Abstract
Autophagy (macroautophagy) is often defined as a degradative process and a tributary of the lysosomal pathway. In this context, autophagy carries out cytoplasmic quality control and nutritional functions by removing defunct or disused organelles, particulate targets and invading microbes, and by bulk digestion of the cytoplasm. However, recent studies indicate that autophagy surprisingly affects multiple secretory pathways. Autophagy participates in extracellular delivery of a number of cytosolic proteins that do not enter the conventional secretory pathway via the Golgi apparatus but are instead unconventionally secreted directly from the cytosol. In mammalian cells, a prototypical example of this manifestation of autophagy is the unconventional secretion of a major proinflammatory cytokine, IL-1β. This review examines the concept of secretory autophagy and compares and contrasts the role of autophagy in the secretion of IL-1α and IL-1β. Although IL-1α and IL-1β have closely related extracellular inflammatory functions, they differ in intracellular activation, secretory mechanisms and how they are affected by autophagy. This example indicates that the role of autophagy in secretion is more complex, at least in mammalian cells, than the simplistic view that autophagosomes provide carriers for unconventional secretion of cytosolic proteins.
Key Words: Autophagy, Alarmins, Inflammasome, Calpain
Introduction: Secretory Autophagy
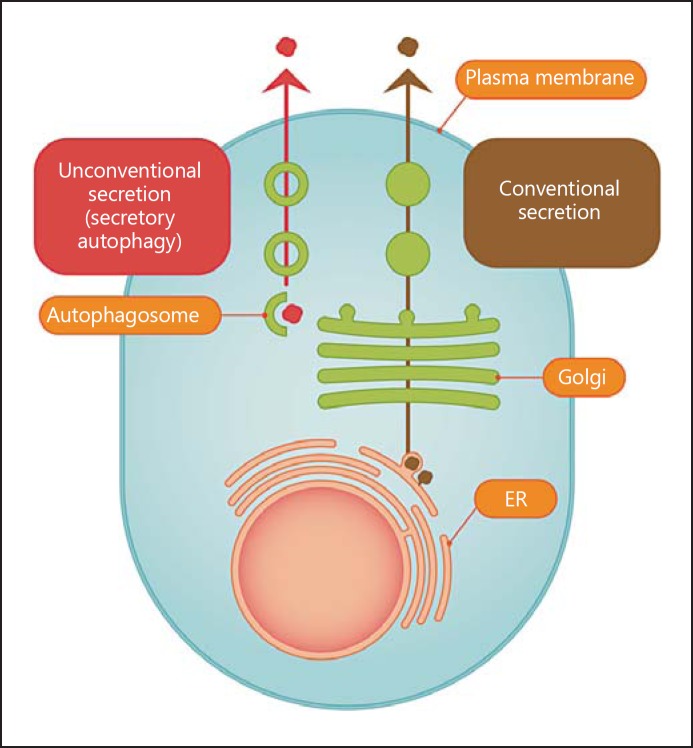
Our current view of protein secretion from eukaryotic cells is dominated by the well-established classic paradigm of conventional secretion (fig. 1, right). This paradigm rests on the extensively characterized biosynthetic pathway utilized by proteins endowed with sorting signals (leader peptides) authorizing them to enter the lumen of the endoplasmic reticulum (ER). From there, they undergo modifications and transport through the Golgi apparatus (G), and are finally secreted upon fusion of post-Golgi carriers with the plasma membrane (PM). A new protein secretion paradigm has been recently defined in yeast [1, 2, 3] as autophagy-based unconventional secretion in eukaryotic cells. In principle, this phenomenon (secretory autophagy; fig. 1, left) may enable any cytosolic protein to be exported from the cell where it may exert either its primary or a cryptic biological function. A direct function of autophagy in unconventional secretion by providing carriers for secretion (fig. 1, left) has been described in yeast, using a combination of genetic and cell biological tools [1, 2, 3]. Secretory autophagy has also been implicated in mammalian cells [4, 5, 6]. At present, there are two principal features defining secretory autophagy: (1) participation of Atg factors and contribution of autophagy as a pathway, and (2) dependence on Golgi reassembly and stacking protein (GRASP; fig. 2) known as GRASP55 and GRASP65 in mammalian cells [7], dGRASP in Drosophila, GrpA in Dictyostelium and Grh1 in yeast [6, 8, 9, 10, 11].
Fig. 1.
Protein secretion: conventional biosynthetic pathway versus unconventional secretion through secretory autophagy. The well-developed paradigm of conventional protein secretion through the ER, Golgi and post-Golgi trafficking (right arrow) versus the process (left arrow) of autophagy-dependent unconventional secretion of cytosolic proteins (secretory autophagy). The proteins destined for conventional secretion enter ER via signal peptides, whereas cytosolic proteins destined for secretory autophagy are sequestered into autophagosomes to be exported from the cell.
Fig. 2.
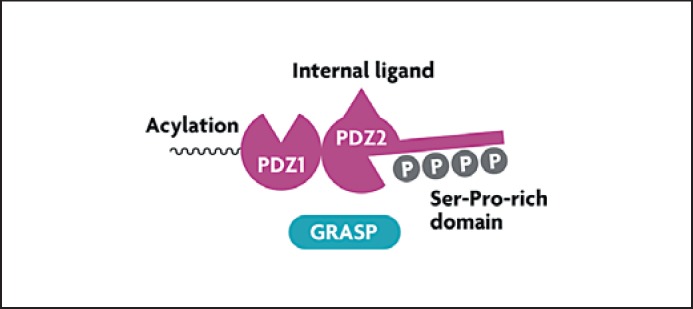
GRASP domains and salient features. GRASP contains two PDZ domains, an internal protein sequence (‘internal ligand’) that binds to one of the PDZ domains allowing homo-oligomerization, N-terminal acylation that facilitates GRASP association with membranes, and a Ser-Pro-rich domain that is a substrate for phosphorylation by Ser/Thr kinases. Phosphorylation modulates GRASP interactions and the oligomerization state.
Degradative Autophagy
Autophagy is currently best known for its role in metabolism as well as a form of organelle and protein turnover and quality control [12, 13]. Canonical degradative autophagy (macroautophagy) allows cells to digest their cytoplasmic components as an endogenous source of nutrients and energy at times of starvation or as a mechanism for the clearance of defective organelles and toxic intracellular aggregates [14]. The canonical autophagy pathway depends on a suite of autophagy-specific factors (termed Atg and followed by a number) responsive to upstream signaling by TOR, AMPK and other inputs [12, 15, 16]. The Atg factors are responsible for the execution of autophagy and the formation of specialized double membrane organelles, termed autophagosomes. The Atg factors organize in several subcomplexes, including the Atg5-Atg12/Atg16 complex that acts as a conceptual equivalent of E3 ligases in the ubiquitin system. It regulates C-terminal lipidation of Atg8 (or its mammalian equivalents LC3s and GABARAPs) with phosphatidylethanolamine essential for autophagosomal membrane growth [12]. Atg8-phosphatidylethanolamine can effect membrane tethering and fusion [17, 18], albeit autophagic organelles are also subject to conventional membrane fusion via SNAREs, the canonical regulators of membrane fusion in most compartments of the eukaryotic cell [19]. The membrane precursors for the formation of autophagosomes originate from transient domains on the ER, termed omegasomes. There are also potential contributions from other compartments such as the plasma membrane, mitochondria and Golgi. To carry out degradative functions, autophagosomes fuse with lysosomes to form autolysosomes where the captured cargo is eventually degraded.
As stated above, autophagy is assumed to represent primarily a catabolic, lysosomal degradative pathway. The notion of autophagy as a purely degradative pathway was recently challenged by the emergence of three independent reports on the role of autophagy in unconventional secretion of Acb1 in yeast [1, 2, 3] and IL-1β and other cargo (e.g. HMGB1 and IL-18) in mammalian cells [4, 5]. These new insights assign a nondegradative function to autophagy manifested as unconventional protein secretion (fig. 1). Furthermore, it has become apparent that autophagy broadly intersects with protein trafficking and secretion to include effects on the constitutive biosynthetic pathway [20], regulated exocytosis [21] and alternative sorting of integral membrane proteins to the plasma membrane [22].
Secretory Autophagy and Unconventional Secretion
The information regarding how autophagy promotes unconventional secretion comes, at present, from a handful of studies [1, 2, 3, 4, 22]. It is most completely defined in yeast, using the yeast prototypical cargo for unconventional secretion, Acb1 [1, 2, 3]. The role of autophagy in unconventional protein secretion and trafficking is a recently recognized function of the autophagic machinery. Not all unconventionally secreted proteins are secreted by the same mechanism. Unconventional secretion is a catch-all term for a spectrum of diverse processes [10]. This collection of often unrelated secretory processes delivers, via unique mechanisms, specific protein cargo localized in the cytosol to the extracellular environment or assists in the trafficking of integral membrane proteins to the plasma membrane without having to pass through the ER-G-PM pathway [10]. Only a subset of unconventional secretion is dependent on autophagy. When autophagy provides specific membrane carriers to execute unconventional secretion, this subset is referred to as autosecretion [5], type III secretion [6] or secretory autophagy, a term preferentially used in this review.
Secretory versus Degradative Autophagy Precursors
Are the precursors for secretory and degradative autophagy related? A membrane structure in yeast, termed compartment for unconventional protein secretion (CUPS) [2] has been identified as the source of organelles or trafficking intermediates for autophagy-based unconventional secretion. CUPS, the term used in yeast, may be related (although this has still to be established) to the mammalian cell omegasome, which acts as a cradle for generating nascent autophagosomes normally associated with degradative autophagy [23]. CUPS forms in the general vicinity of but does not completely coincide with the ER exit sites marked by Sec13 [2]. The organizers of the ER exit sites (Sec12 and Sec16) and COPII components (Sec23 and Sec24, but not Sec13 and Sec31) are required for the canonical autophagosome formation in yeast [24]. Participation of the regulators of the early secretory pathway in autophagy has also been noted in mammalian cells [25, 26]. Starvation induces both CUPS [2] and omegasomes [23]. These structures are morphologically similar, PI3P-positive, and are decorated by autophagy (Atg) factors. As CUPS is reportedly not induced by rapamycin, it has been proposed to be distinct from omegasomes [2]. However, the original description of the omegasome and its role in autophagy along with its PI3P positivity did not report the use of rapamycin and was based solely on the starvation-dependent induction of autophagy [23]. The relationship of CUPS to the well-known preautophagosomal structure (PAS) in yeast also needs to be clarified since Sec23 of COPII is required for autophagy via PAS [24], but its mutation does not interrupt unconventional secretion of Acb1. Similarly, Sec12 matters for autophagy [24] but appears to be dispensable for CUPS formation in yeast [2]. The subtle differences outlined above may be technical but could indicate some subcompartmentalization of domains within precursor for autophagosomal organelles (CUPS, PAS and omegasomes) destined for degradative or secretory autophagy.
Secretory versus Degradative Autophagy: Cargo Selection
How is the cargo destined for secretory autophagy selected and separated from degradative autophagy cargo? Thus far, this has not been explored. Of potential relevance is that CUPS contains Vps23 [2]. Vps23, in mammalian systems known as TSG101, is a member of the endosomal sorting complex required for transport (ESCRT)-I complex associated with multivesicular endosomal sorting but also participating in a number of other trafficking processes. ESCRT components other than Vps23 are however not found in CUPS [2] and Vps23 itself is not necessary for CUPS formation, indicating that Vps23 may be a component of sorting elements associated with CUPS. Of further interest is that VPS23/TSG101, in its sorting function, can bind to ubiquitinated cargo, a typical recognition tag for autophagic adaptors [27]. A role in secretory autophagy for conventional autophagy adaptors, e.g. p62, NBR1, NDP52 and optineurin, remains to be addressed.
GRASP in Unconventional Secretion and Secretory and Canonical Autophagy
The protein named GRASP (fig. 2), with orthologs in yeast [2, 3, 8], metazoans [28] and mammalian cells [29, 30, 31], is the only specific marker thus far known to be essential for unconventional protein secretion [10]. However, how it functions in autophagy-based unconventional secretion is not understood. A recently published study [4, 5] indicates that GRASP55 (one of the two mammalian GRASPs) is also necessary for canonical, degradative autophagy [4]. Thus, the early secretory and degradative autophagosomes may originate from the common ancestral membrane domains, with subsequent divergence and specialization for the secretion or digestion of captured substrates.
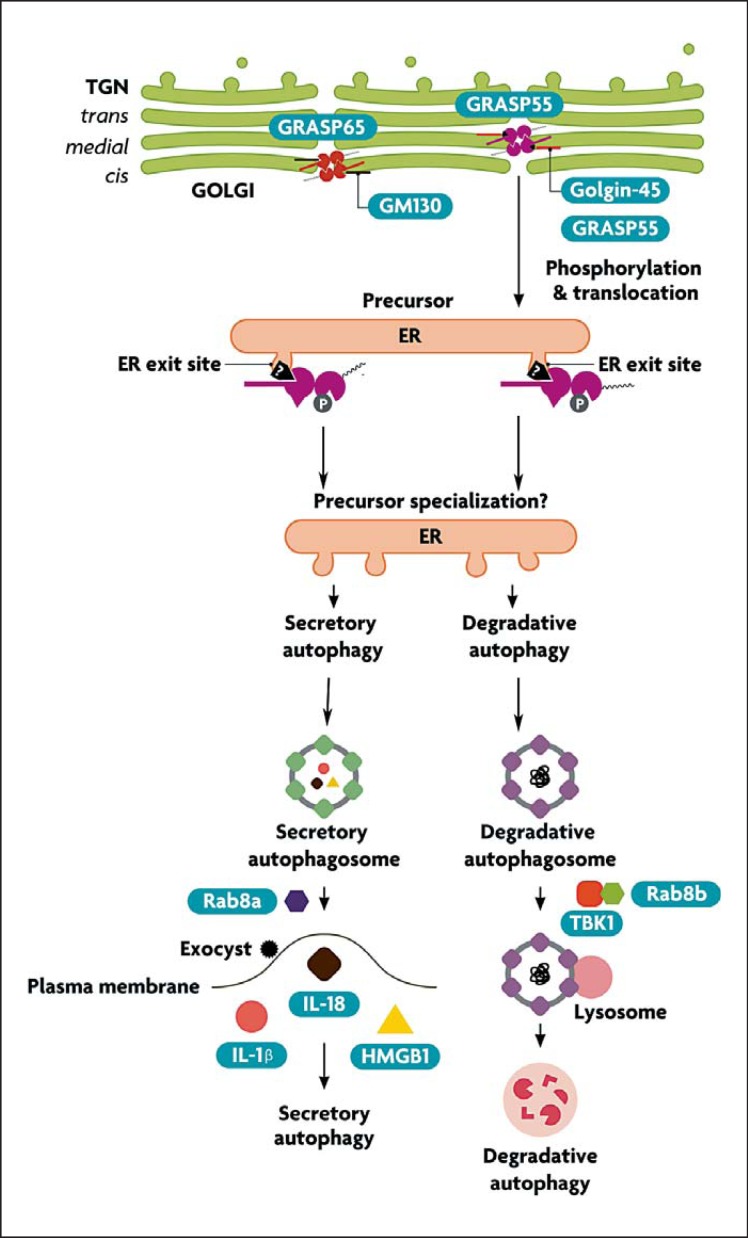
The yeast GRASP (Grh1) redistributes to CUPS. It is not known what signaling events regulate redistribution of the yeast GRASP orthologs to CUPS upon starvation. In mammalian cells, GRASP55 and GRASP65 localize to the Golgi ribbon, where they interact with Golgin-45 and GM130 (fig. 3). GRASP translocation in mammalian cells has been detected [4] and it could mirror the events in yeast, although this has still to be established. There are two types of factors that could be expected to control GRASP55 localization during degradative and secretory autophagy: (1) kinase(s) that may affect the GRASP55 homo-oligomerization state and thus the tethering of adjacent membranes (as in the Golgi ribbon), and (2) compartment-specific interacting partners binding to the PDZ domains of GRASPs, thus tethering them to the appropriate membranes that eventually coalesce. GRASPs have the ability to link membranes through their homotypic interactions between PDZ domains and internal PDZ-binding motifs (‘internal ligand’; see fig. 2). This is morphologically best recognized in the formation, maintenance and disruption of Golgi ribbons, where GRASPs are known to play a role in the linking of Golgi cisternae [30, 31] (fig. 3). The ability of GRASPs to undergo homo-oligomerization depends on the phosphorylation state of their Ser-Pro-rich C-terminal domains and can be disrupted, e.g. during mitosis (causing Golgi dispersal) following complex phosphorylation patterns by several kinases including ERK, CDK1 and PLK1 [30, 31]. In addition to homo-oligomerization via the internal ligand, GRASP55 and GRASP65 interact in the Golgi with Golgin-45 and GM130, respectively. Although phosphorylation of GRASPs can disrupt their homo-oligomerization (resulting in events such as Golgi disruption in mitosis), it is not known whether similar events could lead to GRASP translocation and modulation of interactions with other putative partners involved in secretory autophagy and unconventional secretion in general.
Fig. 3.
Hypothetical model and the proposed putative points of divergence between degradative and secretory autophagy. The steps are discussed in the text.
GRASP carries out its function in conventional autophagy as well, at least in mammalian cells [4]. The yeast GRASP equivalent is important for Atg9 relocalization upon starvation [2], mirroring Vps23 localization to CUPS. The effect of the yeast GRASP ortholog Grh1 on the sole integral membrane autophagy factor (Atg9) indirectly corroborates with the reported role of the mammalian GRASPs (GRAPS55) in control of autophagy initiation [4].
Regulation of Secretory versus Degradative Autophagy
In mammalian cells, there is an involvement in autophagy of two closely related but distinct isoforms of the small GTPase Rab8: Rab8a and Rab8b [4, 32]. There are indications of specialization between Rab8a and Rab8b in secretory versus degradative autophagy (fig. 3). It has been shown that Rab8a, and potentially exocyst components, play a role in secretory autophagy [4]. In contrast, Rab8b affects the maturation stages of degradative autophagy [32].
Rab8b interacts with the Ser/Thr protein kinase TBK-1. TBK-1, in turn, phosphorylates two autophagy adaptors: optineurin [33] and the classic autophagy adaptor p62 (sequestosome 1) [32]. The latter is a key autophagy factor [34] and the founding member of a new subfamily of pattern recognition receptors termed sequestosome-like receptors [27]. This function of TBK-1 is likely superimposed on the tonic level of basal p62 phosphorylation carried out by casein kinase 2 at the same Ser-403 residue [35]. Of note, TBK-1 is a member of the IKK family of kinases, the well-known regulators of innate immunity [36]. IKKs can be either canonical (IKK-α and IKK-β) or atypical/IKK-related kinases (IKKε and TBK-1). IKKα and IKKβ play a role in the induction of autophagy [16]. A role for TBK-1 in autophagy maturation indicates a sequential action in autophagy of the canonical IKKs at the initiating stages and TBK-1 at the completion end of the process. Whether TBK-1 regulates autophagy at the point of divergence between secretory and degradative autophagy (by, for example, promoting the latter) remains to be determined.
Autophagy Role in IL-1β Secretion and Inflammasome Activation
In mammalian cells, several cytosolic proteins are secreted through different pathways of unconventional secretion [6, 10]. The most prominent candidate to be unconventionally secreted via autophagy is the proinflammatory cytokine IL-1β, as proposed [37] immediately following the initial reports on the molecular machinery of secretory autophagy in yeast [1, 3]. However, a number of converging reports focussing on IL-1β from the immunological perspective have shown, through the observations in mice lacking Atg16L1 [38] followed up by mechanistic ex vivo and in vivo analyses [4, 39, 40, 41, 42], that autophagy dampens IL-1β activation by the inflammasome. This negative effect of autophagy on IL-1β activation is a dominant effect of autophagy under steady-state conditions. Nevertheless, a contribution of secretory autophagy to the release of IL-1β from the cells has been detected as a transient autophagy-dependent secretion of IL-1β during the early, acute phase of inflammasome activation [4]. The latter observation supports a role (albeit only transiently detectable) of secretory autophagy in the transport of cytosolic IL-1β from the cells via autophagy-dependent processes.
IL-1β is synthesized as a leaderless cytosolic protein secreted from cells via membranous organelles [43, 44]. Secretory autophagy may assist in these unconventional secretion processes [4]. Once secreted, IL-1β binds to the IL-1 receptor (IL-1RI) to initiate a number of inflammatory processes that contribute to innate immunity but can also cause tissue damage [45]. Pro-IL-1β is processed from its inactive cytosolic precursor by inflammasome, a multiprotein complex responding to microbial products referred to as PAMPs (pathogen-associated molecular patterns) and endogenous molecules signaling damage or danger termed alarmins or DAMPs (damage-associated molecular patterns) [46]. An inflammasome typically consists of three parts: a PAMP- or DAMP-detecting module in the form of a Nod-like receptor (e.g. NLRP3) or an endogenous DNA (released from mitochondria)-detecting module such as AIM2, the hub adaptor protein ASC and caspase-1 that, in turn, enzymatically processes substrates, e.g. pro-IL-1β into mature, biologically active products such as IL-1β. Autophagy inhibits IL-1β activation indirectly, by lowering the endogenous sources, such as reactive oxygen species (ROS) or DAMPs such as mitochondrial DNA released by depolarized or otherwise unkempt mitochondria accumulating in autophagy-deficient cells [39, 40] and recognized by the inflammasome components. Autophagy may also act via the proposed autophagic degradation of inflammasome constituents [41, 42, 47]. In principle, the primary role of autophagy in the inflammasome context is to keep the cellular interior clean of endogenous DAMPs, lest these inflammasome agonists cause sterile inflammation and pathological tissue damage. Autophagy also plays a membrane-trafficking role contributing to acute unconventional secretion of IL-1β, but this contribution is masked under steady-state conditions [4]. Secretory autophagy also contributes to the release of other cytosolic inflammasome substrates, e.g. IL-18 and other alarmins such as high-mobility group protein B1 (HMGB1) [4]. Thus, autophagy plays a dual role in inflammasome activation and secretion of IL-1β and the release of other alarmins from the cell. It is important to recognize this duality as follows: (1) the positive contribution of autophagy to acute delivery of IL-1α and alarmins from a cell is a manifestation of secretory autophagy as a membrane-trafficking process [4], and (2) the tonic suppression of inflammasome activation is a manifestation of degradative autophagy in its function as a quality control process in the cytoplasm.
Is the Role of Autophagy in IL-1α Activation and Secretion Different from the Effects on IL-1β?
Another proinflammatory pathway, linked but distinct from inflammasome activation, is affected by autophagy. This pathway involves IL-1α, a proinflammatory cytokine that shares two features with IL-1β: they are both are cytoplasmic proteins and they both engage the same receptors and downstream signaling following extracellular release and binding to IL-1RI. However, IL-1α is activated and secreted in a manner that differs from pro-IL-1β processing and activation. IL-1α is produced as a cytosolic proform that can be processed by calpain or other proteases [48] and actively exported out of the cell [49, 50] or be passively released upon cell death [51]. IL-1α secretion has been reported to involve both caspase 1-independent and caspase 1-dependent pathways [52]. Caspase 1−/− macrophages stimulated with LPS show a 20–30% reduction in IL-1α secretion compared to controls [53], thus indicating that a major route of IL-1α secretion is independent of caspase 1. Interestingly, despite this divergence, autophagy-defective cells (e.g. Atg5fl/fl LysM-Cre+ macrophages) also generate excess IL-1α. This is consistent with the existence of another major, inflammasome-independent activation pathway [50] that is also augmented in the absence of the cytoplasmic housekeeping functions of autophagy [4, 39, 40, 41, 42]. This pathway shares with the inflammasome-IL-1β the very upstream triggers - e.g. ROS emanating from unkempt mitochondria in autophagy-deficient cells. However, it differs in the lower, execution part of the pathway. ROS can activate calpain [54, 55], which in turn activates IL-1α [50].
The findings with the ROS-calpain axis in IL-1α activation [50] and reports regarding the ROS-RLR signaling [56] expand proinflammatory phenomena to noninflammasome pathways downstream of the accumulation of dysfunctional mitochondria and ROS in autophagy-deficient cells. Other changes with inflammatory consequences have been noted in mice with Atg5-deficient macrophages [57, 58]. It is not known whether IL-1α is a substrate for the alternative secretory pathway, as may be the case for IL-1β [4]. Thus far, only the negative role of autophagy in activation processes upstream of the IL-1α secretion has been studied [50]. Kinetic studies of IL-1α release may help discern a possible positive contribution of autophagy to the physical act of secretion of IL-1α versus the dominant role of autophagy in keeping the upstream activation triggers down.
Concluding Remarks
Thus far, genetic studies support the existence of secretory autophagy. It remains to be defined whether and how secretory autophagy provides specific membrane carriers to capture cytosolic proteins and secrete them outside of the cell. The precise cell biological mechanisms executing secretory autophagy, the adaptors for cargo selection, the switches that separate degradative and secretory autophagy as well as other aspects of vectorial membrane transports and protein cargo sorting have still to be delineated. The intersections of this pathway with the role of GRASP need to be understood. The spectrum of cytosolic proteins externalized by secretory autophagy and the scope of their extracellular functions also remain to be catalogued, in order to appreciate in full the biological and medical significance of this potential pathway.
We caution the reader that the models depicted in the figures of this review are hypothetical and should serve not as a firm summary of what is known but as a speculative starting point and a platform to test and - if need be - disprove these models, in the hope that this will stimulate further research. Regardless of the details, secretory autophagy is an area of investigation that is already expanding the sphere of influence of autophagy from a purely intracytoplasmic domain to the extracellular space. Furthermore, the existence of secretory autophagy argues that autophagy is not only a lysosomal degradative process but may play important biogenesis roles in the extracellular delivery of bioactive molecules. Even if this expanded concept is accepted, a common denominator for both processes will still be the removal of macromolecules and organelles from the cytoplasm, which can occur by degradation or secretion.
Acknowledgments
V.D. acknowledges recent and current support by AI042999, AI069345 and ARRA RC1AI086845 from the National Institutes of Health Colitis Foundation of America CCFA2053 and the Bill and Melinda Gates Grand Challenge Explorations grant. E.F.C. was supported by a supplement 3R01AI042999-13S1. This work was supported in part by the NCRR and the National Center for Advancing Translational Sciences NIH Grant UL1 TR000041.
References
- 1.Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruns C, McCaffery JM, Curwin AJ, Duran JM, Malhotra V. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011;195:979–992. doi: 10.1083/jcb.201106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012;22:397–406. doi: 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabouille C, Malhotra V, Nickel W. Diversity of unconventional protein secretion. J Cell Sci. 2012 doi: 10.1242/jcs.103630. in press. [DOI] [PubMed] [Google Scholar]
- 7.Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabral M, Anjard C, Malhotra V, Loomis WF, Kuspa A. Unconventional secretion of AcbA in Dictyostelium discoideum through a vesicular intermediate. Eukaryot Cell. 2010;9:1009–1017. doi: 10.1128/EC.00337-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 11.Giuliani F, Grieve A, Rabouille C. Unconventional secretion: a stress on GRASP. Curr Opin Cell Biol. 2011;23:498–504. doi: 10.1016/j.ceb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Ann Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 13.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, Rain JC, Ninomiya-Tsuji J, Fuentes JM, Lavandero S, Galluzzi L, Maiuri MC, Kroemer G. Inhibition of autophagy by TAB2 and TAB3. Embo J. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N-termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, Tavare S, Inoki K, Shimizu S. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG. Rescue of deltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011;146:746–760. doi: 10.1016/j.cell.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 26.Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, Lichtenberg M, Menzies FM, Ravikumar B, Imarisio S, Brown S, O'Kane CJ, Rubinsztein DC. α-Synuclein impairs macroautophagy: implications for Parkinson's disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schotman H, Karhinen L, Rabouille C. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev Cell. 2008;14:171–182. doi: 10.1016/j.devcel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Barr FA. Purification and functional interactions of GRASP55 with Rab2. Methods Enzymol. 2005;403:391–401. doi: 10.1016/S0076-6879(05)03034-X. [DOI] [PubMed] [Google Scholar]
- 30.Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarvela T, Linstedt A. Golgi GRASPs: moonlighting membrane tethers. Cell Health Cytoskeleton. 2012;4:37–47. [Google Scholar]
- 32.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, Deretic V. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 36.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Molecular Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 37.Pfeffer SR. Unconventional secretion by autophagosome exocytosis. J Cell Biol. 2010;188:451–452. doi: 10.1083/jcb.201001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 39.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 40.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JA. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubartelli A, Cozzolino F, Talio M, Sitia R. A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J. 1990;9:1503–1510. doi: 10.1002/j.1460-2075.1990.tb08268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 45.Dinarello CA. An IL-1 family member requires caspase-1 processing and signals through the ST2 receptor. Immunity. 2005;23:461–462. doi: 10.1016/j.immuni.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peral de Castro C, Jones SA, Ni Cheallaigh C, Hearnden CA, Williams L, Winter J, Lavelle EC, Mills KH, Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144–4153. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 48.Afonina IS, Tynan GA, Logue SE, Cullen SP, Bots M, Luthi AU, Reeves EP, McElvaney NG, Medema JP, Lavelle EC, Martin SJ. Granzyme B-dependent proteolysis acts as a switch to enhance the proinflammatory activity of IL-1alpha. Mol Cell. 2011;44:265–278. doi: 10.1016/j.molcel.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yazdi AS, Guarda G, Riteau N, Drexler SK, Tardivel A, Couillin I, Tschopp J. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1alpha and IL-1beta. Proc Natl Acad Sci USA. 2010;107:19449–19454. doi: 10.1073/pnas.1008155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castillo E, Dekonenko A, Arko-Mensah J, Mandell M, Dupont N, Jiang S, Delgado-Vargas M, Timmins G, Bhattacharya D, Yang H, Hutt J, Lyson C, Dobos K, Deretic V. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad USA. 2012 doi: 10.1073/pnas.1210500109. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CJ, Kono H, Golenbock D, Reed G, Akira S, Rock KL. Identification of a key pathway required for the sterile inflammatory response triggered by dying cells. Nat Med. 2007;13:851–856. doi: 10.1038/nm1603. [DOI] [PubMed] [Google Scholar]
- 52.Gross O, Yazdi AS, Thomas CJ, Masin M, Heinz LX, Guarda G, Quadroni M, Drexler SK, Tschopp J. Inflammasome activators induce interleukin-1alpha secretion via distinct pathways with differential requirement for the protease function of caspase-1. Immunity. 2012;36:388–400. doi: 10.1016/j.immuni.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 53.Schott WH, Haskell BD, Tse HM, Milton MJ, Piganelli JD, Choisy-Rossi CM, Reifsnyder PC, Chervonsky AV, Leiter EH. Caspase-1 is not required for type 1 diabetes in the NOD mouse. Diabetes. 2004;53:99–104. doi: 10.2337/diabetes.53.1.99. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Arnold JM, Pampillo M, Babwah AV, Peng T. Taurine prevents cardiomyocyte death by inhibiting NADPH oxidase-mediated calpain activation. Free Radic Biol Med. 2009;46:51–61. doi: 10.1016/j.freeradbiomed.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 55.Sharma AK, Rohrer B. Sustained elevation of intracellular cGMP causes oxidative stress triggering calpain-mediated apoptosis in photoreceptor degeneration. Curr Eye Res. 2007;32:259–269. doi: 10.1080/02713680601161238. [DOI] [PubMed] [Google Scholar]
- 56.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWT. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]