaling by Rho Family GTPases | 0.036 |
| LXR/RXR Activation | 0.037 |
| Sonic Hedgehog Signaling | 0.038 |
| Melatonin Signaling | 0.038 |
| Regulation of eIF4 and p70S6K Signaling | 0.040 |
Geneset Analysis
Gene set analysis was also conducted using the transcriptional gene modules described by Banchereau et al.37 Almost two-thirds of the transcriptional modules examined (164/260) exhibited significantly different (p < 0.05) expression patterns upon viral infection. In contrast, only a single transcriptional module, M7.35 (Table 6), varied between high and low antibody responders. Gene set analysis using a subset of the BROAD genesets (those that included the keyword “immune” in their title/functional description) indicated that 200 of the 234 immunology-related genesets were differentially activated upon infection (p<0.05), while geneset 132 (Innate_Immune_Response) was the only geneset with significant differences when comparing high and low antibody responders.
Table 6.
Transcriptional Module M7.35
| Symbol | Description |
|---|---|
| ANKRD22 | Homo sapiens ankyrin repeat domain 22 (ANKRD22), mRNA. |
| CCNA1 | Homo sapiens cyclin A1 (CCNA1), mRNA. |
| CD163 | Homo sapiens CD163 molecule (CD163), transcript variant 1, mRNA. |
| CD177 | Homo sapiens CD177 molecule (CD177), mRNA. |
| CLEC5A | Homo sapiens C-type lectin domain family 5, member A (CLEC5A), mRNA. |
| CREB5 | Homo sapiens cAMP responsive element binding protein 5 (CREB5), transcript variant 4, mRNA. |
| DAAM2 | Homo sapiens dishevelled associated activator of morphogenesis 2 (DAAM2), mRNA. |
| ECHDC3 | Homo sapiens enoyl Coenzyme A hydratase domain containing 3 (ECHDC3), mRNA. |
| GPR84 | Homo sapiens G protein-coupled receptor 84 (GPR84), mRNA. |
| IL1R1 | Homo sapiens interleukin 1 receptor, type I (IL1R1), mRNA. |
| KIAA1026 | Homo sapiens kazrin (KIAA1026), transcript variant B, mRNA. |
| LOC400793 | PREDICTED: Homo sapiens hypothetical LOC400793, transcript variant 2 (LOC400793), mRNA. |
| LOC401233 | Homo sapiens similar to HIV TAT specific factor 1; cofactor required for Tat activation of HIV-1 transcription (LOC401233), mRNA. |
| LOC651612 | PREDICTED: Homo sapiens hypothetical protein LOC651612 (LOC651612), mRNA. |
| METTL7B | Homo sapiens methyltransferase like 7B (METTL7B), mRNA. |
| MYO10 | Homo sapiens myosin X (MYO10), mRNA. |
| OLAH | Homo sapiens oleoyl-ACP hydrolase (OLAH), transcript variant 1, mRNA. |
| ORM1 | Homo sapiens orosomucoid 1 (ORM1), mRNA. |
| ORM2 | Homo sapiens orosomucoid 2 (ORM2), mRNA. |
| PCOLCE2 | Homo sapiens procollagen C-endopeptidase enhancer 2 (PCOLCE2), mRNA. |
| PFKFB2 | Homo sapiens 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 (PFKFB2), transcript variant 2, mRNA. |
| SLC1A3 | Homo sapiens solute carrier family 1 (glial high affinity glutamate transporter), member 3 (SLC1A3), mRNA. |
| SLC2A11 | Homo sapiens solute carrier family 2 (facilitated glucose transporter), member 11 (SLC2A11), transcript variant 3, mRNA. |
| SYN2 | Homo sapiens synapsin II (SYN2), transcript variant IIa, mRNA. |
| TDRD9 | Homo sapiens tudor domain containing 9 (TDRD9), mRNA. |
| TLR2 | Homo sapiens toll-like receptor 2 (TLR2), mRNA. |
| ZDHHC19 | Homo sapiens zinc finger, DHHC-type containing 19 (ZDHHC19), mRNA. |
Viral Gene Expression
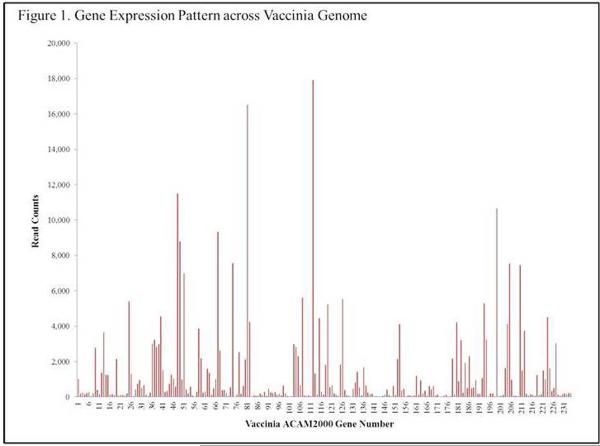
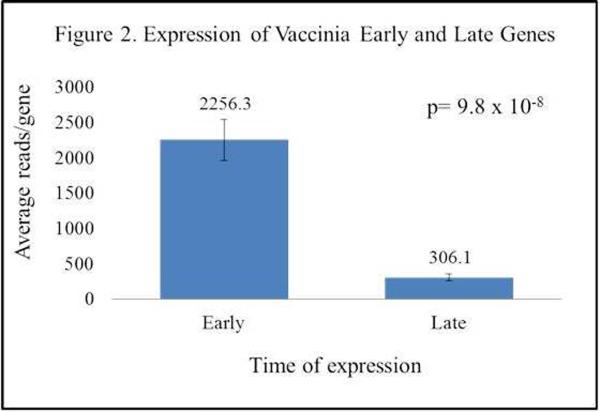
mRNA-Seq analysis provides both host and viral gene expression data, and allowed us to investigate host-pathogen interaction at the gene expression level. Figure 1 illustrates the average read count across all samples (infected read count – uninfected read counts) for each of the ~250 open reading frames (ORFs) of vaccinia virus. Reads were mapped to the ACAM2000 sequence (Genbank: AY313848.1). We detected gene expression of each of the putative viral genes across a wide range of expression levels. Mean viral read counts ranged from 71 for the A38L semaphorin gene to 10,740 for the H5R transcription factor. Closer examination of the data found that genes with the lowest expression values were predominantly expressed late in the viral life cycle, while genes classified as “early” were expressed at significantly higher levels (Figure 2). Vaccinia virus possesses a number of immunomodulatory proteins, almost all of which were detected in our samples. Table 5 illustrates the expression levels of these viral genes. Interestingly, there were no significant differences between viral gene expression in samples from high and low antibody responders.
Figure 1.
Gene Expression Pattern across Vaccinia Genome
Figure 2.
Expression of Vaccinia Early and Late Genes
Table 5.
Immunomodulatory Gene Expression by Vaccinia Virus.
| ACAM Name | COP Name | Function | Median (IQR) # Reads Stim | Median # Reads Unstim |
|---|---|---|---|---|
| 001 | C23L, B29R | Chemokine binding protein | 1,006 (485 – 1,546) | 1 |
| 002 | C22L, B28R | TNFaR | 166 (103 – 223) | 0 |
| 003 | TNFaR | 231 (122 – 305) | 0 | |
| 013 | C12L | Serpin 1,2,3 | 3,640 (2,026 – 5,187) | 7 |
| 014 | C11R | EGF | 1,254 (577 – 1,973) | 2 |
| 015 | C10L | IL-1R antagonist | 1,244 (672 – 1,886) | 2 |
| 018 | apoptosis, host defense mod | 92 (45 – 145) | 0 | |
| 019 | IL-18 binding prot | 2,147 (1,158 – 3,661) | 4 | |
| 031 | C4L | IL 1R antagonist | 502 (241 – 706) | 1 |
| 032 | C4L | IL 1R antagonist | 666 (304 – 882) | 1 |
| 033 | C4L | IL 1R antagonist | 155 (66 – 210) | 0 |
| 037 | N1L | NFkB inh./virokine | 3,230 (1,684 – 4,651) | 4 |
| 042 | M2L | NF-kB Inh | 4,554 (3,176 – 7,382) | 8 |
| 045 | K3L | IFN resist, PKR inh | 335 (215 – 785) | 1 |
| 049 | K7R | Inh IFNb signal | 572 (238 – 750) | 1 |
| 069 | E3L | IFN resist, PKR inh, dsRNA binding | 9,325 (5,180 – 15,798) | 16 |
| 183 | A46R | IL-1 signal inhibitor TLR-like | 4,233 (2,232 – 6,450) | 8 |
| 189 | A52R | TLR/IL-1 signal inhibitor | 486 (243 – 666) | 1 |
| 201 | B8R | IFNg Receptor | 10,664 (6,994 – 14,488) | 16 |
| 206 | B13R | SPI-2, crmA, IL-1 convertase, | 4,110 (2,351 – 5,452) | 5 |
| 207 | B14R | SPI-2, crmA, IL-1 convertase | 7,536 (4,242 – 10,295) | 9 |
| 212 | B19R | secreted IFNa/b receptor | 7,452 (4,629 – 10,948) | 13 |
| 224 | IL 18 binding | 1,235 (700 – 2,270) | 2 | |
| 229 | C12L | SPI-1 | 4,506 (2,566 – 6,306) | 8 |
| 239 | TNFaR like | 210 (140 – 310) | 0 | |
| 240 | TNFaR like | 187 (110 – 223) | 0 | |
| 034 | C3L | Complement Binding | 72 (46 – 134) | 0 |
| 044 | K2L | serpin, SPI-3, host defense modulator | 263 (167 – 513) | 0 |
| 190 | A53R | TNFR Crm C | 538 (279 – 789) | 1 |
| 209 | B16R | IL-1b Receptor | 73 (42 – 125) | 0 |
Discussion
The transcriptional profiles of PBMCs from individuals vaccinated with the Dryvax® smallpox vaccine were assessed after stimulation with vaccinia virus for eight hours. Our primary interest was to examine the early transcriptomic events that occur during innate responses to vaccinia virus in a mixed cell population. Because the subjects tested were taken from those with the two extremes of the humoral immune response after smallpox vaccination we also compared gene expression patterns in individuals with high and low vaccinia-specific neutralizing antibody responses. Transcriptomic analysis of the effect of vaccinia infection on host cells has previously been reported primarily in immortalized cell lines or single cell subsets, and has typically utilized microarray technology. Here we report the use of mixed-cell population PBMCs and NGS technology to assess global gene expression changes (both cellular and viral).
Individual Genes – Stim/Unstim
Previous reports of host gene expression after vaccinia infection indicate a generalized downregulation of gene expression with a few select genes being upregulated. Rubins et al used microarrays to compare the effect of vaccinia and monkeypox infection on gene expression patterns in macrophages, fibroblasts, and HeLa cells and identified clusters of genes involved in innate immunity that were downregulated upon infection with VACV.38 Of the genes that they identified, in our experimental system only CXCL3 and STAT1 were downregulated, while the remaining genes exhibited insignificant changes in expression pattern. This is likely due to the different cell types examined. Rubins et al reported that their three cell types responded to viral infection with distinct differences in the gene expression pattern. Moss et al. used RNA sequencing to simultaneously analyze host and viral gene expression patterns after infection of HeLa cells with VACV.39 Four hours postinfection, they reported that 50–75% of host genes were decreased, while relatively few genes were overexpressed. They also indicated that expression of genes involved in NF-kB signaling, apoptosis, signal transduction, and other ligand-mediated signaling pathways was significantly altered. These results match our findings in a mixed-cell population (Table 1), indicating that general features of vaccinia infection may be shared across cell type. Interestingly, a number of histone genes exhibited strong (2–40 fold) upregulation upon vaccinia infection. This is similar to the results obtained in published reports using monkeypox and rabbitpox.40, 41 The authors speculate that cellular histone proteins may play a role in the organization and compaction of the viral genome.40 Our data support these earlier results; vaccinia virus elicits a similar effect as the other poxviruses. At this point it is too early to determine if this effect is the result of an antiviral host response or necessary for the poxvirus life cycle.
Several groups have shown that monocytes are particularly susceptible to vaccinia infection.42, 43 In our hands, flow cytometry analysis of PBMCs infected with vaccinia virus for eight hours indicate that the vast majority (>85%) of monocytes are infected (data not shown) and our mRNA-Seq results indicated that a large number of monocyte/macrophage related genes (CD14, MPEG1, PDCD1LG2) have significantly altered expression. The CD14 gene produces a surface antigen expressed on macrophages (and to a lesser extent by some granulocytes and dendritic cells)44 that recognizes lipopolysaccharide,45 leading to activation of NF-kB, cytokine secretion, and initiation of inflammatory responses. MPEG1 encodes for a macrophage-specific protein with limited homology to perforin.46 PDCD1LG2 encodes for the programmed cell death 1 ligand 2 protein (a costimulatory molecule essential for T cell proliferation and IFNγ production) was downregulated by vaccinia infection. Importantly, IL18 gene expression was also downregulated upon infection. This cytokine has been shown to play a critical role in cellular responses to poxvirus infection,47 and we have previously reported that single nucleotide polymorphisms in both IL18 and IL18R genes are associated with variations in immune response following smallpox vaccination.19
Individual Genes – Interaction
In the interaction analysis (differential effect of vaccinia stimulation in high and low responders), the KIR2DL3 gene was expressed at a significantly lower level in high responders and was downregulated upon vaccinia stimulation; in contrast, the low responders had higher baseline levels that increased upon viral stimulation. KIR2DL3 is a killer cell Ig-like receptor with two immunoglobulin domains and a longer cytoplasmic tail containing the immune tyrosine-based inhibitory motif (ITIM) that inhibits natural killer (NK) cell lysis of target cells expressing HLA-C alleles. KIR2DL3 has been linked to the resolution of hepatitis C virus infection, and this effect requires expression of both the NK receptor and its HLA-C1 ligand. It is possible that downregulation of this NK receptor in high responders allows for increased killing of vaccinia-infected cells; however, a cause and effect relationship, if one exists, between high responder status and KIR2DL3 expression is unclear and will require additional study. The second gene of interest, TPSD1, encodes for a mast cell serine protease. TPSD1 was expressed at similar levels in both high and low responders, but vaccinia stimulation resulted in increased expression in high responders and the opposite effect in low responders. TPSD1 contains a premature stop codon that leads to the loss of the C terminal regions necessary for optimal catalytic activity, but has been implicated in autoimmune pathology.48, 49 Although TPSD1 is largely inactive, an increase in gene expression may serve as an indicator of mast cell activity. Elevated levels may be due to mast cell recognition of vaccinia through IgE, or may merely be an indirect effect of other immune recognition pathways. The third significant gene, UNC13A, is a phorbol ester receptor similar to protein kinase C that is integral to synaptic vesicle priming.50 UNC13A expression levels were lower to begin with in high responders and decreased 20% upon viral stimulation, while in low responders background levels were high to begin with and exhibited a dramatic decrease (~45%) upon stimulation (p = 2.96 × 10−5). UNC13A has also been shown to interact with Rab37 and control TNFα secretion in activated macrophages;51 however, TNFA mRNA expression did not differ between the high and low responder groups. The high Ab responders secreted greater amounts of TNFα (224.7 pg/ml versus 150.8 pg/ml in low responders), although the difference did not reach statistical significance (p=0.14).
Pathway Analysis
Our pathway analyses indicated several important immune recognition pathways exhibiting differential activation upon viral stimulation, including complement, pattern recognition receptor, dendritic cell maturation pathways, as well as oxidative stress response, TREM1 signaling and FcγR-mediated phagocytosis. In spite of high level transcription of immunomodulatory genes, some innate immune recognition pathways are still activated in response to vaccinia infection, while others are suppressed. These results further illustrate the dynamic tension that exists between host and pathogen. TREM1 encodes for an Ig superfamily receptor expressed on monocytes and neutrophils and is upregulated in response to pathogen associated molecular patterns resulting in monocyte activation, secretion of IL-8, TNFa, MCP-1, and upregulation of adhesion and costimulatory molecules (ITGB1, CD40).52 Flow analysis of PBMCs from these subjects infected with vaccinia virus revealed a strong propensity for vaccinia virus to infect CD14+ monocytes (data not shown). Our transcriptomic data indicate that TREM1 and multiple downstream components of its signaling pathways are decreased upon viral infection, which might impair the immune function of infected monocytes and may lead to a decreased ability to prime adaptive immune responses.
Gene Set Analysis
Our gene set analysis comparing uninfected and infected samples indicated that a large number of transcriptional modules were differentially expressed. These modules were identified from subjects with a variety of immunologic conditions and so it is not surprising that many of the same modules would be affected by a viral infection. With the exception of module M7.35, we did not see significant differences between subjects with robust or weak antibody responses to smallpox vaccine. A potential functional role for this module has not yet been determined; however, several genes within the module are integral to innate and inflammatory responses (IL1R1, TLR2). Other genes include: the monocyte marker CD163; CLEC5A, a C-type lectin that serves as a macrophage recognition receptor for dengue virus stimulating pro-inflammatory responses;53 and the ORM1 and ORM2 proteins involved in transport of lipophilic compounds in the blood.54 The ORM proteins are also thought to regulate immune function during acute phase responses.55 Further examination of the contributions of these genetic elements in host responses to vaccinia infection is warranted and may provide additional insights into host-pathogen interactions. The Broad gene set M3064 (annotated by GO:0045087), containing 23 innate immune response genes, was the only geneset with significantly different expression when comparing high and low antibody responders. This gene set includes several defensins (DEFB1, DEFB118, DEFB127), IL12A and IL12B, as well as receptors involved in NK cell activity (CD1D, CRTAM, and NCR1). It is possible that high antibody responders have more innate responses that more readily recognize and react to vaccinia virus, and that this increased innate activity promotes stronger adaptive responses culminating in higher vaccinia-neutralizing antibody titer.
Viral Gene Expression Analysis
Examination of the expression levels of viral genes indicated robust viral gene expression, predominantly in early genes (Figure 2). We saw a striking similarity of viral gene expression between individuals with high and low immune response to the smallpox vaccine. One possible explanation may be the experimental setup in which subjects' PBMCs were isolated, frozen, thawed, and placed in tissue culture with growth medium for an eight-hour in vitro infection. These conditions abrogate immediate binding by virus-specific serum antibody and may not allow sufficient time for the differential cellular immune reactivity between these two groups to alter the initial and early rounds of viral replication.
Assarsson et al used a genome-tiling array to measure expression kinetics of 223 vaccinia genes after infection of Hela cells and found that a majority of the viral genes were detected at the eight-hour time point.56 Several of the genes that they did not find expressed at any of their studied time points were expressed in our experiments (mean read counts are as follows: WR092 = 41 reads; WR097 = 766 reads; WR145 = 378 reads; WR162= 141 reads; WR206 = 5,022. See Figure 1). Our data indicates that each of the 241 ACAM2000 ORFs were expressed in the stimulated samples and for some viral genes we identified relatively high expression levels (5,000–10,000 readcounts), indicating that strain and, more likely, cell-specific differences can dramatically affect viral gene expression and care must be taken when comparing expression data across studies.
A limitation of this study is the possible dilution of observable effects, given that each individual cell type may respond differently to vaccinia infection. On the other hand, this system more closely matches the in vivo environment during infection, and allows for the myriad cell-cell interactions that will occur during an infection or vaccination event. These interactions, involving either cell-to-cell contact, or mediated through soluble factors, are likely to alter the local microenvironment and the individual cell response to infection. The response of a mixed T cell:macrophage population may not reflect the response of a pure T cell population nor that a pure macrophage population, but, rather, will include at least four components: 1) a T cell response to infection; 2) a macrophage response to infection; 3) a T cell response to infected macrophages; and 4) a macrophage response to infected T cells. However, further studies on purified cell populations such as monocytes (given our findings with multiple monocyte-specific genes), or, alternatively, B cells or Th cells (given the neutralizing antibody titer-based subject selection) will yield valuable additional information. Our subjects were selected based on differences in humoral immunity and are likely to have different numbers of vaccinia-specific memory T and B cells. It is possible that inter-individual differences in the memory lymphocyte pool contribute to the differences seen in this study. We selected an eight hour time-point in order to allow for the development of early innate responses while minimizing the contribution of the memory T or B cells.
A strength of this study is the combined use of both individual analyses and pathway/geneset analyses along with highly sensitive NextGen Sequencing technology. Taken together, these separate analyses can identify individual components of the immune response, and the interaction of multiple signaling components. The geneset-type analyses have the additional benefit of reducing the number of associations to help offset false discovery.
Previous reports of gene expression in the context of vaccinia infection have focused on established cell lines or on primary cultures of single cell types. Our results indicate that numerous innate genes and pathways are activated upon vaccinia infection of a complex mixture of PBMCs. A number of chemokines, cytokines, interferons, and macrophage-associated genes exhibited significant downregulation upon infection. Upregulated genes included histones, IFNβ, IFNγ and heat shock proteins. Our data also indicate that notable differences in gene expression between high and low responders to the smallpox vaccine exist. It is possible that these differences are the result of divergent immunoregulatory processes in high and low responders. Further investigation of the effect of these loci on immune responses to viral vaccines may lead to important findings regarding genetic control of immune responses and the ability to use such information in engineering new vaccine candidates.10, 12, 15
Supplementary Material
Table 2.
Differential effect of vaccinia stimulation on high and low responders
| Gene Name | Gene Description | FC High responders | FC Low responders | p-value | FDR |
|---|---|---|---|---|---|
| KIR2DL3 | Killer cell immunoglobulin-like receptor, long cytoplasmic tail | 0.88 | 1.2 | 1.55E-12 | 3.76E-09 |
| TPSD1 | Tryptase, delta 1 | 1.15 | 0.81 | 1.68E-11 | 3.49E-08 |
| UNC13A | Unc-13 homolog A | 0.83 | 0.62 | 2.96E-05 | 0.05 |
Acknowledgements
We gratefully acknowledge the subjects who participated in this study. We wish to thank the nurses, phlebotomists and study coordinators at both the Naval Health Research Center and the Mayo Clinic – particularly, Dr. Meg Ryan for her time and efforts in subject recruitment, and Shaun Riska, Sumit Middha and Asha Nair for biostatistical and bioinformatics support. This study was supported by the National Institutes of Health contract HHSN266200400065C (AI40065). The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Conflict of Interest/Disclosures
Dr. Poland is the chair of a Safety Evaluation Committee for investigation vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice to Merck & Co. Inc, CSL Biotherapies, Avianax, Sanofi Pasteur, Dynavax, Novartis Vaccines and Therapeutics, and PAXVAX Inc. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. The other authors do not have any conflict of interest. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
Supplementary information is available at http://www.nature.com/gene/index.html.
References
- 1.Fenner F. Smallpox and its eradication. World Health Organization; Geneva: 1988. [Google Scholar]
- 2.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, et al. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 1999;281(22):2127–37. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy RB, Ovsyannikova I, Poland GA. Smallpox vaccines for biodefense. Vaccine. 2009;27(Suppl 4):D73–9. doi: 10.1016/j.vaccine.2009.07.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Giulio DB, Eckburg PB. Human monkeypox: an emerging zoonosis. The Lancet Infect Dis. 2004;4(1):15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trindade GS, Guedes MI, Drumond BP, Mota BE, Abrahao JS, Lobato ZI, et al. Zoonotic vaccinia virus: clinical and immunological characteristics in a naturally infected patient. Clin Infect Dis. 2009;48(3):e37–40. doi: 10.1086/595856. [DOI] [PubMed] [Google Scholar]
- 6.Essajee S, Kaufman HL. Poxvirus vaccines for cancer and HIV therapy. Expert Opinion Biol Ther. 2004;4(4):575–88. doi: 10.1517/14712598.4.4.575. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert PA, McFadden G. Poxvirus cancer therapy. Recent Patents on Anti-infective Drug Disc. 2006;1(3):309–21. doi: 10.2174/157489106778777592. [DOI] [PubMed] [Google Scholar]
- 8.Arlen PM, Skarupa L, Pazdur M, Seetharam M, Tsang KY, Grosenbach DW, et al. Clinical safety of a viral vector based prostate cancer vaccine strategy. J Urol. 2007;178(4 Pt 1):1515–20. doi: 10.1016/j.juro.2007.05.117. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Hadjipanayis AG, Parker MD. Viral vectors for use in the development of biodefense vaccines. Adv Drug Deliv Rev. 2005;57(9):1293–314. doi: 10.1016/j.addr.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Oberg AL, Kennedy RB, Li P, Ovsyannikova IG, Poland GA. Systems biology approaches to new vaccine development. Current Opin Immunol. 2011;23(3):436–43. doi: 10.1016/j.coi.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poland GA, Jacobson RM, Ovsyannikova IG. Trends affecting the future of vaccine development and delivery: the role of demographics, regulatory science, the anti-vaccine movement, and vaccinomics. Vaccine. 2009;27(25–26):3240–4. doi: 10.1016/j.vaccine.2009.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poland GA, Kennedy RB, Ovsyannikova IG. Vaccinomics and personalized vaccinology: is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathog. 2011;7(12):e1002344. doi: 10.1371/journal.ppat.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poland GA, Ovsyannikova IG, Jacobson RM. Personalized vaccines: the emerging field of vaccinomics. Expert Opini Biol Ther. 2008;8(11):1659–67. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poland GA, Ovsyannikova IG, Jacobson RM, Smith DI. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol and Ther. 2007;82(6):653–64. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- 15.Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS. 2011;15(9):625–36. doi: 10.1089/omi.2011.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy R, Pankratz VS, Swanson E, Watson D, Golding H, Poland GA. Statistical approach to estimate vaccinia-specific neutralizing antibody titers using a high-throughput assay. Clin Vacc Immunol : CVI. 2009;16(8):1105–12. doi: 10.1128/CVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, et al. Gender effects on humoral immune responses to smallpox vaccine. Vaccine. 2009 doi: 10.1016/j.vaccine.2009.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Human leukocyte antigen genotypes in the genetic control of adaptive immune responses to smallpox vaccine. J Infect Dis. 2011;203(11):1546–55. doi: 10.1093/infdis/jir167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haralambieva IH, Ovsyannikova IG, Dhiman N, Kennedy RB, O'Byrne M, Pankratz VS, et al. Common SNPs/Haplotypes in IL18R1 and IL18 Genes Are Associated With Variations in Humoral Immunity to Smallpox Vaccination in Caucasians and African Americans. J Infect Dis. 2011;204(3):433–41. doi: 10.1093/infdis/jir268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manischewitz J, King LR, Bleckwenn NA, Shiloach J, Taffs R, Merchlinsky M, et al. Development of a novel vaccinia-neutralization assay based on reporter-gene expression. J Infect Dis. 2003;188(3):440–8. doi: 10.1086/376557. [DOI] [PubMed] [Google Scholar]
- 21.Oberg AL, Bot BM, Grill DE, Poland GA, Therneau TM. Technical and biological variance structure in mRNA-Seq data: life in the real world. BMC Genomics. 2012;13(1):304. doi: 10.1186/1471-2164-13-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullagh P, Nelder JA. Generalized linear models. Chapman and Hall; London, England: 1983. [Google Scholar]
- 23.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–32. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 26.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121–30. [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 28.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23(8):980–7. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- 30.Fridley BL, Jenkins GD, Biernacka JM. Self-contained gene-set analysis of expression data: an evaluation of existing and novel methods. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0012693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher RA. Statistical methods for research workers. 4th edn Oliver and Boyd; Edinburgh etc.: 1932. [Google Scholar]
- 32.Witten DM, Tibshirani R. Testing Significance of Features by Lassoed Principal Components. Ann Appl Stat. 2008;2(3):986–1012. doi: 10.1214/08-AOAS182SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Team RDC. R: A language and environment for statistical computing. 2008. [Google Scholar]
- 35.SAS Institute I . SAS/STAT User's Guide. Version 9 SAS Institute; Cary, NC: 2005. [Google Scholar]
- 36.Mack TM, Noble J, Jr., Thomas DB. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg. 1972;21(2):214–8. doi: 10.4269/ajtmh.1972.21.214. [DOI] [PubMed] [Google Scholar]
- 37.Banchereau R, Jordan-Villegas A, Ardura M, Mejias A, Baldwin N, Xu H, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7(4):e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubins KH, Hensley LE, Relman DA, Brown PO. Stunned silence: gene expression programs in human cells infected with monkeypox or vaccinia virus. PLoS One. 2011;6(1):e15615. doi: 10.1371/journal.pone.0015615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad of Sci USA. 2010;107(25):11513–8. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alkhalil A, Hammamieh R, Hardick J, Ichou MA, Jett M, Ibrahim S. Gene expression profiling of monkeypox virus-infected cells reveals novel interfaces for host-virus interactions. Virol J. 2010;7:173. doi: 10.1186/1743-422X-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology. 2003;315(2):322–34. doi: 10.1016/s0042-6822(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Puig JM, Sanchez L, Roy G, Blasco R. Susceptibility of different leukocyte cell types to Vaccinia virus infection. Virol J. 2004;1:10. doi: 10.1186/1743-422X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Q, Jones B, Hu N, Chang H, Ahmad S, Liu J, et al. Comparative analysis of tropism between canarypox (ALVAC) and vaccinia viruses reveals a more restricted and preferential tropism of ALVAC for human cells of the monocytic lineage. Vaccine. 2006;24(40–41):6376–91. doi: 10.1016/j.vaccine.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Griffin JD, Ritz J, Nadler LM, Schlossman SF. Expression of myeloid differentiation antigens on normal and malignant myeloid cells. J Clin Invest. 1981;68(4):932–41. doi: 10.1172/JCI110348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver J, et al. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4(4):407–14. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 46.Spilsbury K, O'Mara MA, Wu WM, Rowe PB, Symonds G, Takayama Y. Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood. 1995;85(6):1620–9. [PubMed] [Google Scholar]
- 47.Wang Y, Chaudhri G, Jackson RJ, Karupiah G. IL-12p40 and IL-18 play pivotal roles in orchestrating the cell-mediated immune response to a poxvirus infection. J Immunol. 2009;183(5):3324–31. doi: 10.4049/jimmunol.0803985. [DOI] [PubMed] [Google Scholar]
- 48.Pallaoro M, Fejzo MS, Shayesteh L, Blount JL, Caughey GH. Characterization of genes encoding known and novel human mast cell tryptases on chromosome 16p13.3. J Biol Chem. 1999;274(6):3355–62. doi: 10.1074/jbc.274.6.3355. [DOI] [PubMed] [Google Scholar]
- 49.Caughey GH. Mast cell tryptases and chymases in inflammation and host defense. Immunol Rev. 2007;217:141–54. doi: 10.1111/j.1600-065X.2007.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossner S, Fuchsbrunner K, Lange-Dohna C, Hartlage-Rubsamen M, Bigl V, Betz A, et al. Munc13-1-mediated vesicle priming contributes to secretory amyloid precursor protein processing. J Biol Chem. 2004;279(27):27841–4. doi: 10.1074/jbc.C400122200. [DOI] [PubMed] [Google Scholar]
- 51.Mori R, Ikematsu K, Kitaguchi T, Kim SE, Okamoto M, Chiba T, et al. Release of TNF-alpha from macrophages is mediated by small GTPase Rab37. Euro J Immunol. 2011;41(11):3230–9. doi: 10.1002/eji.201141640. [DOI] [PubMed] [Google Scholar]
- 52.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164(10):4991–5. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 53.Chen ST, Lin YL, Huang MT, Wu MF, Cheng SC, Lei HY, et al. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 2008;453(7195):672–6. doi: 10.1038/nature07013. [DOI] [PubMed] [Google Scholar]
- 54.Treuheit MJ, Costello CE, Halsall HB. Analysis of the five glycosylation sites of human alpha 1-acid glycoprotein. Biochem J. 1992;283(Pt 1):105–12. doi: 10.1042/bj2830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fournier T, Medjoubi NN, Porquet D. Alpha-1-acid glycoprotein. Biochim Biophys Acta. 2000;1482(1–2):157–71. doi: 10.1016/s0167-4838(00)00153-9. [DOI] [PubMed] [Google Scholar]
- 56.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, et al. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad of Sci USA. 2008;105(6):2140–5. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.