Abstract
We report the characterization of the diheme cytochrome c peroxidase (CcP) from Shewanella oneidensis (So) using UV/Visible absorbance, Electron Paramagnetic Resonance Spectroscopy, and Michaelis-Menten kinetics. While sequence alignment with other bacterial diheme cytochrome c peroxidases suggests that So CcP may be active in the as-isolated state, we find that So CcP requires reductive activation for full activity, similar to the canonical Pseudomonas-type of bacterial CcP enzyme. Peroxide turnover initiated with oxidized So CcP shows a distinct lag-phase, which we interpret as reductive activation in situ. A simple kinetic model is sufficient to recapitulate the lag-phase behavior of the progress curves and separate the contributions of reductive activation and peroxide turnover. The rates of catalysis and activation differ between MBP-fusion and tag-free So CcP, and also depend on the identity of the electron donor. Combined with Michaelis-Menten analysis these data suggest that So CcP can accommodate electron donor binding in several possible orientations, and that the presence of the MBP tag affects the availability of certain binding sites. To further investigate the structural basis of reductive activation in So CcP we introduced mutations into two different regions of the protein that have been suggested to be important for reductive activation in homologous bacterial CcPs. Mutations in a flexible loop region neighboring the low-potential heme significantly increased the activation rate, confirming the importance of flexible loop regions of the protein in converting the inactive, as-isolated enzyme into the activated form.
Reactive oxygen species such as hydrogen peroxide and superoxide are generated in hypoxic conditions as a result of a leaky or inefficient electron transport chain (1), which may further yield toxic hydroxyl radicals through Fenton chemistry (2). For many gram negative bacteria, it is thought that periplasmic peroxide scavenging is achieved by soluble diheme cytochrome c peroxidases (3). Bacterial diheme cytochrome c peroxidases (BCcPs1) differ from the canonical monoheme peroxidases such as horseradish peroxidase (HRP) (4) and yeast cytochrome c peroxidase (yCcP) (5) in their cofactor content and mechanism. The two heme cofactors in bacterial peroxidase enzymes are covalently bound within two separate cytochrome c-like domains. In all characterized BCcPs, the high-potential (∼ +330 mV vs hydrogen) Met-His ligated heme (“H-heme”) serves as an electron transfer site, while the other, low-potential ‘L-heme’ (around −300mV vs. NHE) serves as the peroxidatic active site (6, 7). In the as-isolated state of the BCcP, both heme sites are in the ferric oxidation state, with the H-heme in a low-spin/high-spin equilibrium and the L-heme in a low-spin state (7). The high-potential heme, which is not present in monoheme peroxidases, may store a second oxidizing equivalent during the catalytic cycle (6), and is hypothesized to mediate electron transfer from electron donor proteins to the peroxidatic heme (8, 9).
Most BCcPs are isolated in a state that is catalytically inactive where both hemes are each in the ferric state, as exemplified by the diheme peroxidase from Pseudomonas aeruginosa (Psa) (10). In the absence of reducing equivalents provided by an electron donor protein or small molecule such as ascorbate, both hemes of the BCcP remain in the ferric state where the active site is bis-His ligated, preventing binding of the peroxide substrate (11), as illustrated in Figure 1. The active form of the enzyme can be achieved by a one-electron reduction termed ‘reductive activation’: reduction of the H-heme results in local conformational changes that cause reorientation of the liganding groups on the low-potential heme, allowing for peroxide to access the active site (Figure 1B, 11, 12, 13). The overall phenomenon of reductive activation may serve a regulatory role, preventing turnover of the peroxidase in the absence of sufficient reducing equivalents provided by periplasmic electron donor proteins.
Figure 1.
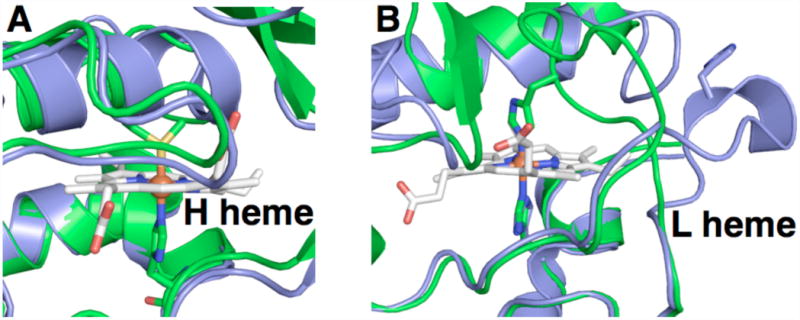
Comparison of the structures of the inactivated (green) versus activated (blue) states of BCcPs, focusing on the residues surrounding the high potential heme (A) and low potential heme (B). Pseudomonas aeruginosa cytochrome c peroxidase as-isolated is shown in green (PDB code = 1EB7); constitutively active cytochrome c peroxidase from Nitrosomonas europaea is shown in blue (PDB code = 1IQC).
The reductive activation process is an essential step in most of the known BCcPs. In addition to the Psa enzyme (10), reductive activation is required for the BCcPs from Paracoccus denitrificans (Pd) (14), Pseudomonas nautica (Psn) (15), Rhodobacter capsulatus (Rc) (16), Pseudomonas stutzeri (Pss) (17) and Geobacter sulfurreducens (Gs) (18). In contrast, diheme peroxidases from Nitrosomonas europaea (Ne) (19) and Methyloccocus casulatus (Bath) (20) do not need to undergo the activation step; the crystal structure of Ne CcP shows that the low-potential heme is pentacoordinate in the as-isolated form (13) (Figure 1B, blue). A sequence alignment of representative BCcP enzymes (Figure 2) shows that the sequences of key loop regions that change conformation during reductive activation differ between Psa-type CcPs that require reductive activation and Ne CcP, which does not. Swapping only two amino acids in one of these loop regions in the Psa-type CcP from Geobacter sulfurreducens generates a constitutively active protein, mimicking the Nitrosomonas-like class of BCcPs (18). Intriguingly, on the basis of sequence the Shewanella oneidensis (So) CcP is highly similar to Ne CcP (60% identity by primary amino acid sequence), and diverges more strongly from Psa-like CcPs (44% identity to Psa CcP). The sequence similarities between Ne and So are particularly striking in the flexible loop regions that have been identified as mediators of the conformational changes in reductive activation. We sought to test the hypothesis that, based on sequence identity, the So CcP should be a member of the Ne-type functional sub-class of BCcPs, which would not require reductive activation.
Figure 2.
Partial sequence alignment of various bacterial CcPs. Gs: Geobacter sulfurreducens, Psa: Pseudomonas auruginosa, So: Shewanella oneidensis, Ne: Nitrosomonas europaea. Heme binding motifs (CXXCH) are marked with purple shading, and the three loop regions implicated in reductive activation are shaded in yellow, blue or orange.
In this report, we characterize the peroxidase activity of a diheme peroxidase from Shewanella oneidensis. We express the So CcP gene (SO2178) recombinantly in E. coli as a maltose-binding protein fusion that has been previously described (21), as well as the presumed redox partner for So CcP, a monoheme cytochrome c5, SO0264 (21). We show, based on EPR, UV/Visible spectroscopy, and steady-state kinetics of recombinant So CcP, that the So CcP is a conventional Psa-type BCcP requiring reductive activation. We find that the as-isolated form of So CcP undergoes rapid reductive activation under in vitro assay conditions, with a rate that depends on the concentration of the electron donor. We present a computational model for the peroxidase activity of as-isolated So CcP that permits the deconvolution of the reductive activation rate and the peroxide turnover rate from the enzymatic progress curves. Following our initial characterization of the So CcP system, we demonstrate that this protein serves as a facile model system for structure/function analysis through further investigation the role of the activation loops of So CcP. We show that mutations in loop 1 and loop 3 regions do not completely inhibit peroxidase activity, but instead show defects in peroxidase turnover and increases in the activation rate.
Materials and Methods
Expression and Purification of Diheme Cytochrome c Peroxidase from Shewanella oneidensis
The gene coding for the Shewanella cytochrome c peroxidase (SO2178) in the parent vector pMKL1 was a kind gift to our laboratory from Drs. Frank Collart and Yuri Londer (Argonne National Labs). The SO2178 gene is expressed as a fusion to the C-terminus of the maltose-binding protein (MBP), with a TEV site between the MBP and So CcP to allow isolation of a tag-free So CcP protein. The MBP protein also includes an N-terminal hexahistidine tag for Ni2+ affinity purification. The So CcP-MBP fusion protein is expressed in E. coli JM109 cells (Strategene). Maximal So CcP expression was achieved by co-transformation with pEC86, a plasmid bearing the cytochrome c maturation cassette (22). Transformed cells were grown in 10 mL starter culture at 37 °C for 8 hours. Starter cultures were resuspended into fresh media and used to inoculate bulk expression cultures containing 1 L of 2× YT (Fisher BioReagents) supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol in a 2 L flask. Bulk expressions were grown at 37 °C for 14 to 16 hours with good aeration (200 rpm) to high optical density (A600 > 2). Induction for 4 hours with 80 mg/L of IPTG yielded pink to brown pellets as an indication of cytochrome c production. Cells were harvested, sonicated in 20mM Tris pH 7.4, 150 mM NaCl, 100 μM EDTA, 0.13 mg/mL lysozyme, clarified by centrifugation, and immediately loaded onto an amylose affinity resin column (NEB) to avoid proteolytic cleavage of the CcP-MBP fusion. For sensitive applications requiring very pure sample, amylose chromatography was followed by size exclusion chromatography on an S-100 column (GE Healthcare prepacked HiPrep™ Sephacryl™). Removal of MBP was achieved by cleavage overnight with TEV protease and subsequent Ni2+-NTA chromatography. So CcP mutants in loop 1 (P75T/H81K/E84Q) and loop 3 (M219Q/F247N) were constructed using the QuikChange Mutagenesis Kit (Stratagene; see Supporting Information), and expressed and purified as MBP fusions as described for the wild-type protein.
Expression and Purification of Recombinant Pseudomonas aeruginosa Azurin
The plasmid pETAz9+ was a kind gift of Michelle McGuirl (UMontana). The plasmid was transformed into E. coli BL21(DE3), and the transformants were grown in 4 mL starter cultures of 2× YT until cloudy. Starter cultures were used to inoculate 1 L cultures of 2× YT with 50 μg/mL kanamycin for bulk expression. The 1L cultures were grown at 37 °C overnight to reach high optical density, and induced in the morning with 1 mM IPTG for 4 hours. The resulting cells were lysed gently by sonication in buffer containing 20 mM Tris pH 8, 20% glucose, 15 μg/mL lysozyme and 5 μg/mL DNase. Clarified lysate was supplemented with 100 μM final concentration of CuSO4, and holo-azurin was loaded on Q-Sepharose equilibrated in 10 mM diethanolamine pH 9. Azurin was eluted with 500mM NaCl in 10 mM diethanolamine pH 9.00. Azurin-containg fractions were reduced with 1 mM dithionite and further purified by size exclusion chromatography (GE Healthcare prepacked HiPrep™ Sephacryl™S-100).
Expression and purification of Shewanella oneidensis cytochrome c5
The Shewanella oneidensis gene SO0264, which codes for cytochrome c5, was synthesized by Genewiz and sub-cloned into pET25b+ (Invitrogen) at the EcoRV restriction site. The resulting plasmid was named pSOc5, and was used for the expression of a tag-free version of the cytochrome bearing the wild-type periplasmic localization sequence at the N-terminus. Recombinant S. oneidensis c5 was expressed in E. coli BL21(DE3) carrying the pEC86 plasmid (22). For over-expression, 1 L of 2× YT media containing 100 μg/mL ampicillin and 25 μg/mL chloramphenicol was inoculated with 5 mL of resuspended overnight BL21(DE3)/pEC86/pSOc5 culture and grown without IPTG induction at 37 °C for 22 hours with moderate shaking (170 rpm). Cell pellets were harvested by centrifugation, resuspended in 80 mL of lysis buffer (20 mM Tris pH 8, 1 mM EDTA, 50 mM NaCl, 0.1 mg/mL lysozyme), and lysed by sonication. Lysate was clarified by centrifugation, the pH of the clarified lysate was adjusted to 4.5 by drop-wise addition of 1 M acetic acid, and precipitated proteins were removed by centrifugation. The resulting pink supernatant was oxidized by addition of equimolar potassium ferricyanide and loaded onto MacroPrep HighS resin (10 mL bed volume, BioRad) equilibrated in 10 mM sodium acetate pH 4.5. Bound proteins were eluted with an 80 mL gradient from 50 to 500 mM NaCl in 10 mM sodium acetate pH 4.5. So cyt c5 eluted in a broad peak centered around 250 mM NaCl, and fractions with purity ratios (A410/A280) of 4.0 or greater were pooled and stored in 15% glycerol at −80°C.
EPR Spectroscopy
X-band (9.62 GHz) EPR spectra were recorded on a Bruker 300 spectrometer equipped with an Oxford ESR-910 liquid helium cryostat. The quantification of all signals is relative to a Cu-EDTA spin standard. For quantification, the spectra were recorded under non-saturating microwave conditions, meaning that the signal intensity was found to be proportional to the square root of the microwave power. The EPR spectrum of the Cu-EDTA standard is recorded daily at a temperature and microwave power of 10K and 2μW. The copper concentration of the standard is determined by atomic absorption. The microwave frequency was calibrated with a frequency counter and the magnetic field with a NMR gaussmeter. The sample temperature was calibrated with a carbon-glass resistor (LakeShore CGR-1-1000) placed at the position of the sample in an NMR tube. A modulation frequency of 100 KHz with 10 G modulation amplitude was used for all EPR experiments. The EPR simulation software was written by one of the authors. The simulations are least-squares fits of the experimental spectra generated with consideration of all intensity factors, which allows computation of simulated spectra for a specified sample concentration. The simulations therefore allow a quantitative determination of protein signal intensities. The Windows software package (SpinCount) is available for general application to any mono- or dinuclear metal complex by contacting M. Hendrich.
Activity Assays
The electronic absorption spectra properties of the CcP-MBP and the tag-free CcP were conducted on a Cary 50 spectrophotometer (Varian). All assays were conducted at 23 °C in assay buffer (5 mM MES, 5 mM HEPES, 10 mM NaCl and 1 mM CaCl2 at pH 6.00) following the protocol previously reported for the P. denitrificans enzyme (14). Horse heart cytochrome c (Sigma), Psa azurin, or So cyt c5 was reduced by treatment with 20 mM sodium L-sodium ascorbate, and excess ascorbate was removed from the electron donor by a PD-10 desalting column (NEB) immediately before the assay. A stock of between 5-10 μM enzyme was prepared in assay buffer. As-isolated proteins were treated with K3Fe(CN)6 to ensure that enzyme was the fully oxidized. Semi-reduced enzyme was prepared by incubation for at least 1 hour in 1 mM sodium L- ascorbate and 10 μM diaminodurol. The oxidation of the reduced horse heart cytochrome c was monitored at 550 nm in the presence of H2O2 using an extinction coefficient difference between the oxidized and reduced protein of (Δε550) 21.5 mM−1cm−1 (23), whereas oxidation of reduced So cyt c5 was monitored at 553 nm using a difference in extinction coefficient of (Δε553) 12.5 ± 2 mM−1cm−1 as determined by comparison to protein concentrations determined by the BCA assay (Thermo Scientific). When azurin was used as the electron donor, the increase in absorbance at 628 nm is monitored using an extinction coefficient of ε628 = 5.0 mM−1cm−1 (24).
Lag-Phase Kinetics Model
A kinetic model for activation of the fully oxidized enzyme was implemented in Python 2.7, and the complete code is given in the Supporting Information. Because the concentration of active, semi-reduced CcP enzyme changes over the course of the experiment, the differential equations that describe the behavior of the system do not have a closed solution. Instead, we implement numerical integration with a time-step of 0.2 s to find an approximate analytical solution to the differential equations. Least-squares optimization was accomplished using the optimize.leastsq function in SciPy.
The amount of CcP converted from the as-isolated, oxidized state to the semi-reduced state during each time step depends on both the fraction of oxidized CcP that is bound to an electron donor and the rate of electron transfer in the bound complex. Based on the observation that even large concentrations of electron donor proteins (> 50 μM) are not saturating in the peroxidase assay, we assume that binding of oxidized CcP to the reduced electron donor (ED) protein has an equilibrium dissociation constant greater than 10 μM, and can be modeled by a rapid equilibrium. The fraction of oxidized CcP in the bound complex is given by:
where [ED]red is the concentration of reduced electron donor and KD is the equilibrium dissociation constant for reduced electron donor binding to oxidized CcP.
During each time step, the concentration of semi-reduced CcP increases by the factor:
where kact is a rate constant with units of s−1 that describes the rate of electron transfer in the bound complex.
During each time step there are two ways for electron donor to become oxidized. The first is donating an electron to activate one molecule of CcP. The second is donating an electron to a molecule of CcP that has been oxidized by peroxide during the catalytic cycle. CcP must be recharged with two electrons during each catalytic cycle (6). The rate of peroxide catalysis depends on [CcP]active, and is first order in terms of the reduced electron donor concentration. To ensure the reaction is zero-th order in peroxide, we use peroxide concentrations in excess of 9 μM, an order of magnitude greater than the hydrogen peroxide Km (see Results). The total change in the concentration of reduced electron donor at each time-step is:
where k1 is the second-order rate constant for the peroxide turnover. Mulitplying k1 by the electron donor concentration gives as first-order rate constant that should match the kcatobs from Michaelis-Menten analysis.
Total absorbance is calculated using extinction coefficients for oxidized and reduced forms of the electron donor. For horse heart cytochrome c, ε(550 nm, oxidized) = 8.4 mM−1cm−1, ε(553 nm, reduced) = 29.9 mM−1cm−1 (23). For Shewanella oneidensis cyt c5, extinction coefficients were experimentally determined by comparison to BCA assay-derived concentrations, ε(553 nm, oxidized) = 5.1 ± 0.5 mM−1cm−1, ε(553 nm, reduced) = 17.6 ± 2 mM−1cm−1.
Following these three calculations, new values for [CcP]active, [CcP]inactive, [ED]ox, and [ED]red are calculated, and used as initial values for the next time-step. For the best fit to the data, we include an additional time-dependent decay of the active So CcP into an inactive state.
Results
Expression and Purification of So CcP as an MBP Fusion
The bacterial diheme peroxidase from Shewanella oneidensis was overexpressed in E. coli JM109 as a soluble MBP fusion. Induction at high optical density (A600 > 1) and in the presence of the c-type cytochrome maturation cassette (ccmABCDEFGH) (22) yields 10-25 mg of fusion protein per liter of culture. Because the presence of a fusion protein can change the activity of the protein of interest (25, 26), we proceeded to investigate whether the recombinant protein behaved like a native BCcP in biochemical assays.
Following removal of the MBP protein by TEV protease and Ni-NTA chromatography, the purity index (A408/A280) was typically around 4.8, matching previously reported values for a bacterial diheme peroxidases isolated from their native source (27). Because all bacterial diheme peroxidases have two hemes and a comparable number of tryptophan residues, a similar purity index suggests that a majority of the protein has the native heme loading. The MBP-fusion protein runs on an SDS-PAGE gel with an apparent molecular weight of 70 kDa, and the tag-free So CcP runs in a diffuse band with an apparent molecular weight of 35 kDa (Figure S1), consistent with the size predicted by the expressed amino acid sequence. Like all other reported BCcPs, both the MBP-fusion and tag-free So CcP proteins run as a dimer in size-exclusion chromatography (Figure S2) (16, 19, 20, 28).
Optical Properties of the CcP-MBP Fusion and Tag-free CcP
The optical absorbance spectra of fully oxidized verses semi-reduced So CcP, both as an MBP-fusion protein and with the MBP tag removed, are shown in Figure 3. The spectral features at wavelengths greater than 300 nm characteristic of heme absorption are virtually identical in the MBP-fusion So CcP verses the tag-free So CcP, suggesting that the heme environments are not substantially affected by the presence of the fusion protein. The fusion protein has a higher absorbance at 280 nm, consistent with the presence of additional tryptophans in the MBP tag. Semi-reduced samples have extremely high absorbance at wavelengths below 300 nm due to the presence of excess sodium L-ascorbate, which is required to keep the sample in the semi-reduced state.
Figure 3.
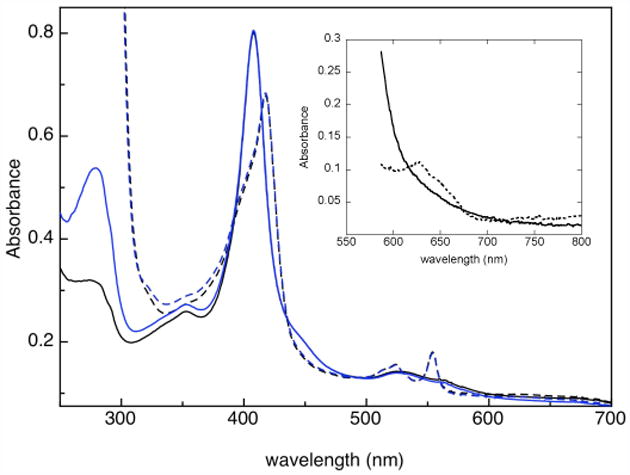
Electronic absorption spectrum of the oxidized (solid, black) and semi-reduced (dashed, black) tag-free So CcP, and oxidized (solid, blue) and semi-reduced (dashed, blue) MBP-fusion So CcP. The inset shows the absorption spectrum of tag-free oxidized and semi-reduced So CcP at 8 times higher concentration.
Fully oxidized So CcP shows a characteristic Soret maximum at 407 nm and broad β- and α-bands at 540 and 553 nm. The fully oxidized enzyme does not display the 640 nm band indicative of high-spin heme (Figure 3, inset). Upon reduction with L-ascorbate, the Soret band maximum shifts to 417 nm and a Soret shoulder around 407 nm persists, the β- and α-bands at 540 and 553 nm dramatically sharpen (consistent with the reduction of the H-heme (29)), and a charge transfer band at around 640 nm, suggestive of a high-spin ferric heme state, is clearly apparent (Figure 3, inset).
EPR Properties of the CcP-MBP and Tag-free CcP
The fully oxidized state of So CcP shows two low-spin ferric species (Figure 4A) in equal amounts. The signal at g = 3.40 is from a highly anisotropic low-spin species observed in all bacterial diheme peroxidases in their as-isolated state (both hemes in the ferric state) and originates from the H-Heme of the protein (17, 19, 30, 31). Species with g = 3.0, 2.21, 1.30 (L1-Heme) originate from the L-Heme and have g-values similar to that of the Psa-type peroxidases (31, 32). The simulation overlaid on the data quantitatively agrees with the total heme concentration of the sample, and from the simulation, the two heme species are found to be present in equal amount. So CcP does not show a high-spin ferric signal near g = 6, which is a minority species in other diheme peroxidases (Figure S3) (17, 19, 29, 30, 33). Upon reduction with ascorbate the H-Heme signal disappears (Figure 4B), suggesting conversion of the ferric H-Heme to an EPR silent low-spin ferrous species as observed in other peroxidases. The signal from L1-Heme loses intensity and a new low-spin heme species with g = 2.87, 2.36, 1.51 (L2-Heme) appears. This change can be attributed to the conversion of a fraction of the low-potential species into the new L2-Heme species. The concentrations of L1- and L2-Heme in the semi-reduced CcP together account for the concentration of low-potential heme before reduction. Table 1 summarizes the various species in the oxidized and semi-reduced enzyme with their amounts obtained from quantitative simulations.
Figure 4.
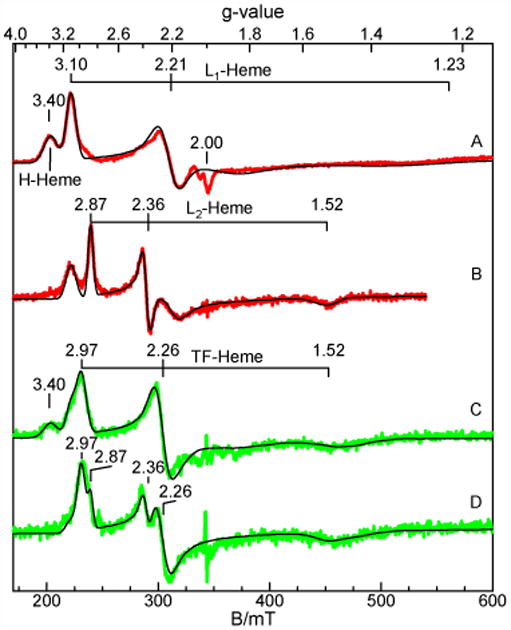
EPR spectra of 0.5 mM CcP in the absence and presence of sodium L-ascorbate with simulations overlaid. (A) So CcP-MBP, (B) Semi-reduced So CcP-MBP, (C) tag-free So CcP, (D) semi-reduced tag-free So CcP. Experiments were conduced with microwave frequency of 9.62 GHz, power of 0.02 mW, and temperature of 7 K.
Table 1.
EPR parameters of the CcP-MBP and tag-free So CcP in the oxidized and semi-reduced states.
| Heme Species | Amount with respect to total Fe (%) | |||
|---|---|---|---|---|
| CcP-MBP | Semi-red CcP-MBP | Tag free CcP | Semi-red Tag free CcP | |
| H-Heme | 50 | 0 | 33 | 0 |
| L1-Heme | 50 | 34 | 33 | 11 |
| L2-Heme | 0 | 20 | 8 | |
| TF-Heme | 0 | 0 | 33 | 33 |
Tag-free So CcP (Figure 4C) shows the same H-Heme species and L1-Heme species as the fusion protein, but both at an overall lower amount (Table 1). In addition, a new feature is observed at g = 2.97. The new signal represents a significant amount of heme and overlaps with the g = 3.10 signal of the L1-Heme. As shown by the simulation on Figure 4C, the spectrum can be accurately simulated with three heme species, using the same parameters as H- and L1-Heme species, and a new heme site with g = 2.97, 2.26, 1.52 (TF-Heme). The simulations indicate that the concentrations of the H- and L1-Heme species are approximately equal, and both lower in amount commensurate with the amount of the TF-Heme present. Thus, the new TF-Heme species appears at the expense of both H- and L1-Heme species. For the semi-reduced tag-free protein (Figure 4D), the signal g = 3.40 vanishes indicating full reduction of the H-Heme. The signal (shoulder) at g = 3.1 decreases and the signal from the L2-Heme is clearly visible at g = 2.36. The simulation shown in Figure 4D use the same parameters for L1-, L2- and TF-Hemes as that of Figure 4B or 4C, except in different amounts. The percentages of species determined from the simulation are given in Table 1. The relative amounts of the L1-Heme and L2-Heme are consistent with that observed from the fusion protein. The TF-Heme species in the semi-reduced sample is not affected by ascorbate addition and the amount of this species remains the same before and after reduction.
Catalytic Properties of So CcP
The activity of So CcP was determined in the presence of micromolar concentrations of various redox partners (Table 2). So cyt c5 is the presumed native electron donor in the periplasm of Shewanella, whereas horse heart cyt c and azurin are electron donors that have been previously used for kinetic analysis of BCcPs, and are included in this study for comparison. We determined kinetic parameters for both MBP-fusion So CcP and the tag-free So CcP. The linear initial rates (for semi-reduced protein) or fastest linear phases (for oxidized protein) at various concentrations of peroxide were used to calculate kinetic parameters using the Michaelis-Menten formalism (Table 2; Figure S4). Because the electron donor proteins bind extremely weakly to So CcP (Km > 10 μM), we could not achieve saturating concentrations in vitro using any of the electron donors studied, and as a result the values reported below are not maximal turnover rates.
Table 2.
Kinetic parameters of both the oxidized and semi-reduced states of So CcP-MBP and tag-free So CcP.
| CcP-MBP | Tag-free CcP | ||||
|---|---|---|---|---|---|
| Electron Donor† | Parameter§ | Semi-red | Oxidized | Semi-red | Oxidized |
| Horse heart cyt c | Km (μM) for H2O2 | 0.13 ± 0.08 | 0.3 ± 0.1 | 0.03 ± 0.01 | 0.17 ± 0.05 |
| kcatobs (s−1) | 35 ± 3 | 13 ± 1 | 1.9 ± 0.2 | 1.3 ± 1.0 | |
| kcatobs/Km (M−1s−1) ×106 | 280 | 46 | 74 | 7.5 | |
| So cyt c5 | Km (μM) for H2O2 | 0.1 ± 0.1 | 0.8 ± 0.1 | 0.3 ± 0.1 | 0.6 ± 0.5 |
| kcatobs (s−1) | 10 ± 1 | 0.8 ± 0.1 | 73 ± 5 | 7 ± 5 | |
| kcatobs/Km (M−1s−1) ×106 | 100 | 1 | 240 | 12 | |
| Azurin | Km (μM) for H2O2 | 0.2 ± 0.2 | 0.3 ± 0.1 | 0.2 ± 0.1 | 0.4 ± 0.1 |
| kcatobs (s−1) | 40 ± 20 | 12 ± 2 | 3.4 ± 0.3 | 1.3 ± 0.1 | |
| kcatobs/Km (M−1s−1) ×106 | 202 | 38 | 17 | 3 | |
Electron donors are horse heart cytochrome c (10 μM), So cyt c5 (4 μM) or azurin (12 μM).
Assays were conducted in 5 mM MES, 5 mM HEPES, 10 mM NaCl, 1 mM CaCl2 (pH 6.0) at 23°C.
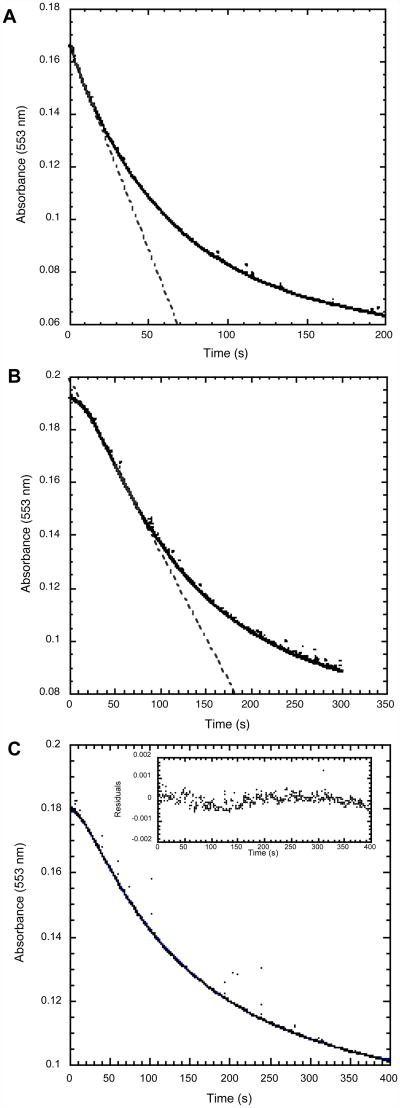
Qualitative inspection of the progress curves reveals that while the oxidation of electron donor occurs in a typical manner for reactions initiated by semi-reduced enzyme (Figure 5A), a distinct lag phase is observed when fully oxidized So CcP is used (Figure 5B). The lag phase reflects the conversion of the oxidized state to the active, semi-reduced state in vitro. In all cases, kcatobs values for the semi-reduced So CcP are faster than the oxidized So CcP. We suspect that this is partially because of a time-dependent deactivation of the CcP during the several minute time course of the experiment. We have seen that for both semi-reduced and oxidized So CcP, the electron donor is not completely oxidized at the endpoint of the experiment if the initial rate is not sufficiently fast (data not shown).
Figure 5.
Comparison of peroxidase catalytic activity using So cyt c5 (4 μM) as the electron donor for (A) semi-reduced tag-free So CcP and (B & C) oxidized tag-free So CcP. Experiments are conducted in 5 mM MES, 5 mM HEPES, 10 mM NaCl, 1 mM CaCl2 (pH 6.0) at 23°C. In A & B, points represent the data and a line represents the linear rate used for Michaelis-Menten analysis. Pane C compares the experimental data (points) to the least-square fit of the data to the kinetic model (line) as described in the text. The inset in (C) shows the residuals of the least-square fit.
Kinetic parameters for So CcP vary depending on the electron donor used or the presence of the MBP tag. For the non-native electron donors horse heart cyt c and azurin, peroxide turnover of the MBP-tagged So CcP is approximately 10 times faster than for the native, tag-free form in the same oxidation state. In contrast, when So cyt c5 is used the opposite is true; peroxide turnover rates for the tag-free protein are 7 to 10 times faster for the tag-free form of the protein than for the MBP-tagged protein, suggesting that the MBP tag affects the rate-limiting step of turnover.
Peroxide binding as assessed by Km values is the same, within error, for almost all combinations of peroxidase and electron donor assayed. A notable exception is that Km values are generally two times weaker for the oxidized form of the enzyme when compared to the semi-reduced form under the same conditions, which may reflect an incomplete activation of the oxidized protein on the time-scale of the in vitro assay.
Comparison of Peroxide Turnover to Other BCcP Family Members
Peroxide turnover kinetics have been reported for numerous members of the BCcP family (Table 3). Among the BCcPs in Table 3, all have estimated or measured Km values for electron donors greater than 50 μM (17, 20, 24, 34). Because peroxide turnover was measured at concentrations much less than the Km, the rate of the reaction is approximately linearly proportional to the electron donor concentration. In this linear range, the peroxide turnover divided by the electron donor concentration (kcatobs/[ED]) gives an effective bimolecular rate constant that can be compared between the BCcPs. The value of kcatobs/[ED] for So CcP is the fastest of all of the BCcPs we surveyed. So CcP also has the tightest peroxide binding, as measured by Km.
Table 3.
Comparison of literature values of the peroxide turnover parameters of various bacterial diheme cytochrome c peroxidases in the semi-reduced state using cytochromes c as an electron donor.
| Organism ‡ | Km (μM) for H2O2 | kcatobs (s−1) | Electron donor (concentration) | kcatobs/[ED] (s−1μM−1)& | Required Pre-reduction? | Reference |
|---|---|---|---|---|---|---|
| Psa | 6 | 118 | Psa cyt 551 (13 μM) | 14 | Yes | (24) |
| Rc* | 33 | 40 | Rc cyt c2 (18 μM) | 2 | Yes | (34) |
| Pss | 1.8 | 88 | Pss cyt c551 (7 μM) | 13 | Yes | (17) |
| Gs | 6.2 | 15.5 | ABTS (3 mM) | Yes | (18) | |
| Mc | 0.5 | 50 | Mc cyt c555 (9 μM) | 5 | No | (20) |
| So | 0.3 | 73 | So cyt c5 (4 μM) | 18 | Yes | This Study |
Abbreviations match those used in the text.
Turnover for as-isolated protein.
This approximation holds because the concentration of electron donor is much less than the estimated Km, therefore the rate is approximately linear with electron donor concentration.
Computational Modeling of Lag-Phase Kinetics
Our simple kinetics model, which considers an equilibrium binding step, an activation step, and a peroxidase turnover step, recapitulates the shape of the peroxide turnover progress curve (Figure 5C). Residuals are very small but nonrandom, suggesting a minor systematic error of unknown origin.
Implementing the computational model requires us to make some educated hypotheses about the So CcP:So cyt c5 system. The model contains two different parameters, the activation rate and the dissociation constant for binding of the electron donor to So CcP, either of which could effect the length of the lag phase. Neither of these parameters has been measured independently for So CcP and So cyt c5. In our model, we are unable to separate the effect of the activation rate constant from that of the equilibrium dissociation constant of the oxidized form of So CcP for the reduced electron donor. We therefore implement the model by keeping the equilibrium dissociation constant the same (7μM) for all cases. This value for KD is in the range of equilibrium binding constants previously reported for BCcPs and monoheme electron donor proteins (9). Differences in the activation rates that we report (kact) reflect either changes in electron transfer rate or differences in binding of the electron donor.
Analysis of lag-phase kinetics using the computational model (Table 4) confirms the results from Michealis-Menten analysis. The MBP-fusion So CcP turns over peroxide at approximately the same rate with either horse heart cyt c or So cyt c5 as the electron donor, whereas the tag-free So CcP turns over peroxide 25 times more rapidly with the native electron donor So cyt c5 when compared to horse heart cyt c. Activation rates are significantly slowed when tag-free So CcP is assayed in the presence of horse heart cyt c as the electron donor. This suggests that horse heart cyt c binds to the tag-free protein either more weakly or in a conformation that gives less efficient electron transfer.
Table 4.
Model-derived kinetic parameters for peroxidase turnover initiated with oxidized So CcP.
| Electron donor | Peroxidase | Activation rate kact (s−1) | Peroxide turnover rate k1 (s−1)* |
|---|---|---|---|
| So cyt c5 | So CcP-MBP | 0.05 ± 0.01 | 1.6 ± 0.4 |
| So CcP | 0.07 ± 0.02 | 7 ± 1 | |
| Horse heart cyt c | So CcP-MBP | 0.08 ± 0.02 | 1.6 ± 0.4 |
| So CcP | 0.02 ± 0.007 | 0.28 ± 0.08 |
Turnover values correspond to 4 μM electron donor, with the KD for electron donor binding to CcP set to 7 μM.
Characterization of Activation Loop Mutants
Additional kinetic analyses were performed for two tag-free So CcP variants containing mutations in two of the three loops that are known to undergo conformational changes upon reductive activation of a Psa-type CcP (depicted in Figure 1): either loop 1 (P75T/H81K/E84Q) or loop 3 (M219Q/F247N). We did not make mutations in the loop 2 region because the amino acid sequence of the So enzyme already matches that of the Ne CcP in all key positions that are different between the two functional sub-classes. The visible absorbance spectra of both So CcP variants in the semi-reduced and oxidized states are identical when compared to spectrum of the wild-type protein in the same oxidation state (data not shown), indicating that the protein variants are folded with hemes in a similar environment as the wild-type enzyme. Despite the location of the mutations (many of which are far from the presumed peroxide binding site), both variants show defects in peroxide turnover (Table 5), suggesting that the wild-type enzyme is highly optimized for a high rate of peroxide turnover. The triple mutant in loop 1, P75T/H81K/E84Q, shows a five-fold decrease in turnover of the semi-reduced state and 20-fold decrease in turnover of the oxidized state when compared to the wild-type protein under identical conditions. The double mutant M219Q/F247N shows a slightly larger defect in peroxide turnover in the semi-reduced state, a 12-fold decrease when compared to wild-type protein under identical conditions. Peroxide binding for the semi-reduced state of the mutants, as measured by Km, is unchanged (within error) for the mutants when compared to wild-type, but both mutants show a slight increase of the Km of the oxidized state for peroxide. The So CcP protein variants also demonstrated lag-phase kinetics for peroxide turnover when initiated by oxidized enzyme. For the loop 1 triple mutant, the activation rate is 7 times faster than wild-type, and the loop 3 double mutant shows a very slight increase in activation rate over the wild-type protein.
Table 5.
Kinetic parameters of both the oxidized and semi-reduced states of tag-free So CcP variants using So c5 as the electron donor.
| Km for H2O2 (μM) | kcatobs (s−1) | kact (s−1) | k1 (s−1)* | |||
|---|---|---|---|---|---|---|
| Loop 1 | P75T/H81K/E84Q | Oxidized | 1.0 ± 0.3 | 0.3 ± 0.1 | 0.6 ± 0.2 | 0.56± 0.04 |
| Semi-reduced | 0.4 ± 0.3 | 13 ± 1 | n.a. | n.a. | ||
| Loop 3 | M219Q/F247N | Oxidized | 1.6 ± 0.2 | 0.5 ± 0.1 | 0.09 ± 0.01 | 0.60 ± 0.04 |
| Semi-reduced | 0.5 ± 0.3 | 5.9 ± 0.5 | n.a. | n.a. | ||
| Wild type | Oxidized | 0.6 ± 0.5 | 7 ± 5 | 0.07 ± 0.02 | 7 ± 1 | |
| Semi-reduced | 0.3 ± 0.1 | 73 ± 5 | n.a. | n.a. | ||
Turnover values correspond to 4 μM electron donor.
The kinetic differences noted for these two constructs were reflected in differences in their EPR characterization: whereas the fully-oxidized wild-type MBP-tagged enzyme reveals two states H (g = 3.37, 2.20, 1.50) and L1 (g= 3.08, 2.26, 1.30) corresponding to the H- and L-hemes, both the loop 1 triple mutant and the loop 3 double mutant additionally reveal significant population of states L2 and TF (Figure S5). Thus, substantial amounts (Table S1) of oxidized MBP-enzymes have already adopted conformations that would be associated activated L-heme (i.e., the L2 state). Consequently, the presence of lag-phase kinetics is at present unclear. The lower activities of the variants do not quantitately correlated with the presence of the TF-heme. If we assume that the TF-heme state is inactive, we would expect at most a factor of two decrease in activity, yet activity is decreased by approximately an order of magnitude. We suspect, but cannot yet substantiate, that the conformational changes observed in the EPR spectra of the oxidized states of the variants reflect changes in access to the hemes, their redox properties, or the ability of specific enzymatic states to inter-convert, as required for catalysis.
Discussion
Optical Spectroscopy of So CcP
The electronic absorption spectra of the as-isolated state of the MBP-fusion and tag-free So CcP both exhibit the Soret maximum at 407 nm characteristic of ferric c-type hemes. Changes to the spectrum upon reduction with ascorbate or dithionite mirror changes that have been seen other BCcPs (15, 16, 18, 19, 29): Reduction of the H-heme with ascorbate causes a red-shift in the Soret maximum to 417 nm and a sharpening of the β- and α-bands at 540 nm and 553 nm, while the ascorbate-reduced spectrum maintains a Soret feature at 407 nm, indicating that one heme remains in the ferric state. Further reduction with dithionite results in the disappearance of the Soret shoulder and an increase in the intensity of the Soret peak at 417 nm (data not shown). Intriguingly, the only significant differences in the optical characteristics of the So enzyme, with respect to canonical CcPs is the absence of a 640 nm band with the fully oxidized enzyme. As observed in the Pd and Psa enzymes, the 640 nm band has been attributed to a high-spin state of the high-potential heme (29,30,33)). Thus, the So enzyme appears to possess a low-spin H-heme. However, the 640 nm band is observed in the semi-reduced protein, indicating the same high-spin state of the oxidized L-heme, in parallel with other BCcP enzymes that require reductive activation to attain activity (29,30,33)).
EPR Characterization of the So CcP
Comparisons of the So enzyme with other bacterial diheme peroxidases by electron paramagnetic resonance spectroscopy (EPR) demonstrates the similarity of So peroxidase to the Psa (30) and Pd (31) peroxidases in both its oxidized and semi-reduced form, as compared to the Ne and Mc peroxidases. In the oxidized state, the gz-value (3.10) of the L-Heme is closer to that of Pd (3.00) than Ne (2.85). In the semi-reduced state of both the So and Pd enzymes, a fraction of the L-heme changes to a new species, whereas no such change occurs for the Ne enzyme. Contrasting the optical spectroscopy, we observe no evidence in the EPR for conversion of heme to the high-spin state as has been previously observed for the Psa enzyme (30). For the So enzyme, the two L-Heme species represent 34% and 20% of the heme in the sample after ascorbate addition. Based on the g-values, the new L2-heme species falls into the imidazolate classification of EPR signals: using the crystal-field parameters of Taylor (35), Δ/λ = 2.736 and v/Δ = 0.762 for L2, while L1 displays analogous parameters that are indicative of neutral imidazole binding (Δ/λ = 2.687 and v/Δ = 0.524). Thus, it appears that upon semi-reduction the axial His for 40% of the L-heme is deprotonated. For all these enzymes, the gz-values of the H-heme are approximately the same, and this heme is reduced with ascorbate in the semi-reduced form. Approximately one-half of the tag-free So CcP protein appears to be the same as the fusion protein. The signals are in the same locations, and upon ascorbate addition, this fraction of the protein undergoes the same spectroscopic changes as that observed for the fusion protein: H-heme vanishes and a fraction of the L-Heme changes to a second species. The spectrum of the tag-free protein, however, has a significant additional new low-spin heme species (TF-heme) with gz = 2.97 relative to the MBP-fusion protein. This TF-heme also appears to be a bis-His heme based on the g-values (with crystal-field parameters that make distinguishing between neutral His and imidazolate character ambiguous). The amount of TF-heme is approximately equal to that of the normal heme species, which suggest that approximately one-half of the protein is in this other state. The addition of ascorbate to the tag-free protein had no effect on the state ascribed to the TF-heme, suggesting that it cannot be reduced by ascorbate.
The molecular basis of the generation of the new TF-heme state is unclear at this time, and because the visible absorbance spectra of tag-free enzyme are nearly identical to the MBP-fusion, it is presumed that the generation of the TF-heme state may be reversible at room temperature. However, these data do imply that there are inherent differences in conformational flexibility in the MBP-tagged verses the tag-free forms of the So CcP enzyme, consistent with the differences seen in the kinetics of peroxide turnover and activation rate in the two forms of the enzyme.
Catalytic behavior of So CcP
We monitored the kinetic parameters of peroxide turnover and activation of So CcP in the presence of several electron donor proteins as well as for MBP-tagged and tag-free versions of the enzyme. It is clear that the kinetic behavior of the MBP-tagged protein differs substantially from the tag-free protein; without the MBP-tag, So cyt c5 gives the highest peroxide turnover, whereas the MBP-tagged protein is most active when horse heart cyt c or azurin are used as electron donors. These data indicate that the relatively large MBP protein has a principle effect on the binding of electron donors.
It is surprising that the peroxide turnover rates of the oxidized protein never reach the rates of the semi-reduced proteins. This may indicate that there is a slow step in reductive activation that is required for maximum turnover efficiency and is not attained during the time course of the in vitro experiments. This is supported by the observation that peroxide binding (Km) differs between oxidized and semi-reduced So CcP. The slow time-dependent decay of activated protein, which was required for optimal fit of the computation model to the lag-phase data, may also contribute to the rate differences. One possible explanation for this deactivation is dissociation of the dimeric So CcP at the low nanomolar concentrations in the in vitro assay (14).
The catalytic parameters we report for peroxide turnover of So CcP in the presence of various electron donors are in the same range as those reported for other BCcPs elsewhere in the literature. So CcP binds peroxide much more tightly, as assayed by Km values, than other representative BCcPs. This may reflect a difference in peroxide sensitivity of components of the Shewanella periplasm when compared with other gram negative bacteria. The values of kcatobs reported in Table 3 are not the maximum values for the BCcP enzymes; maximum peroxide turnover, in the presence of saturating electron donor, will be several times higher. Given the reported concentrations of So c5 isolated from cells grown under low oxygen conditions (34 micromoles of c5 per kilogram of cell pellet; 36), we estimate that the in vivo concentration of So cyt c5 in the periplasm is on the order of 300 μM, suggesting that in vivo rates of peroxide turnover will be much faster than those reported here, where the maximum cyt c5 concentration used is 40 μM (with the relationship between peroxide turnover and electron donor concentration being linear).
Studies with other BCcPs have demonstrated that a variety of small proteins can act as electron donors, and that the peroxide turnover rate of the BCcP depends on the identity of the electron donor (37). This variability is attributed to differences the binding orientation of the various electron donors. In the case of Pd CcP, computational modeling and NMR identified two possible electron donor binding sites: one that was favored by horse heart cytochrome c and a different binding site favored by the native Pd electron donor cytochrome c-550 (8, 9). Our data suggest a similar model for So CcP. For So CcP, peroxide turnover is most efficient for the tag-free protein when the native electron donor So cyt c5 is used, suggesting that So cyt c5 binds to the So CcP surface in a unique mode that is optimized for electron transfer. The lower turnover rates for non-native electron donors may result from binding more weakly to the So CcP surface, or in a slightly different orientation that is less optimal for electron transfer. Moreover, we show that the presence of the MBP-tag favors the turnover of peroxide in the presence of the non-native electron donors azurin and horse heart cyt c. This suggests that the bulky MBP protein either increases the binding affinity of non-native electron donors, or forces the non-native electron donor to bind in a mode that facilitates more rapid electron transfer.
Previous studies of BCcP family members and electron donor proteins have suggested that the electrostatic potential of the protein surfaces allows the native electron donor to select a proper binding site. So cyt c5 has an equal number of positive and negative amino acids, but the arrangement of positive and negative charges results in a substantial dipole moment (0.56 D/atom) (38). Horse heart cytochrome c has an excess of positively charged residues, but the arrangement of charges results in a lower net dipole (0.324 D/atom). This difference in the electrostatic potential of the two electron donors may contribute to the differences in binding mode. We are currently undertaking biophysical experiments to explore the differences in the equilibrium binding between electron donors and various forms of the So CcP protein.
Tag-free vs. MBP-fusion So CcP
The So CcP protein was expressed in E. coli with an N-terminal MBP fusion. This fusion protein system allows purification of the So CcP protein in high yield, an important criterion for use as a model BCcP in biophysical studies. We therefore investigated the potential differences between the So CcP-MBP fusion protein and the tag-free So CcP. Optical spectroscopy failed to show any differences between the heme environment in the fusion protein verses the tag-free protein. However, EPR spectroscopy demonstrates that the tag-free protein has greater conformational flexibility than So CcP-MBP, as half of the tag-free So CcP was trapped in a “TF-heme” state in the low-temperature EPR experiments. The peroxidase turnover data suggests that this additional conformational flexibility facilitates interaction with the native electron donor So cyt c5, resulting in higher rates of peroxide turnover. We conclude that the MBP fusion protein affects the conformational flexibility, but not the underlying fold, of the So CcP protein, demonstrating that fusion proteins should be employed carefully in biochemical and biophysical studies.
Kinetics Modeling and Comparison to Other BCcPs
Previous reports (39) have investigated the lag-phase behavior of the model BCcP family member from Pseudomonas aeruginosa. The authors noted, as we do in this report, that the lag-phase behavior may result from equilibrium binding between oxidized CcP and reduced electron donor followed by electron transfer, which can be modeled using an equilibrium binding constant and an activation rate. The authors reported kinetic parameters (Km = 4 μM and kact = 0.2 s−1) (39) that agree remarkably well with those we have established for So CcP. Our use of a modern computational framework in Python and SciPy allows us to quickly extract the activation and peroxidase rates from the kinetic progress curve. The framework we developed has been used here and in continuing experiments to study the differential effects of mutations on the activation or the peroxide turnover rate.
Effect of Activation Loop Mutations on Activation and Peroxide Turnover
There are few reports of mutagenesis studies of BCcP family members. With the exception of the Geobacter CcP loop 2 mutation (18), reports have been limited to mutation of residues with hypothesized roles in heme ligation, proton-delivery or electron transfer between the hemes (40, 41). In general, these mutational studies have resulted in complete deactivation of the peroxide turnover activity of the enzyme, and full kinetic characterization has been rarely reported (40, 41). In this study, we focus on mutations in flexible regions that are not spcificially implicated in catalysis. These locations can be mapped on to the fully oxidized Psa CcP x-ray crystal structure, as shown in Figure 6, which demonstrates that in the case of the M219Q/F247N double mutation, both positions are well removed from the low-potential catalytic heme.
Figure 6.
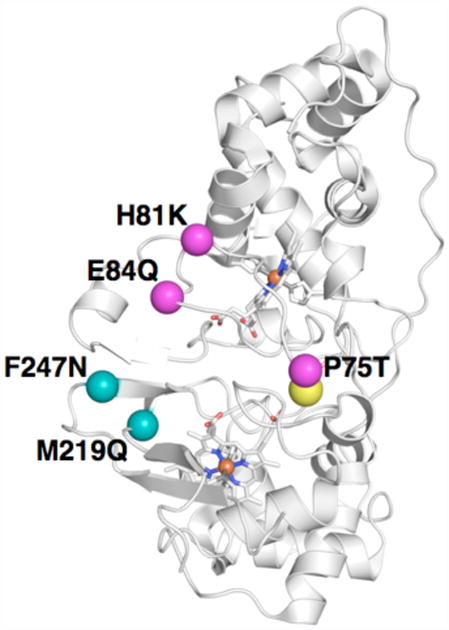
Depiction of the mutations made in either loop 1 or loop 3 in So CcP. Using the Psa fully oxidized structure as a model, locations of the loop 1 triple mutant P75T/H81K/E85Q are indicated as magenta spheres at the alpha carbon position. The loop 3 double mutant positions M219Q and F247N are shown as teal spheres at the predicted alpha carbon positions.
A sequence alignment of Ne and So CcPs shows extremely strong sequence conservation in several loop regions. Based on this sequence conservation and the structural evidence of the importance of these loops in the conformational change required for activation, we predicted that the So CcP would not require reductive activation. However, UV/Visible spectroscopy, EPR spectroscopy, and peroxide turnover indicate that this is not the case. We therefore sought to make mutations in the activation loops of So CcP in an attempt to convert So CcP into a Ne-type BCcP. For the BCcP of Geobacter sulfurreducens, the S134P/V135K double mutant transforms the L-loop (loop 1) into an “always open” state, as determined crystallographically (18). In attempting a similar conversion, we constructed So CcP mutations in loop 1 (P75T/H81K/E84Q) and loop 3 (M219Q/F247N). We did not make mutations in the loop 2 region because the sequences are already nearly identical between So and Ne CcPs; the only differences are in positions otherwise conserved between Psa- and Ne-type BCcPs that are not likely to be important in reductive activation.
We found that neither set of mutations successfully transformed So CcP into a constitutive, Ne-type BCcP. However, the loop 1 triple mutation (P75T/H81K/E84Q) was activated much more quickly than the wild-type protein during the in vitro peroxidase turnover assay. This suggests that the three simultaneous mutations have successfully modulated the structure of loop 1 to favor the active state. Mutations in the loop regions were accompanied by severe decreases in peroxidase activity for the So CcP variants, indicating that the mutations have additional effects on catalytic activity in addition to changes on the equilibrium between the active and inactive states. Interestingly, the M219Q/F247N mutation affects catalysis despite its location near the H-heme, far from the catalytic site. This suggests that the M219Q/F247N mutation also has a subtle impact on the equilibrium between the inactive and active conformations of the protein. Indeed, the EPR characterization of either of the two mutants described here underscore that structural conformations around the H-heme are strongly affected by the sequences of loops 1 and 3. The loop 1 and loop 3 mutants both reveal that multiple heme states can be adopted, while fully oxidized, which presumably negatively impact catalytic competency. And, while the tag-free version of the loop 3 M219Q/F247N mutant appears similar to native enzyme, the L-heme clearly displays slight perturbations in g values, suggesting that the electronic structure of the L-heme is not truly native-like. Together, these data strongly suggest that all of the flexible loop regions identified in other BCcP family members are also important for redox-coupled conformational changes in the So CcP.
Conclusions
This study describes the recombinant expression and characterization of a BCcP family member, the So CcP. UV/Visible spectroscopy and EPR spectroscopy confirm the similarity between So CcP and other Psa-like peroxidases, while EPR also suggests subtle differences in the conformational flexibility of the So CcP when compared to other bacterial peroxidases. Peroxide turnover of So CcP can be monitored using a variety of native and non-native electron donor proteins, and Michaelis-Menten analysis confirms that So CcP catalyzes the turnover of peroxide with catalytic efficiencies similar to other documented BCcPs. The So CcP can be easily purified in high yield from the E. coli system, and sequence variants have proven easy to express and purify, making So CcP an excellent system for mutational study. We hope that the detailed kinetic characterization we present here can serve to initiate extensive mechanistic study of the BCcP family, and more broadly, the mechanism of electron transfer in proteins.
Supplementary Material
Acknowledgments
The authors wish to thank Drs. Frank Collart and Yuri Londer (Argonne), Michelle McGuirl (UMontana) and Mr. Ben Levin for their kind supply of reagents used in this work.
Footnotes
Abbreviations: CcP, cytochrome c peroxidase; BCcP, bacterial cytochrome c peroxidase; MBP, maltose binding protein; HRP, horseradish peroxidase; Psa, Pseudomonas aeruginosa; So, Shewanella oneidensis; Gs, Geobacter sulfurreducens; Ne, Nitrosomonas europaea; Rc,Rhodobacter capsulatus; Pd, Paracoccus denitrificans; Pss, Pseudomonas stutzeri; Psn, Pseudomonas nautica; Mc, Methyloccocus casulatus (Bath); NHE, Normal Hydrogen Electrode; EPR, Electron Paramagnetic Resonance; S-100, Sepharcryl 100.
This work was supported by a National Institutes of Health research grant (R01-GM072663), and the Boston University Undergraduate Research Opportunities Program.
Conflict of Interest Declaration. The authors declare no competing financial interest.
Supporting Information Available. Mutagenesis primers, Python code for least-squares fitting of lag-phase kinetics data and SDS-PAGE gel of So CcP purification as MBP-fusion. This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B, Gutteridge JMC. Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett. 1992;307:108–112. doi: 10.1016/0014-5793(92)80911-y. An update. [DOI] [PubMed] [Google Scholar]
- 3.Van Spanning RJ, De Boer AP, Reijnders WN, Westerhoff HV, Stouthamer AH, Van Der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 4.Hiner A, Raven EL, Thorneley R, Garcia-Canovas F, Rodriguez-Lopez JN. Mechanisms of compound I formation in heme peroxidases. J Inorg Biochem. 2002;91:27–34. doi: 10.1016/s0162-0134(02)00390-2. [DOI] [PubMed] [Google Scholar]
- 5.Finzel BC, Poulos TL, Kraut J. Crystal structure of yeast cytochrome c peroxidase refined at 1.7-A resolution. J Biol Chem. 1984;259:13027–13036. [PubMed] [Google Scholar]
- 6.Ellfolk N, Ronnberg M, Aasa R, Andreasson LE, Vanngard T. Properties and function of the two hemes in Pseudomonas cytochrome c peroxidase. Biochim Biophys Acta. 1983;743:23–30. doi: 10.1016/0167-4838(83)90413-2. [DOI] [PubMed] [Google Scholar]
- 7.Pettigrew GW, Echalier A, Pauleta SR. Structure and mechanism in the bacterial dihaem cytochrome c peroxidases. J Inorg Biochem. 2006;100:551–67. doi: 10.1016/j.jinorgbio.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Pettigrew GW, Prazeres S, Costa C, Palma N, Krippahl L, Moura I, Moura JJ. The structure of an electron transfer complex containing a cytochrome c and a peroxidase. J Biol Chem. 1999;274:11383–9. doi: 10.1074/jbc.274.16.11383. [DOI] [PubMed] [Google Scholar]
- 9.Pettigrew GW, Pauleta SR, Goodhew CF, Cooper A, Nutley M, Jumel K, Harding SE, Costa C, Krippahl L, Moura I, Moura J. Electron transfer complexes of cytochrome c peroxidase from Paracoccus denitrificans containing more than one cytochrome. Biochemistry. 2003;42:11968–11981. doi: 10.1021/bi034829c. [DOI] [PubMed] [Google Scholar]
- 10.Ronnberg M, Ellfolk N. Pseudomonas cytochrome c peroxidase. Initial delay of the peroxidatic reaction Electron transfer properties. Biochim Biophys Acta. 1978;504:60–66. doi: 10.1016/0005-2728(78)90006-3. [DOI] [PubMed] [Google Scholar]
- 11.Echalier A, Goodhew CF, Pettigrew GW, Fulop V. Activation and catalysis of the di-heme cytochrome c peroxidase from Paracoccus pantotrophus. Structure. 2006;14:107–117. doi: 10.1016/j.str.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Echalier A, Brittain T, Wright J, Boycheva S, Mortuza GB, Fulop V, Watmough NJ. Redox-linked structural changes associated with the formation of a catalytically competent form of the diheme cytochrome c peroxidase from Pseudomonas aeruginosa. Biochemistry. 2008;47:1947–1956. doi: 10.1021/bi702064f. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu H, Schuller DJ, Lanzilotta WN, Sundaramoorthy M, Arciero DM, Hooper AB, Poulos TL. Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases. Biochemistry. 2001;40:13483–13490. doi: 10.1021/bi011481h. [DOI] [PubMed] [Google Scholar]
- 14.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura JJG, Moura I. The kinetics of the oxidation of cytochrome c by Paracoccus cytochrome c peroxidase. Biochem J. 1994;300:907–914. doi: 10.1042/bj3000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves T, Besson S, Duarte LC, Pettigrew GW, Girio F, Devreese B, Vandenberghe I, Van Beeumen JJ, Fauque G, Moura I. A cytochrome c peroxidase from Pseudomonas nautica 617 active at high ionic strength: expression, purification and characterization. Biochim Biophys Acta Protein Struct Mol Enzymol. 1999;1434:248–259. doi: 10.1016/s0167-4838(99)00188-0. [DOI] [PubMed] [Google Scholar]
- 16.De Smet L, Pettigrew GW, Van Beeumen JJ. Cloning, overproduction and characterization of cytochrome c peroxidase from the purple phototrophic bacterium Rhodobacter capsulatus. Eur J Biochem. 2001;268:6559–6568. doi: 10.1046/j.0014-2956.2001.02610.x. [DOI] [PubMed] [Google Scholar]
- 17.Timoteo CG, Tavares P, Goodhew CF, Duarte LC, Jumel K, Girio FMF, Harding S, Pettigrew GW, Moura I. Ca2+ and the bacterial peroxidases: the cytochrome c peroxidase from Pseudomonas stutzeri. J Biol Inorg Chem. 2003;8:29–37. doi: 10.1007/s00775-002-0382-y. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann M, Seidel J, Einsle O. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J Mol Biol. 2009;393:951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Arciero D, Hooper AB. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J Biol Chem. 1994;269:11878–11886. [PubMed] [Google Scholar]
- 20.Zahn JA, Arciero DM, Hooper AB, Coats JR, DiSpirito AA. Cytochrome c peroxidase from Methylococcus capsulatus Bath. Arch Microbiol. 1997;168:362–372. doi: 10.1007/s002030050510. [DOI] [PubMed] [Google Scholar]
- 21.Londer YY, Giuliani SE, Peppler T, Collart FR. Addressing Shewanella oneidensis “cytochromome”: the first step towards high-throughput expression of cytochromes c. Protein Expression Purif. 2008;62:128–137. doi: 10.1016/j.pep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Arslan E, Schulz H, Zufferey R, Kunzler P, Thony-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem Biophys Res Commun. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 23.Van Gelder B, Slater EC. The extinction coefficient of cytochrome c. Biochim Biophys Acta. 1962;58:593–5. doi: 10.1016/0006-3002(62)90073-2. [DOI] [PubMed] [Google Scholar]
- 24.Soininen R, Ellfolk N. Pseudomonas cytochrome c peroxidase. IV. Some kinetic properties of the peroxidations reaction, and enzymatic determination of the extinction coefficients of Pseudomonas cytochrome c-551 and azurin. Acta Chem Scand. 1972;26:861–872. doi: 10.3891/acta.chem.scand.26-0861. [DOI] [PubMed] [Google Scholar]
- 25.Mason AB, He QY, Halbrooks PJ, Everse SJ, Gumerov DR, Kaltashov IA, Smith VC, Hewitt J, MacGillivray RTA. Differential effect of a his tag at the N- and C-termini: functional studies with recombinant human serum transferrin. Biochemistry. 2002;41:9448–9454. doi: 10.1021/bi025927l. [DOI] [PubMed] [Google Scholar]
- 26.Song J, Markley JL. Cautionary tail: the presence of an N-terminal tag on dynein light-chain Roadblock/LC7 affects its interaction with a functional partner. Protein Pept Lett. 2007;14:265–268. doi: 10.2174/092986607780090801. [DOI] [PubMed] [Google Scholar]
- 27.Goodhew CF, Wilson IB, Hunter DJ, Pettigrew GW. The cellular location and specificity of bacterial cytochrome c peroxidases. Biochem J. 1990;271:707–712. doi: 10.1042/bj2710707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmour R, Prazeres S, McGinnity DF, Goodhew CF, Moura JJ, Moura I, Pettigrew GW. The affinity and specificity of Ca(2+)-binding sites of cytochrome-c peroxidase from Paracoccus denitrificans. Eur J Biochem. 1995;234:878–86. doi: 10.1111/j.1432-1033.1995.878_a.x. [DOI] [PubMed] [Google Scholar]
- 29.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura I, Moura JJ. Spectroscopic characterization of cytochrome c peroxidase from Paracoccus denitrificans. Biochem J. 1993;294:745–752. doi: 10.1042/bj2940745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foote N, Peterson J, Gadsby P, Greenwood C, Thomson A. Redox-linked spin-state changes in the di-haem cytochrome c-551 peroxidase from Pseudomonas aeruginosa. Biochem J. 1985;230:227–237. doi: 10.1042/bj2300227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prazeres S, Moura JJ, Moura I, Gilmour R, Goodhew CF, Pettigrew GW, Ravi N, Huynh BH. Mossbauer characterization of Paracoccus denitrificans cytochrome c peroxidase. Further evidence for redox and calcium binding-induced heme-heme interaction. J Biol Chem. 1995;270:24264–24269. doi: 10.1074/jbc.270.41.24264. [DOI] [PubMed] [Google Scholar]
- 32.Aasa R, Ellfolk N, Ronnberg M, Vanngard T. Electron paramagnetic resonance studies of Pseudomonas cytochrome c peroxidase. Biochim Biophys Acta. 1981;670:170–175. doi: 10.1016/0005-2795(81)90005-2. [DOI] [PubMed] [Google Scholar]
- 33.Foote N, Peterson J, Gadsby PM, Greenwood C, Thomson AJ. A study of the oxidized form of Pseudomonas aeruginosa cytochrome c-551 peroxidase with the use of magnetic circular dichroism. Biochem J. 1984;223:369–378. doi: 10.1042/bj2230369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu W, De Smet L, Van Driessche G, Bartsch RG, Meyer TE, Cusanovich MA, Van Beeumen J. Characterization of cytochrome c-556 from the purple phototrophic bacterium Rhodobacter capsulatus as a cytochrome-c peroxidase. Eur J Biochem. 1998;258:29–36. doi: 10.1046/j.1432-1327.1998.2580029.x. [DOI] [PubMed] [Google Scholar]
- 35.Taylor C. The EPR of low spin heme complexes: Relation of the tau2g hole model to the directional propertis of the g tensor, and a new method for calculating the ligand field parameters. Biochim Biophys Acta Protein Struct. 1977;491:137–148. doi: 10.1016/0005-2795(77)90049-6. [DOI] [PubMed] [Google Scholar]
- 36.Meyer TE, Tsapin AI, Vandenberghe I, DeSmet L, Frishman D, Nealson KH, Cusanovich MA, vanBeeumen JJ. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS. 2004;8:57–77. doi: 10.1089/153623104773547499. [DOI] [PubMed] [Google Scholar]
- 37.Pauleta SR, Cooper A, Nutley M, Errington N, Harding S, Guerlesquin F, Goodhew CF, Moura I, Moura JJG, Pettigrew GW. A copper protein and a cytochrome bind at the same site on bacterial cytochrome c peroxidase. Biochemistry. 2004;43:14566–14576. doi: 10.1021/bi0485833. [DOI] [PubMed] [Google Scholar]
- 38.Felder CE, Prilusky J, Silman I, Sussman JL. A server and database for dipole moments of proteins. Nucleic Acids Res. 2007;35:W512–521. doi: 10.1093/nar/gkm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foote N, Turner R, Brittain T, Greenwood C. A quantitative model for the mechanism of action of the cytochrome c peroxidase of Pseudomonas aeruginosa. Biochem J. 1992;283:839–843. doi: 10.1042/bj2830839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Smet L, Savvides SN, Van Horen E, Pettigrew G, Van Beeumen JJ. Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases. J Biol Chem. 2006;281:4371–4379. doi: 10.1074/jbc.M509582200. [DOI] [PubMed] [Google Scholar]
- 41.Hsiao HC, Boycheva S, Watmough NJ, Brittain T. Activation of the cytochrome c peroxidase of Pseudomonas aeruginosa. The role of a heme-linked protein loop: a mutagenesis study. J Biol Inorg Chem. 2007;101:1133–1139. doi: 10.1016/j.jinorgbio.2007.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.