Abstract
Catalytic hairpin assembly (CHA) is an enzyme-free amplification method that has previously proven useful in amplifying and transducing signals at the terminus of nucleic acid amplification reactions. Here, for the first time, we engineered CHA be thermostable from 37 °C to 60 °C and in consequence have generalized its application to real-time detection of isothermal amplification reactions. CHA circuits were designed and optimized for both high and low temperature rolling circle amplification (RCA) and strand displacement amplification (SDA). The resulting circuits not only increased the specificity of detection, they also improved sensitivity by as much as 25- to 10,000-fold over comparable real-time detection methods. These methods have been condensed into a set of general rules for the design of thermostable CHA circuits with high signals and low noise.
Isothermal nucleic acid amplification reactions such as nucleic acid sequence-based amplification (NASBA),1 signal-mediated amplification of RNA technology (SMART),2 rolling circle amplification (RCA),3 strand displacement amplification (SDA),4 and loop-mediated isothermal amplification (LAMP)5 may be much more tractable than PCR for the amplification of nucleic acid analytes in point-of-care applications. However, the robust detection of amplicons remains analytically difficult, and in many cases still relies on the detection of final products by methods such as electrophoresis followed by dye staining (using ethidium bromide or SYBR green)6–8 or through monitoring the increase in calcein fluorescence9 or solution turbidity due to pyrophosphate release.10,11 These methods can allow for real-time monitoring of signals but sacrifice specificity because any accumulation of non-specific or parasitic amplicons will yield a false positive signal. While some sequence-specific detection methods have also been developed, such as fluorescent resonance transfer (FRET) probes,12,13 molecular zippers,14 molecular beacons,15,16 and hybridization between DNA-coated gold nanoparticles,17,18 these approaches merely mirror the development of amplicons and provide no opportunity for programming how amplicon signals might be amplified, integrated, and displayed in real-time.
In contrast, non-enzymatic nucleic acid circuits have previously provided a useful means for both amplifying and transducing signals from nucleic acid analytes.19–25 One of these circuits, catalytic hairpin assembly (CHA), has been engineered to yield hundreds-fold catalytic amplification with negligible background and can transduce analyte-binding to a variety of detection modalities, such as fluorescent and electrochemical signals.26–29 Recently, CHA has been used as a specific end-point transducer30 for enzyme-based isothermal amplification reactions, such as LAMP, that sometimes produce parasitic amplicons and result in a high false positive rate.30–32
The further adaptation of such circuits to real-time detection would likely advance their general utility as molecular transducers, just as the use of exonuclease-sensitive (Taqman) probes33 led to great improvements in PCR sensitivities. In particular, the use of CHA transducers in real-time detection schemes may lead to concomitant improvements in the convenience, sensitivity, and programmability of isothermal reactions. However, the development of real-time CHA detection has previously posed several challenges. Many isothermal amplification reactions are carried out at 60 °C or above,34 a temperature that may increase background due to hairpin breathing.35 Therefore, we for the first time engineered a thermostable CHA circuit that could operate at temperatures up to 60 °C without sacrificing performance. This achievement allowed for the use of CHA as a real-time, sequence-specific detector for two isothermal amplification reactions (RCA and SDA). The development of real-time CHA transduction also yielded insights into circuit design that could be immediately applied to the creation of circuits that operate in different reaction conditions.
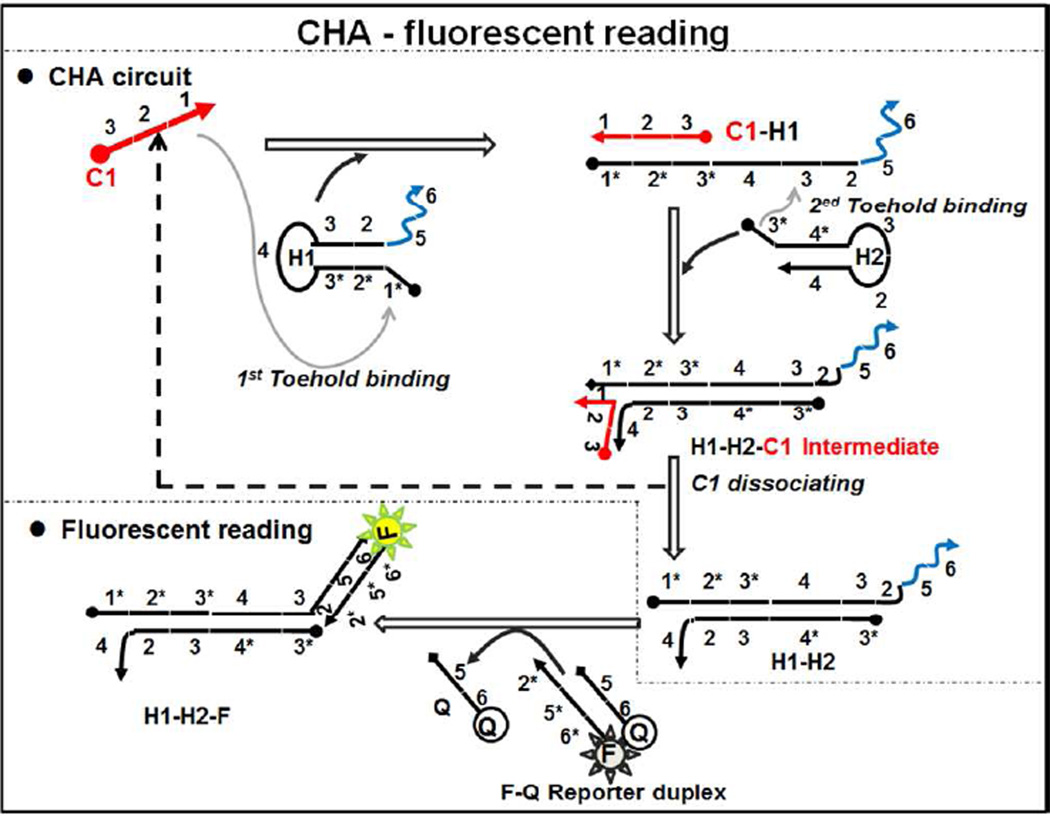
As a first step we investigated whether circuits could be designed to operate at high temperatures. A circuit designed to perform at 37 °C in the presence of 140 mM NaCl and 5 mM KCl at pH 7.5 (hereafter referred to as the TNaK reaction, Table S4) was initially chosen and monitored for performance at higher temperatures. In this circuit (Figure 1), Domain 1 (8-base, Table S1) on C1 (here specified as C1LT, where LT indicates “low temperature”) served as a toehold to bind Domain 1* on H1LT, and initiated a branch migration reaction to open H1LT stem (16-base). Domain 3 (8-base) on H1LT then served as a second toehold to initiate another strand displacement reaction to open H2LT stem (11-base), forming the C1LT-H1LT-H2LT intermediate. C1LT can dissociate from H1LT and catalyze additional hairpin assembly reactions. To monitor the reaction process, H1LT-H2LT initiates strand displacement (via a 7-base toehold) of a Black(R) (QLT) fluorescence quencher from an oligonucleotide-FAM (FLT) signal.
Figure 1.
Scheme of CHA circuit. Complementary domain strands are indicated with an asterisk such that Domain 1 complements Domain 1*.
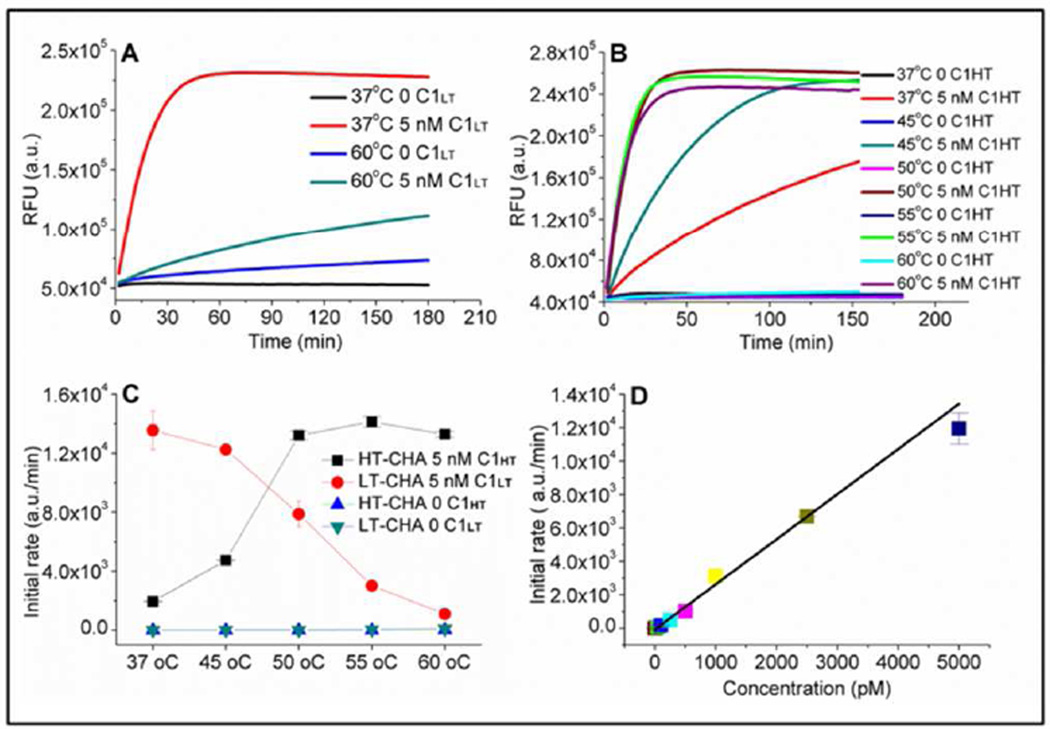
At 37 °C this design showed ca. 50–100 fold amplification within 5–10 hours, and little background leakage (Figure 2A). While the uncatalyzed reaction rate (background leakage rate) remained low below 50 °C (Figures 2C and S1), at 60 °C it increased almost 10-fold (Figure 2A, blue). In parallel, the catalyzed reaction rate decreased sharply with increasing temperature, and was reduced by almost 10-fold at 60 °C (Figure 2A, green). Thus, the circuit designed to function at 37 °C suffered a near 100-fold decrease in performance at 60 °C.
Figure 2.
A) Fluorescence response of LT-CHA at 37 °C and 60 °C, with and without catalyst. B) Fluorescence response of HT-CHA at various temperatures, with and without catalyst. C) Initial rates of LT-CHA and HT-CHA at various temperatures, with and without catalyst, as calculated from the data in A & B. D) Initial rate vs. CHT concentrations for HT-CHA at 60 °C (r2 = 0.999). The sequences and conditions used in these experiments are found in Tables S3 and S4 respectively.
Adaption of the CHA circuit for high-temperature function required improving both the stability of the stem structures (to reduce background) and the binding of the catalyst to the toehold (to increase the catalyzed reaction rate). These conditions could be respectively met by increasing the length of the stem and the length of the toehold. As a basis for design, the predicted free energies for folding and binding at 50 °C (the highest temperature that the original circuit could tolerate well) were used to guide changes in the length and sequence of a modified circuit (Table S2). Three base-pairs were added to the two hairpin stems (H1HT, H2HT, where HT indicates “high temperature”) and to the reporter duplex (FHT-QHT), while the toehold was extended from 8 to 9 nucleotides in length (CHT). A general ‘4-rule’ principle is listed in the supporting information for designing CHA circuits that can function at any temperature. In short, as the desired temperature for circuit function is increased, the lengths of hairpin stems should be extended so as to retain the free energy of base-pairing; the lengths of toeholds should be extended to maintain efficient catalysis; hairpin sequence and length should be optimized to avoid mis-folding and unintended secondary structures; and the lengths of oligonucleotides should not be extended so far as to accumulate excessive impurities during chemical synthesis.
The augmented CHA circuit (HT-CHA) was assayed at various temperatures. At 60 °C it proved to have a catalytic reaction rate as fast as the LT-CHA had at 37 °C. Moreover, it demonstrated 5-fold less background leakage than LT-CHA at 60 °C (Figures 2B and 2C). There was also an excellent linear relationship (r=0.999) between catalyst (C1HT) concentration and the initial rate of the circuit reaction at 60 °C (Figures 2D, S2, and S3). As low as 100 pM C1HT could be discriminated from background (by three standard deviations) within one hour.
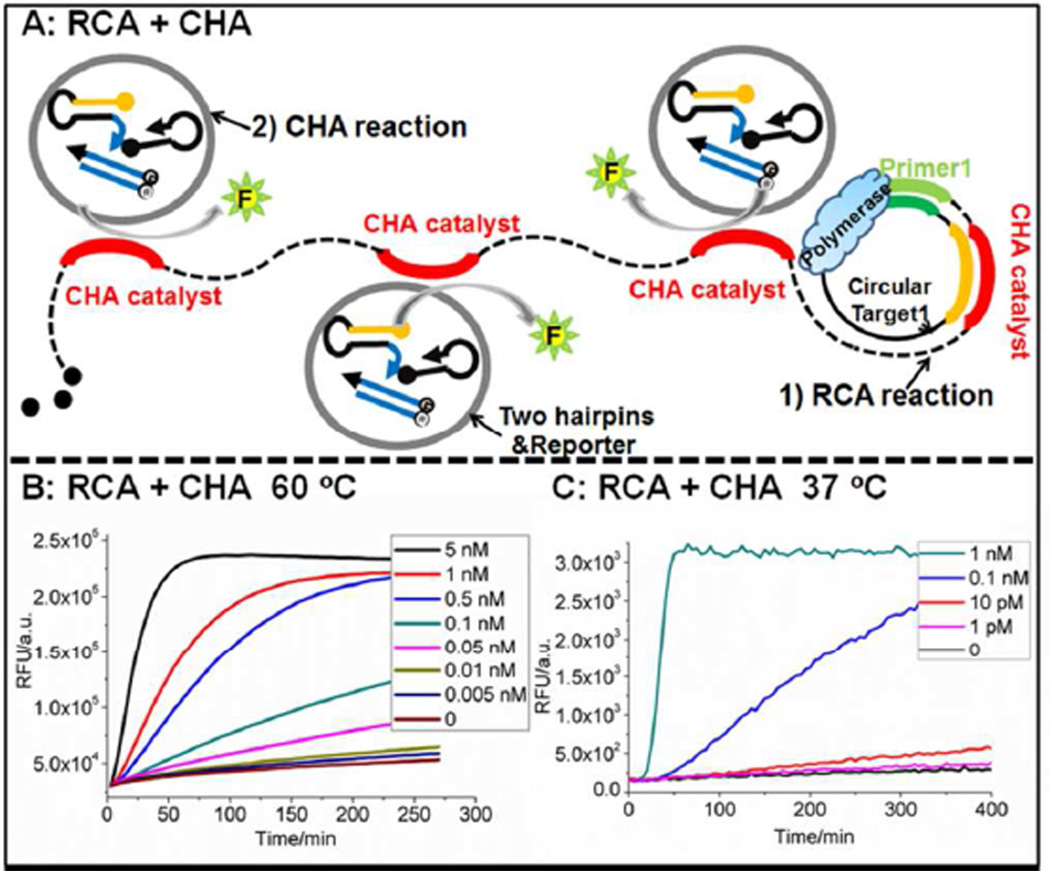
With a functional HT-CHA in hand, the question became whether it could be used as a real-time signal transducer in high-temperature isothermal amplification reactions. A synthetic 77-mer ssDNA containing an antisense template of the CHA catalyst was used to detect rolling circle amplification (RCA) at 60 °C (Figure 3A). Following amplification, each single-stranded monomer should contain the equivalent of the catalyst sequence once, and it will therefore be present thousands of times in each concatemer. The “RCA+CHA” amplification and real-time analysis system was driven by the Bst polymerase at 60 °C in the presence of 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8 (referred to as the ThermoPol reaction, Table S4). Bst polymerase was chosen because it has previously been widely used for high temperature RCA reactions.36–37 The salt concentration in the ThermoPol reaction was lower than that in the TNaK reaction previously used for the design and assay of the HT-CHA circuit. To account for this difference, interactions between the catalyst sequence and Domain 1* of H1 were extended by 1 base-pair, yielding H1EHT (where E denotes the extended toehold). When comparing this redesigned circuit to the original under the new low salt conditions, HT-CHA had a 5-fold decrease in catalytic rate, but EHT-CHA showed only a 2-fold decrease. While EHT-CHA performed better in low salt conditions, there was no noticeable difference in performance between EHT-CHA and HT-CHA in the TNaK reaction, indicating that both toeholds are likely saturated at high salt concentrations.
Figure 3.
A) Scheme for CHA as a real-time detector of linear RCA. B) Concentration dependence of Circular THTRCA of RCA-CHA at 60 °C in the ThermoPol reaction, using EHT-CHA and Bst large fragment polymerase. C) Concentration dependence of Circular TLTRCA of RCA-CHA at 37 °C in the Phi29 reaction, using EHT-CHA and Phi29 polymerase. The sequences and conditions used in these experiments are found in Tables S3 and S4 respectively.
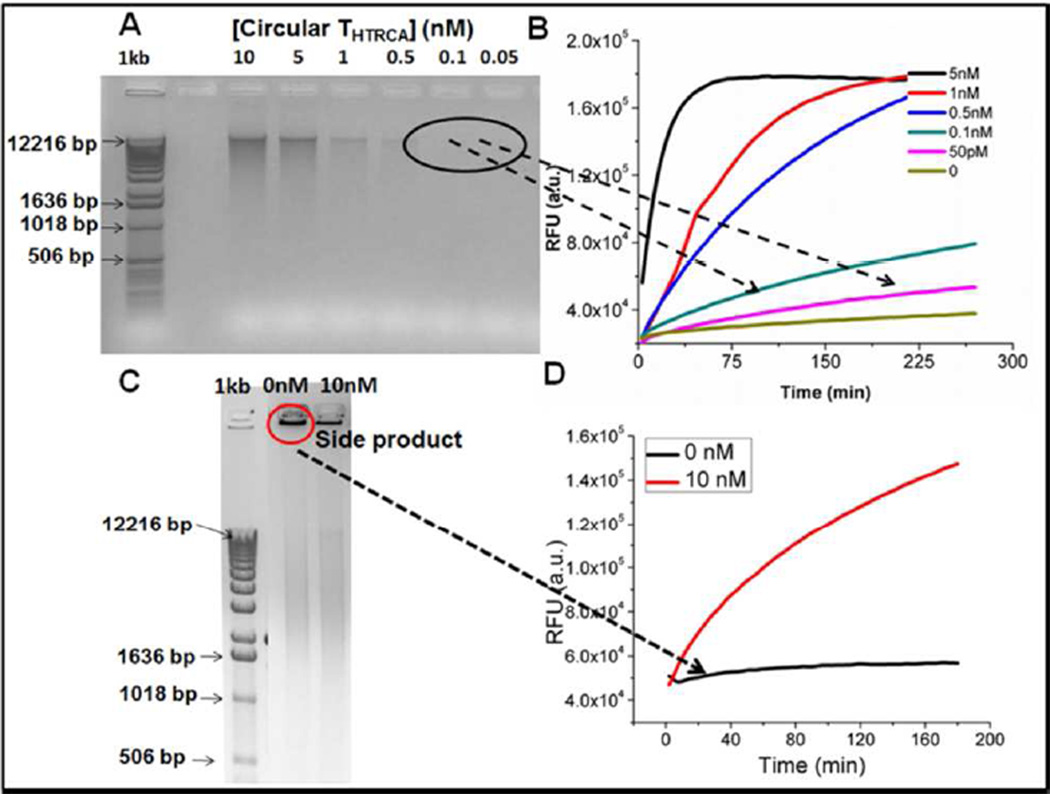
The EHT-CHA was able to monitor RCA in real-time (Figure 3B) and could detect as low as 5 pM circular DNA (Circular THTRCA, 100 amol) within 4.5 hours at 60 °C. As was previously the case for LAMP,30–32 the use of CHA to transduce and report the presence of correct isothermal amplification products leads to much greater specificity of detection than would just monitoring the accumulation of DNA (Figures 4A–4D). While false amplicons accumulate during RCA even in the absence of template, these amplicons are not reported by the EHT-CHA circuit.
Figure 4.
High-temperature CHA transduction is sensitive and specific. A) 1% agarose gel of the product generated from a 2 hour RCA reaction with and without Circular THTRCA. B) The same RCA products from Figure A were diluted 4-fold with EHT-CHA, followed by a CHA reaction in ThermoPol reaction conditions at 60 °C. The concentrations listed are those before dilution. C) 1% agarose gel of the product generated from an overnight RCA reaction with and without Circular THTRCA. D) The same RCA products from Figure C were diluted 4-fold with HT-CHA, followed by a CHA reaction in ThermoPol reaction conditions at 60 °C. The concentrations listed are those before dilution.
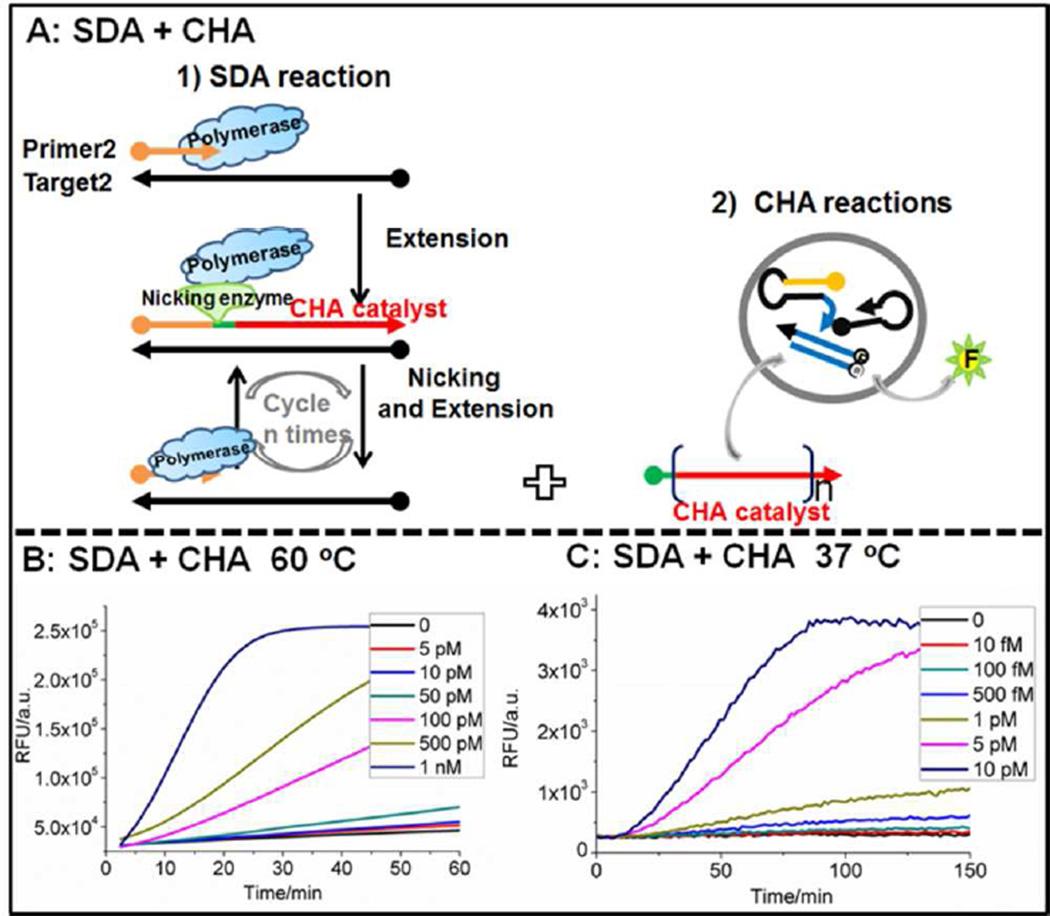
To demonstrate the generality of real-time CHA transduction and detection of isothermal amplification, the same high-temperature CHA circuit was attempted with a different reaction, strand displacement amplification (SDA; Figure 5A). In this instance, primer (PHTSDA) binding initiates an extension reaction by the Bst polymerase. The extended duplex is nicked by the enzyme Nb.BsrDI to generate a single-stranded DNA containing the catalyst sequence. A cycle of extension, nicking, and product dissociation leads to the accumulation of multiple single-stranded DNA catalyst sequences. In this “SDA+CHA” system for real-time analysis, the reaction was carried out in 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9 (referred to as the NEBuffer 2 reaction, Table S4) instead of the ThermoPol reaction conditions so as to improve the coordinate performance of the Bst and Nb.BsrDI enzymes. As low as 5 pM input DNA (100 amol THTSDA) could be detected within one hour (Figure 5B). The performance of HT-CHA and EHT-CHA was similar. As there are almost no real-time sequence-specific detectors for high-temperature linear SDA or RCA, these results potentially open the way for a wider application of nucleic acid circuitry in diagnostics.
Figure 5.
A) Scheme for CHA as a real-time detector of SDA. B) Concentration dependence of Circular THTSDA of SDA-CHA at 60 °C in the NEBuffer 2 reaction using EHT-CHA, Bst large fragment polymerase, and Nb. BsrDI. C) Concentration dependence of Circular TLTSDA of SDA-CHA at 37 °C in the NEBuffer 2 reaction using LT-CHA, Klenow (3’–5’exo-) polymerase, and Nb.BbvCI. The sequences and conditions used in these experiments are found in Tables S3 and S4 respectively.
Given our ability to monitor high-temperature isothermal amplification reactions, the next step was to see if real-time monitoring of lower temperature isothermal amplification reactions might be feasible. This was an especially difficult challenge, since single-stranded products form structures that might occlude binding to the CHA circuits. Once again, CHA was used to try and monitor both RCA and SDA, but at 37 °C. This required another change in the enzymes and hence the reaction conditions used. The RCA reaction uses Phi29 polymerase in 10 mM MgCl2, 10 mM (NH4)2SO4, 4 mM dithiothreitol, pH 7.5 (referred to as the Phi29 reaction, Table S4), while the SDA reaction uses Klenow (3’–5’exo-) polymerase, Nb.BbvCI nicking enzyme, and NEBuffer 2. Both the Phi29 reaction and the NEBuffer 2 reaction have high salt concentrations (10 mM Mg2+) similar to that of the TNaK reaction, and thus the original LT-CHA circuit did not require modification. As shown in Figures 3C and 5C, real-time monitoring of both RCA and SDA reactions was successful. The detection limit for the RCA target (Circular TLTRCA) was 1 pM (20 amol), which is 40-fold lower than a real-time molecular beacon detector.38 Additionally, the detection limit for the SDA target (TLTRCA) was 100 fM (2 amol), which is 25- to 10,000-fold lower than a recently reported real-time detection with molecular beacons39,40 and was better even than electrochemical detectors.41 The improved sensitivities of LT-CHA relative to HT-CHA are not due to increased background with the latter, but may stem from the improved functionality of the “Klenow(3’–5’exo-)-Nb.BbvCI” enzyme combination over “Bst-Nb.BsrDI.”
In conclusion, we have successfully engineered thermostable CHA circuits for the first time, and demonstrated the potential of both high- and low-temperature CHA circuits as real-time monitors of isothermal amplification reactions. Overall, the results strongly support the conclusion that CHA can be readily and rationally adapted to real-time monitoring of a variety of isothermal amplification reactions. It was surprising how purely thermodynamic considerations of hybridization propensities (at different temperatures, in different buffers, and with different enzymes) produced circuit designs that worked almost the first time they were tried. In light of these and others’ results, CHA can now be seen to have a number of advantages as a transducer for amplification reactions, including not only its adaptability to temperature and buffer conditions, but also to different detection modalities28,30 and the incorporation of programmable molecular logic circuits into analytical readouts.26,27,29
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Bill and Melinda Gates Foundation (OPP1028808); the Defense Advanced Research Projects Agency (HR0011-12-2-0001, 5-35830) and the National Institute of Health TR01 (5 R01 AI092839).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting Information includes materials and methods, Figures S1–S3, Table S1–S4, and the design principles of the CHA circuit and HT-CHA circuit. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Compton J. Nature. 1991;350:91. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 2.Wharam SD, Marsh P, Lloyd JS, Ray TD, Mock GA, Assenberg R, McPhee JE, Brown P, Weston A, Cardy DL. Nucleic Acids Res. 2001;29:e54. doi: 10.1093/nar/29.11.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lizardi PM, Huang XH, Zhu ZR, Bray-Ward P, Thomas DC, Ward DC. Nat. Genet. 1998;19:225. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 4.Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Nucleic Acids Res. 1992;20:1691. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter NG, Strunk G. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7937. doi: 10.1073/pnas.91.17.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hataoka Y, Zhang LH, Mori Y, Tomita N, Notomi T, Baba Y. Anal. Chem. 2004;76:3689. doi: 10.1021/ac035032u. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto T, Sonobe T, Hayashi K. J. Clin. Microbiol. 2003;41:2616. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomita N, Mori Y, Kanda H, Notomi T. Nat. Protoc. 2008;3:877. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 10.Mori Y, Nagamine K, Tomita N, Notomi T. Biochem. Biophys. Res. Commun. 2001;289:150. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 11.Mori Y, Kitao M, Tomita N, Notomi T. J. Biochem. Biophys. Methods. 2004;59:145. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Chou PH, Lin YC, Teng PH, Chen CL, Lee PY. J. Virol. Methods. 2011;173:67. doi: 10.1016/j.jviromet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Jung C, Chung JW, Kim UO, Kim MH, Park HG. Anal. Chem. 2010;82:5937. doi: 10.1021/ac100606m. [DOI] [PubMed] [Google Scholar]
- 14.Yi J, Zhang W, Zhang DY. Nucleic Acids Res. 2006;34:e81. doi: 10.1093/nar/gkl261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiman B, van Aarle P, Sillekens P. Mol. Biotechnol. 2002;20:163. doi: 10.1385/MB:20:2:163. [DOI] [PubMed] [Google Scholar]
- 16.Nadeau JG, Pitner JB, Linn CP, Schram JL, Dean CH, Nycz CM. Anal.Biochem. 1999;276:177. doi: 10.1006/abio.1999.4350. [DOI] [PubMed] [Google Scholar]
- 17.Jaroenram W, Arunrut N, Kiatpathomchai W. J. Virol. Method. 2012;186:36. doi: 10.1016/j.jviromet.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Seetang-Nun Y, Jaroenram W, Sriurairatana S, Suebsing R, Kiatpathomchai W. Mol. Cell. Probe. 2013;27:71. doi: 10.1016/j.mcp.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 20.Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15275. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 22.Li BL, Jiang Y, Chen X, Ellington AD. J. Am. Chem. Soc. 2012;134:13918. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang KA, Pei R, Stefanovic D, Stojanovic MN. J. Am. Chem. Soc. 2012;134:1642. doi: 10.1021/ja2084256. [DOI] [PubMed] [Google Scholar]
- 24.Yan H, Zhang XP, Shen ZY, Seeman NC. Nature. 2002;415:62. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 25.Li YG, Tseng YD, Kwon SY, D'Espaux L, Bunch JS, McEuen PL, Luo D. Nature Materials. 2004;3:38. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Ellington AD, Chen X. Nucleic Acids Res. 2011;39:e110. doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng AX, Li J, Wang JR, Song XR, Chen GN, Yang HH. Chem. Commun. 2012;48:3112. doi: 10.1039/c2cc30305a. [DOI] [PubMed] [Google Scholar]
- 28.Li F, Zhang H, Wang Z, Li X, Li X, Le XC. J. Am. Chem. Soc. 2013;135:2443. doi: 10.1021/ja311990w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J, Wang J, Han L, Wang E, Wang J. Chem. Commun. 2011;47:10563. doi: 10.1039/c1cc13973h. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Chen X, Ellington AD. Anal. Chem. 2012;84:8371. doi: 10.1021/ac301944v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paris DH, Imwong M, Faiz AM, Hasan M, Bin Yunus E, Silamut K, Lee SJ, Day NPJ, Dondorp AM. Am. J. Trop. Med. Hyg. 2007;77:972. [PubMed] [Google Scholar]
- 32.Kimura Y, de Hoon MJL, Aoki S, Ishizu Y, Kawai Y, Kogo Y, Daub CO, Lezhava A, Arner E, Hayashizaki Y. Nucleic Acids Res. 2011;39:e59. doi: 10.1093/nar/gkr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris T, Robertson B, Gallagher M. J. Clin. Microbiol. 1996;34:2933. doi: 10.1128/jcm.34.12.2933-2936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craw P, Balachandran W. Lab. Chip. 2012;12:2469. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- 35.Altan-Bonnet G, Libchaber A, Krichevsky O. Phys. Rev. Lett. 2003;90:138101. doi: 10.1103/PhysRevLett.90.138101. [DOI] [PubMed] [Google Scholar]
- 36.Beals TP, Smith JH, Nietupski RM, Lane DJ. BMC Mol. Biol. 2010;11:94. doi: 10.1186/1471-2199-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura T, Nishida K, Uchibayashi K, Ohuchi S. Nucleic Acids Symp. Ser. 2006;50:305. doi: 10.1093/nass/nrl152. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson M, Gullberg M, Dahl F, Szuhai K, Raap AK. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Connolly AR, Trau M. Angew. Chem. Int. Ed. 2010;49:2720. doi: 10.1002/anie.200906992. [DOI] [PubMed] [Google Scholar]
- 40.Tian T, Xiao H, Zhang X, Peng S, Zhang X, Guo S, Wang S, Liu S, Zhou X, Meyers C, Zhou X. Chem. Commun. 2013;49:75. doi: 10.1039/c2cc36728a. [DOI] [PubMed] [Google Scholar]
- 41.Fang X, Zhang H, Zhang F, Jing F, Mao H, Jin Q, Zhao J. Lab. Chip. 2012;12:319. doi: 10.1039/c2lc40384f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.