Abstract
Background
Hematopoietic stem cell (HSC) regulation is highly dependent on interactions with the marrow microenvironment, of which osteogenic cells play a crucial role. While evidence is accumulating for an important role of intrinsic miR-17 in regulating HSCs and HPCs, whether miR-17 signaling pathways are also necessary in the cell-extrinsic control of hematopoiesis hereto remains poorly understood.
Methodology/Principal Findings
Using the immortalized clone with the characteristics of osteoblasts, FBMOB-hTERT, in vitro expansion, long-term culture initiating cell (LTC-IC) and non-obese diabetic/severe combined immunodeficient disease (NOD/SCID) mice repopulating cell (SRC) assay revealed that the ectopic expression of miR-17 partly promoted the ability of FBMOB-hTERT to support human cord blood (CB) CD34+ cell expansion and maintain their multipotency. It also seemed that osteoblastic miR-17 was prone to cause a specific expansion of the erythroid lineage. Conversely, deficient expression of miR-17 partly inhibited the hematopoietic supporting ability of FBMOB-hTERT. We further identified that HIF-1α is responsible for, at least in part, the promoted hematopoietic supporting ability of FBMOB-hTERT caused by miR-17. HIF-1α expression is markedly enhanced in miR-17 overexpressed FBMOB-hTERT upon interaction with CB CD34+ cells compared to other niche associated factors. More interestingly, the specific erythroid lineage expansion of CB CD34+ cells caused by osteoblastic miR-17 was abrogated by HIF-1α knock down.
Conclusion/Significance
Our data demonstrated that CB CD34+ cell expansion can be partly promoted by osteoblastic miR-17, and in particular, ectopic miR-17 can cause a specific expansion of the erythroid lineage through augmenting HIF-1α in osteoblasts.
Introduction
Hematopoietic stem cells (HSCs) are multipotent progenitor cells that give rise to all types of mature blood cells. Tracer studies of transplanted HSCs reveal that they most likely reside in bone cavities specifically adjacent to endosteal bone lined by osteoblast cells [1], [2], [3]. HSCs share an important relationship with osteoblasts and other stromal elements of the bone marrow niche critical to their maintenance and protection [1], [4], [5]. Furthermore, it is now widely accepted that gradients of oxygen from below 1% in hypoxic niches to 6% in the sinusoidal cavity exist within the human bone marrow, which also keeps HSCs in a low proliferative and relatively quiescent state [6], [7], [8]. Proliferating progenitors are distributed in O2-rich areas [9], [10], [11], [12]. In line with these reports, Rankin et al have recently showed that the HIF signaling pathway from osteoblasts play key roles in hematopoiesis [13]. Collectively, this evidence suggests that the interaction between HSCs and osteoblasts, forming specialized hypoxia, is crucial in keeping the HSC pool size in vivo and to prevent exhaustion of HSCs from uncontrolled cell-cycle entry and excessive proliferation.
MicroRNAs (miRNAs) are short non-coding RNAs comprised of 21 to 23 nucleotides in length that post-transcriptionally regulate mRNA expression [14]. Involvement of miRNAs in hematopoiesis is strongly suggested by the position of miRNA genes near translocation breakpoints and by their presence in loci targeted for deletion in human leukemias [15]. Moreover, expression profiling data suggest a major role for miRNAs in the regulation of hematopoietic cell commitment, proliferation, apoptosis, survival and differentiation [16], [17], [18]. Most of the studies that have been performed so far on miRNA expression in hematopoietic stem and progenitor cells focus on hematopoietic lineage differentiation [19], [20], [21]. MiR-17 (also called miR-17-5p), an important member of the miR-17-92 cluster [22], is expressed abundantly in hematopoietic progenitors and promotes hematopoietic cell expansion by targeting sequestosome 1 (sqstm1) regulated pathways in mice [23]. Consistent with this data, expression of miR-17 is detected in human CD34+ cells and is shown to be significantly down-regulated during in vitro differentiation toward mature megakaryocytes, monocytes and monocytopoiesis [17], [24]. Collectively, these examples illustrate a more general role for the autocrine production of miR-17 as a regulator of critical pathways determining normal hematopoietic cell fate and differentiation. While evidence is accumulating for a crucial role of intrinsic miR-17 in regulating HSCs and HPCs, whether miR-17 signaling pathways within the hematopoietic niche, especially in osteoblasts, are also necessary in the cell-extrinsic control of hematopoiesis has not yet been examined. Interestingly, one group recently found that some miRNAs are expressed differently between two stromal cell lines that have distinguishable functional characteristics and gene expression profiles for hematopoiesis, suggesting a potential role for miRNAs in regulating hematopoietic cell migration and niche function [25]. Related to this, two other separate studies described a regulatory role for miRNAs in controlling the expression of hematopoietic niche associated genes in endothelial cells [26], [27].
We have previously reported one immortalized clone with the characteristics of osteoblasts [28], designated as FBMOB-hTERT, derived from human fetal bone marrow stromal cells with retroviral vectors containing the human telomerase catalytic subunit (hTERT) gene [28]. The FBMOB-hTERT cells support the human cord blood (CB) HSCs and HPCs expansion and maintain their self-renewal and multipotency [28]. Using these cells, we found that miR-17 was significantly overexpressed. The ectopic expression of miR-17 partly promoted the ability of FBMOB-hTERT to support human CB CD34+ cell expansion and maintain their self-renewal and multipotency. It is noted that ectopic miR-17 in FBMOB-hTERT preferentially supports a specific expansion of the erythroid lineage. Conversely, miR-17 knockdown in FBMOB-hTERT suppressed the hematopoietic supporting ability of FBMOB-hTERT, in particular the mature erythroid cell growth. We further identified that HIF-1α is responsible for, at least in part, the promoted function of ectopic miR-17 in FBMOB-hTERT on hematopoiesis. The expression of HIF-1α was significantly enhanced in miR-17 overexpressed FBMOB-hTERT upon interaction with CB CD34+ cells compared with other niche associated factors such as KL, SDF-1 and EPO. More interestingly, selective expansion of the erythroid lineage of CB CD34+ cells through osteoblastic miR-17 was abrogated by HIF-1α knock down, demonstrating that HIF-1α was, at least partly, a mediator of miR-17-induced CB CD34+ cell expansion in FBMOB-hTERT.
In summary, in addition to intracellular miR-17, our data raised the possibility that miR-17 was also necessary in the cell-extrinsic control of HSCs and HPCs function, which is, at least in part, through the augmented HIF-1α signal pathways.
Materials and Methods
Cell Cultures
The hTERT-transduced fetal bone marrow osteoblasts (FBMOB-hTERT) [28] and cryopreserved primary bone marrow stromal cells (BMSCs) [28] were thawed and plated in DMEM (Gibco) containing 10% FBS (Gibco) at 37°C supplemented with 5% CO2, as described previously [28]. The Phoenix cell line, obtained from American Type Culture Collection (ATCC) was also cultured in the same medium. Human CB samples were obtained as described previously [28]. Briefly, CB mononuclear cells (MNCs) were isolated by the lymphocyte separation medium (1.077 g/ml) (TBD Biotech, Tianjing, China), and were immunomagnetically enriched for CD34+ cells using the MACS CD34+ Cell Isolation Kit (Miltenyi Biotech Inc., Bergisch Gladbach, Germany) according to the manufacturer’s instructions. The purity of CD34+ cells was about 80%–90%, as determined by flow cytometry (FCM). This study was approved by the Ethics Committee of Peking University. Before the experiments, the subjects were informed of the objectives, requirements and procedures of the experiments. All subjects gave informed written consent to participate in the study.
Construction and Packaging of shRNA Vectors
The two miR-17-specific small hairpin RNAs (17/KD and 17/KD1) and HIF-1α-specific shRNA (HIF1α/KD) oligomers [29] were tested. The details of the shRNA sequences are provided in Table 1 . Sense and antisense oligomers were used to produce double-stranded oligomers, and the oligomers were inserted into the retroviral vector RNAi-pSIREN-RetroQ, which drives shRNA production from the U6 promoter and also contains puromycin resistance (Clontech). Inserts were confirmed by sequencing. If not otherwise mentioned, RNAi-pSIREN-RetroQ vectors containing scrambled target sequences not complementary to any known miRNA were served as controls for 17/KD or HIF1α/KD (CTRL). The Phoenix packaging cell line was co-transfected with the RNAi-pSIREN-RetroQ retroviral plasmid and the viral packaging plasmid by Lipofectanine 2000 (Invitrogen) following the manufacturer’s instructions. Viral supernatants were collected at 48 or 72 hours after transfection and stored at −80°C for future use.
Table 1. Primers used in this study.
| Name | Sequence |
| shRNA-I | 5′-GGATCCGTCAAAGTGCTTACAGTGCAGGTTCAAGAGACCTGCACTGTAAGCACTTTGACTTTTTTGAATTC-3′ |
| shRNA-II | 5′-GGATCCGTGCATCTACTGCAGTGAAGGCTTCAAGAGAGCCTTCACTGCAGTAGATGCACTTTTTTGAATTC-3′ |
| shRNA-III | 5′-GGATCCGCTGGAGACACAATCATATCTTTTCAAGAGAAAGATATGATTGTGTCTCCAGCTTTTTTGAATTC-3′ |
| Scrambled shRNA | 5′ -GGATCCACACGTCCGAACATACTACTTCAAGAGAGTAGTATGTTCGGACGTGTTTTTTTGAATTC-3′ |
| pre-miR-17 | 5′ -CGGATCCAGTTTGAGGTGTTAAT-3′; 5′ -GGAATTCACTATCTGCACTAGATG-3′ |
| Hsa-miR17 | 5′ -AGTGCGTGTCGTGGAGTC-3′; 5′ -GCAAAGTGCTTACAGTGCA-3′ |
| U6 | 5′ -GCTTCGGCAGCACATATACTAAAAT-3′; 5′ -CGCTTCACGAATTTGCGTGTCAT-3′ |
| HIF-1α | 5′ -CCATTAGAAAGCAGTTCCGC-3′; 5′ - TGGGTAGGAGATGGAGATGC -3′ |
| SDF-1 | 5′- AACGCCAAGGTCGTGGTCGTGCTG-3′; 5′ - CACATCTTGAACCTCTTGTTTAAAAGC -3′ |
| KL | 5′- GACAGCTTGACTGATCTTCTGGAC-3′; 5′ - ACTGCTGTCATTCCTAAGGGAGCT -3′ |
| EPO | 5′- GAGAATATCACGACGGGCTG -3′; 5′ - CCACTGACGGCTTTATCCAC -3′ |
| 17α-satellite | 5′- ACGGGATAACTGCACCTAAC -3′; 5′ - CCATAGGAGGGTTCAACTCT -3′ |
Cloning of Human pre-miR-17 Gene, the Derivative Construction and Retroviral Infection
Total DNA was isolated using DNA Mini kit (Watson Biotechnologies, Inc., Shanghai, China) according to the manufacturer’s instructions. Primers for amplifying human pre-miR-17 gene were provided in Table 1 . PCR was carried out following standard protocols. The full-length human pre-miR-17 expression clone was inserted into the retroviral vector, yielding the expression constructs, pCMV-pre-miR-17 (17/OE). If not otherwise mentioned, vectors containing the empty intron sequence served as controls for 17/OE (CTRL). Viral supernatants were produced and stored as described as above.
The retroviral supernatant (1.0 ml) containing indicated retrovirus and polybrene (8ug/ml) was added to culture dishes when indicated cells grew up to 70%–80% confluent. After incubation for 5 hours at 37°C, the transduction medium was replaced by 2.0 ml DMEM, and puromycin (4ug/ml) was added to the cultures 24 hours later. Puromycin resistant clones grew out after 4–5 weeks of culture, and the pool cells were transferred to a fresh dish for expansion.
RNA Extraction and Real-time RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen). Real-time RT-PCR analysis was performed with a Bio-Rad iCycler using the iQSYBER green supermix (Bio-Rad). U6 snRNA (for miRNA) or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (for mRNA) was used as the endogenous control. Standard curves for internal control and tested genes were measured each time to determine the relative level of the respective transcript. The copy number was normalized to endogenous control levels.
Antibodies and Western Blotting
Cells were washed twice with phosphate buffered saline (PBS) and resuspended in RIPA buffer with protease inhibitor, and allowed to lyse on ice for 30 min. After lysis, samples were cleared by centrifuging at 15,000×g for 15 min at 4°C and supernatants were collected. Protein concentrations of supernatants were determined using a BCA (bicinchoninic acid) Protein Assay kit (Pierce). Samples were analyzed by western blotting using standard procedures. Mouse monoclonal GAPDH (Santa Cruz) was used at 1:5000. Rabbit anti-HIF-1α (Santa Cruz) was used at 1:1000. The blots were visualized by an enhanced chemiluminescence (ECL) kit following the manufacturer’s instructions. To ensure equal protein loading, each membrane was probed with the GAPDH antibody.
Luciferase Assay
17/OE FBMOB-hTERT or CTRL cells were seeded in 24-well plates at a density of 1.0×105 cells per well and were allowed to grow for 24 hours before transfection. Afterward, each well was transiently transfected with 900 ng pGL3 of either wild-type or mutant 3′UTR of HIF-1α [30] with a pRLTK vector (100 ng) for normalization of transfection efficiency using lipofectamine 2000. Cell lysates were harvested 36 hours post transfection, and firefly and renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega). The value of relative luciferase activity denotes the firefly luciferase activity normalized to that of renilla for each assay. Three independent experiments were done in triplicate.
Co-culture of Stromal Cells with Human CB CD34+ Cells
Co-culture assay was performed as described previously [28]. Briefly, the indicated stromal cells were irradiated at a dose of 40 Gy, and seeded in 24-well plates (5.0×105/well) overnight. CD34+ cells (2.0×104/well) in Iscove’s Modified Dulbecco’s Medium (IMDM) (Bio-WHITTAKER) were supplemented with 10% FBS, 10−4M 2-mercaptoethanol, 2 mM L-glutamine, 5 mg/ml insulin, 100 U/ml penicillin, and 100 mg/ml streptomycin as well as a cytokine cocktail consisting of Flt ligand (FL; 10 ng/ml), SCF (10 ng/ml), thrombopoietin (TPO, 10 ng/ml), and interleukin (IL)-6 (10 ng/ml), all of which were purchased from Peprotech Inc. After 14 days of culture, non-adherent and adherent hematopoietic cells that were loosely attached to stromal cells were harvested by gentle pipetting, counted, and analyzed for CD34 and CD38 expression by flow cytometry.
Long-term Culture Initiating Cells (LTC-IC) Assay
LTC-IC was analyzed as described previously [28]. Briefly, 1.0×104 CB CD34+ cells were plated into six-well plates containing a nearly confluent, irradiated indicated stromal cell monolayer in LTC medium (Myelo-Cult, StemCell Inc., Vancouver, BC, Canada) along with 10−6M hydrocortisone sodium hemisuccinate (Sigma). The LTC medium contained horse serum, fetal bovine serum, 2-Mercaptoethanol and α-MEM. 50% of the medium was replaced weekly with fresh medium. Both non-adherent and adherent cells were harvested weekly during 5–8 weeks of culture, and were cultured in the complete methylcellulose medium containing 2.8% BSA, 30% FBS (Hyclone), 50 ng/ml SCF, 20 ng/ml IL-3, 20 ng/ml GM-CSF, 20 ng/ml IL-6, and 3 U/ml EPO (Kirin, Tokyo, Japan), at 37°C with 5% CO2. After 14–16 days of cultures, colonies with greater than 50 cells were counted to assess LTC-IC activities. Colony-forming units (CFU-Cs), colony-forming unit-mix (CFU-Mix’s), colony-forming unit-erythrocyte (CFU-Es) and burst-forming unit-erythrocyte (BFU-Es) were counted to calculate the frequency of LTC-IC according to the manufacturer’s instructions (StemCell Inc.).
NOD/SCID Repopulating Cell (SRC) Assay
5.0×104 CB CD34+ cells were co-cultured with indicated stromal cells for 4 weeks, harvested as described above, and injected intravenously (i.v.) into 8-week-old, sublethally irradiated (3.5 Gy) NOD/SCID mice. Positive control mice were transplanted with freshly isolated CD34+ cells. The mice were sacrificed 12 weeks post-transplantation. Bone marrow mononuclear cells were harvested and analyzed by flow cytometry. Human cell repopulation was assessed using anti-human-CD45-FITC (fluorescein isothiocyanate), anti-human-CD34-PE (phycoerythrin), anti-human-CD36-PC7 (PE-cyanin 7) and anti-glycophorin A (GPA)-APC (allophycocyanin) antibodies. Isotype controls were mouse IgG1 conjugated to PE, APC PC7 or FITC. All antibodies were from Becton Dickinson. PCR amplification of the human 17α-satellite gene was employed as a second test for the presence of human cells in the NOD/SCID mice that had received transplants. In cases where secondary transplants were performed, bone marrow cells were prepared from primary recipients as described below, pelleted 4 minutes at 720 g and resuspended at 1.6 to 2.4 ×106/mL in IMDM/10% FBS. Secondary recipients each received 6.5 ×105 human CD34+ cells from primary recipients. All care and handling of animals were performed with the approval of Institutional Authority for Laboratory Animal Care of Peking University.
Statistical Analysis
The results are expressed as the mean ± standard deviation (SD). Statistical comparisons were performed using a two-tailed Student’s t -test.
Results
MiR-17 is Endogenously Expressed in FBMOB-hTERT and Primary BMSCs
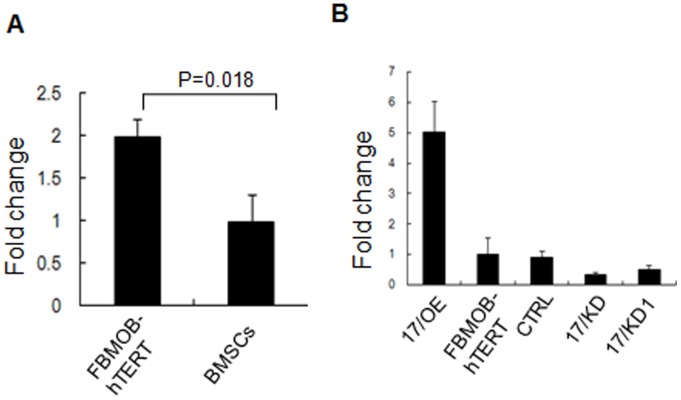
To test our hypothesis that osteoblastic miR-17 may influence hematopoiesis, we first determined the miR-17 expression level in the FBMOB-hTERT and primary bone marrow stromal cells (BMSCs). Real-time RT-PCR assay showed that miR-17 is endogenously expressed in these cells. Compared with primary BMSCs, FBMOB-hTERT expressed a significantly higher level of miR-17 ( Figure 1A ). MiR-17 is also endogenously expressed in cord blood (CB) CD34+ cells (data not shown), which is consistent with the previous reports [16], [23]. The higher expression of miR-17 in FBMOB-hTERT cells was of interest given that such an expression was likely to have hematopoietic functional consequences.
Figure 1. The expression of miR-17 in FBMOB-hTERT cells.
A: The expression level of miR-17 in FBMOB-hTERT cells was evaluated by real-time RT-PCR. Each reaction was performed in triplicate. The data are presented as the ratio of miR-17 levels (relative to U6) in FBMOB-hTERT to that in bone marrow stromal cells (BMSCs). B: Real-time RT-PCR was performed to evaluate the expression level of miR-17 in FBMOB-hTERT cells after retrovirally transduced with vectors for miR-17 overexpression (17/OE), miR-17 knockdown (17/KD or 17/KD1), or control (CTRL). The data are presented as the ratio of miR-17 levels (relative to U6) in 17/OE, FBMOB-hTERT, 17/KD or 17/KD1 to that in CTRL. Each reaction was performed in triplicate.
The Function of osteoblastic miR-17 on Expansion of CB CD34+ Cells
To analyze the function of osteoblastic miR-17 on the expansion of HSCs and HPCs, the miR-17 overexpressing and knockdown models were created using FBMOB-hTERT cells by using retroviral vectors. The miR-17 levels in the FBMOB-hTERT cells transduced with the indicated virus were determined using real-time RT-PCR. We observed obvious up and down-modulation of miR-17 in 17/OE cells and 17/KD cells respectively compared to the CTRL ( Figure 1B ). Based on shRNA influence on miR-17 expression, 17/KD was chosen for further studies.
We first investigated the ability of the osteoblastic miR-17 to expand CB CD34+ cells in vitro. CD34+-enriched CB cells were co-cultured with irradiated 17/OE, 17/KD, or CTRL cells. After 14 days of culturing, the numbers of total cells, CD34+ cells, CD34+CD38− cells, and CD34+CD38+ cells were counted, respectively. As shown in Table 2 , although both CD34+CD38− and CD34+CD38+ cells were expanded dramatically in the presence of FBMOB-hTERT, the 17/OE cells appeared to be more potent than CTRL cells (CD34+CD38− cells: 24.21- versus 18.45-folds) in supporting CD34+CD38− cell expansion. Conversely, deficient expression of miR-17 in FBMOB-hTERT suppressed CD34+CD38− cell expansion (CD34+CD38− cells: 13.27- versus 18.45-folds). These results suggest that miR-17 in FBMOB-hTERT cells can promote CD34+CD38− cell expansion in vitro.
Table 2. Ex vivo expansion of CB CD34+ cells over 14 days.
| 17/OE | CTRL | 17/KD | |
| Total cells | 80.01±10.11 | 82.13±8.18 | 86.29±5.02 |
| CD34+ cells | 30.14±4.23* | 24.65±3.89 | 17.01±5.41* |
| CD34+CD38− cells | 24.21±3.05* | 18.45±2.31 | 13.27±3.11* |
| CD34+CD38+ cells | 92.67±9.01 | 92.16±12.87 | 91.88±7.90 |
The cells were stained with PE-conjugated mAb to CD34 and FITC-conjugated mAb to CD38, and analyzed by flow cytometry. Values indicate the fold increase compared with the initial number of cells (2.0×104/well).The results are given as mean ± standard deviation (SD) (n = 6). *p<0. 05 versus the CTRL cells (Student’s t-test).
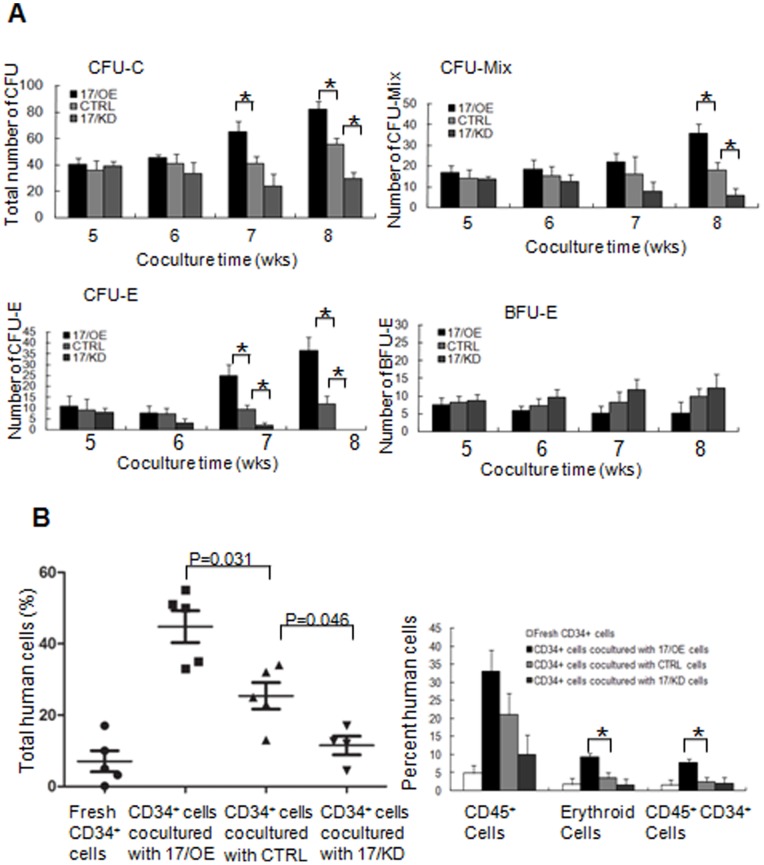
To test the capability of osteoblastic miR-17 in supporting self-renewal and maintaining multipotent differentiation of HSC, we performed a Long-term culture initiating cells (LTC-IC) assay. CB CD34+cells were co-cultured with 17/OE, 17/KD, or CTRL cells in LTC-IC medium for 5–8 weeks, and then subject to a CFU assay. After 14–16 days of culturing, the colonies with more than 50 cells were counted. As shown in Figure 2A , the number of total CFCs from the cells co-cultured with 17/OE cells for 7 or 8 weeks was significantly higher than that of the cells co-cultured with the CTRL. Moreover, after co-cultured for 8 weeks, the number of total CFCs from the cells co-cultured with 17/KD cells became significantly lower compared to the cells co-cultured with the CTRL, suggesting that osteoblastic miR-17 partly supports the LTC-IC activity of HSC. Interestingly, the number of mature erythroid (CFU-E) from the cells co-cultured with 17/OE cells for 7 or 8 weeks was significantly higher than that of the cells co-cultured with the CTRL, whereas only after co-cultured for 8 weeks, the number of total CFU-Mix’s from the cells co-cultured with 17/OE cells was significantly higher than that of the cells co-cultured with the CTRL, suggesting that ectopic miR-17 in FBMOB-hTERT preferentially supports a specific expansion of the erythroid lineage. Knockdown of miR-17 in FBMOB-hTERT cells, on the other hand, resulted in reduced hematopoietic support, which was followed by diminishing CFU output. It is noted that compared to a significant and specific increase or decrease in the number of CFU-E from cells co-cultured with 17/OE or 17/KD cells for 7 or 8 weeks than that from cells co-cultured with the CTRL, the number of immature erythroid (BFU-E) progenitors did not change significantly after osteoblastic miR-17 modulation.
Figure 2. The effect of miR-17 modulation in FBMOB-hTERT cells on CB CD34+ cells.
A: The effect of miR-17 modulation in FBMOB-hTERT cells on long-term culture initiating cells activity of CB CD34+ cells. 1.0×104 CD34+ CB cells were co-cultured with FBMOB-hTERT cells after transduced with vectors for miR-17 overexpression (17/OE), miR-17 knockdown (17/KD), or control (CTRL) for 5-8 weeks and then subject to colony-forming-unit (CFU) assay. After 14-16 days of culture, the colonies, including CFU-Mixs, CFU-E, and BFU-E with greater than 50 cells were counted. The results are expressed as mean ± SD (n = 6). *p<0.05, compared between 17/OE or 17/KD, and CTRL group (Student’s t-test). B: Effect of miR-17 modulation in FBMOB-hTERT cells on repopulation of CB CD34+ cells in non-obese diabetic/severe combined immunodeficient disease (NOD/SCID) mice. 5.0×104 CB CD34+ cells were co-cultured with 17/OE, 17/KD or CTRL, harvested at 4 weeks of culture and then injected intravenously into the sublethally irradiated NOD/SCID mice (n = 6 per group). The mice were sacrificed 12 weeks after transplantation and the mononuclear cells from bone marrow were analyzed for human cells composed of CD45+, CD45–CD36+ and CD36–GPA+ cells and CD45+CD34+ population by flow cytometry. The level of total human cell engraftment was shown in the left panel. P = 0.031 or 0.046, compared between the NOD/SCID mice injected with CD34+ cells co-cultured with 17/OE or 17/KD and those injected with CD34+ cells co-cultured with CTRL (Student’s t-test). The fraction of CD45+, erythroid (CD45−CD36+ and CD36−GPA+) and CD45+CD34+ population among the engrafted human cells was shown in right panel. *p<0.05, compared between the NOD/SCID mice injected with CD34+ cells co-cultured with 17/OE and those injected with CD34+ cells co-cultured with CTRL (Student’s t-test). The significant difference was only analyzed between the mice injected with CD34+ cells co-cultured with 17/OE or 17/KD and those injected with CD34+ cells co-cultured with CTRL.
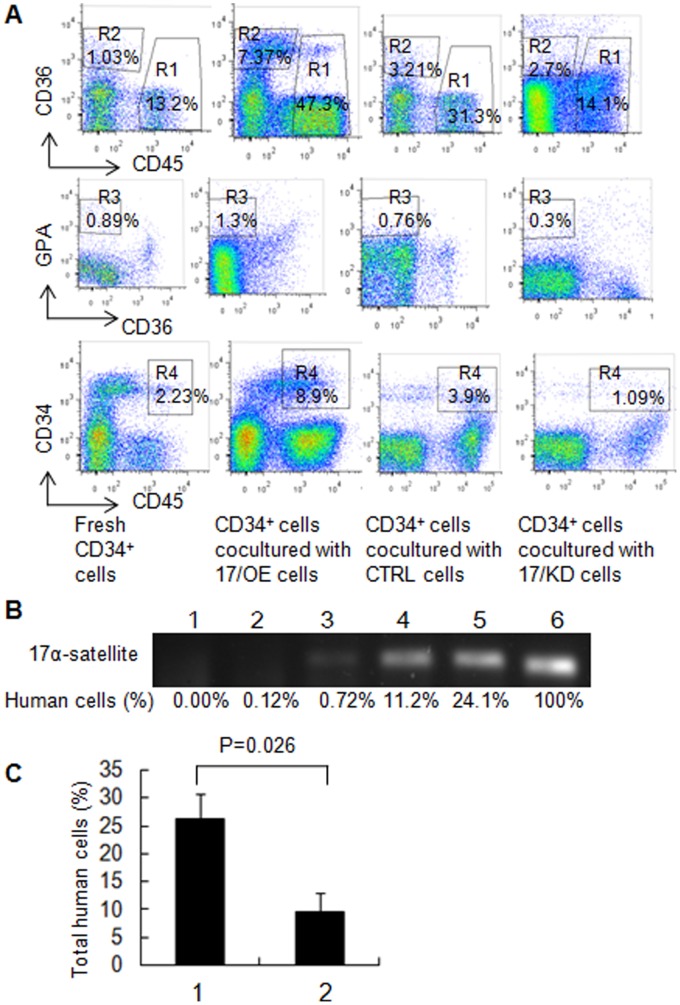
To further support our in vitro expansion and LTC-IC results, we examined the engraftment of CB CD34+ cells after co-cultured with 17/OE, 17/KD, or CTRL cells in NOD/SCID mice. The sublethally irradiated NOD/SCID mice were transplanted with 5.0×104 CB CD34+ cells from the co-cultures with irradiated 17/OE, CTRL, or 17/KD cells for 4 weeks. The level of total human cell engraftment composed of CD45+, CD45–CD36+ and CD36–GPA+ cells, and CD45+CD34+ populations were assessed in the bone marrow mononuclear cells of the graft mice at 12 weeks post-transplant by flow cytometry. At 12 weeks, the degree of total human cells from the mice injected with CD34+ cells co-cultured with 17/OE or 17/KD was significantly higher or lower than that of the mice injected with CD34+ cells co-cultured with CTRL ( Figure 2B left panel ; p = 0.031 and 0.046 respectively). The statistical analysis was only done in comparison between the mice injected with CD34+ cells co-cultured with 17/OE or 17/KD and those injected with CD34+ cells co-cultured with CTRL. We further analyzed the multilineage development from input CD34+ populations. Flow cytometry analysis of the human graft in a representative graft mouse from each group is shown in Figure 3A . There was no significant difference in the percentage of CD45+ cells by comparing the 17/OE or 17/KD group with the corresponding CTRL group ( Figure 2B right panel ). However, we observed a significantly higher percentage of erythroid cells, including CD45–CD36+ and CD36–GPA+ populations in 17/OE group compared to the percentage in the CTRL group. Similarly, compared to the CTRL group, the 17/OE group also showed a significantly higher percentage of CD45+CD34+ cells ( Figure 2B right panel ). The observed multilineage development from input CD34+ populations co-cultured with 17/OE cells coincides with expansion and LTC-IC assays in vitro. Whereas the percentage of CD45+CD34+ cells and erythroid cells in the 17/KD group whose transplants of CD34+ cells had been co-cultured with 17/KD cells showed a tendency, although this was not significant, to be lower than that of mice receiving transplants of CD34+ cells co-cultured with CTRL cells. To further confirm that the human cells determined by flow cytomtry were of human origin, the human-specific 17α-satellite gene was detected by PCR in several representative graft mice, which contained different percentages of human cells. We found that the human 17α-satellite gene could be detected by PCR amplification when the percentage of human cells was over 0.72% ( Figure 3B , lanes 3–6), whereas it was indetectable at a percentage of 0.12% ( Figure 3B , lanes 2). These results partly confirmed our in vitro data and suggest that osteoblastic miR-17 partly promotes the ability of FBMOB-hTERT to maintain the multipotency of CB CD34+ cells in vitro. It also seemed that osteoblastic miR-17 is prone to cause a specific expansion of the erythroid lineage.
Figure 3. Effect of miR-17 modulation on repopulation of CD34+ cells in primary and secondary NOD/SCID mice.
A: Flow cytometry analysis of the human CB CD34+ cell repopulation in a representative primary NOD/SCID mouse after co-cultured with 17/OE, 17/KD or CTRL cells. Fresh CD34+ cells were served as controls. The mononuclear cells from bone marrow harvested from the engrafted NOD/SCID mice were examined by flow cytometry for the assessment of human cells composed of CD45+ cells (R1) and erythroid cells including CD45–CD36+ (R2) and CD36–GPA+ (R3) population and CD45+CD34+ cells (R4). B: The bone marrow mononuclear cells (MNCs) containing the different percentage of human cells (lanes 2-5) from the primary representative engrafted mice were analyzed for human-specific 17α-satellite DNA by PCR. The human-specific 17α-satellite gene was detected when the human cells were over 0.70% (lanes 3–6) whereas it was indetectable at a percentage of 0.12% (lane 2). Lane 1, one mouse without transplants; lane 2, one mouse receiving transplants of fresh CD34+ cells; lane 3, one mouse receiving transplants of CD34+ cells co-cultured with 17/KD cells; lane 4, one mouse receiving transplants of CD34+ cells co-cultured with CTRL cells; lane 5, one mouse receiving transplants of CD34+ cells co-cultured with 17/OE cells; lane 6 indicates positive control (human peripheral blood (PB) MNCs). C: Effect of miR-17 modulation in FBMOB-hTERT cells on repopulation of CB CD34+ cells in secondary NOD/SCID mice. Human cells were analyzed in secondary mice 12 weeks after intravenous injection of bone marrow cells from primary mice, which were injected with CB CD34+ cells co-cultured with 17/OE or CTRL for 4 weeks and sacrificed 8 weeks after transplantation. P = 0.026, compared between 1 and 2 (1, bone marrow mononuclear cells in secondary mice from primary mice injected with CB CD34+ cells co-cultured with 17/OE; 2, bone marrow mononuclear cells in secondary mice from primary mice injected with CB CD34+ cells co-cultured with CTRL) (Student’s t-test).
To further confirm whether or not the human cell fraction in the primary engraftment contained NOD/SCID repopulating cells (SRCs), we collected bone marrow from the primary mice that received a transplant of CD34+ cells co-cultured with 17/OE cells or CTRL cells and transplanted the marrow into sublethally irradiated secondary recipients. In these experiments, the bone marrows (BMs) of six mice from the two groups were transplanted into eight recipients (calculated human CD34+ cells received/mouse was 6.5×105). Human cells in the BM from the secondary recipients consisting of CD45+, CD45–CD36+ and CD36–GPA+ populations and the CD45+CD34+ cells were also analyzed by flow cytometry. While the cells from the two groups could engraft secondary recipients, the percentage of total human cell engrafting the BM from the primary recipient transplanted with CD34+ cells co-cultured with 17/OE, as demonstrated by flow cytometry, was significantly higher than that of the primary recipient transplanted with CD34+ cells co-cultured with the CTRL ( Figure 3C ). Although only a limited number of secondary recipients have been analyzed, these data suggest that the ability of FBMOB-hTERT to maintain the multipotency of CB CD34+ cells in vitro was partly promoted through miR-17 over-expression.
MiR-17 Up-regulates HIF-1α Expression upon Interaction with CB CD34+ Cells
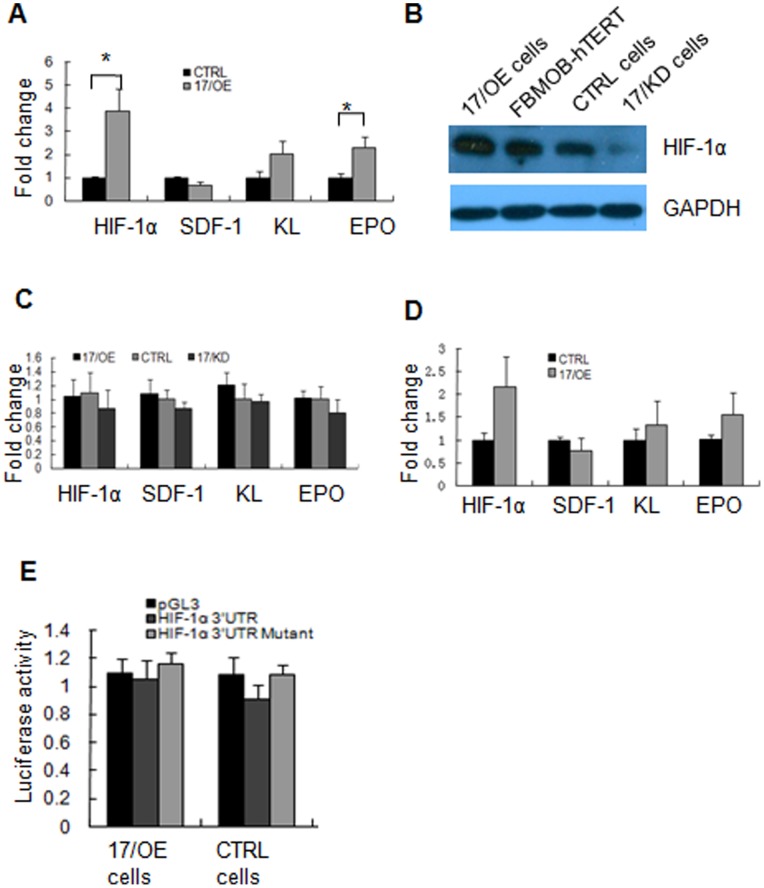
To explore the mechanisms by which miR-17 promotes the function of FBMOB-hTERT in supporting hematopoiesis thus causing a specific expansion of the erythroid lineage, we examined the production of hematopoietic supporting growth factors including the hypoxia-inducible transcription factor (HIF-1α), stromal cell-derived factor (SDF-1), stem cellfactor/c-kit ligand (SCF/KL) and erythropoietin (EPO) by 17/OE and CTRL cells during interaction with CB CD34+ cells. The 17/OE and CTRL cells were co-cultured with or without 1.0×104 CD34+ cells for 24 hours. The adherent co-cultured cells were harvested after washing off the loosely adherent and non-adherent cells, and analyzed by real-time RT-PCR or western-blotting. Among these genes, HIF-1α and EPO were found to be significantly increased in 17/OE cells after interaction with CB CD34+ cells compared to that in CTRL cells ( Figure 4A , about 4 and 2 folds respectively). In contrast, KL exhibited no significant increase and there was almost no change in SDF-1 expression in 17/OE cells after interaction with CB CD34+ cells compared to that in CTRL cells. Protein expression of HIF-1α was also measured by western-blotting. As shown in Figure 4B , the levels of HIF-1α protein were significantly up-regulated in 17/OE cells after co-cultured with CB CD34+ cells compared to that in CTRLs. We also confirmed the up-regulation of HIF-1α in primary BMSCs after ectopic expression of miR-17 upon interaction with CB CD34+ cells. These results suggest that ectopic miR-17 in FBMOB-hTERT augmented the expression of niche associated genes during co-cultured with CB CD34+ cells, which may be responsible for the hematopoietic supporting ability of osteoblastic miR-17.
Figure 4. MiR-17 up-regulates HIF-1α expression upon interaction with CB CD34+ cells.
A: 1.0×106 17/OE or CTRL cells were co-cultured with CB CD34+ cells (1.0×104/well) in six-well plates for 24 hours, and then washed twice to remove loosely adherent and non-adherent cells. The adherent cells were harvested and analyzed by real-time RT-PCR for transcripts of the niche associated genes: HIF-1α, SDF-1, KL and EPO. The results are expressed as mean ± SD (n = 3). *p<0.05, compared between 17/OE cells and CTRL cells (Student’s t-test). B: HIF-1α protein levels were measured by western blotting in 17/OE, 17/KD or CTRL cells after interaction with CB CD34+ cells for 24 hours. C: The transcripts of the niche associated genes: HIF-1α, SDF-1, KL and EPO were analyzed by real-time RT-PCR in 17/OE, 17/KD or CTRL cells. The results are expressed as mean ± SD (n = 3). Without CB CD34+ cell existing, the expression of the indicated niche associated genes was not changed significantly in 17/OE or 17/KD cells comparing with that in CTRL cells. D. The transcripts of the niche associated genes were analyzed by real-time RT-PCR in bone marrow stromal cells (BMSCs) with over-expressed miR-17 and the control cells upon interaction with CB CD34+ cells. The expression of HIF-1α was up-regulated after miR-17 over-expressed whereas the expression of SDF-1, KL and EPO did not changed obviously. The results are expressed as mean ± SD (n = 3). E: Luciferase reporter assays to check whether miR-17 directly target HIF-1α in FBMOB-hTERT. The luciferase activities were not significantly decreased in 17/OE cells compared with that in CTRL cells. The results are given as mean ± standard deviation (SD) (n = 3).
HIF-1α Knock Down Partially Abrogate the Hematopoietic Supporting Ability of Osteoblastic miR-17
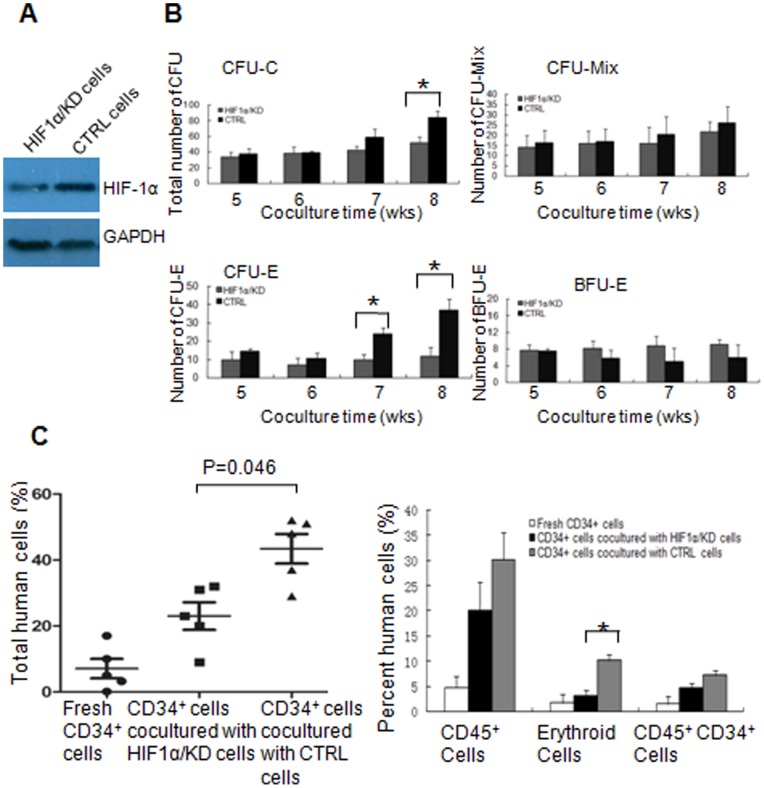
Since ectopic miR-17 in FBMOB-hTERT cells can significantly up-regulate HIF-1α upon interaction with CB CD34+ cells, we examined whether or not the hematopoietic supporting ability of osteoblastic miR-17 is dependent on the augmented HIF-1α activity in FBMOB-hTERT cells. The HIF-1α knockdown model was created using 17/OE FBMOB-hTERT cells by stably expressing HIF-1α shRNA or nonspecific shRNA (termed HIF1α/KD and CTRL respectively) via retroviral transduction. We performed western-blotting to detect HIF-1α protein levels in the retrotransduced 17/OE cells. As shown in Figure 5A , the level of HIF-1α protein was significantly knocked down by HIF-1α shRNA in HIF1α/KD cells when compared to levels in CTRL cells.
Figure 5. HIF-1α knockdown partially abrogates the hematopoietic supporting ability of osteoblastic miR-17.
A: Western blotting was performed to evaluate the expression level of HIF-1α protein in 17/OE cells after transduced with vectors for HIF1α knockdown (HIF1α/KD), or control (CTRL). B: The effect of the HIF1α/KD cells on long-term culture initiating cells activity of CB CD34+ cells. 1.0×104 CD34+ CB cells were co-cultured with HIF1α/KD cells or CTRL for 5-8 weeks and then subject to CFU assay. After 14–16 days of culture, the colonies, including CFU-Mixs, CFU-E, and BFU-E with greater than 50 cells were counted. The results are expressed as mean ± SD (n = 6). *p<0.05, compared between HIF1α/KD and CTRL group (Student’s t-test). C: Effect of HIF1α/KD cells on repopulation of CB CD34+ cells in non-obese diabetic/severe combined immunodeficient disease (NOD/SCID) mice. 5.0×104 CB CD34+ cells were co-cultured with HIF1α/KD or CTRL cells, harvested at 4 weeks of culture and then injected intravenously into the sublethally irradiated NOD/SCID mice (n = 6 per group). The mice were sacrificed 12 weeks after transplantation and the mononuclear cells from bone marrow were analyzed for human cells composed of CD45+, CD45–CD36+ and CD36–GPA+ cells and CD45+CD34+ population by flow cytometry. The level of total human cell engraftment was shown in the left panel. p = 0.046, compared between the mice injected with CD34+ cells co-cultured with HIF1α/KD and those injected with CD34+ cells co-cultured with CTRL (Student’s t-test). The fraction of CD45+, erythroid (CD45−CD36+ and CD45− CD36−GPA+) and CD45+CD34+ cells among the engrafted human cells was shown in the right panel. *p<0.05, compared between the mice injected with CD34+ cells co-cultured with HIF1α/KD and those injected with CD34+ cells co-cultured with CTRL (Student’s t-test). The significant difference was only analyzed between the mice injected with CD34+ cells co-cultured with HIF1α/KD and those injected with CD34+ cells co-cultured with CTRL.
Using the cells above, we further investigated the hematopoietic supporting ability of osteoblastic miR-17 after HIF-1α knock down. The in vitro expansion assay ( Table 3 ) demonstrated that CTRL cells appeared to be more potent than HIF1α/KD cells (CD34+CD38− cells: 24.83- versus 15.65-folds) in supporting CD34+CD38− cell expansion. The LTC-IC ( Figure 5B ) assay showed that, only after co-cultured for 8 weeks, the total number of CFCs from the cells co-cultured with CTRL cells was significantly higher than that of the cells co-cultured with HIF1α/KD cells. There was no significant difference between the number of CFU-Mix’s from the cells co-cultured with CTRL cells, and from the cells co-cultured with HIF1α/KD cells. These suggested that the hematopoietic supporting ability of osteoblastic miR-17 was partially abrogated by HIF-1α knock down. It is interesting that the number of mature erythroid (CFU-Es) from the cells co-cultured with the CTRL for 7 or 8 weeks was significantly higher than that of the cells co-cultured with HIF1α/KD, which further suggested that the specific erythroid lineage expansion of CB CD34+ cells caused by osteoblastic miR-17 was abrogated by HIF-1α knock down. The number of immature erythroid (BFU-E) progenitors from the cells co-cultured with HIF1α/KD cells did not change significantly.
Table 3. Ex vivo expansion of CB CD34+ cells over 14 days.
| HIF1α/KD | CTRL | |
| Total cells | 82.09±7.16 | 80.27±9.03 |
| CD34+ cells | 21.91±4.52* | 30.02±3.99 |
| CD34+CD38− cells | 15.65±4.12* | 24.83±4.03 |
| CD34+CD38+ cells | 92.33±11.30 | 91.98±8.31 |
The effect of ex vivo expansion of HIF1α/KD cells on CB CD34+ cells was assayed according to the methods described above.
p<0. 05 versus CTRL cells (n = 6) (Student’s t-test).
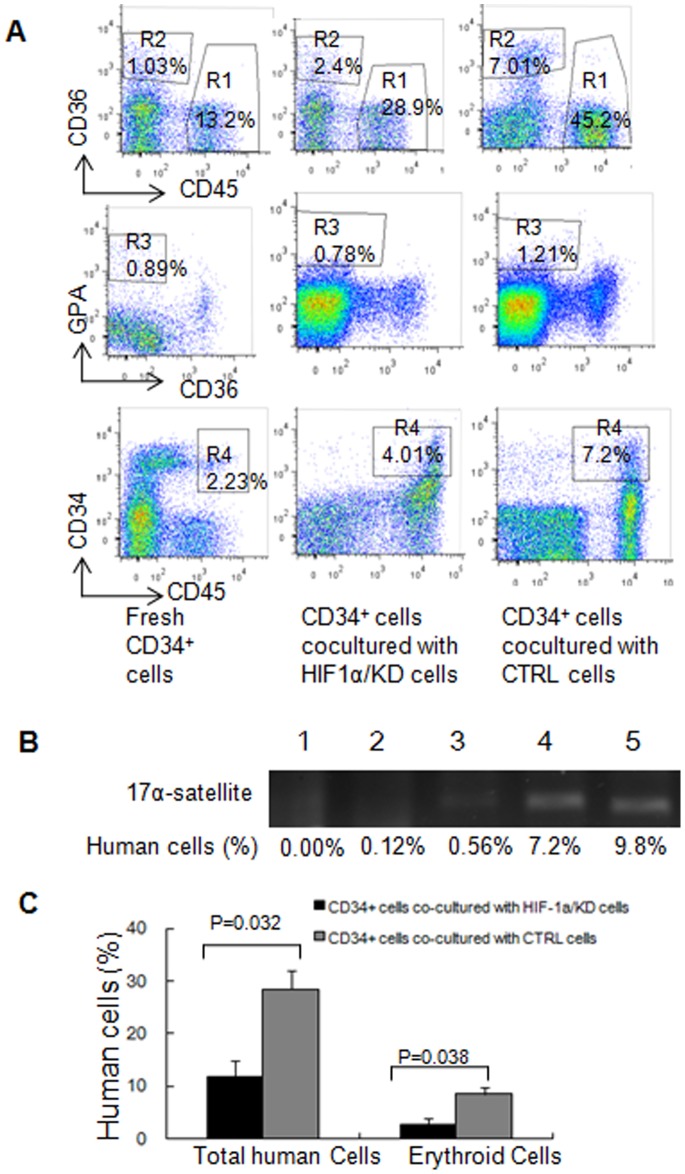
To support the above in vitro expansion and LTC-IC results, we further examined the engraftment of CB CD34+ cells after co-cultured with HIF1α/KD or CTRL cells in NOD/SCID mice ( Figure 5C ). The sublethally irradiated NOD/SCID mice were transplanted with 5.0×104 CB CD34+ cells from the co-cultures with irradiated HIF1α/KD or CTRL for 4 weeks. The level of total human cell engraftment composed of CD45+, CD45–CD36+ and CD36–GPA+ cells, and CD45+CD34+ population were assessed in the bone marrow mononuclear cells of the engrafted mice at 12 weeks post-transplant by flow cytometry. Flow cytometry analysis of the human graft in a representative graft mouse from each group is shown in Figure 6A . At 12 weeks, the degree of total human cells from the mice injected with CD34+ cells co-cultured with HIF1α/KD is significantly lower than that in the mice injected with CD34+ cells co-cultured with the CTRL ( Figure 5C left panel ; p = 0.046). The significant difference was only analyzed between the mice injected with CD34+ cells co-cultured with HIF1α/KD and those injected with CD34+ cells co-cultured with CTRL. We further analyzed the multilineage development from input CD34+ populations. There was no significant difference in the percentage of CD45+ cells ( Figure 5C right panel ) between the two groups; however, we observed a significantly lower percentage of erythroid cells including CD45–CD36+ and CD36–GPA+ population in mice injected with CD34+ cells co-cultured with HIF1α/KD compared to that in mice injected with CD34+ cells co-cultured with the CTRL ( Figure 5C right panel ), which is consistent with the results from the LTC-IC assay in vitro. The level of CD45+CD34+ cells showed no significant difference between the two groups. Human-specific 17α-satellite gene was also detected by PCR in three representative graft mice ( Figure 6B ) to confirm that the human cells determined by flow cytomtry were of human origin. We found that the human 17α-satellite gene could be detected by PCR amplification when the percentage of human cells was more than 0.56% ( Figure 6B , lanes 3–5), whereas it was indetectable at a percentage of 0.12% ( Figure 6B , lanes 2). These results confirmed our in vitro data and demonstrated that the specific erythroid lineage expansion of CB CD34+ cells caused by osteoblastic miR-17 was abrogated by HIF-1α knock down. All these suggested that HIF-1α is, at least partly, a mediator of CB CD34+ cell expansion caused by miR-17 in FBMOB-hTERT cells.
Figure 6. Repopulation of CD34+ cells after co-cultured with HIF1α/KD in primary and secondary NOD/SCID mice.
A: Flow cytometry analysis of the human CD34+ cell repopulation in a representative primary NOD/SCID mouse after co-cultured with HIF1α/KD or CTRL. Fresh CD34+ cells were served as controls. The mononuclear cells from bone marrow harvested from the primary injected NOD/SCID mice were examined by flow cytometry for the assessment of human cells composed of CD45+ cells (R1) and erythroid cells including CD45–CD36+ (R2) and CD36–GPA+ population (R3) and CD45+CD34+ cells (R4). B: The bone marrow mononuclear cells (MNCs) containing the different percentage of human cells (lanes 2-5) from the primary representative engrafted mice were analyzed for human-specific 17α-satellite DNA by PCR. The human-specific 17α-satellite gene was detected when the human cells were over 0.50% (lanes 3-5), whereas it was indetectable at a percentage of 0.12% (lanes 2). Lane 1, one mouse without transplants; lanes 2-3, two mice receiving transplants of fresh CD34+ cells; lane 4, one mouse receiving transplants of CD34+ cells co-cultured with HIF1α/KD cells; lane 5, one mouse receiving transplants of CD34+ cells co-cultured with CTRL cells. C: Effect of HIF-1α knockdown in 17/OE cells on repopulation of CB CD34+ cells in secondary NOD/SCID mice. Human cells were analyzed in secondary mice 12 weeks after intravenous injection of bone marrow cells from primary mice, which were injected with CB CD34+ cells co-cultured with HIF1α/KD or CTRL for 4 weeks and sacrificed 8 weeks after transplantation. The percentage of total human cell or erythroid cells in the secondary BM from the primary recipient transplanted with CD34+ cells co-cultured with HIF1α/KD was significant lower than that from the primary recipient transplanted with CD34+ cells co-cultured with CTRL (p = 0.032 and 0.038 respectively) (Student’s t-test).
To further determine whether the human cell fraction in the primary engraftment contained NOD/SCID repopulating cells (SRCs), we collected bone marrow from the primary mice that received a transplant of CD34+ cells co-cultured with HIF1α/KD or CTRL cells and transplanted the marrow into sublethally irradiated secondary recipients. The BMs of six mice from two groups were transplanted into eight recipients (calculated human CD34+ cells received/mouse was 6.5×105). Human cells in the BM from the secondary recipients consisting of CD45+, CD45–CD36+ and CD36–GPA+ populations and the CD45+CD34+ cells were also analyzed by flow cytometry. The percentage of total human cells or erythroid cells in the secondary BM from the primary recipient transplanted with CD34+ cells co-cultured with HIF1α/KD was significantly lower than that of the primary recipient transplanted with CD34+ cells co-cultured with the CTRL, as demonstrated by flow cytometry ( Figure 6C ). Although only a limited number of secondary recipients have been analyzed, these data suggest that the specific erythroid lineage expansion of CB CD34+ cells caused by osteoblastic miR-17 was abrogated by HIF-1α knock down.
Discussion
Osteogenic cells, lining in endosteal bone, play a crucial role in regulating HSC function [3], [4]. The FBMOB-hTERT cell line without tumorigenicity has the characteristics of osteoblasts [28] and can actively maintain the capacity of self-renewal and multipotency of HSCs and HPCs [28]. In this study, using FBMOB-hTERT cells [28], we identified that miR-17 in FBMOB-hTERT was significantly higher than that in bone marrow stromal cells (BMSCs) ( Figure 1A ), which is possibly responsible for the hematopoietic-supporting characteristic of FBMOB-hTERT. As expected, the expansion and LTC-IC assay ex vivo suggested that miR-17 in FBMOB-hTERT partly promoted the ability of FBMOB-hTERT to support human CB CD34+ cell expansion and maintain their multipotency ( Figure 2A ). It seems that the ability of miR-17 in FBMOB-hTERT to promote CB CD34+ cell expansion requires a significant amount of time. This idea is evidenced in the fact that the number of CFU-Mix from the cells co-cultured with 17/OE or 17/KD was significantly higher or lower than that of the cells co-cultured with CTRL cells only after co-cultured for 8 weeks. Although after co-cultured for 5, 6 or 7 weeks, there was a trend toward a decrease or increase in the number of CFU-Mix from the cells co-cultured with 17/KD or 17/OE compared to that from the cells co-cultured with CTRL cells, statistical analyses of the cohort indicated that it did not meet statistical significance (p>0.05). It is of interest to note that osteoblastic miR-17 seemed to be more prone to support erythroid lineage expansion because the number of mature erythroid (CFU-E) from the cells co-cultured with miR-17 modulated FBMOB-hTERT for 7 weeks was significantly changed in comparison to the cells co-cultured with CTRL cells. However, compared to a significant and specific change in the number of mature erythroid (CFU-E) from cells co-cultured with 17/OE or 17/KD cells for 7 or 8 weeks to that of cells co-cultured with CTRL, the number of immature erythroid (BFU-E) progenitors from CB CD34+ cell did not change significantly after osteoblastic miR-17 modulation. The function of miR-17 on CB CD34+ cells was confirmed by the primary and secondary engraftment assay in NOD/SCID mice. A significantly higher percentage of human CD45+CD34+ cells and erythroid cells (CD45–CD36+ and CD36–GPA+ cells) was observed in the bone marrow of mice transplanted with CD34+ cells co-cultured with 17/OE cells compared to that in the bone marrow of mice transplanted with CD34+ cells co-cultured with CTRL cells. In contrast, the percentage of human CD45+ cells in the bone marrow of mice whose transplants of CD34+ cells had been co-cultured with 17/OE cells showed a tendency, although this was not significant (p>0.05), to be higher than that of mice receiving transplants of CD34+ cells co-cultured with CTRL cells. All these suggested that the ectopic miR-17 signal pathway in FBMOB-hTERT cells may create a niche which can partly promote HSC and HPC expansion and is more suitable for erythroid progenitor differentiation, which subsequently leads to more mature erythroid cells.
The mechanisms underlying the enhanced expansion are largely unclear. One of the mechanisms is likely mediated by a variety of HSC-supporting growth factors, such as HIF-1α, which are constitutively activated by overexpressed miR-17 upon interaction with CB CD34+ cells. In support of this notion, we found that the transcription of HIF-1α was significantly up-regulated in 17/OE cells after co-cultured with CB CD34+ cells ( Figure 4A ). The protein level of HIF-1α was also up-regulated upon interaction with CB CD34+ cells ( Figure 4B ). It seemed that this special environment is vital for the up-regulation of HIF-1a caused by miR-17, because HIF-1α was not changed without the existence of CB CD34+ cells regardless of the level of miR-17 expression ( Figure 4C ). These data suggested that the different expressions of HIF-1a in different culture environments were caused by miR-17. The microenvironment plays a critical role in the regulation of cell fate and subsequent tissue formation [31], [32], [33] and various signaling molecules were subsequently ignited. Taguchi et al [30] suggested that HIF-1α was repressed by miR-17–92 only under a normoxic condition, whereas HIF-1α was robustly induced under hypoxia regardless of the level of miR-17–92 expression [30]. In addition, Jin et al [34] found that miR-17 modulated the diverse effect of canonical Wnt signaling in different microenvironments [34]. On the basis of our results and previous reports, we put forth the idea that different microenvironments lead to different effects of miR-17 and that mechanism is the key point of our further research.
Except for the environment, the intricate and finely tuned relationship between HIF-1α and miR-17 is also likely dependent on cellular context and appears to be promoter-independent in FBMOB-hTERT. After transfecting pGL3 with the 3′UTR of HIF-1α, the luciferase activities were not significantly decreased in 17/OE cells compared to the activities in CTRL cells ( Figure 4E ). The contradiction with the previous report may be due to the different cellular contexts of non-tumor and tumor cells. Additionally, the mRNA level of EPO was also significantly up-regulated in 17/OE, although they were not as high as HIF-1α, which was also dependent upon the interaction with CB CD34+ cells. EPO expression is tightly regulated by developmental, physiological, and cell-type-specific factors during development [35], [36], which can be activated in response to acute anemia or hypoxia to functionally regulate erythropoiesis [13], [37]. Increased EPO by miR-17 may be a feasible explanation for the inclination of 17/OE cells to support the growth of mature erythroid cells. Although the transcript of KL was not significantly altered, further experiments are still needed to explore whether the up-regulated KL, a hematopoietic supporting growth factor [38], is responsible for the promoted hematopoietic supporting ability of 17/OE cells. However, the mRNA levels of SDF-1, which is expressed by the niche cellular components and is important for the migration of HSCs [39], was not altered in 17/OE cells with or without interaction with CB CD34+ cells. Overall, by changing the expression of hematopoietic supporting factors, ectopic expression of miR-17 in osteoblastic cells may create a suitable niche for HSC expansion, in particular the specific expansion of the erythroid lineage.
Of great interest, our experiments on ex vivo expansion, LTC-IC and SRC in vivo assay revealed that selective expansion of the erythroid lineage of CB CD34+ cells through osteoblastic miR-17 was abrogated by HIF-1α knock down ( Figure 5 ). These data suggested that the function of osteoblastic miR-17 on HSCs and HPCs was through, at least in part, the HIF-1α signaling pathway. Although the relationship between miR-17 and HIF-1α is dependent on the environment and cellular context, our data showed a functional link between HIF-1α and miR-17, which has also been demonstrated by other research groups [30], [40].
In summary, our data suggested the potential contribution of miR-17 in bone marrow stem cell niches and an osteoblastic-miR-17-HIF-1α-HSC crosstalk in hematopoietic development. Our study demonstrated that, in addition to regulating the cellular constituents of HSCs and HPCs, miR-17 may also participate in the regulation of hematopoietic microenvironment and be involved in intercellular communications between HSCs and their niche cells. Defining the role of miR-17 in osteoblasts on hematopoiesis raises the possibility that miR-17 may play a key part in regulating the hematopoietic niche. Further characterization of miR-17 and other miRNAs on this field will be particularly important, not only for a better understanding of the detailed mechanisms behind HSC self-renewal and lineage commitment, but also for developing novel and efficient molecular targets to prevent and treat hematopoietic disorders.
Acknowledgments
We thank Dr. Sai-Feng Wang from the Institute of Systems Biomedicine, Peking University Health Science Center, Beijing, China for providing the retroviral vector RNAi-pSIREN-RetroQ. We also thank our colleagues in the Department of Medical Genetics, Peking University for their support during the project.
Funding Statement
This project was supported by the National Natural Science Foundation of China (31201019), Beijing Natural Science Foundation (5122022) and Leading Academic Discipline Project of Beijing Education Bureau (8910026970103004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones DL, Wagers AJ (2008) No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol 9: 11–21. [DOI] [PubMed] [Google Scholar]
- 2. Wilson A, Trumpp A (2006) Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol 6: 93–106. [DOI] [PubMed] [Google Scholar]
- 3. Calvi LM, Adams GB, Weber KWJM, Olson DP, Knight MC, et al. (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841–6. [DOI] [PubMed] [Google Scholar]
- 4. Zhang J, Niu C, Ye L, Huang H, He X, et al. (2003) Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425: 836–41. [DOI] [PubMed] [Google Scholar]
- 5. Arai F, Hirao A, Ohmujra M, Sato H, Matsuoka S, et al. (2004) Tie/Angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118: 149–61. [DOI] [PubMed] [Google Scholar]
- 6. Lekli I, Gurusamy N, Ray D, Tosaki A, Das DK (2009) Redox regulation of stem cell mobilization. Can. J. Physiol. Pharmacol 87: 989–995. [DOI] [PubMed] [Google Scholar]
- 7. Chow DC, Wenning LA, Miller WM, Papoutsakis ET (2001) Modeling pO(2) distributions in the bone marrow hematopoietic compartment. II. Modified Kroghian models. Biophys J 81: 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jang YY, Sharkis SJ (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low oxygenic niche. Blood 110: 3056–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hermitte F, Brunet de la Grange P, Belloc F, Praloran V, Ivanovic Z (2006) Very low O2 concentration (0.1%) favors G0 return of dividing CD34+ cells. Stem Cells 24: 65–73. [DOI] [PubMed] [Google Scholar]
- 10. Roy S, Tripathy M, Mathur N, Jain A, Mukhopadhyay A (2012) Hypoxia improves expansion potential of human cord blood-derived hematopoietic stem cells and marrow repopulation efficiency. Eur J Haematol 88(5): 396–405. [DOI] [PubMed] [Google Scholar]
- 11. Ivanovic Z, Dello Sbarba P, Trimoreau F, Faucher JL, Praloran V (2000) Primitive human HPCs are better maintained and expanded in vitro at 1 percent oxygen than at 20 percent. Transfusion 40: 1482–1488. [DOI] [PubMed] [Google Scholar]
- 12. Danet GH, Pan Y, Luongo JL, Bonnet DA, Simon MC (2003) Expansion of human SCID-repopulating cells under hypoxic conditions. J Clin Invest 112: 126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rankin EB, Wu C, Khatri R, Wilson TL, Andersen R, et al. (2012) The HIF signaling pathway in osteoblasts directly modulates erythropoiesis through the productionof EPO. Cell 149: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartel DP (2004) MicroRNAs: genomics, biogenesis,mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 15. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. ProcNatlAcadSci USA 101: 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merkerova M, Vasikova A, Belickova M, Bruchova H (2009) MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev 19: 17–26. [DOI] [PubMed] [Google Scholar]
- 17. Garzon R, Pichiorri F, Palumbo T, Luliano R, Cimmino A, et al. (2006) MicroRNA fingerprints during human megakaryocytopoiesis. ProcNatlAcadSci USA 103: 5078–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neilson JR, Zheng GX, Burge CB, Sharp PA (2007) Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev 21: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bissels U, Wild S, Tomiuk S, Hafner M, Scheel H, et al. (2011) Combined Characterization of microRNA and mRNA Profiles Delineates Early Differentiation Pathways of CD133(+) and CD34(+) Hematopoietic Stem and Progenitor Cells. Stem Cells 29: 847–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, et al. (2010) MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. ProcNatlAcadSci USA 107: 21505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, et al. (2010) MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. ProcNatlAcadSci USA 107: 14235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tagawa H, Seto M (2005) A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia 19: 2013–2016. [DOI] [PubMed] [Google Scholar]
- 23.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, et al.. (2011) MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood 118: 916–25. Epub 2011 May 31. [DOI] [PMC free article] [PubMed]
- 24. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, et al. (2007) MicroRNAs 17–5p–20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 9: 775–787. [DOI] [PubMed] [Google Scholar]
- 25. Pillai MM, Yang X, Balakrishnan I, Bemis L, Torok-Storb B (2010) MiR-886–3pdown regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS One 5: e14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Solingen C, de Boer HC, Bijkerk R, Monge M, van Oeveren-Rietdijk AM, et al. (2011) MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(+)/Lin(−) progenitor cells inischaemia. Cardiovasc Res 9: 449–55. [DOI] [PubMed] [Google Scholar]
- 27. Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, et al. (2009) Dilivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2: ra81. [DOI] [PubMed] [Google Scholar]
- 28. Yang YX, Miao ZC, Zhang HJ, Wang Y, Gao JX, et al. (2007) Establishment and characterization of a human telomerase catalytic subunit-transduced fetal bonemarrow-derived osteoblastic cell line. Differentiation 75: 24–34. [DOI] [PubMed] [Google Scholar]
- 29. Wagegg M, Gaber T, Lohanatha FL, Hahne M, Strehl C, et al. (2012) Hypoxia promotes osteogenesis but suppresses adipogenesis of human mesenchymal stromalcells in a hypoxia-inducible factor-1 dependent manner. PLoS One 7: e46483 doi: 10.1371/journal.pone.0046483. Epub 2012 Sep 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, et al. (2008) Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res 68: 5540–5. [DOI] [PubMed] [Google Scholar]
- 31. Burdick JA, Vunjak-Novakovic G (2009) Engineered microenvironments for controlled stem cell differentiation. Tissue Eng Part A 15: 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamashita A, Nishikawa S, Rancourt DE (2010) Microenvironment modulates osteogenic cell lineage commitment in differentiated embryonic stem cells. PLoS One 5: e9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu Y, Cai K, Luo Z, Zhang Y, Li L, et al. (2012) Regulation of the differentiation of mesenchymal stem cells in vitro and osteogenesis in vivo by microenvironmental modification of titanium alloy surfaces. Biomaterials 33: 3515–3528. [DOI] [PubMed] [Google Scholar]
- 34. Liu W, Liu Y, Guo T, Hu C, Luo H, et al. (2013) TCF3, a novel positive regulator of osteogenesis, plays a crucial role in miR-17 modulating the diverse effect of canonical Wnt signaling in different microenvironments. Cell Death Dis 4: e539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kapitsinou PP, Liu Q, Unger TL, Rha J, Davidoff O, et al. (2010) Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood 116: 3039–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haase VH (2010) Hypoxic regulation of erythropoiesis and iron metabolism. Am. J. Physiol. Renal Physiol. 299: F1–F13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weidemann A, Kerdiles YM, Knaup KX, Rafie CA, Boutin AT, et al. (2009) The glial cell response is an essential component of hypoxia-induced erythropoiesis in mice. J. Clin. Invest 119: 3373–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takagi S, Saito Y, Hijikata A, Tanaka S, Watanabe T, et al. (2012) Membrane-bound human SCF/KL promotes in vivo human hematopoietic engraftment and myeloid differentiation. Blood 119: 2768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ratajczak MZ, Serwin K, Schneider G (2013) Innate immunity derived factors as external modulators of the CXCL12-CXCR4 axis and their role instem cell homing and mobilization. Theranostica 3: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He M, Wang QY, Yin QQ, Tang J, Lu Y, et al.. (2012) HIF-1α downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ doi:10.1038/cdd.2012.130. [DOI] [PMC free article] [PubMed]