Abstract
Background:
To setup a non-invasive genetic screening method for colorectal cancer, we evaluated the promoter methylation status of secreted frizzled-related protein1 (sfrp1) in stool samples of colorectal cancer with respect to a series of healthy individuals, using methylation-specific polymerase chain reaction.
Materials and Methods:
In stool samples from 25 patients with colorectal cancer and 25 healthy control subjects, isolated DNA was treated with sodium bisulfite and analyzed by methylation-specific polymerase chain reaction with primers specific for methylated or unmethylated promoter sequences of the SFRP1 gene.
Result:
Methylation of the SFRP1 promoter was present in the stool DNA of patients with colorectal cancer. A sensitivity of 52% and specificity of 92% were achieved in the detection of colorectal neoplasia. The difference in methylation status of the SFRP1 promoter between the patients with colorectal neoplasia and the control group was statistically highly significant (P = 0.006).
Conclusions:
The results indicate that this DNA stool test of methylation of the SFRP1 promoter is a sensitive and specific method. It is assumed that the test is potentially useful for the early detection of colorectal cancer.
Keywords: Colorectal cancer, DNA methylation, secreted frizzled-related protein, stool DNA test
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer in the world, and is the second major cause of death from cancer in Europe and in the USA.[1,2] Detection of early disease and precancerous lesions Seems to be a key measure to reducing mortality rate from this disease.[3] CRC can be most effectively treated when diagnosed at an early stage.[4] Early detection can improve prognosis, but the recognition that virtually all CRCs arise from a discrete and accessible precursor lesion raises the prospect that cancer can essentially be prevented with an appropriate screening.[5] Yet, the acceptance of current screening methods is low. Only a minority (14 – 17 %) of average-risk adults older than aged 50 years undergo colonoscopy.[6,7] Fecal occult bleod testing (FOBT) is far more widely used;[8,9] however, tumors without bleeding can remain undetected.[10] The primary goal today is to identify the most sensitive and effective screening approaches that would maximize patient compliance. Colonocytes are continuously shed into the stool. Therefore, an analysis of their molecular and genetic alterations may aid CRC detection.[11] Studies aiming to make some correlation with various SNP have already been done in Isfahan.[12,13,14] However, it is a first attempt in using stool DNA methylation as a molecular marker in CRC detection in Isfahan. In a multicenter study in 2004, stool-based DNA tests were found to be 4 times more effective than FOBT for detecting CRC.[10] In addition, only one stool sample is needed for stool DNA tests compared with 3 samples for FOBT, and compliance and patient acceptance are clearly higher than for colonoscopy.[15,16]
The emergence of molecular stool testing provides a possible user-friendly alternative to conventional methods of CRC screening. One such strategy would be to develop tests for the detection of fecal DNA methylation patterns that will improve the sensitivity of non-invasive screening tests for colorectal neoplasia.[17,18] One of the principal epigenetic mechanisms known to be involved in carcinogenesis is the methylation of the cytosine residues of CpG-rich sequences (CpG islands) located within the promoter regions of genes regulating cell proliferation, apoptosis, and DNA repair. A number of genes have now been shown to be hypermethylated in CRC.[19] Silencing of SFRPs genes which are glycoproteins working as inhibitory modulators of a putative tumorigenic pathway (the Wnt signaling pathway) induced by promoter hypermethylation plays a key role in colorectal tumorigenesis.[20] In 1983, Feinberg and Vogelstein observed altered methylation of genes in colorectal tumors and adenomas.[21,22] In 2002, Suzuki et al. observed frequent promoter hypermethylation and silencing of SFRP genes in CRC and identified them as potential gatekeeper genes whose loss of function occurs early in CRC progression.[23] In present study, we studied the methylation status of SFRP1 gene in stool samples from patients with CRC and healthy individuals, using methylation-specific polymerase chain reaction (MSP), as a potential marker for stool-based early detection and early screening of colorectal cancer.
MATERIALS AND METHODS
Patients and collection of fecal DNA sample
DNA Samples: Human stool samples were collected from 50 individuals including 25 healthy volunteers and 25 patients with colorectal cancer before any treatment. About 5 g stool was collected from each individual. All the samples were collected in dry clean plastic containers. An informed consent was obtained from every subject prior to the study. Stools were collected prior to any preparation for colonoscopy or 4 - 5 days following this procedure. Tumor characteristics such as location, size, and stage, as well as, age, sex and other necessary information were recorded in a questionnaire. The stool specimens were stored at -20°C immediately after collection, to avoid potential enzymatic degradation of nucleic acids, and if longer storage was needed, then transferred to a -70°C.
DNA isolation from fecal samples
Samples were randomly coded before processing to ensure adequate blinding of the clinical information. DNA was isolated from stool samples (250 mg) by use of the QIAamp DNA Stool Mini Kit (Qiagene Germany) according to the manufacturer's protocol.
Bisulfite modification
DNA was chemically modified by sodium bisulfite to convert all unmethylated cytosines to uracils while leaving methyl cytosines unaltered (EpiTect Bisulfite Kit, Qiagen) and eluted in 50 μL of elution buffer.
Methylation specific PCR (MSP)
The bisulfate-modified DNA was used as a template for MSP as described previously.[24] Proper positive and negative controls were included in each batch of PCR reaction. Methylated and unmethylated primer sequences used in this study[25,26] as well as the annealing temperature and, product sizes are given in Table 1. For the MSP, 2 μL of bisulfite-converted DNA was used in each amplification reaction. PCR was performed in a reaction volume of 25 μL consisting of 17.875 μL ddH2O, 2.5 μL 10X PCR buffer, 0.2 mM dNTP mixture, 10 pM of each forward and reverse primers, and 2 units of TaKaRa Taq HS. Thermal cycling profile performed as follow: 95°C for 15 min, followed by 40 cycles at 94°C for 30 sec, specific annealing temperature (62°C for methylated primer pairs and 58°C for unmethylated primer pairs) for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. The MSP products were then analyzed by 2.5% agarose gel electrophoresis.
Table 1.
SFRP1 primers sequences, annealing temperature, and product size for MSP assays

Statistical analysis
Pearson chi-squared test was used to evaluate the association between the methylation status of the SFRP1 promoter in the DNA from all stool samples, as well as to evaluate the association between methylated SFRP1 promoter (positive or negative), tumor location (colon vs. rectum), patient group (control vs. CRC), and demographic variables, such as age and gender. P > 0.05 was considered to be significant. All statistical analyses were performed with the SPSS 13 software package (SPSS Inc., Chicago, IL).
RESULT
Detection of methylated SFRP1 gene in fecal DNA
We assessed methylation status of SFRP1 promoter in fecal DNA from the patients and control groups by MSP reaction. The information of patients is shown in Table 2.
Table 2.
Characteristics of patients

Patients had mean (sd) age of 58 (12.87) years. 35% of cases were female and 65% of patients were male. The most common tumor site was rectum (40%); other sites were cecum (25%), splenic flexture of colon (10%), ascending colon (15%), and hepatic flexture (10%), respectively. All the tumors were invasive adenocarcinoma. Tumor sizes were less than 3 cm in 40%, 3 - 6 cm in 25%, and more than 6 cm in 35%.
Methylation status of SFRP1 promoter for 13 patients was positive. In addition, for 23 subjects in control group, methylation of SFRP1 gene was negative. Based on our results, sensitivity of SFRP1 was 52% and specificity was 92%. Methylation status (positive vs. negative) of SFRP1 gene between CRC and control groups was significantly different (P value = 0.006).
DISCUSSION
The low acceptance of current screening methods has stimulated the search for a non-invasive, highly sensitive screening test. In analyzing various issues of CRC screening and the different screening tests, the following aspects need to be considered: (a), sensitivity and specificity, (b) safety, (c), acceptability, which often determines compliance, (d) cost, (e) efficacy (the extent to which medical interventions improve health under ideal circumstances), and (f) effectiveness, which is important because it indicates the accuracy of detecting and removing precancerous lesions.[27] Although currently colonoscopy is a gold standard procedure for the CRC diagnosis, but exhibits certain disadvantages like high costs, increased risk of perforation and bleeding, difficult preparation for the patients, and the need for sedation.[28] Disadvantages of FOBT are low sensitivity, low specificity, poor compliance, and the need for colonoscopy to confirm a positive test result.[29]
Stool-based DNA hypermethylation testing is a new, non-invasive method of colorectal cancer screening. It is easier to perform and is more sensitive than fecal occult blood testing, only a single stool sample is needed, does not require diet or medication restrictions, and evaluates the whole colon and rectum.[30] Aberrant Wnt signaling pathway is an early molecular event in 90% of CRCs, contributing to the growth, proliferation, and loss of apoptosis of tumor cells.[31] SFRPs are tumor suppressor proteins that contain a domain similar to one of WNT-receptor frizzled proteins (Fz) and may block Wnt signaling either by interacting with Wnt proteins to prevent them from binding to Fz proteins or by forming non-functional complexes with Fz.[32] Epigenetic inactivation of SFRP genes induced by promoter hypermethylation has been shown to play an important role in development of CRC by allowing constitutive WNT signaling [Figure 1].[33,34]
Figure 1.
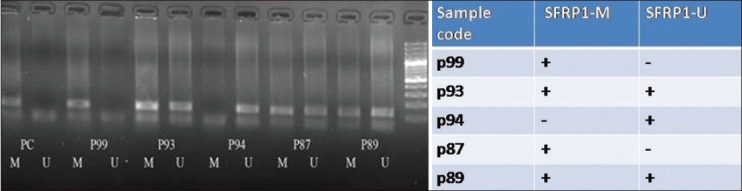
Detection of unmethylated (U) and methylated (M) SFRP1 in stool sample of CRC patients. P:patient, PC: positive control, M:methylated, U:Unmethyl
Detection of tumor-derived DNA alterations in fecal samples is an intriguing new approach with high potential for the non-invasive detection of CRC.[35] Methylation analysis of a number of gene promoters in DNA from fecal samples has been less comprehensively investigated, but has been suggested to be a sensitive diagnostic tool for colorectal tumor.[36] Our test indicated a sensitivity of 52% and specificity of 92% (P < 0.006). The presence of both methylated and unmethylated promoter sequences is in agreement with the heterogeneous mixture of dysplastic, tumor, and normal cells characteristically observed in early stages of carcinogenesis. Recently, Muller and colleagues[35] reported the detection of SFRP1 promoter methylation in the stool DNA of patients with CRC. In a preliminary setup with 10 CRC patients and 13 healthy control subjects, they achieved 90% sensitivity and 77% specificity, and in the fecal DNA from an independent test set of 13 patients with CRC and 13 healthy control subjects, a sensitivity of 77% and a specificity of 77% were obtained. In various stool-based studies using SFRP1 gene, different levels of sensitivity and specificity were obtained, mostly with satisfactory achievements.[36,37]
CONCLUSION
Hypermethylation of SFRP1 gene promoter in feces are novel epigenetic biomarkers of CRC and carried high potential for the remote detection of CRC as an non-invasive screening method.
ACKNOWLEDGMENTS
The authors are grateful to Mr. Mohammad Salehi for his help in statistical analysis.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–92. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Smigal C, Thun MJ. Cancer statistics. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancerstatistics 2003. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 5.Hung KE, Chung DC. Colorectal cancer screening: Today and tomorrow. South Med J. 2006;99:240–9. doi: 10.1097/01.smj.0000203817.89741.29. quiz 247-9. [DOI] [PubMed] [Google Scholar]
- 6.Vernon SW. Participation in colorectal cancer screening: A review. J Natl Cancer Inst. 1997;89:1406–22. doi: 10.1093/jnci/89.19.1406. [DOI] [PubMed] [Google Scholar]
- 7.Schroy PC 3rd, Heeren TC. Patient perceptions of stool-based DNA testing for colorectal cancer screening. AmJ Prev Med. 2005;28:208–14. doi: 10.1016/j.amepre.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: Effectiveness of biennial screening for fecal occult blood. J Natl Cancer Inst. 1999;91:434–7. doi: 10.1093/jnci/91.5.434. [DOI] [PubMed] [Google Scholar]
- 9.Imperiale TF, Ransohoff DF, Itzkowitz SH. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 10.Loktionov AO, Neill IK, Silvester KR. Quantiation of DNA from exfoliated colonocytes isolatedfrom human stool surface as a novel noninvasive screening test for colorectalcancer. Clin Cancer Res. 1998;4:337–42. [PubMed] [Google Scholar]
- 11.Deenadayalu VP, Rex DK. Fecal-based DNA assays: A new, noninvasive approach to colorectal cancer screening. Cleve Clin J Med. 2004;71:497–503. doi: 10.3949/ccjm.71.6.497. [DOI] [PubMed] [Google Scholar]
- 12.Daraei A, Salehi R, Salehi M, Emami MH, Janghorbani M, Mohamadhashem F, et al. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadiccolorectal cancer in Iranian population. Med Oncol. 2012:291044–9. doi: 10.1007/s12032-011-9980-2. [DOI] [PubMed] [Google Scholar]
- 13.Daraei A, Salehi R, Mohamadhashem F. DNA-methyltransferase 3B 39179 G > T polymorphism and risk of sporadic colorectal cancer in a subset of Iranian population. J Res Med Sci. 2011;16:807–13. [PMC free article] [PubMed] [Google Scholar]
- 14.Daraei A, Salehi R, Mohamadhashem F. PTGS2 (COX2) -765G>C gene polymorphism and risk of sporadic colorectal cancer in Iranian population. Mol Biol Rep. 2012;39:5219–24. doi: 10.1007/s11033-011-1319-8. [DOI] [PubMed] [Google Scholar]
- 15.Mandel JS, Church TR, Bond JH. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603–77. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- 16.Ahlquist DA, Shuber AP. Stool screening for colorectalcancer: Evolution from occult blood to molecularmarkers. Clin Chim Acta. 2002;315:157–68. doi: 10.1016/s0009-8981(01)00712-4. [DOI] [PubMed] [Google Scholar]
- 17.Belshaw NJ, Elliott GO, Williams EA, Bradburn DM, Mills SJ, Mathers JC, et al. Use of DNA from human stools to detect aberrant CpG island methylation of genes implicated in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1495–501. [PubMed] [Google Scholar]
- 18.Leung WK, To KF, Man EP, Chan MW, Hui AJ, Ng SS, et al. Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am J Gastroenterol. 2007;102:1070–6. doi: 10.1111/j.1572-0241.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 19.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 20.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg AP, Vogelstein B. Hypomethylation distin-ishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Gabrielson E, Chen W. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 24.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang ZH, LiL H, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol. 2007;13:950–4. doi: 10.3748/wjg.v13.i6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou H, Molina JR, Harrington JJ, Osborn NK, Klatt KK, Romero Y, et al. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barretts esophagus. Int J Cancer. 2005;116:584–91. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 27.Ra Wagner JL. Cost-effectiveness of screening for common cancers. Cancer metastasis Rev. 1997;16:281–94. doi: 10.1023/a:1005800227359. [DOI] [PubMed] [Google Scholar]
- 28.Shane E, Hendon, Jack A. Practices for colon cancer screening division of gastroenterology, University of South Alabama College of Medicine. Mobile AL. USA Keio J Med. 2005;54:179–83. doi: 10.2302/kjm.54.179. [DOI] [PubMed] [Google Scholar]
- 29.Lang CA, Ransohoff DF. Fecal occult blood screening for colorectal cancer. Is mortality reduced by chance selection for screening colonoscopy? J Am Med Assoc. 1994;271:1011–3. [PubMed] [Google Scholar]
- 30.Schroy PC, Heeren Tc. A comparative study of patient perceptions and screening preferences for stool-based DNA testing(SBDNA), fecal occult blood testing (FOBT), or colonoscopy (CS) Gastroenterology. 2003;124:A481. [Google Scholar]
- 31.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 32.Kawano Y, Kypta R. Secreted antagonists of the Wnt signaling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 34.Jubb AM, Bell SM, Quirke P. Methylation and colorectal cancer. J Pathol. 2001;195:111–34. doi: 10.1002/path.923. [DOI] [PubMed] [Google Scholar]
- 35.Muller HM, Oberwalder M, Fiegl H. Methylation changes in fecal DNA: A marker for colorectal cancer screening? Lancet. 2004;363:1283–5. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Bauer M., Croner R.S., Pelz J.O., Lodygin D., Hermeking H., et al. DNA stool test for colorectal cancer: Hypermethylation of the secreted frizzled-related protein-1 gene. Dis. Colon Rectum 50. 2007:1618–26. doi: 10.1007/s10350-007-0286-6. [DOI] [PubMed] [Google Scholar]
- 37.Azuara D, Rodriguez-Moranta F, de Oca J, Soriano-Izquierdo A, Mora J, Guardiola J, et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clinical colorectal cancer. 2010;9:168–76. doi: 10.3816/CCC.2010.n.023. [DOI] [PubMed] [Google Scholar]


