Abstract
The amyloid fold is usually considered a result of protein misfolding. However, a number of studies have recently shown that the amyloid structure is also used in nature for functional purposes. CsgA is the major subunit of Escherichia coli curli, one of the most well-characterized functional amyloids. Here we show, using a highly efficient approach to prepare monomeric CsgA, that in vitro fibrillation of CsgA occurs under a wide variety of environmental conditions and that the resulting fibrils exhibit similar structural features. This highlights how fibrillation is “hardwired” into amyloid that has evolved for structural purposes in a fluctuating extracellular environment and represents a clear contrast to disease-related amyloid formation. Furthermore, we show that CsgA polymerization in vitro is preceded by the formation of thin needlelike protofibrils followed by aggregation of the amyloid fibrils.
Amyloid fibrils are commonly associated with devastating diseases such as Parkinson’s and Alzheimer’s diseases, type II diabetes, and transmissible spongiform encephalopathies such as Creutzfeld-Jacob disease.1–4 Amyloid formation is considered to be the result of misfolding followed by spontaneous polymerization of otherwise soluble protein into biochemically and structurally similar fibrils with a characteristic cross-β structure in which the β-strands are arranged perpendicular to the fiber axis.5–9 Amyloid fibrils examined by TEM or AFM are typically found to be composed of intertwined filaments. The fibrils are of indeterminate length and have a width on the order of 7–20 nm, varying with fibril morphology.4,6
The fact that many structurally unrelated proteins can form amyloid fibrils indicates that the general amyloid fold is governed by protein backbone interactions, while the kinetics and thermodynamics of fibrillation may be modulated by side chain interactions.6,10–13 The β-strands underlying the amyloid structure can arrange in numerous different ways.14 Thus, a given amyloid structure does not necessarily represent a global energetic minimum in the folding landscape as is the case for the native structure of simple globular proteins15,16 but rather represents the most accessible conformational amyloid state under the given experimental or physiological conditions.17–19 Variations in the molecular structures of amyloid fibrils made of the same polypeptide chain are thought to be responsible for the existence of multiple strains of mammalian prion diseases and yeast prion phenotypes.20–25 Furthermore, such polymorphism may produce variations in the toxicity or patterns of deposition of amyloid fibrils in Alzheimer’s and other amyloid diseases.26,27
Despite its common association with diseases, the amyloid structure is also used for beneficial purposes in nature. The curli system in Escherichia coli is one of the best studied systems of functional amyloids. Curli fimbriae are part of the complex extracellular matrix that is essential for biofilm formation, host cell adhesion, and invasion, and they are reported to be important stimulants of the host inflammatory response.28–34
A highly regulated pathway involving two divergently expressed operons is required for curli biogenesis. The csgBA operon encodes curli subunits CsgA and CsgB. csgDEFG encodes proteins essential in the regulation and transportation of CsgA and CsgB to the cell surface.31,35–37 The major curli subunit, CsgA, is secreted to the cell surface as a soluble unstructured protein.38,39 Here, it interacts with membrane-bound CsgB, which nucleates the fibrillation of CsgA. After this initial step, amyloid fibrils are predicted to grow by addition of CsgA to the growing tip.37,39
CsgA and CsgB both contain five imperfect repeats with highly conserved glutamine and asparagine residues that are important for amyloid formation.40 Each repeating unit is predicted to form a strand–loop–strand motif that closely resembles the cross-β fold described for many disease-related amyloids.8,41 It seems reasonable to hypothesize that the presence of specific side chain interactions in and between curli subunits as well as the presence of five repeating units in one protein sequence guides the fibrillation of CsgA toward a single well-defined amyloid form. Here we have evaluated the robustness of the fibrillation process by checking the sensitivity of the final CsgA amyloid structure toward various environmental conditions in vitro. We have investigated the structural features using a broad range of biophysical tools, including Fourier-transform infrared spectroscopy, circular dichroism, X-ray fiber diffraction, and transmission electron microscopy, and conclude that CsgA forms fibrils over a broad swathe of physiologically relevant conditions. This robustness toward changing environmental conditions is consistent with evolutionarily optimized fibrillation properties.
MATERIALS AND METHODS
E. coli MG1665 mutant SM2258,42 used for curli production, was kindly provided by S. Molin (BioCentrum-DTU, Lyngby, Denmark).
Purification of Curli
Curli were isolated by a protocol modified from Collinson et al.43 as described previously by Dueholm et al.44
Purification of CsgA from Curli
Curli were lyophilized, dissolved in 98% formic acid, and lyophilized again. The depolymerized curli were then dissolved in anion exchange (AIEX) binding buffer [8 M urea, 1 mM DTT, and 20 mM bistris (pH 6.5)] and filtered through a 0.22 µm filter. The sample was subjected to ion exchange on a 5 mL HiTrap Q HP column equilibrated with AIEX binding buffer using a flow rate of 5 mL/min and eluted by stepwise increasing the concentration of NaCl (0, 100, 200, 500, and 1000 mM). Fractions containing CsgA were pooled and dialyzed against deionized water for 2 days. This resulted in precipitation of CsgA. The precipitate was collected, resuspended in 98% formic acid, and lyophilized. The lyophilized material was suspended in GdmCl buffer [6 M GdmCl and 20 mM NaPO4 (pH 7.0)]. CsgA monomers were isolated from aggregated material and dimers by gel filtration in GdmCl buffer using a Superdex 200 10/300 column (GE Healthcare). CsgA was desalted into deionized water using a 5 mL HiTrap desalting column right before the protein was used for fibrillation studies. This thorough procedure was required to obtain reproducible fibrillation curves in the fibrillation assay.
Fibrillation Assay
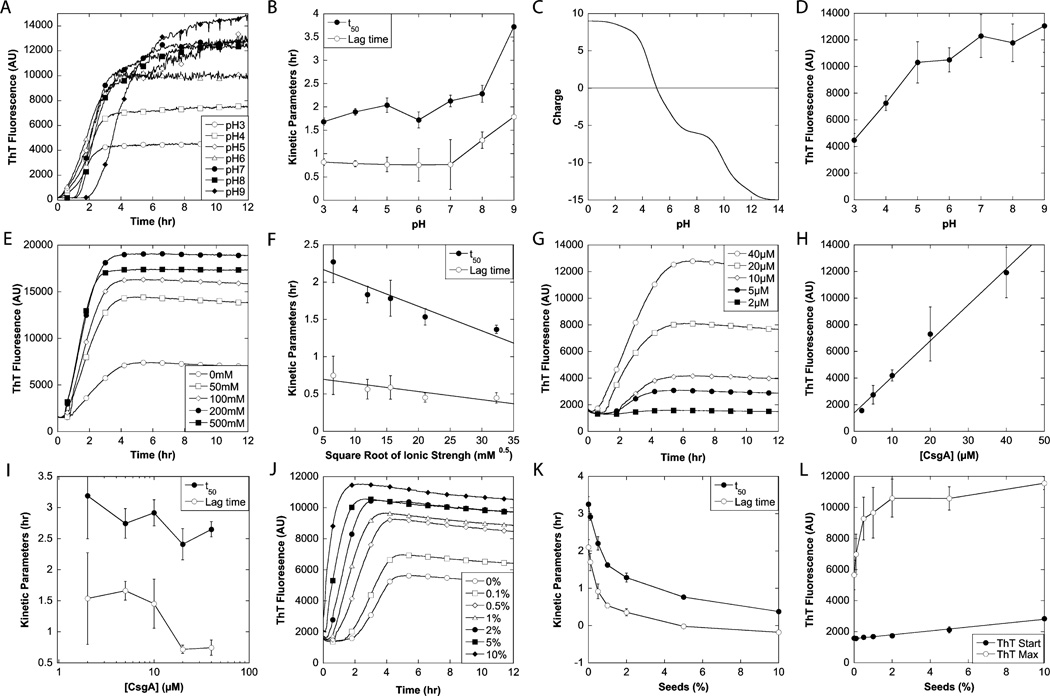
Gel-filtered CsgA in 6 M GdmCl was desalted using a HiTrap desalting column equilibrated with deionized water. Protein concentrations were estimated by the UV absorbance of the peptide bond using the empirical formula of Waddell.45 The protein was diluted in deionized water to 2 times the final concentration and mixed with a premade 2× cocktail of buffer, NaCl or seeds, and 80 µM ThT; 200 µL samples were loaded in a 96-well plate. Immediately afterward, the plate was transferred to a Tecan GENios Pro plate reader and fluorescence was measured using excitation/emission at 448/485 nm (ThT) or 360/485 nm (ANS), and a gain of 60. Measurements were taken by bottom reads every 2 min, and shaking for 30 s (orbital, amplitude of 2.5 mm) was applied between the reads. Reads was integrated for 40 µs. Six fibrillation curves were averaged to reduce the signal-to-noise ratio. For seed preparation, 5 mL of a 1 mg/mL suspension of native curli fibrils was sonicated using a rod sonicator at its maximal effect for 30 s. The following buffers were used: 20 mM citric acid-NaOH for pH 3–5, 20 mM histidine-HCl for pH 6, 20 mM phosphoric acid-NaOH for pH 7, 20 mM Tris-HCl for pH 8, and 20 mM glycine-HCl for pH 9. In all cases, the ionic strength was adjusted to 50 mM by the addition of NaCl. Note that we do not include error bars for the various time profiles in the figures, as this will make it impossible to gain a clear overview of the data. Rather, the quality of the data may be assessed through the error bars of the kinetic parameters in different panels of Figure 1. All these values are based on kinetic parameters from individual fibrillation curves and therefore represent true errors.
Figure 1.
Effect of environmental conditions on the fibrillation of CsgA. (A, E, G, and J) Fibrillation of CsgA followed by ThT at various pH values (A), NaCl concentrations (E), protein concentrations (G), and CsgA fibril seed concentrations (J). The standard conditions for the fibrillations were 20 µM CsgA in 20 mM sodium phosphate (pH 7) without addition of NaCl or seeds. (B, F, I, and K) Effect of pH (B), ionic strength (F), protein concentration (I), and CsgA fibril seeds (concentrations in weight percentage) (K) on the kinetic parameters t50 and lag time. (C) Theoretical overall charge of mature CsgA as a function of pH calculated using CLC DNA Workbench version 5.7.1. (D and E) Effect of pH (C) and protein concentration (H) on the ThT maximum. (L) Effect of seeds on ThT start level and maximum.
Circular Dichroism
Circular dichroism (CD) spectra from 250 to 190 nm were recorded on a Jasco J-810 spectropolarimeter using 0.2 nm steps, a scan speed of 50 nm/min, a bandwidth of 3 nm, and a response time of 2 s. A light path of 0.1 mm was used, and the temperature was kept constant at 20 °C with a thermostatically controlled cell holder. The protein concentration was 0.2 mg/mL, and all spectra were baseline corrected with respect to buffer. To improve the signal-to-noise ratio, five scans were averaged for each sample. The results were expressed as mean residue ellipticity (MRE). All samples except those collected during the fibrillation were sonicated briefly to minimize fibril precipitation.
Fourier-Transform Infrared Spectroscopy
Fourier-transform infrared spectroscopy (FTIR) was conducted using a Tensor 27 (Bruker) FTIR spectrophotometer equipped with a deuterated triglycine sulfate (DTGS) midinfrared detector and a Golden Gate single-reflection diamond attenuated total reflectance (ATR) cell (Specac). Approximately 4 µg of protein was dried on the ATR crystal using dry nitrogen. Spectra were recorded from 4000 to 1000 cm−1 using a nominal resolution of 2 cm−1 and 64 accumulations. Fourier self-deconvolution of the spectra in the amide I region was performed using a built-in apodization function with a deconvolution factor of 2, a noise reduction factor of 0.5, and a Lorentzian line shape in the OPUS 5.5 system (Bruker). Identification of the different components of the amide I region was performed by secondderivative analysis in the OPUS 5.5 system.
Transmission Electron Microscopy
A fibril solution (10–20 µL) was applied to the nickel grid for 30 s. The grids were washed with one drop of glass-distilled water, stained with three drops of 1% phosphotungstic acid (pH 6.9), and blotted dry. Electron microscopy was conducted using a JEOL 1010 TEM instrument at 60 kW. Images were taken using an Olympus KeenView camera. For size determination, a standard grid size replica plate (2160 lines/mm) was used.46
X-ray Fiber Diffraction
Fiber diffraction specimens were prepared on a stretch frame using a suspension of approximately 5 mg/mL fibrillated CsgA and dried at room temperature. Data were collected in house using a Cu Kα rotating-anode source (FR591 Enraf Nonius, Delft, The Netherlands; wavelength of 1.5418 Å), equipped with a MAR Research 345 image plate X-ray detector (345 mm diameter). The sample–detector distance was 400 mm with exposure times of 20–60 min. The images were evaluated using Fiber Fix for Windows version 1.3.1. Bragg distances were measured as the peak maxima from radial-averaged intensity plots.
SDS–PAGE
SDS–PAGE was performed according to the method of Laemmli with modifications by Ames.47,48 Samples were mixed with SDS–PAGE loading buffer with DTT, boiled for 5 min, and analyzed on a 15% SDS–PAGE gel. The concentration of non-amyloid CsgA was estimated from the CsgA monomer band intensity using the ImageJ gel analysis tool (http://rsbweb.nih.gov/ij/). We did not detect significant amounts of CsgA dimer on the gel.
RESULTS
Effects of pH and Ionic Strength
The ability of CsgA to form amyloid fibrils was investigated under a wide range of conditions using thioflavin T (ThT) fluorescence as a probe of amyloid formation. For such experiments, it is essential to employ highly pure CsgA preparations devoid of preformed nuclei and nucleator protein CsgB because artifacts may otherwise arise from seeding reactions. Highly pure monomeric preparations of the Aβ peptide have also been shown to lead to more reproducible fibrillation.49 Accordingly, we developed a procedure for obtaining CsgA in which E. coli curli were depolymerized in formic acid and subsequently separated into different protein components by ion exchange in buffer containing 8 M urea. CsgA fractions were dialyzed against water, lyophilized, and resuspended in formic acid to facilitate CsgA dissociation. CsgA was subsequently purified by gel filtration in a buffer containing 6 M GdnHCl (Figure S1 of the Supporting Information). The fraction containing CsgA monomers was then transferred to water on a desalting column immediately prior to aggregation assays.
CsgA clearly polymerized to ThT-binding aggregates over the entire pH range (pH 3–9) tested (Figure 1A). Polymerization kinetics displayed a sigmoidal time profile, in which a lag time of ≤2 h was followed by a rapid increase in ThT fluorescence, which typically stabilized at a plateau level after ~6 h. The data were highly reproducible and showed overall variations in terms of lag time and end ThT fluorescence of 5– 10%. Both the half-time for fibrillation, t50 (i.e., the time required to reach half the final ThT fluorescence), and the lag time (i.e., the time at which the tangent to the fibrillation curve at t50 intersects with the start ThT level) remained fairly constant from pH 3 to 7 (Figure 1B) but increased at higher pH values. Closer inspection of the first 4 h of the fibrillation revealed that when the pH was below 7, the ThT fluorescence increased without any clear lag phase (Figure S2 of the Supporting Information). This increase was most pronounced around the pI of mature CsgA, which according to the theoretical titration in Figure 1C lies around pH 5–6. Given that ThT can also bind unspecifically to amorphous aggregates and give rise to small increases in fluorescence (ref 44 and references therein), the early stage increase in the magnitude of the ThT signal may reflect rapid and relatively nonspecific aggregation (seen as visible cloudiness in the solution) because of the lack of electrostatic repulsion near the pI of CsgA.
The end point ThT values generally increased with pH (Figure 1D). This could be due to either the pH dependence of ThT binding or fluorescence or genuine changes in fibril structures. Using pH-jump experiments in which acid or base was added to preformed fibrils, it was clear that ThT fluorescence levels in the presence of CsgA fibrils could reversibly and rapidly switch from one level at one pH value to another at another pH level (Figure S3A of the Supporting Information) and switch back again. Given that fibrils are unlikely to reconfigure so rapidly and reversibly from one stable state to another, we consider it most likely that the different ThT fluorescence levels reflect largely pH-dependent effects (in terms of either affinity or fluorescence intensity) of binding to similar fibril structures. Note that there are small variations. Close comparison of the end levels for the pH jumps in Figure S3A of the Supporting Information with the original fibrillation curves at different pH values in Figure 1A shows that the end ThT levels at pH 9 are higher than at pH 7 (Figure 1A), while the pH-jump experiments show that going from pH 7 to 9 leads to a small decrease in fluorescence. The differences are relatively small and within the variation range seen for typical fibrillation experiments.
Overall, it is remarkable that CsgA can fibrillate over such a broad range of pH values spanning the neutral region, given that extreme pH values are often required to induce fibrillation in many other cases.50–52
The effect of ionic strength on the fibrillation of CsgA was examined at pH 7 by varying the NaCl concentration from 0 to 500 mM (Figure 1E). CsgA fibrillation was evident at all NaCl concentrations, and the half-time for fibrillation as well as the lag time decreased in proportion to the square root of the ionic strength (Figure 1F), which indicates a role for Debye–Huckel screening effects in the aggregation process. The end point ThT levels clearly increased with NaCl concentration (Figure 1E). This could in principle reflect changes in ThT binding affinity. Modulation of ThT binding affinity by electrostatic screening has been observed in other contexts.53 However, this did not seem to play a role in our experiments as addition of NaCl to preformed fibrils did not alter the ThT fluorescence (Figure S3B of the Supporting Information), unlike the effect of a change in pH (Figure S3A of the Supporting Information). Another possibility is that different amounts of aggregates are formed at different ionic strengths. This can be ruled out by the observation (see below) that CsgA monomers are completely incorporated into higher-order structures. A final possibility is either a change in fibrillar architecture (in which more densely clumped fibrils will have less accessible surface area for ThT to bind) or the ratio between fibrils and nonfibrillar aggregates, but this is difficult to verify using these data (see the section on electron microscopy below).
Effect of Protein Concentration
The effect of protein concentration on the kinetics of fibrillation was investigated (Figure 1G). A linear relation was found between the protein concentrations and end ThT intensities (Figure 1H), indicating that the same fibrils (with similar ThT binding properties) are formed at different concentrations and the amount scales simply with the amount of precursor monomer. The non-zero intercept reflects background contributions from free ThT. Although the lag time decreased 2–3-fold over a 20-fold increase in protein concentration (Figure 1I), the half-time for fibrillation, t50, remained essentially invariant (Figure 1I), effectively making the time required to reach the end plateau (~6 h) independent of CsgA concentration. Powers and Powers have demonstrated that a concentration-independent t50 corresponds to a situation in which the protein is above the supercritical concentration (scc), so that the fibrillation rate is independent of protein concentration.54 The situation is a little more complex for CsgA, given that the lag time does decrease slightly with concentration. However, the invariance of t50 does suggest that either CsgA has an extremely low scc or the monomer is the active fibrillating nucleus. Such a nucleus could conceivably form if the monomer, which is initially unstructured,55 folds back on itself via the five imperfect repeats to form an elongation-competent monomeric nucleus.
Effect of Seeding
The effect of seeding on the fibrillation kinetics (Figure 1J) was evaluated by replacing a part of the monomeric CsgA with preformed native curli fibril seeds. The lag time decreased markedly with an increasing seed concentration (Figure 1K). In contrast, the elongation phase was fairly constant. Even the lowest seed concentration (0.1%) had a significant effect on fibrillation. This is very good evidence that the monomer CsgA solution contained no or very few seeds. The start value of ThT fluorescence scaled linearly with seed concentration (Figure 1L), consistent with a linear relationship between the amount of seed present and the number of binding sites for ThT. However, a higher seed concentration results in an asymptotic increase in the end ThT level (Figure 1L). A simple but speculative explanation for this is that a large amount of nuclei leads to many small fibrils while a small amount leads to a few large fibrils. These two classes of fibrils may have different ThT binding properties.
Analysis of Fibril Structure
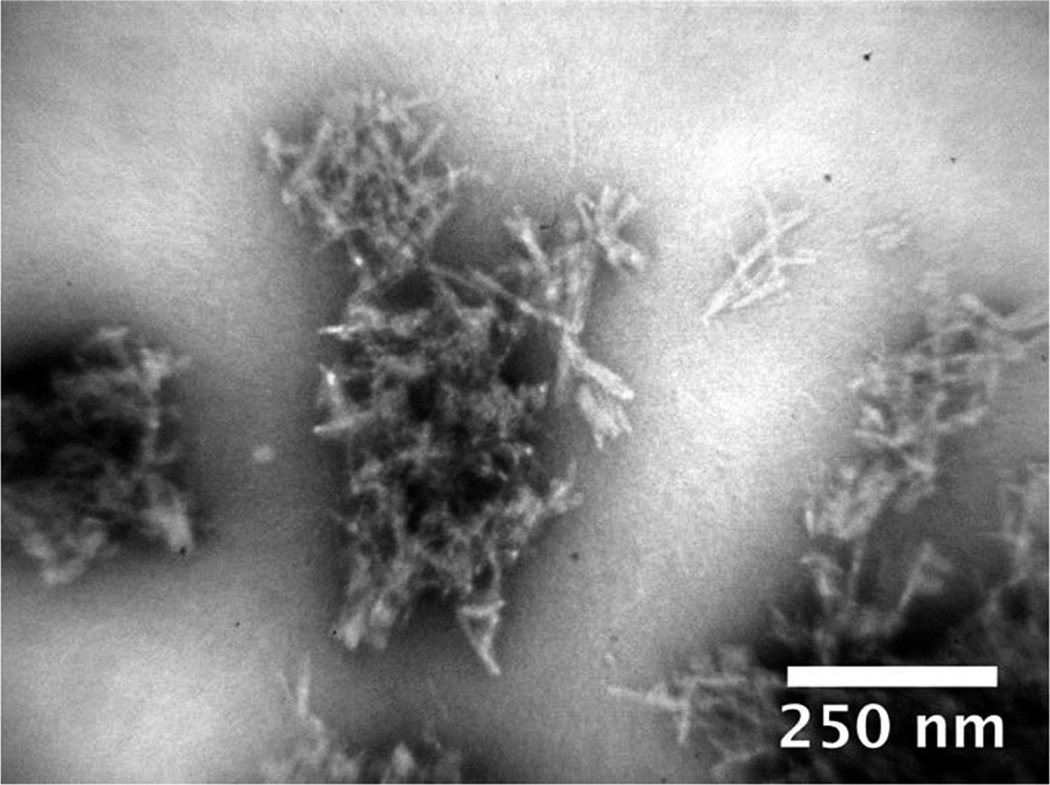
We characterized the structure of the fibrils formed under different conditions. The morphology of fibrils formed at various pH values and NaCl concentrations was examined by transmission electron microscopy (TEM). All samples exhibited similar needlelike fibrils, which aggregated into large clumps (a representative sample is shown in Figure 2). In all samples, we also detected agglomerated structures, which could be interpreted as either dense amorphous aggregates or clumps of tightly associated fibrils and protofibrils. For this reason, it was difficult to quantify the relative amounts of fibrils and potential amorphous aggregates. Samples exposed to sonication prior to TEM still contained significant amounts of these clumps (data not shown), indicating strong adhesive forces linking the aggregates together.
Figure 2.
Morphology of the CsgA fibrils. TEM image of CsgA fibrils formed in 20 mM sodium phosphate (pH 7).
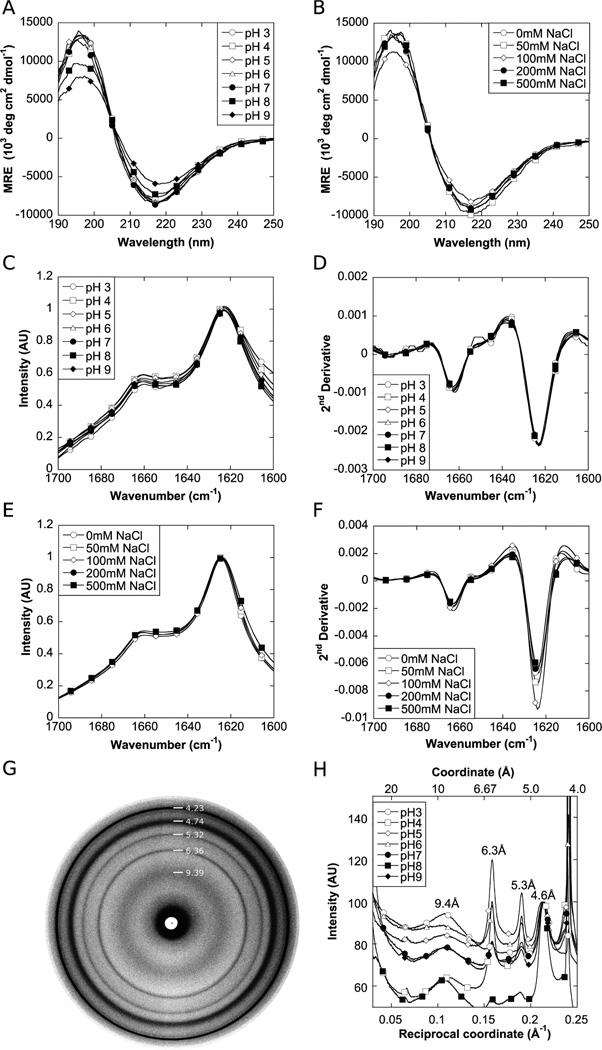
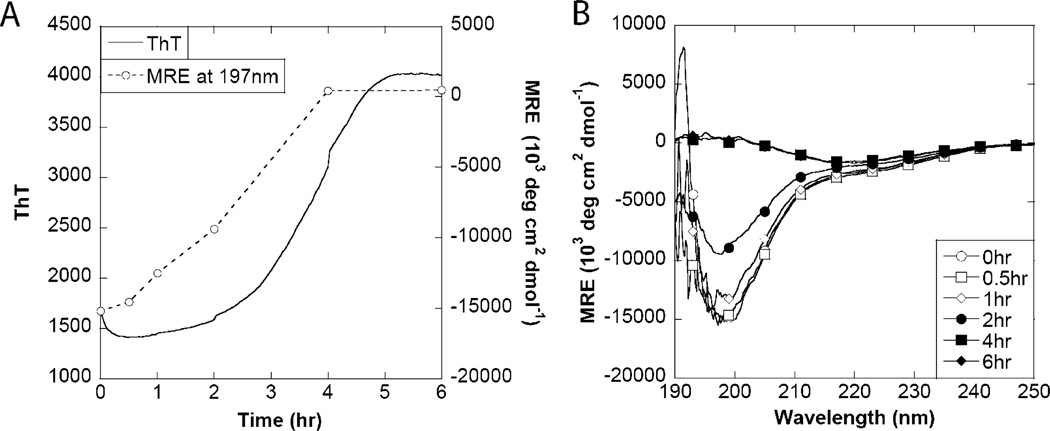
The secondary structure of the fibrils was analyzed by CD and FTIR (Figure 3A – F). All CD spectra exhibited similar strong β-sheet signatures with the characteristic minimum at ~217 nm and maximum at ~197 nm (Figure 3A,B). FTIR also showed very similar spectra for the fibrils with a well-defined peak at 1623 cm−1, indicative of β-sheet in an amyloid-like conformation maintained by very strong hydrogen bonds and a minor shoulder at 1663 cm−1, which indicates β-turns (Figure 3C – F).56–58
Figure 3.
Biophysical characterization of CsgA fibrils formed under various environmental conditions. (A and B) CD spectra of CsgA fibrils formed at various pH values (A) and at pH 7 with varying NaCl concentrations (B). (C and D) FTIR (C) and second-derivative (D) spectra of CsgA fibrils formed at various pH values. (E and F) FTIR (E) and second-derivative (F) spectra of CsgA fibrils formed at pH 7 with varying NaCl concentrations. (G) X-ray fiber diffraction pattern of CsgA fibrils formed at pH 7. (H) Radial averaged X-ray fiber diffraction spectra of fibrils formed at various pH values.
The structure of fibrils formed at various pH values was examined by X-ray fiber diffraction (Figure 3G,H). For all samples, a characteristic Bragg reflection at 4.6 Å was seen, corresponding to the spacing between β-strands in the amyloid fold6 and a reflection at 8–10 Å, corresponding to intersheet spacing.6,59 The latter manifested itself as a broad and rather featureless peak. Similar reflections have been shown for wild-type curli.60 Additional well-defined reflections were seen for all samples at 6.3, 5.3, and 4.2 Å. The structural origin of these reproducible reflections is not clear.
If fibrils are well-aligned, they will show characteristic meridional reflections around 4.8 Å and equatorial reflections at 10 Å, reflecting the amyloid arrangement in which β-strands are orthogonal to the fibril axis, whereas the β-sheets are arranged parallel to the axis. This was not the case for the CsgA fibrils, whose highly aggregative disposition (see the following section and Figure 4) prevented good alignment of the fibers and consequently hindered a more detailed analysis of the structural features of the fibers. Nevertheless, the similarities of the observed reflections suggest little difference in the packing of the CsgA fibers and a common core structure regardless of pH.
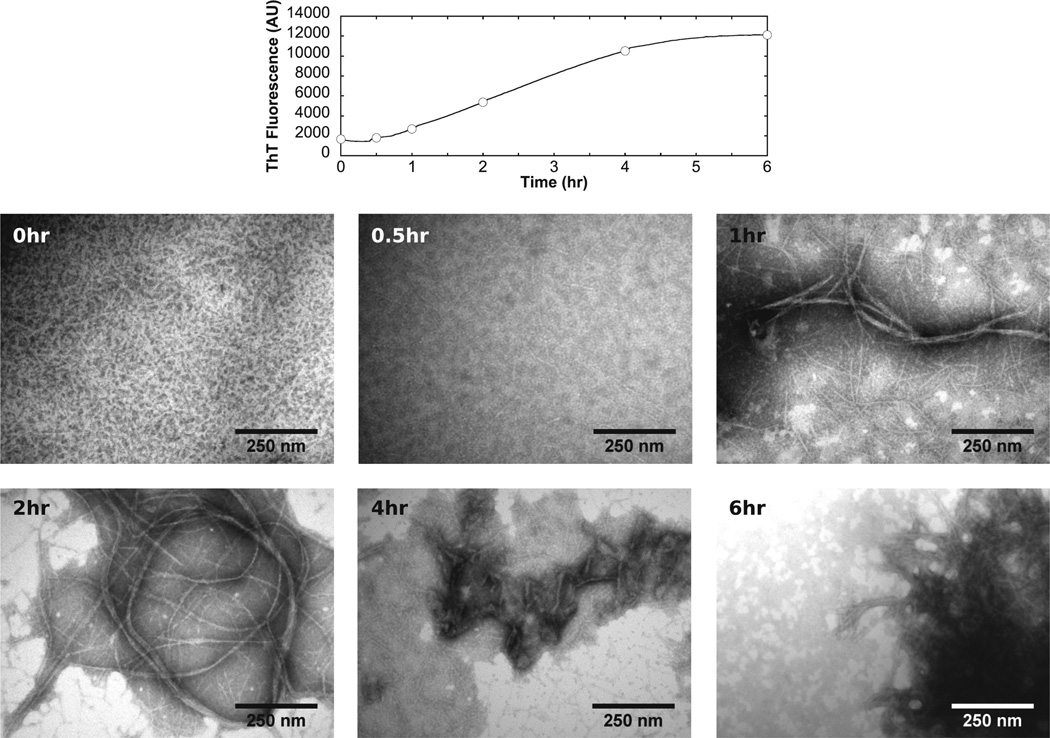
Figure 4.
Fibrillation of CsgA followed by TEM. Samples was collected at various time points during the fibrillation of 40 µM CsgA at pH 7 and examined by TEM. The fibrillation curve measured by ThT is shown at the top, with circles corresponding to the time points shown in the bottom panels.
Fibrillation Followed by TEM
We examined samples collected during the fibrillation process by TEM (Figure 4). Immediately after monomeric CsgA had been mixed with buffer, a few short, thin filaments were observed. After 1 h, larger fibrils were seen to coexist with many thin filaments. At 2 h, the fibrils appeared to have grown even larger at the expense of the small filaments. At 4 h, the fibrils started to aggregate and form larger clumps. A few thin filaments were still present. These filaments were completely absent after 6 h, and only large clumps of fibers were present. Some amorphous aggregates were seen in all samples except for the 0 h sample. Over time, these aggregates appeared to assemble into larger dumps.
Fibrillation of CsgA Followed by ANS Fluorescence
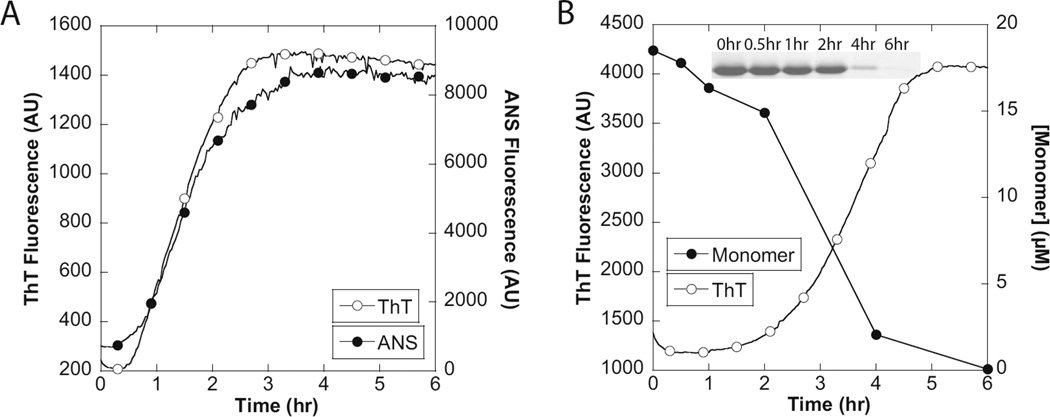
We used the hydrophobic probe 8-anilino-l-naphthalenesulfonate (ANS), whose fluorescence is enhanced by the presence of hydrophobic regions,61–63 to investigate if hydrophobic protofibrils of non-amyloid character were formed prior to the formation of ThT binding amyloid fibrils (Figure 5A). In general, the two probes exhibited similar fibrillation profiles, indicating that aggregation to form ThT-binding fibrils is accompanied by the exposure of contiguous patches of hydrophobic surfaces, but that there is no separate non-ThT binding preaggregate species.
Figure 5.
Fibrillation of CsgA followed by ANS fluorescence and SDS–PAGE. (A) Fibrillation of 20 µM CsgA followed by ThT and ANS fluorescence. (B) Fibrillation of 20 µM CsgA followed by ThT and SDS–PAGE. SDS–PAGE data were expressed as the CsgA monomer concentration calculated from the CsgA monomer band intensity using the ImageJ gel analysis tool (http://rsbweb.nih.gov/ij/).
Fibrillation of CsgA Followed by SDS–PAGE
To investigate if the increase in the magnitude of the ThT signal correlates with an increase in the relative amount of fibrillated CsgA, samples were collected during fibrillation and analyzed by SDS–PAGE (Figure 5B). Fibrillar CsgA cannot be dissolved in SDS–PAGE loading buffer in the absence of formic acid, whereas monomerized CsgA remains soluble in the SDS– PAGE loading buffer and enters the gel. It was found that essentially all CsgA was converted to the fibrillar form and thus that the increase in ThT fluorescence arises from the increase in the percentage of CsgA in the fibril form.
Note that we cannot say anything definite about the existence of oligomeric structures based on this SDS-PAGE analysis. We do not see any higher-order structures on the gel, but if potential oligomeric species are sensitive to SDS, they may not survive as intact species on the gel. However, any such oligomer might be present in only small quantities, as discussed by Wang et al.55 and suggested for many other fibrillating proteins.52
Fibrillation of CsgA Followed by Circular Dichroism
CD showed a clear transition from a random coil structure to one with a high content of β-sheet structure (Figure 6). This transition generally correlated with the increase in ThT fluorescence, although direct correlation to the mean residue ellipticity was problematic because of protein precipitation and settling. Thus, we cannot reach any definite conclusions about the secondary structure content of the needlelike fibrils formed at early stages of aggregation according to TEM (Figure 4); although they are likely to contain regular secondary structure, the absolute CD signal will depend both on the amount of solubilized material and on the fraction of CsgA that forms these structures rather than the original random coil structure. However, the lack of defined structures at 0.5 and 1 h suggests that only a small fraction of CsgA is in the fibrillated state, which is in good agreement with the SDS-PAGE data. TEM is biased toward large and aggregated species and is inappropriate for determining the fraction of protein in a given form.
Figure 6.
Fibrillation of CsgA followed by circular dichroism. (A) Fibrillation of 20 µM CsgA followed by ThT and CD signal intensity at 197 nm. (B) CD wavelength spectra of samples collected during fibrillation.
DISCUSSION
The ability of CsgA monomers to assemble into amyloid-like fibrils was evaluated under a wide range of conditions and clearly shows that the protein has an inherent tendency to fibrillate. This is consistent with fibrillation properties being highly optimized for specific purposes through evolution. Most other proteins require specific circumstances to expose their amyloid forming propensities. In vitro fibrillation is typically promoted by acidic pH, elevated temperatures,11 submicellar concentrations of anionic surfactants,64 or moderate concentrations of denaturants65 or organic solvents.66 Fibrillation by disease-associated proteins under physiological conditions usually requires either aggregation-inducing mutations, long-term exposure to a chemically reactive environment (e.g., dopamine as suggested for α-synuclein in Parkinson’s disease67), or proteolytic cleavage to either destabilize the protein (e.g., gelsolin68) or release peptides (e.g., Aβ or ADan/ ABri69) that fibrillate when removed from their normal physiological context.
The pH may also be a parameter for controlling the formation of functional amyloid. For example, pH changes modulate the solubility of the amyloid form of pmel17 involved in melanin deposition70 and may even be a way to control the storage of peptide hormones as temporary amyloid deposits.71 CsgA fibrils formed at different pH values do show variations in the level of ThT fluorescence that cannot entirely be ascribed to simple pH changes; this is also seen in the fiber diffraction data in which the relative sizes of different diffraction peaks vary. Nevertheless, fibrils form over the entire pH range tested.3–9 Presumably, the robust fibrillation of CsgA reflects the need to ensure reliable fibril formation and integrity under a wide range of extracellular conditions outside the control of E. coli, in contrast to the carefully controlled environment of melanin production and hormone release.
Despite CsgA’s “hardwired” ability to fibrillate, we still observe variations in the fibrillation kinetics depending on the specific conditions. It was found that the kinetic parameters t50 and lag time were shortest near and below the pI of CsgA and longest at higher pH values. This shows that negatively charged residues at higher pH values reduced the propensity of CsgA to form a fibrillation prone nucleus, whereas the positive charges present below the pI have only minor effects. This is not surprising if we investigate the spatial arrangement of the charged residues in Chapman’s proposed β-helical model of CsgA in the curli fibrils,39 which is based on the proposed structure of the AgfA β-helix41 and supported by solid state NMR experiments.60 The negative charges originate from aspartic acid and glutamic acid residues, and these are situated next to Ser1 and around Gln18 in the repeats. According to the proposed structure, hydrogen bonding between these amino acid residues as well as Gln7 and Asn12 of the repeats provides stability to the amyloid structure. Introduction of repulsive charges around these amino acids will therefore reduce the stability of the nucleus. The transition time (i.e., the time taken to reach the ThT maximum from the lag phase) was relatively constant (~3 h) throughout the tested pH range (Figure 1A). This shows that interactions involved in fibril growth are only slightly perturbed by changes in pH. This is in good agreement with an amyloid structure stabilized by pH-independent polar interactions between the side chains of glutamine and asparagine residues. It furthermore shows that charges in the vicinity of Ser1 and Asn12 at most have a minor effect on fibril elongation despite a clear effect on nucleation. It should also be noted that R5, the most aggregation prone repeat of CsgA, does not contain any acidic groups, as is true for R1 and R5 of CsgB.72 It is therefore likely that the repeats directly involved in nucleation have been tailored to withstand changes in the pH of the environment. Charge still plays a role, given that the rates of nucleation and growth both decreased in proportion to the square root of the ionic strength, implying a screening of charged groups in CsgA. However, the effect is modest, given that the lag times and half-times decrease less than 2-fold as we increase the ionic strength to 1 M.
Fibril morphology in TEM images, X-ray fiber diffraction patterns, and secondary structure analysis by FTIR and CD all point toward a similar amyloid structure. This is in agreement with the initial hypothesis that CsgA forms a well-defined amyloid structure because of the stabilizing effect of the repeating units and side chain interactions.
All these fibrils are prepared in vitro, raising the obvious question of the extent to which our findings can be extrapolated to in vivo conditions. In this context, we note that the general biophysical properties of in vitro polymerized CsgA and that of curli fibers are the same. Both structures are rich in β-strand, bind to ThT and Congo red, and have grossly the same appearance by TEM.38,60 Nevertheless, techniques such as solid state NMR may reveal differences in the ultrastructures of in vitro polymerized CsgA and curli fibers. For example, curli in vivo are the result of initial templating by the CsgB nucleator protein. Therefore, we predict that the structure of curli fibers in vivo will reflect properties of CsgB as well as CsgA. Information about the fibrillation properties of CsgA is thus a necessary part of a fuller understanding of the formation of curli fibers.
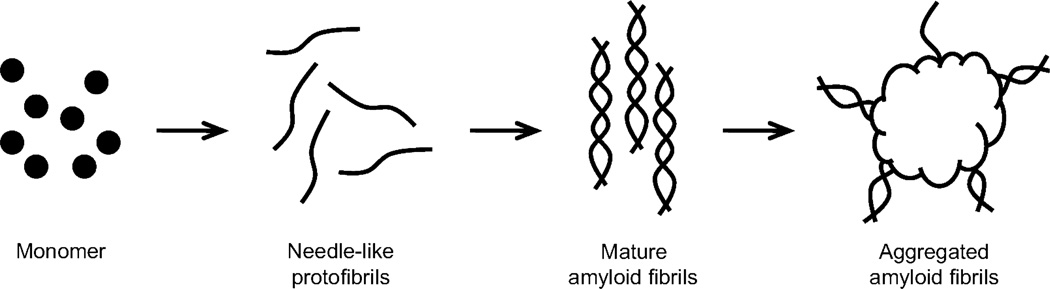
In summary, detailed information about the CsgA fibrillation process was obtained by analyzing samples collected at various times during the fibrillation at pH 7. The samples were analyzed via TEM, ANS binding, CD, and SDS–PAGE. It was found that the fibrillation was initiated by a conversion of the monomers into thin needlelike protofibrils (Figure 7). These protofibrils later converted into hydrophobic amyloid fibrils, which associate to larger aggregates. It is tempting to speculate that the highly aggregative and hydrophobic nature of the CsgA fibrils may be useful in curli-mediated E. coli and Salmonella surface attachment and biofilm formation.
Figure 7.
Proposed CsgA fibrillation mechanism. Monomeric CsgA initially forms thin needlelike protofibrils. These protofibrils rearrange to form amyloid fibrils, which subsequently aggregate into dense clumps.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Zhou Yizhou for constructive comments about the manuscript.
Funding This work is supported by a grant from the Villum Kann Rasmussen Foundation to M.S.D. and D.E.O. D.E.O. is supported by the Danish Research Foundation (inSPIN).
ABBREVIATIONS
- AFM
atomic force microscopy
- AIEX
anion exchange
- ANS
8-anilino-l-naphthalenesulfonate
- ATR
attenuated total reflectance
- CD
circular dichroism
- DTGS
deuterated triglycine sulfate
- FTIR
Fourier-transform infrared spectroscopy
- MRE
mean residue ellipticity
- scc
supercritical concentration
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- t50
half-time for fibrillation
- ThT
thioflavin T
- TEM
transmission electron microscopy
Footnotes
ASSOCIATED CONTENT
Supporting Information
Figures showing purification of CsgA (Figure S1), (2) close-up of the fibrillation time course at the beginning of the reaction (Figure S2), and pH and NaCl jumps conducted with CsgA fibrils (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: The importance of being unfolded. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Knight JD, Miranker AD. Phospholipid catalysis of diabetic amyloid assembly. J. Mol. Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- 3.Volles MJ, Lansbury PT. Vesicle permeabilization by protofibrillar α-synuclein is sensitive to Parkinson’s disease-linked mutations and occurs by a pore-like mechanism. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 4.Krebs MR, Morozova-Roche LA, Daniel K, Robinson CV, Dobson CM. Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci. 2004;13:1933–1938. doi: 10.1110/ps.04707004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 6.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tycko R. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 2004;14:96–103. doi: 10.1016/j.sbi.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Fandrich M, Dobson CM. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen JS, Christensen G, Otzen DE. Modulation of S6 fibrillation by unfolding rates and gatekeeper residues. J. Mol. Biol. 2004;341:575–588. doi: 10.1016/j.jmb.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen JS, Dikov D, Otzen DE. N- and C-terminal hydrophobic patches are involved in fibrillation of glucagon. Biochemistry. 2006;45:14503–14512. doi: 10.1021/bi061228n. [DOI] [PubMed] [Google Scholar]
- 14.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 15.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark PL. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem. Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen JS, Dikov D, Flink JL, Hjuler HA, Christiansen G, Otzen DE. The changing face of glucagon fibrillation: Structural polymorphism and conformational imprinting. J. Mol. Biol. 2006;355:501–523. doi: 10.1016/j.jmb.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 18.Pedersen JS, Otzen DE. Amyloid-a state in many guises: Survival of the fittest fibril fold. Protein Sci. 2008;17:2–10. doi: 10.1110/ps.073127808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen JS, Flink JM, Dikov D, Otzen DE. Sulfates Dramatically Stabilize a Salt-Dependent Type of Glucagon Fibrils. Biophys. J. 2006;90:4181–4194. doi: 10.1529/biophysj.105.070912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 21.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 22.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat. Rev. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 24.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 25.Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 27.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, Vigouret JM, Paganetti P, Walsh DM, Mathews PM, Ghiso J, Staufenbiel M, Walker LC, Jucker M. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 28.Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 30.Johansson C, Nilsson T, Olsen A, Wick MJ. The influence of curli, a MHC-I-binding bacterial surface structure, on macrophage-T cell interactions. FEMS Immunol. Med. Microbiol. 2001;30:21–29. doi: 10.1111/j.1574-695X.2001.tb01545.x. [DOI] [PubMed] [Google Scholar]
- 31.Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. Role of fibronectin in curli-mediated internalization. FEMS Microbiol. Lett. 2002;212:55–58. doi: 10.1111/j.1574-6968.2002.tb11244.x. [DOI] [PubMed] [Google Scholar]
- 33.Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 34.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 35.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli . J. Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 37.Hammar M, Bian Z, Normark S. Nucleatordependent intercellular assembly of adhesive curli organelles in Escherichia coli . Proc. Natl. Acad. Sci. U.S.A. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J. Mol. Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 42.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis . J. Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dueholm MS, Petersen SV, Sønderkaer M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Otzen DE. Functional amyloid in Pseudomonas . Mol. Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- 45.Waddell WJ. A simple ultraviolet spectrophotometric method for the determination of protein. J. Lab. Clin. Med. 1956;48:311–314. [PubMed] [Google Scholar]
- 46.Klemm P, Christiansen G, Kreft B, Marre R, Bergmans H. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J. Bacteriol. 1994;176:2227–2234. doi: 10.1128/jb.176.8.2227-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 48.Ames GF. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J. Biol. Chem. 1974;249:634–644. [PubMed] [Google Scholar]
- 49.Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid β-Protein Aggregation Produces Highly Reproducible Kinetic Data and Occurs by a Two-Phase Process. ACS Chem. Neurosci. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fandrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 52.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–336. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 53.Groenning M, Olsen L, van de Weert M, Flink JM, Frokjaer S, Jorgensen FS. Study on the binding of thioflavin T to β-sheet-rich and non-β-sheet cavities. J. Struct. Biol. 2007;158:358–369. doi: 10.1016/j.jsb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 54.Powers ET, Powers DL. The kinetics of nucleated polymerizations at high concentrations: Amyloid fibril formation near and above the “supercritical concentration”. Biophys. J. 2006;91:122–132. doi: 10.1529/biophysj.105.073767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 57.Gasset M, Baldwin MA, Lloyd DH, Gabriel JM, Holtzman DM, Cohen F, Fletterick R, Prusiner SB. Predicted α-helical regions of the prion protein when synthesized as peptides form amyloid. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10940–10944. doi: 10.1073/pnas.89.22.10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zandomeneghi G, Krebs MR, McCammon MG, Fandrich M. FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. doi: 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 62.Ptitsyn OB, Pain RH, Semisotnov GV, Zerovnik E, Razgulyaev OI. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- 63.Engelhard M, Evans PA. Kinetics of interaction of partially folded proteins with a hydrophobic dye: Evidence that molten globule character is maximal in early folding intermediates. Protein Sci. 1995;4:1553–1562. doi: 10.1002/pro.5560040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Otzen DE, Nesgaard LW, Andersen KK, Hansen JH, Christiansen G, Doe H, Sehgal P. Aggregation of S6 in a quasi-native state by sub-micellar SDS. Biochim. Biophys. Acta. 2008;1784:400–414. doi: 10.1016/j.bbapap.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 65.De Young LR, Dill KA, Fink AL. Aggregation and denaturation of apomyoglobin in aqueous urea solutions. Biochemistry. 1993;32:3877–3886. doi: 10.1021/bi00066a006. [DOI] [PubMed] [Google Scholar]
- 66.Chiti F, Taddei N, Bucciantini M, White P, Ramponi G, Dobson CM. Mutational analysis of the propensity for amyloid formation by a globular protein. EMBO J. 2000;19:1441–1449. doi: 10.1093/emboj/19.7.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rochet J-C, Outeiro TF, Conway KA, Ding TT, Volles MJ, Lashuel HA, Bieganski RM, Lindquist SL, Lansbury PT. Interactions among α-synuclein, dopamine, and biomembranes: Some clues for understanding neurodegeneration in Parkinson’s disease. J. Mol. Neurosci. 2004;23:23–34. doi: 10.1385/jmn:23:1-2:023. [DOI] [PubMed] [Google Scholar]
- 68.Chen CD, Huff ME, Matteson J, Page L, Phillips R, Kelly JW, Balch WE. Furin initiates gelsolin familial amyloidosis in the Golgi through a defect in Ca2+ stabilization. EMBO J. 2001;20:6277–6287. doi: 10.1093/emboj/20.22.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rostagno A, Revesz T, Lashley T, Tomidokoro Y, Magnotti L, Braendgaard H, Plant G, Bojsen-Møller M, Holton J, Frangione B, Ghiso J. Complement Activation in Chromosome 13 Dementias. J. Biol. Chem. 2002;277:49782–49790. doi: 10.1074/jbc.M206448200. [DOI] [PubMed] [Google Scholar]
- 70.McGlinchey RP, Shewmaker F, McPhie P, Monterroso B, Thurber K, Wickner RB. The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U.S.A. 2009 doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.