Abstract
The human intestine houses a dense microbial ecosystem in which the struggle for nutrients creates a continual and dynamic selective force. Host-produced mucus glycans provide a ubiquitous source of carbon and energy for microbial species. Not surprisingly, many gut resident bacteria have become highly adapted to efficiently consume numerous distinct structures present in host glycans. We propose that sophistication in mucus consumption is a trait most likely to be found in gut residents that have co-evolved with hosts, microbes that have adapted to the complexity associated with the host glycan landscape.
Keywords: gut microbiota, host-microbial interaction, microbiome, mucin, polysaccharide utilization
Mucus glycans foster interactions with intestine resident microbes
A complex community of microbes known as the intestinal microbiota populates the distal gut of mammals. In this dense microbial ecosystem, competition for nutrients is fierce and repeated purging of the microbes most poorly adapted to the gut environment has ensured the selection of a community that possesses traits optimized for gut survival. In the human intestinal microbiota, the consumption of diverse classes of carbohydrates is among the most abundantly represented traits (Cantarel et al. 2009; Lozupone et al. 2012). The success of polysaccharide utilizing microbes in the gut reflects the abundance and diversity of the glycan landscape. Complex dietary glycans, such as plant cell-wall polymers (e.g. dietary fiber) pass to the distal gut undigested and represent a major food source for the microbiota. In addition, host-secreted mucus is ubiquitous and glycan-rich.
The classic definition of mucus as a protective layer has recently changed to include important roles in microbiota interaction. Mucus has been viewed as a secretion that functions primarily as an immunological barrier that physically protects the host from the dense adjacent microbial community. Recent insight into the lifestyles of intestine-resident microbes has required an expansion of the original view of mucus to include a role in the sustenance and tethering of gut-resident microbial mutualists (Sonnenburg et al. 2004; Martens et al. 2008; Johansson et al. 2011; Hansson 2012; Figure 1). In this view, the mucus serves not just as a consistent endogenous source of carbohydrates for these microbes, but also as a signpost of proximity to host tissue within a vast and disorienting landscape of otherwise homogenized digesta that lacks informative landmarks.
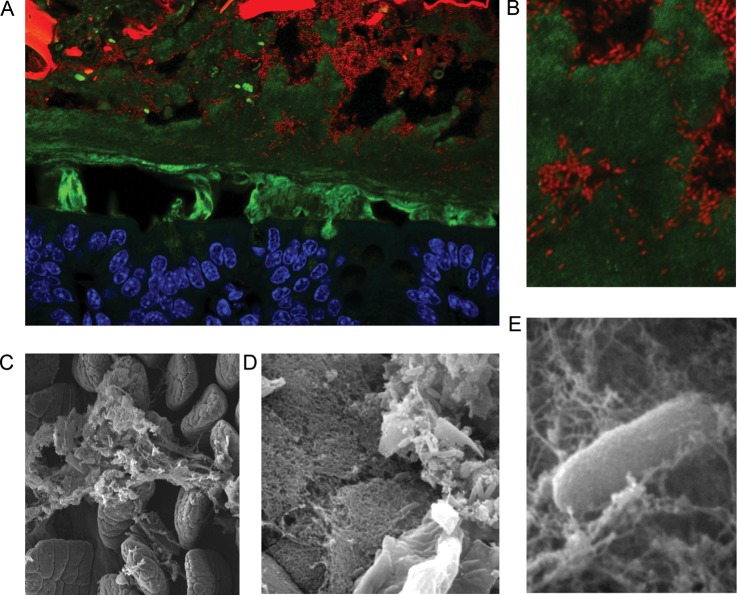
Fig. 1.
Localization of Bacteroides thetaiotaomicron (Bt) to the outer layer of colonic mucus. (A) A cross-section of distal colon of a gnotobiotic mouse colonized with Bt. Cells are stained with DAPI and false colored with blue (host tissue) or red (bacterial cells) and mucus is stained with an anti-MUC2 antibody (green). The small Bt rods occupy discrete microhabitats that include dietary plant material (intense large red and green objects), loose lumenal mucus (diffuse green signal) and the outer loose layer of epithelium-adjacent mucus. (B). Zoomed in the view of Bt embedded within mucus. Scanning electron micrographs of (C) mucus covering intestinal villi of the mouse; (D) Bt embedded in a mucus lattice (middle) overlying epithelial cells (bottom left), and in food particle (upper right), within the gnotobiotic mouse gut (Sonnenburg et al. 2005); (E) zoomed in view of Bt in mucus (Sonnenburg et al. 2005). Bt cells are ∼1 × 5 μm.
Consistent with the role in fostering host interaction with gut resident microbes, genetic alteration of host gut epithelial and mucus glycans in humans and mouse models influences microbial communities and host-microbial interaction. FUT2, known as the secretor locus in humans, encodes a polymorphic α1-2-fucosyltransferase. Non-secretors, which are homozygous for non-functional alleles and lack α1-2-fucose residues in secretory tissues and on mucus, are at increased risk for several diseases including Crohn's disease and celiac disease (McGovern et al. 2010; Franks 2012; Parmar et al. 2012) and have gut communities that are distinct from secretors, who possess the functional fucosyltransferase (Rausch et al. 2011). Loss of core-1 or core-3 derived O-glycans increases intestinal epithelial permeability and susceptibility to colitis; in the case of core-3 loss, colorectal tumor susceptibility is also enhanced (An et al. 2007; Fu et al. 2011). These examples support an important role for host epithelial and mucosal glycans in dictating aspects of microbial community composition and microbiota-host dynamics. Although the potential mechanisms contributing directly or indirectly to these effects are numerous, it is likely that the attachment to and foraging of host-generated carbohydrates is governed by the structural motifs that are present.
Structural nuances of mucin glycans have likely served as a strong selective force in microbiota assembly and maintenance within species and between individuals in a population. The glycan landscape of a mucosal surface such as the gut dictates aspects of microbial community composition and the nature of interaction at the mucosal interface in every gut on the planet. So, although it may seem that these nuances are only of interest to a few expert glycobiologists around the world, countless microbes that inhabit each person's bowel share this appreciation.
Inferring a microbial appreciation of mucus glycan structures
Mucins are cell surface or secreted glycoproteins that help to form the mucosal barrier that lines the gastrointestinal (GI) tract, and therefore, constitute a significant surface area for interaction with gut microbes. The extensive, primarily O-linked glycosylation of mucins contributes to their many physiological roles attributed to the mucosal layer. Among the numerous roles that these proteins play at mucosal surfaces (Corfield et al. 2001; Eckburg et al. 2005; Patsos and Corfield 2009), one is specific to the glycan moieties which promotes the establishment and maintenance of the gut microbiota. Gut microbiota composition varies by host species, region of the GI tract and developmental period, all factors that correspond to differences and dynamics in host mucosal glycan (Nordman et al. 1997; Robbe et al. 2004; Thomsson et al. 2012). Several studies have detailed the refined responses of mucus-consuming bacteria to purified mucus-derived or mucus-like carbohydrates, which further support the importance of mucin glycan structural motifs in bacterial recognition and consumption.
Genomic analysis has expanded the list of genes classified as mucins in humans to over 20 (Corfield et al. 2001; Patsos and Corfield 2009; Moran et al. 2011). A subset of these is specifically expressed in the GI tract. A hallmark of mucins is their extensive glycosylation. Glycosyltranferases and their expression patterns have been manipulated to tease apart the enzymatic contributions to the complex glycosylation pathways as well as explore the biological functions of specific structures (Manzi et al. 2000; Thomsson et al. 2002, 2012; Ismail et al. 2011). Mucins have repeated peptide sequences of serine or threonine O-glycan acceptor sites resulting in a molecule that may be up to 80% carbohydrate by mass. Secreted mucins can form polymeric sheets to generate the mucous layer that lines various epithelial surfaces. These secreted forms are synthesized primarily by specialized goblet cells present along those epithelial tracts including the GI tract. Whether released from mucosal scrapings or from specific purified mucin glycoproteins, a large number of GI mucin O-glycans have been structurally characterized using a combination of methods that include mass-spectrometry and NMR (Nordman et al. 1997; Karlsson, Herrmann, et al. 1997; Karlsson, Nordman, et al. 1997; Larsson et al. 2009, 2011).
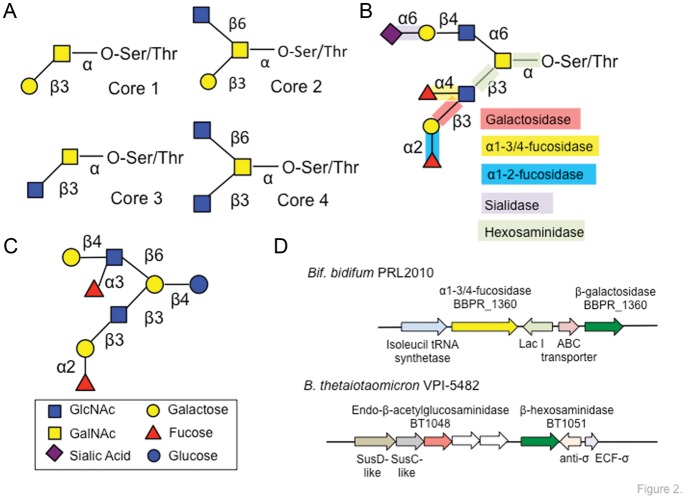
Early structural work on colonic mucins (Podolsky 1985a, b) elucidated that although structural variation exists, glycans from these proteins are mostly variations on general themes. More recently, glycan core structures 1–7 have been identified along the GI tract. Extending off of these core structures (Figure 2A) is often repeating N-acetyllactosamine units and terminating sialic acids and/or fucose residues. Structural variety can be generated between individuals, partly attributed to the expression of blood group epitopes (Hanisch et al. 1993; Karlsson, Herrmann, et al. 1997; Capon et al. 2001; Robbe et al. 2003, 2004). The human adult intestinal mucins have a decreasing gradient of fucosylation and an increasing acidic gradient (i.e. sulfation and sialylation) down the length of the GI tract. Although these gradients are absent in fetal intestinal mucins, the diversity of glycan structures parallels that of adults with more than a 100 structures identified (Robbe-Masselot et al. 2009). Additional variation is introduced by sulfation and acetylation, modifications that cannot only affect the charge state or chemical properties of the mucus, but also require enzymatic removal prior to microbial glycoside hydrolase (GH)-mediated liberation of monosaccharides for fermentation (Figure 2B).
Fig. 2.
Structural features of intestinal mucin glycans and similarities with HMOs. (A) The vast majority of mucin glycans are based on variations of an O-linked core in which N-acetylgalactosamine is attached to either a serine or a threonine residue on the mucin polypeptide backbone. Seven discrete core structures have been associated with gut mucin glycans; the predominant core-1 thru core-4 are shown (Varki et al. 2009). (B) Examples of a mucin O-glycan extended core-4 structure from the ileum (Robbe et al. 2004) and the diverse set of GHs required to liberate monosaccharides from mucin glycans and makes them available for microbial fermentation in the gut. (C) Example of a branched HMO built upon a lactose core; note the similarity to the mucin glycan structure. (D) Prototypic operons within B. bifidum and Bt that encode machinery compatible with mucin glycan use, including multiple GHs.
Structural information for GI mucin glycans is available for non-human mammals such as pig (Karlsson, Nordman, et al. 1997; Nordman et al. 1997), rat (Slomiany et al. 1980; Karlsson, Herrmann, et al. 1997) and mouse (Manzi et al. 2000; Thomsson et al. 2002, 2012; Ismail et al. 2011). These studies illustrate that there are common core structures, extensive structural diversity and tissue specific glycoforms. Differences exist at the level of relative abundance of core types, of neutral, acidic or sulfated glycans and the localization of specific glycan linkages. For example, the murine colonic mucin Muc2 contains more core-1 and -2 type glycans and more neutral glycans than detected on the human colonic MUC2 in addition to differences in fucosylation, sialylation and sulfation (Thomsson et al. 2012). These differences suggest that if gut microbes are well-adapted and have co-evolved with one particular host species, there may be incompatibilities between a symbiont's mucin-recognition/adherence/consumption machinery and glycan landscape nuances of a new host species. These differences are also important to keep in mind when studying gut bacteria in the lab. Much of the available data for the use of mucin by human-derived microbiota strains has been obtained using either porcine mucin (one of the only commercially available sources) during in vitro growths or murine mucin glycans from in vivo experiments in gnotobiotic mice. It is clear from these studies that much can be learned in this heterologous setting, likely due to the strong structural commonalities of mucin glycans in general; however, less clear is the extent to which incompatibilities between gut microbe and host glycans may be maladaptive in a competitive gut ecosystem.
Human milk oligosaccharides (HMOs) are a class of oligosaccharides that are structurally related to mucin glycans and are present in the gut early in life. Similar to mucin glycans, these molecules have structural diversity based on common cores (Figure 2C). The abundance (5–15 g/L) of HMO in milk and the inability of these molecules to be digested and absorbed by infants in the upper GI tract is consistent with these oligosaccharides serving as a natural prebiotic to seed mucus adapted beneficial microbes in the developing infant microbiota, among other possible roles (Miller and McVeagh 1999; Gnoth et al. 2000; Bode 2012; Ruhaak and Lebrilla 2012). Well-described HMO consumption pathways in Bifidobacteria and Bacteroides strains, along with the finding that Bacteroides employ mucus-utilization machinery for consumption of HMOs supports its role as a prebiotic (Sela and Mills 2010; Marcobal et al. 2011). Thirteen core structures are used to classify the HMO glycome and more than a 100 structures of neutral or acidic character have been described. The structural composition varies by individual, secretor status and stage of lactation (Ninonuevo et al. 2006; Wu et al. 2010; Bode 2012). The structural features common between HMO and mucin glycans highlights a potential common importance in the role these molecules play in establishing and maintaining intestine adapted microbes (Urashima et al. 2012; Yu et al. 2013). It is tantalizing to speculate that HMOs have been crafted over the course of evolution to mimic mucin glycan structures, a theory that implies that mucin utilization is a characteristic of beneficial (non-pathogenic) microbes.
Mucus on the menu
The ability of gut resident bacteria to degrade and consume intestinal mucus has been appreciated for over 50 years (Hoskins and Zamcheck 1968; Bayliss and Turner 1982). Over the past decade the molecular and genomic details of microbial interaction with mucin are beginning to come to light (Figure 2D). Molecular studies have primarily focused on two major groups of genes: (i) those encoding proteins that confer adhesion to mucus and (ii) those encoding proteins required to liberate, import and metabolize monosaccharides from the complex glycan structures, such as GHs, sulfatases or proteases. Although the diversity of bacteria that have been studied to date is somewhat limited, the existing information provides a glimpse of themes that are likely to become pervasive for intestinal isolates as new genera and species become the focus of detailed laboratory investigation.
Members of the genera Lactobacillus and Bifidobacterium, and specifically strains used as probiotics, have been the focus of several mucus adhesion studies. In Lactobacillus, surface-associated proteins (e.g. anchored to the cell wall) have been described as adhesion factors (Buck et al. 2005; Velez et al. 2007; Goh and Klaenhammer 2010). Lactobacillus reuteri encodes the mucus-binding protein MUB, which contains a repeated MucBP domain (Roos and Jonsson 2002). Homologs of MUB and MucBP-type domain have been found in multiple Lactobacillus spp. (Boekhorst et al. 2006; MacKenzie et al. 2010) and are implicated in mucin adhesion. Lactobacillus rhamnosus GG contains cell wall bound pili, generally associated with the ability to form biofilms (Kankainen et al. 2009). Interestingly, this strain was originally selected for mucus-binding ability (Gorbach 2000) and a recent comparative genomic study has identified a unique genomic region encoding pilus genes within strain GG (Douillard et al. 2013). 32-Mmubp in Lactobacillus fermentum (Macias-Rodriguez et al. 2009), the mannose-specific adhesin protein in Lactobacillus plantarum (Miyoshi et al. 2006) and the mucus adhesion protein MapA in L. reuteri and L. fermentum (Rojas et al. 2002; Miyoshi et al. 2006) are additional examples of different surface proteins that facilitate adhesion of different Lactobacillus spp. to intestinal mucin.
Diverse extracellular proteins appear to be employed by Bifidobacterium spp. in mucus binding and adherence, although direct functional tests are required to validate several predictions. Functional genomic analysis of Bifidobacterium breve revealed type IVb pilus-type proteins are required to facilitate host-colonization (O'Connell Motherway et al. 2011). Mucin-binding assays with cloned Bifidobacterium bifidum extracellular transaldolase demonstrated that the expression of this extracellular protein was correlated with a mucin-binding phenotype (Gonzalez-Rodriguez et al. 2012). Bifidobacterium longum NCC2705 encodes a protein with high homology to type 2 glycoprotein-binding fimbriae, which are cell-surface filaments that can mediate the adhesion to host mucus (Schell et al. 2002). The use of glycan arrays revealed that family-1 of solute binding proteins encoded in Bifidobacterium longum subsp. infantis genome has an affinity for host glycans (Garrido et al. 2011).
The absence of other information related to mucus adherence in other gut resident microbes is likely due to a lack of study. For example, it is likely that Bacteroides spp. encode novel types of mucus binding machinery, as suggested in recent work on B. fragilis (Huang et al. 2011). Emerging genetic screening tools along with an increasing number of investigators developing (and sharing) methods to investigate mucus adherence promise to push this field rapidly forward in the immediate future.
Genomic and metagenomic sequencing have greatly expanded the number of predicted microbial enzymes that degrade polysaccharides, the GHs and polysaccharide lyases (PL; Cantarel et al. 2009). Based on these activities predicted from the GH/PL profile of a given organism's or ecosystem's sequence, it is possible to infer the polysaccharide degrading potential, and these predictions are fueling hypothesis-driven experiments that aid definitive characterization (Sonnenburg et al. 2010; Martens et al. 2011). Figure 2B shows GH activities that are required for mucin degradation, and co-occurrence of these enzymes in bacterial operons often serve as a signature of mucin-degrading capacity of a microbe. These enzymes are typically exo-hydrolases (release monosaccharides from the non-reducing terminus of the glycan) and are often secreted or cell-surface associated likely in order to facilitate the liberation of terminal monosaccharides from the massive mucin molecules. Microbes adept at mucin degradation often encode sulfatases, acetylases, α-sialidases and α-fucosidases. The removal of these monosaccharides and modifications that often terminate mucin glycan structures is required for further monosaccharide liberation from the extended core structures. The free monosaccharides produced in this de-capping process may be utilized by the bacterium responsible for liberation in either catabolic or biosynthetic (e.g. capsule biosynthesis) metabolism or shed into the ecosystem for consumption by another microbe (Comstock et al. 2000; Troy et al. 2010; Marcobal et al. 2011). Production of α-N-acetylgalactosaminidase that allows the hydrolysis of the common O-glycosidic linkages between galactose β1-3/4-N-acetylgalactosamine and serine or threonine in mucin peptide backbone is another indication of a mucin-adapted microorganism (Ruas-Madiedo et al. 2008). Furthermore, the presence of N-acetyl-β-hexosaminidases, β-galactosidases or lacto-N-biosidases (Wada et al. 2008) is additional activities that are consistent with mucin degradation.
Bacteroides have long been known to utilize a broad spectrum of polysaccharides including mucin glycans (Roberton and Stanley 1982; Macfarlane and Gibson 1991). Studies over the past two decades, largely focused on the model human gut symbiont Bacteroides thetaiotaomicron (Bt) have provided genomic and mechanistic understanding of polysaccharide use by members of this genus (D'Elia and Salyers 1996; Shipman et al. 2000; Xu et al. 2007). Functional genomics and molecular genetics have expanded our insight into how the machinery encoded by a polysaccharide utilization locus (PUL) accomplishes the task of turning an extracellular glycan into an intracellular glycolytic substrate and how a single bacterium can efficiently regulate over 80 loci with differing specificities depending upon nutrient availability (Bjursell et al. 2006; Martens et al. 2008; Sonnenburg et al. 2010; Lynch and Sonnenburg 2012).
PULs are characterized by the presence of homologs of two starch utilization related genes susC and susD (Reeves et al. 1996; Martens et al. 2008) and often encode glycolytic enzymes, sensor regulators and other accessory proteins such as sulfatases and monosaccharide transporters. susC encodes a TonB-dependent transporter involved in importing starch into the periplasm (Reeves et al. 1996), and susD encodes a secreted α-helical starch binding protein associated with the outer Bt membrane (Koropatkin et al. 2008). Numerous Bt PULs involved in mucin glycan consumption have been described in vitro and in vivo and the deletion of five of these loci compromised Bt's transmission from gnotobiotic mouse mother to offspring, an event in which mucin glycan use appears to be critical (Sonnenburg et al. 2005; Bjursell et al. 2006; Martens et al. 2008). It is clear that at least some of these loci respond to discrete structural motifs, like core-1 disaccharide (Martens et al. 2008) consistent with structural and biochemical work on sensor regulators that delineate-specific recognition of host glycans (Lowe et al. 2012). More recently, studies addressing how a PUL-associated sensor regulator enables Bt to prioritize a class of dietary glycans over a mucus carbohydrate provided some insight into the role of mucin glycans in Bt's life in a dynamic gut ecosystem (Lynch and Sonnenburg 2012). Indeed, the ability of Bacteroides to adaptively forage on available carbohydrates and switch between dietary glycans and host mucin based on availability and a substrate priority hierarchy appears to be a hallmark of this genus (Sonnenburg et al. 2005; Xu et al. 2007).
The importance of sulfatase production has also been described for Bt (Dierks et al. 2003; Benjdia et al. 2007, 2011; Carlson et al. 2008). Twenty putative sulfatase genes have been identified in the Bt genome as well as one anaerobic sulfatase-maturing enzyme (anSME) gene required for sulfatases activity. Deletion of anSME results in the loss of Bt sulfatase activity, which is essential for Bt gut colonization and sulfated glycan consumption. Analysis of sequenced Bacteroidetes genomes revealed that anSME and sulfatases are encoded within many species (Benjdia et al. 2011), consistent with earlier studies showing other Bacteroides are able to release sulfate from mucin (Willis et al. 1996).
The ability to metabolize mucin glycans appears to be somewhat restricted within Bifidobacterium (Ruas-Madiedo et al. 2008; Wasilewska et al. 2008), with B. bifidum appearing to be the most adept at this function. Whole-genome sequencing of the B. bifidum strain PRL2010 revealed a large proportion of GHs associated with mucin degradation, most of which are uniquely present in B. bifidum relative to other sequenced strains within this genus (Turroni et al. 2011). Proteomics and transcriptomics reveal the expression of several genes related to mucin breakdown: two exo-α sialidases, a α1-2-l-fucosidase, two α1-3-l-fucosidases, a putative endo-α-N-acetylgalactosaminidase and a lacto-N-biosidase. In addition, the B. bifidum genome encodes four N-acetyl-β-hexosaminidases, four β-galactosidases as well as several putative carbohydrates transporters that may aid in import of the mucin glycan degradation products. Comparative genomic hybridization from different B. bifidum strains links the absence of a specific putative carbohydrate transporter and α1-2-l-fucosidase to reduced growth on mucin-containing media (Turroni et al. 2011).
Akkermansia muciniphila of the Verrucomicrobia phylum is a prominent member of the human microbiota and was isolated in pure culture due to its capacity to grow on mucin (Derrien et al. 2004). The genome sequence from A. muciniphila reveals numerous putative mucin-degradation-related genes. The small size of the Akkermansia genome along with the high abundance of mucin-related genes (predicted 11% of open reading frames encode for mucin-consumption-related functions) suggests that this microorganism has specialized in mucus use, in contrast to many of the mucin-utilizing Bacteroides that exhibit broad polysaccharide degradation potential. The Akkermansia sp. genome encodes eleven N-acetyl-β-hexosaminidases, two α1-2-fucosidases, four sialidases and two β-galactosidases, most of which are characterized by the presence of signal peptide suggesting extracellular functions, explaining its ability to deplete complex glycan structures produced by the host. However, this species lacks the canonical mucus-binding domains involved in the adherence to the intestinal mucus layer, indicating possible novel mucin adhering mechanisms. The depletion of Akkermansia in the mucosa of individual's experiencing IBD compared with healthy controls (Png et al. 2010) is consistent with its adaptation to a non-inflamed mucosal environment.
Multiple other taxa appear to be endowed with mucin glycan utilization abilities (Hoskins and Zamcheck 1968; Salyers et al. 1977; Pultz et al. 2006), and detailed investigations are likely to be highly informative to the extent of convergent functionalities between distantly related bacteria. The role of lateral gene transfer from other gut residents or environmental microbial gene reservoirs in honing and altering the specificity of Bacteroides PULs has been demonstrated for certain dietary polysaccharides (Hehemann et al. 2010, 2012). As the rules governing the evolution of glycan consumption emerge, it will be interesting to see the extent of symmetry between how systems specific for dietary polysaccharides versus those that target host mucin glycans adapt to the respective carbohydrate dynamics.
Sophistication in mucus use: a trait more likely in co-evolved gut residents?
Human gut pathogens interact with host glycans in diverse ways (Moran et al. 2011); however, many appear to be poorly adapted to utilize complex glycans associated with host mucus. For those bacteria that cause disease as a strategy, it is possible that the absence of complex mucus-adapted machinery is due to a pathogenic lifestyle. Host antagonism often results in the immune-mediated eradication of the microbe from either the gut or mucosal sites or reduced host fitness (which translates into reduced microbiota transmission/fitness). Therefore, changes in the host glycan landscape that occur over developmental, spatial or evolutionary windows may be difficult for transiently interacting pathogens to adapt to. Co-adaptation between symbiont and host mucus glycans is supported by the selective interaction between the squid Euprymna scolopes and its light organ symbiont Vibrio harveyi and extends to microbial regulation of glycan structure (Nyholm et al. 2000). For example, the ability of Bt to up-regulate host gut fucosyltransferase as a way to increase the “harvestable” fucose on mucus glycans, an ability that depends upon the bacterium's ability to catabolize that sugar suggests that microbes may be adapted to manipulate and shape the glycan landscape to suit their consumption preferences (Bry et al. 1996; Hooper et al. 1999).
Several examples exist that support a reliance of gut pathogens on commensal GHs for mucus monosaccharide liberation. Salmonella typhimurium and Clostridium difficile, two pathogens common to the human GI tract, possess the ability to catabolize the mucosal monosaccharide sialic acid, but neither encode machinery necessary to harvest sialylated glycan on their own, including the absence of a sialidase. Both pathogens are therefore reliant upon other members of the resident microbiota to liberate this sugar from glycosidic linkage (K. Ng, J. Ferreyra, J. Sonnenburg, manuscript in preparation). Commensal liberated fucose provides an advantage to Campylobacter jejuni in vivo (Stahl et al. 2011). Similarly, there is evidence that vancomycin resistant Enterococci consume mucosal carbohydrates that have been liberated by other commensal microbes (Pultz et al. 2006). Enterohaemorrhagic E. coli has been shown to rely upon microbiota-liberated fucose as a signal to induce virulence and metabolic transcriptional programs in the gut (Pacheco et al. 2012). And detailed analysis of pathogenic and non-pathogenic E. coli strains reveals a capacity to use diverse monosaccharides derived from host glycans despite lacking many of the corresponding GHs (Fabich et al. 2008). Therefore, cross-feeding of commensal carbohydrates appears to be an important theme in enteric pathogen strategy; preliminary genomic analysis of common gut pathogens indicates that many are limited in the GHs necessary for liberation of mucus-derived monosaccharides (S. Smits, A. Marcobal, J. Sonnenburg, unpublished data).
Clearly, many selective pressures shape intestinal mucin glycan structures including interactions with enteric pathogens and mutualistic microbiota residents (Figure 3). Several intestinal mutualists, such as Bacteroides species, have adapted to use carbohydrates present in the distal intestine including dietary plant polysaccharides and host mucus as carbon and energy sources. These non-pathogenic residents have spent eons co-evolving with host-mucus structures. Considering co-evolution between a microbe and host mucus would be expected to result from a long, sustained association, it is probable that sophistication in mucus consumption is associated with bacteria that are unlikely to cause disease. The presence of soluble mucin-like glycans in human milk (i.e. HMO) may represent an attempt to control the rapid and chaotic assembly of a newborn infant's intestinal microbiota. Promoting colonization by beneficial or benign species may be best accomplished by attracting mucus-adapted bacteria. Therefore, an infant may gain a selective advantage if milk oligosaccharides are confined to structures that co-opt extant mucin glycan utilization pathways. Attracting such mucin-adapted resident mutualists, such as Bacteroides spp. that can adaptively forage on dietary glycans, may provide the added benefit of ensuring some stability in microbiota composition as the infant diet transitions to solid food.
Fig. 3.
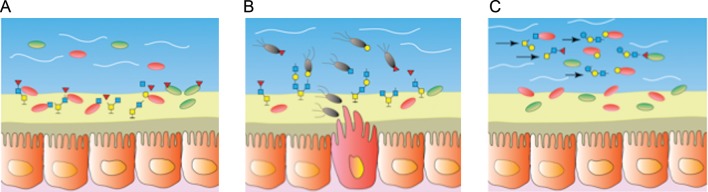
The role of mucus in supporting a gut ecosystem. (A) Mucus supports an adhesive community of microbial mutualists that dine on and embed within the outer loose layer of mucus. Degradation of the structural complexity of mucin glycans is accomplished by this community of highly adapted gut residents. (B) During times of perturbation, efficient partitioning of host mucus resources may be disrupted enabling less adapted strains, like pathogenic species (represented with flagellae) to cross-feed on free monosaccharides. (C) Milk oligosaccharides offer an orally delivered mucin-glycan-like substrate to the distal gut to aid in the nutritional support and establishment of a beneficial community of microbes.
Diet and the extent of microbiota reliance upon mucin: food for thought (and microbes)
It is worthwhile to consider that existing data suggest that decreased dietary plant polysaccharides shifts the burden of microbiota nutritional support to host mucus glycans. This was originally demonstrated in vivo for Bt in the gnotobiotic mouse intestine: depletion of polysaccharides from the host diet resulted in profound shift by the gut symbiont to host mucus consumption (Sonnenburg et al. 2005). Accruing data suggest that decreased fiber or plant material in host diet also leads to decreases in microbiota diversity and shifts in composition consistent with a reliance upon host mucus. For example, in the gut of hibernating ground squirrels in late winter, the microbiota exhibits loss of diversity and enrichment of Bacteroidetes and Akkermansia spp. (Carey et al. 2013). Additionally, microbiotas of the industrialized world have much less diversity compared with the microbiota of traditional societies, which correspond to vastly different lifestyles that include much lower dietary fiber consumption in “Westerners” (De Filippo et al. 2010; Yatsunenko et al. 2012). The increased reliance upon host mucus in the presence of a Western diet has been demonstrated in mice (Mahowald et al. 2009) and microbial breach of colonic mucus is observed in ulcerative colitis (Johansson et al. 2013). So while the ability to utilize complex mucus glycan appears to be a trait of co-evolved gut symbionts and may play an important role in structuring the microbiota during infancy or after microbiota disturbance, the effects of an increasing reliance of gut microbes on host derived carbohydrates for extended periods of time and the relevance to Western diseases is worthy of detailed investigation.
Funding
This work was supported in part by a grant from the NIH (DK085025). J.L.S. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Conflict of interest
None declared.
Abbreviations
anSME, anaerobic sulfatase-maturing enzyme; Bt, Bacteroides thetaiotaomicron; GH, glycoside hydrolase; GI, gastrointestinal; HMO, human milk oligosaccharide; PL, polysaccharide lyase; PUL, polysaccharide utilization locus.
Acknowledgements
We thank Erica Sonnenburg for critical comments on the manuscript.
References
- An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. doi:10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CE, Turner RJ. Examination of organisms associated with mucin in the colon by scanning electron-microscopy. Micron. 1982;13:35–40. [Google Scholar]
- Benjdia A, Leprince J, Guillot A, Vaudry H, Rabot S, Berteau O. Anaerobic sulfatase-maturating enzymes: Radical SAM enzymes able to catalyze in vitro sulfatase post-translational modification. J Am Chem Soc. 2007;129:3462–3463. doi: 10.1021/ja067175e. doi:10.1021/ja067175e. [DOI] [PubMed] [Google Scholar]
- Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem. 2011;286:25973–25982. doi: 10.1074/jbc.M111.228841. doi:10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. doi:10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. doi:10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology. 2006;152:273–280. doi: 10.1099/mic.0.28415-0. doi:10.1099/mic.0.28415-0. [DOI] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. doi:10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Buck BL, Altermann E, Svingerud T, Klaenhammer TR. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2005;71:8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. doi:10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. doi:10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon C, Maes E, Michalski JC, Leffler H, Kim YS. Sd(a)-antigen-like structures carried on core 3 are prominent features of glycans from the mucin of normal human descending colon. Biochem J. 2001;358:657–664. doi: 10.1042/bj3580657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey HV, Walters WA, Knight R. Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am J Physiol Regul Integr Comp Physiol. 2013;304:R33–R42. doi: 10.1152/ajpregu.00387.2012. doi:10.1152/ajpregu.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BL, Ballister ER, Skordalakes E, King DS, Breidenbach MA, Gilmore SA, Berger JM, Bertozzi CR. Function and structure of a prokaryotic formylglycine-generating enzyme. J Biol Chem. 2008;283:20117–20125. doi: 10.1074/jbc.M800217200. doi:10.1074/jbc.M800217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Pantosti A, Kasper DL. Genetic diversity of the capsular polysaccharide C biosynthesis region of Bacteroides fragilis. Infect Immun. 2000;68:6182–6188. doi: 10.1128/iai.68.11.6182-6188.2000. doi:10.1128/IAI.68.11.6182-6188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield AP, Carroll D, Myerscough N, Probert CSJ. Mucins in the gastrointestinal tract in health and disease. Front Biosci. 2001;6:D1321–D1357. doi: 10.2741/corfield. doi:10.2741/Corfield. [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. doi:10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia JN, Salyers AA. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1996;178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. doi:10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Dierks T, Schmidt B, Borissenko LV, Peng JH, Preusser A, Mariappan M, von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C-α-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. doi:10.1016/S0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Jarvinen HM, Kant R, Pietila TE, Randazzo C, Paulin L, Laine PK, Caggia C, von Ossowski I, et al. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol. 2013;79:1923–1933. doi: 10.1128/AEM.03467-12. doi:10.1128/AEM.03467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. doi:10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. doi:10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks I. Gut microbiota: FUT2 genotype influences the gut microbiota in patients with Crohn's disease and healthy individuals. Nat Rev Gastroenterol Hepatol. 2012;9:2–2. doi: 10.1038/nrgastro.2011.237. doi:10.1038/nrgastro.2011.237. [DOI] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T, Johansson MEV, Liu X, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. doi:10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One. 2011;6:e17315–e17315. doi: 10.1371/journal.pone.0017315. doi:10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–3020. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- Goh YJ, Klaenhammer TR. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2010;76:5005–5012. doi: 10.1128/AEM.00030-10. doi:10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rodriguez I, Sanchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol. 2012;78:3992–3998. doi: 10.1128/AEM.08024-11. doi:10.1128/AEM.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbach SL. Probiotics and gastrointestinal health. Am J Gastroenterol. 2000;95:S2–S4. doi: 10.1016/s0002-9270(99)00806-0. doi:10.1016/S0002-9270(99)00806-0. [DOI] [PubMed] [Google Scholar]
- Hanisch FG, Chai WG, Rosankiewicz JR, Lawson AM, Stoll MS, Feizi T. Core-typing of O-linked glycans from human gastric mucins-lack of evidence for the occurrence of the core sequence Gal1-6-GalNAc. Eur J Biochem. 1993;217:645–655. doi: 10.1111/j.1432-1033.1993.tb18288.x. doi:10.1111/j.1432-1033.1993.tb18288.x. [DOI] [PubMed] [Google Scholar]
- Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. doi:10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J-H, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–U123. doi: 10.1038/nature08937. doi:10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci USA. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. doi:10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. doi:10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins LC, Zamcheck N. Bacterial degradation of gastrointestinal mucins. Comparison of mocus constituents in stools of germ-free and conventional rats. Gastroenterology. 1968;54:210. [PubMed] [Google Scholar]
- Huang JY, Lee SM, Mazmanian SK. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe. 2011;17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. doi:10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail MN, Stone EL, Panico M, Lee SH, Luu Y, Ramirez K, Ho SB, Fukuda M, Marth JD, Haslam SM, et al. High-sensitivity O-glycomic analysis of mice deficient in core 2 beta 1,6-N-acetylglucosaminyltransferases. Glycobiology. 2011;21:82–98. doi: 10.1093/glycob/cwq134. doi:10.1093/glycob/cwq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, Hannson GC. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2013 doi: 10.1136/gutjnl-2012-303207. [Epub ahead of print 20 February 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Larsson JMH, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. doi:10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. doi:10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson NG, Herrmann A, Karlsson H, Johansson MEV, Carlstedt I, Hansson GC. The glycosylation of rat intestinal Muc2 mucin varies between rat strains and the small and large intestine—A study of O-linked oligosaccharides by a mass spectrometric approach. J Biol Chem. 1997;272:27025–27034. doi: 10.1074/jbc.272.43.27025. doi:10.1074/jbc.272.43.27025. [DOI] [PubMed] [Google Scholar]
- Karlsson NG, Nordman H, Karlsson H, Carlstedt I, Hansson GC. Glycosylation differences between pig gastric mucin populations: A comparative study of the neutral oligosaccharides using mass spectrometry. Biochem J. 1997;326:911–917. doi: 10.1042/bj3260911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. doi:10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JMH, Karlsson H, Crespo JG, Johansson MEV, Eklund L, Sjovall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–2307. doi: 10.1002/ibd.21625. doi:10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- Larsson JMH, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19:1568–1569. doi: 10.1093/glycob/cwp048. doi:10.1093/glycob/cwp090. [DOI] [PubMed] [Google Scholar]
- Lowe EC, Basle A, Czjzek M, Firbank SJ, Bolam DN. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci USA. 2012;109:7298–7303. doi: 10.1073/pnas.1200479109. doi:10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R. Identifying genomic and metabolic features that can underline early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 2012;22:1974–1984. doi: 10.1101/gr.138198.112. doi:10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JB, Sonnenburg JL. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol. 2012;85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. doi:10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR. Formation of glycoprotein degrading enzymes by Bacteroides fragilis. FEMS Microbiol Lett. 1991;77:289–293. doi: 10.1016/0378-1097(91)90567-t. doi:10.1111/j.1574-6968.1991.tb04363.x. [DOI] [PubMed] [Google Scholar]
- Macias-Rodriguez ME, Zagorec M, Ascencio F, Vazquez-Juarez R, Rojas M. Lactobacillus fermentum BCS87 expresses mucus- and mucin-binding proteins on the cell surface. J Appl Microbiol. 2009;107:1866–1874. doi: 10.1111/j.1365-2672.2009.04368.x. doi:10.1111/j.1365-2672.2009.04368.x. [DOI] [PubMed] [Google Scholar]
- MacKenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, Roos S, Walter J, Juge N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology. 2010;156:3368–3378. doi: 10.1099/mic.0.043265-0. doi:10.1099/mic.0.043265-0. [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. doi:10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzi AE, Norgard-Sumnicht K, Argade S, Marth JD, van Halbeek H, Varki A. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology. 2000;10:669–689. doi: 10.1093/glycob/10.7.669. doi:10.1093/glycob/10.7.669. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. doi:10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. doi:10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001221. doi:10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DPB, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. doi:10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JB, McVeagh P. Human milk oligosaccharides: 130 reasons to breast-feed. Br J Nutr. 1999;82:333–335. doi: 10.1017/s0007114599001567. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Okada S, Uchimura T, Satoh E. A mucus adhesion promoting protein, MapA, mediates the adhesion of Lactobacillus reuteri to Caco-2 human intestinal epithelial cells. Biosci Biotechnol Biochem. 2006;70:1622–1628. doi: 10.1271/bbb.50688. doi:10.1271/bbb.50688. [DOI] [PubMed] [Google Scholar]
- Moran AP, Gupta A, Joshi L. Sweet-talk: Role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut. 2011;60:1412–1425. doi: 10.1136/gut.2010.212704. doi:10.1136/gut.2010.212704. [DOI] [PubMed] [Google Scholar]
- Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. doi:10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Nordman H, Davies JR, Herrmann A, Karlsson NG, Hansson GC, Carlstedt I. Mucus glycoproteins from pig gastric mucose: Identification of different mucin populations from the surface epithelium. Biochem J. 1997;326:903–910. doi: 10.1042/bj3260903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. doi:10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JAM, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108:11217–11222. doi: 10.1073/pnas.1105380108. doi:10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. doi:10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar AS, Lappalainen M, Paavola-Sakki P, Halme L, Farkkila M, Turunen U, Kontula K, Aromaa A, Salomaa V, Peltonen L, et al. Association of celiac disease genes with inflammatory bowel disease in Finnish and Swedish patients. Genes Immun. 2012;13:474–480. doi: 10.1038/gene.2012.21. doi:10.1038/gene.2012.21. [DOI] [PubMed] [Google Scholar]
- Patsos G, Corfield A. Management of the human mucosal defensive barrier: Evidence for glycan legislation. Biol Chem. 2009;390:581–590. doi: 10.1515/BC.2009.052. doi:10.1515/BC.2009.052. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. doi:10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Podolsky DK. Oligosaccharides structures of huma colonic mucin. J Biol Chem. 1985a;260:8264–8271. [PubMed] [Google Scholar]
- Podolsky DK. Oligosaccharides structures of isolated human colonic mucin species. J Biol Chem. 1985b;260:5510–5515. [PubMed] [Google Scholar]
- Pultz NJ, Vesterlund S, Ouwehand AC, Donskey CJ. Adhesion of vancomycin-resistant Enterococcus to human intestinal mucus. Curr Microbiol. 2006;52:221–224. doi: 10.1007/s00284-005-0244-2. doi:10.1007/s00284-005-0244-2. [DOI] [PubMed] [Google Scholar]
- Rausch P, Rehman A, Kuenzel S, Haesler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. doi:10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Delia JN, Frias J, Salyers AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. doi:10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, Michalski JC. Evidence of regio-specific glycosylation in human intestinal mucins—Presence of an acidic gradient along the intestinal tract. J Biol Chem. 2003;278:46337–46348. doi: 10.1074/jbc.M302529200. doi:10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- Robbe-Masselot C, Maes E, Rousset M, Michalski J-C, Capon C. Glycosylation of human fetal mucins: A similar repertoire of O-glycans along the intestinal tract. Glycoconj J. 2009;26:397–413. doi: 10.1007/s10719-008-9186-9. doi:10.1007/s10719-008-9186-9. [DOI] [PubMed] [Google Scholar]
- Roberton AM, Stanley RA. In vitro utilization of mucin by Bacteroides fragilis. Appl Environ Microbiol. 1982;43:325–330. doi: 10.1128/aem.43.2.325-330.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Ascencio F, Conway PL. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl Environ Microbiol. 2002;68:2330–2336. doi: 10.1128/AEM.68.5.2330-2336.2002. doi:10.1128/AEM.68.5.2330-2336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos S, Jonsson H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology. 2002;148:433–442. doi: 10.1099/00221287-148-2-433. [DOI] [PubMed] [Google Scholar]
- Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–1940. doi: 10.1128/AEM.02509-07. doi:10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45:442–451. doi: 10.5483/BMBRep.2012.45.8.161. doi:10.5483/BMBRep.2012.45.8.161. [DOI] [PubMed] [Google Scholar]
- Salyers AA, West SEH, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from human colon. Appl Environ Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. doi:10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Mills DA. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. doi:10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. doi:10.1128/JB.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomiany BL, Murty VLN, Slomiany A. Isolation and characterization of oligosaccharides from rat colonic mucus glycoprotein. J Biol Chem. 1980;255:9719–9723. [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. doi:10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. doi:10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–U1256. doi: 10.1016/j.cell.2010.05.005. doi:10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. L-Fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc Natl Acad Sci USA. 2011;108:7194–7199. doi: 10.1073/pnas.1014125108. doi:10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, Hansson GC. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fuc α1-2 glycosyltransferase. Biochem J. 2002;367:609–616. doi: 10.1042/BJ20020371. doi:10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsson KA, Holmen-Larsson JM, Angstrom J, Johansson MEV, Xia L, Hansson GC. Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1-and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology. 2012;22:1128–1139. doi: 10.1093/glycob/cws083. doi:10.1093/glycob/cws083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy EB, Carey VJ, Kasper DL, Comstock LE. Orientations of the Bacteroides fragilis capsular polysaccharide biosynthesis locus promoters during symbiosis and infection. J Bacteriol. 2010;192:5832–5836. doi: 10.1128/JB.00555-10. doi:10.1128/JB.00555-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F, Milani C, van Sinderen D, Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: An example of possible human-microbe co-evolution. Gut microbes. 2011;2:183–189. doi: 10.4161/gmic.2.3.16105. doi:10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- Urashima T, Asakuma S, Leo F, Fukuda K, Messer M, Oftedal OT. The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv Nutr. 2012;3:473S–482S. doi: 10.3945/an.111.001412. doi:10.3945/an.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings JH, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2nd ed. Cold Spring Harbor (NY): CSHL Press; 2009. O-GalNAc glycans. [PubMed] [Google Scholar]
- Velez MP, De Keersmaecker SCJ, Vanderleyden J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol Lett. 2007;276:140–148. doi: 10.1111/j.1574-6968.2007.00908.x. doi:10.1111/j.1574-6968.2007.00908.x. [DOI] [PubMed] [Google Scholar]
- Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. doi:10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska E, Markiewicz LH, Bielecka M. Stimulation of Bifidobacteria growth by oligofructose and mucin. Polish J Food Nutr Sci. 2008;58:443–449. [Google Scholar]
- Willis CL, Cummings JH, Neale G, Gibson GR. In vitro effects of mucin fermentation on the growth of human colonic sulphate-reducing bacteria. Anaerobe. 1996;2:117–122. doi:10.1006/anae.1996.0015. [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. doi:10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:1574–1586. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z-T, Chen C, Kling DE, Liu B, McCoy JM, Merighi M, Heidtman M, Newburg DS. The principal fucosylated oligosaccharides of human milk exhibit prebiotic properties on cultured infant microbiota. Glycobiology. 2013;23:169–177. doi: 10.1093/glycob/cws138. doi:10.1093/glycob/cws138. [DOI] [PMC free article] [PubMed] [Google Scholar]