Abstract
Bacterial cytochrome c peroxidase (CcP) enzymes are diheme redox proteins that reduce hydrogen peroxide to water. They are canonically characterized by a peroxidatic (called L, for “low reduction potential”) active site heme, and a secondary heme (H, for “high reduction potential”) associated with electron transfer, and an enzymatic activity that exists only when the H-heme is pre-reduced to the FeII oxidation state. The pre-reduction step results in a conformational change at the active site itself, where a histidine-bearing loop will adopt an “open” conformation allowing hydrogen peroxide to bind to the FeIII of the L-heme. Notably, the enzyme from Nitrosomonas europaea does not require pre-reduction. Previously, we have shown that protein film voltammetry (PFV) is a highly useful tool in distinguishing the electrocatalytic mechanisms of the Nitromonas-type of enzyme from other CcPs. Here, we apply PFV to the recently described enzyme from Geobacter sulfurreducens, and the Geobacter S134P/V135K double mutant, which has been shown to be similar to the canonical sub-class of peroxidases and the Nitrosomonas sub-class of enzymes, respectively. Here we find that the wild-type Geobacter CcP is indeed similar electrochemically to the bacterial CcPs that require reductive activation, yet the S134P/V135K mutant shows two phases of electrocatalysis: one that is low in potential, like the wild-type enzyme, and a second, higher-potential phase that has a potential dependent upon substrate binding and pH, yet is at a potential that is very similar to the H-heme. These findings are interpreted in terms of a model where rate-limiting intra-protein electron transfer governs the catalytic performance of the S134P/V135K enzyme.
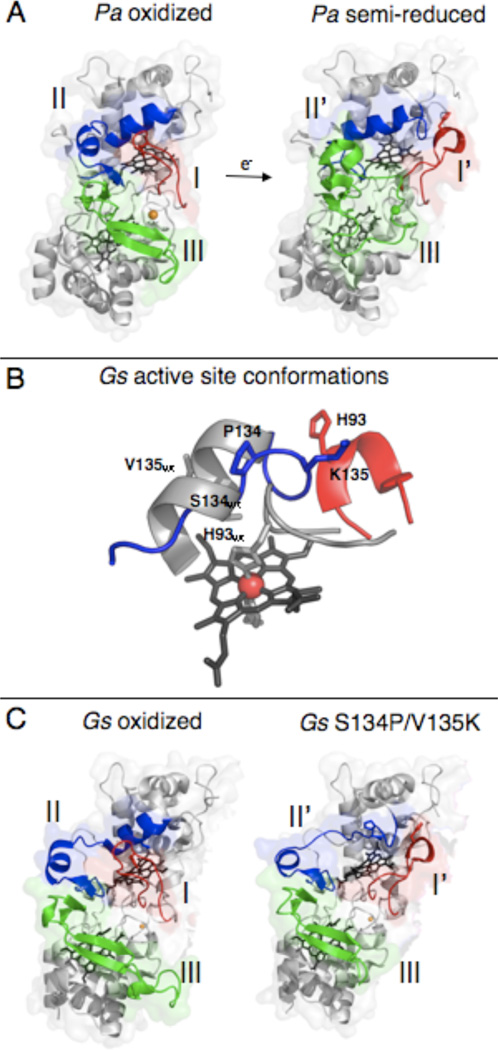
Close investigation of the genome sequence of Geobacter sulfurreducens (Gs), an organism initially classified as a strict anaerobe (1), revealed the presence of enzymes that typically enable a microbe to perform aerobic respiration (2), including homologs to catalase, superoxide dismutase, rubrerythrin, cytochrome c oxidase and heme peroxidases (3). Subsequent assessment of the ability of Geobacter to grow aerobically (4) and studies where Fe(III) citrate is provided as the terminal electron acceptor suggested that CcpA (gene GSU2813), a diheme-containing cytochrome c peroxidase (CcP) (2), is an important component of Geobacter biochemistry, as it is responsible for the facile reduction of H2O2 to water (5). The Gs CcpA gene product has recently been shown to be a highly basic cytochrome c peroxidase that is quite similar to the canonical diheme peroxidases, including the disposition of the two heme cofactors, the observation of a Ca2+ ion bound at the interface of the two heme-bearing domains common to all bacterial CcP enzymes (6–10), and the requirement for pre-reduction of the His/Met ligated high potential heme (H) in order to attain activity at the peroxidatic active site (L) (15). As indicated in Table 1, the majority of bacterial heme peroxidases require reductive activation (5, 6, 13, 18, 19), aside from a small number of CcP enzymes, including that from Nitrosomonas europaea (Ne) (10, 11, 20–22). The overall process of reductive activation begins with the low midpoint potential (11–13) L-heme existing in the oxidized state, with a bis-His ligated coordination environment. Reduction of the high-potential heme (H), by one electron causes a conformational change in the L-loop region containing the distal His ligand, as well as two other loop regions (Figure 1A, where loops are labeled 1 through 3). Figure 1A shows the global conformational changes associated with reductive activation have been revealed for the canonical peroxidase from Pseudomonas aeruginosa (Pa) (16,17) which has been crystallographically characterized in its resting and reductively-activated states (9, 14), where the activated form of the enzyme displays an accessible L-heme ready to bind and react with hydrogen peroxide.
Table 1.
Comparison of the Activity Requirements for Bacterial Cytochrome c Peroxidases
Figure 1.
Crystal structures of members of the canonical class of bacterial cytochrome c peroxidases. (A) Structures of the oxidized P. aeruginosa CcP (top, PDB 1EBY) with loops I (red), II (blue), and III (green) highlighted. The addition of one electron to reduce the high potential heme causes the rearrangement of loops I, II, and III in the semi-reduced protein (bottom, PDB 2VHD). (B) G. sulfurreducens in both the wild-type oxidized (grey) and S134P/V135K mutant (red/blue) enzyme. The mutations to residues S134 and V135 in the wild-type protein has significant effects on the structure surrounding the distal face of the L-heme. This changes the activity requirements of the enzyme seen in solution and PFV analyses. (Figures of the active site of wild-type and S134P/V135K were prepared using Protein Data Bank files 3HQ6 and 3HQ8 respectively (15).) (C) Structures of the wild-type and double mutant S134P/V135K enzymes from G. sulfurreducens. The wild-type oxidized protein (top, PDB 3HQ6) has loop I (red) in the closed position. The S134P and V135K mutations (bottom, PDB 3HQ8) cause only loops I (red) and II (blue) to rearrange. Loop III (green) retains the wild-type conformation and does not resemble the semi-reduced form of the P. aeruginosa enzyme.
As the Ne enzyme does not require activation to achieve full activity, recent work studying the Geobacter CcpA has focused on the structural features that govern the required activation step. In an attempt to mimic the properties of the constitutively active Ne enzyme, mutants were created based on the protein sequence of the Ne CcP. The most notable construct was the S134P/V135K (Gs numbering) double mutant, where both mutations are installed on the second loop region of Gs CcpA. This mutation allows for the enzyme to be constitutively active without the requirement for pre-reduction, and a comparison of the active site crystal structures of the wild-type and double mutant enzymes (Figure 1B) emphasizes the conformational change around the L-heme, as well as the changes in the global protein fold (Figure 1C) that allows for activity without pre-reduction of this enzyme (15).
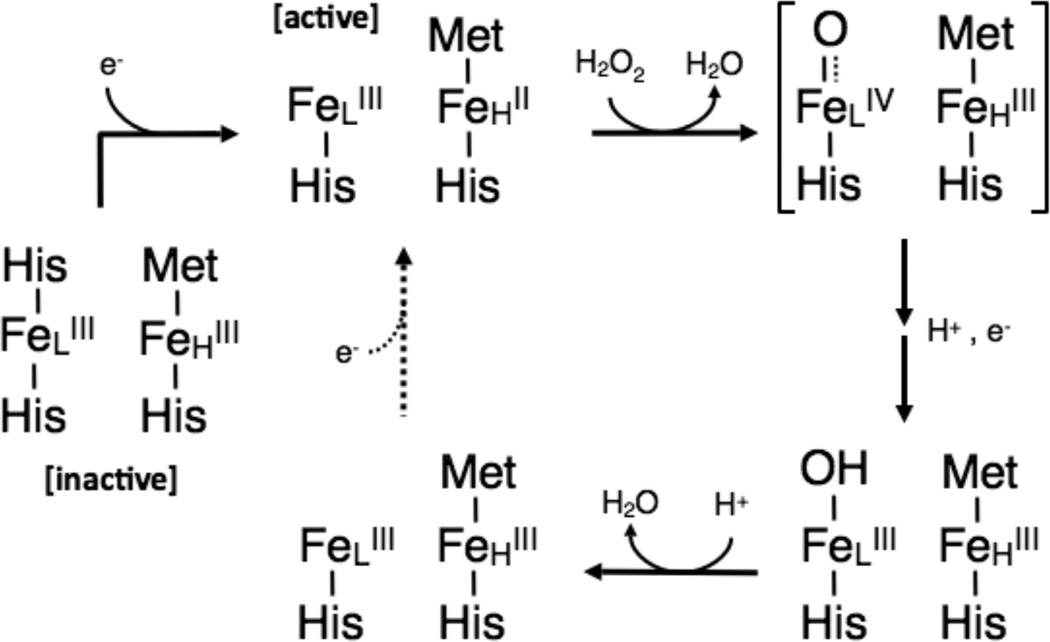
Mechanistically, generation of the reductively activated state of a diheme CcP not only engenders the loop movements that ultimately release the distal His ligand at the active site (7, 9, 14), but also yields the storage of a redox equivalent in the H-heme (Scheme 1). Preparation of the semi-reduced state allows for peroxide to bind, and heterolytic cleavage of the O—O bond causes the generation of a compound-I like intermediate, where the exact nature of the iron-oxygen bond is not known, but where one oxidizing equivalent is hypothesized to be stored at the Fe4+ oxidation state in the L-heme, while the second oxidizing equivalent is found on the H-heme. Subsequent reduction, and proton transfer steps are required, though their ordering is uncertain at present: Another electron is presumably transferred from a redox partner through the H-heme, resulting in the reduction of the L-heme to the ferric state. Reduction of the L-heme is coupled to the uptake of two protons ultimately, resulting in the second molecule of water in the overall reaction; release of the product, along with an additional electron (dotted arrow, Scheme 1) are required to return the enzyme to the active, mixed-valent state. Intermolecular and inter-cofactor electron transfers are important for the catalytic cycle of the canonical subclass of bacterial cytochrome c peroxidases, as their ability to form the active five-coordinate state of the L-heme depends on the ability of electrons to be transferred through the H-heme. While the subtleties of this mechanism are still under investigation, previously we have used protein film voltammetry (PFV) as a mechanistic tool that rapidly illuminates the differences between the peroxidases which do require reductive activation (the Pa enzyme) and those that do not (from Ne). PFV allows for the direct analysis of the redox activity of the enzyme active site during catalysis (24).
Here, we use protein film voltammetry (PFV) as a biophysical tool to assess the similarities and differences between Geobacter CcpA and other members of the bacterial cytochrome c peroxidase family, and to interrogate the S134P/V135K double mutant which appears to behave like the Ne enzyme in solution assays. As described below, we find that CcpA shows electrocatalytic voltammetry that is similar to that observed previously for canonical bacterial CcPs that require redox-poising to attain activity (12, 14), and the S134P/V135K double mutant is indeed distinct from the wild-type enzyme, displaying electrochemical characteristics that shed insight into the potentially rate limiting role of intercofactor (H-heme) electron transfer.
MATERIALS AND METHODS
Protein Expression and Purification
G. sulfurreducens cytochrome c peroxidase was produced in E. coli using the pETSN::CcpA plasmid in combination with the pEC86 plasmid encoding for the c-type heme maturation genes. For the complete expression, mutagenesis, and purification conditions please refer to reference (15).
Electrochemistry
Protein film voltammetry experiments were preformed on a PGSTAT 12 AutoLab (Ecochemie) potentiostat, equipped with FRA and EDC modules. A three-electrode configuration was used in a water-jacketed glass cell. A platinum wire was used as the counter electrode and a saturated calomel reference electrode was used. Potentials are reported here versus a standard hydrogen electrode (SHE). Calomel potentials were corrected by +242 mV. All experiments were done at 0°C unless otherwise noted. A cell solution of 10 mM CHES, HEPES, MES, and TAPS with 100 mM NaCl allowed for a broad range of pH values to be investigated throughout the experiments. When necessary the electrode was rotated with an EG&G electrode rotator. Protein films were generated on pyrolytic edge plane graphite electrodes (PGE). Protein films were grown on electrodes by directly depositing 1 µL of a 300 µM stock protein solution directly onto the electrode surface and incubated for five minutes. Excess protein was rinsed from the electrode surface with either water or the cell solution buffer of a specific pH.
Nonturnover electrochemical signals were generated with the electrochemical cell surrounded by a Faraday cage to eliminate electrochemical noise from the system. To remove oxygen from the buffer argon was gently bubbled through the cell solution. Catalytic electrochemical experiments were preformed in an MBraun Labmaster glovebox in an anaerobic environment. Data were collected with the GPES software package (Ecochemie). Nonturnover signals were analyzed by subtraction of the graphite baseline electrochemical response from the raw data using the SOAS package, courtesy of Dr. Christophe Léger. SOAS was also used for the analysis of the limiting currents and catalytic midpoint potentials of the catalytic voltammograms (25). A linear baseline was subtracted from the cathodic scan of the raw catalytic data to extract kinetic parameters and analyze the catalytic data.
RESULTS
Direct Electrochemistry
We have carried out a series of direct electrochemical experiments to investigate the diheme cytochrome c peroxidase from G. sulfurreducens using protein film voltammetry. Protein samples of either wild-type CcpA or the S134P/V135K mutant were applied directly to the graphite electrode. Attempts at using surfaces other than graphite (e.g. modified gold) were unsuccessful. Films formed on graphite electrodes are stable at a broad range of pH values, ranging from pH 3 to pH 10, and multiple scans can be taken before the protein film needs to be regenerated.
Catalytic PFV Experiments – reduction of H2O2
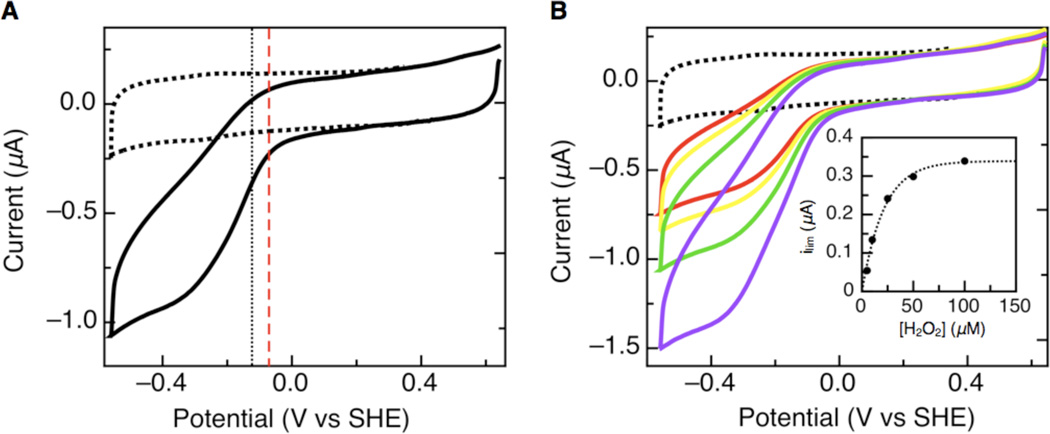
When substrate is added to the electrochemical cell the oxidative and reductive half-scans are transformed into reversible sigmoidal waves. This transformation occurs as the electrode provides undirected electrons to the protein and subsequently to the substrate. The midpoint potential of the transition of the sigmoidal wave represents Ecat, the electrocatalytic potential. A typical catalytic voltammogram for wild-type G. sulfurreducens CcpA at pH 7 is shown in Figure 2A. Based on previous solution experiments for the wild-type G. sulfurreducens CcpA, which requires a reductive activation step, we expected to see Ecat values related to that of the P. aeruginosa CcP, which is the parent member of the canonical family of enzymes requiring a reductive activation step. The electrocatalytic reduction of H2O2 by G. sulfurreducens CcpA, at the electrode surface, has an Ecat = −120 mV vs SHE at pH 7. This is indeed similar to the reduction potential of P. aeruginosa CcP which is −70 mV vs SHE, though shifted more negative by 50 mV (12). Catalytic waves are also used to calculate Michaelis-Menten parameters for the reaction of CcPs with H2O2 by taking the limiting current, ilim, as a measure of enzymatic velocity. At pH 7 and 0°C, with increasing concentrations of H2O2, wild-type CcpA shows a Km = 14 µM (Figure 2B, inset). As expected wild-type G. sulfurreducens CcpA behaves similarly on the graphite electrode surface, as in solution, to the P. aeruginosa enzyme and exhibits similar electrocatalytic properties.
Figure 2.
Electrocatalytic reduction of hydrogen peroxide at a PGE electrode with wild-type G. sulfurreducens CcpA adsorbed onto the graphite electrode surface. (A) Voltammogram collected for wild-type G. sulfurreducens CcpA with 25 µM peroxide in the cell solution. The dotted voltammogram is the baseline before the addition of peroxide. The vertical black dotted line represents the catalytic potentials for wild-type G. sulfurreducens CcpA and the dashed red line shows the P. aeruginosa CcP catalytic potential for comparison. (B) Voltammograms collected for increasing concentrations of peroxide in the cell solution (red = 5 µM, yellow = 10 µM, green = 25 µM, purple = 50 µM). The inset shows the calculation of Km for voltammograms collected at a specific pH. (Experimental conditions: pH 7, scan rate = 20 mV/s, rotation rate = 1000 rpm.)
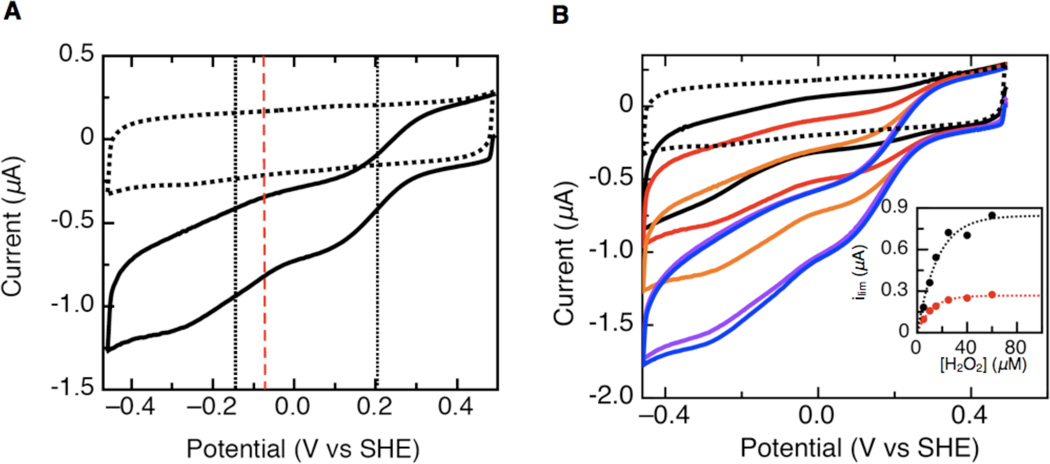
Assessing the catalytic voltammetry of the G. sulfurreducens CcpA S134P/V135K double mutant revealed dramatic changes in the waveshapes (Figure 3), compared to both the wild-type enzyme (Figure 2), and other cytochrome c peroxidases described thus far. Preparing and handling the enzymes in precisely the same manner, the S134P/V135K enzyme did not yield a simple sigmoidal wave like the wild-type protein, instead it resulted in catalytic waves that had not one component, but two (Figure 3A). Upon initial inspection there is a lower potential feature (Ecat,1) very similar to that seen for wild-type: here Ecat,1 is −145 mV at pH 7, versus −120 mV. More importantly, an additional higher potential feature (Ecat,2) is now seen. At pH 7 with 25 µM substrate, Ecat,2 is +200 mV vs SHE. This is the first time that a P. aeruginosa type enzyme has, upon mutation, successfully displayed a shift in Ecat to potentials more closely related to those of the N. europaea CcP than values collected for the P. aeruginosa enzyme. As Figure 3B shows, both components of the catalytic current increase and saturate their intensity with increasing amounts of hydrogen peroxide. For each substrate concentration, the individual contribution of the limiting current for Ecat,2 was determined at a potential of 50 mV, and compared to the overall response of the reductive wave at −400 mV. Thus, we can use these currents as enzymatic velocities to demonstrate Michaelis-Menten kinetics (Figure 3B, inset). Through this treatment, S134P/V135K reveals an apparent Km of 11 µM using the overall current response, the portion of the current that corresponds to Ecat,2 yields a Km = 8 µM, with both being in good agreement with the solution-based measurement of Km of 12 µM (15). The S134P/V135K Ecat,1 and Ecat,2 values are compared with that of P. aeruginosa CcP (red dash) in Figure 3A, while the Ecat value for the constitutively active N. europaea CcP is > 500 mV, and thus is off scale, with respect to the data shown.
Figure 3.
Electrocatalytic reduction of hydrogen peroxide at a PGE electrode with S134P/V135K G. sulfurreducens CcpA adsorbed onto the graphite electrode surface. (A) Voltammogram collected for S134P/V135K with 25 µM peroxide in the cell solution. The dotted voltammogram is the baseline before the addition of peroxide. The black dotted vertical lines represents the catalytic potentials for S134P/V135K while the red dashed line shows the P. aeruginosa CcP (−70 mV) catalytic potential. (B.) Voltammograms collected for increasing concentrations of peroxide in the cell solution. (black = 5 µM, red = 10 µM, orange = 20 µM, purple = 40 µM, blue = 50 µM) The inset shows the calculation of Km for voltammograms collected at a specific pH. (Experimental conditions: pH 7, scan rate = 20 mV/s, rotation rate = 1000 rpm.)
Catalytic PFV Experiments - pH Dependencies
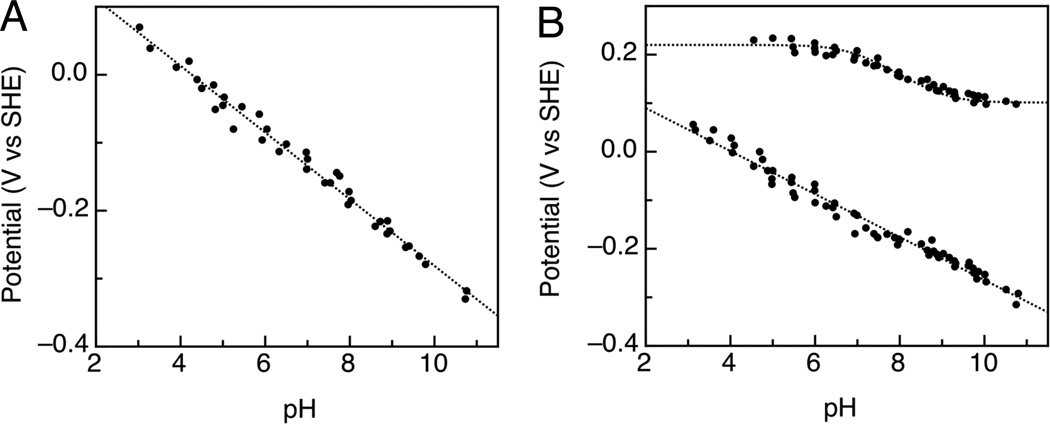
The pH dependence of the Ecat value for the electrocatalytic reduction of hydrogen peroxide by the enzyme can be used to further compare the wild type and S134P/V135K CcpA. Figure 4 shows the pH dependence curves of the Ecat values determined for voltammograms of G. sulfurreducens CcpA and S134P/V135K in the presence of 25 µM substrate (H2O2). The low potential features of both the wild-type and S134P/V135K enzyme display similar pH dependencies having slopes that fit well to the ideal 1e−:1H+ model (slope = −54 mV/pH decade at 0°C). Values calculated from the pH dependence curve for wild-type G. sulfurreducens CcpA (Figure 4A) are pKox < 3 and pKred > 11. S134P/V135K displays one pH dependence curve that is markedly similar to wild-type G. sulfurreducens CcpA and one that is noticeably different (Figure 4B). The double mutant’s Ecat,1 feature gives rise to a curve similar to that of wild-type G. sulfurreducens CcpA, with calculated values similar to those for the wild-type enzyme. The pH dependence curve of the higher potential feature leads to calculated values of pKox = 6.8, pKred = 9.1, Eacid = 220 mV, and Ealk = 100 mV. As shown above, Ecat,2 of the double mutant fits well to the same model as the low potential feature. However the pKox value of the double mutant is >3 pH units greater than that of the low potential feature. The pH dependence curve for the high potential feature also shows an overall higher potential value at each pH unit ranging from a 160 mV to almost a 500 mV difference from the lower potential feature depending on the value of pH. pH invariant regions of the high potential feature also occur at extreme pH values (less than pH 6.5 and greater than pH 9) whereas the low potential feature does not display any distinct pH invariant regions at valued of pH that we were able to probe.
Figure 4.
pH dependence of Ecat determined for voltammograms of wild-type G. sulfurreducens CcpA (A) and S134P/V135K (B) in the presence of 25 µM substrate. The data are fit to a 1e−:1H+ process, possessing the expected slope of −54 mV/pH decade at 0°C.
Catalytic PFV Experiments - H2O2 Concentration Dependencies
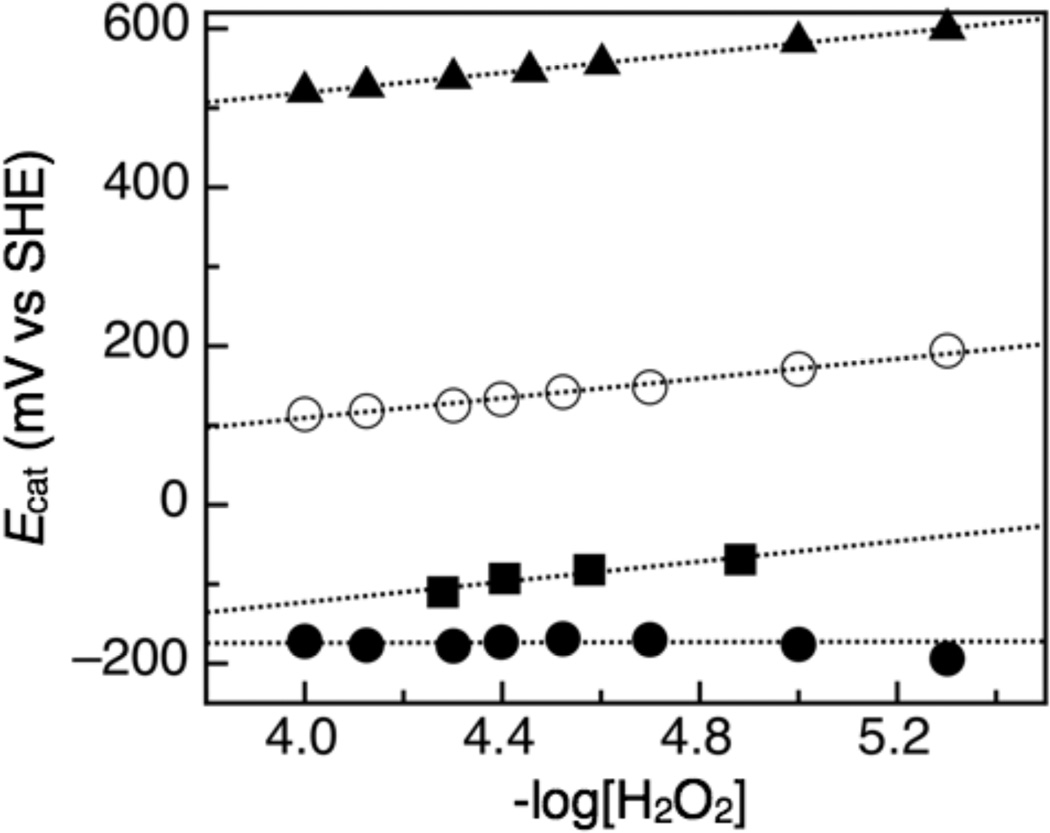
In order to assess the possible role of substrate binding to the catalytic midpoint potentials, the Ecat values were plotted with respect to the substrate concentration (Figure 5). For both wild-type (Ecat) and S134P/V125K (Ecat,1) the dependence upon substrate is essentially non-existent (1 mV per [H2O2] decade). However, the Ecat values of the high potential feature of S134P/V135K show a slope of 62 mV per [H2O2] decade at varying concentrations of peroxide. Comparison of the peroxide dependence for wild type G. sulfurreducens CcpA and S134P/V135K are shown alongside data for the canonical, Pseudomonas subclass of peroxidases (12), as well as the N. europaea enzyme (20), both of which display slopes of peroxide dependence ~ 60 mV/[H2O2] decade.
Figure 5.
Plot of Ecat vs −log[H2O2] for S134P/V135K. The black closed circles represent the low potential feature and the black open circles represent the high potential feature of the G. sulfurreducens S134P/V135K CcpA. The squares represent the P. aeruginosa catalytic feature and the triangles represent the N. europaea catalytic feature.
DISCUSSION
Here we report the electrochemical characterization of the wild-type enzyme and S134P/V135K double mutant of Geobacter sulfurreducens cytochrome c peroxidase, CcpA. The former of these enzymes has been described previously as a homologue of the canonical Pseudomonas-type enzyme, requiring reductive activation of the H-heme in order to engender activity at the active site, while the latter double mutant has been structurally characterized, revealing that the active site has already achieved the necessary open conformation, independent of pre-reduction (15). In the presence of substrate we can compare the two Geobacter enzymes directly, and place them in the context of other bacterial cytochrome c peroxidases. For the first time, mutations of a wild-type cytochrome c peroxidase have transformed a Pseudomonas-like, canonical CcP into an enzyme that has displays electrocatalytic responses suggestive of a Nitrosomonas-like enzyme.
Wild-type G. sulfurreducens CcpA Catalytic Voltammetry
In parallel to our previous work conducted with the Pseudomonas (12) and Nitrosomonas (20) CcP enzymes, we have used PFV at graphite electrodes to observe the redox chemistry of wild-type Gs CcpA. The addition of substrate results in electrocatalytic limiting currents that are related to the kinetics of the enzyme, and analysis of the data shows that catalytic electrochemistry is low in reduction potential, is fast and reversible in a cyclic voltammetry experiment (i.e., no hysteresis), possesses a shape of the electrocatalytic wave that indicates a 1-electron redox process, and yields limiting currents that when treated as enzymatic velocities predict values of Km for substrate and maximal activities as a function of pH, that agree with solution measurements. Thus, in a similar fashion to solution measurements that mark the Gs CcpA as similar to other canonical bacterial peroxidases (6, 12, 18–20), the electrochemical traits we observe here are highly similar to those found for the Pseudmonas enzyme.
With respect to the comparisons with the Pseudmonas enzyme, our many catalytic experiments presented here suggest that the Ecat value of wild-type G. sulfurreducens CcpA corresponds to an active site process, yet a process that has a very low reduction potential of −120 mV (pH 7). This low potential of catalysis makes the Gs CcpA similar to the canonical Pseduomonas sub-class of bacterial CcP enzymes (where Ecat is reported at −70 mV under identical conditions (12)). As with the Pseudmonas enzyme, we find that electrocatalysis of the wild-type Geobacter enzyme is reversible, clearly one-electron in nature, and displays pH dependent behavior indicating a stoichiometry of 1:1 protons to electrons. However, the primary difference between the Geobacter data, and that of the Pseudomonas enzyme is that the midpoint potentials associated with catalysis does not show a dependence upon hydrogen peroxide for the Geobacter enzyme, while the Pseudomonas enzyme does show a peroxide dependence. Thus, while the one-electron redox couple that largely governs electrocatalysis appears to be similar to the FeII/FeIII redox couple of the Pseudomonas enzyme, it cannot be identical.
The nature of the species that contributes to electrocatalysis in the case of the Geobacter enzyme are still uncertain at this time, and may be due to an outer-sphere redox process occurring at the active site, rate limiting electron transfer into active site directly from the electrode, or localized unfolding of the His/Met-ligated H-heme, as has been suggested to occur at some hemoproteins when adsorbed to graphite (36, 37). Indeed, the recent effort of Paes de Sousa and co-workers suggests that the low-potential electrocatalysis displayed by the Pa enzyme is likely due to unfolding of the H heme, and that the penta-coordinate active site is actually “silent” (36). However in the cases of the Pa and Gs enzymes, such a circumstance is unlikely: not only do our studies yield electrochemically determined values of Km in agreement with solution measurements, and display reversible electrocatalysis (whereas all voltammetry in Ref. 36 is marked by dramatic hysteresis), but here it is clear that active site itself cannot be “silent”, as the S134P/V135K mutation transmutes the electrochemical response. It is implausible that this mutation would give rise to such a dramatic change in the voltammetric wave-shape if the electrocatalytic signature were due to the H-heme.
S134P/V135K Catalytic Voltammetry
The S134P/V135K double mutation is the first example of a mutant bacterial CcP that bridges the differential reactivity of the P. aeruginosa-like peroxidases and N. europaea-like enzymes. Whereas the double mutant is constitutively active in solution (15), and the crystal structure of the enzyme revealed that the diferric form of the enzyme displays an open conformation of the active site (Figure 1B), the electrocatalysis displayed in Figure 3 is not akin to that found for the either sub-classes of CcP, showing two distinct phases of the reduction of substrate. For S134P/V135K lower potential electrocatalytic feature (Ecat,1) remains close to values of potential observed for the wild-type enzyme, but the additional, higher-potential feature (Ecat,2) is found to have Ecat = +200 mV vs SHE at pH 7. In a similar fashion to the wild-type enzyme, the kinetics of the S134P/V135K active site can be calculated using electrocatalytic limiting current data. The values of Km calculated for both the low potential and high potential electrocatalytic features, using ilim as a function of substrate (H2O2) reveals values of substrate binding similar to other bacterial cytochrome c peroxidases (6, 12, 15, 18–20), as well as to solution based values for S134P/V135K (15). Thus we again conclude that the enzyme, when adsorbed, displays native-like properties. Given its similarity to the wild-type electrochemical signals, the nature of the lower-potential catalytic component centered at Ecat,1 is most likely to be identical to the wild-type enzyme. However, it is uncertain currently if the electrochemistry associated with Ecat,1 is due to a sub-population of the adsorbed enzyme that behaves like wild-type protein, or if, when adsorbed the mutant behaves like wild-type when lower potentials are applied. While the nature of Ecat,1 is unclear, the second, higher-potential feature (centered at Ecat,2 of +200 mV) is more intriguing due to its permutation from wild-type behavior. Our initial hypothesis was that the PFV analyses of the mutant would likely reveal electrocatalytic behavior similar to the Nitrosomonas enzyme (a high-potential (>500 mV) one-electron wave) and while Ecat,2 is perturbed from the wild-type behavior, is it still ~250 mV lower in potential than the electrocatalytic features of the Nitrosomonas enzyme. Thus, two main questions result from our unanticipated data of the double-mutant: What does the high potential feature centered at Ecat,2 represent?, and Why has the S134P/V135K enzyme not fully achieved Nitrosomonas enzyme-like characteristics?
Electrochemical basis of Ecat,2
Regarding the nature of the catalytic process governing Ecat,2, several possibilities can be excluded. Previously, we have shown electrocatalytic reduction of hydrogen peroxide with the Pseudomonas enzyme, centered at low potentials (−70 mV); where we interpreted the substrate-dependence and the relatively low potential as representing a rate-limiting process of a FeII/FeIII redox couple at the active site L-heme, such as the release of water from L (12). Here, based on the potential of Ecat,2, it seems very unlikely a similar phenomenon is at work, though the potential is located in the range for an FeII/FeIII redox couple. Similarly, the value of potential excludes the involvement of a ferryl (or higher) heme oxidation state: previously with the N. europaea enzyme the high potential value for catalysis (>550 mV) was interpreted to correspond to a proton-coupled species generated after the formation of the FeIV=O:R+· compound I-like intermediate (20). Thus, the remaining possibilities to describe Ecat,2 include the rate-limiting electron transfer into the enzyme from the electrode, or a relay mechanism of rate-limiting electron transfer between the two heme cofactors.
Interfacial electron transfer would be mediated via the His/Met ligated, H-heme, which is known to have a potential spanning a range of +200 to +350 mV in the bacterial CcP enzymes (6, 10, 13). In such a model, electrocatalysis would be governed by the injection of electrons into the enzyme via the H-heme from the electrode, and therefore the response should bear the H-heme pH dependence and presumably be independent of the concentration of substrate. However, the pH dependence of the FeII/FeIII couple of the H-heme is not known currently for the wild-type CcpA. It is quite clear from our data, however that Ecat,2 observed in the Geobacter double mutant variant is dependent on both pH and substrate concentration (Figures 4 and 5). Further assessment of the possible role of limiting interfacial electron transfer requires detection of an electrochemical signal for H in the absence of substrate, such that both the pH-dependence and the interfacial electron transfer rate, k0, can be measured; while such signals have yet to be observed, comparison of k0 to kcat (determined both in solution and for the adsorbed enzyme) will make it clear if interfacial electron transfer is in fact slower than the enzymatic reaction.
Alternately, internal redox relay steps can govern electrocatalysis, even if electron transfer is fast between the enzyme and electrode (26–29). Such behavior has been observed for several enzymes, such as fumarate reductase (27, 30–33), sulfite oxidase (29, 34, 35), and flavocytochrome b2 (29). Efforts by Léger and co-workers allows for the quantitative modeling of such redox relays where a two-electron reaction occurs at an active site, which is mediated by electron transfer via a one-electron redox cofactor (29). In such an electrokinetic model, the precise appearance of the catalytic waves depend upon the relay rates of forward and backward electron transfer steps between the relay and the active-site, as well as redox potentials of the key states involved. Here, application of such a model cannot be achieved: an estimate of + 250 mV can be used for the H-heme relay redox potential, but potentials of key intermediate species associated with catalysis (e.g., the reduction of Compound I and Compound II) are not known. Yet, it is clear that Ecat,2 is quite close to estimates of the H-heme reduction potential, and thus suggests the model described by Léger, where electron relay is occurring at approximately the same rate as catalysis, and the electrocatalytic response takes on the appearance of a one-electron feature at a potential close to that of the relay.
Structural bases for incomplete conversion to an Ne-type enzyme
While it is clear from our electrochemical analysis that the S134P/V135K double mutant has traits that are distinct from the wild-type enzyme and other bacterial CcPs electrochemically characterized to date, the design of the S134P/V135K double mutation was to transform a Pseudomonas-like CcP enzyme to be akin to a Nitrosomonas-type enzyme that does not require reductive activation. Here we have demonstrated that by electrochemical measures, the conversion of the wild-type enzyme has been significant, though not complete. The double mutation has not achieved full conversion of the enzyme to a Nitrosomonas-like enzyme, it is clear that the reorganization of the distal-face of the L-heme attained in the S134P/V135K crystal structure (Figure 1B) is not enough to convert the electrochemical behavior from one type of peroxidase to the other. This difference can be rationalized on the basis of the structural evidence in comparison of the oxidized and semi-reduced forms of the Pseudomonas enzyme, shown in Figure 1A: Upon the reductive activation of CcPs there are three general loop movements that can be seen after reduction of the protein by one electron, and while Figure 1B demonstrates that the distal face of the double mutant has adopted an open conformation, as required for peroxide binding, this conformational change is incomplete and is achieved with motion of only two of the three loops that are required to rearrange as a part of reductive activation. Indeed, one specific region of the S134P/V135K enzyme that does not rearrange with respect to the wild-type enzyme is the area of the third loop region (labeled III/green in Figure 1C) located around the H-heme. Thus, our data suggest that fast electron transfer to and/or from the H-heme may require the conformational changes of Loop 3 and that the “incomplete” conversion of the Gs enzyme may be due to insufficiently reorganized sub-structures of the S134P/V135K mutant. The differences in function of variants of the Geobacter enzyme will be assessed with additional point mutations to the third loop region of the protein, to complete the three loop rearrangement of the enzyme.
CONCLUSIONS
From the electrochemical studies performed on wild-type G. sulfurreducens CcpA and S134P/V135K we now have a better understanding of the catalytic mechanism of this peroxidase as compared with the canonical P. aeruginosa and N. europaea homologues. Distinct from the wild-type that displays a low potential catalytic wave, the S134P/V135K double mutant is the first bacterial CcP to display a shifted electrocatalytic potential (Ecat,2) to more closely resembles the N. europaea sub-class of enzymes. These findings are interpreted in terms of a model where the S135P/V135K mutant mechanism is limited by intra-protein electron transfer. Future studies that combine structural and electrochemical analyses will enable allow for this model to be developed quantitatively, and further our understanding of how loop rearrangements in proximity to both the L-heme and the H-heme can modulate the redox mechanism displayed by bacterial cytochrome c peroxidases.
Scheme 1.
The proposed scheme for activation of resting state P. aeruginosa-type CcP and the catalytic cycle for the reduction of hydrogen peroxide to two molecules of water. The G. sulfurreducens CcpA shows solution-based kinetic parameters that mimic the P. aeruginosa type of CcP. The second electron coupled step responsible for the continuation of the catalytic cycle is shown as a dotted arrow.
Abbreviations
- PFV
protein film voltammetry
- CcP
cytochrome c peroxidase
- SHE
standard hydrogen electrode
- FRA
frequency response analyzer
- ECD
electrochemical detections module
- L-heme
low reduction potential peroxidatic heme
- H-heme
high reduction potential electron transfer heme
- CHES
2-(cyclohexylamino-ethanesulfonic acid
- HEPES
2-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- MES
2-(N-morpholino)ethanesulfonic acid
- TAPS
N-[tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid
- NaCl
sodium chloride
- PGE
pyrolytic edge
- Em
midpoint potential
- Ecat
catalytic midpoint potential
- ilim
limiting current
Footnotes
This work was supported by a National Institutes of Health grant (R01-GM072663) and by Deutsche Forschungsgemeinschaft (Ei-520/1 an IRTG 1478).
REFERENCES CITED
- 1.Caccavo F, Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 1994;60:3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leang C, Adams LA, Chin KJ, Nevin KP, Methe BA, Webster J, Sharma ML, Lovley DR. Adaptation to disruption of the electron transfer pathway for Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 2005;187:5918–5926. doi: 10.1128/JB.187.17.5918-5926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 4.Lin WC, Coppi MV, Lovley DR. Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ. Microbiol. 2004;70:2525–2528. doi: 10.1128/AEM.70.4.2525-2528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunford HB, Stillman JS. On the function and mechanism of action of peroxidases. Coord. Chem. Rev. 1976;19:181–251. [Google Scholar]
- 6.De Smet L, Savvides SN, Van Horen E, Pettigrew G, Van Beeumen JJ. Structural and mutagenesis studies on the cytochrome c peroxidase from Rhodobacter capsulatus provide new insights into structure-function relationships of bacterial di-heme peroxidases. J. Biol. Chem. 2006;281:4371–4379. doi: 10.1074/jbc.M509582200. [DOI] [PubMed] [Google Scholar]
- 7.Dias JM, Alves T, Bonifcaia C, Pereira AS, Trincao, Bourgeois D, Moura I, Romao MJ. Structural Basis for the Mechanism of Ca2+ Activation of the Di-Heme Cytochrome c Peroxidase from Pseudomonas nautica 617. Structure. 2004;12:961–963. doi: 10.1016/j.str.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Echalier A, Goodhew CF, Pettigrew GW, Fulop V. Activation and catalysis of the di-heme cytochrome c peroxidase from Paracoccus pantotrophus. Structure. 2006;14:107–117. doi: 10.1016/j.str.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Fulop V, Ridout CJ, Greenwood C, Hajdu J. Crystal structure of the di-haem cytochrome c peroxidase from Pseudomonas aeruginosa. Structure. 1995;3:1225–1233. doi: 10.1016/s0969-2126(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu H, Schuller DJ, Lanzilotta WN, Sundaramoorthy M, Arciero DM, Hooper AB, Poulos TL. Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases. Biochemistry. 2001;40:13483–13490. doi: 10.1021/bi011481h. [DOI] [PubMed] [Google Scholar]
- 11.Arciero DM, Hooper AB. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J. Biol. Chem. 1994;269:11878–11886. [PubMed] [Google Scholar]
- 12.Becker CF, Watmough NJ, Elliott SJ. Electrochemical evidence for multiple peroxidatic heme states of the diheme cytochrome c peroxidase of Pseudomonas aeruginosa. Biochemistry. 2009;48:87–95. doi: 10.1021/bi801699m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellfolk N, Ronnberg M, Aasa R, Andreasson LE, Vanngard T. Properties and function of the two hemes in Pseudomonas cytochrome c peroxidase. Biochim. Biophys. Acta. 1983;743:23–30. doi: 10.1016/0167-4838(83)90413-2. [DOI] [PubMed] [Google Scholar]
- 14.Echalier A, Brittain T, Wright J, Boycheva S, Mortuza GB, Fulop V, Watmough NJ. Redox-linked structural changes associated with the formation of a catalytically competent form of the diheme cytochrome c peroxidase from Pseudomonas aeruginosa. Biochemistry. 2008;47:1947–1956. doi: 10.1021/bi702064f. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann M, Seidel J, Einsle O. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J. Mol. Biol. 2009;393:951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Ronnberg M, Ellfolk N. Pseudomonas cytochrome c peroxidase. Initial delay of the peroxidatic reaction. Electron transfer properties. Biochim. Biophys. Acta. 1978;504:60–66. doi: 10.1016/0005-2728(78)90006-3. [DOI] [PubMed] [Google Scholar]
- 17.Ronnberg M, Ellfolk N. Heme-linked properties of Pseudomonas cytochrome c peroxidase. Evidence for non-equivalence of the hemes. Biochim. Biophys. Acta. 1979;581:325–333. doi: 10.1016/0005-2795(79)90252-6. [DOI] [PubMed] [Google Scholar]
- 18.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura JJ, Moura I. The kinetics of the oxidation of cytochrome c by Paracoccus cytochrome c peroxidase. Biochem. J. 1994;300:907–914. doi: 10.1042/bj3000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timoteo CG, Tavares P, Goodhew CF, Duarte LC, Jumel K, Girio FM, Harding S, Pettigrew GW, Moura I. Ca2+ and the bacterial peroxidases: the cytochrome c peroxidase from Pseudomonas stutzeri. J. Biol. Inorg. Chem. 2003;8:29–37. doi: 10.1007/s00775-002-0382-y. [DOI] [PubMed] [Google Scholar]
- 20.Bradley AL, Chobot SE, Arciero DM, Hooper AB, Elliott SJ. A distinctive electrocatalytic response from the cytochrome c peroxidase of Nitrosomonas europaea. J. Biol. Chem. 2004;279:13297–13300. doi: 10.1074/jbc.C400026200. [DOI] [PubMed] [Google Scholar]
- 21.Zahn JA, Arciero DM, Hooper AB, Coats JR, DiSpirito AA. Cytochrome c peroxidase from Methylococcus capsulatus Bath. Arch. Microbiol. 1997;168:362–372. doi: 10.1007/s002030050510. [DOI] [PubMed] [Google Scholar]
- 22.Elliott SJ, Bradley AL, Arciero DM, Hooper AB. Protonation and inhibition of Nitrosomonas europaea cytochrome c peroxidase observed with protein film voltammetry. J. Inorg. Biochem. 2007;101:173–179. doi: 10.1016/j.jinorgbio.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Mazumdar A, Chatterjee R, Adak S, Ghosh A, Mondal C, Banerjee RK. Characterization of sheep lacrimal-gland peroxidase and its major physiological electron donor. Biochem. J. 1996;314:413–419. doi: 10.1042/bj3140413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armstrong FA, Heering HA, Hirst J. Reactions of complex metalloproteins studies by protein-film voltammetry. Chemical Society Reviews. 1997;26:169–180. [Google Scholar]
- 25.Fourmond V, Hoke K, Heering HA, Baffert C, Leroux P, Leger C. SOAS: a free software to analyse electrochemical data and other one-dimensional signals. Bioelectrochemistry. 2009;76:141–147. doi: 10.1016/j.bioelechem.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong FA. Recent developments in dynamic electrochemical studies of adsorbed enzymes and their active sites. Curr. Opin. Chem. Biol. 2005;9:110–117. doi: 10.1016/j.cbpa.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Heering HA, Weiner JH, Armstrong FA. Direct Detection and Measurement of Electron Relays in a Multicentered Enzyme: Voltammetry of Electrode-Surface Films of E. coli Fumarate Reductase, an Iron-Sulfue Flavoprotein. J. Am. Chem. Soc. 1997;119:11628–11638. [Google Scholar]
- 28.Leger C, Elliott SJ, Hoke KR, Jeuken LJ, Jones AK, Armstrong FA. Enzyme electrokinetics: using protein film voltammetry to investigate redox enzymes and their mechanisms. Biochemistry. 2003;42:8653–8662. doi: 10.1021/bi034789c. [DOI] [PubMed] [Google Scholar]
- 29.Leger C, Lederer F, Guigliarelli B, Bertrand P. Electron flow in multicenter enzymes: theory, applications, and consequences on the natural design of redox chains. J. Am. Chem. Soc. 2006;128:180–187. doi: 10.1021/ja055275z. [DOI] [PubMed] [Google Scholar]
- 30.Weiner JH, Dickie P. Fumarate reductase of Escherichia coli Elucidation of the covalent-flavin component. J. Biol. Chem. 1979;254:8590–8593. [PubMed] [Google Scholar]
- 31.Blaut M, Whittaker K, Valdovinos A, Ackrell BA, Gunsalus RP, Cecchini G. Fumarate reductase mutants of Escherichia coli that lack covalently bound flavin. J. Biol. Chem. 1989;264:13599–13604. [PubMed] [Google Scholar]
- 32.Manodori A, Cecchini G, Schroder I, Gunsalus RP, Werth MT, Johnson MK. [3Fe-4S] to [4Fe-4S] cluster conversion in Escherichia coli fumarate reductase by site-directed mutagenesis. Biochemistry. 1992;31:2703–2712. doi: 10.1021/bi00125a010. [DOI] [PubMed] [Google Scholar]
- 33.Van Hellemond JJ, Tielens AG. Expression and functional properties of fumarate reductase. Biochem. J. 1994;304:321–331. doi: 10.1042/bj3040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, Enemark JH, Rees DC. Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell. 1997;91:973–983. doi: 10.1016/s0092-8674(00)80488-2. [DOI] [PubMed] [Google Scholar]
- 35.Elliott SJ, McElhaney AE, Feng C, Enemark JH, Armstrong FA. A voltammetric study of interdomain electron transfer within sulfite oxidase. J. Am. Chem. Soc. 2002;124:11612–11613. doi: 10.1021/ja027776f. [DOI] [PubMed] [Google Scholar]
- 36.Paes de Sousa PM, Pauleta SR, Simoes Gonçalves ML, Pettigrew GW, Moura I, Moura JJG, Correia dos Santos MM. Artefacts induced on haem proteins by electrode surfaces. J. Biol. Inorg. Chem. 2011;16:209–215. doi: 10.1007/s00775-010-0717-z. [DOI] [PubMed] [Google Scholar]
- 37.Ye T, Kaur R, Senguen FT, Michel LV, Bren KL, Elliott SJ. Methionine ligand lability of type I cytochromes c: detection of ligand loss using protein film voltammetry. J. Am. Chem. Soc. 2008;130:6682–6683. doi: 10.1021/ja801071n. [DOI] [PMC free article] [PubMed] [Google Scholar]