Abstract
A high throughput screening campaign was conducted to identify small molecules with the ability to inhibit the interaction between the vitamin D receptor (VDR) and steroid receptor coactivator 2. These inhibitors represent novel molecular probes to modulate gene regulation mediated by VDR. The peroxisome proliferator-activated receptor δ (PPARδ) agonist GW0742 was among the identified VDR-coactivator inhibitors and has been characterized herein as a pan nuclear receptor antagonist at concentrations higher than 12.1 µM. The highest antagonist activity for GW0742 was found for VDR and the androgen receptor (AR). Surprisingly, GW0742 behaved as PPAR agonist/antagonist activating transcription at lower concentration and inhibiting this effect at higher concentrations. A unique spectroscopic property of GW0742 was identified as well. In the presence of rhodamine-derived molecules, GW0742+ increased fluorescence intensity and fluorescence polarization at an excitation wavelength of 595 nm and emission wavelength of 615 nm in a dose dependent manner. The GW0742-inhibited NR-coactivator binding resulted in a reduced expression of five different NR target genes in LNCaP cells in the presence of agonist. Especially VDR target genes CYP24A1, IGFBP-3 and TRPV6 were negatively regulated by GW0742. GW0742 is the first VDR ligand inhibitor lacking the secosteroid structure of VDR ligand antagonists. Nevertheless, the VDR-meditated downstream process of cell differentiation was antagonized by GW0742 in HL-60 cells that were pretreated with the endogenous VDR agonist 1,25-dihydroxyvitamin D3.
Keywords: GW0742, vitamin D receptor, androgen receptor, peroxisome proliferator-activated receptor, steroid receptor coactivator 2, fluorescence polarization, high throughput screening, IGFBP-3, CYP24A1, TRPV6, UGT1A1, PSA, BTG2
Nuclear receptor (NR) ligands represent one of the largest class of currently FDA approved drugs for a wide variety of human diseases.1 The 48 identified members of this superfamily of transcription factors share a similar protein structure and bind, in case of the steroid hormone receptors, to endogenous ligands with a similar steroid structure.2 During the past decade, many new synthetic NR ligands with diverse scaffolds have been introduced, especially for the peroxisome proliferation activated receptors (PPARα (3363 ligands), PPARγ (3891 ligands), and PPARδ (1729 ligands).3 Although the structural optimization of NR ligand has been guided by affinity to a given NR, it is generally accepted that ligand selectivity among NRs has to be optimized as well in order to reduce side effects. The discovery that NR–coactivator binding is required for NR-mediated transcription led to a more stringent concept of selectivity for NR ligands.4, 5 NR ligand drug candidates should not only bind selectively to a certain NR but also selectively modulate recruitment of particular nuclear receptor coactivators.6 Many NR ligands with improved selectivity have been recently introduced but the sheer number of NRs and affiliated coactivators that govern their specific gene regulation pattern has complicated the selectivity assessment of these ligands.7
A new concept to selectively modulate NR function involves the development of small molecules that directly inhibit the interaction between NRs and coregulators instead of interacting with the receptor ligand binding pocket to allosterically modulate interactions of receptor with its target proteins.8 The development of an inhibitor selective towards a particular coregulator-NR interaction could be highly useful for the vitamin D receptor (VDR).9 VDR binds to its endogenous ligand, 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3), with high affinity,10 mediating changes in transcription of genes responsible for cell differentiation, proliferation, and calcium homeostasis.11 Based on its biological function, the VDR has been identified as an important pharmaceutical target for the treatment of metabolic disorders, skin diseases, cancer, autoimmune diseases, and cardiovascular diseases.12 VDR, similar to other NRs, contains several functional domains, including a ligand-binding domain (VDR-LBD), which mediates ligand-dependent gene regulation.13 VDR binds DNA as a heterodimer with the retinoid X receptor (RXR).14 In the unliganded state, VDR is associated with corepressor proteins, which repress transcription of VDR target genes.15 In the presence of 1,25-(OH)2D3, the VDR-LBD undergoes a conformational change which prevents corepressor binding and permits interactions with coactivators, resulting in the formation of a multi-protein complex responsible for VDR-mediated transcription.16
VDR agonists have been developed to treat metabolic bone diseases and proliferative skin disorders.12 In contrast to the large number of reported VDR agonists, only a limited number of VDR ligand antagonists have been described with the ability to allosterically inhibit the interactions between VDR and its coactivators.17–24 All of these antagonists are based on the secosteroid structure of 1,25-(OH)2D3. Mita et al. introduced the first reversible small molecule direct inhibitors of VDR–coactivator interactions with moderate NR selectivity.25 The reported compounds inhibited both VDR-mediated and estrogen receptor β-mediated transcription. Recently, we reported the first irreversible direct inhibitors of the VDR–coactivator interaction.26 This new class of compounds selectively inhibited the interaction between VDR and coactivator SRC227 among six other NR–coactivator interactions tested and reduced the expression of VDR target gene TRPV6 in DU-145 prostate cancer cells in the presence of 1,25-(OH)2D3.
Herein, we describe the evaluation of compound GW0742, which was identified during a high throughput screen (HTS) to identify small molecules that inhibit the interaction between VDR and nuclear receptor coactivators 2. GW0742 was introduced by GlaxoSmithKline in 2003 as a highly selective agonist for the peroxisome proliferator activated receptor δ (PPARδ).28 Since then, GW0742 has been investigated in cell-based assays and in vivo to understand the role of PPARδ in hypertension,29, 30 diabetes,31, 32 inflammation,33, 34 obesity,35 and cancer.36–38 Interestingly, PPAR agonists have been shown to inhibit the transcription of VDR target gene CYP24A1 in the presence of 1,25-(OH)2D3.39 The results presented herein show that GW0742, at micromolar concentrations, behaves as an antagonist of VDR and other nuclear receptors.
Experimental Procedures
Reagents
1,25-(OH)2D3 (calcitriol) was purchased from Endotherm, Germany; GW0742 was purchased from Tocris, triiodothyronine (T3), GW7647, Rosiglitazone, dihydrotestosterone (DHT), Bexarotene, and estradiol were purchased from Sigma. LG190178 was synthesized using a published procedure.40
Labeled coactivator Peptides
Peptides, such as SRC2–3 (CLQEKHRILHKLLQNGNSPA),41 SRC2-2 [CEKHKILHRLLQDSS], DRIP2 [CNTKNHPMLMNLLKDNPAQD] were purchased and labeled with cysteine-reactive fluorophores, such as fluorescein maleimides, Texas-Red maleimides and Alexa Fluor 647 maleimides, in DMF/PBS 50:50. After purification by HPLC, the corresponding labeled peptides were dissolved in DMSO and stored at −20°C.
Protein Expression and Purification
The VDR-LBDmt DNA was kindly provided by D. Moras42 and cloned into pMAL-c2X vector (New England Biolabs). A detailed expression and purification protocol for VDR was reported.41 Detailed expression and purification of protocols for PPARγ-LBD, TRβ-LBD, and AR-LBD were reported as well.43
High Throughput FP Assay
The HTS was carried out at the NIH Chemical Genomic Center (NCGS) Bethesda. For details see [AID:504847/pubchem].
VDR–SRC2–3 inhibition confirmation assay
These assays were conducted in 384 well black polystyrene plates (Corning) using a buffer (25 mM PIPES (pH 6.75) 50 mM NaCl, 0.01% NP-40, 2% DMSO, VDR-LBD protein (0.6 µM), LG190178 (5 µM), and Alexa Fluor 647-labeled SRC2–3, Texas Red-labeled SRC2–3, or Fluorescein-labeled SRC2–3 (7.5 nM) and optional mercaptoethanol 1 mM or 100 mM as indicated). Small molecule transfer into 20 µl assay solution was accomplished using a stainless steel pin tool (V&P Scientific), delivering 100 nl of a 10 mM compound solution. Fluorescence polarization was detected after 2h at an excitation/emission wavelength of 650/665 (Alexa Fluor), 595/615 nm (Texas Red), 495/520 (Fluorescein). Three independent experiments were carried out in quadruplicate and data was analyzed using nonlinear regression with variable slope (GraphPrism).
VDR Ligand Competition Assay
Ligand antagonism was determined by using a FP assay (PolarScreen, Invitrogen), which employs a rhodamine-labeled VDR ligand. Two independent experiments were conducted in quadruplicate and data was analyzed using nonlinear regression with variable slope (GraphPrism).
Transcription assays
Briefly, HEK 293T cells (ATTC) were cultured in 75 cm2 flasks using DMEM/High Glucose (Hyclone, #SH3024301), non-essential amino acids, HEPES (10 mM), penicillin and streptomycin, and 10% of dialyzed FBS (Invitrogen,#26400–044). At 70–80 percent confluency, 2 ml of untreated DMEM containing 1.56 µg of NR plasmid, 16 µg of a luciferase reporter gene, Lipofectamine™ LTX (75 µl), and PLUS™ reagent (25 µl) was added. After 16 hours, the cells were harvested with 0.05% Trypsin (3 ml) (Hyclone, #SH3023601), added to 15 ml of DMEM high Glucose (Hyclone, #SH3028401), non-essential amino acids, sodium pyruvate (1 mM), HEPES (10 mM), penicillin and streptomycin, and 2% percent charcoal treated FBS (Invitrogen, #12676–011), and spun down for 2 minutes at 1000 rpm. The cell were resuspended in the same media and plated in sterile cell culture treated black 384 well plates with optical bottom (Nunc, #142761) at a concentration of 15,000 cells per well, which were treated with a 0.25% solution of Matrigel (BD Bioscience, #354234) beforehand. After 2 hours, plated cells were treated with small molecules in vehicle DMSO, followed by a 16 hours incubation time. The following NR agonists were used: 1,25(OH)2D3 (10 nM), GW7647 (30 nM), Rosiglitazone (300 nM), GW0742 (50 nM), DHT (10 nM), Bexarotene (200 nM), T3 (10 nM), and estradiol 10 nM). Transcription was determined using Bright-Glo (Promega). Cell viability was determined using Cell-Titer-Glo (Promega) for identical treated cells. Controls for cell viability were 3-dibutylamino-1-(4-hexyl-phenyl)-propan-1-one (100 µM in DMSO) (positive) and DMSO (negative). Three independent experiments were conducted in quadruplicate and data was analyzed using nonlinear regression with variable slope (GraphPrism).
Western Blot of in vitro binding reactions between SRC2 bearing all three NIDs and VDR-LBD in the presence of GW0742
GST fusion to the SRC2 bearing all three NIDs were expressed in Escherichia coli BL21. Cultures were grown to OD600 = 0.5–0.6 at 22 °C and induced with 0.5 mM isopropyl-D-thiogalactoside for 12 h. The cultures were centrifuged (1000 x g), and bacterial pellets were resuspended in 20 mM TRIS, pH 7.4, 200 mM NaCl, 1 mM NaN3, 0.5M EDTA, 1 mM DTT, protein inhibitors cocktail (Roche) and sonicated. Debris was pelleted by centrifugation (100,000 x g). The supernatant was incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) and washed. Protein on bead was stored with 10% glycerol at –20°C. Each pull-down reaction was carried out in 100 µl buffer (25 mM PIPES (pH 6.75) 50 mM NaCl, 0.01% NP-40, 2% DMSO) using 100 nM calcitriol, 10 µM VDR-LBD-MPB, and GW0742. After 2 hours at rt, 15 µl of SRC2-beads was added to each reaction followed by 30 minutes incubation. The reaction was filtered, washed with buffer (100 µl) and eluted from the bead using a buffer and 10 mM reduced glutathione. Separation was carried out using SDS-PAGE followed Western blotting using standard procedures with anti-MBP (E8032S, New England BioLabs) and anti-mouse IgG-Tr (sc-2781, Santa Cruz).
Semi-quantitative real time PCR
Briefly, LNCaP cells (ATTC) were cultured in 75 cm2 flasks using DMEM/High Glucose (Hyclone, #SH3024301), non-essential amino acids, HEPES (10 mM), penicillin and streptomycin, and 10% dialyzed FBS (Invitrogen, #26400–044). LNCaP cells were incubated at 37 °C with GW0742 (20 µM) in the presence or absence of DHT (10 nM), Rosiglitazone (5 µM), Triiodothyronine (10 nM), or 1,25(OH)2D3 (10 nM) for 18 h in a 6 well plate. Cells of each well were harvested using 0.3 ml of 0.05% Trypsin (Hyclone, #SH3023601) and added to DMEM media (1 mL). The cell suspension was spun down for 2 min at 1000 rpm, media was removed and the cell pellet was resuspended in RTL buffer (RNAeasy kit, Qiagen) with the addition of mercaptoethanol. The cells were lysed using a QIAshredder (Qiagen) and total RNA was isolated using RNAeasy kit (Qiagen). A QuantiFast SYBR Green RT-PCR Kit (Qiagen) was used for the real time PCR following manufacturer’s recommendations. Primers used in these studies are as follows: GAPDH FP 5’-accacagtccatgccatcac-3’, GAPDH RP 5’-tccaccaccctgttgctgta-3’; TRPV6 FP 5’-ACTGTCATTGGGGCTATCATC-3’, TRPV RP 5’-CAGCAGAATCGCATCAGGTC-3’; IGFBP3 FP 5’-CGCCAGCTCCAGGAAATG-3’, IGFBP3 RP 5’- GCATGCCCTTTCTTGATGATG-3’; UGT1A1 FP 5’-GCCATGCAGCCTGGAATT-3', UGT1A1 RP 5'-GGCCTGGGCACGTAGGA-3'; PSA FP 5'-TCTGCCTTTGTCCCCTAGAT-3', PSA RP 5'-AACCTTCATTCCCCAGGACT-3'; BTG2 FP 5'-AGGGTAACGCTGTCTTGTGG-3', BTG2 RP 5'-TACAGTTCCCCAGGTTGAGG-3'; ANGPTL4 FP 5'-AGGGTAACGCTGTCTTGTGG-3', ANGPTL4 FP 5'-TACAGTTCCCCAGGTTGAGG-3'. Real-time rt-PCR was carried out on a Mastercycler (Eppendorf). We used the ΔΔCt method to measure the fold change in gene expression of target genes. Standard errors of mean were calculated from 4 biological independent experiments performed in triplicates.
HL60 differentiation assay
HL60 cells (ATTC) were cultured in 75 cm2 flasks using RPMI-1640 (ATTC, #30–2001), non-essential amino acids, penicillin and streptomycin, and 10% dialyzed FBS (Invitrogen, #26400–044). The cells were treated with 1,25(OH)2D3 (50 nM) or vehicle DMSO and plated at a concentration of 2M cell/ml in 384 well plates with optical bottom (Nunc 142761). Cells were incubated with different concentrations of GW0742 for 4 days at 37°C. 4 µl of a nitro blue tetrazolium chloride (5.4 mg/ml) and phorbol-12-myristate-13-acetate (PMA) (100 µg/ml) was added to each well and incubated for 30 minutes at 37°C. The amount of oxidized nitro blue tetrazolium was quantified at a wavelength of 550 nm applying a 4×4 scan of full well area using the Tecan M1000. Two independent experiments were conducted in quadruplicate and data was analyzed using nonlinear regression with variable slope (GraphPrism).
Results
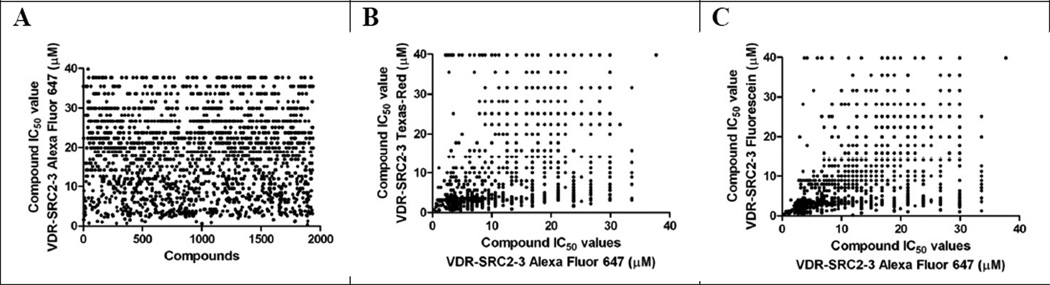
A small molecule screen of about 390,000 compounds at different concentrations was conducted in 1536-well black polystyrene plates using fluorescence polarization (FP) [AID:504847/pubchem]. The HTS identified inhibitors of the interaction between VDR and fluorescently labeled peptide SRC2–3, representing the NR interaction domain of the steroid receptor coactivator 2 (SRC2). The screen was carried out in the presence of VDR-LBD protein (800 nM), VDR agonist LG19017840 (5 µM) and Alexa Fluor 647 labeled coregulator peptide SRC2–3 (7.5 nM). Fluorescence polarization was detected at an emission wavelength at 650 nm and excitation wavelength of 665 nm. 1938 compounds exhibited IC50 values of 40 µM and lower in the primary screen (Figure 1, A). Furthermore, these compounds were validated with two alternative FP assays employing Texas Red-labeled SRC2–3 (excitation/emission 595/615 nm) [AID:602201/pubchem] and Fluorescein-labeled SRC2–3 (excitation/emission 495/520 nm) [AID:602200/pubchem] (Figure 1, B and C).
Figure 1.
IC50 values of 1938 hit compounds using VDR-LBD and SRC2–3 labeled with: A Alexa Fluor 647; B Texas-Red in correlation with Alexa Fluor 647; C Fluorescein in correlation with Alexa Fluor 647.
Further characterization of the 1938 initial hits revealed that 69% of the compounds exhibited IC50 values of 40 µM and lower in the FP assay employing VDR-LBD and Texas Red-labeled SRC2–3 (Figure 1, B). A good correlation between compound activities in this assay and validation assays was observed. 83% of the 1938 hit compounds also exhibited IC50 values of ≤ 40 µM in the FP assay employing Fluorescein-labeled SRC2–3 (Figure 1, C). Also in this case, a good correlation with the assay employing SRC2–3 Alexa Fluor 647 was observed.
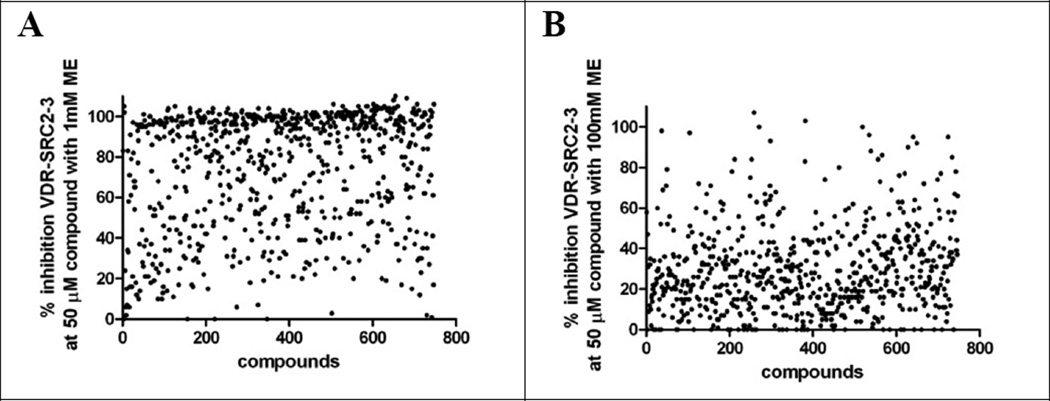
Based on these results, 747 compounds were selected with an emphasis on diverse scaffolds and functionality. In order to delineate compounds that might inhibit coactivator binding by irreversibly reacting with cysteine residues of the VDR-LBD protein, the VDR–SRC2–3 FP assay was carried out in the presence of 1 mM or 100 mM 2-mercaptoethanol (ME). The hypothesis here is that electrophilic compounds are more likely to react in the presence of excess nucleophilic ME than with protein nucleophilic protein residues such as cysteine. We introduced this method for the electrophilic compound DHPPA, which showed reduced thyroid receptor binding in the presence of 100 mM ME.44 The results of both assays are depicted in Figure 2.
Figure 2.
Inhibition for the VDR-SRC2–3 Alexa Fluor 647 interaction by small molecules (50 µM) in the presence of 1 mM or 100 mM of 2-mercaptoethanol (ME).
Interestingly, we observed significant reduction of the majority of compounds to inhibit the interaction between VDR and SRC2–3 in the presence of 100 mM ME in comparison with 1 mM ME (Figure 2, B). This suggests that a large number of our initial hits may inhibit VDR–coactivator interactions via modification of surface cysteine residues. However, activities of a significant minority of compounds were not influenced by a higher concentration of ME (100 mM) and among these was GW0742 (Figure 3, A).
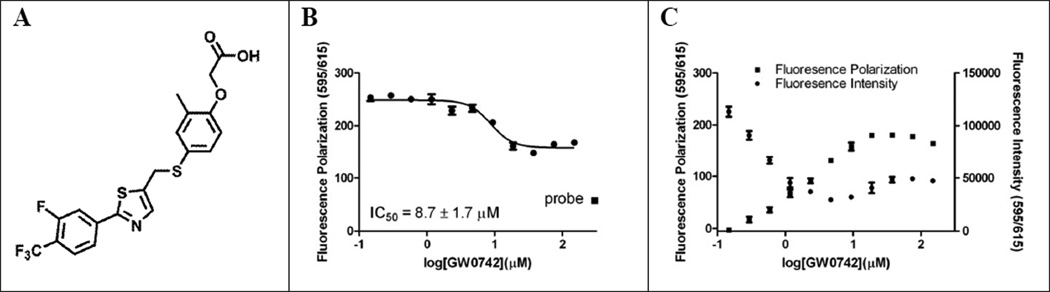
Figure 3.
VDR–GW0742 interactions. A chemical structure of GW0742; B FP VDR ligand competition assay; C Fluorescence interaction between GW0742 and rhodamine-labeled VDR ligand.
The following activities were measured for GW0742 during the HTS campaign: 1. VDR–SRC2–3 inhibition (FP assays) (IC50 value): 14 µM (Alexa Fluor 647), 25.1 µM (Fluorescein), inconclusive (Texas-Red); 2. VDR transcription assay (VDR–GeneBLAzer, Invitrogen): The IC50 value was 26 µM at a maximum response of 30% as defined by the absence of VDR agonist 1,25(OH)2D3 [AID:602202, pubchem]; 3. Toxicity in HEK293T cells (CellTiter-Glo, Promega): 18% cell death at a concentration of 45.8 µM [AID:602204, pubchem].
In order to determine that GW0742 is interacting with VDR and not the coactivator a commercially available VDR ligand binding FP assay was used employing VDR and a rhodamine-labeled VDR ligand (Figure 3, B).
Using increasing concentrations of GW0742, competition of VDR–ligand binding was confirmed with an IC50 value of 8.7 ± 1.7 µM. Interestingly, partial inhibition was observed with respect to 100% free probe (55 mP) measured in the absence of VDR (Figure 3, B (square)). Prompted by this result and by the fact that the FP assay employing Texas-Red SRC2–3 was inconclusive during the HTS campaign, we analyzed the interaction between GW0742 and rhodamine-labeled VDR ligand in the absence of VDR (Figure 3, C). Surprisingly, we found that both fluorescence intensity and fluorescence polarization of the rhodamine-labeled VDR ligand increased at higher concentrations of GW0742. Similar results were found for the Texas-Red labeled SRC2–3 in the presence of GW0742 using the same excitation and emission wavelength (results not shown). GW0742 by itself showed no significant fluorescence under these conditions. Based on these results, we concluded that GW0742 inhibited the interaction between VDR and rhodamine-labeled VDR ligand, but that IC50 values measured with Alexa Fluor 647 labeled SRC2–3 are a more reliable measure of the inhibitory activity of the ligand. No change of fluorescence intensity and fluorescence polarization was detected at an emission wavelength at 650 nm and excitation wavelength of 665 nm for Alexa Fluor 647 labeled SRC2–3 and increasing concentrations of GW0742 (results not shown). In addition, we investigated the interaction between GW0742 and SRC2 using isothermal titration calorimetry (ITC) but no specific interaction was observed (supporting information).
FP assays were carried out to determine the selectivity of GW0742 with respect to its ability to inhibit the interaction between other nuclear receptors and Alexa Fluor 647-labeled coactivator peptides. These included VDR and SRC2–3, thyroid hormone receptor β (TRβ) and SRC2–2, peroxisome proliferation activated receptor γ (PPARγ) and DRIP2, derived from coactivator DRIP20545, and AR and SRC2–3. The results are depicted in Figure 4.
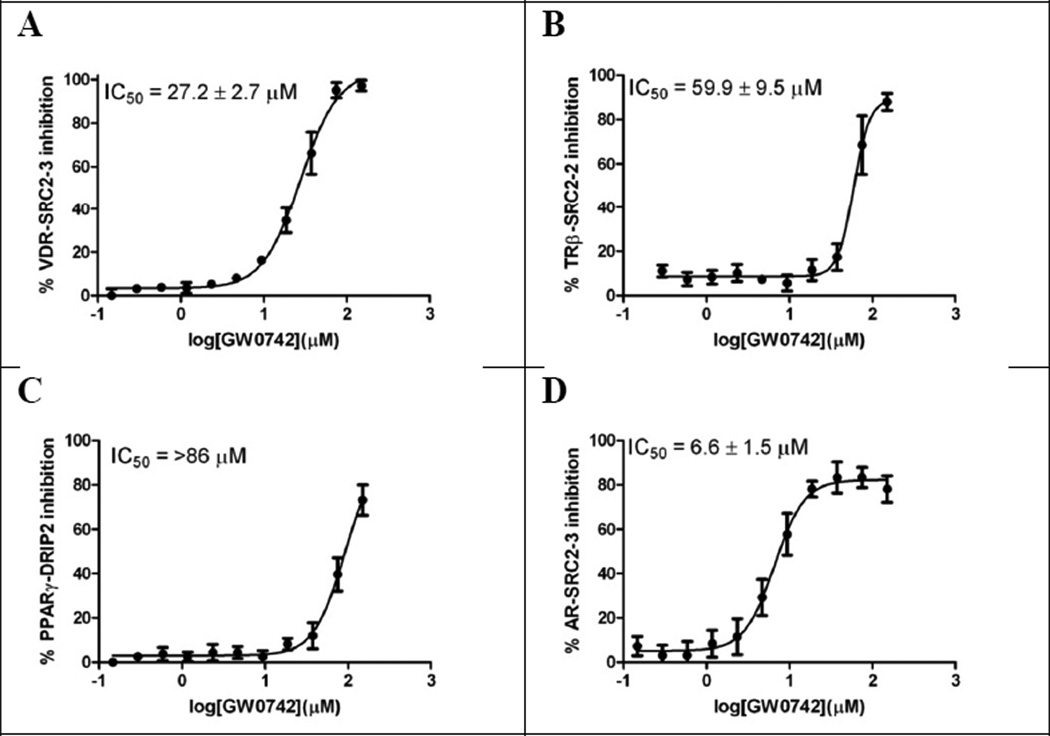
Figure 4.
Summary of nuclear receptor-coactivator inhibition in the presence of GW0742 using FP. A Inhibition of the VDR–SRC2–3 interaction; B Inhibition of the TRβ–SRC2–2 interaction; C Inhibition of the PPARγ–DRIP2 interaction; D Inhibition of the AR–SRC2–3 interaction. The conditions for different NRs were as follows: VDR-LBD (0.8 µM), Alexa Fluor 647 labeled SRC2–3 (7 nM), and LG190178 (2 µM); TRβ-LBD (1 µM), Alexa Fluor 647 labeled SRC2– 2 (7 nM), and triiodothyronine (10 nM); PPARγ-LBD (5 µM), Alexa Fluor 647 labeled DRIP2 (7 nM), and rosiglitazone (5 µM); AR-LBD (5 µM), Alexa Fluor 647 labeled SRC2–3 (7 nM), and dihydrotestosterone (10 nM) were incubated with GW0742 for 2h.
The inhibition of VDR-SRC2–3 binding in the presence of GW0742, demonstrated during the HTS campaign, was confirmed with a calculated IC50 value of 27.2 ± 2.7 µM (Figure 4, A). The ability of GW0742 to disrupt the TR–SRC2–2 and the PPARγ–DRIP2 interactions was modest with IC50 values of 59.9 ± 9.5 µM and >86 µM, respectively (Figure 4, B and C). The strongest GW0742 inhibition was observed for the AR–SRC2–3 interaction with an IC50 value of 6.6 ± 1.5 µM (Figure 4, D).
Many other groups have investigated GW0742 in respect to different nuclear receptor binding using a variety of assays. These include PPARα, PPARγ and PPARδ using cell-based transactivation assays (alkaline phosphatase as reporter enzyme), where GW0742 exhibited agonistic EC50 values of 1.1 ± 0.109 µM (PPARα), 2.0 ± 1.3 µM (PPARγ), and 0.001± 0.002 µM (PPARδ), respectively. Antagonistic activity of GW0742 was also observed in HTS campaigns using a GeneBLAzer assays (Invitrogen) employing the estrogen receptor α (ERα, AID:588513, pubchem), the glucocorticoid receptor (GR, AID:588533, pubchem), PPAR δ (AID:588535, pubchem), PPARγ (AID588537, pubchem), the retinoid X receptor (RxR, AID:588546, pubchem), and TRβ (AID:588547, pubchem). Activity of GW0742 found for the majority of these nuclear receptors was in the micromolar range and exhibited inconclusive dose-response curves in most cases.
Therefore, an array of nuclear receptor mediated transcription assays was used to confirm the pan nuclear receptor–coactivator inhibition caused by high concentrations of GW0742 in the FP assay. For VDR, TRα, and TRβ, HEK293T cells were transfected with nuclear receptor expression vector and luciferase reporter under control of the cognate nuclear receptor response elements or target gene promoters. For all other nuclear receptors, one-hybrid assays employing nuclear receptor LBDs fused to the DNA binding domain (DBD) of GAL4 and a luciferase reporter plasmid driven by five copies of the GAL4 binding site fused to a tk promoter were employed. The assays were carried out in 384-well plates and the amount of luciferase formed was quantified after 18 hours using Bright-Glo. The transcription assays were carried out in the presence and absence of endogenous ligands or synthetic agonists using different concentrations of GW0742. The EC50 and IC50 values are summarized in Table 1.
Table 1.
Evaluation of GW0742 in different nuclear receptor reporter assays.
| Entry | Nuclear Receptor |
Agonist EC50 (µM)11 |
Antagonist IC50 (µM)11 |
|---|---|---|---|
| 1 | VDR | 10 | 14.7 ± 1.51 |
| 2 | PPARα | 1.3 ± 0.3 | 37.4 ± 8.22 |
| 3 | PPARγ | 2.8 ± 0.7 | 20.2 ± 5.43 |
| 4 | PPARδ | 0.0037 ± 0.0014 | 21.6 ± 4.94 |
| 5 | AR | 10 | 12.1 ± 5.35 |
| 6 | RxRα | 10 | 22.9 ± 3.86 |
| 7 | TRα | 10 | 31.4 ± 4.07 |
| 8 | TRβ | 10 | 25.8 ± 5.28 |
| 9 | ERα | 10 | 21.3 ± 7.29 |
1,25(OH)2D3(10nM)
GW7647 (30nM)
Rosiglitazone (300nM)
GW0742 (50nM)
DHT (10nM)
Bexarotene (200nM)
T3 (10nM)
T3 (10nM)
estradiol (10nM)
no activation detected
three independent experiments were conducted in quadruplicate and data was analyzed using nonlinear regression with variable slope (GraphPrism).
As expected, GW0742 was only able to activate transcription mediated by PPARα, PPARγ, and PPARδ among the nuclear receptors tested. The measured EC50 values, 1.3 ± 0.3 µM (PPARα), 2.8 ± 0.7 µM (PPARγ), and 0.0037± 0.0014 µM (PPARδ) (Table 1, entries 2, 3, and 4), were in agreement with previously reported results 28. In contrast, inhibition of transcription was found for all nuclear receptors within an IC50 value range of 12.1 to 37.4 µM. Interestingly, GW0742 exhibited significantly lower IC50 values for VDR and AR relative to other nuclear receptors with IC50 values of 12.1 µM and 14.7 µM, respectively (Table 1, entries 1 and 5). The cell viability in the presence of GW0742 was quantified under the same conditions using CellTiter-Glo (data not shown). At a GW0742 concentration of 37.5 µM, 93 ± 3 % of the cell population was viable and at a concentration of 18.7 µM of GW0742 no toxicity was observed. This observation is in agreement with the HTS data described before.
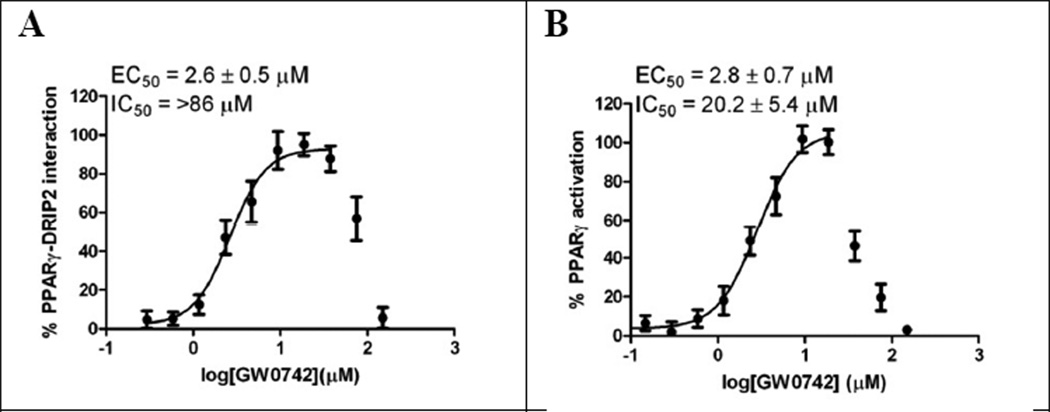
For PPARs, GW0742 exhibited an agonistic effect at lower concentration and an antagonistic effect at higher concentrations as shown for PPARγ for the FP assay (Figure 4) and transcription assay depicted in Figure 5. GW0742 EC50 values determined in both assays are very similar with 2.6 µM (FP assay) and 2.8 µM (transcription), respectively. At GW0742 concentrations of >20 µM, we observed significant PPARγ-mediated transcriptional inhibition (Figure 5, B), whereas significant inhibition between PPARγ-LBD and DRIP2 was observed for GW0742 at higher concentrations (>86 µM).
Figure 5.
Agonistic and antagonist effect of GW0742 with respect to PPARγ. A FP assay using PPARγ-LBD and Alexa Fluor 647-labeled DRIP2; B One-hybrid transcription assay employing PPARγ.
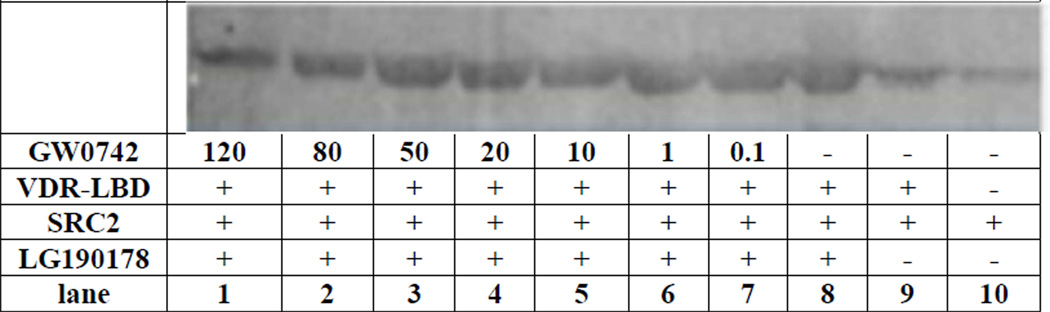
GW0742 also inhibited the interaction between VDR-LBD and an SRC2 fragment bearing three nuclear interaction domains in a western pull-down assay (Figure 6). Control experiments indicated that SRC2 binds to VDR in the presence of VDR ligand LG190178 (Figure 6, lane 8) but not in the absence of ligand (Figure 6, lane 9). The faint bands in the absence of ligand and VDR were caused by nonspecific binding of VDR antibody to SRC2 beads. The VDR–SRC2 interaction was blocked in a dose dependent manner by GW0742 (Figure 8, lane 1–7). Although significant inhibition of the VDR–SRC2 interaction was observed at 80–120 µM GW0742, residual bands could still be detected partially due to unspecific binding of the VDR antibody to SRC2 beads as observed for the controls.
Figure 6.
Inhibition of the VDR–SRC2 interaction by GW0742 analyzed by a Western pull-down assay.
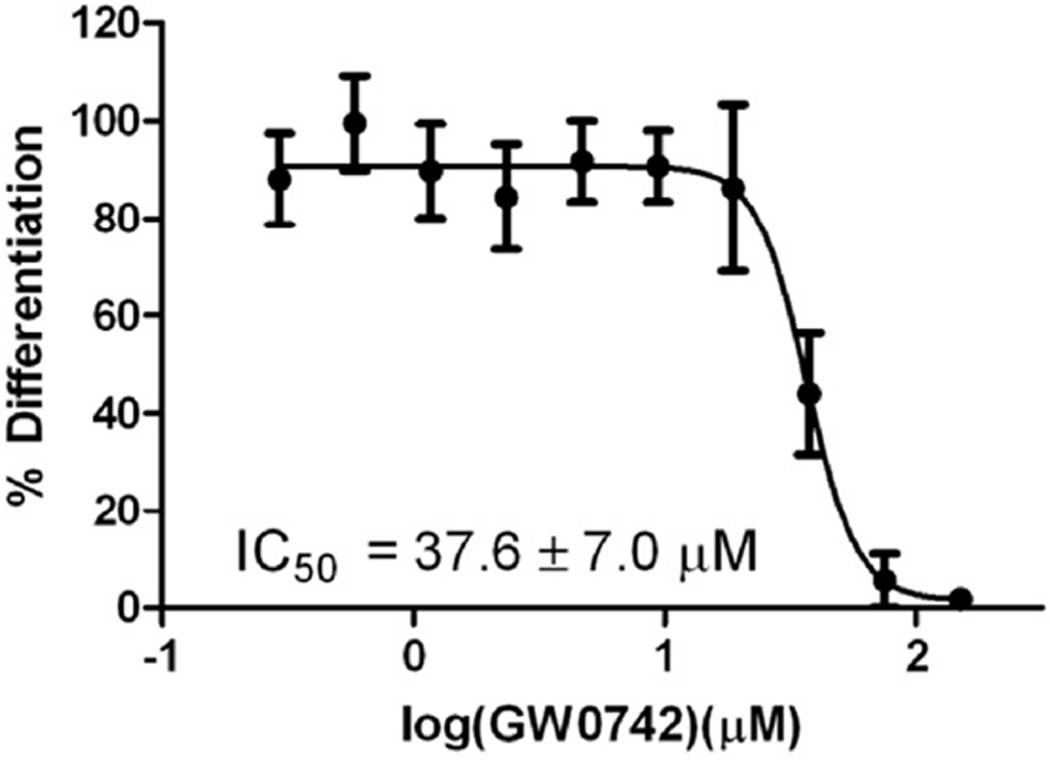
Figure 8.
Inhibition of differentiation of HL-60 cells induced by 1,25(OH)2D3 (50 nM) in the presence of GW0742.
LNCaP cancer cells express different nuclear receptors including AR46, PPARγ47, TRβ48 and VDR49. Furthermore, steroid receptor coactivator 1, 2 and 3 and DRIP205, essential for transcriptional activation, are expressed in these cells.50, 51 Based on these reports, we investigated the ability of GW0742 to inhibit nuclear receptor transcriptional activity by quantifying the RNA levels of different target genes in the presence and absence of endogenous or synthetic ligands (Figure 7).
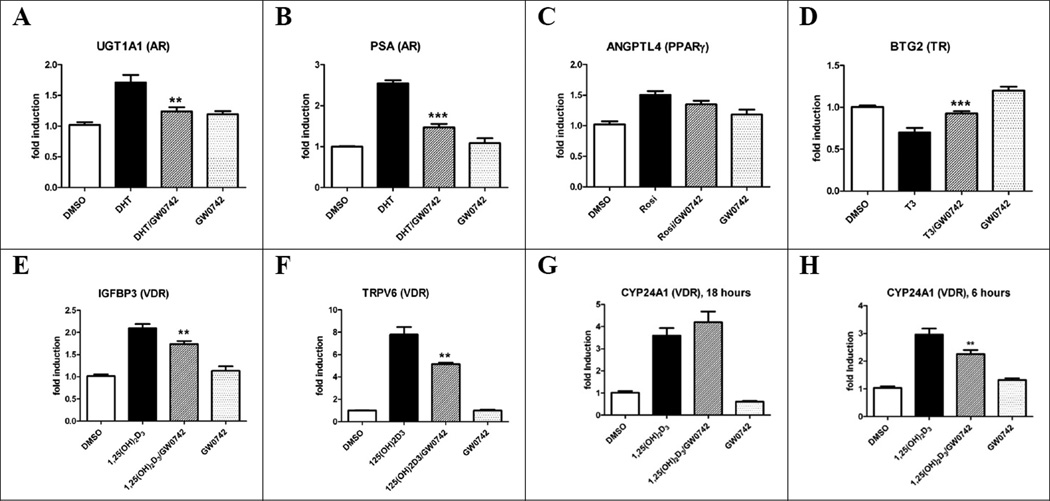
Figure 7.
Gene regulation by GW0742 (20 µM, or 50 µM for H) in LNCaP cells after 18 hours (or as indicated) in the presence and absence of NR ligands. A and B DHT (10 nM); C Rosiglitazone (5 µM); D Triiodothyronine (10 nM); E-H 1,25(OH)2D3 (10 nM). Standard errors of mean were calculated from four biological independent experiments performed in triplicates. Stars represent P<0.05 (*), P<0.01 (**), P<0.001 (***) (Student’s t-test).
For AR, expression of UDP glucuronosyltransferase 1 family polypeptide A cluster (UGT1A1)52 and the prostate-specific antigene (PSA)53 were upregulated in the presence of DHT (Figure 7, A and B). LNCaP cells treated with DHT (10 nM) and 20 µM GW0742 showed a significant inhibitory effect. PPARγ target gene angiopoietin-related protein 4 (FIAF/ANGPTL4)54 was induced by rosiglitazone (5 µM) and this response was unaffected by GW0742 (20 µM) (Figure 7, C). TR target gene B-cell translocation gene 2 (BTG2)55 was down-regulated in the presence of T3 as shown in Figure 7, D. This effect was reversed by GW0742 (20 µM) confirming the inhibitory action of GW0742 upon TR demonstrated in other assays. Finally, VDR target genes IGFBP-356, 57 and TRPV658, 59 (Figure 7, E and F) were strongly induced by 1,25(OH)2D3 (10 nM) and GW0742 selectively inhibited this effect. For VDR target gene CYP24A1,60 we observed an induction at 6 hours and 18 hours in the presence of 1,25(OH)2D3. Interestingly, only at the earlier time point of 6 hours and a concentration of 50 µM GW0742 a significant inhibition of this induction was determined (Figure, 7, G and H).
As mentioned earlier, VDR governs expression of genes responsible for differentiation of human promyelocyte HL-60 cell into monocytes by 1,25(OH)2D3.61, 62 VDR antagonists with a secosteroid structure inhibit this effect.18,19,22 The established differentiation assay employs nitro blue tetrazolium chloride to colorimetrically quantify the amount of cellular superoxide in the presence of phorbol-12-myristate-13-acetate (PMA) that function as a stimulant of the respiratory burst.63 HL-60 cells were incubated for 4 days with and without 1,25(OH)2D3 (10 nM) and GW0742 in the a dose dependent manner. The results are shown in Figure 8.
A strong HL-60 cell differentiation was observed in the presence of 1,25(OH)2D3. A significant reduction of the 1,25(OH)2D3-induced differentiation was observed for GW0742 at concentration of higher than 37.5 µM. Significant HL-60 cell death was only observed for a GW0742 concentration of 150 µM.
Discussion
This study reports a successful HTS campaign to identify new inhibitors of VDR–coactivator interaction. A novel strategy to delineate potential irreversible inhibitors, which are likely to react with nucleophilic protein residue, from a pool of hit compounds was introduced by executing a secondary screen in the presence of a high concentration of the nucleophile ME. A concentration of 100 mM ME was essential to significantly reduce the number of hit compounds.
GW0742, a PPARδ agonist, was identified among the VDR–coactivator inhibitors that were not influenced by ME. Interestingly, GW0742 exhibited characteristics of a pan nuclear receptor antagonist at concentrations higher than 12.1 µM, as demonstrated for biochemical FP assays and cell-based transcription assays. While GW0742 inhibited activity of a large number of nuclear receptors, albeit at micromolar concentrations, it inhibited VDR and AR activity most effectively. Interestingly, GW0742 also behaved as an agonist/antagonist for PPARα, PPARγ, and PPARδ, activating transcription at low concentrations and inhibiting this effect at higher concentrations.
The mechanism by which GW0742 inhibits activity of VDR and other nuclear receptors is not absolutely clear. Unexpectedly, we found that GW0742 inhibits the interaction between VDR and its small molecule ligand. While ligand competition appears incomplete, we suspect that our ligand binding assay underestimates the effectiveness of GW0742 competition because of an unexpected spectroscopic property of GW0742 discovered in this study. In the presence of rhodamine derived compounds an increase of fluorescence intensity was observed at higher GW0742 concentrations using an excitation wavelength of 595 nm and emission wavelength of 615 nm. The interaction between GW0742 and rhodamine compound also increase fluorescence polarization at the same excitation and emission wavelength. This unusual spectroscopic behavior of GW0742 might partly mask its ability to compete with the fluorescent VDR agonist and could also represent an underlying reason for a number of false positive FP HTS hits. Thus, this data is suggestive of a conventional antagonist mode of action, in which GW0742 interacts with the VDR ligand binding site and allosterically disrupts the interaction between VDR and SRC2.
Results with PPARs, in which low concentrations of GW0742 display agonist behaviors and high concentrations inhibit this effect, suggest that other mechanisms may be at play. Similar behaviors have been reported for 4-hydroxytamoxifen (HT) in regard to estrogen receptors (ERs). A crystal structure of ERβ showed one HT molecule bound to the ligand binding site and another at the coactivator binding site.64 As a result, ER was activated at low HT concentrations by ligand-induced conformational changes that allowed coactivator recruitment but ER-coactivator interaction was inhibited at higher HT concentrations by competition for ER coactivator binding.65 A similar mechanism might be one explanation for the agonist/antagonist behavior of GW0742 in regard to PPAR binding. There are other possibilities; PPARs possess large ligand binding pockets that can accommodate more than one ligand.66 Thus, high GW0742 concentrations could result in unusual PPAR/ligand stoichiometries that could trigger inactive receptor conformations. Clarification of this issue will require further investigation.
Regardless of the mechanism of action, our studies revealed that GW0742 is effective in cell culture models. The therapeutic ratio (efficacy: toxicity) for GW0742 is relative small for VDR-mediated processes (5:1) but very large for PPARδ (20000:1). The downstream effect of nuclear receptor–coactivator inhibition by GW0742 resulted in the suppression of VDR genes IGFBP-3 and TRPV6 in LNCaP cells in the presence of 1,25(OH)2D3. Other than in the prostate, the proteins encoded by these genes are highly expressed in the placenta.67 Therefore, GW0742 treatments might be problematic during pregnancy. As observed by other groups, the classic VDR target gene CYP24A1 was moderately upregulated by 1,25(OH)2D3 in LNCaP cells.49 The inhibition of CYP24A1 transcription induced by 50 µM GW0742 at only the earlier time point is likely due to different expression levels of CYP24A1 over time, which has been shown in PC-3 cells.68 In addition, we found the inhibition of transcription of AR target genes UGT1A1 and PSA in the presence of GW0742 in LNCaP when stimulated with DHT. The proteins encoded by these genes are found in increased levels in patients with prostate cancer and prostatitis. Thus, GW0742 might have beneficial effects for both conditions. BTG2, a TRβ target gene, encodes the coregulator BTG2, which has been shown to inhibit cell cycle progression at the G1 checkpoint by repressing cyclin D1 transcription.69 In the presence of T3, BTG2 is down-regulated in LNCaP cells but the T3 suppression is inhibited in the presence of 20 µM GW0742. The regulation for PPAR target gene ANGPL4, which plays a significant role in fat metabolism, was inconclusive at the GW0742 concentration tested. ANGPTL4 activation in the presence of Rosiglitazone and GW0742 was reported for HT29 colon cancer cells although at a GW0742 concentration of 1 µM.70 Thus, high local concentrations of GW0742 have a strong influence on the gene regulation mediated by PPARs and other nuclear receptors. The activation of transcription by 1,25(OH)2D3 induces differentiation of HL-60 cells, a process which was shown to be inhibited by GW0742 in a dose dependent manner. Overall, we have demonstrated the interactions between GW0742 and different NRs with diverse transcriptional and biological effects underlining the possibility that GW0742 might share a privileged scaffold recognized by many members of the NR superfamily. In respect to VDR, GW0742 is the first ligand antagonist lacking a secosteroid structure, which inhibits the transcriptional activation of VDR target genes in the presence of 1,25(OH)2D3. Because of the fact that the three dimensional folding of VDR and resulting interactions with coregulators are highly depended on the structure of the bound ligand,71 we expect that GW0742 will exhibit different biological responses than current secosteroid VDR antagonist. We are currently in the process to develop new VDR-selective antagonists based on GW0742 in order to determine the downstream effect of VDR–coactivator inhibition without influencing PPAR signaling.
Supplementary Material
Acknowledgment
We would like to thank Robin Goy and Nicholas Nassif for the production of VDR protein.
Funding.
This work was supported by the University of Wisconsin-Milwaukee [LAA], the UWM Research Growth Initiative (RGI grant 2012) [LAA], the NIH R03DA031090 [LAA], the UWM Research Foundation (Catalyst grant), the Lynde and Harry Bradley Foundation [LAA], the Richard and Ethel Herzfeld Foundation [LAA], and the Molecular Libraries Initiative of the National Institutes of Health Roadmap for Medical Research (U54MH084681) [GB, AS, AJ, AY, DM].
Abbreviations
- VDR
vitamin D receptor
- NR
nuclear hormone receptor
- FP
fluorescence polarization
- SRC2
steroid receptor coactivator 2
- SRC2–3
third NR interaction domain of the steroid receptor coactivator 2 peptide
- TRPV6
transient receptor potential vanilloid type 6 gene
- CYP24A1
1,25-dihydroxyvitamin D3 24-hydroxylase
- 1,25-(OH)2D3
1,25-dihydroxyvitamin D3, DBD, DNA binding domain
- LBD
ligand-binding domain
- RxR
retinoid X receptor
- HTS
high throughput screening
- DMSO
dimethyl sulfoxide
- HEK293T
human embryonic kidney cells
- AR
androgen receptor
- ERα
estrogen receptor alpha
- TRβ
thyroid receptor beta
- PPARα
peroxisome proliferator-activated receptor α
- PPARγ
peroxisome proliferator-activated receptor γ, PPARδ, peroxisome proliferator-activated receptor δ
- DRIP205
VDRR-interacting protein 205
- GST
glutathione-S-transferase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- CYP24A1
1,25-dihydroxyvitamin D3 24-hydroxylase gene
- NCGC
NIH Chemical Genomic Center
- ME
mercaptoethanol
- T3
triiodothyronine
- DHT
dihydrotestosterone
- GAL4
yeast transcription activator protein GAL4
- HT
4-hydroxytamoxifen
- UGT1A1
UDP glucuronosyltransferase 1 family polypeptide A cluster
- PSA
prostate-specific antigene
- ANGPTL4
angiopoietin-related protein 4
- BTG2
B-cell translocation gene 2
- IGFBP-3
insulinlike growth factor-binding protein
- HL-60
human promyelocytic leukemia cell
- PMA
phorbol-12-myristate-13-acetate
- DMF
dimethylforamide
- PBS
Phosphate-buffered saline
- HPLC
high pressure liquid chromatography
- PIPES
piperazine-N,N’-bis(2-ethanesulfonic acid
- NP-40
Tergitol-type NP-40
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- FBS
Fetal bovine serum
- DMEM
Dulbecco’s Modification of Eagle’s Medium
- NID
nuclear receptor interaction domain
- EDTA
Ethylenediaminetetraacetic acid
- DTT
Dithiothreitol
- MBP
maltose binding protein
- LNCaP
human prostate cancer cells
- PMA
phorbol-12-myristate-13-acetate
Footnotes
Supporting Information Available.
The absence of binding between GW0742 and SRC2 was determined using ITC. This material is available free of charge via the Internet at http:/pubs.acs.org.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- 3.The Binding Database. ChemAxon. 2013 [Google Scholar]
- 4.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 5.Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna NJ, O'Malley BW. Nuclear receptors, coregulators, ligands, and selective receptor modulators: making sense of the patchwork quilt. Ann N Y Acad Sci. 2001;949:3–5. doi: 10.1111/j.1749-6632.2001.tb03997.x. [DOI] [PubMed] [Google Scholar]
- 7.O'Malley BW, Malovannaya A, Qin J. Minireview: Nuclear Receptor and Coregulator Proteomics--2012 and Beyond. Mol Endocrinol. 2012 doi: 10.1210/me.2012-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore TW, Katzenellenbogen JA. Inhibitors of Nuclear Hormone Receptor/Coactivator Interactions. Annu Rep Med Chem. 2009;44:443–457. [Google Scholar]
- 9.Bikle DD, Teichert A, Arnold LA, Uchida Y, Elias PM, Oda Y. Differential regulation of epidermal function by VDR coactivators. J Steroid Biochem Mol Biol. 2010;121:308–313. doi: 10.1016/j.jsbmb.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumbaugh PF, Haussler MR. 1 Alpha,25-dihydroxycholecalciferol receptors in intestine. I. Association of 1 alpha,25-dihydroxycholecalciferol with intestinal mucosa chromatin. J Biol Chem. 1974;249:1251–1257. [PubMed] [Google Scholar]
- 11.Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2:203–216. doi: 10.1023/a:1010062929140. [DOI] [PubMed] [Google Scholar]
- 12.Feldman D, Pike JW, Glorieux FH. Vitamin D. second ed. 1–2. Elsevier: Burlington; 2005. [Google Scholar]
- 13.Yamada S, Shimizu M, Yamamoto K. Vitamin D receptor. Endocr Dev. 2003;6:50–63. doi: 10.1159/000072769. [DOI] [PubMed] [Google Scholar]
- 14.Toell A, Polly P, Carlberg C. All natural DR3-type vitamin D response elements show a similar functionality in vitro. Biochem J. 2000;352(Pt 2):301–309. [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JY, Son YL, Lee YC. Involvement of SMRT corepressor in transcriptional repression by the vitamin D receptor. Mol Endocrinol. 2009;23:251–264. doi: 10.1210/me.2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rachez C, Freedman LP. Mechanisms of gene regulation by vitamin D(3) receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 17.Lamblin M, Spingarn R, Wang TT, Burger MC, Dabbas B, Moitessier N, White JH, Gleason JL. An o-aminoanilide analogue of 1alpha,25-dihydroxyvitamin D(3) functions as a strong vitamin D receptor antagonist. J Med Chem. 2010;53:7461–7465. doi: 10.1021/jm1007159. [DOI] [PubMed] [Google Scholar]
- 18.Kato Y, Nakano Y, Sano H, Tanatani A, Kobayashi H, Shimazawa R, Koshino H, Hashimoto Y, Nagasawa K. Synthesis of 1alpha,25-dihydroxyvitamin D3-26,23-lactams (DLAMs), a novel series of 1 alpha,25-dihydroxyvitamin D3 antagonist. Bioorg Med Chem Lett. 2004;14:2579–2583. doi: 10.1016/j.bmcl.2004.02.076. [DOI] [PubMed] [Google Scholar]
- 19.Inaba Y, Yoshimoto N, Sakamaki Y, Nakabayashi M, Ikura T, Tamamura H, Ito N, Shimizu M, Yamamoto K. A new class of vitamin D analogues that induce structural rearrangement of the ligand-binding pocket of the receptor. J Med Chem. 2009;52:1438–1449. doi: 10.1021/jm8014348. [DOI] [PubMed] [Google Scholar]
- 20.Nakabayashi M, Yamada S, Yoshimoto N, Tanaka T, Igarashi M, Ikura T, Ito N, Makishima M, Tokiwa H, DeLuca HF, Shimizu M. Crystal structures of rat vitamin D receptor bound to adamantyl vitamin D analogs: structural basis for vitamin D receptor antagonism and partial agonism. J Med Chem. 2008;51:5320–5329. doi: 10.1021/jm8004477. [DOI] [PubMed] [Google Scholar]
- 21.Saito N, Kittaka A. Highly potent vitamin D receptor antagonists: design, synthesis, and biological evaluation. Chembiochem. 2006;7:1479–1490. doi: 10.1002/cbic.200600054. [DOI] [PubMed] [Google Scholar]
- 22.Cho K, Uneuchi F, Kato-Nakamura Y, Namekawa J, Ishizuka S, Takenouchi K, Nagasawa K. Structure-activity relationship studies on vitamin D lactam derivatives as vitamin D receptor antagonist. Bioorg Med Chem Lett. 2008;18:4287–4290. doi: 10.1016/j.bmcl.2008.06.095. [DOI] [PubMed] [Google Scholar]
- 23.Herdick M, Steinmeyer A, Carlberg C. Antagonistic action of a 25-carboxylic ester analogue of 1alpha, 25-dihydroxyvitamin D3 is mediated by a lack of ligand-induced vitamin D receptor interaction with coactivators. J Biol Chem. 2000;275:16506–16512. doi: 10.1074/jbc.M910000199. [DOI] [PubMed] [Google Scholar]
- 24.Ishizuka S, Kurihara N, Miura D, Takenouchi K, Cornish J, Cundy T, Reddy SV, Roodman GD. Vitamin D antagonist, TEI-9647, inhibits osteoclast formation induced by 1alpha,25-dihydroxyvitamin D3 from pagetic bone marrow cells. J Steroid Biochem Mol Biol. 2004;89–90:331–334. doi: 10.1016/j.jsbmb.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 25.Mita Y, Dodo K, Noguchi-Yachide T, Miyachi H, Makishima M, Hashimoto Y, Ishikawa M. LXXLL peptide mimetics as inhibitors of the interaction of vitamin D receptor with coactivators. Bioorganic & Medicinal Chemistry Letters. 2010;20:1712–1717. doi: 10.1016/j.bmcl.2010.01.079. [DOI] [PubMed] [Google Scholar]
- 26.Nandhikonda P, Lynt WZ, McCallum MM, Ara T, Baranowski AM, Yuan NY, Pearson D, Bikle DD, Guy RK, Arnold LA. Discovery of the first irreversible small molecule inhibitors of the interaction between the vitamin D receptor and coactivators. J Med Chem. 2012;55:4640–4651. doi: 10.1021/jm300460c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, Lambert MH, Willson TM, Oliver WR, Jr, Sternbach DD. Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPARdelta)--synthesis and biological activity. Bioorg Med Chem Lett. 2003;13:1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 29.Harrington LS, Moreno L, Reed A, Wort SJ, Desvergne B, Garland C, Zhao L, Mitchell JA. The PPARbeta/delta agonist GW0742 relaxes pulmonary vessels and limits right heart hypertrophy in rats with hypoxia-induced pulmonary hypertension. PLoS One. 2010;5:e9526. doi: 10.1371/journal.pone.0009526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarzuelo MJ, Jimenez R, Galindo P, Sanchez M, Nieto A, Romero M, Quintela AM, Lopez-Sepulveda R, Gomez-Guzman M, Bailon E, Rodriguez-Gomez I, Zarzuelo A, Galvez J, Tamargo J, Perez-Vizcaino F, Duarte J. Antihypertensive effects of peroxisome proliferator-activated receptor-beta activation in spontaneously hypertensive rats. Hypertension. 2011;58:733–743. doi: 10.1161/HYPERTENSIONAHA.111.174490. [DOI] [PubMed] [Google Scholar]
- 31.Maria Quintela A, Jimenez R, Gomez-Guzman M, Jose Zarzuelo M, Galindo P, Sanchez M, Vargas F, Cogolludo A, Tamargo J, Perez-Vizcaino F, Duarte J. Activation of peroxisome proliferator-activated receptor-beta/-delta (PPARbeta/delta) prevents endothelial dysfunction in type 1 diabetic rats. Free Radic Biol Med. 2012 doi: 10.1016/j.freeradbiomed.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 32.Matsushita Y, Ogawa D, Wada J, Yamamoto N, Shikata K, Sato C, Tachibana H, Toyota N, Makino H. Activation of peroxisome proliferator-activated receptor delta inhibits streptozotocin-induced diabetic nephropathy through anti-inflammatory mechanisms in mice. Diabetes. 2011;60:960–968. doi: 10.2337/db10-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galuppo M, Di Paola R, Mazzon E, Esposito E, Paterniti I, Kapoor A, Thiemermann C, Cuzzocrea S. GW0742, a high affinity PPAR-beta/delta agonist reduces lung inflammation induced by bleomycin instillation in mice. Int J Immunopathol Pharmacol. 2010;23:1033–1046. doi: 10.1177/039463201002300408. [DOI] [PubMed] [Google Scholar]
- 34.Zingarelli B, Piraino G, Hake PW, O’Connor M, Denenberg A, Fan H, Cook JA. Peroxisome proliferator-activated receptor {delta} regulates inflammation via NF-{kappa}B signaling in polymicrobial sepsis. Am J Pathol. 2010;177:1834–1847. doi: 10.2353/ajpath.2010.091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington WW, C SB, J GW, N OM, J GB, D CL, W RO, M CL, D MI. The Effect of PPARalpha, PPARdelta, PPARgamma, and PPARpan Agonists on Body Weight, Body Mass, and Serum Lipid Profiles in Diet-Induced Obese AKR/J Mice. PPAR Res. 2007;2007:97125. doi: 10.1155/2007/97125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bility MT, Devlin-Durante MK, Blazanin N, Glick AB, Ward JM, Kang BH, Kennett MJ, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor beta/delta (PPAR beta/delta) inhibits chemically induced skin tumorigenesis. Carcinogenesis 29. 2008:2406–2414. doi: 10.1093/carcin/bgn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollingshead HE, Killins RL, Borland MG, Girroir EE, Billin AN, Willson TM, Sharma AK, Amin S, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) ligands do not potentiate growth of human cancer cell lines. Carcinogenesis. 2007;28:2641–2649. doi: 10.1093/carcin/bgm183. [DOI] [PubMed] [Google Scholar]
- 38.Marin HE, Peraza MA, Billin AN, Willson TM, Ward JM, Kennett MJ, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor beta inhibits colon carcinogenesis. Cancer Res. 2006;66:4394–4401. doi: 10.1158/0008-5472.CAN-05-4277. [DOI] [PubMed] [Google Scholar]
- 39.Sertznig P, Dunlop T, Seifert M, Tilgen W, Reichrath J. Cross-talk between vitamin D receptor (VDR)- and peroxisome proliferator-activated receptor (PPAR)-signaling in melanoma cells. Anticancer Res. 2009;29:3647–3658. [PubMed] [Google Scholar]
- 40.Boehm MF, Fitzgerald P, Zou A, Elgort MG, Bischoff ED, Mere L, Mais DE, Bissonnette RP, Heyman RA, Nadzan AM, Reichman M, Allegretto EA. Novel nonsecosteroidal vitamin D mimics exert VDR-modulating activities with less calcium mobilization than 1,25-dihydroxyvitamin D3. Chem Biol. 1999;6:265–275. doi: 10.1016/S1074-5521(99)80072-6. [DOI] [PubMed] [Google Scholar]
- 41.Teichert A, Arnold LA, Otieno S, Oda Y, Augustinaite I, Geistlinger TR, Kriwacki RW, Guy RK, Bikle DD. Quantification of the vitamin D receptor-coregulator interaction. Biochemistry. 2009;48:1454–1461. doi: 10.1021/bi801874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5:173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 43.Hwang JY, Huang WW, Arnold LA, Huang RL, Attia RR, Connelly M, Wichterman J, Zhu FY, Augustinaite I, Austin CP, Inglese J, Johnson RL, Guy RK. Methylsulfonylnitrobenzoates, a New Class of Irreversible Inhibitors of the Interaction of the Thyroid Hormone Receptor and Its Obligate Coactivators That Functionally Antagonizes Thyroid Hormone. Journal of Biological Chemistry. 2011:286. doi: 10.1074/jbc.M110.200436. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estebanez-Perpina E, Arnold LA, Jouravel N, Togashi M, Blethrow J, Mar E, Nguyen P, Phillips KJ, Baxter JD, Webb P, Guy RK, Fletterick RJ. Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor. Mol Endocrinol. 2007;21:2919–2928. doi: 10.1210/me.2007-0174. [DOI] [PubMed] [Google Scholar]
- 45.Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43:1809–1818. [PubMed] [Google Scholar]
- 47.Mueller E, Smith M, Sarraf P, Kroll T, Aiyer A, Kaufman DS, Oh W, Demetri G, Figg WD, Zhou XP, Eng C, Spiegelman BM, Kantoff PW. Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proc Natl Acad Sci U S A. 2000;97:10990–10995. doi: 10.1073/pnas.180329197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh ML, Juang HH. Cell growth effects of triiodothyronine and expression of thyroid hormone receptor in prostate carcinoma cells. J Androl. 2005;26:422–428. doi: 10.2164/jandrol.04162. [DOI] [PubMed] [Google Scholar]
- 49.Skowronski RJ, Peehl DM, Feldman D. Vitamin D and prostate cancer: 1,25 dihydroxyvitamin D3 receptors and actions in human prostate cancer cell lines. Endocrinology. 1993;132:1952–1960. doi: 10.1210/endo.132.5.7682937. [DOI] [PubMed] [Google Scholar]
- 50.Linja MJ, Porkka KP, Kang Z, Savinainen KJ, Janne OA, Tammela TL, Vessella RL, Palvimo JJ, Visakorpi T. Expression of androgen receptor coregulators in prostate cancer. Clin Cancer Res. 2004;10:1032–1040. doi: 10.1158/1078-0432.ccr-0990-3. [DOI] [PubMed] [Google Scholar]
- 51.Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 2007;67:4034–4041. doi: 10.1158/0008-5472.CAN-06-3039. [DOI] [PubMed] [Google Scholar]
- 52.Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, Ijichi N, Ouchi Y, Shirahige K, Aburatani H, Inoue S. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- 53.Riegman PH, Vlietstra RJ, van der Korput JA, Brinkmann AO, Trapman J. The promoter of the prostate-specific antigen gene contains a functional androgen responsive element. Mol Endocrinol. 1991;5:1921–1930. doi: 10.1210/mend-5-12-1921. [DOI] [PubMed] [Google Scholar]
- 54.Mandard S, Zandbergen F, Tan NS, Escher P, Patsouris D, Koenig W, Kleemann R, Bakker A, Veenman F, Wahli W, Muller M, Kersten S. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J Biol Chem. 2004;279:34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 55.Tsui KH, Hsieh WC, Lin MH, Chang PL, Juang HH. Triiodothyronine modulates cell proliferation of human prostatic carcinoma cells by downregulation of the B-cell translocation gene 2. Prostate. 2008;68:610–619. doi: 10.1002/pros.20725. [DOI] [PubMed] [Google Scholar]
- 56.Colston KW, Perks CM, Xie SP, Holly JM. Growth inhibition of both MCF-7 and Hs578T human breast cancer cell lines by vitamin D analogues is associated with increased expression of insulin-like growth factor binding protein-3. J Mol Endocrinol. 1998;20:157–162. doi: 10.1677/jme.0.0200157. [DOI] [PubMed] [Google Scholar]
- 57.Peng L, Malloy PJ, Feldman D. Identification of a functional vitamin D response element in the human insulin-like growth factor binding protein-3 promoter. Mol Endocrinol. 2004;18:1109–1119. doi: 10.1210/me.2003-0344. [DOI] [PubMed] [Google Scholar]
- 58.Hoenderop JG, van der Kemp AW, Hartog A, van de Graaf SF, van Os CH, Willems PH, Bindels RJ. Molecular identification of the apical Ca2+ channel in 1, 25-dihydroxyvitamin D3-responsive epithelia. J Biol Chem. 1999;274:8375–8378. doi: 10.1074/jbc.274.13.8375. [DOI] [PubMed] [Google Scholar]
- 59.Meyer MB, Watanuki M, Kim S, Shevde NK, Pike JW. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20:1447–1461. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- 60.Chen KS, DeLuca HF. Cloning of the human 1 alpha,25-dihydroxyvitamin D-3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 61.Miyaura C, Abe E, Kuribayashi T, Tanaka H, Konno K, Nishii Y, Suda T. 1 alpha,25-Dihydroxyvitamin D3 induces differentiation of human myeloid leukemia cells. Biochem Biophys Res Commun. 1981;102:937–943. doi: 10.1016/0006-291x(81)91628-4. [DOI] [PubMed] [Google Scholar]
- 62.Mangelsdorf DJ, Koeffler HP, Donaldson CA, Pike JW, Haussler MR. 1,25-Dihydroxyvitamin D3-induced differentiation in a human promyelocytic leukemia cell line (HL-60): receptor-mediated maturation to macrophage-like cells. J Cell Biol. 1984;98:391–398. doi: 10.1083/jcb.98.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J Exp Med. 1979;149:969–974. doi: 10.1084/jem.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Chirgadze NY, Briggs SL, Khan S, Jensen EV, Burris TP. A second binding site for hydroxytamoxifen within the coactivator-binding groove of estrogen receptor beta. Proc Natl Acad Sci U S A. 2006;103:9908–9911. doi: 10.1073/pnas.0510596103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kojetin DJ, Burris TP, Jensen EV, Khan SA. Implications of the binding of tamoxifen to the coactivator recognition site of the estrogen receptor. Endocr Relat Cancer. 2008;15:851–870. doi: 10.1677/ERC-07-0281. [DOI] [PubMed] [Google Scholar]
- 66.Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, Jr, McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han VK, Bassett N, Walton J, Challis JR. The expression of insulinlike growth factor (IGF) and IGF-binding protein (IGFBP) genes in the human placenta and membranes: evidence for IGF-IGFBP interactions at the feto-maternal interface. J Clin Endocrinol Metab. 1996;81:2680–2693. doi: 10.1210/jcem.81.7.8675597. [DOI] [PubMed] [Google Scholar]
- 68.Khanim FL, Gommersall LM, Wood VH, Smith KL, Montalvo L, O’Neill LP, Xu Y, Peehl DM, Stewart PM, Turner BM, Campbell MJ. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23:6712–6725. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 69.Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, Augusti-Tocco G, Mattei E, Lakshmana MK, Krizhanovsky V, Reeves SA, Giovannoni R, Castano F, Servadio A, Ben-Arie N, Tirone F. Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J Neurosci. 2004;24:3355–3369. doi: 10.1523/JNEUROSCI.3860-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aronsson L, Huang Y, Parini P, Korach-Andre M, Hakansson J, Gustafsson JA, Pettersson S, Arulampalam V, Rafter J. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4) PLoS One. 2010;5 doi: 10.1371/journal.pone.0013087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlberg C, Molnar F, Mourino A. Vitamin D receptor ligands: the impact of crystal structures. Expert Opin Ther Pat. 2012;22:417–435. doi: 10.1517/13543776.2012.673590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.