Abstract
Clostridium cellulolyticum, a mesophilic anaerobic bacterium, produces highly active enzymatic complexes called cellulosomes. This strain was already shown to bind to cellulose, however the molecular mechanism(s) involved is not known. In this context we focused on the gene named hycP, encoding a 250-kDa protein of unknown function, containing a Family-3 Carbohydrate Binding Module (CBM3) along with 23 hyaline repeat modules (HYR modules). In the microbial kingdom the gene hycP is only found in C. cellulolyticum and the very close strain recently sequenced Clostridium sp BNL1100. Its presence in C. cellulolyticum guided us to analyze its function and its putative role in adhesion of the cells to cellulose. The CBM3 of HycP was shown to bind to crystalline cellulose and was assigned to the CBM3b subfamily. No hydrolytic activity on cellulose was found with a mini-protein displaying representative domains of HycP. A C. cellulolyticum inactivated hycP mutant strain was constructed, and we found that HycP is neither involved in binding of the cells to cellulose nor that the protein has an obvious role in cell growth on cellulose. We also characterized the role of the cellulosome scaffolding protein CipC in adhesion of C. cellulolyticum to cellulose, since cellulosome scaffolding protein has been proposed to mediate binding of other cellulolytic bacteria to cellulose. A second mutant was constructed, where cipC was inactivated. We unexpectedly found that CipC is only partly involved in binding of C. cellulolyticum to cellulose. Other mechanisms for cellulose adhesion may therefore exist in C. cellulolyticum. In addition, no cellulosomal protuberances were observed at the cellular surface of C. cellulolyticum, what is in contrast to reports from several other cellulosomes producing strains. These findings may suggest that C. cellulolyticum has no dedicated molecular mechanism to aggregate the cellulosomes at the cellular surface.
Introduction
Cellulose, a major polysaccharide on earth, is a linear polymer of glucose organized in a regular crystalline arrangement and forming insoluble linear microfibrils. In plant cell walls, these fibrils are surrounded by a complex matrix made up of other polysaccharides as hemicellulose or pectin [1], [2]. Several cellulolytic microorganisms carry out efficient deconstruction of crystalline cellulose and other polysaccharides of the plant cell wall. Among them, Clostridium cellulolyticum, a mesophilic anaerobic bacterium, produces highly active extracellular enzymatic complexes called cellulosomes together with free enzymes. In this cellulolytic strain, cellulosomes are made up of a non enzymatic scaffolding protein called CipC, composed of a CBM3, two hydrophilic modules (×2) whose function remains unknown and eight type I cohesins [3]. The cohesins bind with high affinity to the dockerin modules typically borne by the cellulosomal enzymes, thus leading to cellulosomes assembly [4]. Cellulosomal or free plant cell wall degrading enzymes display catalytic modules classified into three distinct groups in the CAZY database: the glycoside hydrolase, the pectate lyase, and the carbohydrate esterase group (http://www.cazy.org/ [5]).
Cellulolytic bacteria were early reported to bind to cellulose [6], [7], [8]. The adherence to their substrate is expected to bring them several competitive advantages: (i) the enzymes are secreted closer to the substrate, avoiding their diffusion in the extracellular medium, (ii) the hydrolysis products are released in the vicinity of the bacterium and can be directly consumed, thus limiting their diffusion and decreasing the feedback inhibition of the hydrolytic enzymes [8], [9]. Recently the cellulolytic bacterium Clostridium thermocellum was shown to form biofilm on cellulose [10], [11]. Cellulose was found to be significantly degraded in the biofilm area, compared to the areas without biofilm, highlighting the importance of cell adherence for cellulolytic activity.
In C. thermocellum, cellulosomes were shown to mediate cell binding to cellulose through the CBM3 borne by the cellulosomal scaffolding protein, CipA [6], [12], [13]. CipA contains a type II dockerin which interacts with type II cohesins hosted by 3 other non catalytic proteins OlpB, Orf2p, and SbdA [14], [15], [16]. These latter proteins are bound to cell surface through their Surface Layer Homology (SLH) modules. At the cell surface, cellulosomes form protuberances which can be observed using scanning electron microscopy [13]. These ultra-structures are missing at the surface of the non adherent C. thermocellum AD2 strain, which is no longer able to attach cellulosomes to the cell surface [6], [17]. In other cellulolytic species, such as Clostridium cellulovorans, Acetivibrio cellulolyticus, and Bacteroides cellulosolvens, similar ultrastructures were also observed [13], [18]. The cellulosomes were therefore hypothesized to be implicated in cellulose adherence process in these strains [12]. Molecular evidence supports this hypothesis: in the genome of A. cellulolyticus, and B. cellulosolvens genes encoding cell surface proteins were discovered which may mediate anchorage of the cellulosome scaffolding protein [12]. The anchorage would be done through type II cohesin/dockerin interactions as it was observed for C. thermocellum. C. cellulovorans lacks type II dockerin in the scaffolding protein CbpA. In this strain, cell binding to cellulose may be mediated by the cellulosomal enzyme Eng5E [19]. This protein may anchor cellulosomes to the cell surface thanks to the presence of a C-terminal type I dockerin and N-terminal Surface Layer Homology domains (SLH). In addition, hydrophilic modules of the scaffolding protein CbpA were shown to bind to C. cellulovorans cell wall fractions and were proposed to help to maintain the cellulosomes at the cell surface [20]. Thus in these species, the scaffolding protein of the cellulosomes seems to be directly or indirectly involved in cell adhesion to cellulose.
Clostridium cellulolyticum was shown to bind to cellulose [7], but in contrast to the cellulolytic species described above, the factors involved in this process have not yet been elucidated. In addition to the scaffolding protein CipC, the genome of C. cellulolyticum encodes 8 other putative CBM3-containing proteins [21]. Among them, seven were predicted to contain Family-9 glycoside hydrolase catalytic modules and are expected to be cellulases. They may be incorporated within cellulosomes since all of them bear a dockerin module. The eighth CBM3-containing putative protein is the product of the gene located at the locus Ccel_1491. Annotation of this gene in NCBI database indicates that the corresponding protein contains a CBM3a, similar to the CBM3 of CipC which is known to bind strongly to crystalline cellulose [22]. Moreover, it is a very large protein of 250 kDa, of unknown function, and for which computational analysis failed to predict any catalytic-, dockerin-, or cohesin-module [21]. The presence of such a protein in C. cellulolyticum prompted us to analyze its function and its putative role in cell adhesion to cellulose as well as that of the scaffolding protein CipC, since cellulosomal scaffolding proteins were proposed to mediate cell adhesion in several other cellulolytic bacteria.
Results
Bioinformatic analysis of the protein encoded by the gene at the locus Ccel_1491
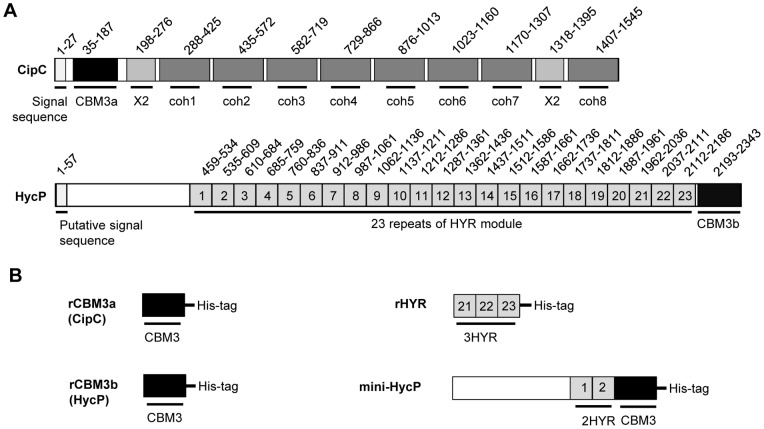
The structural organizations deduced from bioinformatic analysis of the product of the gene at the locus Ccel_1491 and of CipC are presented in figure 1. CipC is a well described protein of 160 kDa which contains eight cohesins, and a CBM3 at the N-terminus. It is a secreted protein and its precursor harbors a typical gram positive signal peptide. In contrast, the newly identified product of the gene present at the locus Ccel_1491 is predicted either as a secreted protein with a putative 58 amino acids long signal peptide or as a membrane protein with a transmembrane helix located between amino acids 34 and 51. This 250-kDa protein exhibits 23 copies of a hyaline repeat module (HYR) and a CBM3 at the C-terminus. For convenience, the product of the gene at locus Ccel_1491 will be named HycP for HYR modules and CBM3 containing protein. At the N-terminus of HycP, a region of about 250 amino-acids does not match with any other classified domain except with a bacterial Ig-like domain family 3 (BID_1 in SMART database), with a very weak E-value. This domain is usually found in bacterial cell surface proteins.
Figure 1. Structural organization of the proteins used in this study.
A. Structural organization of CipC and HycP from Clostridium cellulolyticum. Numbers above the protein correspond to the starting and final amino acids of each module in the full length molecule. The numbers in HycP indicate the order of the HYR modules starting from the N-terminus. HYR modules were identified using PFAM, SMART and by manual search (see alignments in data S1). B. Modular organization of recombinant proteins produced in the present study. The numbers of the HYR modules in recombinant proteins correspond to the same HYR modules in the wild-type protein.
HycP is composed of 23 copies of HYR modules which account for nearly 75% of its sequence. Each HYR module is about 75 amino-acids long; an alignment of these modules is presented in data S1. The HYR modules were initially discovered in the hyalin protein found in the echinoderm extra-embryonic matrix and are responsible for the recognition of this protein by its cell surface receptor [23]. The hyalin protein contains exclusively this type of repeated modules. HYR modules belong to the immunoglobuline like fold, like the Fn3 domain [24]. They are found in eukaryotes as well as in prokaryotes where they are detected in some surface proteins, associated with Family-18 glycoside hydrolase modules (chitinase) or as part of hypothetical proteins. Their function in these proteins is unknown [24]. For HycP no obvious function can be deduced from a structural organization analysis neither did the genetic environment give further clues, since the gene encoding HycP is framed upstream and downstream by genes of unknown function.
Classification and characterization of HycP CBM3
HycP contains a CBM3 found at the N-terminus of the protein. The CBM3 family is sub divided in several subfamilies. The CBM3 found in CipC belongs to the CBM3a subfamily as those in other scaffolding proteins [25], [26]. A sequence search in the NCBI data bank for microbial proteins sharing similarity with the CBM3 of HycP provided a list of proteins containing CBM3a or CBM3b. The two highest scores were obtained with the CBM3b of the exoglucanase CelY from Clostridium stercorarium (accession number gi|1708082) and the CBM3a of the scaffolding protein CipA from C. thermocellum (accession number gi|2554721). In order to further analyze the sequence of HycP CBM3, an alignment with several known CBM3a and 3b sequences was performed (figure 2). As it was formerly shown, the presence or absence of a short 4′ β-strand allows discrimination between CBM3a and CBM3b, respectively [26]. This strand holds a tyrosine which is one of the conserved amino-acids important for the binding of the CBM3a to the planar crystalline cellulose [27], [28], [29]. In contrast to the CBM3b, HycP CBM3 contains an additional stretch of about 8 amino-acids. This stretch lacks the conserved tyrosine found in the 4′ β-strand of the CBM3a, and does not contain any aromatic residues. Remarkably a conserved histidine found in all CBM3a in the 4 β-strand, is replaced by an aromatic amino acid in the HycP CBM3, as observed in the case of the CBM3b subfamily (fig 2). These observations lead us to propose the classification of the CBM3 from HycP in the CBM3b rather than in the CBM3a subfamily.
Figure 2. Amino-acid sequence alignment of CBM3a and CBM3b from various CBM3a or CBM3b containing proteins.
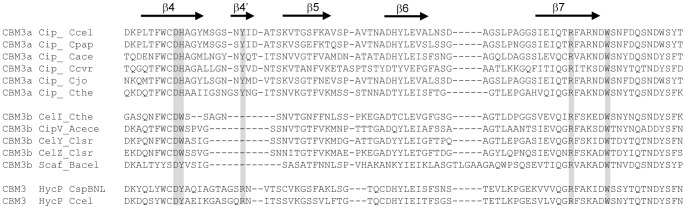
Alignment has been performed using ClustalW2. It is focused on residues considered to participate in planar interaction with cellulose, highlighted in grey box. Regions of secondary structure are marked with an arrow and labeled as in the structure of the CBM3a of the scaffolding protein CipA from Clostridium thermocellum (Tormo 1996). CBM amino-acid sequence aligned (accession numbers codes in parentheses) are: Cip_Ccel(YP_002505087) and HycP_Ccel (YP_002505824) from Clostridium cellulolyticum; HycP_CspBNL (YP_005147316) from Clostridium sp. BNL1100; Cip_Cpap (ZP_08194681) from Clostridium papyrosolvens; Cip_Cace (NP_347546) from Clostridium acetobutylicum; Cip_Ccvr (ZP_07630535) from Clostridium cellulovorans; Cip_Cjo (BAA32429) from Clostridium josui; Cip_Cthe (ZP_14248391) and CelI_Cthe (AAA20892) from Clostridium thermocellum; CipV_Acece (AAF06064) from Acetovibrio cellulolyticus; CelY (YP_007373484) and CelZ (CAA39010) from Clostridium stercorarium; Scaf_Bacel (AAG01230) from Bacteroides cellulosolvens.
Both CBM3a and 3b are known to bind to crystalline cellulose. We analyzed the cellulose binding capacity of the newly discovered CBM3b and compared it with that of the well known CBM3a from the scaffoldin CipC. Recombinant CBMs, referred to as rCBM3a and rCBM3b for respective proteins CipC and HycP (figure 1B), were fused to a polyhistidine tag at the C-terminus, produced in Escherichia coli, purified and used for binding assays. Binding capacities were first investigated on crystalline cellulose and straw. rCBM3a was shown to bind strongly to both crystalline cellulose and straw, whereas rCBM3b seems to have higher affinity for cellulose than for straw (figure 3). We measured the dissociation constant for both CBMs on different cellulosic substrates (Table 1). Both modules bind to phosphoric acid swollen cellulose (PASC) with the same affinity, but the rCBM3b displays approximately 10 times lower affinity on all tested crystalline cellulose (Sigmacell, Avicel and BMCC) compared to the rCBM3a. Nevertheless our results indicate that the CBM3b is functional and is able to bind to crystalline cellulose, but with lower KD values ranging from 10−5 to 10−6 M.
Figure 3. Interactions of rCBM3a and rCBM3b with straw and crystalline cellulose.
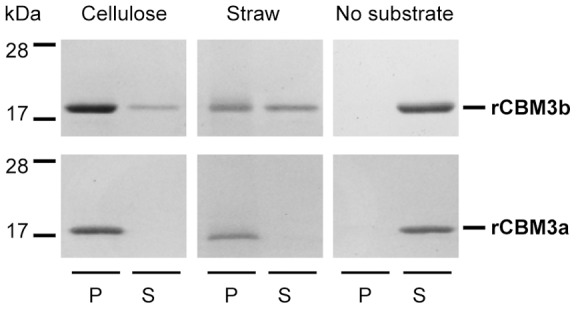
Recombinant proteins were mixed with substrates during one hour. After centrifugation the bound proteins found in the pellet (P), and the unbound proteins present in the supernatant (S), were analyzed by SDS-PAGE.
Table 1. Dissociation constants of rCBM3a and rCBM3b to cellulosic substrates.
| KD (M) | ||
| Substrate | rCBM3a (CipC) | rCBM3b (HycP) |
| BMCC | 2.49E−8 | 9.5E−6 |
| Avicel | 8.017E−7 | 1.16E−5 |
| Sigmacell | 4.33E−7 | 8.83E−6 |
| PAS-cellulose | 8.51E−7 | 7.47E−7 |
HycP enzymatic assays
The HycP protein contains 23 HYR modules along with a functional CBM3b. As CBM3b-containing proteins are often cellulases, we explored if HycP has a catalytic activity towards various cellulosic substrates. In order to facilitate these tests we produced a shortened form, called mini-HycP in E. coli which is composed of the first 409 amino-acids found after the predicted signal sequence cleavage site (the part of the protein which does not match with any conserved domains), the two first HYR modules (which were not reported to display catalytic activity) and the CBM3b (figure 1B). Mini-HycP also contains a C-terminal His-tag to facilitate its purification. Activity assays were performed on straw, crystalline cellulose (Avicel), phosphoric acid swollen cellulose, and Carboxy-Methyl Cellulose (CMC) by measuring the quantity of reducing sugars released. Under our experimental conditions (37°C, pH 6), we were not able to detect any activity of the mini-HycP on any of these substrates (data not shown).
Construction of hycP and cipC C. cellulolyticum mutant strains
Our results showed that HycP has a functional CBM3b but no enzymatic activity toward cellulosic substrates. We therefore explored the possibility that the function of HycP is to induce the binding of C. cellulolyticum to cellulose, since the protein contains a cellulose binding module, as well as numerous HYR modules which are found in many cell surface proteins [24]. Furthermore bioinformatic analyses of the sequence predicted a putative transmembrane helix at the N-terminus. In order to verify this hypothesis, we constructed a mutant strain from C. cellulolyticum in which the hycP gene was inactivated using the ClosTron technique developed by Heap and co-workers [30]. As scaffolding proteins are reported to be involved in cell adhesion to cellulose in several cellulosome-producing bacteria, we decided to inactivate the cipC gene as well, using the same technique. The intron was designed to target the very beginning of cipC in the DNA region encoding the CBM3a module, in order to prevent any production of a truncated CipC form of the protein that would still display the cellulose binding module (figure 1A). Two mutant strains were thus constructed, MTLcipC and MTLhycP.
Analysis of the genomic DNA of both strains by PCR and southern blot confirmed the genetic localization of the mutations and the presence of only one insertion in the chromosome (data not shown). In the strain MTLcipC, the pMTL007cipC vector was cured but not in the strain MTLhycP where the pMTL007hycP persisted in all the tested clones obtained from two transformation events, even after many replicates. In order to detect HycP in the different strains we used rabbit antibodies raised against rHyr, a purified recombinant protein produced in E coli and containing the three C-terminal HYR modules fused to a C-terminal His-tag (Fig 1B). Analysis of both, whole cells and cellobiose culture supernatant of each mutant, indicated that the proteins of interest were absent, while in wild-type cells bands corresponding to proteins of about 280 kDa and 160 kDa were detected using anti-HYR or anti-CipC CBM3a antisera, respectively (fig 4 A and B lanes 1 and 2). In addition, we observed that in the wild-type strain, HycP is more abundant in the supernatant than in the cell fraction, thus indicating that this protein is mainly secreted. The same observation was made concerning CipC what in this case is consistent with its typical gram positive signal sequence. The presence of proteins HycP and CipC in the cellular fraction of the wild-type strain may either be due to their production in the cell prior to secretion, or to a putative association with the cell wall.
Figure 4. Detection of HycP CipC and Cel48F in different Clostridium cellulolyticum strains.
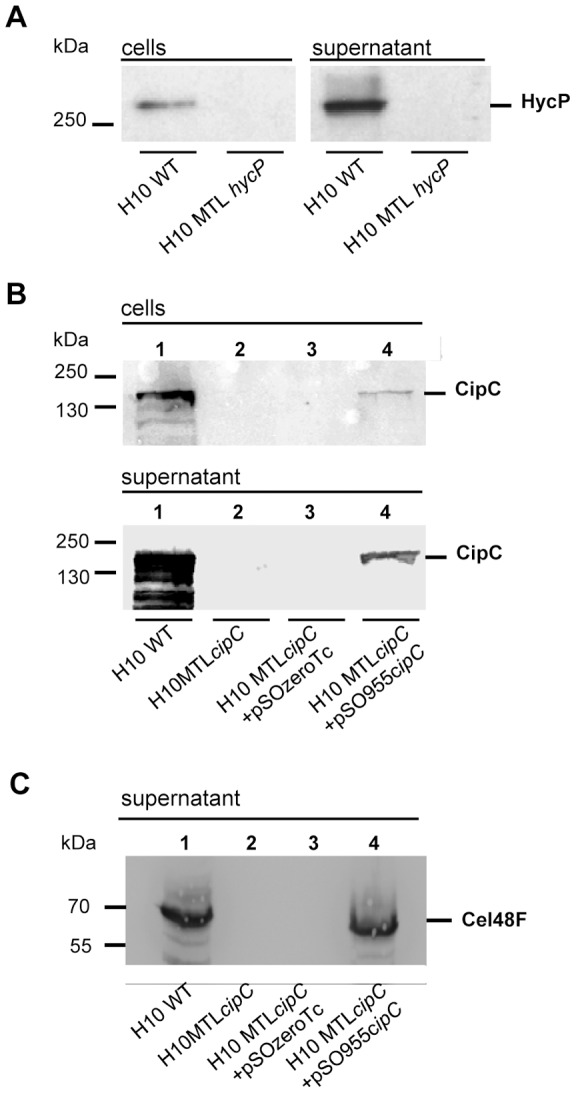
Different C. cellulolyticum strains were studied: wild-type strain, MTLhycP, MTLcipC, MTLcipC(pSOS955cipC) and MTLcipC(pSOSzero-Tc) strains. Aliquot was taken from a culture of these strains at the exponential growth phase on cellobiose substrate, and centrifuged to separate the cells and the supernatant. Cell fraction and 10% TCA precipitated supernatant fraction corresponding to the same culture volume were subjected to SDS-PAGE. After transfer onto nitrocellulose membranes, membranes were probed with antibodies directed against HYR modules from HycP (Panel A), or CipC (Panel B), or Cel48F (Panel C).
Cell binding to cellulose and growth analysis of the MTLhycP mutant strain
In order to test the MTLhycP mutant strain for its ability to bind to cellulose we used a spectrophotometric adhesion test. It indicated that 95% of wild-type C. cellulolyticum cells cultured in cellobiose, bind to filter paper cellulose (figure 5A). In presence of BSA, which reduces the unspecific binding, still 80% of C. cellulolyticum cells bind to cellulose whereas only 10% were found to bind to nitrocellulose, which is a chemically modified cellulose. For comparison Clostridium perfringens, a human pathogen unable to grow on cellulosic substrates, showed only 20 % of adherent cells on filter paper, thus confirming the specificity of the test. Subsequent adhesion tests were all performed in presence of BSA in order to study specific binding. Observations by scanning electron microscopy (SEM) of the filter paper after cell adhesion confirmed the presence of bound cells (data not shown).
Figure 5. Cell adherence to cellulosic substrates.
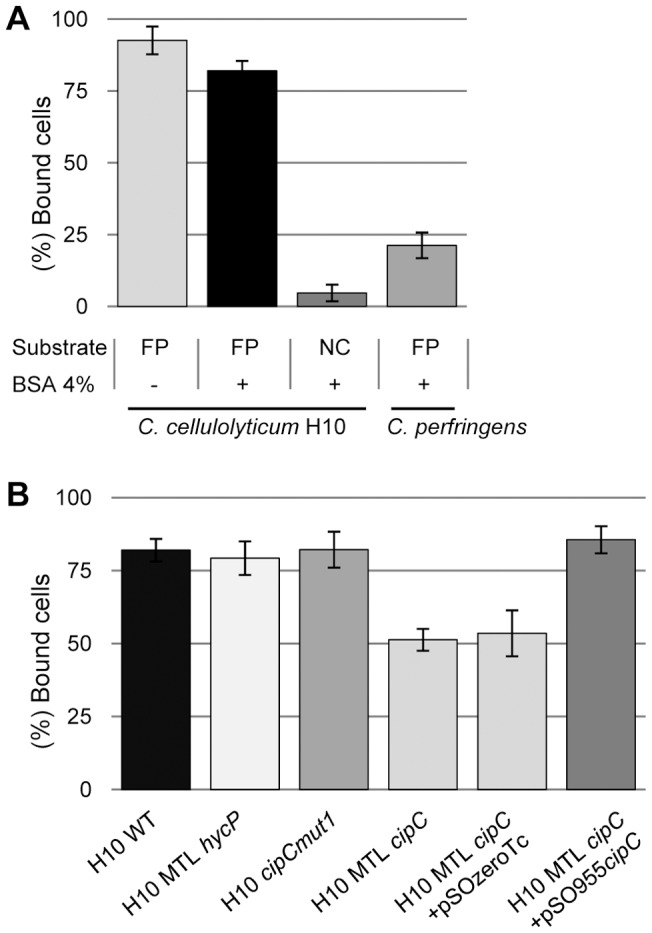
Cells cultured on cellobiose were incubated one hour in anaerobic conditions with a strip of insoluble substrate. Binding percentage is calculated from the level of unbound cells measured in the supernatant by spectrophotometry (optical density at 450 nm) compared to the optical density value of an assay where no insoluble substrate was added. (A) Clostridium cellulolyticum is incubated on filter paper (FP) or on nitrocellulose (NC) strips with or without BSA saturation and compared to the binding level of Clostridium perfringens to BSA saturated filter paper. (B) Cellulose binding capacity of C. cellulolyticum wild-type strain, MTLhycP, MTLcipC, MTLcipC(pSOS955cipC), MTLcipC(pSOSzero-Tc), and cipCmut1 mutant strains. Experiments were performed in triplicates, on at least three independent experiments and two isolated clones for MTLhycP, MTLcipC, MTLcipC(pSOS955cipC) and MTLcipC(pSOSzero-Tc).
The MTLhycP mutant strain was assayed for its binding capacity to cellulose. The results indicated that MTLhycP mutant cells bind to cellulose at the same level as the wild-type strain, thereby demonstrating that HycP has no obvious role in cell binding to cellulose (fig 5B). To further analyze the role of HycP, we measured the growth of the MTLhycP mutant strain in various conditions and compared them with the wild-type strain. On cellobiose rich medium, generation time of MTLhycP mutant was 25% lower than that of wild-type strain, indicating that when HycP is not produced and not secreted, the fitness of C. cellulolyticum on soluble sugars is enhanced. When using insoluble cellulose as the substrate, no significant difference was observed whatever medium (rich or minimal medium) or crystalline cellulose (Avicel or Sigmacell) were used (fig 6).
Figure 6. Growth of Clostridium cellulolyticum wild-type and MTLhycP strains on cellulose.
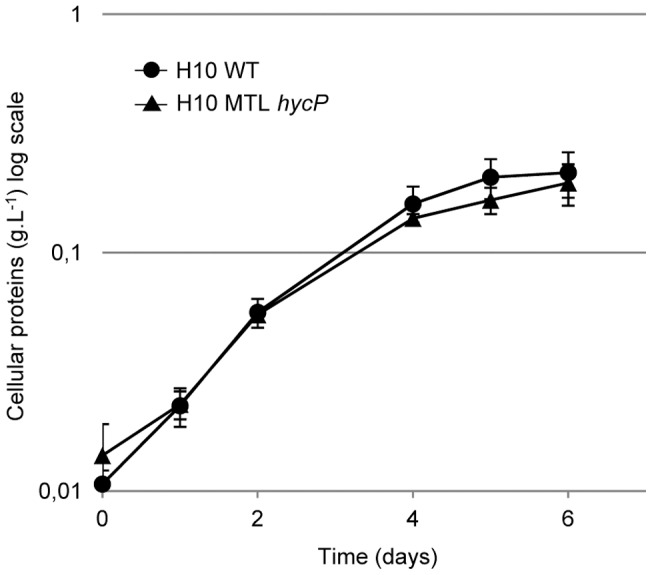
Both strains were grown on rich medium containing 5 g.L−1 Sigmacell. Growth was monitored by measuring total protein content. Experiment was performed in duplicates. Growth performed in other condition as minimal medium containing Sigmacell or Avicel did not show any differences between both strains.
Cell binding to cellulose and analysis of the MTLcipC mutant strain
CipC is the first gene of an operon containing 12 genes which encode mainly glycosyl hydrolases (cel48F, cel8C, cel9G, cel9E, orfX, cel9H, cel9J, man5K, cel9M, rgl11Y, cel5N), directly involved in plant cell wall degradation [31], [32]. We used two different cipC mutant strains: the new MTLcipC mutant constructed in the present study, and a spontaneous mutant cipCmut1 which was formerly characterized [31]. This mutant strain contains an insertion sequence at the 3′ extremity of the cipC gene leading to the production of a truncated CipC. The presence of the insertion sequence in the cipC gene induced a polar effect which caused the abolishment of the expression of all other genes localized in the operon downstream of cipC [31]. We analysed the binding capacities of both strains. We observed that cipCmut1 binds to cellulose at the same level as the wild-type strain. As this strain produces none of the cellulases encoded by the operon cip-cel, our observation might indicate that these proteins do not participate in cell binding. In the MTLcipC mutant strains, only 50% of the cells bound to cellulose. We observed that in this strain, already the second gene downstream cipC, namely cel48F, was not expressed, suggesting the occurrence of the same polar effect as in the cipCmut1 strain (fig 4C, lane 1 and 2). The difference between MTLcipC and cipCmut1 strains is therefore that the MTLcipC mutant strain does not produce any CipC, whereas the cipCmut1 mutant strain still produces a small amount of a truncated form of CipC with the N-terminal CBM3a [31]. This may explain their difference in cellulose adherence and suggests the involvement of CipC, through its CBM3a, in cell binding to cellulose.
In order to validate the involvement of CipC in the phenotype of the mutant constructed in the present study, we complemented MTLcipC strain using a replicative vector (pSOS955cipC). It allows the expression of the cipC gene under control of a constitutive promoter which has been shown to be functional in C. cellulolyticum [33], [34], [35]. A control strain MTLcipC(pSOSzero-Tc) containing the same vector but without the expression cassette, was also constructed. Both strains were analyzed for their CipC content by western blot, along with the wild-type strain. As expected, the complemented strain MTLcipC(pSOS955cipC) produced CipC. However the production of Cel48F was also detected, suggesting that a homologous recombination event occurred between the cipC copy present in the chromosome and the one of the vector, restoring the expression of the operon (fig 4B and C, lanes 3 and 4). Recombination events occurred in all clones obtained after two independent transformation events. Despite this observation, we measured the capacity of the complemented and the control strain to bind to cellulose. The MTLcipC(pSOS955cipC) complemented strain showed equal binding capacity as the wild-type strain, in contrast to the control strain MTLcipC(pSOSzero-Tc), indicating that the complementation restores the fully adherent phenotype (fig 5B). In summary these data strongly suggest that CipC participates in binding of C. cellulolyticum to cellulose while HycP does not.
Observation of the cell surface
Some cellulolytic bacteria are able to form cellulosomal protuberances at their surface which participate in cell adhesion on cellulose and are composed of cellulosomes. The presence of these protuberances has never been shown for C. cellulolyticum. As shown above, in C. cellulolyticum CipC is involved in its adhesion to cellulose. To detect if on the surface of C. cellulolyticum also protuberances are formed, we observed the cell surface of C. cellulolyticum during the growth on filter paper using SEM and compared it to the surface of C. thermocellum grown on the same substrate. Wild-type C. cellulolyticum cell surfaces were entirely smooth and lacked ultra-structural protuberances, in contrast to C. thermocellum whose cell surfaces displayed many protuberances (fig 7).
Figure 7. SEM observation of Clostridium cellulolyticum and Clostridium thermocellum grown on filter paper.
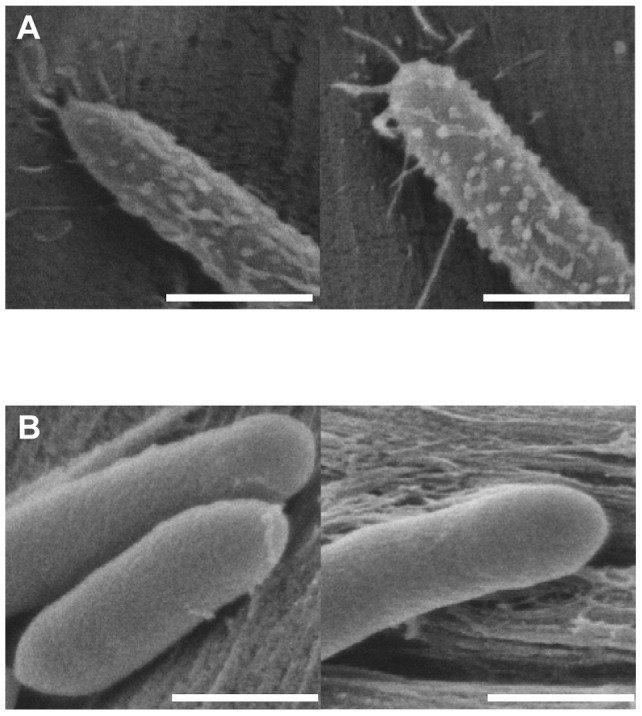
Clostridium thermocellum (A) and Clostridium cellulolyticum (B) were grown on filter paper. Pictures are representative of two independent experiments. Bar represents 500 nm.
Discussion
The CBM3 family contains several subtypes, among them the CBM3a and the CBM3b are known to bind strongly to crystalline cellulose [22], [26], [27], [36]. In the present study, we analyzed the properties of the new HycP CBM3 and compared them to those of the well known CipC CBM3a (Table 2). We showed that HycP contains a functional CBM3 that we classified within the CBM3b subtype according to its amino-acids sequence features. Both rCBM3a and rCBM3b were shown to bind to cellulose and straw as it was previously shown for other CBM3a and b [26]. Determined dissociation constants for rCBM3a with these substrates are consistent with previous data obtained from a recombinant miniCipC protein containing the first three modules of CipC, except for the interaction with PAS cellulose [22]. This difference may be explained by the use of different PAS cellulose preparations in both studies, or by the influence of the surrounding domains present in miniCipC, compared to rCBM3a. We showed that rCBM3b exhibits an overall reduced affinity for crystalline cellulose compared to rCBM3a. A plausible explanation for the difference between both CBM is that in CBM3b a stretch of about 3–4 amino-acids replaces the 4′ β-strand containing a conserved tyrosine in the CBM3a. This latter aromatic amino-acid is involved in one of the important stacking interactions between CBM3a and planar crystalline cellulose [27], [28], [29]. In CBM3b from HycP, no aromatic acid is present in this stretch which might be the cause of its weaker interaction for crystalline cellulose.
Table 2. Summary of the compared properties of CipC and HycP.
| Property | CipC | HycP |
| Modules found | CBM3a | CBM3b |
| Cohesins | HYR modules | |
| X2 | ||
| Preferred binding substrate | Crystalline celllulose | Amorphous cellulose |
| Hydrolytic activity | No | Not detected |
| Role in cell adherence to cellulose | Partial | No |
| Role in cell growth on cellulose | Yes | Not detected |
The presence of the large HycP protein composed of 23 repeats of the HYR module with unknown function, together with a functional CBM3b, raises the question of its function in C. cellulolyticum. HYR domains were initially discovered in eukaryotes but are also found in prokaryotes where they are inserted in some surface proteins, associated with some glycoside hydrolase modules (chitinases) or are part of hypothetical proteins [24]. In the bacterial kingdom, proteins containing multiple HYR modules like HycP are mostly found in marine or freshwater environment microorganisms, where their function is again unknown. The only protein predicted to contain a similar domain organization as HycP, i.e. many HYR domains associated to a CBM3, is found in Clostridium sp BNL1100. This strain is very close to C. cellulolyticum and was isolated from corn stover [37]. Both HYR domain containing proteins share 78% sequence identity. The search for other proteins containing HYR module(s) accompanied with a CBM in the NCBI data base resulted in three proteins: a 390-kDa protein from A. cellulolyticus CD2 (accession number ZP_09466191.1) which is predicted to be composed of a peptidase_C11 domain in the N-terminal part followed by two HYR modules with a CBM3 at the C-terminus, and two putative xylanases which both contain a CBM4_9 and a Family-10 glycoside hydrolase module (accession number gi|147830786, Clavibacter michiganensis subsp. michiganensis NCPPB 382; accession number YP_001360820, Kineococcus radiotolerans SRS30216). No HYR module containing protein is found in other described cellulolytic clostridia as C. thermocellum, C. cellulovorans or C. papyrosolvens. HycP is the only HYR modules containing protein in C. cellullolyticum. The function of the HYR domains in any of these bacterial proteins is unknown.
We explored the possible role of HycP in the light of the function of CBM3-containing proteins found in many other cellulolytic bacteria. CBM3b is usually associated with a Family-9 glycoside hydrolase module in cellulases [26]. Enzymatic assays performed on mini-HycP did not show any glycoside hydrolase activity towards cellulosic substrates, suggesting that the whole protein HycP is devoid of hydrolytic activity on cellulose. This is consistent with the low sequence similarity of the molecule with any known catalytic module. Another function reported for the CBM3-containing proteins is to sense the substrate as it was described in C. thermocellum. Membrane sensor proteins displaying a CBM3 and an anti-sigma factor domain were reported to trigger expression of genes related to the cellulolytic system in the presence of cellulose [38]–[39]. Direct involvement of HycP in the carbohydrate sensing process is however unlikely, since HycP is mainly secreted. In addition, neither the growth on cellulose nor the composition of the cellulosomes are altered when the protein is missing (data not shown). These data strongly suggest that HycP has no direct or indirect role in carbohydrate-sensing. The third function of CBM3-containing proteins is to mediate binding of the whole cell to cellulose. This interaction is established by cellulosomal scaffolding proteins which may contain CBM3a or CBM3b [6], [12], [19], [20]. Our results indicate that HycP is not involved in cell binding to cellulose since no differences were observed in cell adherence to cellulose or growth on cellulose between MTLhycP mutant and wild-type strains. Altogether these results suggest that the protein is not essential for cellulose hydrolysis, and its function remains unclear. The gene encoding HycP is only found in C. cellulolyticum and the related BNL1100 strain, suggesting a recent evolution of both strains, which may result of an adaptation to their specific environment. Similar to the Fn3 domain, HYR modules belong to the immunoglobuline-like fold [24]. It has been reported that Fn3 domains may modify the cellulose surface helping hydrolysis by the cellulases bearing this module [40]. It is possible that HYR modules displays this property, and the association of 23 HYR modules together with a CBM3b found in HycP may further enhance cellulose surface modification. This putative benefit is not observed when C. cellulolyticum is grown on cellulose, but we observed that the secretion of HycP seems to hamper fitness of C. cellulolyticum wild-type strain on soluble sugars. Indeed, the generation time of the mutant MTLhycP strain grown on cellobiose is reduced by 25% compared to wild-type. The persistence of the gene hycP through the evolution of C. cellulolyticum, suggests that this protein brings a benefit, putatively through an ancillary function, which may be useful in specific environments encountered by the bacterium and which has yet to be identified.
The putative involvement of CipC in cell adhesion of Clostridium cellulolyticum to cellulose was addressed in the present study. The scaffolding cellulosomal protein has been reported to be involved in cell adherence of several cellulosomes-producing bacteria [6], [12], [19], [20]. In C. thermocellum, the AD2 mutant failed to attach the cellulosomes at the cell surface and consequently to bind to cellulose [6], [17]. This mutant was found to lack the cellulosomal protuberances observed in the wild-type at the cell surface, highlighting the link between cellulosomal protuberances and adhesion of the cells to cellulose. In contrast in C. cellulolyticum, no protuberances were observed at the surface of C. cellulolyticum wild-type strain and CipC is only partly involved in cell binding to cellulose. It is worth noting that in other cellulolytic bacteria as B. cellulosolvens, A. cellulolyticus or C. cellulovorans, protuberances were also observed, and in all these strains, a molecular mechanism is proposed to tether the cellulosomes to the cell surface [12], [18], [19]. Analysis of the C. cellulolyticum genome failed to identify genes that encode any putative cellulosome cell surface anchoring proteins homologous to EngE from C. cellulovorans, or any predictable cellulosome cell surface anchoring adaptator protein. The lack of cell surface protuberances supports the possibility that, in C. cellulolyticum, no specific mechanism is devoted to the anchorage of cellulosomes to the cell surface. Since no protuberances are observed, the part of the adherence found to be due to CipC in our experiments may occur through other mechanisms. The hydrophilic modules (X2) of the scaffolding protein may exhibit some affinity for the peptidoglycan as suggested for C. cellulovorans [20]. Another possibility is that during the secretion process of the large CipC protein, the CBM3a module may be transiently exposed at the cell surface, allowing its participation in adherence of the cells to cellulose.
The lack of protuberances and the fact that CipC is only partly involved in the mechanism of adhesion to cellulose, suggests that other mechanisms may participate in cell binding to cellulose. Other mechanisms as bacterial glycocalyx or pili were found to be important for bacterial cell adhesion to cellulose [8]. Filamentous fibrillar appendages are reported to be important factors for adhesive properties of bacteria, biofilm formation and colonization. Two types of pilus are described in gram positives bacteria. The first is covalently linked to the peptidoglycan via the action of a sortase which recognizes a LPXTG motif in the protein [41]–[42]. And the second is the gram-negative-like type IV pilus [43], [44], [45]. It is not known whether C. cellulolyticum displays a surface glycocalyx, and no gene encoding sortase and any LPXTG motif containing protein could be found in the C. cellulolyticum genome sequence. But the C. cellulolyticum genome was reported to encode putative type IV pilus components [46]. As demonstrated in the case of Ruminococcus albus, this kind of pilus may also be involved in adhesion of C. cellulolyticum to cellulose [43], [44]. Other cell surface proteins containing SLH modules and CBM may also be involved in cell binding to cellulose, as it was suggested in Caldicellosiruptor saccharolyticus [47]. In C. cellulolyticum it was previously reported that genes encode proteins containing some SLH module(s), together with one or two CBM belonging to families, 9, 17, or 28, reported to bind to cellulosic substrates [21]. Proteins containing these CBMs may therefore participate in the adhesion mechanism(s) of C. cellulolyticum to cellulose. The role of type IV pilus, and other SLH and CBM containing proteins, in adherence of C. cellulolyticum to cellulose will be investigated in the future.
Materials and Methods
Bacterial strains, plasmids, and media
Escherichia coli DH5α (Life Technologies), E. coli BL21(DE3) (Life Technologies), and E. coli SG13009(pREP4) were grown at 37°C in Luria-Bertani medium supplemented with appropriate antibiotics (100 µg.ml−1 of ampicillin, 50 µg. ml−1 of kanamycin). C. cellulolyticum H10 ATCC 35319 [48] and mutants were grown anaerobically at 32°C on basal medium [49] supplemented with either 2 g.L−1 cellobiose (Sigma-Aldrich) or 5 g.L−1 cellulose, Sigmacell 20, (Sigma-Aldrich) or Avicel microcrystalline cellulose (PH101, Fluka, Buchs, Switzerland). When necessary, thiamphenicol (5 µg.ml−1), erythromycin (2.5 µg.ml−1), or tetracyclin (5 µg.ml−1) were added to the medium. Colonies of recombinant C. cellulolyticum strains carrying mutation in their chromosomes were isolated under the anaerobic atmosphere of a glove box (N2-H2, 95∶5 [vol/vol]), on solid basal medium supplemented with 2 g.L−1 of cellobiose, 15 g.L−1 of agar, and 2.5 µg of erythromycin, supplemented with tetracycline (5 µg.ml−1) when necessary for experiments using the complemented strains. Plates were incubated in anaerobic jars under 2×105 Pa of an N2-CO2 (80∶20 [vol/vol]) atmosphere.
Clostridium perfringens (strain CIP 60.61, Institut Pasteur, France) was anaerobically grown at 37°C in standard TGY medium.
Clostridium thermocellum DSM wild-type strain was grown anaerobically at 60°C in previously described medium [6].
Vectors and strains used in this study are reported in Table 3. The expression plasmid pET22b (Novagen) was used for the production in E. coli of the recombinant rCBM3b module, the recombinant protein rHyr, corresponding to the three last HYR modules of the HycP, and the recombinant rCBM3a of CipC in E. coli. pET28a was used for the production of the mini-HycP in E. coli. A derivative of pMTL007 was used for inactivation of hycP or cipC genes in C. cellulolyticum. pSOScipC, pSOS954, pSOSzero-Tc were used for complementation of the C. cellulolyticum mutant strain [23], [33], [35].
Table 3. Bacterial strains and vectors used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
| E. coli DH5a | F− endA1 hsdR17(rK− mK+) supE44 thi-1 λ gyrA96 relA1Δ(lacZYA argF) U169 (Φ80 lacZ ΔM15) recA | Roche Diagnostics |
| E. coli SG13009(pREP4) | F− his pyrD Δlon-100 rpsL (pREP4) | Qiagen |
| E. coli BL21(DE3) | F− ompT hsdS (rB− mB−) gal dcm (DE3) | Novagen |
| C. cellulolyticum H10 | Wild-type, ATCC35519 DSM 5812 | DSMZ |
| C. cellulolyticum MTLhycP | ATCC35319, hycP: intron, Ermr containing the vector pMTL007hycP | This study |
| C. cellulolyticum MTLcipC | ATCC35319 derivative, cipC: intron, Ermr | This study |
| C. thermocellum | Wild-type DSM | DSM [6] |
| C. perfringens | Strain CIP 60.61 | Institut Pasteur |
| pET22b+ | E. coli expression vector; Apr | Novagen |
| pET28a | E. coli expression vector; Kanr | Novagen |
| pETHyr | pET22b+ derivative carrying the735-bp NdeI-XhoI fragment encoding the last three HYR modules of HycP | This study |
| pETCBM3b | pET22b+ derivative carrying the 468-bp NdeI-XhoI fragment encoding the CBM3b module of HycP | This study |
| pETCBM3a | pET22b+ derivative carrying the NdeI-XhoI fragment encoding the CBM3a module of CipC | This study |
| pETMiniHycP | pET28a derivative carrying the 2135-bp NcoI-XhoI fragment encoding the MiniHycP | This study |
| pMTL007 | E. coli/Clostridium shuttle vector (ColE1, pCB102)Ll.ltrBintron (ermBtdRAM2) under the control of Pfac, ltrA; Cmr/Tmr | [30] |
| pMTL007cipC | pMTL007 derivative targeting cipC (locus Ccel_0728) | This study |
| pMTL007hycP | pMTL007 derivative targeting hycP (locus Ccel_1491) | This study |
| pSOSzero-Tc | E. coli/Clostridium shuttle vector (ColE1, pIM13); Apr,Tcr | [33] |
| pSOS954 | E. coli/Clostridium shuttle vector (ColE1, pIM13); Pthl carrying a -35 mutated box expression cassette from C. acetobutylicum, Apr,Ermr | [35] |
| pSOS955 | pSOSzero-Tc derivative carrying SalI-SalI expression cassette from C. acetobutylicum from pSOS954, Apr,Tcr | This study |
| pSOScipC | E. coli/Clostridium shuttle vector (ColE1, pIM13) carrying cipC gene under the control of the mutated Pthl, Apr,Ermr | [31] |
| pSOS955cipC | pSOS955 derivative carrying 4677-bp BamHI-SwaI fragment from pSOScipC, Apr,Tcr | This study |
Apr, ampicilline resistance; Ermr, erythromycin resistance; Kanr, kanamycine resistance; Cmr/Tmr, chloramphenicol/thiamphenicol resistance; Tcr, tetracycline resistance.
Growth measurements
Growth on cellobiose-supplemented basal medium was followed by monitoring optical density at 450 nm over time. When cultured on 5 g.L−1 Sigmacell, growth measurements were based on protein content measurement as described previously [49].
Construction of cipC and hycP mutations in Clostridium cellulolyticum
Gene inactivation in C. cellulolyticum was performed using the ClosTron technology as described by Heap et al., 2007 with minor modifications [30]. The integration sites in the target genes and the primers used to retarget the Ll.LtrB intron in the pMTL007 (IBS, EBS1d and EBS2, see data S2) were generated by the free Perutka algorithm implemented at http://ClosTron.com. Antisens intron integrations were chosen at position 116|117 for cipC and 829|830 for hycP downstream of the start codon. Specific cipC and hycP target primers IBS, EBS1d and EBS2 and the universal primer EBS universal were used to produce a fragment by overlapping PCR using pMTL007 as the matrix. The fragments were subsequently digested by BsrGI and HindIII and cloned in pMTL007 similarly digested. The retargeted resulting vectors were called pMTL007cipC and pMTL007hycp.
The vectors were methylated in vitro with MspI prior to be transferred in C. cellulolyticum by electro-transformation as previously described [50], [51]. The transformed cells were selected using thiamphenicol. Induction of the intron integration was performed by incubation of cells with 3 mM IPTG, and the mutated clones were selected using erythromycin. Clones mutated in cipC and hycP genes were called MTLcipC and MTLhycP, respectively.
Complementation of MTLcipC mutant
For MTLcipC complementation we used the cipC gene previously cloned in an erythromycin resistant pSOScipC vector [31]. As the MTLcipC mutated strain already contains erythromycin resistance brought by the mutation in the genome, we used the tetracycline resistant vector pSOSzero-Tc previously constructed [33]. This vector was digested using SalI and ligated with the expression cassette obtained from pSOS954 digested by the same enzymes [35]. The resulting E. coli-C. cellulolyticum shuttle expression vector called pSOS955 was then digested with BamHI and EheI, and ligated with the cipC gene excised from pSOScipC using BamHI and SwaI. The strain SG13009 (pREP4) was used as the recipient strain for transformation. The resulting vector was called pSOS955cipC. The vectors pSOS955cipC and pSOSzeroTc were transferred in MTLcipC strain thereby generating the cipC complemented strain MTLcipC (pSOS955cipC) and the control strain MTLcipC (pSOSzero-Tc), respectively.
Cloning of the genes encoding rCBM3a, rCBM3b, rHyr and mini-HycP in E. coli
All primers used in this study are presented in data S2. rCBM3a is designed to fuse the CBM3a from cipC (from amino-acid 27 to 187) in frame with a sequence of 6 histidine residues at its C-terminus. The pET-CBM3a was obtained by PCR on the genomic DNA of C. cellulolyticum using the forward CBM3aNdef and reverse CBM3aXhoIR primers, respectively. The amplicon was subsequently digested with NdeI and XhoI and cloned in a NdeI-XhoI linearized pET22b(+) thereby generating pET-CBM3a.
The 468 bp region of the hycP that encodes the CBM3 (from amino-acid 2187 to 2343) was amplified by PCR using the primers CBMHyCPNdeD and CBMHyCPXhoR whereas the 760 bp region of the gene hycP that encodes the three last HYR modules (from the amino-acid 1950 to 2192) was amplified by PCR using the oligonucleotides HyrNdeD and HyrXhoR. These primers introduced NdeI and XhoI sites upstream and downstream of the coding sequence, respectively. One ATG initiation codon was present in the forward primer. Amplicons were digested by NdeI and XhoI, and cloned in a similarly digested pET22b(+) vector. The resulting vectors pET-CBM3b and pET-Hyr contained the coding sequence for the rCBM3b and rHyr proteins fused in frame with a sequence encoding six histidine residues at their C-terminus, respectively.
Mini-HycP was designed to fuse the region starting from the amino-acid 58 to 609 to the region 2190 to 2343, in frame with a sequence encoding six histidine residues at its C- terminus. The gene encoding the Mini-HycP was generated by overlapping PCR performed on genomic DNA from C. cellulolyticum: the first PCR generated a 1681bp fragment using 1491_175NcoID and 1491_1830HyrR primers and the second one generated a 492bp-fragment using 1491_6559CBMD and CBMHyCPXhoR primer. The final amplicon was generated by mixing the two overlapping PCR fragments, and extended using primers 1491_175NcoID and CBMHyCPXhoR. The final amplicon was digested with NcoI and XhoI and cloned in a NcoI – XhoI linearized pET28a thereby generating the pET-miniHycP.
Plasmid pET-CBM3b, pET-Hyr, pET-miniHycP, pET-CBM3a were used to transform the BL21(DE3) strain to produce the corresponding recombinant proteins.
Production and purification of the recombinant proteins
Recombinant E. coli BL21(DE3) were grown at 37°C with shaking to an optical density at 600 nm of 1.0, Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 200 µM, and the cultures were incubated overnight under shaking at 25°C except for BL21(DE3)(pET-mini-HycP) strain for which induction of the heterologous gene expression was performed at 20°C. The cells were then harvested by centrifugation for 15 min at 6000 g and broken in a French press. After centrifugation of the crude extract (10 min, 4°C, 10000 g) the His-tagged proteins present in the supernatant were loaded on a column of Ni-nitrilotriacetic acid superflow resin (Qiagen, Hilden, Germany) equilibrated with 20 mM Tris-HCl (pH 8), and eluted using the same buffer supplemented with 60 mM imidazole. After concentration by ultrafiltration (Vivaspin 20, 10 kDa cutoff, Sartorius, Germany), the proteins were further purified by an anion exchange chromatography (Hi-trap Q-sepharose, GE Healthcare, Buckinghamshire, UK). The fractions of interest were pooled, dialyzed, and concentrated in 20 mM Tris-HCl (pH 8) by ultrafiltration (Vivaspin 20, 10 kDa cutoff, Sartorius, Germany). The absorbance at 280 nm was measured and the protein concentration was determined using their specific extinction coefficient. The purified recombinant rHyr protein was sent to Eurogentec France for polyclonal antibody production using the speedy 28-days protocol.
PAGE and Western blot analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using a vertical electrophoresis system. Gels were stained with Coomassie blue or were electroblotted onto nitrocellulose membranes (Hybond-ECL, GE Healthcare, Buckinghamshire, UK). Membranes were probed with polyclonal rabbit antibodies raised against rHyr protein, or CipC [25]. Primary antibodies were detected using anti-rabbit horseradish peroxidase conjugate (Promega, Madison, WI) and a chemiluminescent substrate (Millipore, Billerica, MA). When necessary, samples were precipitated by 10% ice-cold TCA (v/v), and the pellet was washed twice with acetone, dried and solubilized in loading SDS-PAGE buffer.
Protein binding assays
Binding of protein to polysaccharides were examined by incubating 40 µg of protein with 10 mg of Avicel microcrystalline cellulose (PH101, Fluka, Buchs, Switzerland), or hatched straw (Valagro, Poitiers, France) in 20 mM phosphate buffer (pH 7.0) in a 250 µl final volume during 1 hour at 4°C under gentle shaking. After centrifugation the pellet was washed twice with the same buffer and a sample of the pellet fraction (bound proteins) and of the supernatant (unbound proteins) were analyzed by SDS-PAGE.
Binding constants were determined as formerly described [22]. Binding constants were determined for rCBMs incubated with Avicel PH101, Sigmacell 20 (Sigma), Phosphoric Acid Swollen cellulose (PASC), bacterial microcrystalline cellulose (BMCC) or hatched straw (Valagro, Poitiers, France). PASC was obtained from Avicel PH101 as previously described [52], BMCC and hatched straw were obtained as previously described [53], [54] respectively.
Cell adherence assay
Binding assays protocol was based on the previously described protocol with modifications [55]. In a glovebox, C. cellulolyticum cells at exponential growth phase were mixed with rich medium buffer to reach an optical density of 0.5 at 450 nm. A volume of 2 mL of cell suspension was transferred in 15 mL Hungate tubes with a strip of filter paper, or nitrocellulose (80×10 mm), saturated or not 1 hour at room temperature with 4% BSA. Tubes were then incubated 1 hour with gentle agitation and optical density at 450 nm from supernatant was measured. Adhesion percentage was deduced from optical density measurement of an assay compared with a control where no filter paper or nitrocellulose was added. The reported values presented are the mean of 3 triplicates performed in at least 3 independent experiments.
Scanning electron microscopy
SEM experiments were performed on filter paper after 3 days of growth with C. cellulolyticum or 1 day with C. thermocellum. A piece of the filter paper was incubated with 2.5% glutaraldhehyde in PBS buffer for 30 minutes. Samples are then washed in distilled water and incubated with osmium tetroxyde (4%) for 20 minutes, washed and then gently incubated with 5 ethanol baths containing increasing concentration of ethanol, from 50% to 100%, for 10 minutes each. Filter paper was then incubated two minutes with a 50∶50 [vol/vol] solution of ethanol and hexamethyldisilazane (HMDS) and then 100% HMDS until complete evaporation, and kept dried for gold/palladium alloy coating. Samples were observed in the next few hours using a scanning electron microscope JSM 6320F (Jeol), at the CINaM microscopy service (Centre Interdisciplinaire de Nanosciences de Marseille, CNRS, Marseille).
Protein sequence analysis
Amino acid sequences were compared with those in the NCBI database using the BLAST program (http://blast.ncbi.nlm.nih.gov.gate1.inist.fr/Blast.cgi) [56]. Predictions of domains from amino acids sequences were performed using the Simple Modular Architecture Tool (SMART) (http://smart.embl-heidelberg.de/) [57] and the PFAM protein families database (http://pfam.sanger. ac.uk) [58]. Prediction of signal peptide cleavage sites and transmembrane segments was performed using SIGNALP V4.0 program (http://www. cbs.dtu.dk/services/SignalP) [59] PREDIction of Signal peptide tool (http://www.predisi.de/index.html) [60] and TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM/) [61]. Multiple sequence alignments were performed with the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) [62].
Supporting Information
Amino-acid sequence alignment of HYR modules identified in HycP. Sequences alignment has been performed using ClustalW2. Stars and grey box indicate identical residues; double dot, strongly similar residues; simple dot, weakly similar residues. Sequence of HYR modules were delimited and numbered as shown in figure 1A.
(TIF)
Primer sequences used in the present study.
(TIF)
Acknowledgments
We acknowledge Professor Nigel N. Minton and Mr John T. Heap (University of Nottingham, UK) as creators of the transferred material pMTL007. We thank Pascale de Philip and Hamza Celik for fruitful discussions. We also thank Damien Chaudanson for expert technical assistance for the SEM work at the CINaM.
Funding Statement
The research was supported by a fellowship grant to PHF from the French Ministère de l'enseignement supérieur et de la recherche, and funding from the Centre National de la Recherche Scientifique and Aix-Marseille Université. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- 2. McCann MC, Carpita NC (2008) Designing the deconstruction of plant cell walls. Curr Opin Plant Biol 11: 314–320. [DOI] [PubMed] [Google Scholar]
- 3.Tardif C, Bélaïch A, Fierobe HP, Pagès S, de Philip P, et al. (2006) Clostridium cellulolyticum: cellulosomes and cellulolysis. In Kataeva I editor. Cellulosome. Nova Sciences Publishers Inc. 221–259.
- 4. Fierobe HP, Pagès S, Bélaïch A, Champ S, Lexa D, et al. (1999) Cellulosome from Clostridium cellulolyticum: molecular study of the Dockerin/Cohesin interaction. Biochemistry 38: 12822–12832. [DOI] [PubMed] [Google Scholar]
- 5. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bayer EA, Kenig R, Lamed R (1983) Adherence of Clostridium thermocellum to cellulose. J Bacteriol 156: 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gelhaye E, Petitdemange H, Gay R (1993) Adhesion and growth rate of Clostridium cellulolyticum ATCC 35319 on crystalline cellulose. J Bacteriol. 175: 3452–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miron J, Ben-Ghedalia D, Morrison M (2001) Invited review: adhesion mechanisms of rumen cellulolytic bacteria. J Dairy Sci. 84: 1294–1309. [DOI] [PubMed] [Google Scholar]
- 9. Lu Y, Zhang YH, Lynd LR (2006) Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum . Proc Natl Acad Sci U S A 103: 16165–161659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZW, Lee SH, Elkins JG, Morrell-Falvey JL (2011) Spatial and temporal dynamics of cellulose degradation and biofilm formation by Caldicellulosiruptor obsidiansis and Clostridium thermocellum. AMB Express. doi: 10.1186/2191–0855–1–30. [DOI] [PMC free article] [PubMed]
- 11. Dumitrache A, Wolfaardt G, Allen G, Liss SN, Lynd LR (2013) Form and function of Clostridium thermocellum biofilms. Appl Environ Microbiol 79: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bayer EA, Lamed R, White BA, Flint HJ (2008) From cellulosomes to cellulosomics. Chem Rec 8: 364–377. [DOI] [PubMed] [Google Scholar]
- 13. Lamed R, Naimark J, Morgenstern E, Bayer EA (1987) Specialized cell surface structures in cellulolytic bacteria. J Bacteriol 169: 3792–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujino T, Béguin P, Aubert JP (1993) Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol 175: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lemaire M, Ohayon H, Gounon P, Fujino T, Béguin P (1995) OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol 1995 177: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leibovitz E, Ohayon H, Gounon P, Béguin P (1997) Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J Bacteriol 179: 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dror TW, Rolider A, Bayer EA, Lamed R, Shoham Y (2003) Regulation of expression of scaffoldin-related genes in Clostridium thermocellum . J Bacteriol 185: 5109–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blair BG, Anderson KL (1998) Comparison of staining techniques for scanning electron microscopic detection of ultrastructural protuberances on cellulolytic bacteria. Biotech Histochem 73: 107–113. [DOI] [PubMed] [Google Scholar]
- 19. Kosugi A, Murashima K, Tamaru Y, Doi RH (2002) Cell-surface-anchoring role of N-terminal surface layer homology domains of Clostridium cellulovorans EngE. J Bacteriol 184: 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kosugi A, Amano Y, Murashima K, Doi RH (2004) Hydrophilic domains of scaffolding protein CbpA promote glycosyl hydrolase activity and localization of cellulosomes to the cell surface of Clostridium cellulovorans . J Bacteriol 186: 6351–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blouzard JC, Coutinho PM, Fierobe HP, Henrissat B, Lignon S, et al. (2010) Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 10: 541–554. [DOI] [PubMed] [Google Scholar]
- 22. Pagès S, Gal L, Bélaïch A, Gaudin C, Tardif C, et al. (1997) Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J Bacteriol 179: 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wessel GM, Berg L, Adelson DL, Cannon G, McClay DR (1998) A molecular analysis of hyalin-a substrate for cell adhesion in the hyaline layer of the sea urchin embryo. Dev Biol 193: 115–126. [DOI] [PubMed] [Google Scholar]
- 24. Callebaut I, Gilgès D, Vigon I, Mornon JP (2000) HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci 9: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagès S, Bélaïch A, Fierobe HP, Tardif C, Gaudin C, et al. (1999) Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol 181: 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jindou S, Xu Q, Kenig R, Shulman M, Shoham Y, et al. (2006) Novel architecture of family-9 glycoside hydrolases identified in cellulosomal enzymes of Acetivibrio cellulolyticus and Clostridium thermocellum. FEMS Microbiol Lett. 254: 308–316. [DOI] [PubMed] [Google Scholar]
- 27. Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, et al. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J 15: 5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 28. Yaniv O, Shimon LJ, Bayer EA, Lamed R, Frolow F (2011) Scaffoldin-borne family 3b carbohydrate-binding module from the cellulosome of Bacteroides cellulosolvens: structural diversity and significance of calcium for carbohydrate binding. Acta Crystallogr D Biol Crystallogr 67: 506–515. [DOI] [PubMed] [Google Scholar]
- 29. Yaniv O, Halfon Y, Shimon LJ, Bayer EA, Lamed R, et al. (2012) Structure of CBM3b of the major cellulosomal scaffoldin subunit ScaA from Acetivibrio cellulolyticus . Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP (2007) The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods. 70: 452–464. [DOI] [PubMed] [Google Scholar]
- 31. Maamar H, Valette O, Fierobe HP, Bélaich A, Bélaich JP, et al. (2004) Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol Microbiol 51: 589–598. [DOI] [PubMed] [Google Scholar]
- 32. Maamar H, Abdou L, Boileau C, Valette O, Tardif C (2006) Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum . J Bacteriol 188: 2614–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Celik H, Blouzard JC, Voigt B, Becher D, Trotter V, et al. (2013) A Two-Component System (XydS/R) Controls the Expression of Genes Encoding CBM6-Containing Proteins in Response to Straw in Clostridium cellulolyticum. PLoS One. 8: e56063 doi:10.1371/journal.pone.0056063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perret S, Casalot L, Fierobe HP, Tardif C, Sabathe F, et al. (2004) Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J Bacteriol 186: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perret S, Bélaich A, Fierobe HP, Bélaich JP, Tardif C (2004) Towards Designer Cellulosomes in Clostridia: Mannanase Enrichment of the Cellulosomes Produced by Clostridium cellulolyticum. J Bacteriol. 186: 6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li LL, Taghavi S, Izquierdo JA, van der Lelie D (2012) Complete genome sequence of Clostridium sp. strain BNL1100, a cellulolytic mesophile isolated from corn stover. J Bacteriol 194: 6982–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kahel-Raifer H, Jindou S, Bahari L, Nataf Y, Shoham Y, et al. (2010) The unique set of putative membrane-associated anti-sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol Lett. 308: 84–93. [DOI] [PubMed] [Google Scholar]
- 39. Nataf Y, Bahari L, Kahel-Raifer H, Borovok I, Lamed R, et al. (2010) Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc Natl Acad Sci U S A 107: 18646–18651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kataeva IA, Seidel RD 3rd, Shah A, West LT, Li XL, et al (2002) The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl Environ Microbiol 68: 4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Proft T, Baker EN (2009) Pili in Gram-negative and Gram-positive bacteria-structure, assembly and their role in disease. Cell Mol Life Sci. 66: 613–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Danne C, Dramsi S (2012) Pili of gram-positive bacteria: roles in host colonization. Res Microbiol. 163: 645–658. [DOI] [PubMed] [Google Scholar]
- 43. Pegden RS, Larson MA, Grant RJ, Morrison M (1998) Adherence of the Gram-Positive Bacterium Ruminococcus albus to Cellulose and Identification of a Novel Form of Cellulose-Binding Protein Which Belongs to the Pil Family of Proteins. J Bacteriol 180: 5921–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rakotoarivonina H, Jubelin G, Hebraud M, Gaillard-Martinie B, Forano E, et al. (2002) Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148: 1871–1880. [DOI] [PubMed] [Google Scholar]
- 45. Varga JJ, Nguyen V, O'Brien DK, Rodgers K, Walker RA, et al. (2006) Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol. 62: 680–694. [DOI] [PubMed] [Google Scholar]
- 46. Imam S, Chen Z, Roos DS, Pohlschröder M (2011) Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One 6: e28919 doi:10.1371/journal.pone.0028919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ozdemir I, Blumer-Schuette SE, Kelly RM (2012) S-layer homology domain proteins Csac_0678 and Csac_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus . Appl Environ Microbiol 78: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petitdemange E, Caillet F, Giallo J, Gaudin C (1984) Clostridium cellulolyticum sp. nov., a cellulolytic mesophilespecies from decayed grass. Int J Sys Bacteriol 34: 155–159. [Google Scholar]
- 49. Giallo J, Gaudin C, Belaich JP, Petitdemange E, Caillet-Mangin F (1983) Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl Environ Microbiol 45: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jennert KC, Tardif C, Young DI, Young M (2000) Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology. 146: 3071–3080. [DOI] [PubMed] [Google Scholar]
- 51. Tardif C, Maamar H, Balfin M, Belaich JP (2001) Electrotransformation studies in Clostridium cellulolyticum. . J Ind Microbiol Biotechnol 27: 271–274. [DOI] [PubMed] [Google Scholar]
- 52. Walseth CS (1952) Tech. Assoc. Pulp Pap. Ind. 35: 228–233. [Google Scholar]
- 53. Väljamäe P, Sild V, Nutt A, Pettersson G, Johansson G (1999) Eur J Biochem. 266: 327–334. [DOI] [PubMed] [Google Scholar]
- 54. Fierobe HP, Mingardon F, Mechaly A, Bélaïch A, Rincon MT, et al. (2005) Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem 280: 16325–16334. [DOI] [PubMed] [Google Scholar]
- 55. Gelhaye E, Claude B, Caillez C, Burle S, Petitdemange H (1992) Multulayer adhesion to filter paper of two mesophilic cellulolytic clostridia. Curr Microbiol 25: 307–311. [Google Scholar]
- 56. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, et al. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32: D142–D144 doi:10.1093/nar/gkh088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. (2010) The Pfam protein families database: Nucleic Acids Research. 38: D211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 60. Menne KM, Hermjakob H, Apweiler R (2000) A comparison of signal sequence prediction methods using a test set of signal peptides. Bioinformatics 16: 741–742. [DOI] [PubMed] [Google Scholar]
- 61. Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a Hidden Markov Model: Application to Complete Genomes. J Mol Biol 30: 567–580. [DOI] [PubMed] [Google Scholar]
- 62. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino-acid sequence alignment of HYR modules identified in HycP. Sequences alignment has been performed using ClustalW2. Stars and grey box indicate identical residues; double dot, strongly similar residues; simple dot, weakly similar residues. Sequence of HYR modules were delimited and numbered as shown in figure 1A.
(TIF)
Primer sequences used in the present study.
(TIF)