Summary
Recent studies show that bioactive lipids are important regulators for stem cell survival and differentiation. The sphingolipid ceramide and its derivative, sphingosine-1-phosphate (S1P), can act synergistically on embryonic stem (ES) cell differentiation. We show here simple methods to analyze sphingolipids in differentiating ES cells and to use ceramide and S1P analogs for the guided differentiation of mouse ES cells toward neuronal and glial lineage.
Keywords: Ceramide, sphingolipid, sphingosine-1-phosphate, neuroprogenitor, oligodendrocyte precursor, apoptosis, teratoma
1. Introduction
The use of differentiating stem cells, regardless of being derived from embryonic stem (ES) cells or induced pluripotent stem (iPS) cells is hampered by the persistence of residual pluripotent stem (rPS) cells in the culture of differentiating stem cells. Even though, these cells can be present in very low abundance (<1:106) they may impose a significant risk of teratoma formation when used for transplantation. Previous studies showed that stem cell transplantation can lead to the formation of teratomas even if similar protocols were used for the differentiation and transplantation of ES cells (1–19). Teratomas are stem cell-derived tumors that are fatal if they occur in the brain or heart. The potential for teratoma formation is thus a major safety concern when using larger numbers of ES or iPS cell-derived cells (16). We have introduced for the first time a novel strategy to get rid of residual pluripotent stem (rPS) cells that could form teratomas from stem cells grafts (1, 20). This strategy is based on the use of sphingolipids for guided stem cell differentiation, which promotes different lineage specification depending on the combination of lipid effectors used.
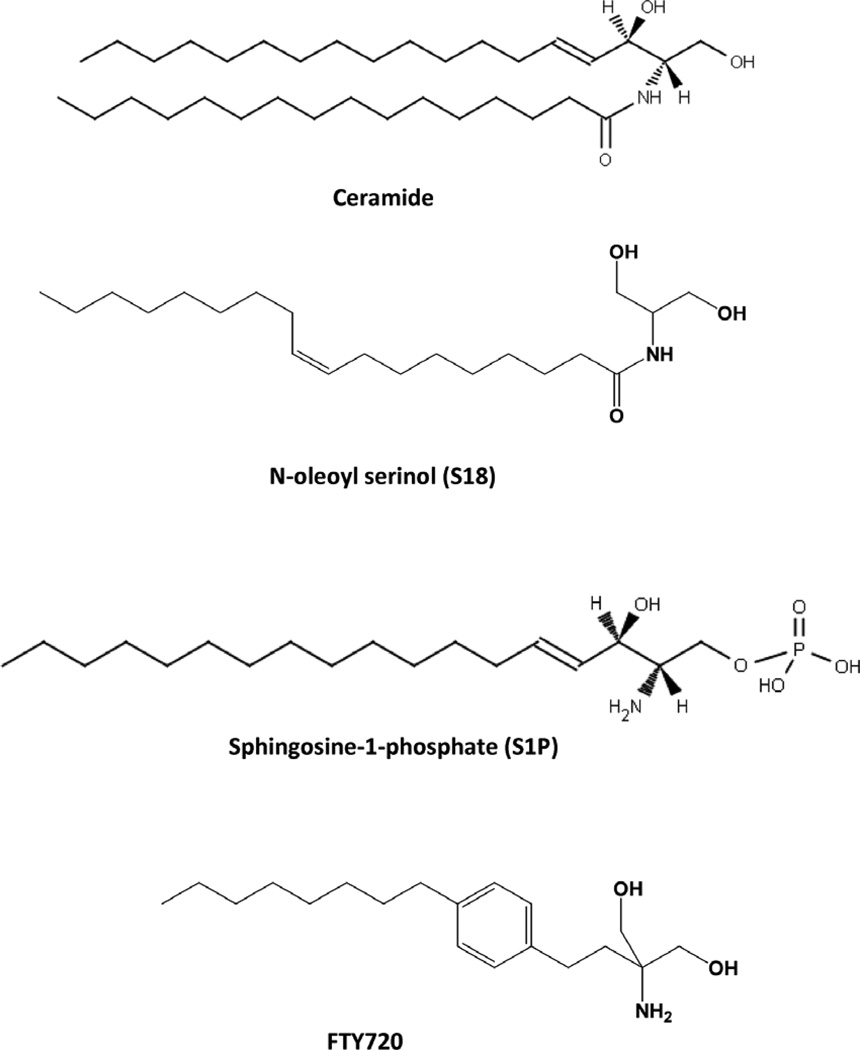
Our studies have shown that rPS cells co-express the pluripotency transcription factor Oct-4 and prostate apoptosis response 4 (PAR-4), a protein that makes rPS cells sensitive toward ceramide and its analogs (1). PAR-4 is an endogenous inhibitor of atypical PKC (aPKC) when associated with ceramide or the novel ceramide analog N-oleoyl serinol (S18, Fig. 1). S18 was for the first time synthesized and its association with aPKC analyzed in our laboratory (1, 20–25). If PAR-4 expression is low S18 or ceramide bind and activate aPKC. Recently, aPKC activation and downstream activation of NF-κB has been found to be critical for neural differentiation of ES cells (26). Therefore, ceramide or S18 can promote neural differentiation via activation of aPKC and its target protein NF-κB (21, 25). However, if PAR-4 expression is high ceramide or S18-associated aPKC binds to PAR-4, which inhibits the enzyme and down-regulates NF-κB-dependent stem cell survival.
Figure 1.
Structure of the sphingolipids and sphingolipid analogs used for stem cell differentiation
We have shown that this mechanism leads to ceramide or S18-induced apoptosis in PAR-4 expressing rPS cells while it promotes neuronal differentiation of PAR-4 (-/low) NPs (1, 25, 27, 28). We have also shown that the ceramide derivative S1P and its pro-drug analog FTY720 promote cell survival and differentiation of NPs toward oligodendroglial lineage because these cells express the S1P receptor S1P1 (20, 29). In particular useful is the combined administration of ceramide/S18 and S1P/FTY720 to in vitro differentiating ES cells (Fig. 1). Since rPS cells express PAR-4 (but not S1P1) and NPs express S1P1 (but not PAR-4) ceramide/S18 eliminates rPS cells and S1P/FTY720 promotes oligodendroglial differentiation of the surviving NPs. Following protocols using sphingolipids and their analogs, ES cells can be differentiated to neuronal or oligodendroglial lineage providing cells for in vitro and in vivo studies.
2. Materials
2.1. Media for the cultivation and differentiation of mouse ES cells (calculated for 100 ml of medium)
FM10 (Feeder cell medium)
89 ml of DMEM (with L-glutamine and sodium pyruvate)
10 ml of heat-inactivated FBS
1 ml 100X stock of penicillin/streptomycin/amphotericin B (fungizone)
(see Note 1)
KSR15 (ES cell medium for cells grown on feeders)
81 ml of Knockout-DMEM
15 ml of Knockout Serum Replacement (KSR)
1 ml of 100x L-glutamine (200 mM)
1 ml of 100x Non-essential amino acids
1 ml of 100x penicillin/streptomycin/amphotericin B
100 µl of ESGRO (LIF)
180 µl of 2-mercaptoethanol
ES15 (ES cell medium for cells grown feeder-free)
81 ml of Knockout-DMEM
15 ml of heat-inactivated ES qualified FBS
1 ml of 100x L-glutamine (200 mM)
1 ml of 100x penicillin/streptomycin/amphotericin B
1 ml of 100x Non-essential amino acids
100 µl of ESGRO (LIF)
180 µl of 2-mercaptoethanol
EB1 (Suspension EB medium)
87 ml of Knockout-DMEM
10 ml of heat-inactivated ES-qualified FBS
1 ml of 100x penicillin/streptomycin/amphotericin B
1 ml of 100x L-glutamine (200 mM)
1 ml of 100x Non-essential amino acids
EB2 (Attached EB medium)
96 ml of DMEM/F12 50/50
1 ml of 100x penicillin/streptomycin/amphotericin B
1 ml of 100x L-glutamine (200 mM)
1 ml of 100x Non-essential amino acids
1 ml of N-2 supplement (100x)
NP (NP medium)
95.5 ml of DMEM/F12 50/50
1 ml of 100x penicillin/streptomycin/amphotericin B
1 ml of 100x L-glutamine (200 mM)
1 ml of 100x Non-essential amino acids
1 ml of N-2 supplement (100x)
500 µl of basic fibroblast growth factor (FGF-2) stock (see Note 2)
Differentiation medium
91.75 ml Neurobasal medium
5 ml of heat-inactivated ES-qualified FBS
1 ml of 100x penicillin/streptomycin/amphotericin B
250 µl of L-glutamine (200 mM stock)
2 ml of 50x B27 supplement
(see Note 3)
Trypsinization
0.25% trypsin/0.2% EDTA in PBS (see Note 4)
Freeze medium
Knockout DMEM with 20% heat-inactivated ES cell-qualified FBS and 10% DMSO
Gelatin coating solution
Dissolve 2 g of gelatin, 300 Bloom in 100 ml of deionized water and autoclave. Gelatin should be completely dissolved after being autoclaved. The 2% gelatin stock solution can be kept refrigerated until further use. For gelatin coating dilute stock solution 1:20 in sterile water and incubate tissue culture dishes for 2 h at room temperature. Then, remove solution and let dishes dry in the hood for 2 h.
3.2. Solutions and reagents for lipid analysis
(Important: see Note 5 for precautionary measures to avoid toxic or hazardous conditions)
Reagents for Folch extraction of lipids
CHCl3/CH3OH (2:1, vol:vol)
Running solvent for TLC
CHCl3/CH3OH (95:1, vol:vol)
Staining solution for lipid detection on TLC
3% cupric acetate in 5% phosphoric acid
3. Methods
3.1. Propagation and differentiation of mouse embryonic stem cells
Overview
In vitro neuronal differentiation of mouse ES cells (ES-J1, ES-D3) followed a serum deprivation protocol as described previously (27, 28, 30–33).
Coat a 100 mm tissue culture dish with 0.1% sterile gelatin solution (freshly prepared from 2% stock) by incubation for 2 h at room temperature. Remove the solution and dry for 2 h in hood with lid only partially covering the dish. Rinse once with FM10 medium.
Seed the dish with 3 × 106 irradiated mouse embryonic feeder fibroblasts (MEFs). Alternatively, feeder fibroblasts mitotically inactivated with mitomycin c can also be used. Cultivate the fibroblasts for 2 days in 10 ml FM10 medium. Mitotically inactivated MEFs are available from commercial sources.
3.2. Propagation of undifferentiated ES cells on feeder fibroblasts
Thaw frozen ES cells and suspend cells in 10 ml freshly prepared KSR15 medium. Spin cells down at 200xg for 5 min. Resuspend cells in 20 ml of KSR15 and plate them on top of the feeder fibroblasts. Do not completely remove supernatant after centrifugation to minimize loss of cells.
After one day of cultivation, replace medium with 20 ml of fresh KSR15. At this stage, the ES cells have attached to the feeder cells, although they may not be clearly visible (small rounded cells on flat fibroblasts).
After another day of cultivation (day 3 after seeding of the ES cells), replace medium again with 20 ml of fresh KSR15.
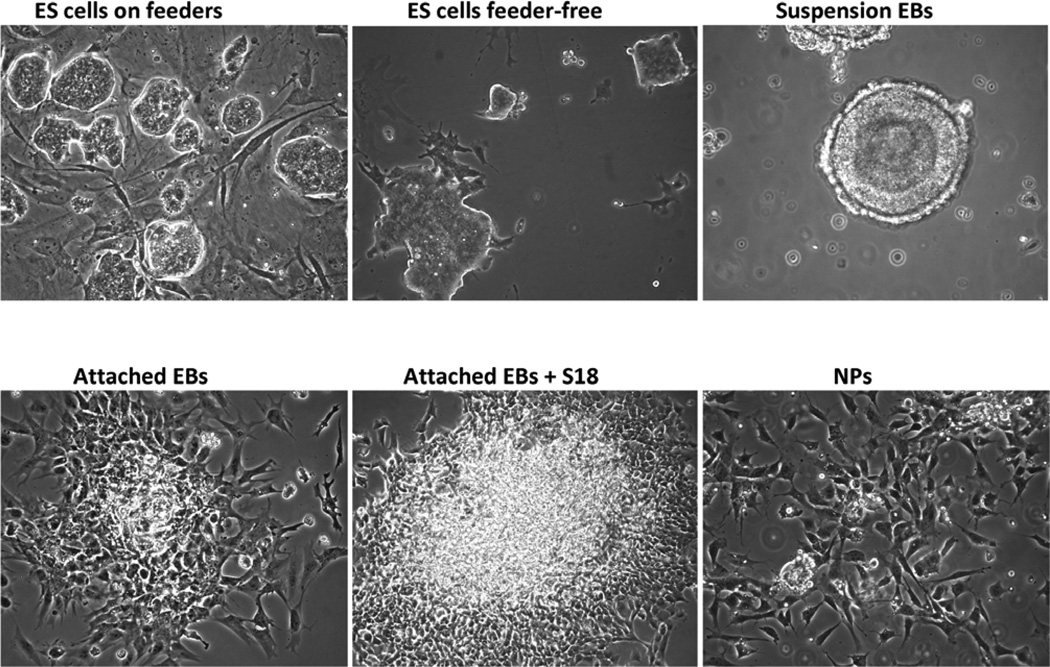
On the fourth day after seeding on the fibroblasts, colonies of ES cells should be clearly visible (Fig. 2). If not change medium again and cultivate for another day. When colonies reach a size as shown in Fig. 2 proceed to feeder-free culture.
Figure 2.
Different stages of mouse ES cell differentiation
3.3. Removal of feeder fibroblasts and propagation of ES cells in feeder-free culture
Prepare four gelatin-coated 100 mm tissue culture dishes without feeder cells.
Rinse the ES cells on the feeders with 10 ml sterile PBS.
Add 4 ml of 0.25% trypsin/EDTA and incubate cells for 5 min at 37 °C.
Immediately add 10 ml of pre-warmed KSR15 and pipette up and down 3 times. Try to rid the ES cell suspension of most of feeder cell patches.
Pellet the cells by centrifugation at 200xg for 5 min.
Resuspend cells in 20 ml of KSR15 and plate them onto a tissue culture dish NOT coated with gelatin. Incubate for 2 h at 37 °C. The feeders will attach while most of the ES cells will stay in suspension.
Take the medium with the ES cells off the plate and transfer it to a gelatin-coated tissue culture dish.
On the next day, change medium to 20 ml of ES15 medium. Change medium every day for another two days. DO NOT let the ES cell colonies exceed 50–70% of the dish surface (Fig. 2). Once cells become too dense, the edges of the colonies start to look ragged. If this happens cells become prematurely differentiated, which will compromise their pluripotency.
3.4. Preparation of suspension embryoid bodies (EBs)
Trypsinize and freeze cells that are not wanted for the preparation of EBs (see freeze medium).
Trypsinize the residual dishes with 0.25% trypsin/EDTA for 2 min. Cells should detach, but still form aggregates. Immediately add 10 ml of EB1 medium to neutralize the trypsin. Then transfer cell aggregates to a 50 ml plastic tube using a 25 ml pipette. Rinse dish with another 10 ml of EB1 medium and combine cell suspensions. Centrifuge for 5 min at 50–100xg or let the cell aggregates settle without centrifugation. Remove medium supernatant and gently wash cell aggregates with 10 ml fresh EB1 medium. Resuspend cell aggregates in 30 ml of EB1 medium and seed them on bacterial dishes. It is important not to use tissue culture dishes to avoid premature attachment of suspension EBs.
Change medium and inspect the cell aggregates every day for the next 3–5 days. The cell aggregates should round up and first form a clearly distinguishable epithelial outer cell layer (primitive endoderm), followed by the formation of basal lamina and an inner cell layer (primitive ectoderm) surrounding a cavity (Fig. 2). This cavity is developmentally equivalent to the pro-amniotic cavity of mammalian embryos. To minimize loss of EBs during medium change tilt the dish and collect the EBs at one side of the dish. DO NOT attempt to completely remove the old medium, but leave some medium behind to maintain the EB suspension.
3.5. Preparation of attached EBs and cultivation of EB-derived cells (EBCs)
Transfer the suspension EBs to tissue culture dishes. Coating is not necessary. Typically, the suspension EBs from one bacterial dish are sufficient to yield one tissue culture dish with attached EBs. Let the EBs attach overnight (Fig. 2).
Change medium to EB2 medium. Keep replacing media daily for 5–7 days. Cells will migrate out of the attached EB. These cells are termed EB-derived cells or EBCs. After sufficient EBCs have been migrated out of the attached EBs, cells can be treated with ceramide analogs or S1P/S1P analogs (see section 3.8). Cells can also be used to generate neuroprogenitors (NPs) prior to drug treatment (see section 3.8).
3.6. Induction of neural differentiation and cultivation of neuroprogenitors (NPs)
Coat tissue culture dishes with p-ornithin laminin from commercially available stock solutions (200 µl p-ornithin or 100 µl laminin in 100 ml of sterile water) by first incubating with p-ornithin and then laminin each for one hour at 37 C. In between and after the two coating steps rinse with sterile water, then store the coated dishes in PBS until use (DO NOT extend storage for more than one day).
Dissociate EBCs with 2 ml of 0.05–0.25% trypsin/EDTA for 5 min at 37 °C. Immediately neutralize the trypsin with 10 ml of EB1 medium then harvest cells by centrifugation at 200xg for 5 min. The dissociation of attached EBs is a critical step that determines the yield of viable NPs (Fig. 2). Harvest cells by centrifugation, resuspend again in NP medium and seed cells on p-ornithin laminin-coated tissue culture dishes or coated cover slips if immunocytochemistry is to be performed (see Note 6).
Cultivate cells for 3–6 days in NP medium, change medium every day. NPs will form rosettes. After 24–48 h in NP medium, cells can be treated with ceramide or S1P analogs as described in section 2. (see Note 7).
Harvest cells by trypsinization with 0.05% trypsin/EDTA. Neutralize trypsin with soybean trypsin inhibitor. Cells can now be either re-plated onto fresh p-ornithin laminin-coated dishes and differentiated or used for transplantation.
3.7. Terminal differentiation to neurons and glia
Incubate replated NPs (treated or not treated with ceramide or S1P analogs) with differentiation medium for up to 10 days.
Assess differentiation lineage by using the respective markers for neuronal, astrocytic, and oligodendroglial differentiation (see section 3.10. for immunocytochemistry protocol).
3.8. Treatment with ceramide and S1P analogs
1. Treatment of attached EBs
1. Two to three days after attachment of suspension EBs to the tissue culture dish, the culture can be incubated with ceramide, S1P, ceramide or S1P analogs, or a combination of these compounds (Figs. 1 and 2). Typically, the following concentrations are useful to eliminate residual pluripotent cells or to direct lineage specification of differentiating cells in the EBs (see Note 8):
N-acetyl sphingosine (C2 ceramide): 5–20 µM*
N-palmitoyl sphingosine (C16 ceramide): 0.5–2.0 µM**
N-oleoyl serinol (S18): 50–100 µM
Sphingosine-1-phosphate (S1P): 0.2–1.0 µM***
FTY720: 0.1–0.5 µM****
* C2 ceramide is water-soluble in the given concentration range, however, may have non-specific and lytic effects. If possible the use of C16 ceramide or S18 is preferred.
** C16 ceramide is not water-soluble in the given concentration range, unless prepared as a 1000x stock solution in ethanol supplemented with 2% dodecane. When using a 1:1000 dilution of this stock, controls with respective final concentration of ethanol and dodecane have to be prepared. If possible the use of the water-soluble S18 is preferred.
*** S1P is only partially water-soluble and is to be prepared as 1000x stock in 1 mg/ml bovine serum albumin/PBS.
**** FTY720 is water-soluble in the given concentration range, however, prolonged exposure at higher concentration may down-regulate S1P1 receptor expression. At 300 nM for 48 h, S1P1 expression was clearly detectable in OPCs (29).
2. After treatment, EBs and EBCs can be dissociated by trypsinization and re-plated to generate NPs or used for transplantation.
2. Treatment of EBCs and NPs
Dissociate cells from attached EBs by trypsinization and re-plate as described in section 3.6. These cells may have been treated with the above-mentioned compounds or not. However, DO NOT continue or start treatment in the first 24 hrs after seeding of the cells. Typically, EBCs will first attached and recover before treatment starts. Treatment is performed with compounds and dosages given in section 3.8.
After 2–3 days of drug treatment, NPs can be used for transplantation or further differentiated as described in section 3.7.
3. Treatment of other stages of ES cell differentiation
Various stages of ES cell differentiation, including undifferentiated ES cells on feeders, feeder-free ES cells, and suspension EBs have been treated with ceramide or ceramide analogs. The outcomes are not as defined with respect to lineage specification as achievable when later stages of ES cell differentiation are used for treatment. Therefore, treatments of earlier differentiation stages are not included in this protocol. However, a brief summary of treatment outcomes will be discussed to provide the opportunity of using protocols published in other studies. For example, Salli et al. (2009) describe the use of nano-liposomal C6 ceramide for the cultivation of undifferentiated ES cells in feeder-free culture (34). Krishnamurthy et al. (2007) describe the use of C16 ceramide or S18 for the restoration of primitive ectoderm in suspension EBs (35).
3.9. Determination of ceramide and S1P using sphingolipid analysis
(Important: see Note 5 for precautionary measures to avoid toxic or hazardous conditions)
1. Sphingolipid extraction
Resuspend EBCs or NPs in 500 µl PBS and add 2.5 ml of CHCl3/CH3OH (2:1, vol/vol) in a Pyrex glass tube capped with Teflon liner. Vortex for 30 seconds, then sonicate for 30 min in an ultrasonication bath (e.g., Branson) or shake for 1 h at room temperature (see Notes 5 and 9)
Centrifuge tube at low speed for 20 min. Two phases and the interphase should be clearly visible. The upper phase contains (partially) water-soluble lipids such as gangliosides and a major portion of phospholipids including S1P. If a comprehensive analysis is desired the upper phase should be removed and kept separately. The lower phase contains ceramide and neutral lipids and should be carefully recovered without disturbing the interphase. The interphase contains denatured protein which should be kept and air dried. After solubilization in SDS sample buffer, this protein fraction can be used for protein quantification, SDS-PAGE, or other methods of protein analysis.
Evaporate the organic solvent from the lower phase with a stream of nitrogen gas. Alternatively, organic solvent can be removed using a rotation evaporator.
Dissolve the lipid residue in CHCl3/CH3OH (1:1, vol/vol) and store at −20 °C until further analysis. Typically, lipids extracted from 10–50 mg cells are re-dissolved in 100–500 µl organic solvent to allow for analysis by TLC.
2. Sphingolipid analysis by thin layer chromatography (TLC)
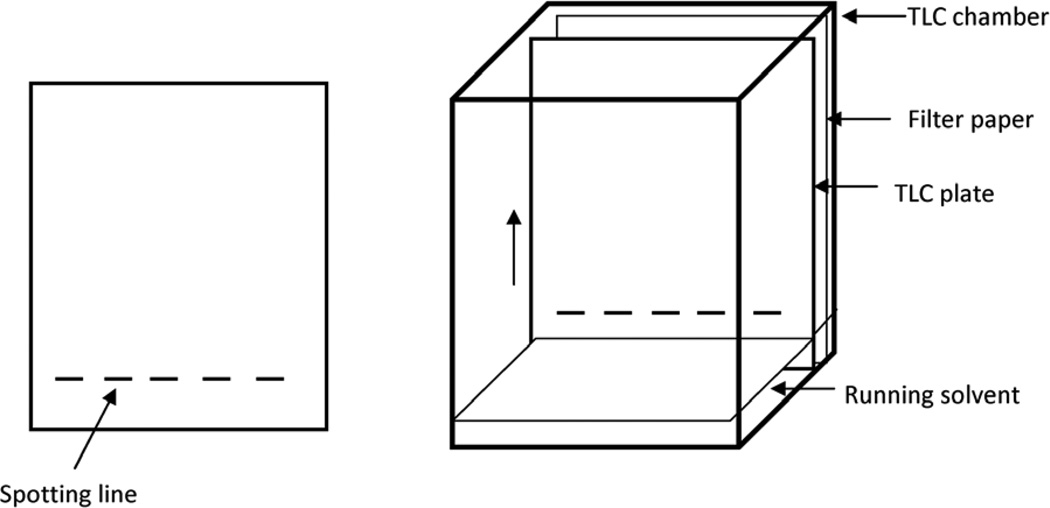
Prepare chromatography chamber (alternatively, a glass beaker can be used) and fill it to about 1/2 inch (1 cm) with the running solvent CHCl3/CH3OH (95:5, vol/vol). For equilibration of the chamber atmosphere with organic solvent a filter paper is placed at the wall of the chamber and saturated with organic solvent all the way up to the rim of the chamber (Fig. 3). To avoid loss of organic solvent the chamber has to be covered with a thick glass plate and a weight placed on top of the plate. Alternatively, aluminum foil can be used to cover the chamber or beaker and secured with a wrapped-around rubber band.
Cut a TLC plate using a glass cutter to fit into the chromatography chamber (alternatively, pre-cut plates are commercially available). Draw spotting lines on the silica gel coat of the plate at about 1 inch (2 cm) from the bottom edge of the plate using a soft pencil as shown in Fig. 3. Each line should be about 1/8−1/4 inch (2–6 mm) in width and 1/4 (5–6 mm) apart from each other. Be careful not to hurt the surface of the silica gel.
Apply the sample (lipids in organic solvent) and lipid standards with a Hamilton syringe or glass tube. The application should be steady and repeatedly on top of the spotting line. Alternatively, pipette tips can be used, although they tend to form drops that hamper a steady application. Be careful not to directly touch the silica gel surface. The amount of lipid to be applied depends on the normalization method. Typically, a lipid amount equivalent to equal amounts of protein or lipid phosphate is applied to each lane on the plate (see Note 10).
After application of the samples and standards, the plate is placed into the chromatography chamber without letting the running solvent get in immediate contact with the applied lipids (Fig. 3). Also ensure that the application lines are dry before placing the plate into the chamber. The running solvent will be soaked into the silica gel and migrate from the bottom to the top (arrow in Fig. 3). The lipids will dissolve into the migrating solvent and co-migrate according to their solubility in organic solvents. More apolar lipids will migrate faster leading to a separation of the lipid mixture into individual lipid species. After the running solvent has reached the top of the plate, the TLC is complete and the plate will be taken out of the chamber for air drying and subsequent staining of the lipids.
Staining of lipid species on TLC plate. (important: see Note 5 for precautionary measures). Spray TLC staining reagent on the surface of the plate until it is wet. Be careful not to inhale the vapors of the spraying reagent. Alternatively, the plate can be briefly dipped into a bath with staining reagent and the back of the glass plate wiped down with a tissue before heating the plate. The plate will be heated (silica gel surface up!) for 5–15 min at 150–180 °C until the charring reaction leads to the appearance of dark bands. Once standards and sample lipids are visible, the plate is removed from the heater for cooling at room temperature. The plate should be scanned immediately because bands will fade over time. For short term storage wrap the plate into aluminum foil and store it at a secure and dark place.
Figure 3.
Assembly for TLC
(see Note 11 for limitations of the staining reaction)
3.10. Immunocytochemistry for lipids and lipid receptors
(see cautionary note 12 for fixation reagents and note 5 for other precautionary measures)
Fix cells or sections with freshly prepared 4% p-formadehyde in PBS for 20 min at room temperature. Wash twice with PBS.
If only surface staining is desired cells or sections do not need to be permeabilized. However, if immunocytochemistry for lipids or proteins inside of cells is desired permebilization can be helpful. For example, a mild permeabilization with 0.2% Triton X-100 in PBS for 5 min at room temperature has not been shown to significantly remove lipids from the sample, but it may alter the distribution of particular lipids in cellular membranes (36). (see Note 13)
Block binding to non-specific antigens by incubation of the sample with 3% ovalbumin/5% donkey serum (secondary antibodies raised in donkey are used) in PBS for 60 min at room temperature or 37 °C. Wash once with PBS.
Incubate with primary antibody diluted in 0.1% ovalbumin/PBS. The proper dilution will need to be tested. Typically, final antibody concentrations of 5 µg/ml will yield excellent results. The antibody reaction can be performed for 2 h at 37 °C, 4 h at room temperature, or overnight at 4 °C (recommended).
Wash sample three times for 10 min with PBS.
Incubate with secondary antibody as described for the first antibody. However, shorten incubation time at room temperature to 2 h and avoid overnight incubation with the secondary antibody. Then wash sample three times for 10 min with PBS. Embed in mounting medium. Note that additional staining for nuclei with Hoechst 33258 dye (1 µg/ml) can be included into the first washing step. After drying, the sample is ready for fluorescence microscopy.
Additional staining reactions such as TUNEL or BrdU assays can be included prior to antibody staining following protocols provided by the manufacturers.
Acknowledgement
This study was in part supported by the NIH grants R01AG034389 and R01NS046835, and a March of Dimes grant 6-FY08-322.
Footnotes
If L-glutamine or sodium pyruvate are not included in the medium they have to added separately to a final concentration of 2 mM or 1 mM, respectively.
Dissolve 10 µg FGF-2 in 100 µl sterile PBS, then add 10 µl of this solution to 500 µl of 0.1% BSA to generate aliquots that can be kept at −20 °C.
To eliminate astrocytes differentiation medium can be supplemented with 20 µM Ara C.
It is recommended to be diluted 1:5 when using for plating of EBCs or NPs; alternatively, non-enzymatic dissociation solution can be used instead of trypsin.
CAUTION: Lipid extraction and analysis requires the use of organic solvents that are toxic for the liver and the nervous system. Therefore, all procedures involving organic solvents must be carried out in a chemical fume hood. The use of organic solvents produces waste that has to be properly disposed of following institutionally enforced regulations and protocols. When handling organic solvents, glass pipettes and glass containers have to be used. The reagent for staining of lipids on TLC contains caustic and acidic components. Therefore, extra care and protection is required such as double gloves and face shield. Do not inhale the spraying reagent or organic vapors. The staining reaction uses a heater in the chemical fume hood which requires that organic solvents are placed in safe distance from the heating device to avoid a fire hazard. It is also recommended to handle the hot TLC plate with heat-resistant gloves or tools to avoid burns.
Prolonged exposure to trypsin can damage the cells and dramatically reduce survival of EBCs after re-plating. Therefore, milder methods of dissociation such as non-enzymatic dissociation solutions should be tested. It is also critical to add EB1 as soon as dissociation is visible. However, DO NOT keep cells for extended time in EB1 medium because it will induce premature differentiation otherwise. Remove medium and wash cells with NP medium.
If the initial survival of EBCs or NPs is low double or triple the concentration of FGF-2 in NP medium.
Incubation with ceramide or ceramide analogs will eliminate rPS cells and enhance neuronal lineage specification, while the addition of S1P or S1P analogs will direct differentiation toward oligodendroglial lineage.
The use of pipette tips should be avoided and glass pipettes used instead. However, if glass pipettes for small volumes are not available pipette tips (NOT plastic pipettes) can be of short term use. In this case, it is advisable to equilibrate the atmosphere within the tip and the pipettor by repeatedly pipetting up and down organic solvent before using it with the sample.
To detect ceramide lipid equivalent to 200–300 µg protein (2–3 mg or 3–5×106 cells) will yield a detectable band using the staining method described in this protocol. Application of an amount of 0.5–1 µg of lipid standard is recommended.
The methods described in this protocol are not suitable for staining of low abundance lipids such as S1P. Further, these methods have to be modified if lipids with a different range of polarity (e.g., phospholipids) are to be analyzed. Many of these modified methods will use radioactive labeling of lipid precursors, which will impose additional hazards. For a more comprehensive lipid analysis it is recommended to use the commercially available service of sphingolipidomics core facilities. In this case, it is also recommended to contact the core facility prior to sample preparation to receive the proper protocol to be used for the sample analysis.
The following methods are only applicable to cultivated cells or cryofixed tissue sections. Paraffin sections cannot be used for immunocytochemistry because organic solvents will dissolve and remove the lipids from the sample. Therefore, it is also not possible to use organic solvents such as methanol or acetone for fixation. The recommended fixation reagent is 4% p-formaldehyde in PBS, which should be prepared fresh. However, frozen stock can also be used for a short period of time. Formaldehyde is a toxic reagent and any steps of the fixation procedure handling open containers should be performed in a chemical fume hood.
It is recommended to perform immunocytochemistry with or without permeabilization and compare the results. In the case that permeabilization cannot be avoided, it is advisable to first perform immunocytochemistry for the lipid without permeabilization, then fix the cells again, and perform the permeabilization afterwards. Proteins inside of the cells can then be immunodetected performing a second round of incubation with the respective primary and secondary antibodies.
References
- 1.Bieberich E, Silva J, Wang G, Krishnamurthy K, Condie BG. Selective apoptosis of pluripotent mouse and human stem cells by novel ceramide analogues prevents teratoma formation and enriches for neural precursors in ES cell-derived neural transplants. J Cell Biol. 2004;167:723–734. doi: 10.1083/jcb.200405144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yanai J, Doetchman T, Laufer N, Maslaton J, Mor-Yosef S, Safran A, Shani M, Sofer D. Embryonic cultures but not embryos transplanted to the mouse's brain grow rapidly without immunosuppression. Int J Neurosci. 1995;81:21–26. doi: 10.3109/00207459509015295. [DOI] [PubMed] [Google Scholar]
- 3.Wakitani S, Takaoka K, Hattori T, Miyazawa N, Iwanaga T, Takeda S, Watanabe TK, Tanigami A. Embryonic stem cells injected into the mouse knee joint form teratomas and subsequently destroy the joint. Rheumatology (Oxford) 2003;42:162–165. doi: 10.1093/rheumatology/keg024. [DOI] [PubMed] [Google Scholar]
- 4.Teramoto K, Hara Y, Kumashiro Y, Chinzei R, Tanaka Y, Shimizu-Saito K, Asahina K, Teraoka H, Arii S. Teratoma formation and hepatocyte differentiation in mouse liver transplanted with mouse embryonic stem cell-derived embryoid bodies. Transplant Proc. 2005;37:285–286. doi: 10.1016/j.transproceed.2004.12.120. [DOI] [PubMed] [Google Scholar]
- 5.Swijnenburg RJ, Tanaka M, Vogel H, Baker J, Kofidis T, Gunawan F, Lebl DR, Caffarelli AD, de Bruin JL, Fedoseyeva EV, Robbins RC. Embryonic stem cell immunogenicity increases upon differentiation after transplantation into ischemic myocardium. Circulation. 2005;112:I166–I172. doi: 10.1161/CIRCULATIONAHA.104.525824. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Pernaute R, Studer L, Ferrari D, Perrier A, Lee H, Vinuela A, Isacson O. Long-term survival of dopamine neurons derived from parthenogenetic primate embryonic stem cells (cyno-1) after transplantation. Stem Cells. 2005;23:914–922. doi: 10.1634/stemcells.2004-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, Gu Y, Ishii M, Fujimiya M, Qi M, Nakamura N, Yoshikawa T, Sumi S, Inoue K. In vivo functioning and transplantable mature pancreatic islet-like cell clusters differentiated from embryonic stem cell. Pancreas. 2003;27:e34–e41. doi: 10.1097/00006676-200308000-00021. [DOI] [PubMed] [Google Scholar]
- 8.Fujikawa T, Oh SH, Pi L, Hatch HM, Shupe T, Petersen BE. Teratoma formation leads to failure of treatment for type I diabetes using embryonic stem cell-derived insulin-producing cells. Am J Pathol. 2005;166:1781–1791. doi: 10.1016/S0002-9440(10)62488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong SP, Tsang KS, Chan AB, Lu G, Poon WS, Li K, Baum LW, Ng HK. Trophism of neural progenitor cells to embryonic stem cells: neural induction and transplantation in a mouse ischemic stroke model. J Neurosci Res. 2007;85:1851–1862. doi: 10.1002/jnr.21319. [DOI] [PubMed] [Google Scholar]
- 10.Choi D, Oh HJ, Chang UJ, Koo SK, Jiang JX, Hwang SY, Lee JD, Yeoh GC, Shin HS, Lee JS, Oh B. In vivo differentiation of mouse embryonic stem cells into hepatocytes. Cell Transplant. 2002;11:359–368. [PubMed] [Google Scholar]
- 11.Bielby RC, Boccaccini AR, Polak JM, Buttery LD. In vitro differentiation and in vivo mineralization of osteogenic cells derived from human embryonic stem cells. Tissue Eng. 2004;10:1518–1525. doi: 10.1089/ten.2004.10.1518. [DOI] [PubMed] [Google Scholar]
- 12.Arnhold S, Klein H, Semkova I, Addicks K, Schraermeyer U. Neurally selected embryonic stem cells induce tumor formation after long-term survival following engraftment into the subretinal space. Invest Ophthalmol Vis Sci. 2004;45:4251–4255. doi: 10.1167/iovs.03-1108. [DOI] [PubMed] [Google Scholar]
- 13.Baker M. Stem cells: Fast and furious. Nature. 2009;458:962–965. doi: 10.1038/458962a. [DOI] [PubMed] [Google Scholar]
- 14.Leor J, Gerecht S, Cohen S, Miller L, Holbova R, Ziskind A, Shachar M, Feinberg MS, Guetta E, Itskovitz-Eldor J. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93:1278–1284. doi: 10.1136/hrt.2006.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum B, Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Cancer Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- 16.Lee AS, Tang C, Cao F, Xie X, van der Bogt K, Hwang A, Connolly AJ, Robbins RC, Wu JC. Effects of cell number on teratoma formation by human embryonic stem cells. Cell Cycle. 2009;8:2608–2612. doi: 10.4161/cc.8.16.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong CY, Gauthaman K, Bongso A. Teratomas from pluripotent stem cells: A clinical hurdle. J Cell Biochem. 2010;111:769–781. doi: 10.1002/jcb.22775. [DOI] [PubMed] [Google Scholar]
- 18.Kuznetsov S, Cherman N, Gehron Robey P. In Vivo Bone Formation by Progeny of Human Embryonic Stem Cells. Stem Cells Dev. 2010;20:269–287. doi: 10.1089/scd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang NK, Tosi J, Kasanuki JM, Chou CL, Kong J, Parmalee N, Wert KJ, Allikmets R, Lai CC, Chien CL, Nagasaki T, Lin CS, Tsang SH. Transplantation of reprogrammed embryonic stem cells improves visual function in a mouse model for retinitis pigmentosa. Transplantation. 2010;89:911–919. doi: 10.1097/TP.0b013e3181d45a61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bieberich E. Smart drugs for smarter stem cells: making SENSe (sphingolipid-enhanced neural stem cells) of ceramide. Neurosignals. 2008;16:124–139. doi: 10.1159/000111558. [DOI] [PubMed] [Google Scholar]
- 21.Bieberich E. Ceramide signaling in cancer and stem cells. Future Lipidol. 2008;3:273–300. doi: 10.2217/17460875.3.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieberich E, Hu B, Silva J, MacKinnon S, Yu RK, Fillmore H, Broaddus WC, Ottenbrite RM. Synthesis and characterization of novel ceramide analogs for induction of apoptosis in human cancer cells. Cancer Lett. 2002;181:55–64. doi: 10.1016/s0304-3835(02)00049-6. [DOI] [PubMed] [Google Scholar]
- 23.Bieberich E, Kawaguchi T, Yu RK. N-acylated serinol is a novel ceramide mimic inducing apoptosis in neuroblastoma cells. J Biol Chem. 2000;275:177–181. doi: 10.1074/jbc.275.1.177. [DOI] [PubMed] [Google Scholar]
- 24.Wang G, Krishnamurthy K, Umapathy NS, Verin AD, Bieberich E. The carboxyl-terminal domain of atypical protein kinase Czeta binds to ceramide and regulates junction formation in epithelial cells. J Biol Chem. 2009;284:14469–14475. doi: 10.1074/jbc.M808909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Silva J, Krishnamurthy K, Tran E, Condie BG, Bieberich E. Direct binding to ceramide activates protein kinase Czeta before the formation of a pro-apoptotic complex with PAR-4 in differentiating stem cells. J Biol Chem. 2005;280:26415–26424. doi: 10.1074/jbc.M501492200. [DOI] [PubMed] [Google Scholar]
- 26.Dutta D, Ray S, Home P, Larson M, Wolfe MW, Paul S. Self Renewal vs. Lineage Commitment of Embryonic Stem Cells: Protein Kinase C Signaling Shifts the Balance. Stem Cells. 2011 doi: 10.1002/stem.605. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieberich E, MacKinnon S, Silva J, Noggle S, Condie BG. Regulation of cell death in mitotic neural progenitor cells by asymmetric distribution of prostate apoptosis response 4 (PAR-4) and simultaneous elevation of endogenous ceramide. J Cell Biol. 2003;162:469–479. doi: 10.1083/jcb.200212067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bieberich E, MacKinnon S, Silva J, Yu RK. Regulation of apoptosis during neuronal differentiation by ceramide and b-series complex gangliosides. J Biol Chem. 2001;276:44396–44404. doi: 10.1074/jbc.M107239200. [DOI] [PubMed] [Google Scholar]
- 29.Bieberich E. There is More to a Lipid than just Being a Fat: Sphingolipid-Guided Differentiation of Oligodendroglial Lineage from Embryonic Stem Cells. Neurochem Res. 2010 doi: 10.1007/s11064-010-0338-5. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock CR, Wetherington JP, Lambert NA, Condie BG. Neuronal differentiation of cryopreserved neural progenitor cells derived from mouse embryonic stem cells. Biochem Biophys Res Commun. 2000;271:418–421. doi: 10.1006/bbrc.2000.2631. [DOI] [PubMed] [Google Scholar]
- 31.Westmoreland JJ, Hancock CR, Condie BG. Neuronal development of embryonic stem cells: a model of GABAergic neuron differentiation. Biochem Biophys Res Commun. 2001;284:674–680. doi: 10.1006/bbrc.2001.5031. [DOI] [PubMed] [Google Scholar]
- 32.Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, McKay RD. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- 33.Roach L, McNelsh JD. Methods for the Isolation and Maintenance of Murine Embryonic Stem Cells. In: Turksen K, editor. Methods in Molecular Biology. Embryonic Stem Cells. Vol. 183. New Jersey: Humana Press; 2002. pp. 1–16. [DOI] [PubMed] [Google Scholar]
- 34.Salli U, Fox TE, Carkaci-Salli N, Sharma A, Robertson GP, Kester M, Vrana KE. Propagation of undifferentiated human embryonic stem cells with nano-liposomal ceramide. Stem Cells Dev. 2009;18:55–65. doi: 10.1089/scd.2007.0271. [DOI] [PubMed] [Google Scholar]
- 35.Krishnamurthy K, Wang G, Silva J, Condie BG, Bieberich E. Ceramide Regulates Atypical PKC{zeta}/{lambda}-mediated Cell Polarity in Primitive Ectoderm Cells: A NOVEL FUNCTION OF SPHINGOLIPIDS IN MORPHOGENESIS. J Biol Chem. 2007;282:3379–3390. doi: 10.1074/jbc.M607779200. [DOI] [PubMed] [Google Scholar]
- 36.Krishnamurthy K, Dasgupta S, Bieberich E. Development and characterization of a novel anti-ceramide antibody. J Lipid Res. 2007;48:968–975. doi: 10.1194/jlr.D600043-JLR200. [DOI] [PubMed] [Google Scholar]