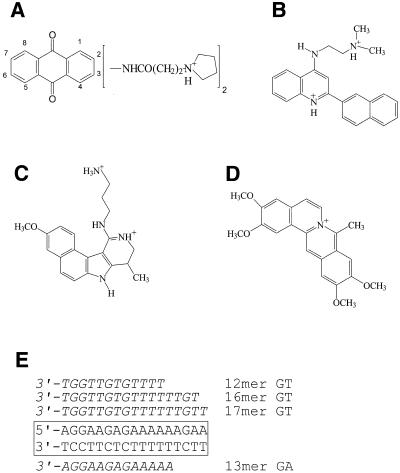
Abstract
We have used DNase I footprinting to examine the interaction of several triplex-binding ligands with antiparallel TG- and AG-containing triplexes. We find that although a 17mer TG-containing oligonucleotide on its own fails to produce a footprint at concentrations as high as 30 µM, this interaction can be stabilised by several ligands. Within a series of disubstituted amidoanthraquinones we find that the 2,7- regioisomer affords the best stabilisation of this TG triplex, though the 1,8- isomer also stabilises this interaction to some extent. By contrast the 1,5- and 2,6- regioisomers show no interaction with TG triplexes. Similar studies with a 13mer AG-containing oligonucleotide show the opposite pattern of stabilisation: the 2,6- and 1,5- isomers stabilise this triplex, but the 2,7- and 1,8-compounds do not. The polycyclic compound BePI strongly stabilises TG- but not AG-containing triplexes, while a substituted naphthylquinoline interacts with both antiparallel triplex motifs.
INTRODUCTION
Intermolecular DNA triplexes are formed by the binding of oligonucleotides in the major groove of duplex DNA (1–3). This interaction is sequence-specific and involves the formation of hydrogen bonds between the third strand bases and substituents on the purine bases of the duplex target. Two triplex motifs have been described in which the third strand runs either parallel or antiparallel to the purine strand of the duplex. Parallel triplexes are characterised by the formation of T·AT and C+·GC triplets, though other triplets including G·GC have also been described. Antiparallel triplexes consist of G·GC, A·AT and T·AT triplets.
Although these triplexes form with high specificity they are less stable than their duplex counterparts, largely as a result of charge repulsion between the phosphodiester backbones. Several strategies have been employed to increase the strength of association (3). One strategy is to develop ligands which bind preferentially to triplex compared to duplex DNA (4). A number of such ligands have been characterised including benzopyridoindole derivatives (such as BePI, Fig. 1C) (4–9), coralyne (Fig. 1D) (10–12), substituted naphthylquinolines (Fig. 1B) (13–16) and a series of bis-substituted amidoanthraquinones (Fig. 1A) (17–19). Molecular modelling studies with the disubstituted anthraquinones suggest that they can traverse the DNA triplex, positioning positively charged side groups in different DNA grooves (19). Studies with parallel (CT-containing) triplexes have shown that the stabilising activity of these compounds depends on the position of the substituents and that they stabilise these triplexes in the order 2,7 > 1,8 = 1,5 > 2,6 (19).
Figure 1.
Chemical structures of the triplex-binding ligands and the interaction of the triplex-forming oligonucleotides with their target site. (A) Bis-amidoanthraquinones, (B) naphthylquinoline, (C) BePI, (D) coralyne. (E) The sequence of the 17mer oligopurine tract in tyrT(34-59) (boxed) and sequences of the TG- and AG-containing triplex-forming oligonucleotides.
There have been fewer studies on the interaction of stabilising ligands with antiparallel triplexes. The naphthylquinoline ligand has been shown to stabilise antiparallel triplexes formed with the oligonucleotides G5T5, T5G5, and A5G5, (16), while low concentrations of BePI (0.5 µM) have been shown to stabilise GT-containing triplexes which have been designed to bind in the antiparallel but not the parallel orientation (5).
One advantage of antiparallel triplexes is that bases do not need be protonated, in contrast to the parallel arrangement in which protonation of cytosine (at pH <6.0) is essential for generating the C+·GC triplet. However, within antiparallel triplexes G·GC and A·AT are structurally very different from T·AT (20), though the limited structural data suggest that A·AT and G·GC are more similar to each other (21,22). In contrast T·AT and C+·GC triplets are nearly isomorphous, though the crystal structure of a DNA triplex reveals that there are minor differences in the backbone positions of different triplets (23). As a result, backbone distortions must occur in the third strand at steps between different triplets. This may account for the observations that antiparallel triplexes are often less stable than their parallel counterparts (24,25), though some sequences generate very stable complexes (26,27). An additional destabilising factor is that they are often dominated by the G·GC triplet (28), and G-rich oligonucleotides can fold into competing higher-order structures (29,30). Furthermore AG-containing oligonucleotides may form other self-associated structures (31,32). There is therefore a need to find methods for improving the stability of these antiparallel complexes.
In this paper we have examined the ability of a series of disubstituted amidoanthraquinones to stabilise the triplexes formed with TG- and AG-containing oligonucleotides. Since antiparallel triplexes are less stable than parallel ones, we have used longer oligonucleotides than in our previous work (19). These results with the anthraquinones are compared with BePI, coralyne and naphthylquinoline.
MATERIALS AND METHODS
Chemicals and enzymes
Oligonucleotides were purchased from Oswel DNA Service (Southampton, UK). These were stored in water at –20°C and diluted to working conditions immediately before use. The various disubstituted amidoanthraquinones were synthesised as their water-soluble addition salts using published procedures (33,34). All the derivatives contained a pyrrolidine end group to ensure that the amine substituents are present in the N-protonated form. The naphthylquinoline triplex-binding ligand was a gift from Dr L.Strekowski (Department of Chemistry, Georgia State University, Atlanta, GA). This was stored at a stock concentration of 20 mM in dimethylsulphoxide (DMSO). Coralyne and BePI were purchased from Sigma and stored at –20°C at a concentration of 10 mM in DMSO.
DNA fragments
TyrT(43-59) is a modification of the original tyrT DNA sequence which contains a 17 base oligopurine tract between positions 43 and 59 (25). The sequence of this region is shown in Figure 1E. The radiolabelled DNA fragment was prepared by digesting the plasmid with EcoRI and AvaI and was labelled at the 3′-end of the EcoRI site using reverse transcriptase and [α-32P]dATP. The labelled 110 bp DNA fragment was separated from the remainder of the plasmid DNA on an 8% (w/v) non-denaturing polyacrylamide gel. The isolated DNA was dissolved in 10 mM Tris–HCl pH 7.5 containing 0.1 mM EDTA to give ∼10–20 c.p.s./µl as determined on a hand-held Geiger counter (<10 nM). For quantitative footprinting experiments the absolute DNA concentration is not important as long as it is lower than the dissociation constant of the DNA-binding compound.
DNase I footprinting
Radiolabelled DNA (1.5 µl) was mixed with 1.5 l oligonucleotide and 1.5 µl triplex-binding ligand. The ligand and oligonucleotide were both dissolved in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MgCl2 (for TG triplexes) or 10 mM MnCl2 for AG triplexes. The concentrations refer to conditions in the final reaction mixture. These mixtures were heated to 65°C for 3 min to destabilise any competing oligonucleotide structures and then equilibrated at 20°C for at least 2 h. The samples were digested by adding 2 µl DNase I (typically 0.01 U ml–1) dissolved in 20 mM NaCl containing 2 mM MgCl2 and 2 mM MnCl2. The reaction was stopped after 1 min by adding 5 µl 80% formamide containing 10 mM EDTA, 10 mM NaOH and 0.1% (w/v) bromophenol blue.
Gel electrophoresis
The products of digestion were separated on 9% polyacrylamide gels containing 8 M urea. Samples were heated to 100°C for 3 min, before rapidly cooling on ice and loading onto the gel. Polyacrylamide gels (40 cm long, 0.3 mm thick) were run at 1500 V for ∼2 h and then fixed in 10% (v/v) acetic acid. These were transferred to Whatman 3MM paper and dried under vacuum at 80°C. The dried gels were either exposed to autoradiography at –70°C using an intensifying screen, or were subjected to phosphorimaging using a Molecular Dynamics STORM PhosphorImager.
Quantitative analysis
The intensity of bands within each footprint was estimated using ImageQuant software (v.4.2A). These were normalised by comparison with a region for which DNase I cleavage was not affected. Footprinting plots (35) were constructed from these data and C50 values, indicating the oligonucleotide concentration which reduces the band intensity by 50%, were calculated by fitting a simple binding curve to the data (19).
The relative affinities of ligands for DNA triplexes can be calculated from these C50 values using the equation 1/ C50 = L/(KT × KL) + 1/KT, where KT is the dissociation constant of the triplex in the absence of added ligand and KL is the triplex–ligand dissociation constant (19). However, this detailed analysis makes several assumptions about the interaction, particularly that each triplex is stabilised by interaction with a single ligand molecule, an assumption which is likely to be correct for the triplex-forming oligonucleotides used in this study. This analysis also requires that we know the dissociation constant for the oligonucleotide–duplex interaction in the absence of added ligand (KT), which cannot be obtained in the present studies since the oligonucleotides do not produce footprints without addition of the stabilising ligands. Because of the limitations in this analysis we have simply used the C50 values to provide an estimate of the relative binding affinities of different ligands.
RESULTS
TG-containing oligonucleotides
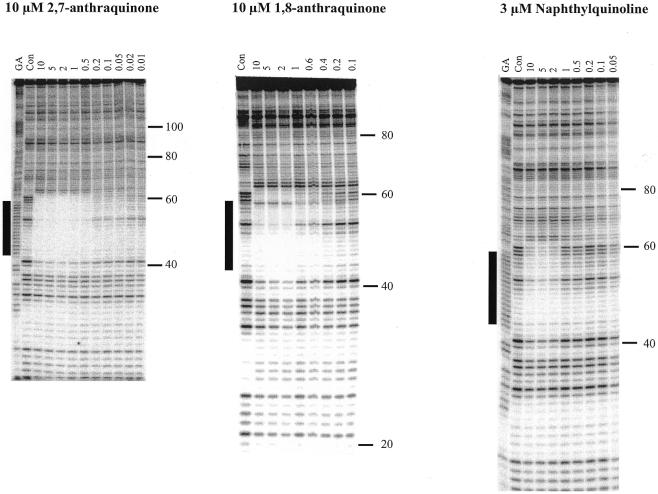
Bis-substituted anthraquinones. Three TG-containing oligonucleotides of 17, 16 and 12 bases were designed to bind to different regions of the 17mer duplex oligopurine tract in an antiparallel orientation, as shown in Figure 1E. Figure 2 shows DNase I footprinting experiments with 10 µM of these oligonucleotides in the presence or absence of 10 µM of several triplex-binding ligands. These experiments were carried out at pH 7.0 in 10 mM Tris–HCl, containing 50 mM NaCl and 10 mM MgCl2. The left portion of Figure 2 shows that 10 µM of the oligonucleotides alone does not affect the cleavage patterns, suggesting that these antiparallel triplexes are not stable, as is frequently observed for triplexes which contain mainly T·AT triplets (24,25). The remainder of Figure 2 shows the effects of 10 µM of the triplex-binding ligands on the triplexes formed with these three oligonucleotides. In the presence of 10 µM naphthylquinoline triplex-binding ligand (Fig.1B) all three oligonucleotides generate footprints, which cover the expected target sites and continue for a few bases 3′ (below) the binding sites. In particular it can be seen that the 16 and 17mers protect the bands at positions 40 and 41, while the 12mer, which binds higher up the target site, does not. There are very marked differences between the effects of the various disubstituted anthraquinones. The 2,6- and 1,5-disubstituted anthraquinones do not induce a footprint with any of the oligonucleotides, whereas clear footprints are formed in the presence of 10 µM of the 2,7- and 1,8-disubstituted anthraquinones. The 2,7-disubstituted compound induces footprints with all three oligonucleotides, while the 1,8- analogue gives clear footprints with the 17 and 16mers but fails to generate a footprint with the 12mer oligonucleotide. This suggests that this ligand binds less well to these triplexes than the 2,7-anthraquinone. The effect of each of these compounds is examined in more detail in the experiments described below.
Figure 2.
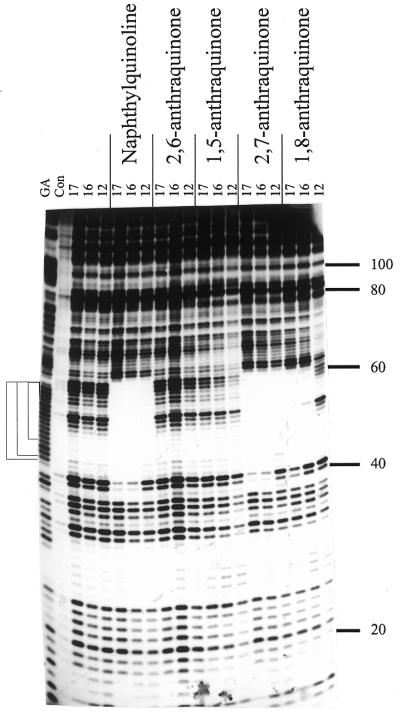
DNase I footprints for the interaction of the various TG-oligonucleotides (10 µM) in the presence of 10 µM of the triplex-binding ligands. The length of the oligonucleotide is shown at the top of each gel lane. The track labelled ‘Con’ shows DNase I digestion of the duplex DNA in the absence of oligonucleotide or ligand. Note that this lane is underdigested in this figure, but the cleavage pattern is identical to the adjacent lanes containing oligonucleotides with no added ligand. ‘GA’ represents a Maxam–Gilbert marker specific for purines. The experiments were performed in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MgCl2. The numbers on the right-hand side indicate the sequence position and are numbered as in Brown et al. (25). The square brackets show the position of the expected target sites.
The first panel of Figure 3 shows the effect of 10 µM of the 2,7-disubstituted anthraquinone on the footprints produced by different concentrations of the 17mer TG oligonucleotide. In this quantitative experiment a clear footprint can be seen which persists to an oligonucleotide concentration of ∼0.2 µM. This footprint extends beyond the triplex-binding site at both the 5′ (upper) and 3′ (lower) ends by 2 or 3 bases, protecting the bands at positions 60–62 and 40–41. Quantitative analysis of the concentration dependence of this footprint yielded a C50 value of 0.14 ± 0.04 µM (Table 1). This value is the oligonucleotide concentration which reduces the intensity of bands in the footprint by 50%. In the presence of lower concentrations of the ligand (data not shown) higher oligonucleotide concentrations are required to generate a footprint; the C50 value in the presence of 3 µM ligand is 0.36 ± 0.07 µM (Table 1). As expected, shorter oligonucleotides are stabilised to a lesser extent by the ligand and quantitative experiments with the 12mer TG oligonucleotide yielded a C50 value of 0.86 ± 0.20 µM in the presence of 10 µM of the 2,7-disubstituted compound.
Figure 3.
DNase I digestion patterns showing the interaction of the 17mer TG-containing oligonucleotide with its target site in tyrT(43-59) in the presence of triplex-binding ligands. Oligonucleotide concentrations (µM) are shown at the top of each gel lane. The track labelled ‘con’ shows DNase I digestion of the duplex DNA in the absence of oligonucleotide or ligand. ‘GA’ represents a Maxam–Gilbert marker specific for purines. The experiments were performed in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MgCl2. The numbers on the right-hand side of each panel indicate the sequence position and are numbered as in Brown et al. (25). The filled boxes show the position of the target site.
Table 1. C50 values (µM) for the interaction of oligonucleotides with their triplex target sites in the presence of various triplex-binding ligands.
| Ligand |
Concentration (µM) |
C50 (mM) |
| 17mer TG |
|
|
| 2,7 amidoanthraquinone |
10 |
0.14 ± 0.04 |
| |
3 |
0.36 ± 0.07 |
| 1,8 amidoanthraquinone |
10 |
0.61 ± 0.23 |
| Naphthylquinoline |
10 |
0.18 ± 0.09 |
| |
3 |
1.4 ± 0.4 |
| BePI |
3 |
.12 ± 0.05 |
| |
1 |
0.24 ± 0.08 |
| |
0.5 |
0.28 ± 0.04 |
| |
|
|
| 12mer TG |
|
|
| 2,7 amidoanthraquinone |
10 |
0.86 ± 0.20 |
| |
|
|
| 13mer AG |
|
|
| 2,6 amidoanthraquinone |
10 |
7.3 ± 1.7 |
| 1,5 amidoanthraquinone |
10 |
1.2 ± 0.3 |
| Naphthylquinoline | 10 | 13.4 ± 3.6 |
The values were determined from quantitative analysis of the footprinting patterns.
The central panel of Figure 3 shows similar footprinting experiments with the 17mer oligonucleotide in the presence of 10 µM of the 1,8-disubstituted anthraquinone. A clear footprint is evident which covers the entire target site and which persists to an oligonucleotide concentration of 2 µM (the band at position 58 is an artifact which corresponds to a stable secondary structure). Although this ligand has stabilised the antiparallel triplex, it is less potent than the 2,7- derivative. Quantitative analysis of this footprinting pattern yielded a C50 value of 0.61 ± 0.23 µM. In contrast, the 2,6- and 1,5-disubstituted anthraquinones failed to stabilise this TG-containing triplex, as shown in Figure 2.
Naphthylquinoline derivative. The third panel of Figure 3 shows the effect of 3 µM naphthylquinoline triplex-binding ligand on the concentration dependence of the footprints produced by the 17mer TG-containing oligonucleotide. This footprint persists to an oligonucleotide concentration of ∼2 µM. Quantitative analysis of these data yields a C50 value of 1.4 ± 0.4 µM which is presented together with the value obtained with 10 µM ligand in Table 1. Although it is clear that this ligand stabilises antiparallel TG-containing triplexes, the resulting complexes are still less stable than those formed with parallel (CT-containing) oligonucleotides.
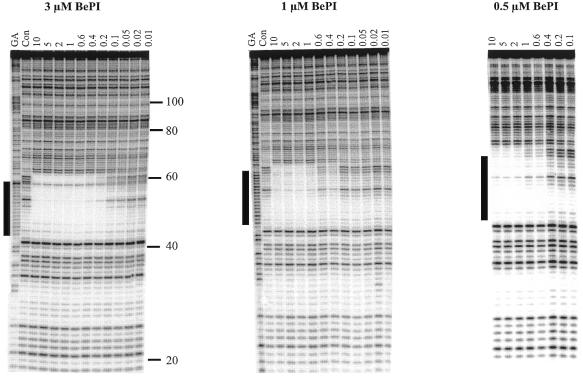
Coralyne and BePI. These two ligands are well known as triplex stabilisers, though in most studies they have been used with parallel triplexes. Coralyne (10 µM) failed to generate a footprint with 10 µM of the 17mer TG-containing oligonucleotide, suggesting that this ligand is not effective in stabilising antiparallel triplexes. In contrast, BePI had a pronounced effect on triplex stability. Figure 4 shows DNase I digestion patterns with different concentrations of the 17mer TG-containing oligonucleotide in the presence of 3 µM (left-hand panel), 1 µM (centre panel) and 0.5 µM BePI (right-hand panel). In the presence of 3 µM BePI a clear footprint is evident which persists to an oligonucleotide concentration of 0.2 µM. This footprint covers the entire target site and is very similar to that generated in the presence of the other ligands. Quantitative analysis of these footprints yields a C50 value of 0.12 ± 0.05 µM, similar to that determined with 10 µM of the naphthylquinoline. Clear footprints are also evident with 1 and 0.5 µM BePI, which yield C50 values of 0.24 ± 0.08 and 0.28 ± 0.03 M, respectively (Table1).
Figure 4.
DNase I digestion patterns showing the interaction of the 17mer TG-containing oligonucleotide with its target site in tyrT(43-59) in the presence of different concentrations of BePI. Oligonucleotide concentrations (µM) are shown at the top of each gel lane. The track labelled ‘Con’ shows DNase I digestion of the duplex DNA in the absence of oligonucleotide or ligand. ‘GA’ represents a Maxam–Gilbert marker specific for purines. The experiments were performed in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MgCl2. The numbers on the right-hand side of the first panel indicate the sequence position and are numbered as in Brown et al. (25). The filled boxes show the position of the target site.
AG-containing oligonucleotides. A 17mer AG oligonucleotide was designed to bind to the oligopurine tract of tyrT(43-59) in an antiparallel orientation. Early footprinting experiments with this oligonucleotide failed to show any interaction with the duplex in the presence of 10 mM MgCl2. Since AG-containing triplexes have previously been shown to be stabilised by manganese (24,36) this was replaced by MnCl2 in subsequent experiments. In this buffer footprinting experiments with the 17mer AG oligonucleotide showed attenuated cleavage within the target site at the highest concentration but did not produce complete protection even with 30 µM oligonucleotide. In order to obtain a clearer indication of the effects of added ligands we chose to work with a 13mer GA-containing oligonucleotide (5′-AAAAAGAGAAGGA), which does not affect the DNase I cleavage pattern even at concentrations as high as 30 µM (data not shown). In contrast to the TG-containing oligonucleotides, addition of 10 µM of either the 2,7- or 1,8-disubstituted anthraquinones failed to generate a footprint with this oligonucleotide. However, both the 2,6- and 1,5-disubstituted anthraquinones stabilised triplex formation when present in 10 µM concentrations.
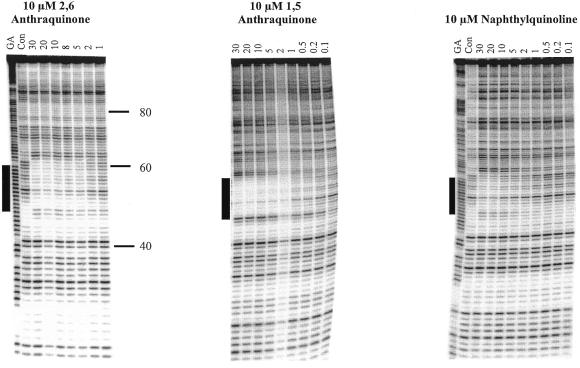
The first panel of Figure 5 shows the effects of 10 µM of the 2,6-disubstituted anthraquinone on the footprints generated with the 13mer AG-containing oligonucleotide. These experiments were conducted at pH 7.0 in 10 mM Tris–HCl, 50 mM NaCl and 10 mM MnCl2. It can be seen that in the presence of the ligand the intensity of bands within the target area is attenuated at an oligonucleotide concentration of 5 µM and above, though the footprint is not complete even at 30 µM oligonucleotide. In the presence of 10 µM of the 1,5-disubstituted anthraquinone (centre panel) the footprint extends to an oligonucleotide concentration of 2 µM. In this instance a strong enhancement is evident at position 48, corresponding to the triplex–duplex junction at the 3′-end of the target site. A similar but weaker enhancement can also be seen with the 2,6-disubstituted compound. Quantitative analysis of these footprints yielded C50 values of 7.3 ± 1.7 and 1.2 ± 0.3 M in the presence of 10 µM of the 2,6- and 1,5- derivatives, respectively (Table 1). The right-hand panel of Figure 5 shows the footprinting pattern on addition of 10 µM naphthylquinoline. In the presence of this ligand the intensity of bands within the target site is attenuated at oligonucleotide concentrations above 10 µM, and this is accompanied by enhanced cleavage at positions 47 and 48. Quantitative analysis of these footprints yielded a C50 value of 13.3 ± 3.6 µM (Table 1). In contrast, neither coralyne or BePI potentiated the interaction with the 13mer AG oligonucleotide, showing no interaction even with 30 µM oligonucleotide and 10 µM ligand.
Figure 5.
DNase I digestion patterns showing the interaction of the 13mer AG-containing oligonucleotide with its target site in tyrT(43-59) in the presence of triplex-binding ligands. Oligonucleotide concentrations (µM) are shown at the top of each gel lane. Tracks labelled ‘Con’ show DNase I digestion of the duplex DNA in the absence of oligonucleotide or ligand. ‘GA’ represents a Maxam–Gilbert marker specific for purines. The experiments were performed in 10 mM Tris–HCl pH 7.0 containing 50 mM NaCl and 10 mM MnCl2. The numbers on the right-hand side of the first panel indicate the sequence position and are numbered as in Brown et al. (25). The filled boxes show the position of the target site.
DISCUSSION
The results presented in this paper demonstrate that there are pronounced differences in the ability of ligands to stabilise AG- and TG-containing triplexes. The 1,8- and 2,7-disubstituted anthraquinones stabilise TG- but not AG-containing triplexes, while the 2,6- and 1,5- regioisomers show the opposite selectivity. BePI is arguably the best ligand for stabilising TG-containing triplexes but shows no activity against AG-triplexes. By contrast the naphthylquinoline ligand interacts with both structures, though it generates more stable TG-containing triplexes.
We suggest that the different effects of the various ligands on AG- and TG-containing triplexes arise from the different structures adopted by these triplexes. Antiparallel T·AT and A·AT triplets have different structures, and neither of these triplets is isomorphous with the G·GC triplet (37–40), though G·GC and A·AT may be more similar to each other (21,22). As a result there must be backbone distortions at steps between different triplets, generating grooves of different shapes and sizes, with varying amounts of overlap between adjacent base triplets.
In general, triplexes generated with TG-containing oligonucleotides have been reported to have a lower intrinsic stability than their AG-containing counterparts (41,42). However, in the present study we find that AG triplexes form weaker complexes. They also do not completely inhibit DNase I cleavage, with bands still evident within the footprints at high oligonucleotide concentrations. Although it is possible that this reflects their weak binding it may be that these AG-containing triplexes are still substrates for DNase I. Since DNase I cuts from the minor groove, while triplex-forming oligonucleotides bind in the major groove, there is no a priori reason why triplexes must abolish DNase I cleavage. In contrast visual inspection of the cleavage patterns suggests that both CT (parallel) and TG (antiparallel) triplexes abolish DNase I cleavage. This difference probably reflects changes in the size and hence accessibilities of the minor grooves in these triplexes.
The simplest explanation for the results presented in this paper is that the ligands function by binding to triplex DNA. However, we cannot rule out the possibility that at least part of their activity is attributable to effects on the self-association properties of the AG- and TG-containing third strands. AG- and TG-containing oligonucleotides are known to adopt self-associated structures (31,32) that compete for triplex formation and thereby reduce the apparent triplex-binding constants. Any ligand which interferes with these self-structures will thereby promote triplex formation.
Disubstituted anthraquinones
All the disubstituted amidoanthraquinone regioisomers examined in this paper stabilise parallel triplexes, and there is only a 6-fold difference in their apparent binding constants. A very different pattern is seen with the antiparallel triplexes. Triplexes formed by TG-containing oligonucleotides are stabilised only by the 2,7- and 1,8-disubstituted anthraquinones and not by their 1,5- and 2,6- regioisomers. In addition the 2,7- and 1,8- compounds have different affinities for TG-containing triplexes: 3 µM of the 2,7-compound produces a C50 value of 0.36 ± 0.07 M, while 10 µM of the 1,8-disubstituted anthraquinone only induces a C50 value of 0.61 ± 0.23 M. The greater effect of the 2,7-compound is evident in that this compound alone is able to stabilise the interaction with the 12mer TG oligonucleotide.
The triplexes formed with the AG-containing oligonucleotides show the opposite order of stabilisation by the various anthraquinones. The 2,7- and 1,8-anthraquinones have no detectable effect on the stability of AG triplexes. The 1,5-disubstituted anthraquinone produces a C50 value of 1.2 ± 0.3 µM with 10 M ligand and appears to be about six times more potent than the 2,6-disubstituted anthraquinone which produces a C50 value of 7.3 ± 1.7 µM.
The observed differences in the abilities of the anthraquinones to stabilise antiparallel triplexes can be attributed to ligand–base stacking differences which depend, in part, on the position of the side chains. Since antiparallel triplets are not isomorphous with each other, backbone distortion will be present within the oligonucleotide at steps between the different triplets. These structural changes are likely to differ according to the neighbouring bases, and will therefore require different ligand structures to stabilise them.
Naphthylquinoline
Unlike the disubstituted anthraquinones, which selectively stabilise either AG- or TG-containing antiparallel triplexes, the naphthylquinoline ligand stabilises both forms, though by differing amounts. The 17mer TG-containing oligonucleotide shows the greatest stabilisation, with a C50 value of 0.18 ± 0.09 µM in the presence of 10 µM ligand. This is very similar to the stabilisation afforded by the 2,7-disubstituted anthraquinone, and is considerably better than the 1,8- compound. With the AG-containing oligonucleotide a weak triplex footprint is induced in the presence of 10 µM naphthylquinoline. In this case both the 2,6- and 1,5-disubstituted anthraquinones are more potent at stabilising this triplex.
The ability of the naphthylquinoline to stabilise both forms of antiparallel triplex may be attributed to the flexibility in its structure and to the small size of its chromophore available for base stacking compared to the anthraquinones. The bond between the ring groups allows free rotation, thereby enabling the ligand to adjust its conformation so as to obtain the best position for stabilisation.
BePI
BePI is one of the best ligands for stabilising parallel (CT-containing) triplexes, and is also the best ligand for stabilising antiparallel TG-containing triplexes. Previous studies have shown that BePI does not stabilise parallel GT-containing triplexes (5). The presence of only 0.5 µM of this ligand produces footprints with the 17mer TG-oligonucleotide with an apparent C50 of 0.28 ± 0.04 M, lower than that for any other ligand at this concentration. It is worth considering whether BePI discriminates between the TG, GT or TT steps in this triplex. The 17mer TG triplex has seven possible intercalation sites between adjacent T·AT triplets, which reduces to a maximum of five if neighbour exclusion applies. The observation that BePI has such a strong effect on this triplex suggests that more than five ligand molecules may be bound to the triplex, implying that it does not discriminate between different triplet steps. In contrast to its high affinity for TC- and TG-containing triplexes BePI does not stabilise the triplex generated with the 13mer AG oligonucleotide.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grants from the Cancer Research Campaign.
References
- 1.Soyfer V.N. and Potoman,V.N. (1996) Triple-Helical Nucleic Acids. Springer-Verlag, New York.
- 2.Thuong N.T. and Hélène,C. (1993) Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angew. Chem. Int. Ed. Engl., 32, 666–690. [Google Scholar]
- 3.Fox K.R. (2000) Targeting DNA with triplexes. Curr. Med. Chem., 7, 17–37. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.L., Duval-Valentin,G., Nguyen,C.H., Perrouault,L., Faucon,B., Rougée,M., Montenay-Garestier,T., Nisagni,E. and Hélène,C. (1992). Triple-helix specific ligands. Science, 256, 1681–1684. [DOI] [PubMed] [Google Scholar]
- 5.Escudé C., Sun,J.S., Nguyen,C.H., Bisagni,E., Garestier,T. and Hélène,C. (1996) Ligand-induced formation of triple helices with antiparallel third strands containing G and T. Biochemistry, 35, 5735–5740. [DOI] [PubMed] [Google Scholar]
- 6.Marchand C., Bailly,C., Nguyen,C.H., Bisagni,E., Garestier,T., Hélène,C. and Waring,M.J. (1996). Stabilization of triple helical DNA by a benzopyridoquinoxaline intercalator. Biochemistry, 35, 5022–5032. [DOI] [PubMed] [Google Scholar]
- 7.Escudé C., Nguyen,C.H., Kukreti,S., Janin,Y., Sun,J.S., Bisagni,E., Garestier,T. and Hélène,C. (1998). Rational design of a triple helix-specific intercalating ligand. Proc. Natl Acad. Sci. USA, 95, 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen C.H., Marchand,C., Delage,S., Sun,J.S., Garestier,T., Hélène,C. and Bisagni,E. (1998). Synthesis of 13H-benzo[6,7]- and 13H-benzo[4,5]indolo[3,2-c]quinolines: a new series of potent specific ligands for triplex DNA. J. Am. Chem. Soc., 120, 2501–2507. [Google Scholar]
- 9.Schmitt P., Nguyen,C.H., Sun,J.S., Grierson,D.S., Bisagni,E., Garestier,T. and Hélène,C. (2000). 13H-benzo[6-7]indolo[3,2-c]quinolines (B[6,7]IQ): optimization of their DNA triplex-specific stabilization properties. Chem. Commun., 2000, 763–764. [Google Scholar]
- 10.Lee J.S., Latimer,L.J.P. and Hampel,K.J. (1993) Coralyne binds tightly to both T·A·T– and C·G·C+-containing DNA triplexes. Biochemistry, 32, 5591–5597. [DOI] [PubMed] [Google Scholar]
- 11.Latimer L.J.P., Payton,N., Forsyth,G. and Lee,J.S. (1995). The binding of analogues of coralyne and related heterocyclics to DNA triplexes. Biochem. Cell Biol., 73, 11–18. [DOI] [PubMed] [Google Scholar]
- 12.Moraru-Allen A.A., Cassidy,S., Alvarez,J.L.A., Fox,K.R., Brown,T. and Lane,A.N. (1997) Coralyne has a preference for intercalation between TA.T triples in intramolecular DNA triple helices. Nucleic Acids Res., 25, 1890–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler S.P., Strekowski,L., Wilson,W.D. and Fox,K.R. (1995) Footprinting studies on ligands which stabilise DNA triplexes. Biochemistry, 34, 7234–7242. [DOI] [PubMed] [Google Scholar]
- 14.Wilson W.D., Tanious,F.A., Mizan,S., Yao,S., Kiselyov,A.S., Zon,G. and Strekowski,L. (1993) DNA triple-helix specific intercalators as antigene enhancers. Biochemistry, 32, 10614–10621. [DOI] [PubMed] [Google Scholar]
- 15.Cassidy S.A., Strekowski,L., Wilson,W.D. and Fox,K.R. (1994) Effect of a triplex-binding ligand on parallel and antiparallel DNA triple helices using short unmodified and acridine-linked oligonucleotides. Biochemistry, 33, 15338–15347. [DOI] [PubMed] [Google Scholar]
- 16.Cassidy S.A., Strekowski,L. and Fox,K.R. (1996) DNA sequence specificity of a naphthylquinoline triple helix-binding ligand. Nucleic Acids Res., 24, 4133–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox K.R., Polucci,P., Jenkins,T.C. and Neidle,S. (1995) A molecular anchor for stabilizing triple-helical DNA. Proc. Natl Acad. Sci. USA, 92, 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kan Y.Z., Armitage,B. and Schuster,G.B. (1997) Selective stabilization of triplex DNA by anthraquinone sulfonamide derivatives. Biochemistry, 36, 1461–1466. [DOI] [PubMed] [Google Scholar]
- 19.Keppler M.D., Read,M.A., Perry,P.J., Trent,J.O., Jenkins,T.C., Reszka,A.P., Neidle,S. and Fox,K.R. (1999) Stabilization of DNA triple helices by a series of mono- and disubstituted amidoanthraquinones. Eur. J. Biochem., 263, 817–825. [DOI] [PubMed] [Google Scholar]
- 20.Giovannangeli C., Rougée,M., Garestier,T., Thuong,N.T. and Hélène,C. (1992) Triple-helix formation by oligonucleotides containing the three bases thymine, cytosine and guanine. Proc. Natl Acad. Sci. USA, 89, 8631–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radhakrishnan I., de los Santos,C. and Patel,D.J. (1993) Nuclear magnetic resonance structural studies of A·AT base triple alignments in intramolecular purine·purine·pyrimidine DNA triplexes in solution. J. Mol. Biol., 234, 188–197. [DOI] [PubMed] [Google Scholar]
- 22.Ji J., Hogan,M.E. and Xao,X.L. (1996) Solution structure of an antiparallel purine motif triplex containing a T·CG pyrimidine base triple. Structure, 4, 425–435. [DOI] [PubMed] [Google Scholar]
- 23.Rhee S., Han,Z.-j., Liu,K., Miles,H.T. and Davies,D.R. (1999) Structure of a triple helical DNA with a triplex-duplex junction. Biochemistry, 38, 16810–16815. [DOI] [PubMed] [Google Scholar]
- 24.Chandler S.P. and Fox,K.R. (1996) Specificity of antiparallel DNA triple helix formation. Biochemistry, 35, 15038–15048. [DOI] [PubMed] [Google Scholar]
- 25.Brown P.M., Madden,C.A. and Fox,K.R. (1998) Triple helix formation at different positions on nucleosomal DNA. Biochemistry, 37, 16139–16151. [DOI] [PubMed] [Google Scholar]
- 26.Svinarchuk F., Bertrand,J.-R. and Malvy,C. (1994) A short purine oligonucleotide forms a highly stable triple helix with the promoter of the murine c-pim-1 proto-oncogene. Nucleic Acids Res., 22, 3742–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svinarchuk F., Paoletti,J. and Malvy,C. (1995) An unusually stable purine(purine–pyrimidine) short triplex. The third strand stabilizes double-stranded DNA. J. Biol. Chem., 270, 14068–14071. [DOI] [PubMed] [Google Scholar]
- 28.Fox K.R. (1994) Formation of DNA triple helices incorporating blocks of G·GC and T·AT triplets using short acridine-linked oligonucleotides. Nucleic Acids Res., 22, 2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng A.-J. and van Dyke,M.W. (1993) Monovalent cation effects on intermolecular purine–purine–pyrimidine triple-helix formation. Nucleic Acids Res., 21, 5630–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivas W.M. and Maher,L.J. (1995). Overcoming potassium-mediated triplex inhibition. Nucleic Acids Res., 23, 1936–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noonberg S.B., Francois,J.-C., Praseuth,D., Guieyesse-Peugot,A.-L., Lacoste,J., Garestier,T. and Hélène,C. (1995) Triplex formation with α anomers of purine-rich and pyrimidine-rich oligodeoxynucleotides. Nucleic Acids Res., 23, 4042–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noonberg S.B., Francoise,J.C., Garestier,T. and Hélène,C. (1995) Effect of competing self-structure on triplex formation with purine-rich oligodeoxynucleotides containing GA repeats. Nucleic Acids Res., 23, 1956–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry P.J., Gowan,S.M., Reszka,A.P., Polucci,P., Jenkins,T.C., Kelland,L.R. and Neidle,S. (1998) 1,4- and 2,6-disubstituted amidoanthracene-9,10-dione derivatives as inhibitors of human telomerase. J. Med. Chem., 41, 3253–3260. [DOI] [PubMed] [Google Scholar]
- 34.Perry P.J., Reszka,A.P., Wood,A.A., Read,M.A., Gowan,S.M., Dosanjh,H.S., Trent,J.O., Jenkins,T.C., Kelland,L.R. and Neidle,S. (1998) Human telomerase inhibition by regioisomeric disubstituted amidoanthracene-9,10-diones. J. Med. Chem., 41, 4873–4884. [DOI] [PubMed] [Google Scholar]
- 35.Dabrowiak J.C. and Goodisman,J. (1989) Quantitative footprinting analysis of drug–DNA interactions. In Kallenbach,N.R. (ed.), Chemistry and Physics of DNA–Ligand Interactions. Adenine Press, New York, pp.143–174.
- 36.Malkov V.A., Voloshin,O.N., Soyfer,V.N. and Frank-Kamenetskii,M.D. (1993) Cation and sequence effects on stability of intermolecular pyrimidine–purine–purine triplex. Nucleic Acids Res., 21, 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishnan I., de los Santos,C. and Patel,D.J. (1991). Nuclear Magnetic Resonance structural studies of intramolecular purine·purine·pyrimidine DNA triplexes in solution. J. Mol. Biol., 221, 1403–1418. [PubMed] [Google Scholar]
- 38.Radhakrishnan I., de los Santos,C. and Patel,D.J. (1993) Nuclear magnetic resonance structural studies of A⋅AT base triple alignments in intramolecular purine·purine·pyrimidine DNA triplexes in solution. J. Mol. Biol., 234, 188–197. [DOI] [PubMed] [Google Scholar]
- 39.Radhakrishnan I. and Patel,D.J. (1993) Solution structure of a purine·purine·pyrimidine DNA triplex containing G⋅GC and T⋅AT triples. Structure, 1, 135–152. [DOI] [PubMed] [Google Scholar]
- 40.Radhakrishnan I. and Patel,D.J. (1994). Hydration sites in purine⋅purine⋅pyrimidine and pyrimidine·pyrimidine·purine DNA triplexes in aqueous solution. Structure, 2, 395–405. [DOI] [PubMed] [Google Scholar]
- 41.Roy C. (1993) Inhibition of gene transcription by purine rich triplex forming oligodeoxyribonucleotides. Nucleic Acids Res., 21, 2845–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faucon B., Mergny,J.L. and Hélène,C. (1996) Effect of third strand composition on triple helix formation: purine versus pyrimidine oligodeoxynucleotides. Nucleic Acids Res., 24, 3181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]