Abstract
Aim
To study prostate cancer zonal differences in TMPRSS2-ERG gene rearrangement.
Methods and Results
We examined 136 well-characterized dominant anterior prostatic tumors, including 61 transition zone (TZ) and 75 anterior peripheral zone (PZ) lesions, defined using strict anatomic considerations. TMPRSS2-ERG FISH and ERG protein immunohistochemistry were performed on tissue microarrays. FISH results, available for 56 TZ and 71 anterior PZ samples, were correlated with ERG staining and TZ-associated “clear cell” histology. Fewer TZ cancers (4/56; 7%) were rearranged than anterior PZ cancers (18/71; 25%) [p=0.009]; deletion was the sole mechanism of TZ cancer rearrangement. ERG protein overexpression was present in 4% (2/56; both FISH +) and 30% (21/71; 17 FISH +) of TZ and anterior PZ tumors. “Clear cell” histology was present in 21/56 (38%) TZ and 8/71 (11%) anterior PZ tumors. 7% of cancers with and 21% without this histology had rearrangement, regardless of zonal origin.
Conclusions
TMPRSS2-ERG rearrangement occurs in dominant TZ and anterior PZ prostate cancers, with all rearranged TZ cancers in this cohort showing deletion. ERG immunohistochemistry demonstrated excellent sensitivity (86%) and specificity (96%) for TMPRSS2-ERG rearrangement. TMPRSS2-ERG fusion is rare in TZ tumors and present at a low frequency in tumors displaying “clear cell” histology.
Keywords: TMPRSS2-ERG rearrangement, transition zone, prostate, cancer, immunohistochemistry
Introduction
Fusion of androgen-regulated transmembrane protease serine 2 (TMPRSS2) to the erythroblastosis virus E26 transforming sequences (ETS) transcription factor gene family members, most commonly to v-ets erythroblastosis virus E26 oncogene homolog (ERG), is an early event in prostate cancer and plays an important role in prostate carcinogenesis (1–5). Since the original report by Tomlins et al in 2005 (6), multiple independent studies have validated the frequency of the TMPRSS2-ETS fusion in prostate cancer, with the dominant rearrangement – TMPRSS2-ERG fusion – being reported in 30–70% of hospital-based surgical cohorts and in a lower frequency (15–30%) of population-based cohorts (1–2, 7–13).
Whereas some studies have suggested association of TMPRSS2-ERG rearrangement with adverse clinicopathologic prognostic variables (2, 8, 10, 14), others have not found this association or have even reported a negative correlation of the rearrangement with prognosis (9, 15). ERG gene overexpression by RT-PCR has also been shown to have a protective effect and associations with lower grade and pathologic stage, negative surgical margins and longer recurrence free survival (7, 16–17).
Prostate cancer arising from different zones has been shown to have morphologic and molecular differences, suggesting different biological mechanisms underlying the development and progression of carcinoma arising in the transition (TZ) v. peripheral (PZ) zone. While TZ tumors may be of larger volume and associated with higher serum PSA values than PZ tumors, most reports have maintained that TZ tumors show lower Gleason scores, a more indolent course and overall more favorable prognosis (18–23). Morphologically, TZ cancers more frequently display “clear-cell” histology than PZ tumors (19, 24–26). Differences in gene expression profiles have also been reported between tumors of different zones (27–28), although molecular mechanisms resulting in different biology have not been delineated.
Compared with reports on TMPRSS2-ERG and outcomes, relatively few studies have addressed TMPRSS2-ERG gene rearrangements in TZ prostate cancer and none have compared fusion status in dominant TZ cancers with that in dominant anterior PZ tumors. The aim of this study therefore, was to analyze a large cohort of TZ and anterior PZ cancers for TMPRSS2-ERG rearrangement to determine zonal incidence and to correlate with the presence of “clear cell” histology. We further compared the utility of a fluorescence in situ hybridization (FISH)-based assay to the ERG antibody immunohistochemistry (IHC) assay for detection of the rearrangement and the overexpression of ERG protein in these cohorts.
Materials and Methods
Cohort Selection
This study was conducted with approval from the Institutional Review Board of Memorial Sloan Kettering Cancer Center. We utilized cases from a well-annotated group of dominant anterior TZ and PZ tumors, based on whole-mounted, entirely-submitted prostates, described in detail in a previously published report from our group (29). Importantly, zonal origin was strictly defined by anatomic considerations, including variation in constitution of the anterior prostate from apex to base – specifically the abundance of anterior PZ tissue in the apical prostate, the so-called ‘apical horns’ – as well as relationship of tumor to the anterior fibromuscular stoma (30). Our previous study demonstrated no significant differences in pathologic features between dominant tumors of TZ and anterior PZ origin with regard to Gleason score, incidence of extraprostatic extension and surgical margin status.
Tissue microarrays (TMA) were constructed from 136 tumors (61 TZ and 75 anterior PZ). Hematoxylin and eosin (H&E) slides of the prostatectomy specimens were reviewed by at least two urological pathologists and slides containing tumor were marked and matched with corresponding paraffin blocks. 0.6 mm tissue cores were punched in triplicate from locations randomly selected within the marked tumor areas and mounted in blank recipient blocks using an automated tissue microarrayer (Beecher Instruments Inc.).
FISH for detection of gene rearrangement status
Fluorescence in situ hybridization was performed as previously described. In brief, a 3-color probe set was used, prepared by combining BAC clones for 3’ ERG (orange), 3’ TMPRSS2 (red) and 5’ TMPRSS2 (green) (all labeled by nick translation using dUTPs supplied by Abbott Molecular Inc., Des Plaines, IL). The tissue microarray sections were dewaxed, pretreated in hot 10mM sodium citrate, followed by pepsin-HCl digestion, after which hybridization, washing and fluorescence detection were performed according to standard procedures (31), including staining with 4’,6-diamidino-2-phenylindole.
Image Analysis
Samples were analyzed using a Metafer MetaCyte automated scanning system (MetaSystems Group Inc, Waltham, MA) and Isis 5.0 FISH imaging software. The hybridized slides were scanned at 5x, and the resulting composite segmented using the MetaCyte tissue microarray tool. Segmentation generated a position list corresponding to each available core, linking slide location to subsequent high-resolution FISH images. Evaluation and analysis of the cases was performed by a urologic pathologist (AG). A minimum of 100 cancer cells were evaluated for each case, whenever possible.
ERG Immunohistochemistry
IHC was conducted using a rabbit monoclonal antibody against the C-terminus of ERG. Positive and negative controls were prostatic carcinomas with known rearrangement status. After deparaffinization, 4 um sections were dehydrated and rinsed in distilled water. Following antigen retrieval with Tris/EDTA buffer (pH 9.0) in a steamer and cooling in EDTA buffer, TMA slides were quenched in 1% hydrogen peroxide in distilled water, blocked in 5% Goat Serum in 2% BSA-PBS and incubated with 1° ERG antibody (Clone EPR3864; Epitomics, Burlingame, CA; dilution 1:250) overnight at 4°C in a humidified chamber. Following 3 washes in Tris-buffered saline with 0.1% Tween-20, labeling was detected using Streptavidin-Biotin secondary Ab and DAB as a chromogen. TMA slides were counterstained with Hematoxylin, washed, dehydrated, cleared and mounted. Only nuclear staining was considered positive.
“Clear cell” histology
The so-called “clear cell” histology, first described by McNeal and colleagues (24, 26), included well-differentiated glands of variable size and contour, composed of tall cuboidal to columnar cells with basally-oriented nuclei, clear to pale pink cytoplasm and occasional eosinophilic luminal secretions. All 136 tumors in this series were evaluated for these morphologic features on the TMA cores. Tumors were considered positive if two or more of the three cores displayed “clear cell” histology. Fisher’s exact test was used to test for a difference in the incidence of TMPRSS2-ERG rearrangement among tumors with and without this histology.
Statistical methods
The associations between tumor zonal origin and deletion or translocation of TMPRSS2-ERG and copy number increase, as well as the association between clear cell histology/tumor zonal origin and deletion or translocation of TMPRSS2-ERG were assessed using Fisher’s exact test.
Results
A total of 9 samples were excluded from these analyses; FISH could not be evaluated in 7 tumors due to technical failure of the assay, while ERG IHC could not be evaluated in 2 tumors due to loss of tissue on TMA sections. FISH results were available for 56 TZ and 71 anterior PZ samples. A summary of the FISH and IHC results is detailed in Table 1. Within each given case, all tumor-bearing cores showed the same results for both FISH and IHC assays. Deletion or translocation of TMPRSS2-ERG was identified in 4 of 56 (7%) TZ samples compared with 18 of 71 (25%) anterior PZ samples (p=0.009). A similar percentage of rearranged tumors – 3 of 4 (75%) TZ and 13 of 18 (72%) anterior PZ – had TMA-core-specific Gleason scores of 6, with the remainder showing Gleason score 7. No case had a Gleason score of 8–10, corresponding well to the GS distribution of our published cohort (29) from which these cases were drawn. Copy number increase was noted in a similar proportion of carcinomas from both zones (TZ: 11% v. anterior PZ: 10%; p>0.9).
Table 1.
TMPRSS2-ERG Rearrangement Results by FISH and ERG Immunohistochemistry
| Transition Zone Tumors [n=56]* | Peripheral Zone Tumors [n=71]* | |||
|---|---|---|---|---|
| FISH (+) [n=4] | FISH (-) [n=52] | FISH (+) [n=18] | FISH (-) [n=53] | |
| ERG IHC (+) [n=23] | 2 | 0 | 17 | 4 |
| ERG IHC (-) [n=104] | 2 | 52 | 1 | 49 |
The number of TZ tumors in this table reflects those for which both FISH and ERG IHC results were available.
FISH = fluorescence in situ hybridization; IHC = immunohistochemistry
In all 4 TZ cancers with TMPRSS2-ERG gene fusion, the mechanism of rearrangement was through deletion. In contrast, 61% of the rearranged anterior PZ tumors in this study had deletion as the mechanism of rearrangement with the remaining rearranged through translocation, an incidence similar to previously reported rates in the prostate cancer literature. No cancers showed concomitant copy number increase with rearrangement.
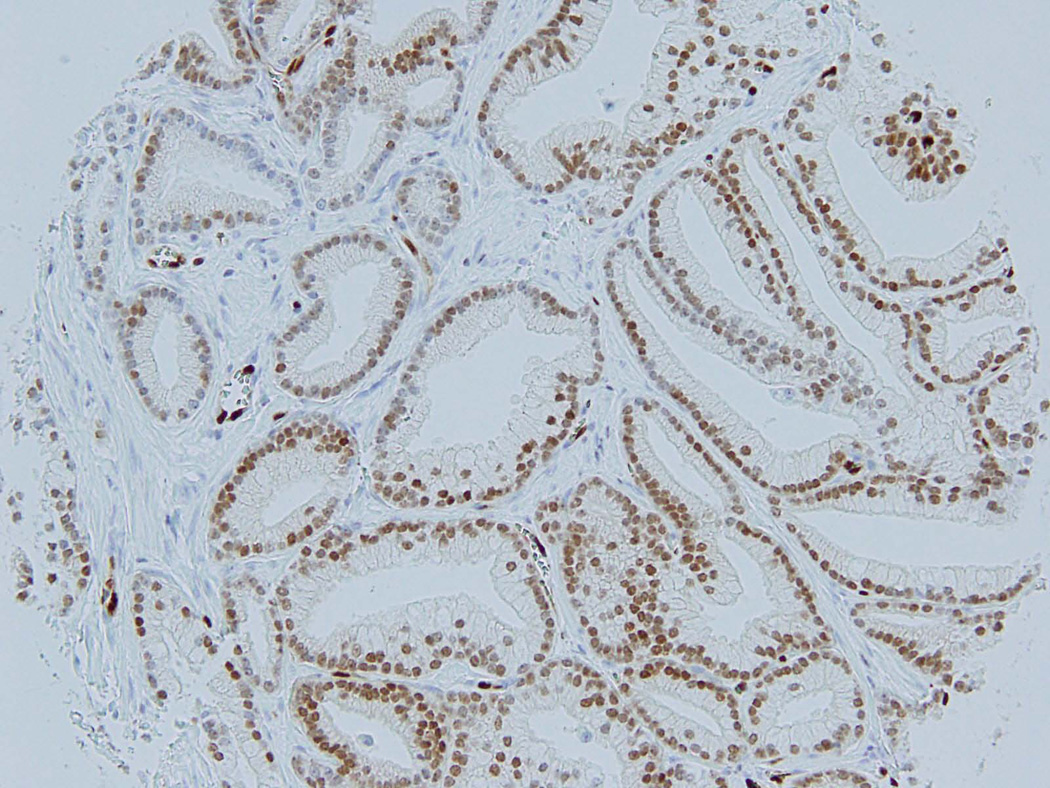
Among the four FISH positive TZ tumors, 2 were IHC positive (Figure 1A–C) and 2 were negative. None of the 52 FISH negative TZ cases was positive by IHC. Of the 18 anterior PZ tumors positive by FISH, 17 were also positive by IHC, while one case was negative. Four additional FISH negative anterior PZ cases were positive by IHC, making the incidence of ERG protein expression in anterior PZ tumors 30%, compared to 25% by FISH. Overall, ERG IHC demonstrated 86% sensitivity and 96% specificity for TMPRSS2-ERG rearrangement as determined by FISH.
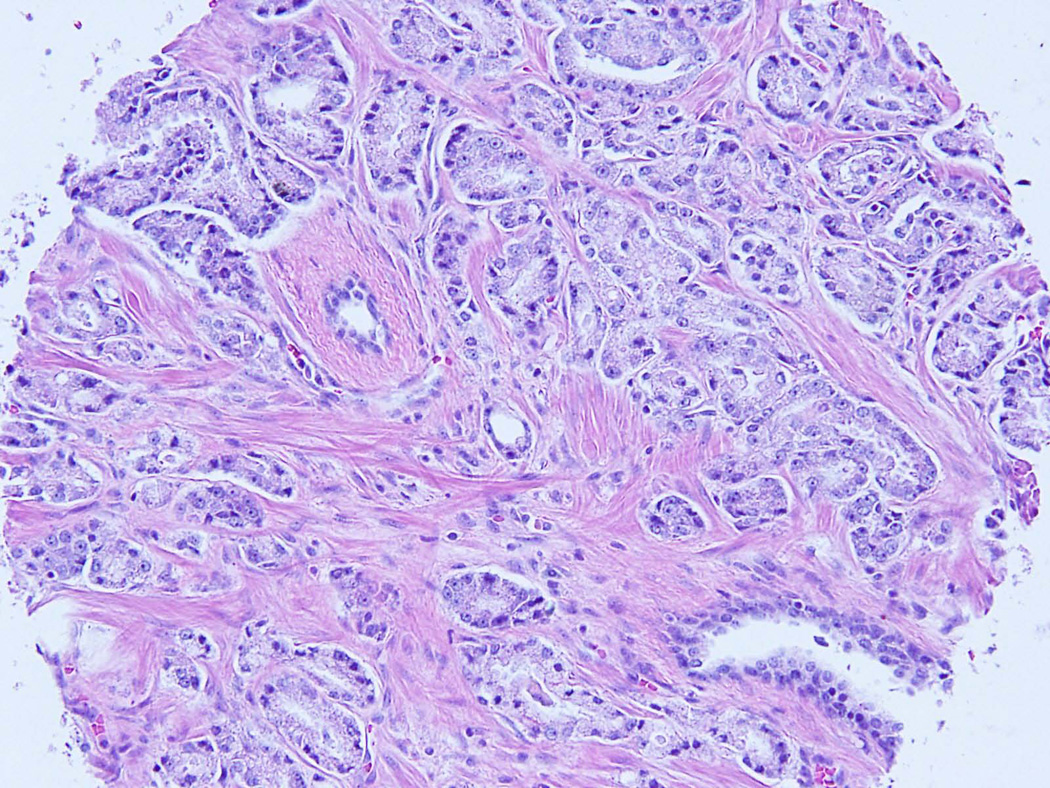
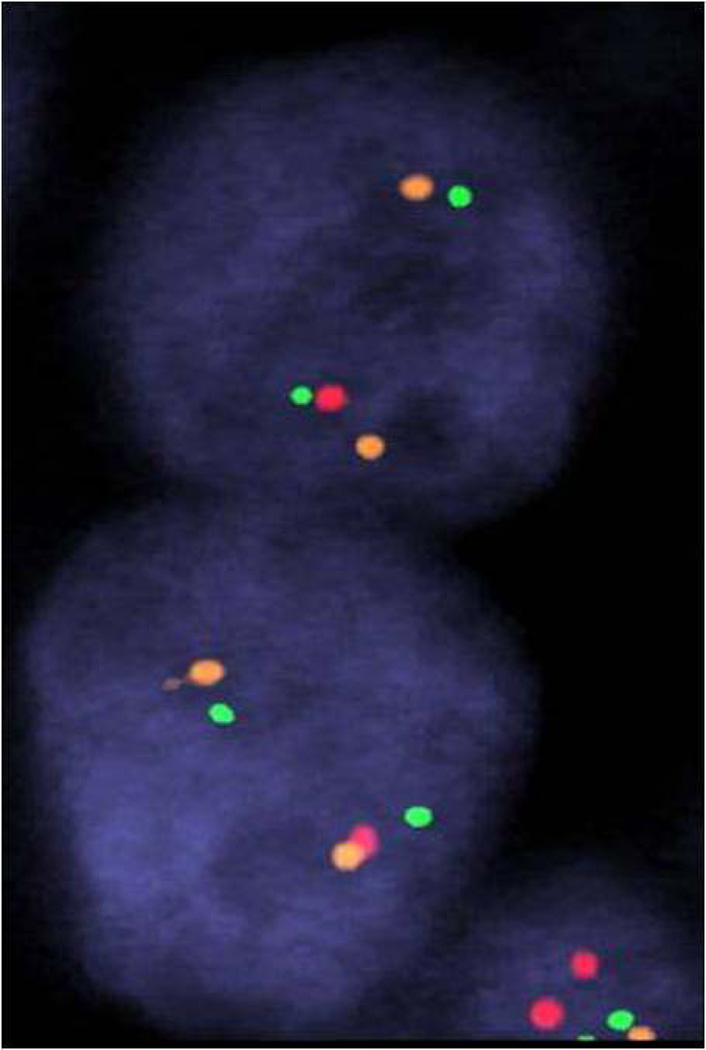
Figure 1.
Tumor of transition zone origin on tissue microarray core – H&E (A) and corresponding positive ERG immunohistochemistry (B); FISH showing TMPRSS2:ERG rearrangement by deletion: one orange-green doublet (3’ERG and 5’TMPRSS2 probes) and loss of corresponding read signal (3’TMPRSS2 probe) in one allele and a wild-type second allele.
Evaluating for “clear cell” histology on TMA cores, TZ tumors were more likely to have this morphology (21/56; 38%) [Figure 2] than were anterior PZ tumors (8/71; 11%) in this cohort (p=0.001). 7% and 21% of samples with and without this specific histologic feature were found to have deletion or translocation of TMPRSS2-ERG, respectively, regardless of zonal origin (p=0.10).
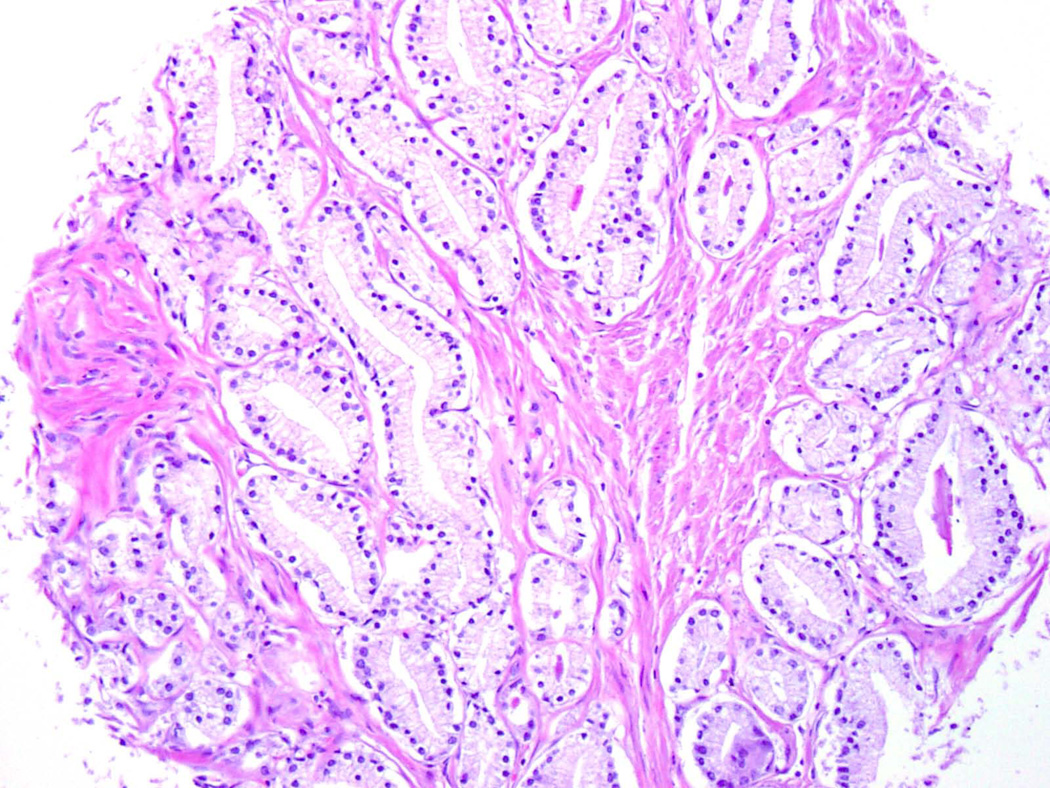
Figure 2.
“Clear cell” histology on tissue microarray core: note clear to pale pink cytoplasm, basally-oriented nuclei and occasional dense pink secretions/crystalloids.
Discussion
Dominant anterior prostatic tumors now represent 20–25% of all prostate cancers in RP specimens (29, 32). Despite more aggressive PSA screening protocols and the standardization of extended needle biopsy templates over the last two decades, the diagnosis of anterior tumors, which are less likely to be palpable on DRE and poorly visualized on imaging (18, 33–34), remains challenging. Most studies that address differences in tumor characteristics by zonal anatomy have considered PZ tumors that are not limited to the anterior portion of the prostate. In such series, TZ tumors have been found to have a larger volume and higher PSA level, yet lower Gleason scores, than PZ tumors (18–20, 22–23). Shannon et al have found that TZ tumors were more likely to be organ confined (23), and Greene et al have reported that TZ tumors have a lower rate of extraprostatic extension as well as a significantly lower rate of biochemical recurrence than PZ tumors of similar volume and grade (19–20).
Interestingly, molecular differences between TZ and PZ tumors have also been reported, with cDNA microarray based gene expression profiling techniques highlighting differences in gene expression levels (27–28). TZ tumors have been reported to have lower proliferation rates, microvessel density and expression levels of p53 and bcl-2 (35) than PZ tumors, suggesting inherent biological differences between cancers arising in these zones. However, many of these differences were no longer significant when controlling for tumor grade (27–28, 35), indicating that more robust clinicopathologic and molecular analyses may be necessary to explain the clinical differences observed.
More recently, our group has characterized in detail a large cohort [n=197] of dominant anterior prostate cancers and found the majority (49%) to be of anterior PZ origin and a smaller number (36%) to be of TZ origin. The remaining minority was split between tumors equally involving both zones (7%) and tumors in which zone of origin could not be determined (8%). No significant differences in Gleason scores, extraprostatic extension or overall surgical margin positivity rate were observed when comparing TZ and anterior PZ tumors (29). Given these conflicting findings, long term clinical outcomes along with molecular analyses are warranted to assess whether true differences in biology and behavior exist between tumors of TZ and anterior PZ origin.
Rearrangements between TMPRSS2 and various members of the ETS transcription factor family, specifically ERG, are now well-recognized as a common and recurrent genetic event in prostate cancer (2, 6, 9). Most radical prostatectomy-based series have reported incidences of TMPRSS2-ERG fusion varying from 30–70% of prostate cancer. The prognostic significance of the rearrangement has been extensively studied, yet conflicting associations with outcome – some showing correlation with adverse clinico-pathologic features and others not finding this association (2, 8–10, 14–15) – have been reported. While heterogeneity of gene rearrangement among tumor nodules in multifocal prostate cancer has been noted (3–4, 36), zonal variation in the incidence TMPRSS2-ERG rearrangement has only been more recently studied (37–40).
In the current study of dominant anterior tumors, FISH assay revealed significantly fewer TZ cancers with the rearrangement (7%) when compared with anterior PZ cancers (25%). These findings are similar to those reported previously in TZ cohorts. Guo et al were the first to report on TZ and PZ tumor foci in 30 prostatectomies, in which the selected tumors represented the largest focus in each zone. In 24 cases of their cases, the largest tumor was located in the TZ. They found TMPRSS2-ERG rearrangement in 43% of PZ tumor foci, yet none of the TZ tumor foci (37). However, in response to a comment by Bismar and Trpkov noting some of the manuscript’s limitations, Guo et al acknowledged that their study was not designed to evaluate the heterogeneity of TMPRSS2-ERG fusion among all tumor foci of all zones (38). Subsequently, and more analogous to our TZ cohort, Falzarano et al evaluated tissue microarrays from 62 cases with dominant TZ tumors of which 46 also showed a PZ tumor focus. Among interpretable cases, 12% (7/59 interpretable cases) of TZ tumors and 34% (12/35 interpretable cases) of PZ tumors revealed TMPRSS2-ERG rearrangement (39). Interestingly, low incidence (13% of 47 patients) of rearrangement has also been reported for incidentally-detected TZ cancers in transurethral resection specimens for benign prostatic hyperplasia and cystoprostatectomy for urothelial carcinoma (40–41). As some have reported fundamental differences between posterior and anterior prostate, including density of innervation (a common route of extraprostatic spread) in these regions (42), the current study is the first to compare ERG rearrangement status in dominant anterior TZ and PZ cancers with comparable pathologic features (29).
Due to the unique location of the two partner genes (TMPRSS2 and ERG) on chromosome 21, two mechanisms of rearrangement have been reported. Deletion is more common, accounting for about two-thirds of cases, while the remainder displays translocation (1–2, 8). Akin to these results, Falzarano et al found that deletion accounted for slightly more than half of the positive tumors in their series of dominant TZ tumors (39). In contrast, we observed only deletion in the four rearranged dominant TZ tumors. Some controversy exists as to the prognostic import of this mode of rearrangement. In a prior publication from our group, we reported that although the presence of deletion alone does not confer a worse prognosis, the occurrence of copy number increase of deleted segments (‘CNID’) was significantly associated with worse clinicopathological features (1, 9). However, none of the rearranged TZ tumors (or anterior PZ tumors) in our series showed concomitant copy number increase with rearrangement. The small number of TZ cases with rearrangement in these studies warrants further large-scale investigation of the mode of fusion in TZ cancers and its relationship to prognosis.
This is also the first study focused on TZ tumors to evaluate ERG expression by immunohistochemical assay. Herein we show a slightly higher incidence of rearranged anterior PZ cancers compared with FISH (30% v. 25%). Conversely, the IHC antibody failed to detect ERG overexpression in two of four FISH positive TZ cancers. We and others (43–48) have previously found excellent correlation between ERG gene rearrangement and truncated ERG protein expression, as well as a high specificity of the antibody for staining prostate cancer cells. However in a small number of cases, discordant results (positive for TMPRSS2-ERG rearrangement by FISH and negative for ERG expression by IHC or vice versa) have been observed (43, 45–47). Regarding this phenomenon, it is first important to highlight that the present study utilized a monoclonal antibody against the C-terminus of ERG, which was significantly more sensitive than an antibody against the N terminus in a recent study (49). Beyond that, a series of possible explanations have been proposed for discordant scenarios. For FISH positive/IHC negative cases: a) rearrangement may lead to expression of a highly truncated protein (e.g. transcripts exclusively involving exons 1–3 or 5–7) lacking target binding sites of the ERG antibody (43) or b) defective/decreased androgen signaling might affect TMPRSS2 (an androgen-regulated element) resulting in lack of ERG overexpression (45, 50). For FISH negative/IHC positive cases: a) technical failure of the FISH assay may result in undetectable rearrangement (45), b) ERG may be overexpressed due to a mechanism other than rearrangement (43, 47), and c) the entire ERG locus may be inserted into a genomically active region, which does not result in a positive break-apart FISH assay but leads to transcript/protein overexpression (47).
Interestingly, the incidence of rearrangement in anterior PZ tumors in this study, detected by both FISH and IHC assay, is lower than that previously reported in radical prostatectomy-based series. Although one might postulate that this reflects biologic differences between anterior PZ tumors and the more common posterior PZ tumors (from which prior cohorts were undoubtedly derived), another possibility exists. Multiple authors have reported a small percentage of cases in which intrafocal heterogeneity is present – that is, tumor cells within the same nodule showing diverse ERG status (41, 51). This may be especially relevant in studies on tissue microarrays in which different, often random areas, within a tumor mass are sampled.
In our series “clear cell” histology assessed on TMA cores was present in 11% of anterior PZ tumors and 38% of TZ tumors, concordant with prior studies showing that this morphology is more frequently seen in TZ tumors (26, 51). Notably however, 7% of tumors with and 21% without this morphology harbored the rearrangement, regardless of zone of origin. Taken together with previous data showing that “clear cell” histology: a) is present non-focally (>25%) in up to 35% of PZ tumors with any such morphology (52) and b) does not predict the presence of TZ cancer at radical prostatectomy when seen on biopsy (53), the current ERG expression data further support the lack of utility of “clear cell” histology in distinguishing zonal origin of prostate cancer.
In this cohort of dominant anterior prostatic tumors, we have shown that rearrangement of the TMPRSS2 and ERG genes, as detected by both the immunohistochemical assay for ERG protein as well as a 3-color break-apart FISH assay, is significantly less common in tumors of transition zone origin than in those of anterior peripheral zone origin. Our finding of deletion as the sole mechanism of rearrangement in transition zone cancers is intriguing and merits further investigation. Taken together, these findings may add to the existing information on and further our understanding of the biology/genetic differences inherent to tumors arising from these different anatomic zones.
Acknowledgments
Supported in part by a Specialized Program of Research Excellence (SPORE) Grant (2P50-CA92629-11) from the National Cancer Institute.
References
- 1.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2007;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 3.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 4.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostatic adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 5.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2-ERG fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Darnel AD, LaFargue CJ, Vollmer RT, Corcos J, Bismar TA. TMPRSS2-ERG fusion is frequently observed in Gleason pattern 3 prostate cancer in a Canadian cohort. Cancer Bio Ther. 2009;8:125–130. doi: 10.4161/cbt.8.2.7134. [DOI] [PubMed] [Google Scholar]
- 8.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 9.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosquera JM, Mehra R, Regan MM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin. Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam RK, Sugar L, Yang W, et al. Expression of TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 13.Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusion between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissue. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Cai Y, Ren C, Ittmann M, et al. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 15.Toubaji A, Albadine R, Meeker AK, et al. Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas. Mod Pathol. 2011 Nov;24(11):1511–1520. doi: 10.1038/modpathol.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovics G, Liu A, Shaheduzzaman S, et al. Frequent expression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 17.Fine SW, Gopalan A, Leversha MA, et al. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010 Oct;23(10):1325–1333. doi: 10.1038/modpathol.2010.120. Epub 2010 Jun 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene DR, Fitzpatrick JM, Scardino PT. Anatomy of the prostate and distribution of early prostate cancer. Semin Surg Oncol. 1995;11:9–22. doi: 10.1002/ssu.2980110104. [DOI] [PubMed] [Google Scholar]
- 19.Greene DR, Wheeler TM, Egawa S, Dunn JK, Scardino PT. A comparison of the morphological features of cancer arising in the transition zone and in the peripheral zone of the prostate. J Urol. 1991;146:1069–1076. doi: 10.1016/s0022-5347(17)38003-5. [DOI] [PubMed] [Google Scholar]
- 20.Greene DR, Wheeler TM, Egawa S, Weaver RP, Scardino PT. Relationship between clinical stage and histological zone of origin in early prostate cancer: morphometric analysis. Br J Urol. 1991;68:499–509. doi: 10.1111/j.1464-410x.1991.tb15394.x. [DOI] [PubMed] [Google Scholar]
- 21.Grignon DJ, Sakr WA. Zonal origin of prostatic adenocarcinoma: are there biologic differences between transition zone and peripheral zone adenocarcinomas of the prostate gland? J Cell Biochem Suppl. 1994;19:267–269. [PubMed] [Google Scholar]
- 22.Noguchi M, Stamey TA, McNeal JE, Yemoto CE. An analysis of 148 consecutive transition zone cancers: clinical and histologic characteristics. J Urol. 2000;163:1751–1755. [PubMed] [Google Scholar]
- 23.Shannon BA, McNeal JE, Cohen RJ. Transition zone carcinoma of the prostate gland: a common indolent tumour type that occasionally manifests aggressive behavior. Pathology. 2003;35:467–471. doi: 10.1080/00313020310001619154. [DOI] [PubMed] [Google Scholar]
- 24.McNeal JE. Normal histology of the prostate. Am J Surg Pathol. 1988;12:619–633. doi: 10.1097/00000478-198808000-00003. [DOI] [PubMed] [Google Scholar]
- 25.McNeal JE. Cancer volume and site of origin of adenocarcinoma in the prostate: relationship to local and distant spread. Hum Pathol. 1992;23:258–266. doi: 10.1016/0046-8177(92)90106-d. [DOI] [PubMed] [Google Scholar]
- 26.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma: correlation with histologic pattern and direction of spread. Am J Surg Pathol. 1988;12:897–906. doi: 10.1097/00000478-198812000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Noel EE, Ragavan N, Walsh MJ, et al. Differential gene expression in the peripheral zone compared to the transition zone of the human prostate gland. Prostate Cancer Prostatic Dis. 2008;11(2):173–180. doi: 10.1038/sj.pcan.4500997. Epub 2007 Jul 24. [DOI] [PubMed] [Google Scholar]
- 28.van der Heul-Nieuwenhuijsen L, Hendriksen PJ, van der Kwast TH, Jenster G. Gene expression profiling of the human prostate zones. BJU Int. 2006 Oct;98(4):886–897. doi: 10.1111/j.1464-410X.2006.06427.x. [DOI] [PubMed] [Google Scholar]
- 29.Al-Ahmadie HA, Tickoo SK, Olgac S, et al. Anterior-predominant prostatic tumors: zone of origin and pathologic outcomes at radical prostatectomy. Am J Surg Pathol. 2008;32:229–235. doi: 10.1097/PAS.0b013e31812f7b27. [DOI] [PubMed] [Google Scholar]
- 30.Fine SW, Al-Ahmadie HA, Gopalan A, Tickoo SK, Scardino PT, Reuter VE. Anatomy of the anterior prostate and extraprostatic space: a contemporary surgical pathology analysis. Adv Anat Pathol. 2007;14:401–407. doi: 10.1097/PAP.0b013e3181597a9c. [DOI] [PubMed] [Google Scholar]
- 31.Leversha MA. Mapping of genomic clones by fluorescence in situ hybridization. Methods Mol Biol. 2001;175:109–127. doi: 10.1385/1-59259-235-X:109. [DOI] [PubMed] [Google Scholar]
- 32.Bott SRJ, Young MPA, Kellett MJ, et al. Anterior prostate cancer: is it more difficult to diagnose? BJU Int. 2002;89:886–889. doi: 10.1046/j.1464-410x.2002.02796.x. [DOI] [PubMed] [Google Scholar]
- 33.Terris MK, Freiha FS, McNeal JE, Stamey TA. Efficacy of transrectal ultrasound for identification of clinically undetected prostate cancer. J Urol. 1991;146:78–83. doi: 10.1016/s0022-5347(17)37718-2. [DOI] [PubMed] [Google Scholar]
- 34.Zakian KL, Eberhardt S, Hricak H, et al. Transition zone prostate cancer: metabolic characteristics at 1H MR spectroscopic imaging: initial results. Radiology. 2003;229:241–247. doi: 10.1148/radiol.2291021383. [DOI] [PubMed] [Google Scholar]
- 35.Erbersdobler A, Fritz H, Schnöger S, et al. Tumour grade, proliferation, apoptosis, microvessel density, p53, and bcl-2 in prostate cancers: differences between tumours located in the transition zone and in the peripheral zone. Eur Urol. 2002 Jan;41(1):40–46. doi: 10.1016/s0302-2838(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 36.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo CC, Zuo G, Cao D, Troncoso P, Czerniak BA. Prostate cancer of transition zone origin lacks TMPRSS2-ERG gene fusion. Mod Pathol. 2009;22:866–871. doi: 10.1038/modpathol.2009.57. [DOI] [PubMed] [Google Scholar]
- 38.Bismar TA, Trpkov K. TMPRSS2-ERG gene fusion in transition zone prostate cancer. Mod Pathol. 2010;23:1040–1041. doi: 10.1038/modpathol.2010.89. [DOI] [PubMed] [Google Scholar]
- 39.Falzarano SM, Navas M, Simmerman K, et al. ERG rearrangement is present in a subset of transition zone prostatic tumors. Mod Pathol. 2010;23:1499–1506. doi: 10.1038/modpathol.2010.150. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Yoshimoto M, Trpkov K, et al. Detection of ERG gene rearrangements and PTEN deletions in unsuspected prostate cancer of the transition zone. Cancer Biol Ther. 2011;11:562–566. doi: 10.4161/cbt.11.6.14376. [DOI] [PubMed] [Google Scholar]
- 41.Braun M, Scheble VJ, Menon R, et al. Relevance of cohort design for studying the frequency of the ERG rearrangement in prostate cancer. Histopathology. 2011;58:1028–1036. doi: 10.1111/j.1365-2559.2011.03862.x. [DOI] [PubMed] [Google Scholar]
- 42.Powell MS, Li R, Dai H, Sayeeduddin M, Wheeler TM, Ayala GE. Neuroanatomy of the normal prostate. The Prostate. 2005;65:52–57. doi: 10.1002/pros.20245. [DOI] [PubMed] [Google Scholar]
- 43.Braun M, Goltz D, Shaikhibrahim Z, et al. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer-a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012 Jun;15(2):165–169. doi: 10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- 44.Chaux A, Albadine R, Toubaji A, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–1020. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falzarano SM, Zhou M, Carver P, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;49:441–447. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 46.Gopalan A, Dudas M, Leversha MA, et al. Evaluation of a novel ERG antibody in prostate cancer and correlation with TMPRSS2-ERG gene rearrangement status by FISH, ACGH and ERG mRNA expression. Mod Pathol. 2011;24(Suppl.1):194A. [Google Scholar]
- 47.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minner S, Luebke AM, Kluth M, et al. High level of Ets-related gene expression has high specificity for prostate cancer: a tissue microarray study of 11483 cancers. Histopathology. 2012;61:445–453. doi: 10.1111/j.1365-2559.2012.04240.x. [DOI] [PubMed] [Google Scholar]
- 49.Mehra R, Lonigro R, Brummell B, et al. Antibody based detection of ERG gene fusions in prostate cancer: an immunohistochemical study comparing c- and n-terminus ERG antibodies. Mod Pathol. 2012;25(Suppl.2):240A. [Google Scholar]
- 50.Dobi A, Furusato B, Shaheduzzaman S, et al. ERG expression levels in prostate tumors reflect functional status of the androgen receptor (AR) as a consequence of fusion of ERG with AR regulated gene promoter. Open Cancer J. 2010;3:101–108. [Google Scholar]
- 51.Falzarano SM, Zhou M, Hernandez AV, Klein EA, Rubin MA, Magi-Galluzzi C. Single focus prostate cancer: pathological features and ERG fusion status. J Urol. 2011;185:489–494. doi: 10.1016/j.juro.2010.09.093. [DOI] [PubMed] [Google Scholar]
- 52.Garcia JJ, Al-Ahmadie HA, Gopalan A, et al. Do prostatic transition zone tumors have a distinct morphology? Am J Surg Pathol. 2008;32:1709–1714. doi: 10.1097/PAS.0b013e318172ee97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haarer CF, Gopalan A, Tickoo SK, et al. Prostatic transition zone directed needle biopsies uncommonly sample clinically relevant transition zone tumors. J Urol. 2009;182:1337–1341. doi: 10.1016/j.juro.2009.06.042. [DOI] [PubMed] [Google Scholar]