Abstract
Nonsense mutations are usually predicted to function as null alleles due to premature termination of protein translation. However, nonsense mutations in the DMD gene, encoding the dystrophin protein, have been associated with both the severe Duchenne Muscular Dystrophy (DMD) and milder Becker Muscular Dystrophy (BMD) phenotypes. In a large survey, we identified 243 unique nonsense mutations in the DMD gene, and for 210 of these we could establish definitive phenotypes. We analyzed the reading frame predicted by exons flanking those in which nonsense mutations were found, and present evidence that nonsense mutations resulting in BMD likely do so by inducing exon skipping, confirming that exonic point mutations affecting exon definition have played a significant role in determining phenotype. We present a new model based on the combination of exon definition and intronic splicing regulatory elements for the selective association of BMD nonsense mutations with a subset of DMD exons prone to mutation-induced exon skipping.
Keywords: DMD, exon skipping, nonsense mutations, Becker muscular dystrophy, splicing motifs, dystrophin
INTRODUCTION
The most common form of muscular dystrophy includes the more severe Duchenne Muscular Dystrophy (DMD; MIM# 310200) and the milder Becker Muscular Dystrophy (BMD; MIM# 300376). Both DMD and BMD are caused by mutations in the ~2.4 Mb DMD gene, the largest known human gene spanned by 79 exons encoding dystrophin, an important cytoskeletal protein. The major determinant of disease severity is whether the mutation results in an mRNA that maintains an open reading frame that allows translation of a functional amino- and carboxy-terminus, with little impact due to the size of the central rod domain deletion(Monaco, et al., 1988). Due to this exceptional feature, antisense-mediated exon skipping is currently one of the more promising therapeutic approaches for DMD (Popplewell, et al., 2010; van Deutekom, et al., 2007) with the induced skipping of central rod domain exons restoring the open reading frame and converting a severe DMD into a milder BMD phenotype. The dispensability of central rod domain exons and the enormous size of the gene provide an unusual context for studying the in vivo effects of exonic variation on mutation-induced exon skipping events.
Nonsense mutations in the DMD gene account for approximately 15% of dystrophinopathy patients (Dent, et al., 2005). Nonsense mutations are expected to result in premature termination of protein translation, and therefore be associated with severe DMD phenotype. However, previous reports have described nonsense mutations that result in significant levels of altered exon splicing and a milder BMD phenotype (Shiga, et al., 1997; Tuffery-Giraud, et al., 2005). One early model suggested that a milder phenotype might result from variable read-through of premature stop codons due to the specific stop codon sequence; however, sequence context effects are apparently not correlated with dystrophinopathy phenotype (Howard, et al., 2004). In contrast, in several of these cases the point mutation results in exclusion of the nonsense mutation-containing exon due to exon skipping (Disset, et al., 2006; Fajkusova, et al., 2001; Ginjaar, et al., 2000; Melis, et al., 1998; Nishiyama, et al., 2008; Shiga, et al., 1997), leading to a model in which point mutations result in either the critical disruption of exonic splicing enhancer (ESE) or creation of exonic splicing suppressor (ESS) motifs in order to account for the exon skipping event. These point mutations are remarkable in that they induce levels of dystrophin restoration similar to those sought by therapeutic approaches to antisense-mediated exon skipping.
In order to investigate the generality of these observations, we analyzed the reading frame of the exons surrounding nonsense mutations identified in a large mutational screen of over 1111 dystrophinopathy mutations (Flanigan, et al., 2009b). We note that the influence of nonsense mutations on exon skipping is significant, as among the 210 patients with nonsense mutations a correlation exists between the phenotype and the reading frame of exons flanking a point mutation-containing exon. The presence of BMD-associated nonsense mutations within exons flanked by exons that can be spliced together to maintain an open reading frame (which we term ‘in-frame’ exons) is consistent with a point mutation-induced exon skipping mechanism. However, the exon distribution of BMD-associated nonsense mutations suggests that there is only a distinct subset of in-frame exons that are prone to restoring significant levels of dystrophin expression. By analysis of multiple exon definition metrics including splice donor/acceptor site strength and exonic splicing enhancer/silencer density, we have developed evidence for a model in which the integrated strength of exon definition elements is a major determinant of whether point mutation-induced exon skipping (and attenuation of the phenotype) is likely to occur.
METHODS
Subjects
As described in detail elsewhere (Flanigan, et al., 2009b) patients were derived from two cohorts. One group was ascertained among patients in the United Dystrophinopathy Project (UDP) and were selected by strict diagnostic criteria which included either (1) clinical features consistent with DMD or BMD and an X-linked family history; or (2) muscle biopsy showing alteration in dystrophin expression by immunofluorescence, immunohistochemistry, or immunoblot; or (3) a mutation in the DMD gene previously detected by clinical testing. After informed consent was obtained (under IRB-approved protocols), blood samples were obtained for DNA analysis; patients were examined, and data was extracted from clinical records for inclusion in the UDP database. Phenotypes in the UDP data set were determined using the directive of “best clinical diagnosis”, based upon an expert clinical diagnosis combining available information regarding clinical presentation features, family history, protein expression, and categorized into one of three diagnostic classes: DMD, BMD, or Intermediate Muscular Dystrophy (IMD). Complete details regarding categorization are available elsewhere (Flanigan, et al., 2009b). A second group consisted of samples sent for analysis in the clinical testing laboratory. For clinical samples, we accepted the referring physician’s clinical diagnosis, and sought confirmatory diagnostic information from family history or muscle biopsy criteria wherever possible.
Mutation Analysis
Mutation analysis is discussed in detail elsewhere (Flanigan, et al., 2009b). Briefly, SCAIP testing, with sequencing of the entire coding region, flanking intronic sequence, and untranslated regions, was performed as previously described (Flanigan, et al., 2003). In selected patients, mRNA was isolated from archived muscle biopsy tissue, and reverse transcription PCR for cDNA sequencing was performed, using conditions and primers published elsewhere (Roberts, et al., 1991). Nucleotide positions were determined according to the reference DMD sequence used for mutation analysis (GenBank accession number NM_004006.2); Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Quantitative Real-time Polymerase Chain Reaction
Real-time quantitative PCR was performed using cDNA on an ABI PRISM® 7900HT Sequence Detection System and results were analyzed using SDS 2.1 software. Total RNA was extracted from muscle sections using Trizol (Invitrogen) according to the manufacturer’s recommended protocol. cDNA was synthesized using Superscript III first strand synthesis kit (Invitrogen) with random hexamer primers. Maxima ™ SYBR Green qPCR Master Mix (2X) (Fermentas) was used for all qPCR reactions. Primers were designed spanning the junctions between exons 30/31 (wild type) and exons 30/32 (exon 31 skipping) forms of the DMD transcript. A common reverse primer in exon 33 was used for both reactions. The wild type DMD transcript, exon 31-skipped DMD transcript, and a housekeeping gene (GAPDH) were amplified in triplicate reactions from both control and patient cDNA. Results were obtained using the comparative CT (ΔΔ CT) method by normalizing the DMD levels to GAPDH, and setting the control wild type DMD transcript value at 100%.
Immunofluorescence
Snap frozen muscle was sectioned to 7 micrometer sections and stained without fixation. Sections were blocked one hour in TBS, 1% BSA, 5% normal goat serum, then incubated two hours with one of three monoclonal antibodies to dystrophin(Nguyen and Morris, 1993) (1:5 dilution): MANDYS16 (Clone No. 1B12; a gift of Dr. G.E. Morris), specific to exons 27/28 at amino acids 1226–1245; MANDYS1 (Clone No. 3B7; a gift of Dr. G.E. Morris), specific to exons 31/32 at amino acids 1431–1505; and Dys2 (clone Dy8/6C5, Novacastra), specific to exons 77–79 at amino acids 3668–3684. Alexa 488 goat anti mouse IgG (Molecular Probes) was used at 1:1000 for secondary detection and images were captured using Olympus Fluoview laser scanning confocal microscopy at 20x magnification.
Splice site motifs, exon definition metrics and statistical analyses
In silico analysis of the effect of point mutations on predicted exonic splice enhancers and silencers in dystrophin exon sequences (NM_004006.2) used the Human Splicing Finder (HSF) (http://www.umd.be/HSF/) tool (Desmet, et al., 2009). This software tool was also used to calculate exonic splice enhancer (ESE) density for the 79 exons in NM_004006.2 using the ESE Finder matrices for SRp40, SC35, SF2/ASF and SRp55 proteins (Cartegni and Krainer, 2002; Smith, et al., 2006), Rescue-ESE hexamers (Fairbrother, et al., 2004), PESE and PESS 8-mers (Zhang and Chasin, 2004), Tra2b, 9G8 and hnRNP A1 binding sites using HSF matrices (Desmet, et al., 2009), exonic splice silencers (Sironi, et al., 2004). The hexamer lists, hexESE and hexESS, were from consolidated lists of PESE and Rescue-ESE motifs, and PESS and FAS-hex3 motifs, as previously described (Ke, et al., 2008). The intronic splicing regulatory (ISR) motifs were a list of 18 conserved pentamers (Table 1 from (Suyama, et al., 2010)) discovered in the flanking introns of skipped exons from a systematic search for pentamer motifs in the flanking introns of known cassette exons (1736 skipped exons in 1473 genes) by using comparative analysis of mammalian genome alignments (Suyama, et al., 2010). The strengths of 3’ and 5’ splice junctions and splice site sequence motifs in the DMD gene were scored using the splice site models and software available at: http://genes.mit.edu/burgelab/maxentl (Yeo and Burge, 2004). The R package wilcox.test (R Development Core Team, 2008) was used for the Mann-Whitney-Wilcoxon test of the null hypothesis that the splice site and exon definition motifs in different exon groups are from identical populations. DMD exons 10 through 64 (based on NM_004006.2) were used for calculations involving rod domain exons.
Table 1.
Association of splice site strength and intron/exon size with exon frame and BMD phenotypes
| In-Frame | Out-of- Frame |
In-Frame (BMD)a |
In-Frame (DMD)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Mean | SD | Mean | SD | p -value |
Mean | SD | Mean | SD | p -value |
| 5’ss strength (MaxEnt) |
7.47 | 1.85 | 8.74 | 1.50 | 0.002 | 7.29 | 2.93 | 7.53 | 1.47 | 0.75 |
| 5’ss strength (MDD) |
11.74 | 2.06 | 12.68 | 2.05 | 0.05 | 10.94 | 2.41 | 11.99 | 1.93 | 0.15 |
| 5’ss strength (MM) |
6.72 | 1.81 | 8.02 | 1.65 | 0.003 | 6.24 | 3.05 | 6.86 | 1.30 | 0.78 |
| 5’ss strength (WMM) |
7.12 | 1.76 | 8.50 | 1.84 | 0.004 | 6.89 | 1.56 | 7.18 | 1.84 | 0.32 |
| 3’ss strength (MaxEnt) |
7.69 | 2.40 | 8.23 | 2.76 | 0.24 | 5.92 | 2.60 | 8.22 | 2.12 | 0.02 |
| 3’ss strength (MM) |
8.06 | 2.53 | 8.61 | 3.22 | 0.17 | 6.10 | 2.66 | 8.66 | 2.22 | 0.02 |
| 3’ss strength (WMM) |
7.38 | 3.81 | 9.08 | 4.83 | 0.06 | 5.08 | 4.24 | 8.08 | 3.47 | 0.06 |
| Upstream intron size (kb) |
16.53 | 29.24 | 36.76 | 52.59 | 0.005 | 7.13 | 7.83 | 8.39 | 10.10 | 0.44 |
| Downstream intron size (kb) |
14.62 | 19.12 | 34.66 | 51.09 | 0.02 | 8.56 | 9.27 | 9.07 | 10.22 | 0.46 |
| Exon size (nts.) | 151 | 33 | 155 | 53 | 0.40 | 150 | 25 | 151 | 36 | 0.42 |
Rod domain in-frame exons: 25, 27, 29, 31, 37, 38, 40
Rod domain in-frame exons: 23, 24, 26, 28, 30, 32, 33, 34, 35, 36, 39, 41, 42
RESULTS
In our survey of 1111 patients, we identified 210 definitively phenotyped non-carrier patients with nonsense mutations (Supp. Table S1). Among these, we noted the significant number who have BMD (n = 30) or Intermediate Muscular Dystrophy (IMD) (n=4), rather than DMD (n =176). In order to investigate the generality of the conclusion that BMD-associated nonsense mutations are due to altered splicing, we first analyzed the reading frame of the exons surrounding each nonsense mutation-containing exon. We postulated that nonsense mutations resulting in a Becker phenotype would more frequently occur in exons where mutation-induced exon skipping would follow the "reading frame rule" (Monaco, et al., 1988), in which single and multi-exon deletions resulting in an in-frame mRNA are more likely to be associated with BMD. Herein we describe as “in-frame” those exons with flanking exons that would be predicted, when spliced together, to result in the maintenance of an open reading frame; conversely, exons flanking an “out-of-frame” exon would be predicted to splice into an mRNA in which the reading frame is disrupted. We excluded one BMD nonsense mutation from our analysis: c.9G>A (p.Trp3X) is found within exon 1 (Flanigan, et al., 2009a), and therefore cannot be said to have a flanking exon reading frame. This mutation results in translation of a significant amount of an amino-terminal truncated dystrophin protein from alternate initiation sites encoded in exon 6 (Gurvich, et al., 2009). It may therefore be considered to result in an open reading frame through the carboxy-terminus, but was nevertheless excluded from the analysis of flanking reading frame.
Exonic distribution of BMD versus DMD nonsense mutations
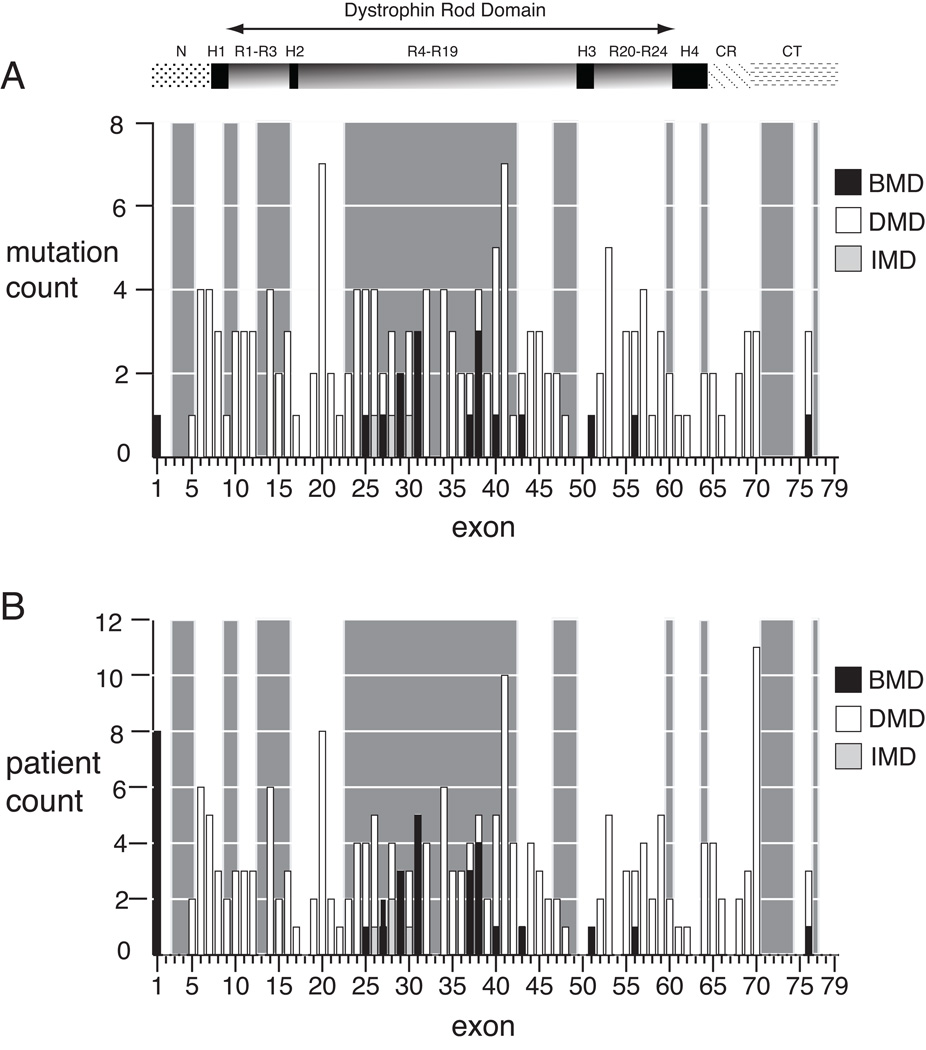
Figure 1A shows the distribution of 160 unique nonsense mutation sites identified in this study among 60 exons, while Figure 1B shows this distribution by patient count from the 210 phenotyped patients. The reading frame of flanking exons is also indicated by the light grey shading of in-frame (skippable) exons (Figure 1). Only one phenotypic discrepancy was found among kindreds: the exon 26 nonsense mutation c.3500C>G was associated with DMD in two boys from one kindred, and with IMD in another unrelated boy (both are represented in Figure 1A). In 202 patients, a flanking reading frame can be established (the exceptions are c.9G>A patients in exon 1). Among those in whom mutation-induced exon skipping is predicted to result in an out-of-frame transcript (n=97), 4% had BMD (n=4) and 96% had DMD or IMD (n=93). In contrast, among patients whose mutations have an in-frame flanking exon context (n=105), 17% had BMD (n=18), and 83% had DMD or IMD (n=87). The difference in proportions of patients with BMD in the out-of-frame versus in-frame subgroups is significant (p =.003 by Pearson’s chi-squared test). These results suggest that in the clinical setting of BMD due to a nonsense mutation, the exonic sequence context is nearly always in-frame (18/22). Notably, the presence of an in-frame exon context is not by itself predictive of BMD, as most (87 of 105, 83%) nonsense mutations found in that context result in DMD or IMD. However, our results suggest that an in-frame context for a nonsense mutation results in a 4-fold higher chance of BMD than a nonsense mutation in the out-of-frame context.
Figure 1.
Exon distribution of dystrophin nonsense mutations by phenotype. A) The number of mutations per exon for unique mutation sites (N=161) is shown for Duchenne (open boxes), intermediate (light boxes), and Becker (dark boxes) phenotypes. Exons with in-frame flanking exon context are highlighted by the light grey background shading, and for each exon within the shaded region, single exon skipping will result in bypass of the truncating mutation. Only a single mutation site (c.3500) is counted twice, due to a discrepant phenotype between kindreds, as discussed in the text. B) The number of mutations per exon by patient (N=210). The exon location of dystrophin protein domains are shown above the graphs: NH2 terminus (N), rod domain spectrin repeats 1 through 24 (R), rod domain hinges 1 through 4 (H), cysteine-rich domain (CR), carboxyl terminus (CT).
BMD mutations occur in a subset of in-frame exons
To examine the general properties of in-frame versus out-of-frame rod domain exons (exons 10 through 64), and in particular, the in-frame exons resulting in BMD, we calculated splice site strengths using algorithms developed by Burge and colleagues, including maximum entropy (MaxEnt), multiple dependence decomposition (MDD), first order Markov model (MM), and a weight matrix model (WMM) (Yeo and Burge, 2004). In-frame dystrophin exons have significantly weaker splice donor site (5’ss) strengths than out-of-frame exons as calculated by each algorithm (Table 1, 5’ss MaxEnt p = 0.002, MDD p=0.05, MM p=0.003, WMM p=0.004). This observation is in agreement with a prior observation finding weaker 5’ss signals in both dystrophin and utrophin in-frame versus out-of-frame exons calculated from consensus values or free energy binding parameters for U1 snRNA (Pozzoli, et al., 2002). We then compared the subset of in-frame rod domain dystrophin exons that contained at least one BMD nonsense mutation (exons 25, 27, 29, 31, 37, 38 and 40), and observed that this subset of in-frame exons has, on average, weaker splice acceptor site (3’ss) strengths compared to non-BMD in-frame exons (Table 1, 3’ss MaxEnt p = 0.02, MM p=0.02). Thus, exons with in-frame BMD nonsense mutations have the weakest aggregate splice site signals of all the DMD exons, as measured by the mean difference between aggregate MaxEnt scores for 3’ss and 5’ss strengths: 13.21 vs. 15.75 (in-frame BMD vs. in-frame DMD/IMD, p =0.02) and 13.21 vs. 16.97 (in-frame BMD vs. out-of-frame exons, p = 0.003).
Exon definition properties of the BMD exon subset
Next, we examined exonic splicing enhancer (ESE), exonic splicing silencer (ESS) and flanking intronic splicing regulatory (ISR) motifs using the mean density of motifs (Table 2). Densities were calculated for SR protein binding site motifs (ESEfinder), hexamer sequences that occur more frequently in exons with weak splice sites (Rescue-ESE), octamer sequencers computationally predicted to occur more frequently in exons (PESE octamers), exon-identity elements (EIE density), ESE motifs for Tra2b and 9G8 binding, ESS motifs predicted from pseudoexons (Sironi Silencer density) and hnRNP motifs (hnRNP A1 density). Also tested were consolidated lists of PESE and Rescue-ESE motifs (hexESE), and PESS and FAS-hex3 motifs (hexESS) that were previously described (Ke, et al., 2008). The mean density of intronic splicing regulatory (ISR) motifs (Suyama, et al., 2010) was calculated for 300 nt. intervals flanking each exon, excluding the splice acceptor and donor sites. Opposite trends are evident for differences in ESE density from in-frame vs. out-of-frame exons and in-frame BMD vs. in-frame DMD exons (Table 2). In-frame exons show increased densities of hexESE motifs versus out-of frame exons (0.26 versus 0.22 motifs/bp, p = 0.02), with similar trends for Rescue-ESE (0.20 versus 0.16 motifs/bp, p = 0.02) and PESE densities (0.10 versus 0.08 motifs/bp, p = 0.07). In contrast, in-frame exons containing BMD mutations show decreased ESE densities (motifs/bp) versus in-frame non-BMD exons: 0.23 versus 0.28 for hexESE, 0.17 versus 0.21 for Rescue-ESE hexamers and 0.07 versus 0.10 (p = 0.03) for PESE octamers. The flanking introns of in-frame exons containing BMD mutations also show increased intronic splicing regulatory densities (Table 2, ISR) versus in-frame non-BMD exons: 0.030 versus 0.021 (p = 0.02). We further tested for enrichment of the most significant ISR motifs identified by analysis of conserved pentamers flanking known cassette exons (Suyama, et al., 2010). These four pentamers (UGCAU, GCAUG, ACUAA and CUAAC) correspond to the hexamer motifs: UGCAUG, which has been identified as the binding site for the Fox-1 proteins (Jin, et al., 2003) and ACUAAC, which is the motif for RNA-binding proteins of the Quaking-like QKI and STAR family (Galarneau and Richard, 2005) and has been recently identified as specifically enriched in muscle-regulated alternative splicing (Sugnet, et al., 2006). These Fox-1 and QKI motifs are enriched in flanking introns of in-frame BMD exons (0.007 motifs/bp) versus in-frame non-BMD exons (0.004 motifs/bp, p = 0.02), and specifically enriched in the 5’ flanking introns of BMD exons (0.01 versus 0.003 motifs/bp, p = 0.02).
Table 2.
Association of ESE/ESS sites and intronic splicing regulatory motifs with exon frame and BMD phenotypes
| In-Frame | Out-of- Frame |
In-Frame (BMD)a |
In-Frame (DMD)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Meanc | SD | Mean | SD | p -value |
Mean | SD | Mean | SD | p -value |
| ESE density (hexESE hexamers) |
0.26 | 0.06 | 0.22 | 0.08 | 0.02 | 0.23 | 0.04 | 0.28 | 0.06 | 0.08 |
| ESE density (RESCUE-ESE hexamers) |
0.20 | 0.06 | 0.16 | 0.07 | 0.02 | 0.17 | 0.04 | 0.21 | 0.06 | 0.14 |
| ESE density (PESE octamers) |
0.10 | 0.03 | 0.08 | 0.03 | 0.07 | 0.07 | 0.03 | 0.10 | 0.03 | 0.03 |
| ESE density (ESEfinder SR motifs) |
0.13 | 0.04 | 0.14 | 0.04 | 0.08 | 0.12 | 0.04 | 0.13 | 0.03 | 0.29 |
| ESE density (EIE) |
0.42 | 0.07 | 0.42 | 0.08 | 0.77 | 0.41 | 0.07 | 0.43 | 0.07 | 0.59 |
| ESE density (Tra2b 9G8 motifs) |
0.15 | 0.03 | 0.14 | 0.04 | 0.39 | 0.14 | 0.03 | 0.15 | 0.03 | 0.35 |
| ESS density (hexESS hexamers) |
0.04 | 0.03 | 0.04 | 0.03 | 0.69 | 0.03 | 0.01 | 0.04 | 0.03 | 0.98 |
| ESS density (Sironi Silencer motifs) |
0.12 | 0.03 | 0.12 | 0.02 | 0.74 | 0.13 | 0.01 | 0.12 | 0.03 | 0.12 |
| ESS density (hnRNP A1 motifs) |
0.06 | 0.02 | 0.05 | 0.02 | 0.51 | 0.07 | 0.01 | 0.05 | 0.02 | 0.07 |
| ISR density (Suyama et al.) |
0.024 | 0.007 | 0.022 | 0.007 | 0.08 | 0.030 | 0.010 | 0.021 | 0.006 | 0.02 |
Rod domain in-frame exons: 25, 27, 29, 31, 37, 38, 40
Rod domain in-frame : 23, 24, 26, 28, 30, 32, 33, 34, 35, 36, 39, 41, 42
density is calculated as sites per base pair
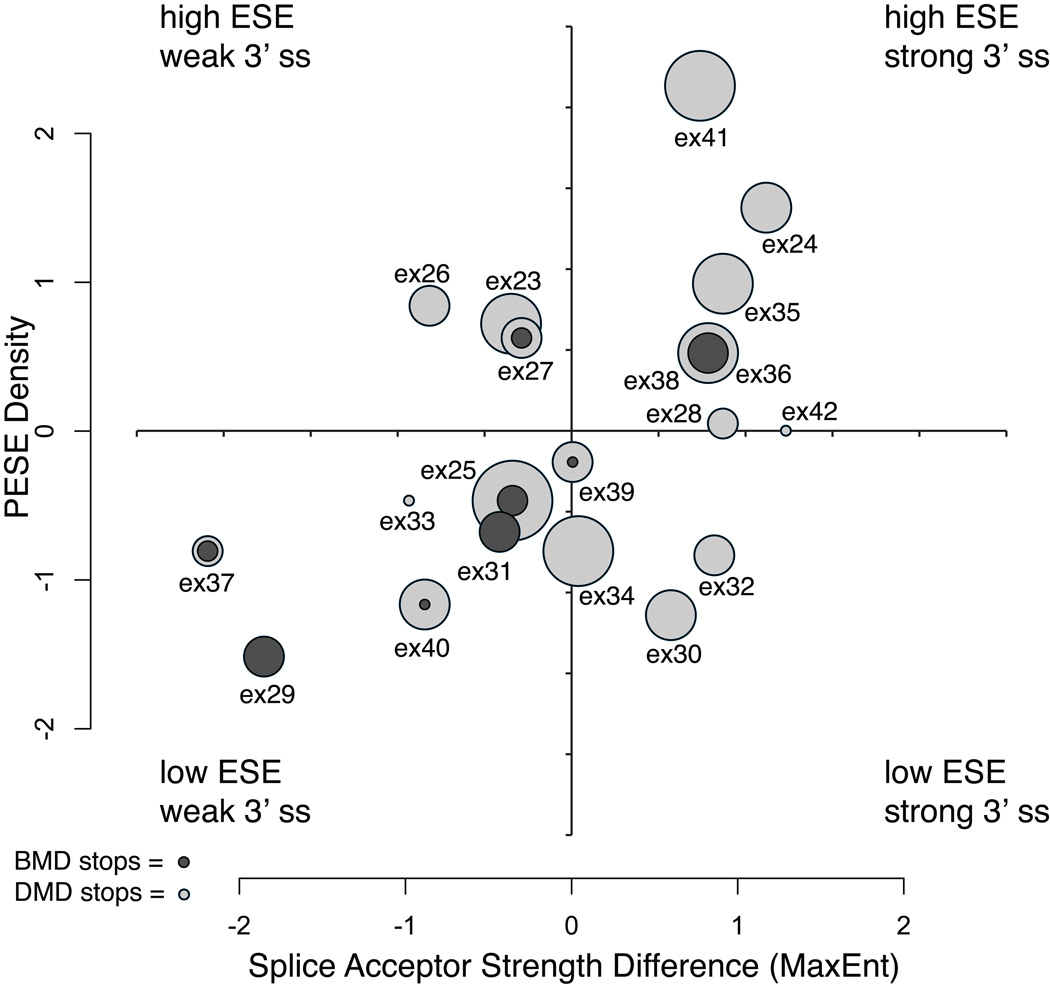
Exons 23 through 42 comprise a contiguous block of in-frame exons, encoding portions of the central rod domain. We further tested for this block of in-frame exons whether BMD exon 3’ss strengths were weaker than the 3’ ss strength in the next distal exon, as might be expected from splice acceptor site competition with the downstream 3’ exon (Nogues, et al., 2003; Habara, et al., 2008). We observed that the 3’ss MaxEnt strength was significantly less for BMD exons versus their next distal exon than for DMD exons versus their next distal exon (BMD mean MaxEnt difference = −2.19 versus DMD exon MaxEnt difference = +1.79; Mann-Whitney-Wilcoxon test, p = 0.01). We then examined the BMD/DMD mutation ratio within each exon comparing their competing 3’ss strength difference versus PESE density (Figure 2). Exon 29 has exclusively BMD mutations, and it has the lowest PESE density and a relatively weak competing 3’ss MaxEnt strength. Five out of six exons in the low PESE, weak 3’ss quadrant have BMD mutations with a combined ratio of 14 BMD to 11 DMD mutations, while only one of seven exons in the high PESE, strong 3’ss quadrant have BMD mutations with a combined ratio of 4 BMD to 27 DMD mutations (Figure 2). Exon 38 also has predominantly BMD mutations but has a relatively strong competing 3’ss strength and high PESE density, suggesting factors not captured by these two metrics, such as ISR motifs, may also contribute to the observed distribution.
Figure 2.
Exonic splicing enhancer density versus relative splice acceptor site strength for in-frame exons 23 through 42. Normalized scores (mean = 0, variance = 1) are plotted for PESE density and the relative 3’ss strength calculated as the difference between 3’ss scores of the mutation-containing exon and the next distal 3’ss. The circles represent individual exons, where the radius is equal to the total number of mutation sites observed for that exon. The nested ratio of BMD (dark gray) versus DMD (light gray) mutations is indicated for each exon.
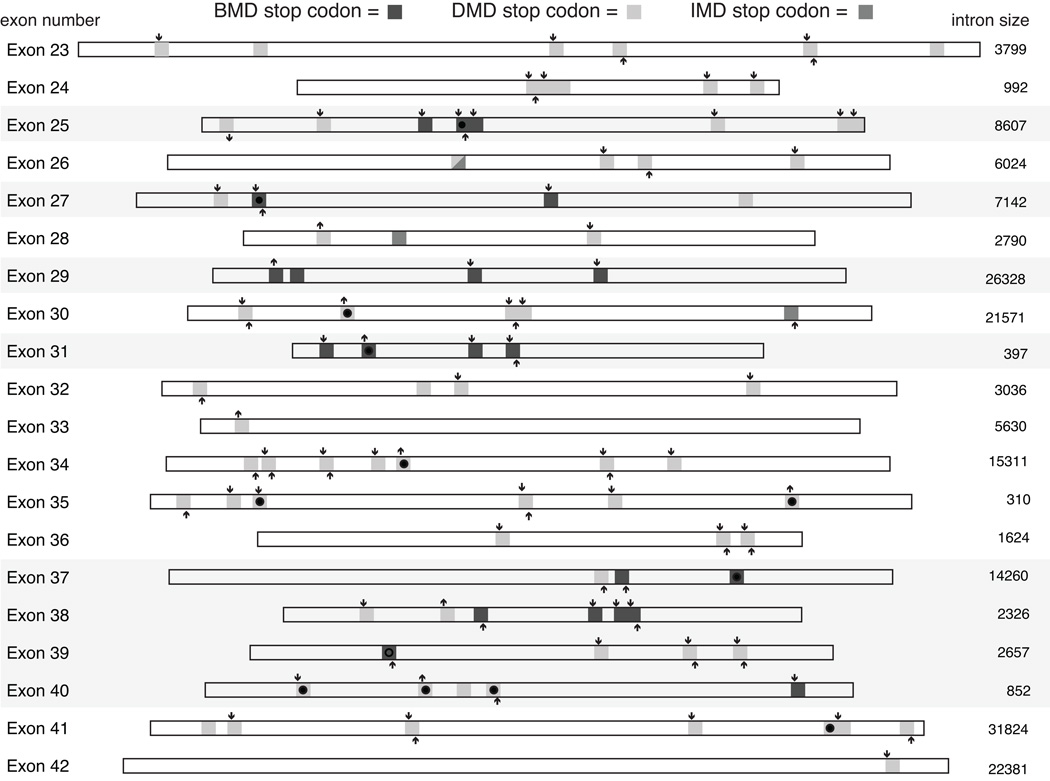
These trends suggest that the subset of in-frame exons containing BMD nonsense mutations tend to have both weaker competing splice acceptor sites, lower ESE densities and higher intronic splicing regulatory (ISR) densities than in-frame exons without these mutations. However, while a weak exon definition context may favor a BMD phenotype, not all nonsense mutations within these exons result in a BMD phenotype. Figure 3 shows the intra-exon locations of nonsense mutations found in exons 23 through 42, and the effect of individual mutations on the disruption and/or creation hexESE or hexESS motifs. Mutations in both phenotypic classes disrupt hexESE motifs and create hexESS motifs, with 1.86 versus 1.46 ESE sites disrupted and 0.76 versus 0.55 ESS sites created for BMD versus DMD mutation, respectively. The differences in either ESE disruption or ESS creation for BMD versus DMD mutations were not significant (t test for ESE disruption, p = 0.43; ESS creation, p = 0.32), confirming that ESE disruption and/or ESS creation in isolation is not sufficient to explain the observed pattern of BMD versus DMD mutations.
Figure 3.
The positions of BMD, DMD and IMD nonsense mutations for in-frame exons 23 through 42. The exon locations of BMD mutations are indicated by dark gray squares, DMD mutations by light gray squares and IMD mutations by medium gray squares. Creation or ablation of an ESE hexamer is indicated by an up or down arrow, respectively, above the mutation site, while creation or ablation of an ESS site by an up or down arrow below the site. The ESE and ESS hexamer set were the merged lists of PESE, Rescue-ESE, PESS and FAS-hex3 sequences previously described (Ke, et al., 2008). Overlap of mutations between this study and other studies (see Supp. Table S1 for list of mutations shown) is shown within each rectangle by no symbol (this study only), filled circle (this study and other studies), and an open circle (other studies only). Exon sizes are drawn to scale and flanking intron sizes are indicated in base pairs.
Within the BMD mutation-containing exons, 21 sites are associated with a BMD phenotype, while 17 sites are associated with a DMD phenotype. We therefore asked whether the effect of these mutations on ESE disruption or ESS creation is associated with a BMD versus DMD outcome within this subset of weakly defined exons. In addition to the difference in hexESE disruption or hexESS creation, we asked whether the effects of these mutations on ESE and ESS motifs predicted using a variety of algorithms in the Human Splicing Finder tool(Desmet, et al., 2009) could detect trends within these BMD exons (Supp. Table S2). In this comparison, nonsense mutations that result in BMD were not associated with ESE or ESS disruption/creation motifs (Supp. Table S2) compared to DMD nonsense mutations using the HSF classification schemes(Desmet, et al., 2009). No significant differences were found for ESE disruption using hexESE, ESE Finder, Rescue-ESE, PESE, EIE, or ESE_HSF matrices or ESS creation using hexESS, Sironi silencers, PESS or hnRNP A1 motfis. Although trends are evident from this analysis suggesting that creation of PESS and hnRNP A1 motifs may be enriched in BMD exons (Supp. Table S2), detailed bioinformatic and experimental analysis of individual mutations may be required to demonstrate specific effects of ESE or ESS disruption/creation.
Molecular confirmation of predicted exon 31 skipping
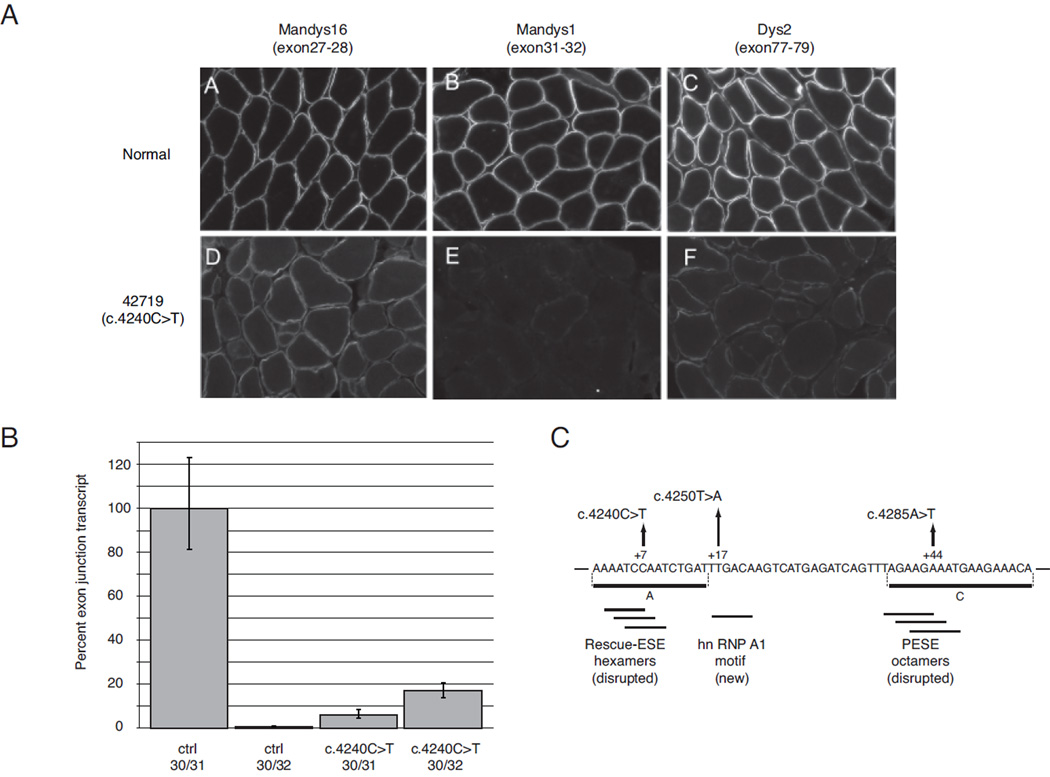
Our dataset allowed the testing of several predictions made from prior in vitro analysis of the exon 31 c.4250T>A nonsense mutation. This mutation has been extensively studied at the biochemical level (Disset, et al., 2006) where it creates a novel ESS sequence capable of binding the splicing repressor protein hnRNP A1 in vitro, as well as potentially disrupting a composite exonic regulatory element of splicing. In that study, deletion analysis of exon 31 minigenes in a mouse muscle C2C12 cell assay predicted the location of two positive splicing regulatory sequences, shown as regions A (nt +1 to +15) and C (nt +39 to + 56) in Figure 4C that flank the c.4250T>A mutation. In our study, two novel BMD nonsense mutations were found in each of these regions, and disrupted predicted ESE sites, including overlapping Rescue-ESE hexamers (c.4240C>T) and overlapping PESE octamers (c.4285A>T).
Figure 4.
RNA and protein level analysis of exon 31 skipping in BMD patient 42719 (c.4240C>T). A) Immunofluorescent analysis of tissue sections with dystrophin antibodies with epitopes encoded by exons 27–28 for Mandys16, exons 31–32 for Mandys1, and exons 77–79 for Dys2. Positive staining indicates presence of the amino acids encoded in epitope-containing exons in the synthesized dystrophin protein; images represent projections of z-series stacks of confocal sections. B) Comparative CT (ΔΔ CT) analysis of exon 30–31 junction and exon 30–32 junction levels from total RNA extracted from muscle sections. C) Schematic diagram of exon 31 shows the location of BMD mutations: p.Gln1414X (c.4240C>T), p.Leu1417X (c.4250T>A), and p.Lys1429X (c.4285A>T). Coordinates are from the intron 30 – exon 31 junction, and horizontal lines below the sequence show predicted ESE/ESS motifs. Horizontal lines A and C are putative ESE regions predicted by Disset et al. (Disset, et al., 2006).
In order to confirm the in vitro prediction that exon 31 c.4240C>T should induce skipping and account for the milder disease, we examined muscle biopsy tissue from a patient with BMD and the c.4240C>T mutation both with quantitative RT-PCR and with a panel of anti-dystrophin antibodies. Immunofluoresence levels of dystrophin (Figure 4A) show uniformly positive, albeit diminished, staining with the antibody Mandys16 (directed toward an epitope encoded by exons 27–28), as well as patchier staining with the antibody Dys2 (directed toward the C-terminus). In contrast, staining with the antibody Mandys1 (an epitope encoded by exons 31–32) is absent, suggesting the absence of peptide sequence translated from exon 31. Relative quantification of exon 31 skipping by real-time PCR is shown in Figure 4B, where a common PCR primer in exon 33 was used with either an exon 30/31 or 30/32 junction primer. The level of exon 30/32 junction product increased 24-fold in muscle from the c.4240C>T patient relative to a normal control, consistent with mutation-induced exon 31 skipping. The exon 30/31 junction product is also substantially decreased relative to normal levels, perhaps indicative of nonsense-mediated decay of the mRNA transcript containing the nonsense mutation.
DISCUSSION
The restricted number of exons in our survey in which nonsense mutations were found to be associated with BMD suggests that critical ESE disruption or ESS creation is not the sole determinant of nonsense mutation-associated exon skipping, as previous ESE-disruption models have postulated (Cartegni and Krainer, 2002; Liu, et al., 2001; Pagani, et al., 2003; Shiga, et al., 1997; Tran, et al., 2006; Zatkova, et al., 2004). Rather, the effects of these single point mutations are dependent on weak intrinsic exon definition elements. Previous reports have identified BMD nonsense mutations associated with exon skipping in the DMD muscle isoform for exon 25 (c.3328G>T) (Fajkusova, et al., 2001), exon 27 (c.3631G>T) (Shiga, et al., 1997), exon 29 (c.3940C>T) (Ginjaar, et al., 2000), exon 31 (c.4250T>A) (Disset, et al., 2006), exon 37 (c.5260G>T) (Hamed, et al., 2005), exon 38 (c.5404C>T) (Janssen, et al., 2005) and exon 72 (c.10304C>A) (Melis, et al., 1998). In this study, only two mutations (exon 31 c.4250T>A and exon 38 c.5404C>T) were found at sites previously described as BMD nonsense mutations despite the large number of patients surveyed, suggesting that target saturation for this class of mutation has not been reached. However, 10 out of 13 BMD nonsense mutations described here occurred in a subset of in-frame exons (25, 27, 29, 31, 37 and 38) that substantially overlap the exons containing these previously described mutations. Additional support for this subset of in-frame exons comes from a recent survey of point mutations described in the UMD-DMD database, which included BMD phenotypes for patients with nonsense mutations in exons 25, 29, 31, 37, 38 and 39 (Tuffery-Giraud, et al., 2009). We observed that patients with nonsense mutations in exons 29 and 31 exclusively have a BMD phenotype, suggesting that these exons have properties that makes them the most prone to nonsense mutation-induced exon skipping. Our observations therefore support a model that prefaces the critical splicing enhancer-disruption / suppressor-creation model with a requirement for multiple cis-acting combinatorial signals leading to intrinsically weak exon definition.
Our data suggest that one key aspect of this exon definition property is a weak splice acceptor (3’ss) site and low exonic splicing enhancer density. The observation that in-frame dystrophin exons have significantly weaker splice donor (5’ss) sites than out-of-frame exons has been made previously using related 5’ss metrics, and appears to be an evolutionarily conserved property of both vertebrate dystrophin and utrophin exons (Pozzoli, et al., 2002). Our analysis indicates that this evolutionary trend in DMD exon definition may be offset by a concomitant increase in ESE density for in-frame exons. Compensation by ESE creation through synonymous substitutions may be one mechanism to offset the observed weakened splice donor strength. Recent genome-wide analysis with human, chimp and macaque sequences comparing the rate of synonymous change in constitutive exons versus intronic change, found evidence that synonymous substitutions tend to create ESE and disrupt ESS motifs (Ke, et al., 2008). They further postulated that positive selection for ESE creation and ESS disruption reflects compensatory evolution working to maintain constitutive exon definition. Using simulated random mutation within human exons, that study also observed that randomly placed mutations tend to disrupt ESE and create ESS motifs. Our observation that both BMD and DMD mutations also tend to disrupt ESE and create ESS motifs, with only a subtle enrichment for ESS creation observed in the BMD class, is in agreement with their simulated data for random mutation. This suggests that without any evolutionary constraint to maintain splicing efficiency, ESE disruption and ESS creation is an expected, commonly found result for point mutations in constitutive human exons.
With disruption and creation of ESE/ESS motifs as the expected outcome, the focus remains on why only a subset of point mutations lead to high level restoration of dystrophin in Becker patients. It is thought that residual levels of ~30% dystrophin are required to achieve the mild phenotype observed in BMD patients (Neri, et al., 2007), and the mRNA level of exon skipping from our single patient with the exon 31 c.4240C>T mutation supports this threshold of dystrophin production. The c.4240C>T and the exon 31 c.4284T>A mutation confirmed a prediction based on in vitro deletion analysis that these regions contain critical ESE motifs that bind Tra2b and SR proteins, and disruption of these sites should result in exon skipping and BMD phenotypes (Disset, et al., 2006). However, for both exon 31 and exon 29, our data also supports that all observed point mutations result in BMD phenotypes. In exon 38 from a BMD patient, a four base deletion has been shown to induce high levels of exon skipping using a transient in vivo assay(Tran, et al., 2006). Further definition of the disrupted site by site-directed mutagenesis suggested that a single point mutation within the deletion could result in the same degree of exon skipping and supported a model of critical ESE disruption. Although exon 38 is not defined as particularly weak using the relative 3’ss and PESE density metrics, we observed point mutations along the length of exon 38 that result in BMD phenotypes.
We also note that exons 37 and 38 have the highest densities (0.02 and 0.017 motifs/bp) of the Fox-1 and QKI binding motifs (UGCAU, GCAUG, ACUAA and CUAAC) observed in the 5’ flanking introns of exon 23 to exon 42. The Fox-1 binding sequence, UGCAUG, is a key element for the regulation of alternative splicing (Brudno, et al., 2001; Lim and Sharp, 1998; Modafferi and Black, 1997) and the Fox-1 RNA recognition motif is conserved from C. elegans to vertebrates (Jin, et al., 2003). Fox-1 binding at GCAUG motifs in the 5’ flanking intron of exon 9 from the human mitochondrial ATP synthase g-subunit gene induces muscle-specific exon skipping(Jin, et al., 2003). The QKI and STAR family of RNA-binding proteins that recognize ACUAAC motifs are also highly conserved alternative splicing regulators (Ohno, et al., 2008). Recent computational analysis indicates that ACUAAC motifs are prominently involved in cassette exon inclusion in mouse muscle transcripts (Barash, et al., 2010). These results suggest that ESE disruption or ESS creation is not sufficient, and suggests that the prerequisite factor influencing the degree of exon skipping is a combination of intrinsic weak exon definition elements. Our results indicate that intronic motifs for RNA binding proteins previously characterized as controlling developmental or tissue-specific cassette exon splicing are also important for mutation-induced exon skipping of constitutive exons in human skeletal muscle.
The 24-fold increase in the exon 30/32 junction fragment observed by qRT-PCR analysis from our patient with the exon 31 c.4240C>T mutation suggests that a large increase over basal level exon skipping is caused by this single point mutation. In vitro splicing assays for internal exon skipping support a general finding that exon inclusion responds incrementally, and in a linearly fashion, to the addition of ESE elements into artificial transcripts (Zhang, et al., 2009). However, the high variability for exon inclusion seen among exons having the same proportion of enhancers in these in vitro experiments and the magnitude of the in vivo effects of single point mutations contradicts a simple model in which the presence of an enhancer/silencer sequence adds to the probability of binding an activator/inhibitor protein which in turn leads to a proportional increase/decrease in splicing. Our observations argue against an Achilles’ heel model of critical ESE disruption/ESS creation, and suggest that a ‘house of cards’ model may better describe the balancing of multiple exon definition elements that ultimately lead to the magnitude of exon skipping observed in BMD patients.
Current views of co-transcriptional splicing of exons favor a kinetic view that suggests splicing is completed prior to release of the nascent mRNA from the chromatin template (Pandya-Jones and Black, 2009). Recent in vivo measurements of transcription and splicing rates of utrophin mRNA suggests that transcription is fast (3.8 kb/min), and that splicing is rapidly completed within minutes after the synthesis of adjacent exons (Singh and Padgett, 2009). For the dystrophin gene, this suggests that the decision to include an exon containing a nonsense-mutation may also involve kinetic interactions between the transcribing RNA polymerase II complex and the assembly of spliceosomal components on nascent exons prior to their juxtaposition for splicing. It is interesting to note that the 3’ introns flanking exons 29 and 31, the two exons most prone to BMD nonsense mutations, are drastically different in size: 26.3 kb versus 0.4 kb. The kinetic view postulates that exon 29 has an ~6 min. advantage over exon 31 in attempting to preempt an exon skipping event. Analysis of the rate and natural order of intron removal for these two exons, and for exons 23 through 42 in general, may shed light on the kinetics of exon inclusion versus skipping and the consequences of point mutations.
Our experimental confirmation is limited due to a limited supply of archived patient tissue (no other biopsies were available), and further mRNA analysis of patient tissue will be required. Our results do not explain the few (n=4) BMD patients with nonsense mutations in out-of-frame flanking contexts (exons 43, 51, 56 and 76). The exon 51 c.7401_7402delGGinsAT mutation has been previously shown to create a cryptic splice site (Winnard, et al., 1992). The exon 76 c.10888C>T mutation may be an example of a truncating mutation beyond exon 70 that results in mild phenotypes if the transcript escapes nonsense-mediated decay and produces a truncated dystrophin capable of rescuing the muscle phenotype (Kerr, et al., 2001). In the other cases, it is possible that multiple exon skipping occurs or alternate splice sites are activated within the exon, such that the reading frame of the mRNA is ultimately preserved. This hypothesis will also require analysis of muscle mRNA, which is not presently available. Meanwhile, the intrinsic “skippability” of an exon, based upon the exon definition metrics, may be of relevance to the design of oligonucleotides for exon-skipping therapies. Previous correlations of the efficiency of antisense-induced exon skipping of DMD exons has found that the strength of the splice acceptor sites of poorly skippable exons were significantly higher than those of exons that could be skipped at medium or high efficiency (Aartsma-Rus and van Ommen, 2007; Wilton, et al., 2007). Further studies of the basal and mutation-induced level of exon skipping may help model the kinetics of in vivo splicing reactions on an actively transcribing DMD gene. Recent genome-wide analysis of cassette exons that are alternatively spliced in tissue-dependent and independent patterns suggest that large data sets can be used to define multiple distinct cis-acting elements, that when combined together, form a probabilistic model for prediction of a given splice pattern (Barash, et al., 2010). Our study suggests that similar combinations of cis-acting elements define constitutive exons, and the subset that we have defined as susceptible to mutation-induced exon skipping may be using similar features that distinguish cassette exons. The kinetic effect of co-transcriptional splicing and the effect of mRNA secondary structure on the binding of spliceosomal components or RNA binding proteins are not integrated into our observations. Further analysis of basal-level exon skipping in normal and diseased muscle samples will be required to address these parameters.
The observation that weakly defined DMD exons are susceptible to point mutation-induced exon skipping may have general implications for predicting the functional consequences of rare exonic sequence variants. Exonic variants that result in non-synonymous substitutions are often classified based on their computationally predicted effect at the protein level, and synonymous variants are often considered neutral. Our results suggest that analyses such as assigning functional consequences and clinical significance to unknown point mutations or pooling rare exonic variants to gain statistical power in tests of association of rare variants with common disease would benefit from a predictive ‘splicing code’ for exonic point mutations. Aside from predictive capabilities, a splicing code for exonic point mutations may eventually help refine patient candidacy for clinical trials of mutation suppression agents. Our study benefited from the relationship of the residual level of truncated dystrophin and the amelioration of the muscle phenotype. Additional approaches, such as high-throughput analysis of exon skipping in patient-derived tissues or cell lines, will be required to extend these observations to a majority of exons. Our results do not suggest that definitive prognostic information can currently be provided to patients and their families based upon which exon contains a nonsense mutation. However, they reinforce the notion that genotype alone cannot predict phenotype, and that clinicians should be particularly cautious of predicting phenotype with nonsense mutations found in an in-frame intronic flanking context.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank all referring physicians; thank A. Bringard and J. Tyce for administrative assistance; and to acknowledge the technical assistance of L. Zhao, T. Tuohy, O. Gurvich, B. Duval, C. Hamil, M. Mahmoud, and A. Aoyagi. The authors also gratefully acknowledge the gift of the monoclonal antibodies MANDYS1 and MANDYS16 from Prof. Glenn Morris (Wolfson Centre for Inherited Neuromuscular Disease, Oswestry, UK). This work is supported by the National Institute of Neurologic Diseases and Stroke (R01 NS043264 [KMF, MTH, RBW]; the National Center for Research Resources (M01-RR00064, to the University of Utah, Dr. L. Betz, P.I).
United Dystrophinopathy Project: Other Investigators and Members
The University of Utah, Salt Lake City, Utah: Mark Bromberg, MD, PhD; Kathy Swoboda, MD; Eduard Gappmaier, DPT; Lynne Kerr, MD, PhD; Kim Hart, MS; Cybil Moural, MS; Kate Hak, BS; Lahdan Heidarian, BS.
Nationwide Medical Center, Columbus, Ohio: Laurent Viollet, PhD; Chelsea Rankin, BS; Cheryl Wall, RN; Wendy King, PT; Susan Gailey, MS
Washington University, St. Louis, Missouri: Glenn Lopate, MD; Paul Golumbek MD, PhD; Jeanine Schierbecker MHS, PT; Betsy Malkus MHS, PT; Renee Renna, RN; and Catherine Siener MHS, PT
University of Iowa, Iowa City, Iowa: Carrie Stephan, RN; Karla Laubenthal, PT, MS, PCS; Kris Baldwin, LPT
Children’s Hospital/University of Pennsylvania, Philadelphia, Pennsylvania: Livija Medne, MS; Allan M. Glanzman, PT, DPT, PCS, ATP; Jean Flickinger, RPT
Cincinnati Children’s Hospital, Cincinnati, Ohio: Paula Morehart, RN; Amy Meyer, PT
University of Minnesota, Minneapolis, Minnesota: Cameron E. Naughton; Marcia Margolis, PT, ATP
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
REFERENCES
- Aartsma-Rus A, van Ommen GJ. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13(10):1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465(7294):53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29(11):2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Dent KM, Dunn DM, von Niederhausern AC, Aoyagi AT, Kerr L, Bromberg MB, Hart KJ, Tuohy T, White S, den Dunnen JT and others. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am J Med Genet A. 2005;134(3):295–298. doi: 10.1002/ajmg.a.30617. [DOI] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet. 2006;15(6):999–1013. doi: 10.1093/hmg/ddl015. [DOI] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32(Web Server issue):W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajkusova L, Lukas Z, Tvrdikova M, Kuhrova V, Hajek J, Fajkus J. Novel dystrophin mutations revealed by analysis of dystrophin mRNA: alternative splicing suppresses the phenotypic effect of a nonsense mutation. Neuromuscul Disord. 2001;11(2):133–138. doi: 10.1016/s0960-8966(00)00169-3. [DOI] [PubMed] [Google Scholar]
- Flanigan KM, Dunn DM, von Niederhausern A, Howard MT, Mendell J, Connolly A, Saunders C, Modrcin A, Dasouki M, Comi GP and others. DMD Trp3X nonsense mutation associated with a founder effect in North American families with mild Becker muscular dystrophy. Neuromuscul Disord. 2009a;19(11):743–748. doi: 10.1016/j.nmd.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM and others. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009b;30(12):1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72(4):931–939. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12(8):691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- Ginjaar IB, Kneppers AL, v d Meulen JD, Anderson LV, Bremmer-Bout M, van Deutekom JC, Weegenaar J, den Dunnen JT, Bakker E. Dystrophin nonsense mutation induces different levels of exon 29 skipping and leads to variable phenotypes within one BMD family. Eur J Hum Genet. 2000;8(10):793–796. doi: 10.1038/sj.ejhg.5200535. [DOI] [PubMed] [Google Scholar]
- Gurvich OL, Maiti B, Weiss RB, Aggarwal G, Howard MT, Flanigan KM. DMD exon 1 truncating point mutations: amelioration of phenotype by alternative translation initiation in exon 6. Hum Mutat. 2009;30(4):633–640. doi: 10.1002/humu.20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habara Y, Doshita M, Hirozawa S, Yokono Y, Yagi M, Takeshima Y, Matsuo M. A strong exonic splicing enhancer in dystrophin exon 19 achieve proper splicing without an upstream polypyrimidine tract. J Biochem. 2008;143(3):303–310. doi: 10.1093/jb/mvm227. [DOI] [PubMed] [Google Scholar]
- Hamed S, Sutherland-Smith A, Gorospe J, Kendrick-Jones J, Hoffman E. DNA sequence analysis for structure/function and mutation studies in Becker muscular dystrophy. Clin Genet. 2005;68(1):69–79. doi: 10.1111/j.1399-0004.2005.00455.x. [DOI] [PubMed] [Google Scholar]
- Howard MT, Anderson CB, Fass U, Khatri S, Gesteland RF, Atkins JF, Flanigan KM. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol. 2004;55(3):422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005;6(1):29–35. doi: 10.1007/s10048-004-0204-1. [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22(4):905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Zhang XH, Chasin LA. Positive selection acting on splicing motifs reflects compensatory evolution. Genome Res. 2008;18(4):533–543. doi: 10.1101/gr.070268.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TP, Sewry CA, Robb SA, Roberts RG. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet. 2001;109(4):402–407. doi: 10.1007/s004390100598. [DOI] [PubMed] [Google Scholar]
- Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18(7):3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27(1):55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- Melis MA, Muntoni F, Cau M, Loi D, Puddu A, Boccone L, Mateddu A, Cianchetti C, Cao A. Novel nonsense mutation (C-->A nt 10512) in exon 72 of dystrophin gene leading to exon skipping in a patient with a mild dystrophinopathy. (Suppl 1).Hum Mutat. 1998:S137–S138. doi: 10.1002/humu.1380110146. [DOI] [PubMed] [Google Scholar]
- Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17(11):6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2(1):90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Neri M, Torelli S, Brown S, Ugo I, Sabatelli P, Merlini L, Spitali P, Rimessi P, Gualandi F, Sewry C and others. Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human. Neuromuscul Disord. 2007;17(11–12):913–918. doi: 10.1016/j.nmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Nguyen TM, Morris GE. Use of epitope libraries to identify exon-specific monoclonal antibodies for characterization of altered dystrophins in muscular dystrophy. Am J Hum Genet. 1993;52(6):1057–1066. [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Takeshima Y, Zhang Z, Habara Y, Tran TH, Yagi M, Matsuo M. Dystrophin nonsense mutations can generate alternative rescue transcripts in lymphocytes. Ann Hum Genet. 2008;72(Pt 6):717–724. doi: 10.1111/j.1469-1809.2008.00468.x. [DOI] [PubMed] [Google Scholar]
- Nogues G, Munoz MJ, Kornblihtt AR. Influence of polymerase II processivity on alternative splicing depends on splice site strength. J Biol Chem. 2003;278(52):52166–52171. doi: 10.1074/jbc.M309156200. [DOI] [PubMed] [Google Scholar]
- Ohno G, Hagiwara M, Kuroyanagi H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 2008;22(3):360–374. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Stuani C, Tzetis M, Kanavakis E, Efthymiadou A, Doudounakis S, Casals T, Baralle FE. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet. 2003;12(10):1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15(10):1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popplewell LJ, Adkin C, Arechavala-Gomeza V, Aartsma-Rus A, de Winter CL, Wilton SD, Morgan JE, Muntoni F, Graham IR, Dickson G. Comparative analysis of antisense oligonucleotide sequences targeting exon 53 of the human DMD gene: Implications for future clinical trials. Neuromuscul Disord. 2010;20(2):102–110. doi: 10.1016/j.nmd.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Pozzoli U, Sironi M, Cagliani R, Comi GP, Bardoni A, Bresolin N. Comparative analysis of the human dystrophin and utrophin gene structures. Genetics. 2002;160(2):793–798. doi: 10.1093/genetics/160.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Roberts RG, Barby TF, Manners E, Bobrow M, Bentley DR. Direct detection of dystrophin gene rearrangements by analysis of dystrophin mRNA in peripheral blood lymphocytes. Am J Hum Genet. 1991;49(2):298–310. [PMC free article] [PubMed] [Google Scholar]
- Shiga N, Takeshima Y, Sakamoto H, Inoue K, Yokota Y, Yokoyama M, Matsuo M. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest. 1997;100(9):2204–2210. doi: 10.1172/JCI119757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16(11):1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, Giorda R, Pozzoli U. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32(5):1783–1791. doi: 10.1093/nar/gkh341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15(16):2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D and others. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2(1):e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Harrington ED, Vinokourova S, von Knebel Doeberitz M, Ohara O, Bork P. A network of conserved co-occurring motifs for the regulation of alternative splicing. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran VK, Takeshima Y, Zhang Z, Yagi M, Nishiyama A, Habara Y, Matsuo M. Splicing analysis disclosed a determinant single nucleotide for exon skipping caused by a novel intraexonic four-nucleotide deletion in the dystrophin gene. J Med Genet. 2006;43(12):924–930. doi: 10.1136/jmg.2006.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L, Moizard MP, Bernard R, Cossee M, Boisseau P and others. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Saquet C, Thorel D, Disset A, Rivier F, Malcolm S, Claustres M. Mutation spectrum leading to an attenuated phenotype in dystrophinopathies. Eur J Hum Genet. 2005;13(12):1254–1260. doi: 10.1038/sj.ejhg.5201478. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM and others. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Fall AM, Harding PL, McClorey G, Coleman C, Fletcher S. Antisense oligonucleotide-induced exon skipping across the human dystrophin gene transcript. Mol Ther. 2007;15(7):1288–1296. doi: 10.1038/sj.mt.6300095. [DOI] [PubMed] [Google Scholar]
- Winnard AV, Jia-Hsu Y, Gibbs RA, Mendell JR, Burghes AH. Identification of a 2 base pair nonsense mutation causing a cryptic splice site in a DMD patient. Hum Mol Genet. 1992;1(8):645–646. doi: 10.1093/hmg/1.8.645. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2–3):377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zatkova A, Messiaen L, Vandenbroucke I, Wieser R, Fonatsch C, Krainer AR, Wimmer K. Disruption of exonic splicing enhancer elements is the principal cause of exon skipping associated with seven nonsense or missense alleles of NF1. Hum Mutat. 2004;24(6):491–501. doi: 10.1002/humu.20103. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Arias MA, Ke S, Chasin LA. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009;15(3):367–376. doi: 10.1261/rna.1498509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18(11):1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.