Abstract
Background/Aims
The aim of this study was to investigate whether genetic variations at positions -1082, -819, and -592 in the interleukin (IL)-10 promoter affect IL-10 production in children with irritable bowel syndrome (IBS).
Methods
Ninety-four children with IBS and 102 children as healthy controls (HCs) were enrolled. Genomic DNA was extracted, and IL-10 -1082, -819, and -592 polymorphisms were detected by direct sequencing from all participants. Peripheral blood mononuclear cells (PBMCs) from 46 IBS children and 38 HCs were isolated and cultured with and without 5 ng/mL Escherichia coli lipopolysaccharide (LPS). IL-10 levels in the culture supernatants were measured by enzyme-linked immunosorbent assay.
Results
There were no significant differences in the distribution of IL-10 -1082, -819, and -592 polymorphisms or in the allele and haplotype frequencies between IBS children and HCs. PBMCs from children with IBS had significantly lower IL-10 levels after LPS stimulation than PBMCs from HCs (p=0.011); however, LPS-induced IL-10 levels in PBMCs with different genotypes of -819 and -592 polymorphisms were not significantly different between IBS patients and HCs.
Conclusions
Although significantly lower LPS-induced IL-10 production by PBMCs was noted, it is unlikely that IL-10 production was fully genetically determined in our IBS children. ClinicalTrials.gov identifier: NCT01131442.
Keywords: Irritable bowel syndrome, Child, Interleukin-10, Interleukin-10 gene polymorphisms
INTRODUCTION
Irritable bowel syndrome (IBS) is a common functional gastrointestinal disorder that mainly affects children >5 years of age and adolescents.1 It is defined as having abdominal pain, bloating, and changes in bowel habits (e.g., diarrhea, constipation, or mixed) in the absence of any organic or structural abnormality. Various etiologies like visceral hyperalgesia, disturbance of brain-gut interaction, autonomic and hormonal events, genetic and environmental factors, food sensitivity, postinfectious sequelae, and psychosocial disturbances have been implicated,2-8 but the precise pathophysiology remains unclear.
Postinfectious IBS introduces a role for the immune activation in the development of IBS symptoms.8 Postinflammatory changes in the gut may produce chronic alterations of the immune system, and altered cytokine profiles in IBS patients have been shown in some studies.9-14 Our previous study also found that children with IBS tend to produce lower amounts of the anti-inflammatory cytokine interleukin (IL)-10 at baseline and after Escherichia coli lipopolysaccharide (LPS) stimulation, implying that defects in immune modulation may contribute to IBS in children.15
Evidences demonstrate that changes in the genetic make-up or expression of cytokines play a critical role in the inflammatory response in the gut.10,16,17 Although a genetic component is suspected, unambiguous susceptibility genes have not been identified so far. Since the capacity for cytokine production can be genetically determined and is mainly related to genetic variations in the promoter region,18 further study that measures cytokine profiles and explores cytokine gene polymorphisms in parallel will be beneficial in this field.
Several polymorphic sites within the IL-10 gene promoter region have been described, including three biallelic polymorphisms at positions -1082 (base G to A, db SNP. rs1800896), -819 (base C to T, db SNP. rs1800871), and -592 (base C to A, db SNP. rs 1800872) from the transcription start site, are known to regulate the capacity to produce IL-10.19 The IL-10 -819 C and T alleles are completely in linkage disequilibrium with the IL-10 -592 C and A alleles, respectively.19,20
The aim of this study was to investigate if genetic variations at positions -1082, -819, and -592 in the IL-10 promoter affect IL-10 production and predispose to the development of IBS.
MATERIALS AND METHODS
1. Study population
Ninety-four children with IBS (49 females and 45 males; age, 5- to 18-year-old) and 102 healthy children as controls (61 females and 41 males; age, 2- to 18-year-old) were enrolled between November 2008 and February 2011. Patients were recruited consecutively from the outpatient Clinic of the Department of Pediatric Gastroenterology at Chang Gung Memorial Hospitals in Keelung and Taoyuan, whereas healthy volunteers were recruited through advertisements. All of the study participants were Han Chinese.
All patients had chronic or relapsing symptoms of IBS consistent with the Rome II criteria.21 The symptoms were present for at least 3 months. Patients were further categorized based on their symptoms and predominant stool patterns.22 Patients with more than three bowel movements per day and loose/watery stool consistency were categorized as diarrhea-predominant IBS (D-IBS, n=32), while those with fewer than three bowel movements per week and hard or lumpy stools were categorized as constipation-predominant IBS (C-IBS, n=33) and those with an alternating bowel pattern were categorized as mixed IBS (M-IBS, n=29).
A comprehensive diagnostic work-up, including hemogram, biochemistry, abdominal sonography, and serial stool testing, were conducted to exclude acute infections or any evidence of structural anomaly that may cause the symptoms.
The hospital's Human Ethics Committee (Institutional Review Board) approved the study and all of the participants provided informed consent.
2. DNA extraction
Genomic DNA was extracted from a 1 mL sample of whole blood from 94 IBS patients and 102 healthy controls and collected into tripotassium ethylenediaminetetraacetic acid sterile tubes. Extraction was performed using QIAamp DNA Blood Mini Kit (QIAGEN Inc., Valencia, CA, USA) according to the manufacturer's instructions.
3. IL-10 genotyping
The three biallelic IL-10 promoter polymorphisms (-1082, -819, and -592) were detected by polymerase chain reaction (PCR) using common primers (forward, 5'-ATC CAA GAC AAC ACT ACT AA-3'; reverse, 5'-TAA ATA TCC TCA AAG TTC C-3'). These primers yielded an amplicon 587 bp in size (-1115 to -528) containing the above polymorphisms. The amplification process was performed in 20 µL containing 1 µL of template DNA, PCR master mix 10 µL (RBC SensiZyme® HotstartTaq Premix; RBC Bioscience, Taipei, Taiwan), MgCl2 (2.5 mmol/L) 1 µL, each primer 2 µL, and free water 4 µL. The parameters for thermocycling were as follows: denaturation at 95℃ for 10 minutes, followed by 30 denaturation cycles at 95℃ for 30 seconds; annealing at 55.5℃ for 30 seconds; and extension at 72℃ for 1 minute. This was followed by a final extension at 72℃ for 5 minutes and then storage at 4℃. After confirming the final products by electrophoresis on agarose gels (3%), all of the PCR products were sequenced using ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The nucleotide polymorphisms at positions -1082, -819, and -592 were read directly.
4. Isolation of peripheral blood mononuclear cells
Blood samples (8 to 10 mL) from 46 (D-IBS, n=20; C-IBS, n=12; and M-IBS, n=14) of the 94 IBS children and 38 of the 102 healthy controls (HCs) were taken upon enrollment and those who used probiotics, antibiotics, analgesics, or immunosuppressive drugs within the past month were not included. In addition, subjects with recent infections, major allergic diseases, food intolerance, or psychiatric disorders were excluded. Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats on Lymphoprep (Nycomed, Oslo, Norway) gradients by centrifugation. After washing, the cells were resuspended at a concentration of 1×106 cells/mL in Roswell Park Memorial Institute 1640 medium containing 10% heat-inactivated fetal bovine serum.
5. Cell cultures
The concentration of PBMCs was adjusted to 106 cells/mL in complete medium and then transferred to 24-well plates. Some were stimulated with E. coli LPS 5 ng/mL (SIGMA L4391; Sigma, St. Louis, MO, USA), while other were not. Duplicate cultures were prepared and incubated for 24 hours at 37℃ in a humidified 5% CO2 atmosphere. The supernatants were collected, pooled, and stored at -20℃ until analysis by enzyme-linked immunosorbent assay (ELISA).
6. ELISA
Concentrations of IL-10 in the culture supernatants were determined using commercially available kits according to the manufacturer's instructions (R&D Systems, Minneapolis, MN, USA: IL-10, D1000B). Optical density was measured at wavelength of 450 nm and reference wavelength of 590 nm. The values were correlated linearly with cytokine standards. The sensitivity limit of the assay was 5 pg/mL.
7. Statistical analysis
Genotype and allele frequencies of each IL-10 polymorphism in the IBS patients and controls were compared by chi-square test. One-way analysis of variance (ANOVA) was performed to compare baseline and 5 ng/mL LPS-stimulated IL-10 levels between the IBS patients, subgroups, and HCs. Owing to the small numbers of subjects in different genotypes of IL-10 promoter polymorphisms and the sampling distribution was not normal, nonparametric test was used to compare baseline and 5 ng/mL LPS-stimulated IL-10 levels at each IL-10 genotype of polymorphisms at positions -1082, -819, and -592 in IBS patients, HCs, and between the two groups. A p<0.05 was considered statistically significant. Box plots indicated the median values at the 25th and 75th percentiles, and the error bars indicated the 10th and 90th percentiles. All statistical calculations were performed using the SPSS version 17.0 for Windows (SPSS Inc., Chicago, IL, USA).
RESULTS
1. Demographic data and polymorphisms of the IL-10 promoter
Sex, mean age, genotype, and allele and haplotype frequencies in the IL-10 gene promoter were analyzed (Table 1). Ninety-four children with IBS (49 females and 45 males; age range, 5 to 18 years; mean age, 14.2±3.1 years) and 102 HCs (61 females and 41 males; age range, 2 to 18 years; mean age, 12.3±5.1 years) were enrolled. At the -1082 locus, the A/A genotype was highly predominant (89.5% in IBS; 93.1% in HCs), while the G allele (either A/G or G/G) was rare. The -819 T allele (IL-10 -819 C/T and T/T) and -592 A allele (IL-10 -592 C/A and A/A) were more prevalent (-819 and -592 in linkage disequilibrium) (Table 1) than the -819 C allele and -592 C allele, respectively, in both the IBS and HC groups (67% vs 33% and 71.6% vs 28.4%, respectively).
Table 1.
Demographic Data and Frequencies of IL-10 Promoter Genotype, Allele, and Haplotype in IBS Children and HCs
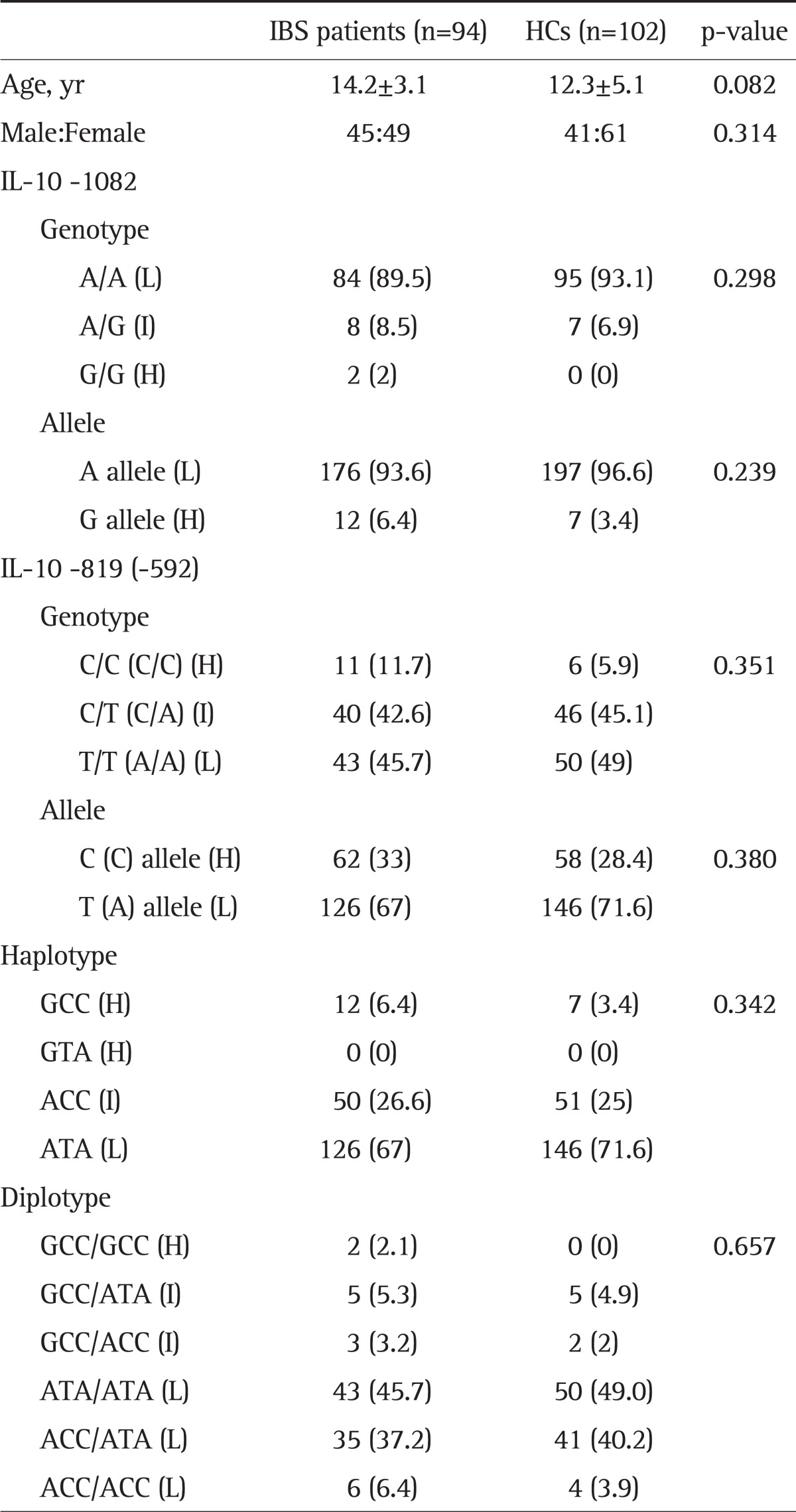
Data are presented as mean±SD or number (%).
IL, interleukin; IBS, irritable bowel syndrome; HC, healthy control; H, IL-10 high production; I, IL-10 intermediate production; L, IL-10 low production.
In this study, the ATA haplotype of IL-10 was more prevalent than the other haplotypes (67% in IBS and 71.6% in HCs). ATA/ATA was the most common diplotype (45.7% in IBS and 49% in HCs), followed by the ACC/ATA haplotype (37.2% in IBS and 40.2% in HCs). There were no differences in genotype, allele, or haplotype frequencies in the three IL-10 polymorphisms between the IBS and control groups.
2. Baseline and E. coli LPS-induced cytokine production
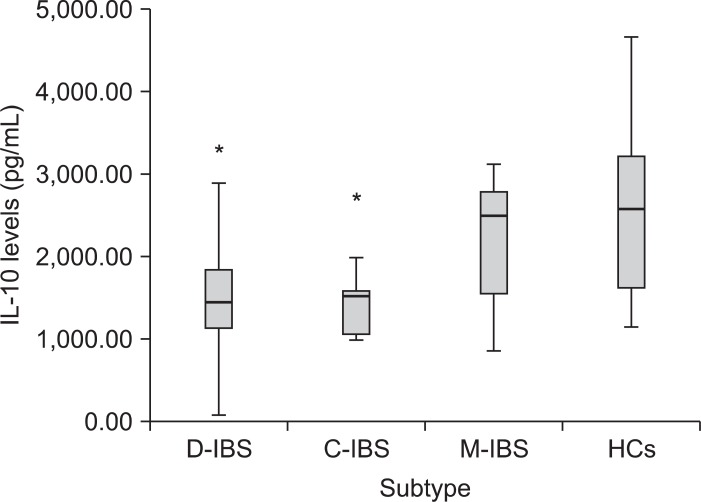
IBS patients had lower baseline IL-10 levels compared with HCs (54.1±36.50 pg/mL vs 114.57±44.10 pg/mL, p=0.292). After stimulation with 5 ng/mL E. coli LPS, the IBS group had significantly lower IL-10 levels compared with the HC group (1,755.26±167.92 pg/mL vs 2,521.16±239.98 pg/mL, p=0.011). Analysis of the three IBS subgroups showed lower baseline IL-10 levels compared with HCs (data not shown). The IL-10 levels were significantly lower under LPS-stimulated condition (5 ng/mL) in the diarrhea and constipation subgroups than in the HCs (p<0.05) (Fig. 1). However, there was no difference among the patient subgroups.
Fig. 1.
Box plot of interleukin (IL)-10 levels following 5 ng/mL lipopolysaccharide stimulation in irritable bowel syndrome (IBS) subtype and healthy controls diarrhea-predominant IBS [D-IBS]: 1,532.68±870.22 vs. constipation-predominant IBS [C-IBS]: 1,428.39±413.66 vs. mixed IBS [M-IBS]: 2,159.44±265.48 vs. HCs [healthy controls]: 2,521.16±1,046.06 pg/mL).
*Statistically significant, p<0.05.
3. Association of IL-10 promoter polymorphism with IL-10 production
To investigate whether genetic variations at positions -1082, -819, and -592 in the IL-10 promoter affected IL-10 production, LPS-induced IL-10 levels were analyzed in different genotypes of polymorphisms in the IBS patients and HCs separately (Table 2). Genotype -1082 A/A was highly predominant (91% in the IBS group and 100% in the HC group) whereas other genotypes at the -1082 loci were rare. Although there were significantly lower IL-10 levels at -1082 A/A in the IBS group compared with HCs (1,709.54±820.04 vs 2,521.16±239.98, respectively; p=0.011), whether IL-10 -1082 A/A affected IL-10 production was inconclusive because few -1082 A/G and G/G genotype were found both in the IBS and HCs group and the association with IL-10 production couldn't be well analyzed.
Table 2.
IL-10 Promoter Polymorphisms and LPS-Induced IL-10 Levels in Children with IBS and HCs
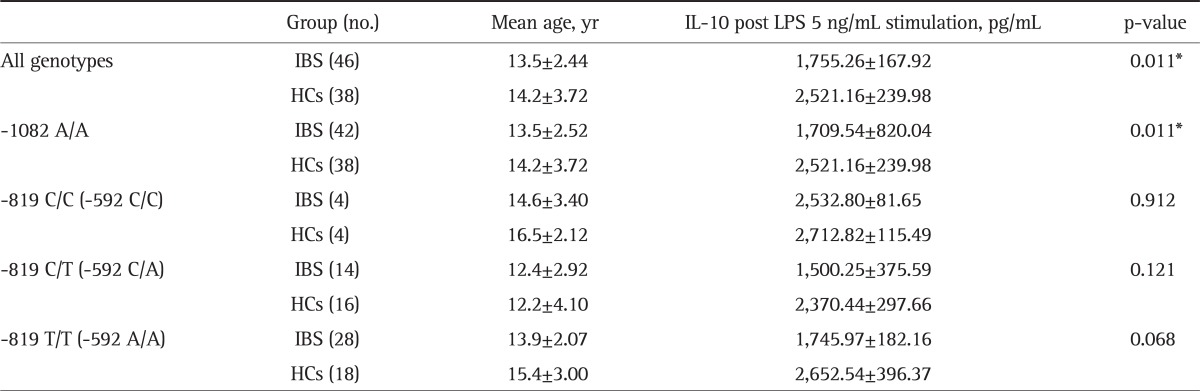
IL, interleukin; LPS, lipopolysaccharide; IBS, irritable bowel syndrome; HC, healthy control.
*Statistically significant, p<0.05.
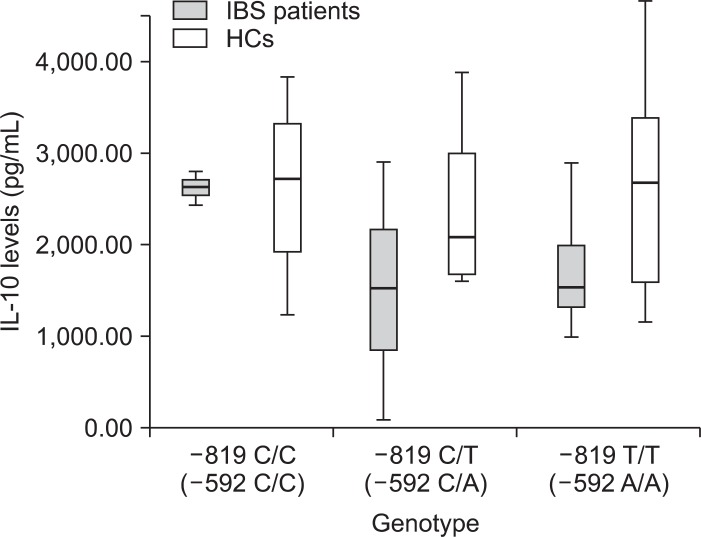
In genotype -819 T/T and -592 A/A, LPS induced IL-10 levels in IBS patients were much lower compared with HCs, approaching statistical significance (p=0.068) (Table 2). In general, PBMCs of the IBS patients tended to produce lower IL-10 levels after stimulation with 5 ng/mL LPS compared with the HC group in different genotypes of -819 and -592 polymorphisms (Fig. 2). However, there were no significantly different LPS-induced IL-10 levels between the IBS patients and HCs (p=0.101).
Fig. 2.
Box plot showing that peripheral blood mononuclear cells (PBMCs) from irritable bowel syndrome (IBS) patients tend to produce lower interleukin (IL)-10 levels after stimulation with 5 ng/mL lipopolysaccharide compared to PBMCs from the healthy controls (HCs) group in different genotypes of -819 and -592 polymorphisms. There was no significant difference between the IBS patients and HC group (p=0.101).
DISCUSSION
Cytokines are involved in the control of gastrointestinal motility and visceral sensitivity.12 They are also important modulators of intestinal immune responses and inflammatory reactions.23-25 Several studies have investigated systemic and mucosal cytokine profiles in IBS revealing evidence of disturbances in the balance of proinflammatory and anti-inflammatory cytokines.9,14,15,26,27
IL-10 is considered to be an anti-inflammatory cytokine that inhibits the production of several other cytokines including interferon-γ, IL-2, IL-4, IL-6, and tumor necrosis factor-α (TNF-α).24 It also regulates B cell proliferation and differentiation and exhibits immunoregulatory activity.28 Reduced IL-10 levels in IBS patients have been mentioned in several literatures.11,12,27,29 Those individuals predisposed to producing lower amounts of IL-10 may be at a higher risk of developing IBS symptoms.
To explore IL-10 production capacity, PBMCs from both IBS group and HCs were stimulated with 5 ng/mL E. coli LPS and determined the IL-10 production of these cells. PBMCs may exhibit similar responses with regard to proliferation and cytokine secretion as those by lamina propria mononuclear cells when they are exposed to bacterial antigens, such as Helicobacter pylori.30 It is worth noting that H. pylori has a weaker stimulatory effect than other intestinal bacteria such as E. coli or Salmonella,31 and it has been reported that the release of cytokines from PBMCs in response to a potent bacterial endotoxin can significantly exceed lamina propria mononuclear cell-mediated cytokine levels.32
In our study, PBMCs from the IBS group produced significantly lower IL-10 levels after stimulation with 5 ng/mL E. coli LPS compared with PBMCs from HCs (1,755.3±167.92 pg/mL vs 2,521.2±239.98 pg/mL; p=0.011). This suggested that defects in immune modulation might contribute to occurrence of IBS in children. Factors that regulate the expression of IL-10 may be involved in the pathogenesis of IBS. In this regard, genetic predisposition such as single nucleotide polymorphisms may be important and further study is required.
IL-10 possesses a highly polymorphic promoter, with variations at -1082, -819, and -592 that have been studied and implicated in regulating the rates of IL-10 gene transcription. The IL-10 A allele at -1082 and T allele at -819 have been associated with low IL-10 production.29,33 The -1082 G, -819 C, and -592 C (GCC) alleles have also been associated with elevated levels of IL-10,34 while ACC and ATA haplotypes exhibit intermediate and low IL-10 gene transcription, respectively.33 In the present study, the prevalences of low production alleles (-1082 A allele and -819 T allele) and haplotype (ATA) are similar in both the IBS group and HCs without statistical significance.
Cytokine gene polymorphisms in individual susceptibility to certain diseases have been documented,35-37 and have been evaluated in adult IBS patients. A genetic predisposition to lower anti-inflammatory cytokine production in IBS patients can mean that control of the inflammatory response may be compromised in some individuals. However, the results are inconsistent.12 Gonsalkorale et al.16 found that IBS patients had a significantly lower frequency of the high-producer genotype for IL-10 (-1082 G/G) compared to controls. Conversely, Barkhordari et al.24,38 found that the low-producer IL-10 (-1082 A/A) had a lower frequency in IBS patients compared to controls, while van der Veek et al.17 found that IL-10 genotypes were similarly distributed in the IBS patients and the controls. Combined high TNF-α and low IL-10 producer (-1082 A/A) genotypes are considerably more frequent in the IBS patients. Lee et al.39 studied IL-10 (-1082 G/A) and TNF-α (-308 G/A) gene polymorphisms and found the genotype and allele distribution were similar in both IBS and control groups.
To date, there is no literature analyzing the association between IL-10 gene polymorphisms and IL-10 production in children with IBS. Whether decreased IL-10 levels in IBS children are primarily determined by cytokine genetic polymorphism or by other mechanisms is not clear and our study is probably a pilot study in this regard. However, LPS-induced IL-10 production by PBMCs is not different between the IBS patients and HCs in -819 and -592 polymorphisms, indicating that lower IL-10 production is not fully genetically determined.
The conflicting results between the current and previous studies regarding the role of IL-10 gene polymorphisms in IBS may be due to several factors. First, it is difficult to ascertain the magnitude of the effects of genetic polymorphisms on disease susceptibility because of the existence of IL-10 homologues. Different IL-10 binding receptors are likely to complicate the determination of IL-10 expression levels in vitro.40 Moreover, insufficiently large sample sizes (especially the IBS subgroups), the younger ages of the study subjects (≤18 years of age), gene-gene and gene-environment interactions, and ethnic differences may contribute to these discrepancies. In this study, the lack of association between IL-10 promoter polymorphisms and lower IL-10 levels in children with IBS implies that different combinations of factors affect the regulation of cytokine production. Genetic variations may affect only a part of the cytokine profile changes that precipitate IBS.
In conclusion, children with IBS tend to produce lower amounts of the anti-inflammatory cytokine IL-10 at baseline and after LPS stimulation. However, there are no significant associations between IL-10 promoter polymorphisms and IL-10 levels in these children. The inherited component and genetic variations in children with IBS require further investigation.
ACKNOWLEDGEMENTS
Members of the PATCH Study Group are J.L.H. (study coordinator), M.C.H., T.C.Y., M.H.T., S.L.L., S.H.L., K.W.Y., W.I.L., L.S.O., L.C.C. (principle investigators). The authors thank the study subjects and their parents for their participation, as well as Chang Gung Memorial Hospital, Keelung for the financial support (CMRPG 290271 and 2B0041).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Wyllie R. Recurrent abdominal pain of childhood. In: Kliegman R, Nelson WE, editors. Nelson textbook of pediatrics. 18th ed. Philadelphia: Saunders; 2007. p. 1627. [Google Scholar]
- 2.Song GH, Venkatraman V, Ho KY, Chee MW, Yeoh KG, Wilder-Smith CH. Cortical effects of anticipation and endogenous modulation of visceral pain assessed by functional brain MRI in irritable bowel syndrome patients and healthy controls. Pain. 2006;126:79–90. doi: 10.1016/j.pain.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum Reprod Update. 2005;11:411–423. doi: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- 4.Locke GR, 3rd, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ., 3rd Familial association in adults with functional gastrointestinal disorders. Mayo Clin Proc. 2000;75:907–912. doi: 10.4065/75.9.907. [DOI] [PubMed] [Google Scholar]
- 5.Morris-Yates A, Talley NJ, Boyce PM, Nandurkar S, Andrews G. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol. 1998;93:1311–1317. doi: 10.1111/j.1572-0241.1998.440_j.x. [DOI] [PubMed] [Google Scholar]
- 6.Solmaz M, Kavuk I, Sayar K. Psychological factors in the irritable bowel syndrome. Eur J Med Res. 2003;8:549–556. [PubMed] [Google Scholar]
- 7.Koloski NA, Talley NJ, Boyce PM. A history of abuse in community subjects with irritable bowel syndrome and functional dyspepsia: the role of other psychosocial variables. Digestion. 2005;72:86–96. doi: 10.1159/000087722. [DOI] [PubMed] [Google Scholar]
- 8.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979–1988. doi: 10.1053/j.gastro.2009.02.074. [DOI] [PubMed] [Google Scholar]
- 9.Dinan TG, Clarke G, Quigley EM, et al. Enhanced cholinergic-mediated increase in the pro-inflammatory cytokine IL-6 in irritable bowel syndrome: role of muscarinic receptors. Am J Gastroenterol. 2008;103:2570–2576. doi: 10.1111/j.1572-0241.2008.01871.x. [DOI] [PubMed] [Google Scholar]
- 10.Collins SM. Dysregulation of peripheral cytokine production in irritable bowel syndrome. Am J Gastroenterol. 2005;100:2517–2518. doi: 10.1111/j.1572-0241.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 11.Macsharry J, O'Mahony L, Fanning A, et al. Mucosal cytokine imbalance in irritable bowel syndrome. Scand J Gastroenterol. 2008;43:1467–1476. doi: 10.1080/00365520802276127. [DOI] [PubMed] [Google Scholar]
- 12.Bashashati M, Rezaei N, Andrews CN, et al. Cytokines and irritable bowel syndrome: where do we stand? Cytokine. 2012;57:201–209. doi: 10.1016/j.cyto.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz-Lucas M, Saz-Peiró P, Sebastián-Domingo JJ. Irritable bowel syndrome immune hypothesis. Part two: the role of cytokines. Rev Esp Enferm Dig. 2010;102:711–717. doi: 10.4321/s1130-01082010001200006. [DOI] [PubMed] [Google Scholar]
- 14.Liebregts T, Adam B, Bredack C, et al. Immune activation in patients with irritable bowel syndrome. Gastroenterology. 2007;132:913–920. doi: 10.1053/j.gastro.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 15.Hua MC, Lai MW, Kuo ML, Yao TC, Huang JL, Chen SM. Decreased interleukin-10 secretion by peripheral blood mononuclear cells in children with irritable bowel syndrome. J Pediatr Gastroenterol Nutr. 2011;52:376–381. doi: 10.1097/MPG.0b013e3181fd9816. [DOI] [PubMed] [Google Scholar]
- 16.Gonsalkorale WM, Perrey C, Pravica V, Whorwell PJ, Hutchinson IV. Interleukin 10 genotypes in irritable bowel syndrome: evidence for an inflammatory component? Gut. 2003;52:91–93. doi: 10.1136/gut.52.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Veek PP, van den Berg M, de Kroon YE, Verspaget HW, Masclee AA. Role of tumor necrosis factor-alpha and interleukin-10 gene polymorphisms in irritable bowel syndrome. Am J Gastroenterol. 2005;100:2510–2516. doi: 10.1111/j.1572-0241.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 18.Mosmann TR. Properties and functions of interleukin-10. Adv Immunol. 1994;56:1–26. [PubMed] [Google Scholar]
- 19.Perrey C, Pravica V, Sinnott PJ, Hutchinson IV. Genotyping for polymorphisms in interferon-gamma, interleukin-10, transforming growth factor-beta 1 and tumour necrosis factor-alpha genes: a technical report. Transpl Immunol. 1998;6:193–197. doi: 10.1016/s0966-3274(98)80045-2. [DOI] [PubMed] [Google Scholar]
- 20.Eskdale J, Keijsers V, Huizinga T, Gallagher G. Microsatellite alleles and single nucleotide polymorphisms (SNP) combine to form four major haplotype families at the human interleukin-10 (IL-10) locus. Genes Immun. 1999;1:151–155. doi: 10.1038/sj.gene.6363656. [DOI] [PubMed] [Google Scholar]
- 21.Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Muller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45(Suppl 2):II43–II47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Park MI, Camilleri M. Genetics and genotypes in irritable bowel syndrome: implications for diagnosis and treatment. Gastroenterol Clin North Am. 2005;34:305–317. doi: 10.1016/j.gtc.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Barkhordari E, Rezaei N, Mahmoudi M, et al. T-helper 1, T-helper 2, and T-regulatory cytokines gene polymorphisms in irritable bowel syndrome. Inflammation. 2010;33:281–286. doi: 10.1007/s10753-010-9183-6. [DOI] [PubMed] [Google Scholar]
- 25.Sartor RB. Cytokines in intestinal inflammation: pathophysiological and clinical considerations. Gastroenterology. 1994;106:533–539. doi: 10.1016/0016-5085(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 26.Drossman DA, Camilleri M, Mayer EA, Whitehead WE. AGA technical review on irritable bowel syndrome. Gastroenterology. 2002;123:2108–2131. doi: 10.1053/gast.2002.37095. [DOI] [PubMed] [Google Scholar]
- 27.O'Mahony L, McCarthy J, Kelly P, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 28.Mocellin S, Panelli MC, Wang E, Nagorsen D, Marincola FM. The dual role of IL-10. Trends Immunol. 2003;24:36–43. doi: 10.1016/s1471-4906(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 29.Kindt S, Van Oudenhove L, Broekaert D, et al. Immune dysfunction in patients with functional gastrointestinal disorders. Neurogastroenterol Motil. 2009;21:389–398. doi: 10.1111/j.1365-2982.2008.01220.x. [DOI] [PubMed] [Google Scholar]
- 30.Windle HJ, Ang YS, Athie-Morales V, McManus R, Kelleher D. Human peripheral and gastric lymphocyte responses to Helicobacter pylori NapA and AphC differ in infected and uninfected individuals. Gut. 2005;54:25–32. doi: 10.1136/gut.2003.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birkholz S, Knipp U, Nietzki C, Adamek RJ, Opferkuch W. Immunological activity of lipopolysaccharide of Helicobacter pylori on human peripheral mononuclear blood cells in comparison to lipopolysaccharides of other intestinal bacteria. FEMS Immunol Med Microbiol. 1993;6:317–324. doi: 10.1111/j.1574-695X.1993.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 32.Sperber K, Silverstein L, Brusco C, Yoon C, Mullin GE, Mayer L. Cytokine secretion induced by superantigens in peripheral blood mononuclear cells, lamina propria lymphocytes, and intraepithelial lymphocytes. Clin Diagn Lab Immunol. 1995;2:473–477. doi: 10.1128/cdli.2.4.473-477.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 34.Koss K, Satsangi J, Fanning GC, Welsh KI, Jewell DP. Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: differential effects on production and allele frequencies. Genes Immun. 2000;1:185–190. doi: 10.1038/sj.gene.6363657. [DOI] [PubMed] [Google Scholar]
- 35.Mahdaviani SA, Rezaei N, Moradi B, Dorkhosh S, Amirzargar AA, Movahedi M. Proinflammatory cytokine gene polymorphisms among Iranian patients with asthma. J Clin Immunol. 2009;29:57–62. doi: 10.1007/s10875-008-9232-1. [DOI] [PubMed] [Google Scholar]
- 36.Movahedi M, Mahdaviani SA, Rezaei N, Moradi B, Dorkhosh S, Amirzargar AA. IL-10, TGF-beta, IL-2, IL-12, and IFN-gamma cytokine gene polymorphisms in asthma. J Asthma. 2008;45:790–794. doi: 10.1080/02770900802207261. [DOI] [PubMed] [Google Scholar]
- 37.Rezaei N, Aghamohammadi A, Shakiba Y, et al. Cytokine gene polymorphisms in common variable immunodeficiency. Int Arch Allergy Immunol. 2009;150:1–7. doi: 10.1159/000210374. [DOI] [PubMed] [Google Scholar]
- 38.Barkhordari E, Rezaei N, Ansaripour B, et al. Proinflammatory cytokine gene polymorphisms in irritable bowel syndrome. J Clin Immunol. 2010;30:74–79. doi: 10.1007/s10875-009-9342-4. [DOI] [PubMed] [Google Scholar]
- 39.Lee HJ, Lee SY, Choi JE, et al. G protein beta3 subunit, interleukin-10, and tumor necrosis factor-alpha gene polymorphisms in Koreans with irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:758–763. doi: 10.1111/j.1365-2982.2010.01496.x. [DOI] [PubMed] [Google Scholar]
- 40.Fickenscher H, Hör S, Küpers H, Knappe A, Wittmann S, Sticht H. The interleukin-10 family of cytokines. Trends Immunol. 2002;23:89–96. doi: 10.1016/s1471-4906(01)02149-4. [DOI] [PubMed] [Google Scholar]