Abstract
Nano-scale complexes of recombinant silk molecules containing tumor homing peptides (THPs) with DNA were designed as less-cytotoxic and highly target-specific gene carriers. We report the preparation and study of these nano-scale silk-based ionic complexes containing pDNA that are able to home specifically to tumorigenic cells due to the presence of THPs, namely, F3 and Lyp1. Genetically engineered silk proteins (MaSp1 monomer) containing poly(L-lysine) domains to interact with pDNA and the THP to bind to specific tumorigenic cells for target-specific pDNA delivery were prepared using Escherichia coli. This process was followed by in-vitro transfection experiments into MDA-MB-435 melanoma cells, highly metastatic human breast tumor MDA-MB-231 cells, and non-tumorigenic MCF-10A breast epithelial cells. The silk-poly(L-lysine) block copolymer containing Lyp1 (ML-Lyp1) showed significant differences from silk-poly(L-lysine) block copolymer containing F3 (ML-F3) in cytotoxicity to MCF10A cells. ML-F3 was the most useful candidate for target delivery into tumorigenic cells. The target specificity of the pDNA complexes to tumorigenic cells was regulated by specific adsorption processes on the cell surface, based on field emission scanning electron microscopy observations.
Keywords: gene delivery, nanocomplex, recombinant protein, spider silk, tumor-homing peptide
Introduction
Silk proteins have been used successfully in the biomedical field as sutures for decades, and also explored as biomaterials for cell culture and tissue engineering, achieving Food and Drug Administration approval for such expanded utility because of their excellent mechanical properties, versatility in processing and biocompatibility.[1,2] Further, the degradation products from the beta-sheet containing silk proteins from the activity of alpha-chymotrypsin showed no significant cytotoxicity in-vitro to neuron cells.[3] On the basis of the biocompatibility of silk proteins, a variety of silk-based biomaterials, including sutures, scaffolds, films, coating, tubes and microspheres have been investigated for biomedical applications.[1,4] Additionally, silk-based gene delivery systems have recently been reported to exhibit biodegradability, biocompatibility, high transfection efficiency, and DNase resistance.[5-8]
Non-viral gene delivery systems using cationic liposomes or chemical synthetic polymers such as polyethyleneimine, the standard in many applications for gene delivery, have improved in terms of transfection efficiency, cell-binding, and endosomal release, resulting in attention as safe alternatives to viral vectors.[9-11] Nevertheless, cytotoxicity and target specificity remain as key challenges to overcome in lipid and synthetic polymer-based gene delivery systems. An ideal non-viral gene vector would be biocompatible, biodegradable, non-cytotoxic, efficient, and can be designed to target specific cell types. These are challenging design goals to meet with liposomes or chemically synthesized polymers, in part due to the limits of chemical synthesis.
In previous studies, we reported new silk-based ionic complexes containing plasmid DNA (pDNA) that were able to home specifically to tumor cells.[12] Silk-based block copolymers were bioengineered with both poly(L-lysine) domains to interact with pDNA and the tumor-homing peptide (THP) to bind specific tumor cells for target-specific pDNA delivery.[12] F3 peptides have been reported to show target specificity and home to nucleolin, which is expressed at the surface of tumor angiogenic endothelial cells, and bind specifically to MDA-MB-435 cells.[13,14] The silk-based block copolymers showed relatively high target specificity and transfection efficiency.[12] For the practical use of silk-based gene delivery system, however, further improvements in transfection efficiency are needed while target specificity is maintained. The content of F3 THP in the silk-based block copolymers was approximately 12 mol%, based on the ratio of the molecular weight of F3 THP (3.4 kDa) and the overall silk-based block copolymer (28.8 kDa).
In the present study, silk-based ionic pDNA complexes with a higher content of THP (25 mol%, 3.4 kDa/13.6 kDa) were designed to enhance specificity and efficiency to home tumor cells in comparison to our previous report.[12] The focus was on how a higher-content of THPs, F3 and Lyp1, with silk-based gene delivery systems enhanced transfection efficiency and specificity to tumor cells. Lyp1 peptide, which is a different type of THP from F3, binds specifically to tumor lymphatics in certain tumors but not to the lymphatics of normal tissues, and Lyp 1 induces cell death in cultured MDA-MB-435 cells.[15,16] To further enhance transfection efficiency, the monomeric silk sequence (MaSp1 monomer) was used in the new designs instead of silk 6mer to prepare smaller pDNA complexes (approximately 90 nm).[12] The rationale for this change was that the diameter of adenovirus, approximately 90 nm, was expected for gene delivery system.[17] Genetically engineered silk block copolymers containing poly(L-lysine) and the THPs were biosynthesized using Escherichia coli system. pDNA complexes of the silk block copolymers were prepared (Figure 1), followed by in-vitro and in-vivo transfection experiments into MDA-MB-435 melanoma cells and highly metastatic human breast tumor MDA-MB-231 cells. Non-tumorigenic MCF-10A breast epithelial cells were used as a control cell line for in-vitro tumor-specific delivery studies.
Figure 1.
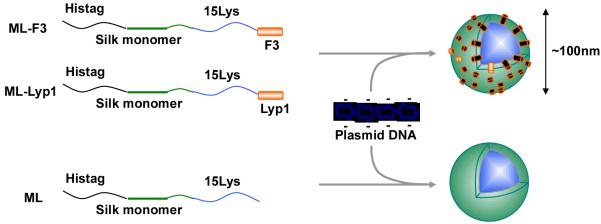
Schematic of formation of the pDNA complexes of the recombinant silk block copolymers. ML-F3: Silk monomer-15 lysines-F3 tumor homing peptide block copolymer. ML-Lyp1: Silk monomer-15 lysines-Lyp1 tumor homing peptide block copolymer. ML: Silk monomer-15 lysines block copolymer.
Experimental Part
Design and Cloning of Recombinant Silk Sequence
The spider silk unit was selected based on the consensus repeat (SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT) derived from the native sequence of the dragline protein MaSp1 sequence from the spider Nephila clavipes (Accession P19837). The silk monomer containing one contiguous copy of this repeat was developed through the transfer of cloned inserts to pET-30a, which had been modified with a linker carrying the restriction sites NheI and SpeI according to our previously published procedures.[18] The sequences of the synthetic oligonucleotides encoding 15 lysine residues were as follows: Lys-a: 5′-CTAGCAAGAAAAAGAAAAAAAAGAAAAAAAAGAAAAAGAAAAAAAAGAAAA-3′, Lys-b: 5′-CTAGTTTTCTTTTTTTTCTTTTTCTTTTTTTTCTTTTTTTTCTTTTTCTTG-3′. The restriction sites for SpeI are italicized. Lys-a and Lys-b are complementary oligonucleotides which were annealed to form double stranded DNA. The double stranded DNA was ligated into pET30-Silk monomer to generate pET30-Silk monomer-15lys by DNA ligase (New England Biolabs Inc, Ipswich, MA).[5] The sequences of the synthetic oligonucleotides encoding F3 and Lyp1 residues were as follows: F3-a: 5′-CTAGCAAAGATGAACCGCAGCGCCGCAGCGCGCGCCTGAGCGCGAAACCGGCGC CGCCGAAACCGGAACCGAAACCGAAAAAAGCGCCGGCGAAAAAAA-3′, F3-b: 5′-CTAGTTTTTTTCGCCGGCGCTTTTTTCGGTTTCGGTTCCGGTTTCGGCGGCGCCGGT TTCGCGCTCAGGCGCGCGCTGCGGCGCTGCGGTTCATCTTTG-3′, Lyp1-a: 5′-CTAGCTGCGGCAACAAACGCACCCGCGGCTGCA-3′, Lyp1-b: 5′-CTAGTGCAGCCGCGGGTGCGTTTGTTGCCGCAG-3′. The restriction sites for SpeI are italicized. F3-a and F3-b as well as Lyp1-a and Lyp1-b are complementary oligonucleotides which were annealed to form double stranded DNA, and subsequently the double stranded DNAs of the sequences were ligated into pET30-Silk monomer-15lys to generate pET30-Silk monomer-15lys-F3, pET30-Silk monomer-15lys-Lyp1(Figure 1). Amino acid sequences of these silk polymers are shown in Figure 2.
Figure 2.

Amino acid sequences of the recombinant silk monomer proteins with 15 lysines and tumor-homing peptides (THPs). Bold: tumor-homing peptide sequences. Underline: representative monomer of spider silk sequence.
Protein Expression and Purification
The constructs pET30-Silk monomer-15lys-F3, pET30-Silk monomer-15lys-Lyp1 and pET30-Silk monomer-15lys were used to transform E. coli strain RY-3041, and the expression and purification of the proteins was carried out by methods reported previously.[6] SDS-polyacrylamide gel electrophoresis (PAGE) was performed using 4-12% precast NuPage Bis-Tris gels (Invitrogen, Carlsbad, CA). The gel was stained with Colloidal blue (Invitrogen, Carlsbad, CA). Purified samples were extensively dialyzed against acetic buffer and then Milli-Q water. For dialysis, Spectra/Por Biotech Cellulose Ester Dialysis Membranes with MWCO of 100-500 Da (Spectrum Laboratories Inc, Rancho Dominguez, CA) were used.
Preparation and Characterization of the Complex
pDNA encoding Firefly Luciferase (Luc, 7041 bp) was amplified in competent E. coli DH5α (Invitrogen) and purified using EndoFree Plasmid Maxi Kits (Qiagen, Hilden, Germany). The complexes of the recombinant silk proteins with pDNA were prepared as reported previously.[6] Briefly, a solution containing silk protein (0.1 mg/mL) was mixed with the pDNA solution (370 μg/mL) at various N/P ratios. Here, N/P ratio refers to the ratio of number of amines/ phosphates from pDNA. The mixture of recombinant silk and pDNA was incubated at room temperature (~20°C) overnight prior to characterization. The pDNA complex solution (around 70 μL) was added to ultra pure water (450 μL, Invitrogen) and then used as a sample for zeta potential and size measurement. Zeta potential and zeta deviation of samples were measured three times by a zeta potential meter (Zetasizer Nano-ZS, Malvern Instruments Ltd, Worcestershire, UK), and the average data were obtained using Dispersion Technology Software version 5.03 (Malvern Instruments Ltd). The pDNA complexes on cells and tissue culture plates were observed by the Supra55VP Field Emission Scanning Electron Microscope (FESEM) (Carl Zeiss Inc., Oberkochen, Germany) that allows surface examination down to nanometer scales. The samples and cells were fixed and hydrated using 2% glutaraldehyde solution. The bottom of the tissue culture plate well with the samples and cells was cut and sputter coated with a Pt/Pd layer (Cressington 208HR) to ensure the sample conductivity necessary for imaging. The FESEM was operated under high vacuum at 10 keV energy electron beam.
Cell Culture, in-vitro Transfection and Cell Viability
The MDA-MB-435 melanoma cell line, MDA-MB-231 human breast tumor cell line and MCF10A non-tumorigenic mammary breast epithelial cells were used to investigate target specificity of the pDNA complexes. All the cell lines were obtained from the American Type Culture Collection. Cultures were grown to confluence using media consisting of Dulbecco’s Modified Eagle Medium (DMEM) and 10% FBS for MDA-MB-435 and MDA-MB-231 cells. The medium for MCF10A was composed of DMEM/F12, 5% horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 0.1 μg/mL cholera toxin and 10 μg/mL insulin. The cultures were detached from their substrates using 0.25% trypsin (Invitrogen), and then replated in the 24-multiwell plate at a density of 10,000 cells/well. Before transfection experiments, the cells were washed with PBS to remove antibiotic and insulin. Media for the in-vitro transfection experiments was consisting DMEM and 10% FBS. pDNA (1.2 μg) and recombinant silk (appropriate amount) complexes were added into each well. After incubation of the cells for 6 h at 37°C, the media was exchanged to the media for each cell lines without pDNA complexes. After another incubation for 72 h, to evaluate luciferase gene expression quantitatively, a Luciferase assay (Promega, Madison, WI) was performed (n=8). The amount of protein in each well was determined using a BCA protein assay (Pierce Biotechnology, Rockford, IL), and then the relative light units (output) / weight of protein (RLU/mg) was obtained. Lipofectamine 2000 (Invitrogen) was used as a positive control vector. For cell viability, the three types of cells (8,000 cells/well) were seeded into the 96-wells plates containing the pDNA complexes and cultured for 48 h in the media (100 μL) used in the transfection experiment. Cytotoxicity to the cells of the pDNA complexes was characterized by standard 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, Madison, WI) according to the manufacturer’s instructions (n=8).
Statistical Analysis
Statistical differences in cell transfection efficiency and cell viability were determined by unpaired t-test with a two-tailed distribution and differences were considered statistically significant at p<0.05. The data in the cell transfection efficiency and cell viability experiments are expressed as means ± standard deviation.
Results and Discussion
Expression and Purification of Recombinant Silk Protein
The recombinant silk proteins containing poly(L-lysine) (15 a.a.) and THPs, F3 (ML-F3) or Lyp1 (ML-Lyp1), were expressed in E. coli and purified with Ni-NTA chromatography. The domain structure and amino acid sequence of the spider silk variants generated with poly(L-lysine) and THP are shown in Figure 1 and Figure 2. Yields of the recombinant silk proteins were approximately 0.8 mg/L after purification and dialysis. The purified proteins were analyzed by SDS-PAGE and stained with Colloidal blue to evaluate molecular weight and purity (Figure 3). Silk monomer-poly(L-lysine) (15 a.a.) block copolymer (ML), ML-Lyp1 and ML-F3 showed major bands corresponding to a molecular weight of approximately 11 kDa, 12kDa and 14kDa, respectively, almost identical to the theoretical molecular weights (monoisotopic mass) of 7,846.58, 11,119.61 and 13,557.15 Da, respectively, considering that silk-based polymers do not run true to size on SDS-PAGE gels, due to the hydrophobic nature of the protein.[18] The results confirmed that the bioengineered proteins were the expected recombinant silk proteins.
Figure 3.
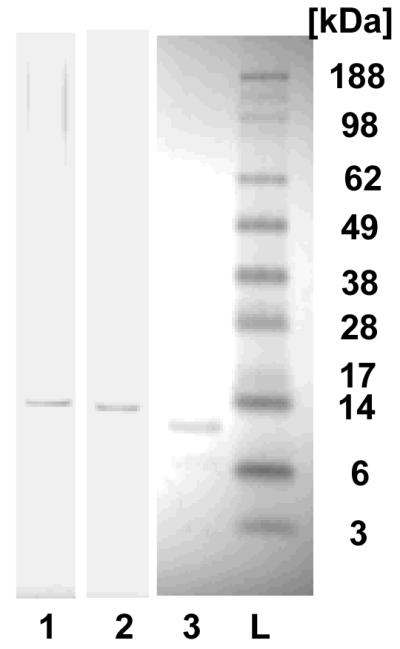
SDS-PAGE of the Silk-polylysine-tumor-homing peptide block copolymers. Lane 1: ML-F3, lane 2: ML-Lyp1, lane 3: ML, and L: molecular weight marker (ladder).
Nanoparticle Formation without pDNA
The ML solutions after the purification with different concentrations were characterized by DLS and zeta-nanosizer (Table 1). The sizes and polydispersity indexes (PDIs) of ML demonstrated formation of ML nanoparticles in the solution with different concentrations raging from 5 to 100 μg/mL. The hydrodynamic diameter of ML nanoparticles decreased from approximately 200 to 80 nm with a decrease in concentration of ML, whereas the zeta potential increased due to positively charged poly(L-lysine) sequence of ML molecules. The smallest average hydrodynamic diameter of ML nanoparticles in the present study was around 80 nm, which is an optimal diameter for gene delivery, at a concentration of 5 μg/mL.
Table 1.
Sizes and zeta potentials of silk monomer-poly(L-lysine) (15 a.a.) block copolymer (ML) with different concentrations.
| Conc. μg/mL | Sizes and PDI a), nm | Zeta potential and deviation, mV |
|---|---|---|
| 100 | 197.0 (0.451) | 0.637 (3.85) |
| 50 | 129.6 (0.349) | 1.35 (3.92) |
| 25 | 155.0 (0.216) | 1.95 (4.31) |
| 10 | 163.4 (0.402) | 7.39 (5.45) |
| 5 | 78.82 (0.433) | 6.51 (5.21) |
polydispersity index.
Preparation and Characterization of pDNA Complexes
Ionic complex formation with pDNA encoding luciferase as a reporter gene and the recombinant silk proteins (ML-F3, ML-Lyp1 and ML) were characterized with different N/P ratios (the ratio of number of amines/ phosphates from pDNA) by zeta-nanosizer and FESEM. The hydrodynamic diameter and zeta potential of the pDNA complexes of the recombinant silks with different N/P ratio are listed in Table 2. The average diameters of the complexes decreased with an increase in N/P ratio, and the pDNA complexes prepared at an N/P 5 demonstrated almost the same values as the complexes at an N/P 10. The zeta potential of pDNA complexes increased with N/P ratio, because of positively charged poly(L-lysine) sequences. Based on these sizes and zeta potentials as well as previous studies,[5,6,8] the most suitable pDNA complexes for in-vitro transfection were determined to be those prepared at an N/P 5 for these recombinant proteins, because of smaller and less negatively charged complexes. In the case of silk 6mer-poly(L-lysine) (n=30) block copolymer, N/P 2 was the best to prepare small and homogeneous ion complexes with pDNA for in-vitro and in-vivo gene delivery.[6,8] More of the silk monomer was required to form homogeneous pDNA complexes as compared to the recombinant proteins containing silk-6mer sequences. The complexes of ML, ML-Lyp1 and ML-F3 prepared at an N/P 5 were 128.5, 251.2 and 220.2 nm in average diameter and −2.04 ± 3.49, −32.9 ± 8.66 and −8.49 ± 2.92 mV in zeta potential, respectively (Table 2). The morphology of the pDNA complexes prepared at an N/P 5 was also observed by FESEM, indicating that the complexes exhibited homogeneous globular morphology and their diameters were less than 100 nm (Figure 4). In the case of FESEM observations, pDNA complexes of silk polymers may be smaller in diameter due to the lower molecular mobility at the surface after fixation and hydration using 2% glutaraldehyde.
Table 2.
Sizes and zeta potentials of the pDNA complexes prepared in this study.
| Sizes and PDI a), nm | Zeta potential and deviation, mV | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| ML-F3 | ML-Lyp1 | ML | ML-F3 | ML-Lyp1 | ML | |
| 2714 | 599.3 | 511.9 | −27.4 | −36.0 | −38.3 | |
| N/P 0.1 | (0.612) | (0.856) | (0.755) | (6.67) | (3.36) | (5.37) |
| 310.0 | 537.6 | 206.1 | −23.0 | −33.4 | −21.3 | |
| N/P 1 | (0.659) | (0.582) | (0.354) | (3.25) | (10.1) | (8.45) |
| 237.3 | 266.9 | 136.0 | −22.5 | −32.3 | −9.36 | |
| N/P 2 | (0.412) | (0.475) | (0.452) | (3.22) | (5.44) | (2.65) |
| 220.2 | 251.2 | 128.5 | −8.49 | −32.9 | −2.04 | |
| N/P 5 | (0.200) | (0.429) | (0.641) | (2.92) | (8.66) | (3.49) |
| 213 | 274.1 | 124.0 | 0.298 | −28.2 | −1.41 | |
| N/P 10 | (0.744) | (0.981) | (0.841) | (3.89) | (9.56) | (6.29) |
| Silk polymer only | 78.8 | 198.3 | 197.0 | 0.902 | −38.5 | 0.637 |
| without pDNA | (0.521) | (0.517) | (0.451) | (3.22) | (7.98) | (3.85) |
polydispersity index.
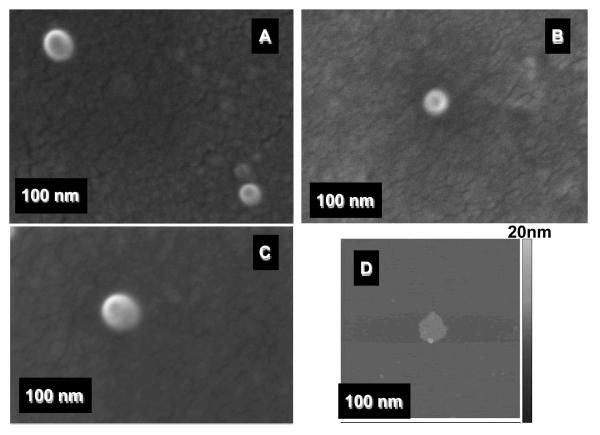
Figure 4.
FESEM images of the pDNA complexes with ML-F3 (A), ML-Lyp1 (B), and ML (C) prepared at an N/P 5. AFM height images of the pDNA complexes with ML-F3 (D). Each scale bar represents 100 nm.
Cytotoxicity of Recombinant Silk Block Copolymers
The cytotoxicity of the pDNA complexes with an N/P ratio of 5 for ML-F3, ML-Lyp1 and ML was determined using the standard MTS assay with MDA-MB-435 melanoma cells, MDA-MB-231 human breast tumor cells and MCF10A mammary breast epithelial cells, which represents two tumorigenic and one healthy cell system. Figure 5 shows the cytotoxicities of the pDNA complexes for 48 h. ML-F3 and ML showed no cytotoxicity to the three types of cells, whereas ML-Lyp1 demonstrated lower cell viability to MDA-MB-435 melanoma cells and MDA-MB-231 human breast tumor cells in comparison to ML-F3 and ML. This is because that Lyp1 peptides bind specifically to tumor lymphatics in certain tumors but not to the lymphatics of normal tissues, as well as induces cell death in cultured tumorigenic cells such as MDA-MB-435 cells.[15,16] Lyp1-conjugated nanoconjugates with a diameter of around 90 nm for targeting drug delivery to lymphatics have been previously shown to exhibit a four-fold increase in-vitro transfection efficiency to BXPC-3 cells as well as an eight-fold increase in-vivo transfection into metastasized lymph nodes.[19] Thus, Lyp1 peptide can be used not only as a tumor-homing peptide but also as a therapeutic peptide to reduce tumor cell numbers. The cell viability of ML-Lyp1 against MCF10A was also lower than those of ML-F3 and ML, suggesting that ML-F3 is less cytotoxic to healthy cells and a more useful candidate as an in-vivo and in-vitro gene delivery carrier in comparison to ML-Lyp1.
Figure 5.
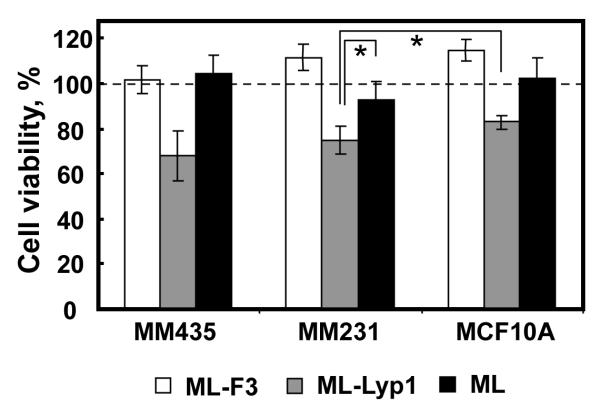
Viability of MDA-MB-435, MDA-MB-231, and MCF10A cells with the pDNA complexes of ML-F3, ML-Lyp1, and ML for 48 h. *Significant difference between two groups at p< 0.05.
In-Vitro Transfection into Three Types of Cells
To evaluate the effects of the THPs on target specificity of the silk-based gene delivery system, in-vitro transfection experiments were carried out using MDA-MB-435, MDA-MB-231 and MCF10A cells. MDA-MB-435 cells had been considered to be breast cancer cells but were recently reported to show melanoma-like properties.[20] Nevertheless, F3 peptides were reported to bind MDA-MB-435 and MDA-MB-231 via nucleolin or nucleolin-related proteins, according to the literature.[13,14] MDA-MB-231 human breast tumor cells were used as a representative model for breast cancer cells. In contrast to these two cell lines, MCF10A non-tumorigenic mammary breast epithelial cells were used as a negative control to evaluate target specificity.
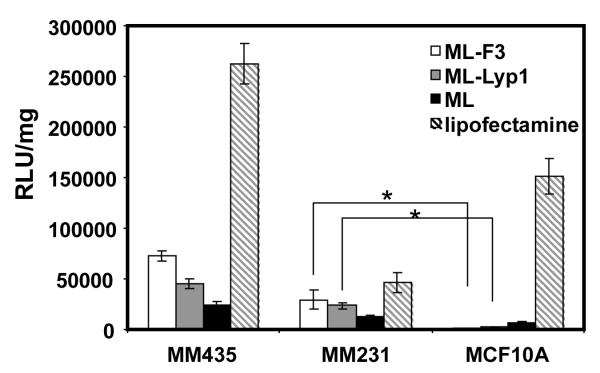
Figure 6 shows the transfection efficiencies to MDA-MB-435, MDA-MB-231, and MCF10A cells for pDNA complexes of ML-F3, ML-Lyp1, ML (N/P 5) and the transfection regent Lipofectamine 2000. ML and Lipofectamine were used as controls. ML, which was composed of silk monomer and polylysine (15a.a.), showed useful transfection to all three cell lines. The pDNA complexes of ML-F3 and ML-Lyp1 exhibited transfection to only MDA-MB-435 and MDA-MB-231 cells, whereas they did not demonstrate useful transfection efficiency to MCF10A non-tumorigenic mammary breast epithelial cells, indicating that the target specificity of the present silk-based gene delivery systems is significantly improved by the additions of THP into the monomeric silk and polylysine sequences. The improvement in target specificity by an addition of THP was reported previously.[12] Here, we discuss transfection specificity (%) to tumor cells, which is defined as follows: [transfection specificity (%) to tumor cells] = [transfection efficiency to tumor cells] / [transfection efficiency to healthy cells] × 100 (%). The transfection specificities of silk 6mer-30 lysines-F3 tumor-homing peptide block copolymer were calculated to be 2,920% to MDA-MB-435 cells (MDA-MB-435: 49100 RLU/mg / MCF10A: 1680 RLU/mg × 100) and 162% to MDA-MB-231 cells (MDA-MB-231: 2720 RLU/mg / MCF10A: 1680 RLU/mg × 100), according to the previous report.[12] Compared to silk 6mer-30 lysines-F3 tumor-homing peptide block copolymer, the present data with ML-F3 showed higher transfection specificity, namely 10,600% to MDA-MB-435 cells (MDA-MB-435: 72500 RLU/mg / MCF10A: 685 RLU/mg × 100) and 4,280% to MDA-MB-231 cells (MDA-MB-231: 29300 RLU/mg / MCF10A: 685 RLU/mg × 100), indicating that higher-content F3 in the silk-based gene delivery systems, like the present system, are capable of enhancing transfection specificity to tumor cells. Further, F3 is a better tumor homing peptide for target delivery to MDA-MB-435 and MDA-MB-231 cells, based on the previous and present results.[12]
Figure 6.
In-vitro transfection results in loading pDNA complexes of ML-F3, ML-Lyp1, and ML into MDA-MB-435, MDA-MB-231, and MCF10A cells. Lipofectamine 2000 was used as a control.
pDNA Complexes on the Cell Surfaces
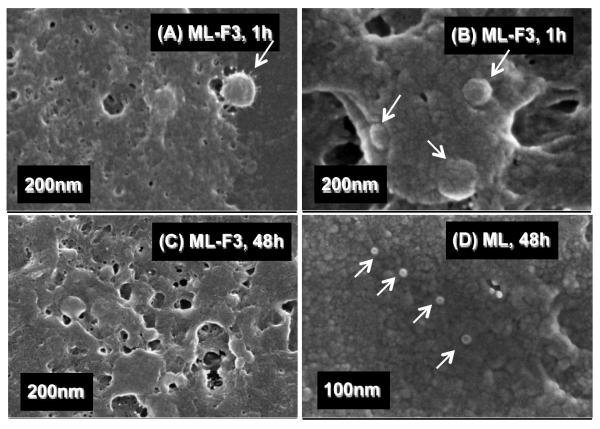
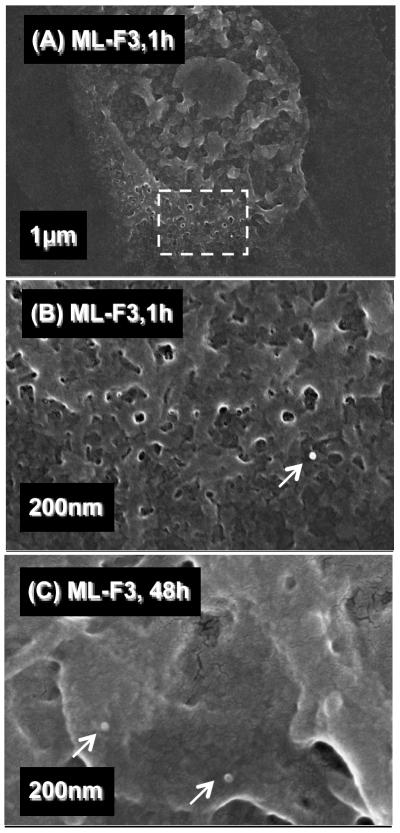
The pDNA complexes fixed at the cell surfaces during the transfection experiments were observed by FESEM. The pDNA complexes of ML-F3, which showed the highest in-vitro transfection specificity and efficiency to MDA-MB-231 cells, were observed on the surface of MDA-MB-231 cells just before (Figure 7A) and during the endocytosis (Figure 7B) after one hour of transfection. These pDNA complexes appeared to be covered with cellular matrices. Almost no pDNA complexes of ML-F3 were observed at the surface of MDA-MB-231 cells after 48 hours of transfection (Figure 7C). In contrast, the pDNA complexes of ML were still observed on the surface of MDA-MB-231 cells after 48 hours of transfection (Figure 7D), indicating that MDA-MB-231 cells do not preferentially uptake the pDNA complexes of ML but those of ML-F3 do. Based on these observations, F3 tumor homing peptides work as ligand molecules to interact with tumorigenic cells. Figure 8A shows the pDNA complexes of ML-F3 at the surface of MCF10A non-tumorigenic mammary breast epithelial cells. Figure 8B, an enlargement of Figure 8A, shows only one complex at the cell surface after incubation of 1 h. Additionally, after 48 h of incubation, the pDNA complexes were still observed at the surface of MCF10A cells (Figure 8C), implying that the non-tumorigenic breast cells do not preferentially uptake the complexes of ML-F3 within a few days. The tumor-target specificity was therefore found to be originating from the adsorption process of the pDNA complexes with THPs on the cell surface. The pDNA complexes of ML-F3 are shown to be significantly specific to tumorigenic cells and useful candidates of tumor-target gene delivery systems. Future studies in-vivo will be required to validate the in-vitro observations reported here.
Figure 7.
FESEM images of the pDNA complexes with ML-F3 on MDA-MB-231 cells at 1 h (A,B) and 48 h (C) after injection of the complexes. White arrows denote the pDNA complexes.
Figure 8.
FESEM images of the pDNA complexes with ML-F3 on MCF10A cells at 1 h (A,B) and 48 h (C) after injection of the complexes. (B) An enlargement of dotted line area in (A). White arrows denote the pDNA complexes.
Conclusion
The present study demonstrates that pDNA complexes of recombinant silk proteins with a relatively high content of THP (25 mol%, 3.4 kDa/13.6 kDa), which are globular and approximately 100 nm in diameter, show significant target specificity to tumorigenic cells by additions of F3 and Lyp1. Lyp1 is significantly cytotoxic to MCF10A cells, and ML-F3 is therefore considered to be a less cytotoxic candidate for target delivery into tumorigenic cells. The mechanism of the specificity of the pDNA complexes to cells is adsorption of the complexes on the cell surface, according to FESEM. Considering previous reports that showed controlled release and relatively high DNase resistance of spider silk-based gene delivery systems, as well as the target specificity reported in the present study,[8,12] the bioengineered silk delivery systems can serve as a versatile and useful new platform polymer for not only non-viral gene delivery but also for drug delivery.
Acknowledgements
This work has been supported by grants from the NIH (P41 Tissue Engineering Resource Center, EB002520) and the NSF (DK). KN was supported by a JSPS Postdoctoral Fellowship for Research Abroad. SEM experiments were performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. CNS is part of the Faculty of Arts and Sciences at Harvard University.
Footnotes
Present address: K. Numata, Enzyme Research Team, RIKEN, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan.
References
- [1].Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- [2].Wong Po Foo C, Kaplan DL. Adv Drug Deliv Rev. 2002;54:1131. doi: 10.1016/s0169-409x(02)00061-3. [DOI] [PubMed] [Google Scholar]
- [3].Numata K, Cebe P, Kaplan DL. Biomaterials. 2010;31:2926. doi: 10.1016/j.biomaterials.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang Y, Kim HJ, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2006;27:6064. doi: 10.1016/j.biomaterials.2006.07.008. [DOI] [PubMed] [Google Scholar]
- [5].Numata K, Subramanian B, Currie HA, Kaplan DL. Biomaterials. 2009;30:5775. doi: 10.1016/j.biomaterials.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Numata K, Hamasaki J, Subramanian B, Kaplan DL. J Control Release. 2010 doi: 10.1016/j.jconrel.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Numata K, Kaplan DL. Adv Drug Deliv Rev. 2010;62:1497. doi: 10.1016/j.addr.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Numata K, Kaplan DL. Biomacromolecules. 2010;11:3189. doi: 10.1021/bm101055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Edelstein ML, Abedi MR, Wixon J. J Gene Med. 2007;9:833. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- [10].Edelstein ML, Abedi MR, Wixon J, Edelstein RM. J Gene Med. 2004;6:597. doi: 10.1002/jgm.619. [DOI] [PubMed] [Google Scholar]
- [11].Lundstrom K. Trends Biotechnol. 2003;21:117. doi: 10.1016/S0167-7799(02)00042-2. [DOI] [PubMed] [Google Scholar]
- [12].Numata K, Reagan MR, Goldstein RH, M. R, Kaplan D. Bioconjugate Chem. doi: 10.1021/bc200170u. in press (DOI: 10.1021/bc200170u) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E. J Cell Biol. 2003;163:871. doi: 10.1083/jcb.200304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Porkka K, Laakkonen P, Hoffman JA, Bernasconi M, Ruoslahti E. Proc Natl Acad Sci U S A. 2002;99:7444. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Laakkonen P, Akerman ME, Biliran H, Yang M, Ferrer F, Karpanen T, Hoffman RM, Ruoslahti E. Proc Natl Acad Sci U S A. 2004;101:9381. doi: 10.1073/pnas.0403317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. Nat Med. 2002;8:751. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- [17].Harrison SC. Science. 2010;329:1026. doi: 10.1126/science.1194922. [DOI] [PubMed] [Google Scholar]
- [18].Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Biochemistry. 1995;34:10879. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- [19].Luo G, Yu X, Jin C, Yang F, Fu D, Long J, Xu J, Zhan C, Lu W. Int J Pharm. 2010;385:150. doi: 10.1016/j.ijpharm.2009.10.014. [DOI] [PubMed] [Google Scholar]
- [20].Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. Breast Cancer Res Treat. 2007;104:13. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]