Abstract
Background
Pteridine metabolic pathway is unusual features of Leishmania, which is necessary for the growth of parasite. Leishmania has evolved a complex and versatile pteridine salvage network which has the ability of scavenging a wide area of the conjugated and unconjugated pteridines especially folate and biopterin. In this study, we focus on the inhibition of ptr1 gene expression.
Methods
L. major ptr1 gene was cloned into pcDNA3 and digested using KpnI and BamHI. The gene was subcloned so that antisense will transcribe and called pcDNA-rPTR. Leishmania major was cultured and late logarithmic-phase promastigotes were harvested. The promastigotes were divided into two groups. One group was transfected with 50 µg of pcDNA-rPTR, whereas the other group was transfected with pcDNA3. Transfected cells were cultured and plated onto semi-solid media. Mouse pritonean macrophages were transfected using pcDNA-rPTR–tansfected promastigotes. Western blotting was performed on mouse transfected pritonean macrophages and extracts from transfected promastigotes of L. major using a L. major ptr1 antibody raised in rabbits.
Results
The PTR1 protein was not expressed in pcDNA-rPTR– tansfected promastigotes and mouse macrophage transfected with pcDNA-rPTR– tansfected promastigotes.
Conclusion
This approach might be used to study the pteridine salvage pathway in Leishmania or to assess the possibility of using gene expression inhibition in the treatment of leishmaniasis.
Keywords: Pteridine reductase 1, Antisense, Leishmania, Gene regulation, Inhibition
Introduction
Pteridine metabolic pathway is unusual features of Leishmania, which is necessary for the growth of parasite. Different parts of pteridine pathway are the target of chemotherapeutic. In order to infect the mammalian host cells pteridine auxotroph, Leishmania absolutely needs an exogenous source of pterin (1–3). Leishmania has evolved a complex and versatile pteridine salvage network which has the ability of scavenging a wide area of the conjugated and unconjugated pteridines especially folate and biopterin (4). Folate and biopterin in fully reduced tetrahydro forms, H4-folate and H4-biopterin, act as co-factors. In Leishmania and mammalian cells, NADPH-dependent enzyme dihydrofolate reductase (DHFR) is the agent for the production of H4-folate from folate and dihydrofolate (5). In Leishmania and other protozoa, DHFR exists as a bifunctional enzyme and joines to thymidylate synthase (DHFR-TS) (6). In the de novo biosynthesis of thymidylate in Leishmania, the main role of H4-folate is as essential co-factor (7).
In mammalian cells, H4-biopterin is synthesized through de novo or salvaged through DHFR-mediated reduction of H2-biopterin (8). While Leishmania lacks de novo biopterin synthetic pathway, DHFR-TS shows no response to biopterin or H2biopterin. Instead, reduced biopterin is generated through the pteridine reductase 1 (ptr1), which sequentially reduces oxidized biopterin to H2-biopterin and then to H4-biopterin (2, 3, 9, 10). Ptr1 is as a gene within the Leishmania H region. Over exposure of ptr1 by gene amplification or DNA transfixion, confers methotrexate (MTX) resistance (3, 11). The predicted ptr1 protein showed homology to a large family of aldo/keto reductases and short-chain dehydrogenases, including several enzymes involved in pteridine metabolism, such as sepiapterin reductase (3) and dihydropteridine reductase (DHPR) (12). Leishmania needs reduced form of folate and biopterin for growth where as the current available anti-pteridines do not have much promising results clinically against leishmaniasis, even though were proved to be effective against other protozoan infectious which justifies research in this field (13).
In this study, ptr1 gene expression was inhibited to study the pteridine salvage pathway in Leishmania or to assess the possibility of using gene expression inhibition in the treatment of leishmaniasis.
Materials and Methods
DNA extraction and gene amplification
Leishmania major was grown in NNN medium and subcultured in RPMI1640 enriched with 10% fetal bovine serum. DNA extraction was done on Leishmania promastigotes harvested at late logarithmic phase. A set of primers (PTR F, 5′-GGA TCC ATG ACT GCT CCG ACC-3′; PTR R, 5′-GGT ACC TCA GGC CCG GGT AAG-3′) was designed based on L. major ptr1 sequence (GenBank Accession No. L01699) with BamHI and KpnI restriction sites established on the 5′-ends of the forward and reverse primers, respectively. The ptr1 coding region was amplified from genomic DNA and the PCR product was ligated to a 3′ T-tailed, EcoRV-digested pBluescript and sequenced.
Construction of the antisense ptr1 gene
Recombinant pBluescript containing L. major ptr1 gene (accession no. EF113119) was digested with KpnI and BamHI enzymes and purified using a Fermentas DNA purification kit (Cat. No. k0513) and then subcloned into pcDNA3 digested with KpnI and BamHI. The recombinant plasmid was transformed into E. coli TOP10 strain. Since ptr1 gene was cloned antiparallel to the sense, this expression plasmid is referred to pcDNA-rPTR or antisense.
Transfection of Leishmania promastigotes
Leishmania major was cultured in liquid medium 199 (Sigma, UK, Dorset, England) supplemented with 10% defined heat-inactivated fetal bovine serum (Biosera, France), 10 mM adenine (Sigma, UK, Dorset, England), 40 mM HEPES (Sigma, UK, Dorset, England), 0.25% hemin (Sigma, UK, Dorset, England), 100 mg/ml streptomycin (Biosera, France) and 100 IU/ ml penicillin (Biosera, France). Late logarithmic phase L. major promastigotes were harvested by centrifugation at 1500g for 10 min and adjusted to 5 × 107/ml in ice-cold transfection buffer (14–17). The promastigotes were divided into two groups: one group was transfected with 50 µg of pcDNA-rPTR (antisense), whereas the other group was transfected with only pcDNA3 using a BioRad Gene Pulser at 450 V and 450 µF capacitance as described previously (15–17). Transfected promastigotes were cultured for 48 h in drug-free medium 199 and subsequently plated onto semi-solid medium 199 containing 40 µg/ml G418 (Sigma, UK, Dorset, England) as a selective antibiotic because pcDNA3 was containing the neomycin resistance gene therefore this antibiotic was used to determine the transfection (14, 18).
Inhibition of ptr1 gene expression in transfected promastigotes by antisense
After two weeks, single colonies were transferred into medium 199 supplemented with 40 µg/mL neomycin (14, 18). Transfected promastigotes were mass cultured and collected by centrifugation at 3,000 g for 10 min and washed twice using 1x TBS (150 mM NaCl, 10 mM Tris pH 7.5). Cells were resuspended in lysis buffer (50 mM Tris, 10% glycerol, 0.1% Triton X-100, 1 mM PMSF) and sonicated, then the lysate was centrifuged at 1,000 g for 10 min and the supernatant was used for analysis. Western blot analysis was performed as described previously (19). Briefly, 106 of each pcDNA-rPTR- transfected L. major promastigotes and pcDNA3-transfected L. major promastigotes were harvested and lysed using sonication. Protein samples were separated on 10% SDS-PAGE, transferred to nitrocellulose membranes and incubated at 37 °C in the presence of a 1:500 dilution of rabbit anti-PTR1 antibody (20) for 1 hour. Then, the membrane was incubated at 37 °C for one hour using horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:5000) as the secondary antibody. Antibody binding was visualized using diaminobenzidine (DAB) (19).
Inhibition of ptr1 gene expression in amastigotes containing pcDNA-rPTR (antisense)
Macrophages were isolated from 4-6 weeks BALB/c mice by peritoneal lavage using chilled RPMI 1640 media (Biosera, France) containing 10% fetal bovine serum and allowed the macrophages to adhere to the surface of flasks for overnight incubation at 37°C in humidified 5% CO2. No adherent macrophages were washed off using Hanks solution. Adherent macrophages were incubated separately with two groups of transfectant parasites (pcDNA-rPTR- transfected L. major promastigotes and pcDNA3-transfected L. major promastigotes) and incubated at 37 °C in humidified 5% CO2 for 2 hours. Unbound parasites were removed by washing with Hanks solution and added RPMI1640 medium containing 10% fetal bovine serum and incubated at 37°C in humidified 5% CO2 for 3 days and the medium was changed every 24 h. Transfected macrophages were separated by scraper and harvested by centrifugation and then lysed by lysis buffer was added and incubated on ice for 30 min. SDS-PAGE and Western blot analysis was performed as described previously (19). Briefly, cells lysate (protein samples) were prepared and separated on 10% SDS-PAGE and electrophoretically transferred onto nitrocellulose membrane and analyzed using a rabbit anti-PTR1 antibody at a 1:1000 dilution as the primary antibody and Goat anti-rabbit IgG HRP conjugation at a 1:5000 dilution as secondary antibody and detected by colorimetry using diamino benzoic acid and H2O2.
Results
Construction of the antisense ptr1 gene
Leishmania major ptr1 gene was cloned into pBluescript, and digested with Bam HI and KpnI (Fig. 1), and subcloned into pcDNA3 as antisense (pcDNA-rPTR). To confirm the identity of the recombinant plasmid, pcDNA-rPTR was digested using restriction enzymes KpnI and BamHI and then an 808bp insert and a 5.4-kb vector fragment was observed using electrophoresis (Fig. 2).
Fig. 1.
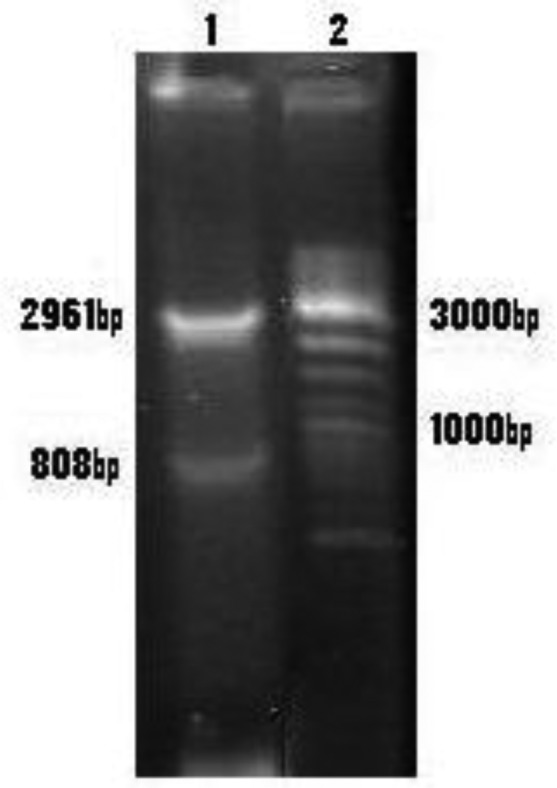
Electrophoresis of digested pBSC-ptr using enzyme/ Lane 1: Digested pBSC-ptr by KpnI and BamHI/Lane 2: 100-bp DNA ladder marker
Fig. 2.
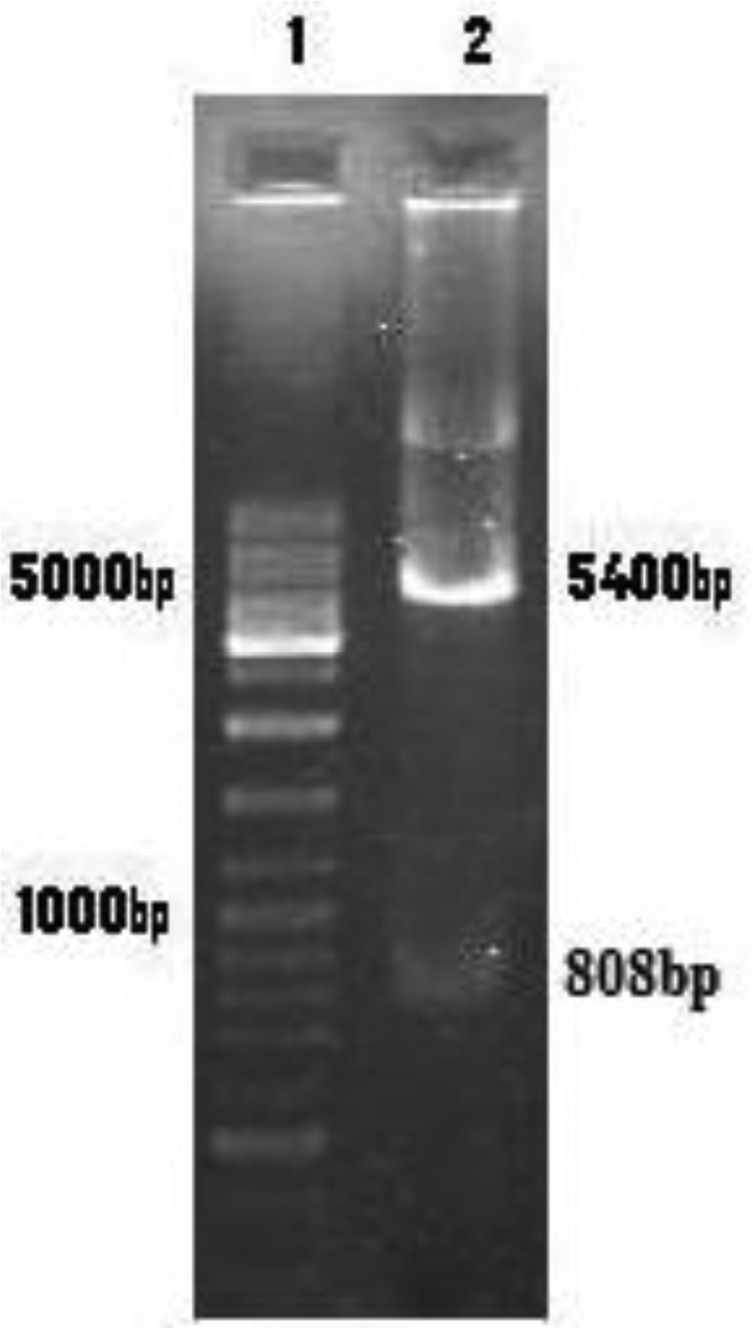
Identification of recombinant plasmid pcDNA-rPTR using enzyme digestion. Lane 1:100-bp DNA ladder marker. Lane 2: pcDNA-rPTR digested with KpnI and BamHI
Promastigotes after transfection
Twenty-four hours after transfection, some dead promastigotes were observed. After selection with 40 µg/mL G418, untransfected promastigotes were dead, and transfected promastigotes were survived. The cell clones were isolated at 10-14 days after G418 selection.
Inhibition of ptr1 expression by antisense in promastigotes
The PTR1 protein was expressed in pcDNA3-transfected promastigotes (Fig. 3, lane 1), but not expressed in pcDNA-rPTR–transfected promastigotes (Fig. 3, lane 2), indicating that the gene was inhibited by the antisense construct.
Fig. 3.
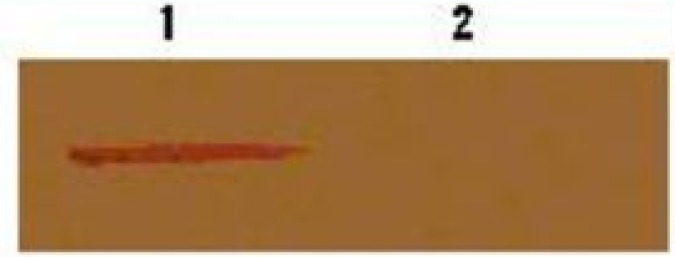
Western blot analysis to demonstrate lack of expression of PTR1 protein in antisense-transfected promastigotes
Lane 1: Lysate of pcDNA3-transfected promastigotes
Lane 2: Lysate of pcDNA-rPTR-transfected promastigotes
Inhibition of ptr1 expression by antisense in amastigotes
The results indicated that ptr1 expression in amastigotes containing pcDNA- rPTR (antisense) was inhibited but inhibition was not seen in amastigotes containing pCDNA3 (Fig. 4).
Fig. 4.

Analysis of expression protein by Western blotting
Lane 1: Infected macrophages by amastigotes containing pcDNA3 /Lane 2: Extract macrophages Lane 3: Infected macrophages by amastigotes containing antisense (pcDNA-rPTR)
Discussion
The properties of ptr1 suggest several mechanisms to explain how its overproduction creates MTX resistance. Because ptr1 binds MTX tightly, it may sequester MTX from DHFR-TS (10). In addition, ptr1 exhibits a broad specificity for pteridine substrates and reduces folate to the H2- and H4 forms (21). Ptr1 may mediate MTX resistance through its ability to reduce folate to generate H2 folate, which is known to be extremely effective in relieving the inhibition of DHFR-TS by MTX in vitro (22, 23). Deletion of the ptr1 gene is lethal to the promastigote in the presence of MTX (5). This gene has also been used to determine the species of Leishmania (24).
In this study, similar to others (25) pcDNA-rPTR was prepared and confirmed; the vector contains the ptr1 gene oriented antiparallel to the sense.
Leishmania major promastigotes were transfected separately with pcDNA-rPTR and pcDNA3, and lysates were examined by western blot analysis, which showed that the expression of the ptr1 gene was inhibited by pcDNA-rPTR successfully. The current data is similar to that generated by Chen et al., in which full-length antisense RNA was used to inhibit the expression of gp63 gene in Leishmania amazonensis (25). Although the targeted gene deletion in L. major GP63 was reported by others (15, 26), the present data demonstrated that inhibition occurs in the cytosol, as Dumas et al. reported in the study using cytosolic antisense RNA to regulate the expression of noncoding RNA in amastigotes (27). Small nucleolar RNA (snoRNA) genes might be silenced in L. major, Leptomonas collosoma and Trypanosoma brucei (28). Silencing is achieved in Leptomonas collosoma and L. major by expressing an antisense transcript complementary to the snoRNA gene, resulting in the accumulation of small interfering RNA (siRNA), the siRNA then eliminates the mature snoRNA (28). Other scientists have used antisense RNA to inhibit beta-tubulin synthesis in Leishmania donovani amastigote, as well as a mini-exon sequence to inhibit amastigote growth (29–33).
Conclusion
In contrast to the previous studies, this work is the first report on inhibition of Leishmania ptr1 by a full-length antisense construct. The results demonstrated that ptr1 antisense RNA might be efficiently block ptr1 mRNA and protein expression in L. major promastigotes and amastigotes which might be used as a model for gene inhibition of Leishmania. This approach might be used to study other infectious diseases and cancer cells when a single gene is involved.
Acknowledgment
This study was supported by the EMGEN (Eastern Mediterranean Health Genomics & Biotechnology Network) through project number 20091. The study was carried out in Cellular and Molecular Biology Research Center, Shahid Beheshti University of Medical Sciences, and the authors would like to thank the personnel and the director of the center. The authors declare that there is no conflict of interest.
References
- 1.Ouellette M, Drummelsmith J, Elfadidi A, Kundig C, Richard D, Roy G. Pterin transport and metabolism in Leishmania and related trypanosomatid parasites. Int J Parasitol. 2002;32(4):385–398. doi: 10.1016/s0020-7519(01)00346-0. [DOI] [PubMed] [Google Scholar]
- 2.Scott DA, Coombs GH, Sanderson BE. Folate utilisation by Leishmania species and the identification of intracellular derivatives and folate- metabolizing enzymes. Mol Biochem Parasitol. 1987;23(2):139–149. doi: 10.1016/0166-6851(87)90149-6. [DOI] [PubMed] [Google Scholar]
- 3.Callahan HL, Beverley SM. A member of the aldoketo reductase family confers methotrexate resistance in Leishmania . J Boil Chem. 1992;267(34):24165–24168. [PubMed] [Google Scholar]
- 4.Cunningham ML, Beverley SM. Pteridine salvage throughout the Leishmania infectious cycle: implications for antifolate chemotherapy. Mol Biochem Parasitol. 2001;113(2):199–213. doi: 10.1016/s0166-6851(01)00213-4. [DOI] [PubMed] [Google Scholar]
- 5.Nare B, Uuba J, Hardy L, Beverley SM. New approach to Leishmania chemotherapy: pteridine reductase 1 (ptr1) as a target and modulator of antifolate sensitivity. Parasitol. 1997;114(Suppl):S101–110. [PubMed] [Google Scholar]
- 6.Hardy LW, Matthews W, Nare B, Beverley SM. Biochemical and genetic tests for inhibitors of Leishmania pteridine pathways. Exp Parasitol. 1997;87(3):158–170. [PubMed] [Google Scholar]
- 7.Beverley SM. Gene amplification in Leishmania . Annu Rev Microbiol. 1991;45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- 8.Nichol CA, Smith GK, Duch DS. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Ann Rev Biochem. 1985;54:729–764. doi: 10.1146/annurev.bi.54.070185.003501. [DOI] [PubMed] [Google Scholar]
- 9.Beck J, Ullman B. Biopterin conversion to reduced folates by Leishmania donovani promastigotes. Mol Biochem Parasitol. 1991;49(1):21. doi: 10.1016/0166-6851(91)90126-q. [DOI] [PubMed] [Google Scholar]
- 10.Bello AR, Nare B, Freedman D, Hardy L, Beverley SM. PTR1: A reductase mediating salvage of oxidized pteridines and methotrexate resistance in the protozoan parasite Leishmania major . Proc Natl Acad Sci USA. 1994;91(24):11442–11446. doi: 10.1073/pnas.91.24.11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania . EMBO J. 1992;11(10):3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiteley JM, Xuong NH, Varughese KI. Is dihydropteridine reductase an anomalous dihydrofolate reductase, a flavin-like enzyme, or a short-chain dehydrogenase? Adv Exp Med Biol. 1993;338:115–121. doi: 10.1007/978-1-4615-2960-6_23. [DOI] [PubMed] [Google Scholar]
- 13.Gourley DG, Luba J, Hardy LW, Beverley SM, Hunter WN. Crystallization of recombinant Leishmania major pteridine reductase 1 (ptr1) Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 9):1608–1610. doi: 10.1107/s0907444999008999. [DOI] [PubMed] [Google Scholar]
- 14.Kapler GM, Coburn CM, Beverley SM. Stable transfection of the human parasite Leishmania major delineates a 30 – kilo base region sufficient for extra chromosomal replication and expression. Mol Cell Biol. 1990;10(3):1084. doi: 10.1128/mcb.10.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi PB, Sacks DL, Modi G, McMaster WR. Targeted gene deletion of Leishmania major genes encoding developmental stage – specific leishmanolysin (Gp63) Mol Microbiol. 1998;27(3):519–530. doi: 10.1046/j.1365-2958.1998.00689.x. [DOI] [PubMed] [Google Scholar]
- 16.Beverley SM, Clayton CE. Transfection of Leishmania and Trypanosome brucei by electroporation. Methods Mol Biol. 1993;21:333. doi: 10.1385/0-89603-239-6:333. [DOI] [PubMed] [Google Scholar]
- 17.Kelly JM, Ward HM, Miles MA, Kendall G. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania . Nucleic Acids Res. 1992;20(15):3963–3969. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rangarajan D, Gokool S, McCrossan MV, Smith DF. The gene B protein localizes to the surface of Leishmania major parasites in the absence of metacyclic stage lipophosphoglycan. J Cell Sci. 1995;108(Pt 11):3359–3366. doi: 10.1242/jcs.108.11.3359. [DOI] [PubMed] [Google Scholar]
- 19.Shewry PR, Fido RJ. Protein blotting, Principels and Applications. In: Ralph Rapley, John M.Walker., editors. Molecular Biomethods handbook. USA: Published by Humana; 1998. pp. 435–444. Press. [Google Scholar]
- 20.Kheirandish F, Bandehpour M, Haghighi A, Mahboudi F, Mohebali M, Mosaffa N, Kazemi B. Molecular cloning and expression of Iranian Leishmania major pteridine reductase 1. Iranian J Parasitol. 2008;3(2):1–9. [PMC free article] [PubMed] [Google Scholar]
- 21.Robello C, Navarro P, Castanys S, Gamarro F. A pteridine reductase gene ptr1 contiguous to a P- glycoprotein confers resistance to antifolates in Trypanosoma cruzi . Mol Biochem Parasitol. 1997;90(2):525–535. doi: 10.1016/s0166-6851(97)00207-7. [DOI] [PubMed] [Google Scholar]
- 22.White JC. Reversal of methotrexate binding to dihydrofolate reductase by dihydrofolate. Studies with pure enzyme and computer modeling using network thermodynamics. J Bio Chem. 1979;254(21):10889–10895. [PubMed] [Google Scholar]
- 23.El Fadili A, Kündig C, Roy G, Ouellette M. Inactivation of the Leishmania tarentolae pterin transporter (BT1) and reductase (ptr1) genes leads to viable parasites with changes in folate metabolism and hypersensitivity to the antifolate methotrexate. J Biol Chem. 2004;279(18):18575–18582. doi: 10.1074/jbc.M400652200. [DOI] [PubMed] [Google Scholar]
- 24.Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, Ouellette M. Unresponsiveness to Glucantime Treatment in Iranian Cutaneous Leishmaniasis due to Drug-Resistant Leishmania tropica Parasites. PLoS Med. 2006;3:e162. doi: 10.1371/journal.pmed.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen DQ, Kolli BK, Yadava N, Lu HG, Gilman-Sachs A, Peterson DA, Chang KP. Episomal expression of specific sense and antisense mRNAs in Leishmania amazonensis: modulation of gp63 level in promastigotes and their infection of macrophages in vitro. Infect Immun. 2000;68(1):80–86. doi: 10.1128/iai.68.1.80-86.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi PB, Kelly BL, Kamhawi S, Sacks DL, McMaster WR. Targeted gene deletion in Leishmania major identifies leishmanolysin (GP63) as a virulence factor. Mol Biochem Parasitol. 2002;120(1):33–40. doi: 10.1016/s0166-6851(01)00432-7. [DOI] [PubMed] [Google Scholar]
- 27.Dumas C, Chow C, Muller M, Papadopoulou B. A novel class of developmentally regulated noncoding RNAs in Leishmania . Eukaryot Cell. 2006;5(12):2033–2046. doi: 10.1128/EC.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang XH, Liu Q, Michaeli S. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc Natl Acad Sci USA. 2003;100(13):7521–7526. doi: 10.1073/pnas.1332001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta D, Adhya S, Basu MK. The effect of beta-tubulin-specific antisense oligonucleotide encapsulated in different cationic liposomes on the suppression (correction of suppression) of intracellular L. donovani parasites in vitro. J Biochem. 2002;132(1):23–27. doi: 10.1093/oxfordjournals.jbchem.a003194. [DOI] [PubMed] [Google Scholar]
- 30.Compagno D, Toulme JJ. Antisense effects of oligonucleotides complementary to the hairpin of the Leishmania mini-exon RNA. Nucleosides Nucleotides. 1999;18(6-7):1701–1704. doi: 10.1080/07328319908044827. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty R, Dasgupta D, Adhya S, Basu MK. Cationic liposome-encapsulated antisense oligonucleotide mediates efficient killing of intracellular Leishmania . Biochem J. 1999;340(Pt 2):393–396. [PMC free article] [PubMed] [Google Scholar]
- 32.Compango D, Lampe JN, Bourge C, Kutyavin IV, Yurchenko L, Lukhtanov EA, Gorn VV, Gamper JR, Toulme JJ. Antisense oligonucleotides containing modified bases inhibit in vitro translation of Leishmania amazonensis mRNAs by invading the mini-exon hairpin. J Biol Chem. 1999;274(12):8191–8198. doi: 10.1074/jbc.274.12.8191. [DOI] [PubMed] [Google Scholar]
- 33.Pascolo E, Blonskim C, Shire D, Toulme JJ. Antisense effect of oligodeoxynucleotides complementary to the mini-exon sequence of the protozoan parasite Leishmania amazonensis . Biochimie. 1993;75(1-2):43–47. doi: 10.1016/0300-9084(93)90023-l. [DOI] [PubMed] [Google Scholar]


