Abstract
Background
The protozoan parasite Toxoplasma gondii can infect any warm blooded nucleated cells. One of the ways for human infection is ingestion of oocysts directly from soil or via infected fruits or vegetables. To survey the potential role of T. gondii oocyst in soil samples, the present study was conducted in Tehran City, Iran.
Methods
A total of 150 soil samples were collected around rubbish dumps, children's play ground, parks and public places. Oocysts recovery was performed by sodium nitrate flotation method on soil samples. For molecular detection, PCR reaction targeting B1 gene was performed and then, the positive results were confirmed using repetitive 529 bp DNA fragment in other PCR reaction. Finally, the positive samples were genotyped at the SAG2 locus.
Results
Toxoplasma DNA was found in 13 soil samples. After genotyping and RFLP analysis in SAG2 locus, nine positive samples were revealed type III, one positive sample was type I whereas three samples revealed mixed infection (type, I & III).
Conclusion
The predominant genotype in Tehran soil samples is type III.
Keywords: Toxoplasma gondii, SAG2, Soil, Genotype, Iran
Introduction
Toxoplasma gondii is a widely distributed coccidian parasite that can infect a wide range of animals and humans. It is over 100 years since the discovery of the parasite in 1908 and now it is used extensively as a model for cell biology of apicomplexan organisms (1, 2). This coccidian parasite is the causative agent of toxoplasmosis, one of the most prevalent parasitic infectious diseases in animals and humans (3). Transmission of this parasite occurs by consumption of raw or undercooked meat containing tissue cyst or by ingestion of mature oocysts from environmental sources such as soil, water, fruits and vegetables (4). It is estimated that 15% to 85% of human population in the world are chronically infected with T. gondii (2). Toxoplasmosis is mainly asymptomatic in immunocompetent individuals, but in immunocompromised patients such as HIV and organ transplantation patients manifestation of clinical signs can be Life threatening (5). Despite of the worldwide distribution of this parasite, there is only one species (T. gondii) that causes toxoplasmosis (1). Prevalence of this parasite had been shown to be up to 50% in Iran, which is verified from different parts of the country (6).
Toxoplasma oocysts are resistant to environmental conditions and may remain infective for more than one year in different types of soils (4, 7).
Soil contamination with oocysts is related to distribution of infected cat feces in environment. Areas such as gardens, park and around rubbish dump are main places that cats may excrete feces in soil (8).
According to the different methods of characterization such as restriction fragment length polymorphism (RFLP), isoenzyme electrophoresis and random amplified polymorphism, T. gondii strains classified into three clonal lineages (genotypes I, II and III) and some atypical genotypes (9–12). It was revealed that three lineages of this parasite have less than 1% difference in genomic level (13). Several genetic markers are available to identify genotypes of T. gondii isolates, that the polymorphic surface antigen two (SAG2) is one of the locuses used for differentiation of these three clonal lineages (12, 14).
Genetic analysis of T. gondii infection in soil and other environmental resources is of importance to comprehend the epidemiology, patterns of transmission and clonal diversity of the parasite in different parts of the world. One of the studies conducted to environmental contamination with this parasite is the survey of Lass et al. in Poland, that he detected T. gondii oocysts in soil samples and confirmed it by molecular methods (15).
The present study was performed to identify T. gondii oocysts in soil samples from Tehran, Iran by molecular method and genotyping of positive samples in SAG2 locus by endonuclease enzymes.
Materials and Methods
Collection of soil samples
One hundred and fifty soil samples were collected from September 2008 to March 2009 from different parts of Tehran city, such as parks, public places, children's play ground and areas around rubbish dumps. Each sample was weighted about 300 gram which was collected from 3 cm of ground depth. Soil samples were dried at laboratory temperature for 48 hours, sieved and concentrated with modified sodium nitrate flotation as described previously (16).
Toxoplasma gondii control standard strains
Three strains were obtained from School of Public Health, Tehran University of Medical Sciences. Tachyzoites of T. gondii RH strain (type I), tissue cysts of Tehran strain (type II) that was previously isolated from human lymphadenitis (17), and tachyzoites of a virulent strain of T. gondii with unknown genotype which is maintain by serial intrapretoneal passages in Department of Parasitology in Tehran University of Medical Science. The strain is introduced as U strain in here.
The tachyzoites were collected from peritoneal cavity of BALB/c mice that were infected three days earlier.
Tissue cysts of Tehran strain (type II) was obtained from brain of BALB/c mice that were injected with bradyzoites of the strain two months earlier.
DNA Extraction
DNA extraction was performed with the commercial genomic mini kit (A & A Biotechnology, Gdynia, Poland) according to manufacturer's instructions. From each samples 100 µl of DNA was eluted and stored at -20°C until use.
Detection of Toxoplasma gondii oocyst by PCR
The target of PCR was the 199 bp fragment of the highly conserved 35 fold repetitive B1 gene (AF179871). For PCR reaction, a pair of primer Toxo1 (5’ GGA ACT GCA TCC GTT CAT GAG 3’) and Toxo2 (5’ TCT TTA AAG CGT TCG TGG TC 3’) were used. The PCR was performed according to Schwab et al. (18). To confirm the results, all positive samples were also examined by another pair of primer Toxo-F (5’ AGG CGA GGG TGA GGA TGA 3’) and Toxo-R (5’ TCG TCT CGT CTG GAT CGC AT 3’). These primers are specific for a 200 to 300 fold repetitive fragment of 134 bp (AF 146527) (19, 20). In this study Taq DNA PreMix (Accupower™, BioNeer, South Korea) was used. With the final reaction volume of 25 µl, the amplification was performed by 10 min at 95°C initial step, followed by 30 cycles: denaturation for 5 s at 95° C, annealing for 10 s at 60° and extension for15 s at 72°C. PCR products were analyzed by electrophoresis on 2% agarose gel and stained with ethidium bromide.
Genotyping and RFLP
For nested-PCR reaction, outer and inner primers for SAG2 locus (3’ and 5’ end) that were previously designed by Howe et al. (12), were used (Table 1).
Table 1.
Names and sequences of the polymerase chain reaction primer pairs used for Nested-PCR
| PCR Reaction | Primer name and sequence | |
|---|---|---|
| 5’ SAG2 primary PCR | SAG2 F4 | 5’-GCTACCTCGAACAGGAACAC-3’ |
| SAG2 R4 | 5’-GCATCAACAGTCTTCGTTGC-3’ | |
| 3’ SAG2 primary PCR | SAG2 F3 | 5’-TCTGTTCTCCGAAGTGACTCC-3’ |
| SAG2 R3 | 5’-TCAAAGCGTGCATTATCGC-3’ | |
| 5’ SAG2 secondary PCR | SAG2 F | 5’-GAAATGTTTCAGGTTGCTGC-3’ |
| SAG2 R2 | 5’-GCAAGAGCGAACTTGAACAC-3’ | |
| 3’ SAG2 secondary PCR | SAG2 F2 | 5’-ATTCTCATGCCTCCGCTTC-3’ |
| SAG2 R | 5’-AACGTTTCACGAAGGCACAC-3’ |
Length of selected fragments was 241-bp and 221-bp in 5’ end and 3’ end, respectively. The protocol for temperature cycling was used as described by Aspinal et al. (21). In order to distinguish T. gondii genotypes, restriction enzymes were used as described previously (12, 22, 23). Two restriction enzymes were selected for RFLP, Sau3aI and HhaI (Fermentas, Germany). Sau3aI enzyme was used for digestion of 5’ end of amplification products that distinguished allele 3 (genotype III) from alleles I and II (genotypes I & II). HhaI enzyme was used for digestion of 3’ end of amplification products that distinguished allele II from types I and III (Fig. 1). For RFLP procedure, 10 µl of nested-PCR products of the 5’ and 3’ ends were digested using 3U of Sau3aI and HhaI restriction enzymes in separate reactions with a total volume of 30 µl at 37° C. Then the fragments were analyzed by 3% agarose gel electrophoresis.
Fig. 1.
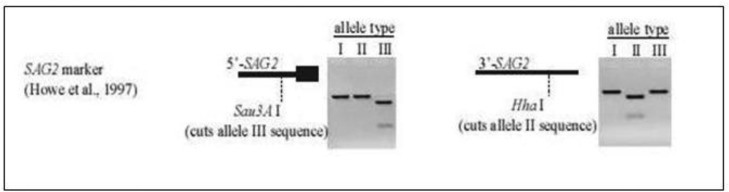
Schematic image for function of restriction enzymes on SAG2 locus (Howe et al. (24)
Sequencing
After nested-PCR, all the positive samples were sequenced by BioNeer Lab (South Korea).
Results
Identification of T. gondii oocysts in soil samples
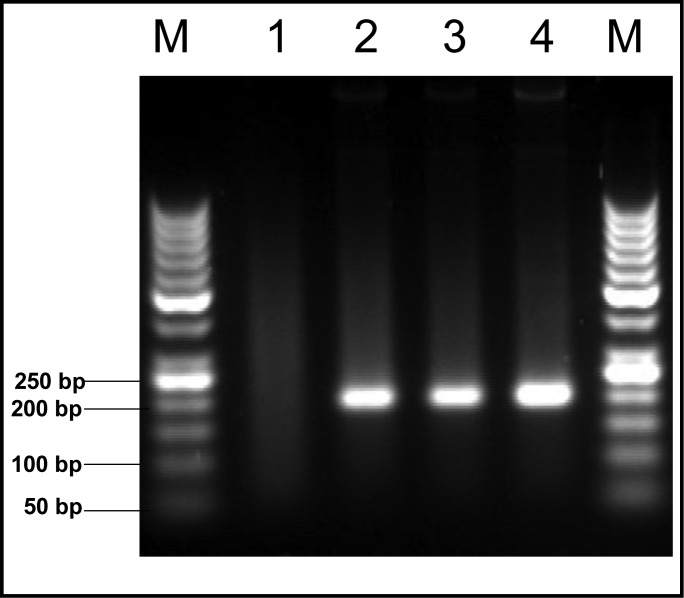
From 150 soil samples that examined by two pairs of primer in two steps of PCR, 13 samples (8.7%) were positive. In the first step, a 194-bp fragment of B1 gene was amplified (Fig. 2). In the second step, all positive samples were examined by other pairs of primers. The length of fragment in this step was 134-bp from 200 to 300 fold-repetitive elements (AF146527) (Fig. 3). All soil samples were examined twice in each PCR reaction.
Fig. 2.
B1 gene amplification products (194 bp) of T. gondii on agarose 2%. Lane M, molecular weight marker 50 bp (Fermentas); Lane 1, Negetive control; Lane 4, positive control (RH strain); Lane 2-3 positive soil samples
Fig. 3.
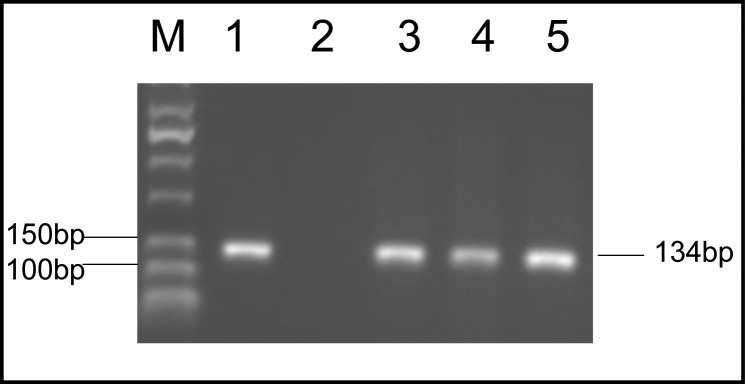
Amplification of 134-bp fragment T. gondii from 200 to 300 folds repetitive 529 bp DNA elements on agarose 2%. Lane M, molecular weight marker 50 bp; Lane 1, positive control (RH strain); Lane 2, Negetive control; Lane 3-5, positive soil samples
Genotyping of positive samples
All of the 13 positive soil samples were examined by nested-PCR at 5’ and 3’ end of SAG2 locus by four pairs of specific primers.
These samples showed a 241-bp amplified band in 5’ end of SAG2 locus and a 221-bp amplified band in 3’ end, respectively. After nested-PCR the amplified fragments were used for RFLP. Genotyping of 13 positive soil samples showed that 9 were type III (69%), 3 were mixed of type I and III (23%) and 1 of the samples was type I (8%) (Figs. 4, 5, 6). The pattern of digestion for RH strain and Tehran strain were corresponded to genotypes I and II, respectively. In addition, genotype of U strain was identified type I. All the steps, including nested-PCR and RFLP were performed twice.
Fig. 4.
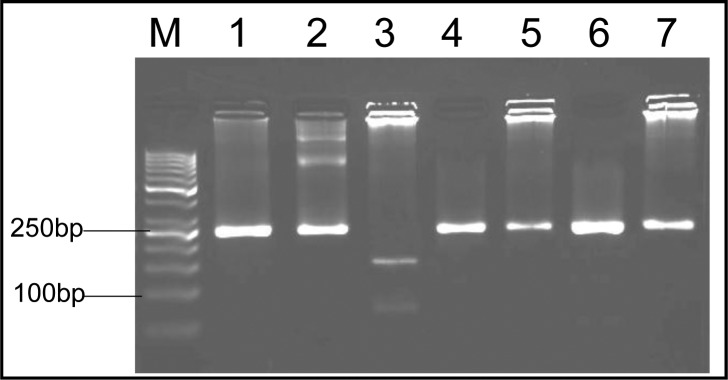
Nested-PCR amplification of SAG2 locus at 3’ end. Lane M, molecular weight marker (50 bp); Lane 1-3, positive controls (RH, unknown and Tehran strains, respectively); Lanes 4-7, soil samples
Fig. 5.
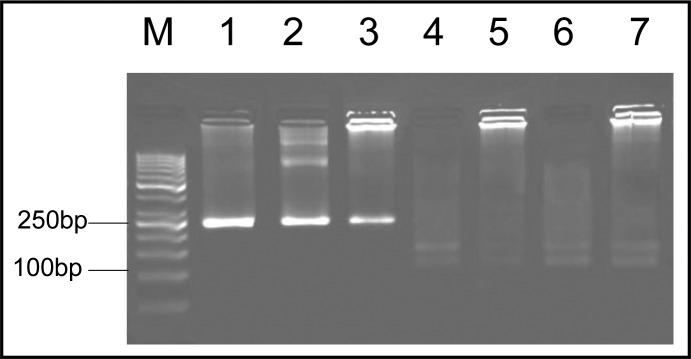
Nested-PCR amplification of SAG2 locus at 5’ end. Lane M, molecular weight marker (50 bp); Lane 1-2, positive controls (RH and Tehran strains, respectively); Lane 3, unknown strain (genotype I); Lanes 4-7, soil samples (genotype III)
Fig. 6.
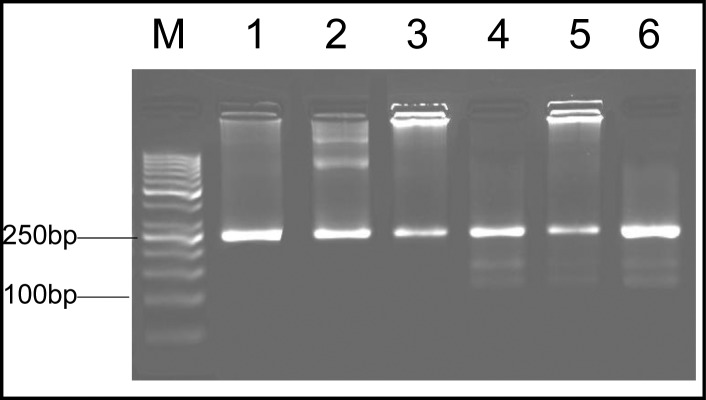
Nested-PCR amplification of SAG2 locus at 5’ end. Lane M, molecular weight marker (50 bp); Lane 1-2, positive controls (RH and Tehran strains, respectively); Lane 3, soil sample (genotype I); Lane 4-6, soil samples (mix genotype of I and III)
Results of sequencing
The products of the first step of nested-PCR were selected for sequencing and sent to BioNeer Company (South Korea). Sequences were aligned using DNASIS MAX software (Version 3.00; Hitachi, Yokohama, Japan). The nucleotide sequences alignment of samples that had shown type III, revealed the 99-100% homology with other reported sequences in GenBank. The nucleotide sequences data from this paper were submitted in the DDBJ/EMBL/Gene Bank nucleotide sequence databases at accession numbers of AB667972-AB667975.
Discussion
There is little information about the presence of T. gondii oocyst in the soil. In the epidemiological point of view, soil is a large and important source of T. gondii infection (4, 24, 25). It is obvious that the abundance of stray cats have a main role in environmental contamination with T. gondii oocysts. The number of oocysts shed by naturally infected domestic cats is largely unknown and both young (< 6 mo) and older (>6 mo) cats have been found shedding oocysts in nature (2). Toxoplasma oocysts are resistance to environmental factors, and remain in soil up to one year (4). Unfortunately, detection of low amounts of oocysts in soil samples is difficult. According to the report of Lelu et al., factors such as dry or humidity of soil had no significant effect on oocyst recovery (26). On the other hand, Toxopalsma oocyst has a resistant wall consisted of several layers, which causes difficult DNA extraction.
Toxoplasma gondii strains classified into three genotypes including I, II, III and some atypical genotypes (9–12). It is obvious that the prevalence of three genotypes is variable in different parts of the world. For example in the study of Howe and Sibley, type III was more common in animals than in human toxoplasmosis (27). Dubey et al. showed the predominance of type I in free- ranging chickens from Brazil (28). In another study, the predominant genotype was type III in free- ranging chickens and ducks from Egypt (29). Genotype I predomination was shown in the study of soil samples from three cities in Poland (15). It seems that ecological and geographical conditions could lead to the different prevalence of these types in different regions.
The present study was performed to investigate T. gondii genotypes in soil samples from Tehran, Iran for the first time. From 13 positive samples, 9 were type III (69%), 3 were mixed of type I and III (23%) and 1 of the samples was type I (8%). Zia-Ali et al. found the type III as dominant genotype in bird hosts in Iran (30); also he showed the distribution of 70% of type III and 30% of type II in animals (31). Behzadi et al. had reported 85.7% (18/22) of human and mice isolates infected with type II (32). Although the resources of our sampling are different from Zia-Ali et al., the results of these two studies are correlated.
Conclusion
Toxoplasma gondii detection and genotyping with molecular methods such as multilocus PCR-RFLP is possible directly in soil samples. Type III is predominant in Tehran soil samples. More studies regarding to determination of the clonal patterns of T. gondii isolates in soil samples by several markers are recommended.
Acknowledgments
We would like to show our appreciation to the cooperation of all staff of Parasitology and Mycology Department, School of Medicine, Tehran University of Medical Sciences. This research is a part of Ph.D Dissertation supported by Tehran University of Medical Sciences (Grant No: P/760). The authors declare that there is no conflict of interest.
References
- 1.Dubey JP. The history of Toxoplasma gondii-the first 100 years. J Eukaryot Microbiol. 2008;55(6):467–75. doi: 10.1111/j.1550-7408.2008.00345.x. [DOI] [PubMed] [Google Scholar]
- 2.Dubey JP. Toxoplasmosis of animals and humans. 2nd ed. Boca Raton: CRC Press; 2010. [Google Scholar]
- 3.Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev. 2012;25(2):264–96. doi: 10.1128/CMR.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumetre A, Darde ML. How to detect Toxoplasma gondii oocysts in environmental samples? FEMS Microbiol Rev. 2003;27(5):651–61. doi: 10.1016/S0168-6445(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 5.Weiss LM, Dubey JP. Toxoplasmosis: A history of clinical observations. Int J Parasitol. 2009;39(8):895–901. doi: 10.1016/j.ijpara.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asmar M, Amirkhani A, Piazak N, Hovanesian A, Kooloobandi A, Etessami R. Toxoplasmosis in Iran, results of a seroepidemiological investigation. Bull Soc Path Ex. 1997;90:19–21. [PubMed] [Google Scholar]
- 7.Frenkel JK, Dubey JP. Effects of freezing on the viability of Toxoplasma oocysts. J Parasitol. 1973;59(3):587–8. [PubMed] [Google Scholar]
- 8.Afonso E, Lemoine M, Poulle ML, Ravat MC, Romand S, Thulliez P, Villena I, Aubert D, Rabilloud M, Riche B, Gilot-Fromont El. Spatial distribution of soil contamination by Toxoplasma gondii in relation to cat defecation behaviour in an urban area. Int J Parasitol. 2008;38(8-9):1017–23. doi: 10.1016/j.ijpara.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Cristina N, Darde ML, Boudin C, Tavernier G, Pestre-Alexandre M, Ambroise-Thomas P. A DNA fingerprinting method for individual characterization of Toxoplasma gondii strains: combination with isoenzymatic characters for determination of linkage groups. Parasitol Res. 1995;81(1):32–7. doi: 10.1007/BF00932414. [DOI] [PubMed] [Google Scholar]
- 10.Darde ML, Bouteille B, Pestre-Alexandre M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiological implications. J Parasitol. 1992;78(5):786–94. [PubMed] [Google Scholar]
- 11.Guo ZG, Johnson AM. Genetic characterization of Toxoplasma gondii strains by random amplified polymorphic DNA polymerase chain reaction. Parasitology. 1995;111(Pt2):127–32. doi: 10.1017/s0031182000064866. [DOI] [PubMed] [Google Scholar]
- 12.Howe DK, Honore S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35(6):1411–4. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature. 1992;359(6390):82–5. doi: 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- 14.Fazaeli A, Ebrahimzadeh A. A new perspective on and re-assessment of SAG2 locus as the tool for genetic analysis of Toxoplasma gondii isolates. Parasitol Res. 2007;101(1):99–104. doi: 10.1007/s00436-006-0449-8. [DOI] [PubMed] [Google Scholar]
- 15.Lass A, Pietkiewicz H, Modzelewska E, Dumetre A, Szostakowska B, Myjak P. Detection of Toxoplasma gondii oocysts in environmental soil samples using molecular methods. Eur J Clin Microbiol Infect Dis. 2009;28(6):599–605. doi: 10.1007/s10096-008-0681-5. [DOI] [PubMed] [Google Scholar]
- 16.Mizgajska-Wiktor H. Recommended method for recovery of Toxocara and other geohelminth eggs from soil. Wiad Parazytol. 2005;51(1):21–2. [PubMed] [Google Scholar]
- 17.Ghorbani M, Samii AH. Toxoplasmic lymphadenitis in Iran. J Trop Med Hyg. 1973;76(7):158–60. [PubMed] [Google Scholar]
- 18.Schwab KJ, McDevitt JJ. Development of PCR enzyme immunoassay oligoprobe detection method for Toxoplasma gondii oocyst, incorporating PCR controls. Appl Environ Microbiol. 2003;69:5819–25. doi: 10.1128/AEM.69.10.5819-5825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassaing S, Bessieres MH, Berry A, Berrebi A, Fabre R, Magnaval JF. Comparison between two amplification sets for molecular diagnosis of toxoplasmosis by real-time PCR. J Clin Microbiol. 2006;44(3):720–4. doi: 10.1128/JCM.44.3.720-724.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homan WL, Vercammen M, De Braekeleer J, Verschueren H. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol. 2000;30(1):69–75. doi: 10.1016/s0020-7519(99)00170-8. [DOI] [PubMed] [Google Scholar]
- 21.Aspinall TV, Guy EC, Roberts KE, Joynson DH, Hyde JE, Sims PF. Molecular evidence for multiple Toxoplasma gondii infections in individual patients in England and Wales: public health implications. Int J Parasitol. 2003;33(1):97–103. doi: 10.1016/s0020-7519(02)00230-8. [DOI] [PubMed] [Google Scholar]
- 22.Aspinall TV, Marlee D, Hyde JE, Sims PF. Prevalence of Toxoplasma gondii in commercial meat products as monitored by polymerase chain reaction--food for thought? Int J Parasitol. 2002;32(9):1193–9. doi: 10.1016/s0020-7519(02)00070-x. [DOI] [PubMed] [Google Scholar]
- 23.Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol. 2006;36(7):841–8. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Alvarado-Esquivel C, Estrada-Martinez S, Pizarro-Villalobos H, Arce-Quinones M, Liesenfeld O, Dubey JP. Seroepidemiology of Toxoplasma gondii infection in general population in a northern Mexican city. J Parasitol. 2010;97(1):40–3. doi: 10.1645/GE-2612.1. [DOI] [PubMed] [Google Scholar]
- 25.Dabritz HA, Conrad PA. Cats and Toxoplasma: implications for public health. Zoonoses Public Health. 2010;57(1):34–52. doi: 10.1111/j.1863-2378.2009.01273.x. [DOI] [PubMed] [Google Scholar]
- 26.Lelu M, Gilot-Fromont E, Aubert D, Richaume A, Afonso E, Dupuis E, et al. Development of a sensitive method for Toxoplasma gondii oocyst extraction in soil. Vet Parasitol. 2011;183(1-2):59–67. doi: 10.1016/j.vetpar.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–66. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- 28.Dubey JP, Graham DH, Blackston CR, Lehmann T, Gennari SM, Ragozo AMA, et al. Biological and genetic characterisation of Toxoplasma gondii isolated from chicken (Gallus domesticus) from Sao Paulo Brazil. Int J Parasitol. 2002;32:99–105. doi: 10.1016/s0020-7519(01)00364-2. [DOI] [PubMed] [Google Scholar]
- 29.Dubey JP, Graham DH, Dahl E, Hilali M, El-Ghaysh A, Sreekumar C, et al. Isolation and molecular characterization of Toxoplasma gondii from chickens and ducks from Egypt. Vet Parasitol. 2003;114:89–95. doi: 10.1016/s0304-4017(03)00133-x. [DOI] [PubMed] [Google Scholar]
- 30.Zia-Ali N, Keshavarz-Valian H, Rezaian M, Khoramizade MR, Kazemi B, Fazaeli A, Darde M. Molecular characterization of Toxoplasma gondii from bird hosts. Iranian J Publ Health. 2005;34(3):27–30. [Google Scholar]
- 31.Zia-Ali N, Fazaeli A, Khoramizadeh M, Ajzenberg D, Darde M, Keshavarz-Valian H. Isolation and molecular characterization of Toxoplasma gondii strains from different hosts in Iran. Parasitol Res. 2007;101(1):111–5. doi: 10.1007/s00436-007-0461-7. [DOI] [PubMed] [Google Scholar]
- 32.Behzadi R, Roohvand F, Razavi MR, Hovanessian A, Assmar M. Genotyping of Toxoplasma gondii strains isolated from patients and mice by PCR-RFLP assay. Iranian J Biotechnol. 2003;1(2):82–6. [Google Scholar]