Abstract
Background:
Giardia duodenalis is one of the most prevalent intestinal parasites of human. It also infects a wide range of mammals. Two genotype of G.duodenalis (A and B) were commonly reported among humans with different frequency of distribution in different geographical locations. This work was conducted to discriminate genotypes of Giardia duodenalis human isolates in Isfahan city using PCR- RFLP. This is the first molecular study on human isolates of G.duodenalis in the area.
Methods:
Samples were collected from different health centers of Isfahan city during June 2011 and February 2012. From 175 Giardia positive stool samples 67 specimens were selected randomly. Cysts of Giardia positive samples were concentrated by flotation sucrose. Extraction of genomic DNA from trophozoite and cysts was performed using QIAamp Stool Mini kit with a modified protocol. PCR- RFLP method was used to amplify a fragment of 458bp at the glutamate dehydrogenase locus, and restriction enzymes BspLI and RsaI differentiated human genotypes A and B and their subgroups.
Results:
PCR – RFLP assay of 67 isolates showed 40(59.7%) isolates as Genotype A group II, 23(34.32%) samples as Genotype B Group III and two (2.98%) sample as Genotype B group IV. Mixed genotype of (AII and B) was detected only in two isolates (2.98%).
Conclusions:
PCR – RFLP assay targeting gdh locus is a sensitive tool and discriminates genotypes, sub genotypes and mixed type of G.duodenalis. Results of our study suggest both anthroponotic and zoonotic origins for the infections respectively.
Keywords: Giardia, gdh, polymerase chain reaction - restriction fragment length polymorphism, Isfahan
INTRODUCTION
Giardia species are intestinal parasites infecting a wide range of vertebrates. The genus comprises 6 species, namely, G. agilis, G. ardeae, G. microti, G. muris, G. psittaci, and G. duodenalis. These species are distinguished on the basis of their morphology.[1] Giardia duodenalis (synonyms: G. intestinalis, G. lamblia), which is one of the most common intestinal parasites of humans,[2] also infects other mammals, including pets and livestock.[2,3,4,5]
Because of the high prevalence of the parasite in humans, domestic animals, and wildlife, it is of public health and veterinary health importance.[2] It has worldwide distribution infecting an estimated 2.8 × 108 people each year. Symptomatic infections have been reported for 200 million people in Asia, Africa, and Latin America with an incidence rate of 500,000 cases per year.[3] It is believed that in the developing countries of the world giardiasis is responsible for 2.5 million diarrhea-associated deaths and nutritional deficiencies children.[1,6] Malnutrition is assumed to be a result of malabsorption caused by G.duodenalis. In symptomatic giardiasis, malabsorption has been seen for 50% of patients. A negative effect of giardiasis on growth and weight gain has been commonly reported. G.duodenalis affects growth and cognitive functions of infected children. Reduction in growth probably via malnutrition was reported in asymptomatic cases as well.[2]
Human infections are caused by G. duodenalis, the species which has also zoonotic potential[2] and is now considered as complex species that comprises at least 7 distinct genetic groups (A–G).[7] Molecular studies on human isolates from different geographic areas show that in almost all cases only assemblages A and B of G. duodenalis are associated with human infections. The prevalence of the mentioned genotypes varies from country to country and even between different geographic areas in a country.[3,4,6,8] The remaining genetic groups (C–G) seem to be host specific.[3] Having host specificity and genetic characteristics, some of previously used species names are suggested for different genotypes of G. duodenalis. Thus for assemblage A the name G. duodenalis, and for assemblages B, E, F, and G the names G. enterica, G. bovis, G. cati, and G. simodi are proposed, respectively. The proposed names for genotypes C and D is G. canis.[2]
A variety of molecular techniques, including Multiplex PCR,[9] PCR-Restriction Fragment Length Polymorphism,[4] real-time PCR,[10] and sequence analysis of housekeeping genes have proved to be used as tools for discrimination of all assemblages and to provide powerful tools for understanding molecular epidemiology, infection sources, and zoonotic potential of human giardiasis.[2,4,5,11] Molecular studies show subgroups (I–VIII) for assemblage A and subgroups (I–VI) for assemblage B.[4]
The current study was conducted to determine G.duodenalis genotypes and subgenotypes in Isfahan, Central Iran, as there is no previous data in this regard. PCR-RFLP method was performed based on gdh, as it is suited for direct typing of Giardia in crude specimens and successfully determines subgenotypes of human isolates.
MATERIALS AND METHODS
Collection of Isolates
In this study, 175 Giardia-positive samples were collected from health centers of Isfahan city during June 2011 to February 2012. After direct observation by microscope, the samples were stored at 4°C without any preservative. Cysts of 67 Giardia-positive specimens were concentrated from the specimens by flotation on 4 layers (0.5, 0.75, 1, and 1.5 M) and single-layer (0.85 M) sucrose. Some of the fresh specimens with a high number of cysts were cultured on TYI-S-33 medium.[12] Purified cysts and cultured trophozoites were stored at -20°C until further examination.
Extraction of DNA
Extraction of genomic DNA was performed from concentrated cysts of 65 and cultured trophozoites of 2 isolates. The trophozoite's DNA was extracted using QIAamp DNA Stool Mini Kit (QIAgen Company, Germany) according to the manufacturer's instruction and used as template for PCR assay. Extraction of genomic DNA from cysts of Giardia was performed as follows: Approximately 200 μL of concentrated sample were used for extraction. Glass beads (cat:G9268-Sigma) and a 500 μL of the lysis buffer (NaCl 50 mM, Tris–HCl 50 mM, EDTA 50 mM, and SDS 1%) were added to the sample and vortexed for 10 min; after repeated freezing and thawing, 20 μL of proteinase K (20 mg/mL), and SDS 10% (1/20 of total volume) were added. The mixture was incubated overnight at 56°C. The incubated mixture was used for DNA extraction by QIAamp DNA Stool Mini Kit. The extracted DNA samples were stored at -20°C for PCR assay.
PCR Amplification and RFLP Analysis
The gdh gene was amplified using a single PCR with the modified GDHF (5’ TCAACGTCAACCGCGGCTTCCT 3’) and GDHR (5’ GTTGTCCTTGCACATCTCC 3’) primers as previously described.[4,5] The reaction mixture for PCR amplification contained 5 μL of 10× buffer (CinnaGen, Iran), 1.5 mM of MgCl2(CinnaGen, Iran), 0.2 mM of each dNTPs, 1 U of Taq polymerase (CinnaGen, Iran), 20 pmole of each primer and 5–10 μL of extracted template DNA. Amplification of extracted DNA was performed using Eppendorf (Germany) thermal cycler in the following conditions: One cycle of 94°C for 5 min (initial denaturation), followed by 35 cycles of 20 s at 94°C, 45 s at 60.5°C, 45 s at 72°C, and a final extension at 72°C for 5 min. The DNA sample extracted from cultured trophozoites was used as the positive control and distilled water was used as the negative control. Electrophoresis of PCR products was performed on 1.5 % (W/V) of agarose gel stained with DNA green viewer.
For the digestion of gdh 458 bp fragment by the enzyme BspLI, 7 μL of PCR product was digested in a total of 20 μL reaction mixture containing 10 U of BspLI (Fermentase EU) and 2 μL of restriction buffer (Tango buffer fermentase), the reaction mixture was incubated at 37°C for 1 h. BspLI discriminates between groups I and II of assemblage A and assemblage B. The PCR products of isolates, identified as genotype B by BspLI, were digested by RsaI for differentiation between groups III and IV of assemblage B as follows: 7 μL of PCR product was added in a mixture of 1 μL (10 U) of RsaI (Fermentase EU), 2 μL of restriction buffer (Tango buffer fermentase) and 10 μL of distilled water. The reaction mixture was incubated at 37°C for 2 h. The digested fragments were fractionated on 3% agarose gel stained by DNA green viewer.
RESULTS
From 67 isolates selected for molecular analysis, 5 isolates were cultured on TYI-S-33, of which 2 isolates were successfully cultivated and were used as positive control for PCR assay. DNA was successfully extracted (100%) from all samples. A 458 bp fragment of gdh locus was amplified for all the extracted DNA samples [Figure 1]. The results showed that using QIAamp DNA extraction kit is a conventional method for DNA extraction from Giardia cysts.
Figure 1.
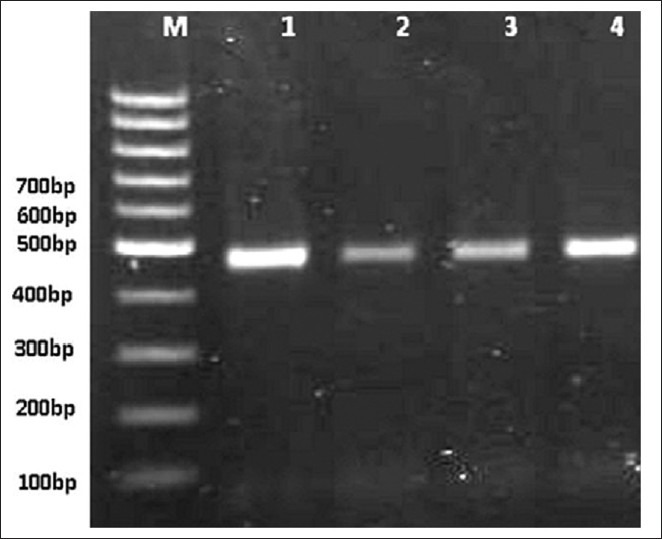
PCR amplification of Giardia duodenalis gdh on 1/5% agarose gel stained with DNA green viewer. (Lane M: 100 bp gene ruler (fermentase); and lanes 1–3: polymerase chain reaction products from examined samples; lane 4: positive control.)
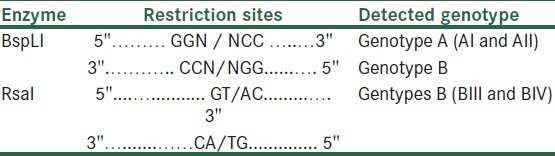
After digestion with enzymes, the amplified fragments were cut and restricted fragments were seen on 3% agarose gel, the restriction sites of enzymes are shown in Table 1.
Table 1.
Restriction sites of digestive enzymes BspLI and RsaI and Genotypes / sub genotypes of Giardia duodenalis, differentiated after digestion of a 458bp fragment
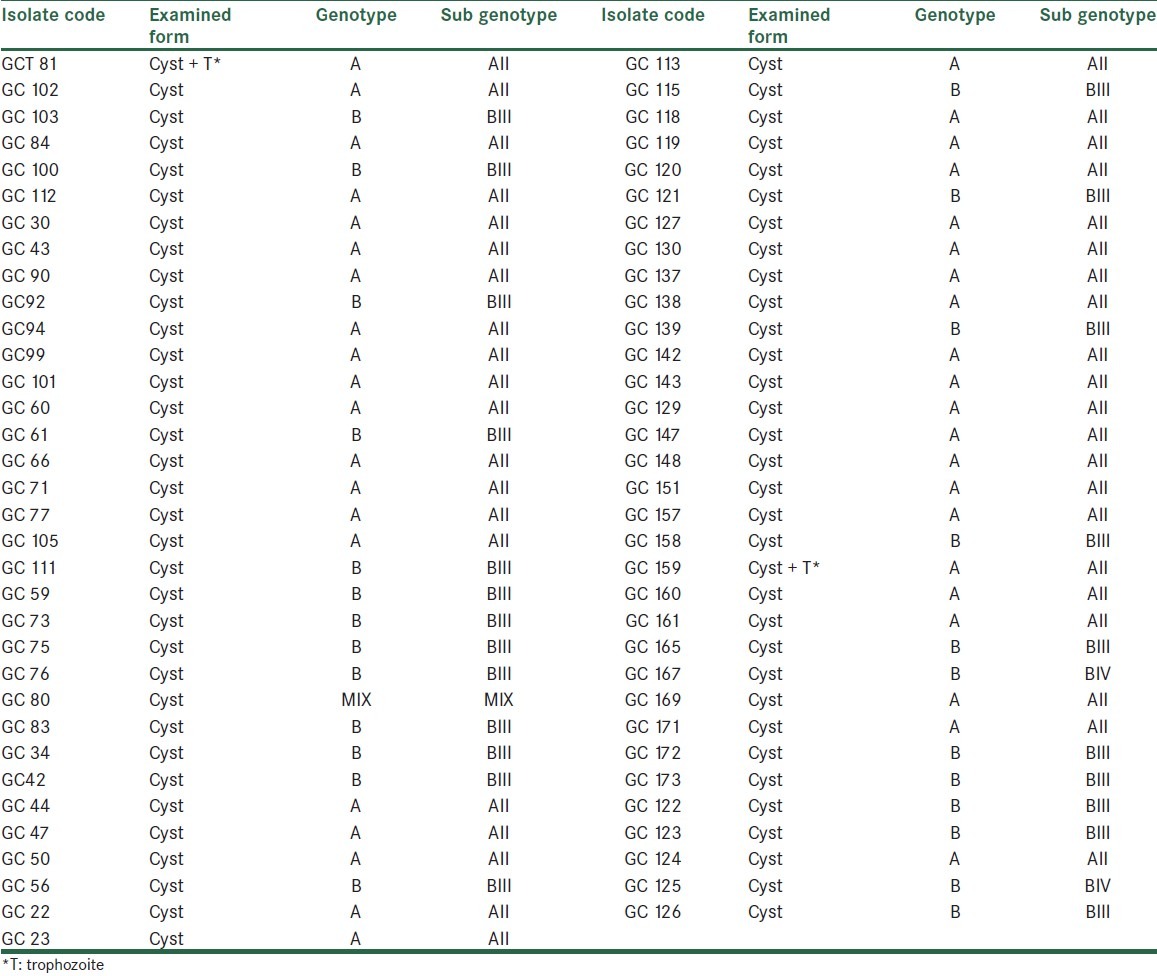
Some of the restricted fragments are diagnostic and determine genotypes and subgenotypes of G.duodenalis [Table 2]. The digested fragments by restriction enzyme BspLI revealed that more isolates were Genotype A group II [Figure 2]. No samples showed the restriction profile of AI. The digested products of the enzyme RsaI showed 2 different patterns of subgenotypes BIII and BIV, most of which were related to the first group [Figure 3]. Resulting from the present research, 40 (59.70%) of 67 specimens were determined as assemblage A group II, 23 (34.32%) isolates as assemblage B group III, 2 isolates (2.98%) as assemblage B group IV, and 2 isolates (2.98%) as a mixed form of genotype A group II and genotype B [Figure 4] [Table 3].
Table 2.
Diagnostic fragments sizes (bp) and genotyping profiles of Giardia duodenalis at the gdh locus when digested by enzymes BspLI and RsaI
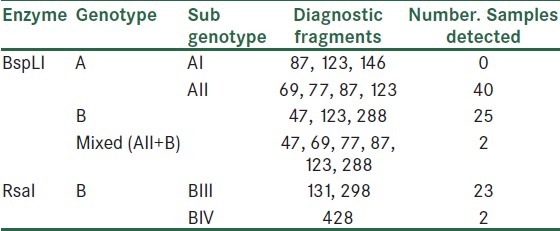
Figure 2.
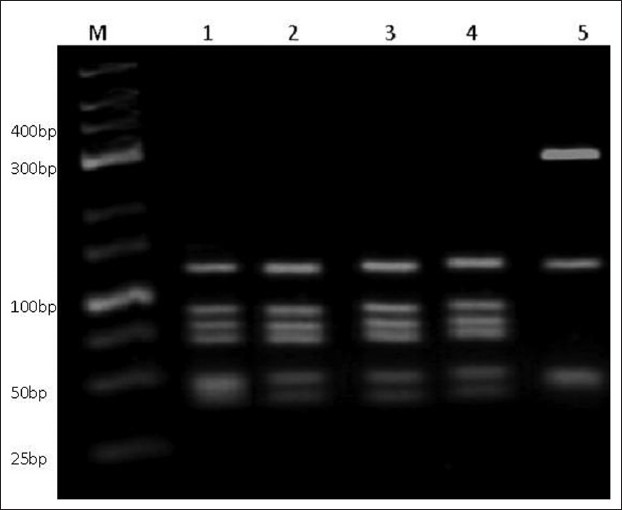
Enzymatic digestion of Giardia duodenalis gdh gene. Polymerase chain reaction products were restricted by BspLI and shown on DNA green viewer stained 3% high resolution agarose gel. (Lane M: 25 bp ladder (fermentase); lane 1-4: G. duodenalisgenotype A group II; and lane 5: G. duodenalisgenotype B.)
Figure 3.
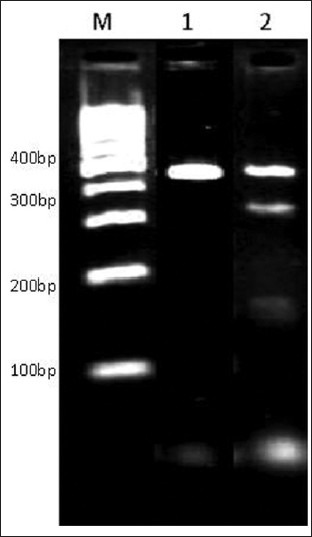
PCR product of G.duodenalis gdh digested by RsaI showing sub genotypes of B group. Lane M ladder 100bp (Fermentase), lane 1genotype B group III, lane 2 genotype B group IV
Figure 4.
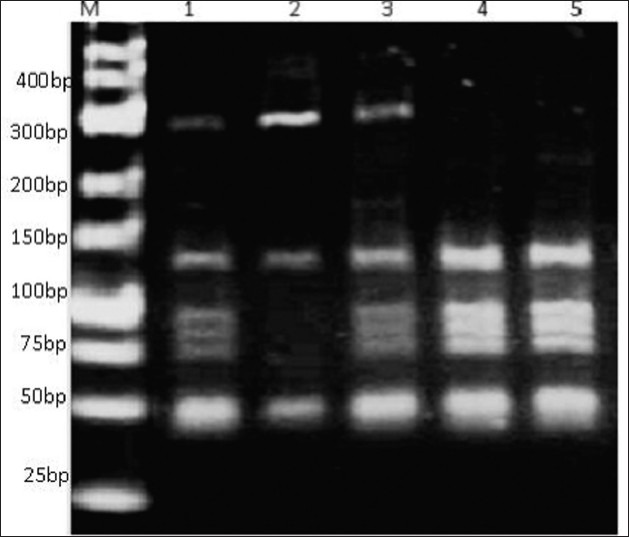
BspLI digestion of G. duodenalis gdh gene. PCR products showing mixed genotypes. Lane M ladder 25bp Gene ruler (Fermentase), lanes 1 and 3 show mixed genotypes.
Table 3.
Genotypes and sub genotypes of G. duodenalis identified by PCR- RFLP method at the gdh locus using restrictions enzymes BspLI and RsaI
DISCUSSION
G. duodenalis is an important and prevalent cause of parasitic gastroenteritis and diarrheal out breaks worldwide.[6,13] The parasite is found to be the most prevalent intestinal protozoan[14] and is taken into WHO's Neglected Diseases Initiative recently.[15] Its direct and easy life cycle, robust cysts that resist water chlorination and environmental conditions, contamination of food, water, and fomites; and its zoonotic potential, make it a common human parasite[2,6] The prevalence of human giardiasis in Iran is reported to be 10.9% and it is the most prevalent intestinal parasite in this country.[16] Genetic heterogeneity of the parasite determined by several molecular studies reveals that G.duodenalis is a complex species consisting of several genotypes.[2,4,7] The distribution of genotypes varies in human and animal hosts; and also it differs for various geographic areas[2,4,6,17,18,19]
Understanding the host range of different genotypes of G. duodenalis, zoonotic potential and cross-species transmission; and environmental factors involved in the exposure of parasite are important aspects of molecular epidemiology of giardiasis.[2] Numerous samples of G. duodenalis collected from humans and animals in various geographic locations have been typed, and the occurrences of the same genotype in human and animals has been reported.[20] Such data indicate the zoonotic potential of the parasite; therefore, most experts agree that G.duodenalis genotypes (especially genotypes A and B) are potentially zoonotic.[2] Human-infecting genotypes (A and B) have different frequencies of distribution in different geographic locations. Many molecular studies consider genotype A as dominant human-infecting assemblage, whereas other studies show a high prevalence of genotype B in humans.[2,4,6] Molecular studies on human isolates in Iran targeting gdh and tpi have shown the existence of genotypes A and B with different frequencies of distribution. Most of the studies show assemblage A as the predominant genotype. In Tabriz, Fallah et al. detected 54.8% genotype A, 41.9% genotype B, and 3.2% mixed genotype out of 34 samples. In a study conducted by Babaei et al.[4] on 38 human isolates in Tehran, 33 (87%) samples were found as G. duodenalis assemblage A group II, 3 (7.8%) samples as assemblage B group III, and 2 (5.2%) samples as of mixed genotypes. Genotyping of G. duodenalis was also performed by Zia-Ali et al.[8] on 30 samples in Kerman, genotype A subgroup II was found in 18 (60%), subgroup AI in 5 (16/6%) samples, and genotype B group III in 7 (23/4%) samples. As the prevalence of zoonotic assemblages varies in the world and also in Iran, there is no data to show the genotypes of G. duodenalis human isolates in Isfahan, Central Iran.
We used PCR-RFLP as it is a sensitive tool that is capable of distinguishing human isolates of G. duodenalis at the genotype and subgenotype level. This method has been used before and discriminated all genotypes of G. duodenalis successfully.[4,5,8]
The extraction of DNA from cysts of the parasite eliminates the difficult stages of parasite cultivation and selective growth of parasite in culture media. We used QIAamp DNA extraction Mini kit with a modified protocol in which glass beads have been used and freeze-thawing was performed. The protocol had a result of 100% successful DNA extraction[4] where only 44% of cysts had a successful growth by an effective method of axenic culture.[12]
In our study, we examined 67 isolates of G. duodenalis from human feces. Our results show genotypes A and B, which were found by many researchers in Iran and other countries. In our study, more isolates were typed as genotype A group II followed by genotype B group III. The higher rate of genotype A in Isfahan is similar to many previous studies. A molecular study in Mexico reported that all the studied isolates were of genotype A. The same results were obtained from studies performed in South Korea and Italy. The results of our study are similar to previous studies in Kerman and East Azerbaijan, but differ with the study conducted in Tehran.
Previous studies have shown that subgenotypes AI, BIII, and BIV have more zoonotic potential.
The higher rate of assemblage A group II reveals an anthroponotic origin of the infection. A considerable number of genotype B in the present study shows a zoonotic transmission route as well.[4] As assemblage B was detected in many waterborne outbreaks of Giardia, contamination of water with animal feces in the area of study is possible. Mixed genotypes of AII and assemblage B was found only in 2 (2/98%) isolates. As previous studies have shown a relationship between mixed infections with multiple sources of infection and complex life cycle of parasites, complex life cycle and multiple source for the parasite is rarely possible in the area of study.
CONCLUSION
In conclusion, molecular studies on human Giardia isolates and discrimination of genotypes is a useful way to know the transmission route and effective prevention methods of giardiasis. Recently, with the advances of molecular typing tools, the epidemiology of giardiasis is systematically addressed. As a final result of our study both anthroponotic and zoonotic origin for the parasite is suggested. Because of the zoonotic potential of G.duodenalis and detection of both genotypes in human samples, further studies on human and animal samples are suggested. Screening of the water sources (water supply and surface water), sewage system and food chain will also help understanding the molecular epidemiology of infection in the area.
ACKNOWLEDGMNTS
We thank the staff of parasitology section of Navab Safavi, Mahdieh, Shahid e Aval Health Networks and St. Al-Zahra Hospital laboratory. The Research Deputy of Isfahan University of Medical Sciences is thanked for providing financial support for this research.
Footnotes
Source of Support: Financially supported by Vice Chancellery of Isfahan University of Medical sciences
Conflict of Interest: None declared
REFERENCES
- 1.Gelanew T, Lalle M, Hailu A, Pozio E, Caccio SM. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 2007;102:92–9. doi: 10.1016/j.actatropica.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and Giardiasis. Clin Microbiol Rev. 2011;24:110–40. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Babaei Z, Oormazdi H, Akhlaghi L, Rezaie S, Razmjou E, Soltani-Arabshahi SK, et al. Molecular characterization of the Iranian Isolates of Giardia lamblia: application of the glutamate dehydrogenase gene. Iran J Publ Health. 2008;37:75–82. [Google Scholar]
- 5.Read CM, Monis PT, Thompson A. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR–RFLP. Infect Genet Evol. 2004;4:125–30. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Fallah E, Nahavandi KH, Jamali R, Mahdavi B, Asgharzadeh M. Genetic characterization of Giardia intestinalis strains from patients having sporadic Giardiasis by PCR assay. J Med Sci. 2008;8:310–5. [Google Scholar]
- 7.Coradi C, David D, Sequeira O, Ribolla R, Carvalho C, Guimaraes G. Genotyping of Brazilian Giardia duodenalis human axenic isolates. J Venom Anim Toxins Incl Trop Dis. 2011;17:353–357. [Google Scholar]
- 8.Etamadi S, Zia-Ali N, Babaie Z, Fasihi Harandi M, Zia–Ali A, Salari Z, et al. The correlation between clinical signs and genotypes of Giardia duodenalis isolated from patients with Giardiasis in Kerman city. J Kerman Univ Med Sci. 2011;18:330–8. [Google Scholar]
- 9.Eligio-Gracia L, Cortes-Campos A, Jimenez-Cardoso E. Classification of Giardia intestinalis isolates by multiple polymerase chain reaction. Parasitol Res. 2008;103:797–800. doi: 10.1007/s00436-008-1042-0. [DOI] [PubMed] [Google Scholar]
- 10.Almeida A, Pozio E, Caccio SM. Genotyping of Giardia duodenalis cysts by new real-time PCR assays for detection of mixed infections in human samples. Appl Environ Microbiol. 2010;76:1895–901. doi: 10.1128/AEM.02305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, et al. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl Trop Dis. 2011;5:e1262. doi: 10.1371/journal.pntd.0001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousefi HA. Pure culture method: Giardia lamblia from different stool samples. J Res Med Sci. 2000;5:78–81. [Google Scholar]
- 13.Kim J, Shin MH, Song KJ, Park SJ. Evaluation of alpha tubulin as an antigenic and molecular probe to detect Giardia lamblia. Korean J Parasitol. 2009;47:287–91. doi: 10.3347/kjp.2009.47.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston AR, Gillespie TR, Rwego IB, McLachlan T, Kent AD, Goldberg T. Molecular epidemiology of cross-species Giardia duodenalis transmission in Western Uganda. PLoS Negl Trop Dis. 2010;4(e683):1–7. doi: 10.1371/journal.pntd.0000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savioli L, Smith H, Thompson A. Giardia and Cryptosporidium join the Neglected Diseases Initiative. Trends Parasitol. 2006;22:203–8. doi: 10.1016/j.pt.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Sayyari A, Imanzadeh F, Bagheri Yazdi SA, Karami H, Yaghoobi M. Prevalence of intestinal parasitic infections in the Islamic Republic of Iran. East Mediterr Health J. 2005;11:377–83. [PubMed] [Google Scholar]
- 17.Traub RJ, Inpankaew T, Reid SA, Sutthikornchai C, Sukthana Y, Robertson ID, et al. Transmission cycles of Giardia duodenalis in dogs and humans in temple communities in Bangkok- a critical evaluation of its prevalence using three diagnostic tests in the field in the absence of a gold standard. Acta Trop. 2009;111:125–32. doi: 10.1016/j.actatropica.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Helmy M, Abdel-Fattah H, Rashed L. Real time PCR/RFLP assay to detect Giardia intestinalis genotypes in human isolates with diarrhea in Egypt. J Parasitol. 2009;95:1–5. doi: 10.1645/GE-1670.1. [DOI] [PubMed] [Google Scholar]
- 19.Cooper MA, Serling CR, Gilman RH, Cama V, Wrtega Y, Adam RD. Molecular analysis of houshold transmission of Giardia lamblia in a highly endemic community of Peru. J Infect Dis. 2010;202:1713–21. doi: 10.1086/657142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monis PT, Thompson RC. Cryptosporidium and Giardia zoonoses: Fact or fiction? Infect Genet Evol. 2003;3:233–44. doi: 10.1016/j.meegid.2003.08.003. [DOI] [PubMed] [Google Scholar]


