Abstract
Background
Prenatal exposure to ethanol elicits a range of neuro-developmental abnormalities, microcephaly to behavioral deficits. Impaired protein synthesis has been connected to pathogenesis of ethanol-induced brain damage and abnormal neuron development. However, mechanisms underlying these impairments of protein synthesis are not known. In this study, we illustrate the effects of ethanol on programmed cell death protein 4 (PDCD4), a tumor and translation repressor.
Methods
Primary cortical neurons (PCNs) were treated with ethanol for 2.5mg/ml and 4mg/ml for different time points (4h to 24h) and PDCD4 expression was detected by Western blotting. Protein synthesis was determined using [35S] methionine incorporation assay. Methyl cap pull down assay was performed to establish the effect of ETOH on association of eIF4A with capped mRNA. Luciferase assay was performed to determine the in vivo translation. A 2-day acute 5-dose binge model with ethanol (4 g/kg body wt, 25% v/v) was performed in Sprague Dawley rats at 12h intervals and analyzed for PDCD4, eukaryotic initiation factor 4A (eIF4A) and eIF4A-methyl cap association.
Results
Ethanol increased PDCD4 expression in a time- and dose-dependent manner in PCNs which inhibited the association of eIF4A with methyl-cap. Ethanol and ectopic PDCD4 expression suppressed in vivo translation in PCNs and RNAi targeting of PDCD4 blocked the inhibitory effect of ethanol on protein synthesis. In utero exposure of pregnant rats to ethanol resulted in a significant increase of PDCD4 protein in fetal cerebral cortex along with inhibition of methyl-cap associated eIF4A, compared to iso-caloric controls. Increased PDCD4 also occurred in pooled fractions of remaining brain regions.
Conclusions
Our data for the first time, illustrate that PDCD4 mediates inhibitory effects of ethanol on protein synthesis in PCNs and developing brain.
Keywords: Ethanol, protein synthesis, PDCD4, primary cortical neurons, eIF4A, cerebral cortex
Introduction
Fetal alcohol syndrome (FAS) expresses developmental aberrations in cerebral cortex and often severe memory, learning and behavior abnormalities (Mattson et al., 1996; Roebuck et al., 1999) all of which are largely dependent on either global or local protein synthesis. Arrays of physiological phenomena such as growth, differentiation, or programmed cell death are governed by protein synthesis (Morley et al., 2005). Gestational exposure to alcohol impairs protein synthesis, protein turnover and proteasome activity in developing brain and fetal cortical neurons (Gutala et al., 2004; Rawat, 1985). Protein synthesis is obligatory in neurons engendering long-term synaptic plasticity (Klan and Deverm, 2004; Pfeiffer and Huber, 2006), lengthening of spines (Vanderklish and Edelman, 2002), transcription independent forms of memory (Sutton et al., 2001). Notably, alcohol-induced deficiency in protein synthesis-dependent anomalies in utero is not typically restored postnatally by “catch up” phenomena even when dietary protein intake is adequate. This underscores the physiological importance of translational regulation underlying developmental abnormalities elicited by prenatal alcohol exposure.
Protein translation is a precisely orchestrated and energy consuming process (Klan and Deverm, 2004; Proud, 2007). mRNA predominantly undergoes translation by cap-dependent scanning mechanisms (Gingras et al., 1999), while 10% of cellular mRNAs are translated by internal ribosomal entry site (IRES) mechanisms (Komar and Hatzoglou, 2005). In general, cap-dependent translation initiation (key rate-limiting step) commences with binding of the eIF4F complex to 5’ cap of mRNA (Pestova et al., 1996; Sonenberg, 2008) leading to assembly of ribosomes to mRNA start codon (AUG). In order for preinitiation complex to locate the first AUG, the complex secondary structure in 5’ UTR must be relaxed by eIF4A, an RNA helicase. Mutations or sequestration of eIF4A impede translation initiation (Pause and Sonenberg, 1992; Yang et al., 2003a). A transformation suppressor protein, PDCD4 interacts with eIFs, eIF4A and eIF4G (Göke et al., 2002; Kang et al., 2002) and inhibits helicase activity of eIF4A thereby cap-dependent translation (Yang et al., 2003a). PDCD4 regulates several essential processes ranging from embryonic development and metamorphosis to normal tissue turnover (Lankat-Buttgereit and Göke, 2009). Since PDCD4 exerts its action chiefly at the translational level, it is a promising target for controlling cancer cell progression (Göke et al., 2004), tumor cell invasion and metastasis (Leupold et al., 2007) and cellular differentiation (Ozpolat et al., 2007). Yet, the availability and modulation of certain factors involved in translation cannot solely influence translation of all mRNAs equally. This warrants a greater understanding of eukaryotic protein synthesis. The data presented in this manuscript confirm that ETOH upregulates PDCD4 expression in a cultured neuron model and in an animal binge model. We also report that ETOH-induced PDCD4 sequesters eIF4A and prevents the in vivo translation thereby protein synthesis in PCNs. Further, PDCD4 silencing reversed ETOH-induced impairment in protein synthesis in cerebral cortical neurons. Consequently, our studies identified for the first time that “PDCD4” could be a novel therapeutic target for mitigation of fetal abnormalities associated with ETOH exposure.
Experimental Procedures
Materials
Smart Pool siRNA against PDCD4 and non-targeting siRNA pool were purchased from Dharmacon Inc., (Lafayette, CO). Antibodies for PDCD4 (Rockland Immunochemcials Inc., Gilbertsville, PA), luciferase, lamin-B, GAPDH (Santa Cruz Biotechnologies, Santa Cruz, CA), eIF4A (Cell Signaling Technology, Beverly, MA), actin, tubulin were from Sigma-Aldrich (St. Louis, MO). 7-methyl-GTP Sepharose-4B beads were obtained from GE Healthcare Biosciences (Piscataway, NJ). 35S Methionine was bought from PerkinElmer Life Sciences (Boston, MA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO). pcDNA-Luc and pcDNA-SL Luc was provided by Dr. Nancy Colburn (Bethesda, MD). pcDNA3-HA-PDCD4 expression construct was a kind gift from Dr. Michele Pagano (New York, NY).
Primary Cortical Neuron (PCN) Cultures
PCNs were prepared from E16-E17 Sprague Dawley rats as described earlier (Ramachandran et al., 2003). Fetal cortices were mechanically dissociated in HBSS and cells suspended in MEM containing 10% FBS and 10% HS. Neurons were maintained in a humidified atmosphere of 95% air and 5% CO2 on poly-D-lysine coated plates. After 1DIV, cultures were enriched for neurons by inhibiting astrocytic growth with 5-fluoro-2'-deoxy uridine (4mg/ml) and uridine (10mg/mL) containing 10% HS supplemented MEM media. Following 48h, fresh media was added and PCNs were treated on 4th and 5th days of culture for transfection experiments and ETOH exposure, respectively. This is a well-established primary neuronal culture system from our laboratory reproducibly yielding ~95% enriched neurons (Narasimhan et al., 2011; Rathinam et al., 2006).
In Vivo Model
Sprague Dawley rats were intubated with ethanol (4 g/kg body wt, 25% v/v) at 12 h intervals on days 16 and 17 of gestation. On day 18, 1 h before sacrifice, a final dose was administered. This 2-day acute ethanol exposure regimen is a very well established animal model that mimics an alcohol binge in humans (Henderson et al., 1995). Pair-fed, weight matched control dams received isocaloric dextrose. All animals were maintained in accordance with Institutional Animal Care and Use Committee-approved procedures. Both isocaloric dextrose administered control and ethanol-fed dams had full access to standard laboratory chow and water ad libitum, whereas pair-fed controls had full access to water but received the weight of chow consumed by the corresponding ethanol dam during the previous 24 h period. The gestational age of the pair-fed control and ethanol rats were staggered by a day, in order to ensure that animals from the pair-fed control received chow at the same stage of gestation as did the corresponding ethanol-treated dams. At the end of treatment, animals were sacrificed; Blood alcohol levels were determined using Analox AM1 analyzer. Fetal brain cortices and rest of the brain were carefully isolated and stored in −80° C until use.
Ethanol treatment
On 5 DIV, PCNs were treated with ETOH (2.5 mg/mL media, 4 mg/mL media) which corresponds to a final concentration of ~54mM and 86mM respectively for 4h, 8h, 12h, or 24h in an incubator pre-saturated with ETOH for 24h to maintain ETOH concentrations (monitored using Analox AM1 alcohol analyzer) in the culture media by placing a beaker filled with ETOH (Narasimhan et al., 2011). Control cells were maintained in the normal incubator. ETOH levels used in the in vitro experiments were clinically relevant, which were at or below those used in other studies to elicit a range of neurotoxic responses in various mouse and rat models (Dong et al., 2011; Narasimhan et al., 2011; Olney et al., 2002; Rathinam et al., 2006).
Reverse transcription-PCR
1.5 µg of total RNA divested of genomic DNA was reverse transcribed by Quantitect reverse transcription kit (Qiagen). 1/10th of cDNA was used in PCR reaction using primers spanning Exon-1 and Exon-2 region of rat PDCD4 (For: 5'-actcggagaactttcagtacc-3'; Rev: 5'-tggagcaaagtagagtggt-3'). After an initial denaturation at 95°C/3 min, PCR was performed for 35 cycles under following conditions: 95°C/30 s, 53°C/30 s, 72°C/60 s, with an extension at 72°C/5 min. The PCR product was run on 1% agarose gel, visualized and photographed using UVP gel documentation system.
Immunoblotting
PCNs, cerebral cortices, or rest of the brain were lysed in RIPA at 4°C, sonicated and centrifuged at 15,000 g for 15 min at 4°C. Equal amounts of protein were separated by SDS-PAGE, transferred onto PVDF membrane and immunoblotted with PDCD4, eIF4A, luciferase, GAPDH, tubulin, actin as previously described (Narasimhan et al., 2011). Both GAPDH and tubulin were used to normalize loading in in vivo samples. The intensity of PDCD4, eIF4A, eIF4G bands were quantified using Image J and normalized to GAPDH.
Cytosolic and nuclear protein fractionation
Neurons seeded at a density of 9 × 106 cells/100mm petri dish were treated with ETOH as mentioned above. At the end of the experiment, nuclear and cytosolic protein fractions were isolated according to manufacturer’s instructions (Pierce, IL). Following protein estimation, equal amounts of protein were used for immunoblotting experiment as described above. Purity of the extracts was confirmed by immunoblotting for Lamin B1 (nuclear) and actin, carbonic anhydrase-II (CA-II), GAPDH (cytosol).
Small interfering RNA (siRNA) transfection
100 nM of siGenome smartpool mix of four PDCD4 specific siRNAs, or non-targeting siRNA pool were transfected into 4DIV neurons using siPort Amine (Ambion). PDCD4 silencing was observed by Western analysis after 24h and 48h. For protein synthesis determinations, 24h post-transfection of siRNA, the cells were exposed to ETOH for additional 24hs and processed for 35S methionine incorporation.
Protein synthesis measurement
Protein synthesis measurements were as previously described (Mahimainathan et al., 2006). Control and ETOH treated PCNs were labeled with 10 µCi/ml [35S] methionine for the terminal 2hs. Cells were washed in PBS, lysed in RIPA and 20 µg of protein was spotted onto 3-mm Whatman filter paper. Filters were washed thrice by boiling for 1 min in 10% TCA containing 0.1g/l methionine. The incorporation of [35S]-aminoacid into TCA-insoluble cellular components was quantified using a Beckman scintillation counter.
Methyl cap pull down assay
Association of eIF4A with the cap mRNA-eIF4F complex was assessed using 7-methyl-GTP Sepharose beads which simulate cap mRNA pool in vivo (Pyronett et al., 2001). 5 DIV PCNs were treated with ETOH for 24hs and lysed in buffer A (50mM Tris-HCl at pH 7.4, 50mM KCl, 1mM EDTA, 0.5% NP-40, and protease inhibitors). Equal amount of protein were used in pull down assays using 7-Methyl-GTP Sepharose-4B beads for 4h at 4°C. Sepharose associated eIF4A depleted supernatant was collected and assessed for PDCD4-eIF4A and PDCD4-eIF4G association following PDCD4 immunoprecipitation. The resin was washed thrice with buffer A and immunoblotted for eIF4A and eIF4G along with PDCD4 immunoprecipitates.
In vivo translation assay
This technique determines in PCNs, if the reporter RNAs in question are synthesized and translated into protein products. 4 DIV PCNs were transfected with either pcDNA-Luc (non-structured) or pcDNA-SL Luc (stem loop structured) using fugene HD to assess in vivo translation. pcDNA-SL Luc reporter construct was engineered to generate an mRNA with 97 nucleotides downstream of the 5’ cap end and an internal 24-bp stem-loop structure that was introduced in the 5’ leader sequence of a luciferase gene. Stem loop in pcDNA-SL Luc was introduced to evaluate the role of eIF4A’s ability to unwind the loop and determine the efficiency of translation process (Gray and Hentze, 1994; Yang et al., 2004). The non-structured pcDNA-Luc reporter construct lacks stem loop. Further, the inclusion of secondary structure to the 5′UTR of a luciferase mRNA (pcDNA-SL Luc) enables the assessment of indirect susceptibility of translational process to PDCD4 or any other mode that can inactivate eIF4A helicase (Fig. s1). 24h post-transfection, cells were treated with or without ETOH (4.0mg/ml) for 24h. In a parallel experiment, pcDNA3-HA-PDCD4 expression plasmid was co-transfected with pcDNA- Luc or pcDNA-SL Luc for 48h. Luciferase activity was determined using Dual Luciferase assay system (Promega, WI) in a Glomax (20/20) luminometer and normalized to protein concentration. Equal amounts of protein were immunoblotted for luciferase and GAPDH. One step RT-PCR was performed as above using luciferase primers (For: 5'-ctgcatccggctatgaagagatacg-3'; Rev: 5'-cccaactgcaactccgataaataacgc-3'); GAPDH primers (For: 5'-agacagccgcatcttcttgt-3'; Rev: 5'-tactcagcaccagcatcacc-3').
Statistical analysis
Data are presented as mean ± s.e.m. Statistical differences were determined using one-way ANOVA followed by Student-Newman-Keuls post-hoc analysis when experiments involved more than two groups. Student’s t-test was used for experiments involving only two groups. P<0.05 was considered as statistically significant.
Results
Ethanol inhibits protein synthesis in PCNs. PDCD4, a translation repressor is expressed in fetal brain cortex and PCNs
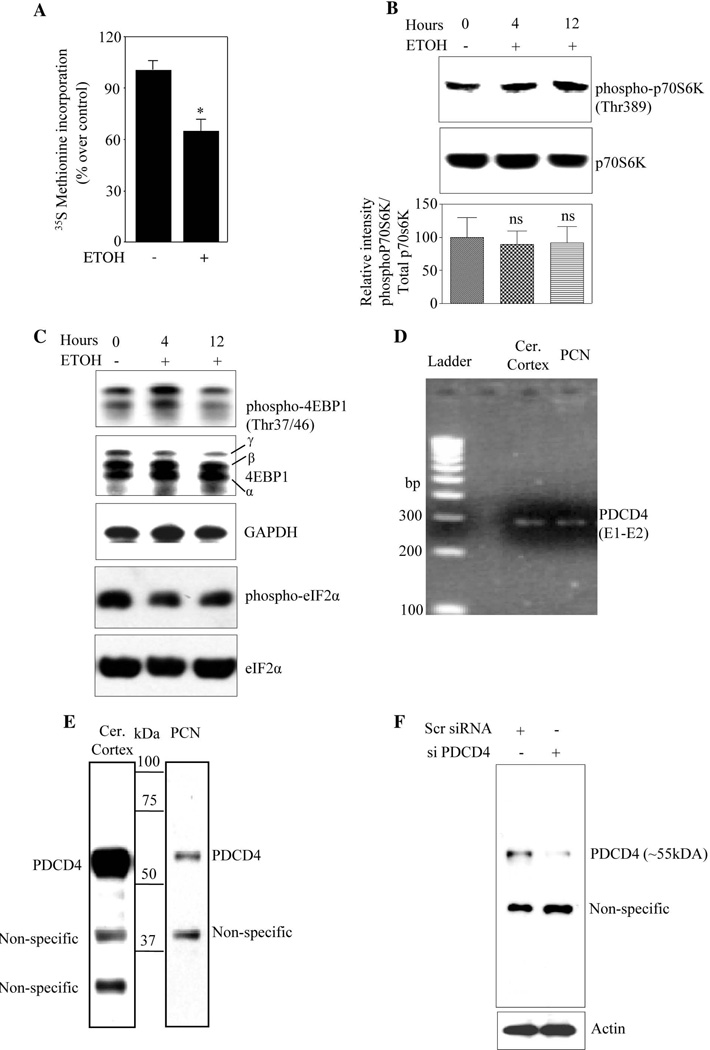
Impaired protein synthesis is associated with pathogenesis of alcohol-induced brain damage (Bonner et al., 2003). However, mechanisms underlying this are poorly understood. We therefore determined ETOH-induced changes in protein synthesis by measuring [35S]-methionine incorporation into proteins in PCNs. ETOH exposure for 24h resulted in 40% reduction in incorporation of [35S]-methionine in TCA precipitates indicating that ETOH inhibits de novo protein synthesis (Fig. 1A). Protein translation is regulated either at initiation or elongation steps and the initiation phase is the predominant rate-limiting step in cap-dependent protein synthesis (Miron and Sonenberg, 2001; Thornton et al., 2003). ETOH could potentially influence several key events that trigger translation initiation viz. phosphorylation of p70S6K, 4EBP1 and eIF2α. ETOH exerted little or no significant changes in phosphorylation levels of these factors in PCNs (Fig. 1B and Fig.1C). Therefore, we posit that ETOH may exert its effects on protein synthesis by blocking eIF4A- associated events. Yang et al. (2003a) reported that PDCD4 suppresses cap dependent translation by binding to eIF4A and inhibiting its helicase activity. Therefore, we determined whether PDCD4 transcript and protein are expressed in fetal brain cerebral cortices and PCNs. RT-PCR analysis using primers spanning exon 1 & 2 of PDCD4 yielded a 291 bp product as predicted (Fig. 1D). Immunoblot analysis showed a major ~55kDa band corresponding to the molecular mass of PDCD4 protein in both cerebral cortex (CC) and PCNs (Fig. 1E). As we detected multiple bands in immunoblot besides the predicted ~55kDa specific PDCD4, we further confirmed the authenticity of bands by employing siRNA knockdown in cultured fetal PCNs. Our results showed that ~55kDa immunoreactive band was specifically suppressed following PDCD4 siRNA treatment, while the other immunoreactive band remained unchanged (Fig. 1F). Together, these data confirm that the immunoreactive band at or around ~55kDa is indeed PDCD4 and that PDCD4 is expressed in fetal rat brain cortex and PCNs.
Figure 1. Ethanol inhibits protein synthesis in PCNs. Rat brain and fetal cortical neurons express PDCD4.
(A) De novo protein synthesis measurement by [35S Methionine] incorporation into proteins in rat PCNs following treatment with ETOH for 24 h. The amount of [35S] radioactivity in the TCA precipitating material was measured and the percentage incorporation of [35S] radioactivity over control was represented. (B) 50µg of protein from untreated and ETOH treated PCN lysates were analyzed using immunoblotting for the extent of phosphorylation in p70S6K at Thr389 using rabbit phospho specific antibody phospho-p70S6K (Thr389) as phosphorylation of Thr389 most closely correlates with p70 kinase activity in vivo. The same blot was stripped and reprobed with total p70S6K which was used as loading control. The bottom panel represents Threonine389 phospho-p70S6K immunoreactivity that was normalized to total p70S6K (p70S6K) and was graphed as percent control. In panel B, ns indicates no significance when compared with untreated control as determined by one way ANOVA. (C) Western analysis was performed using 50µg of protein from untreated and ETOH treated PCN lysates to analyze the extent of phosphorylation in 4EBP1 (Thr37/46) and eIF2α using rabbit phospho specific antibodies. The same blot membrane was stripped and reprobed with corresponding total antibodies that served as loading control. (D) Total RNA isolated from rat brain (RB) and PCN was subjected to one step RT PCR analysis with primers spanning exon1 and exon2 region amplifying a 291 bp product of PDCD4. (E) Western analysis using rabbit anti-PDCD4 polyclonal antibody was performed on total protein lysates from the rat fetal cerebral cortex (75µg) and rat primary fetal cortical neurons (25µg). (F) PCNs were transfected with either non-targeting scrambled siRNA or smart pool mix of four PDCD4 using siPORT amine. Cells were lysed 48 h post transfection and immunoblot analysed with anti-PDCD4 and β-actin.
Ethanol induces PDCD4 expression in PCNs
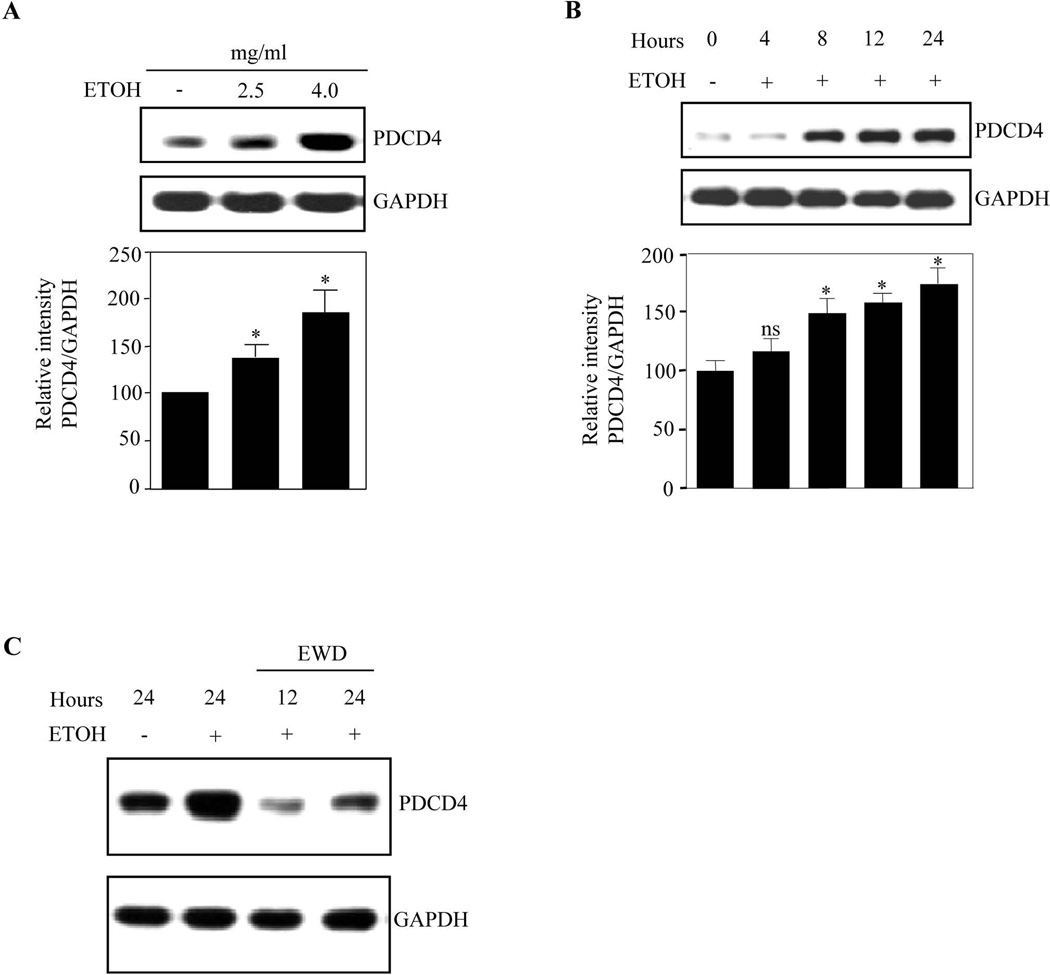
We next determined whether ETOH could modulate PDCD4 which subsequently affects translation by interacting with eIF4A helicase. PCNs treated with 2.5 mg/ml and 4 mg/ml ETOH for 24h showed a ~1.5 fold and ~1.7 fold increase in PDCD4 expression normalized to GAPDH (Fig. 2A). PCNs treated with 4mg/ml ETOH for different time points showed a steady increase in PDCD4 protein (~50%) during first 8h which maximized to 75% at 24h (Fig. 2B). These data indicate a positive correlation between ETOH exposure and PDCD4 increases. To confirm whether PDCD4 upregulation is an ETOH-specific effect, we performed withdrawal experiments in PCNs by replacing ETOH containing culture media with normal media for 12h and 24h after initial 24h of ETOH exposure. ETOH withdrawal reduced PDCD4 expression as early as 12h which tends towards control at 24h (Fig. 2C), suggesting that PDCD4 regulation is alcohol-specific. It is unclear how PDCD4 expression is lowered upon ETOH withdrawal. Nevertheless, these data illustrate that ethanol exposure to cortical neurons upregulates PDCD4 expression and could interfere with downstream events.
Figure 2. PDCD4 expression was strongly induced in PCN in response to ETOH.
(A) PCNs were treated with ETOH 2.5mg/ml and 4mg/ml for 24 h. PDCD4 protein expression was determined by immunoblot analyses. Immunoblotting with anti-GAPDH was used as loading control. Lower panel depicts densitometric scanning analysis ratio of PDCD4 to GAPDH. Quantification data represents mean ± s.e.m, n=3. (B) PCNs were treated with 4mg/ml ETOH for different time points as indicated. Western analysis for PDCD4 and GAPDH was performed. Lower panel indicate the relative intensity of PDCD4 normalized to GAPDH (mean ± s.e.m, n=3). (C) 5 DIV PCNs were treated with ETOH (4mg/ml) for 24 h and the culture media containing ETOH were replaced with normal culture media for additional 12 h and 24 h. Equal amount of lysates were analyzed for PDCD4 and GAPDH protein expression. One way ANOVA was performed to establish statistical significance. In A, B * - P<0.05, when compared with untreated controls; ns-not significant compared with untreated control.
Ethanol promotes both cytoplasmic and nuclear expression of PDCD4
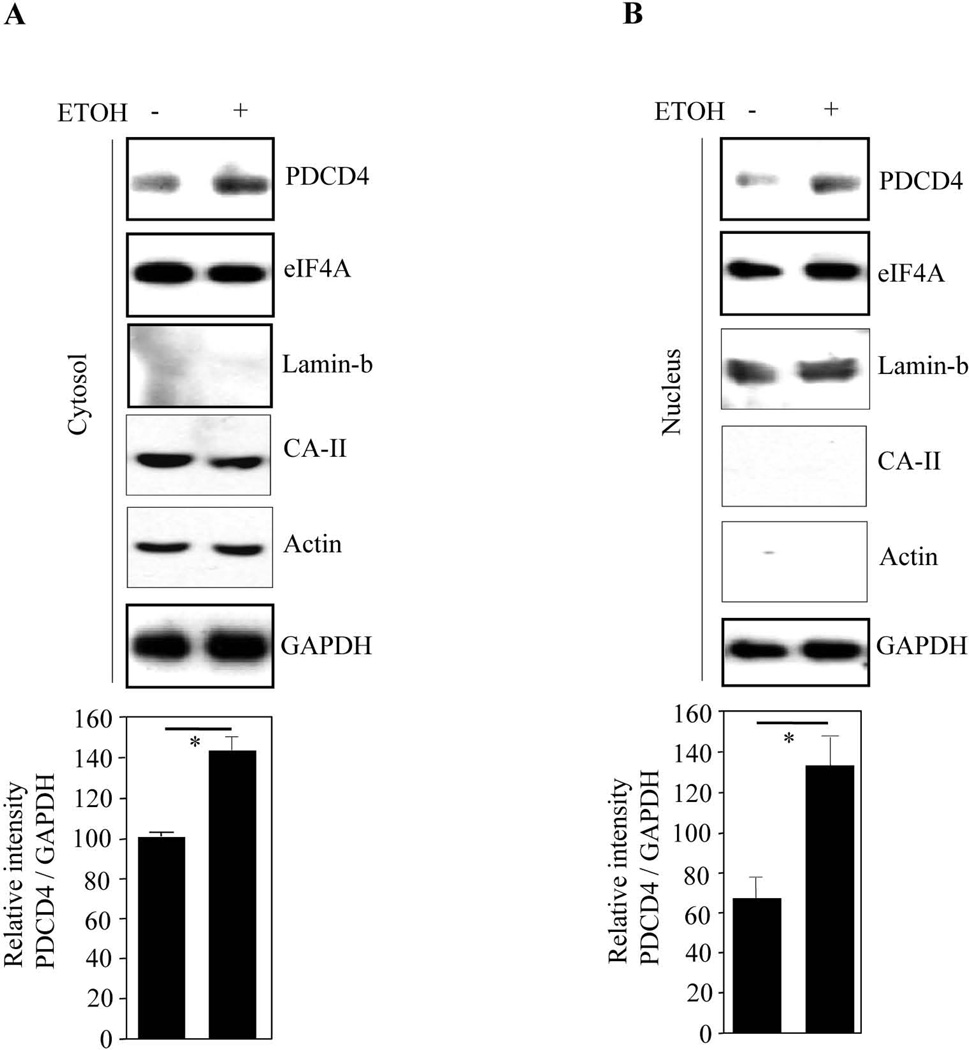
Depending on growth state and cell type, PDCD4 localizes in cytoplasm and inhibits eIF4A-dependent translation. Therefore, we assessed cytoplasmic and nuclear PDCD4 expression following ETOH treatment. Immunoblot analysis showed that ETOH increased both cytosolic PDCD4 (~1.5 fold) (Fig. 3A) and nuclear PDCD4 (~2 fold) (Fig. 3B). We observed GAPDH expression in both the fractions which was unchanged upon ETOH exposure (Fig. 3B) and GAPDH is reported to be both nuclear as well as cytosolic in viable PCNs (Sawa et al., 1997). Purity of the cytosolic fraction was further confirmed by two other specific markers, actin and carbonic anydrase-II (CA-II). We did not observe any compensatory elevation of eIF4A levels. This is the first report where PDCD4 has been shown to localize in both the compartments in post-mitotic neurons, while the ETOH induced (a) cytosolic PDCD4 could impair protein translation events and (b) the nuclear increase could interfere with RNA metabolism and events related to prosurvival (Böhm et al., 2003; Palamarchuk et al., 2005; Yoshinaga et al., 1999).
Figure 3. Ethanol increases cytoplasmic and nuclear content of PDCD4.
Western analysis using PDCD4, eIF4A, GAPDH, actin, CA-II and lamin b1 polyclonal antibodies was performed on cytosolic extracts (a) and nuclear extracts (b) from control and ETOH exposed PCNs. In both A & B, the bottom panel depicts the densitometric scanning ratio of PDCD4/ GAPDH intensities. Student’s t-test determined the significance of treatment. * - represents P<0.05 vs untreated controls (mean ± s.e.m, n=3).
ETOH inhibits eIF4A association with methyl cap mRNA by inducing PDCD4-eIF4A sequestration
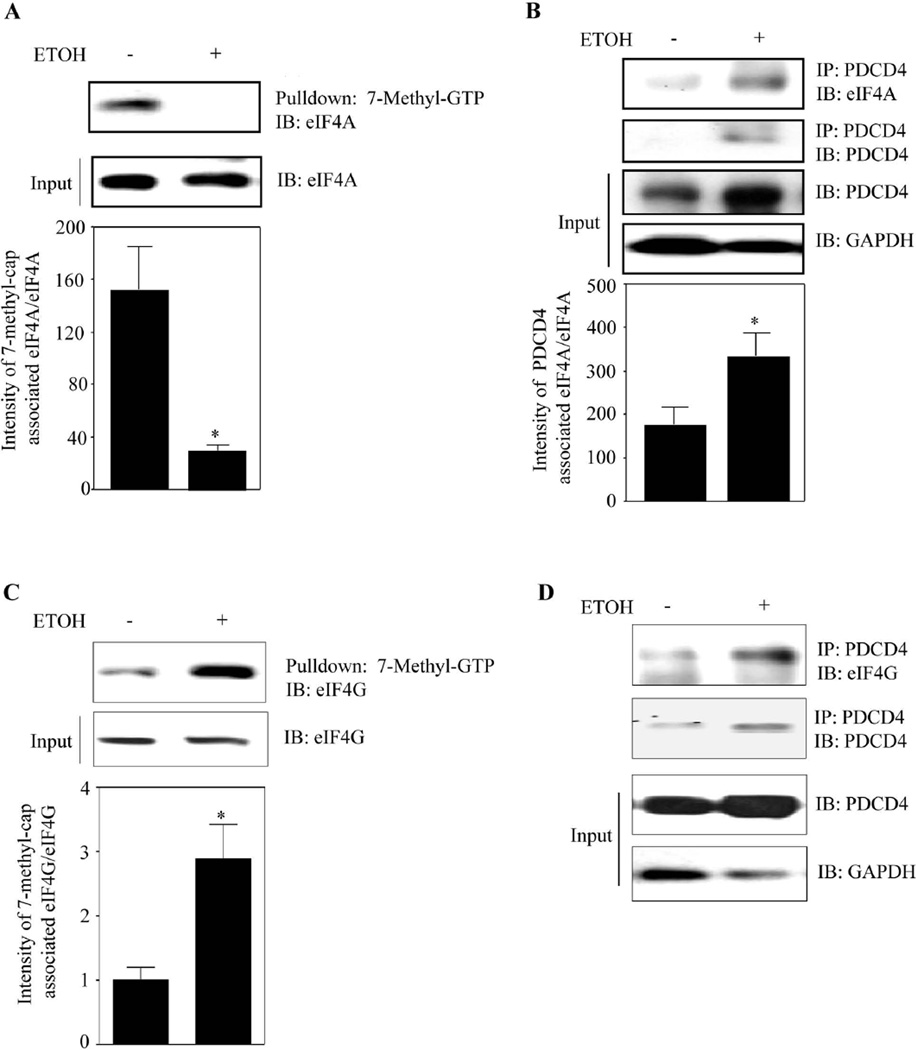
Translation initiation requires binding of methyl-cap of mRNA to eIF4E and unwinding by eIF4A. We next determined whether ETOH influences the association of eIF4A with capped mRNA using a pull-down assay involving 7-methyl-GTP Sepharose-4B beads (mimic of capped mRNA) and assessed for cap-associated eIF4A by immunoblotting. A dramatic reduction in eIF4A associated with 7-methyl-GTP was observed in 24h ETOH treated lysates (~4 fold; Fig. 4A). Importantly, this reduction did not result from decreased eIF4A expression, as eIF4A levels remain unchanged in both control and ETOH treatment (Fig. 4A) suggesting that eIF4A association with capped mRNA could be inhibited by ETOH. Since negligible eIF4A-7-methyl cap analog binding was observed, we next determined if it's sequestration by PDCD4 is increased by ETOH. To evaluate this, supernatant depleted of cap-associated eIF4A was immunoprecipitated with anti-PDCD4 and immunoblotted for eIF4A. ETOH significantly (p<0.05) enhanced PDCD4 binding to eIF4A (Fig. 4B). The supernatants devoid of methyl cap-eIF4A displayed higher levels of PDCD4 and as expected, PDCD4 levels in ETOH-treated samples were increased (Fig. 4B).
Figure 4. ETOH induced PDCD4 sequesters and inhibits eIF4A association with methyl cap mRNA.
(A) PCNs treated with or without ETOH for 24 h were used in a pull-down assay with 7-methyl-GTP Sepharose 4B beads which simulates cap mRNA in vitro. 600µg of protein was used to assess the cap-associated eIF4A by immunoblotting. Lower panel depicts the quantification data and Student’s t test was performed to determine the significance of treatment. (B) Methyl cap associated eIF4A depleted supernatants from methyl cap assay were immunoprecipitated with anti-PDCD4 and immunoblotted with antibodies to eIF4A or PDCD4. Equal loading of input samples was confirmed by measuring GAPDH expression. Lower panel represent the densitometric scanning ratio of PDCD4/ GAPDH intensities (mean ± s.e.m, n=6). (C) Methyl cap assay was performed as in Fig. 4A and the cap-associated eIF4G was assessed by immunoblotting. Lower panel is the quantification data of cap-associated eIF4G to direct eIF4G and statistical significance was assessed by Student’s t test. (D) Supernatant from Fig. 4C was immunoprecipitated with anti-PDCD4 and immunoblotted with eIF4G, PDCD4 and GAPDH. In panels A–C, Student’s t test was performed to determine the significance of treatment. * - P<0.05, when compared with untreated controls.
Scaffolding protein, eIF4G assembles the translation initiation complex constituting eIF4E, eIF4A, PABP1 and others. Hence, we assessed whether the altered association of eIF4A-methyl cap is a result of perturbation in eIF4G binding to methyl cap. Fig. 4C demonstrates that ethanol did not prevent eIF4G binding with cap mimic and in fact, the binding was significantly (p<0.05) increased. Notably, the endogenous levels of eIF4G were unaltered (Fig. 4C). The reason for increased eIF4G association to cap analog under ETOH exposed condition could be to compensate the loss of eIF4A to cap structure (Fig. 4A; Ln 1 vs Ln 2). In spite of increased eIF4G-methyl cap association, we detected increased association of eIF4G to PDCD4 in the methyl cap-depleted supernatant. Taken together, our findings suggests that (a) ETOH-induced inhibition of translation initiation is not a result of perturbing eIF4G’s association with methyl cap and (b) ETOH induced translation suppression appears to be a highly specific event related to unavailability of eIF4A for the capped mRNA structure which is prompted by PDCD4 sequestration of eIF4A.
ETOH and forced expression of PDCD4 inhibits translation in PCNs
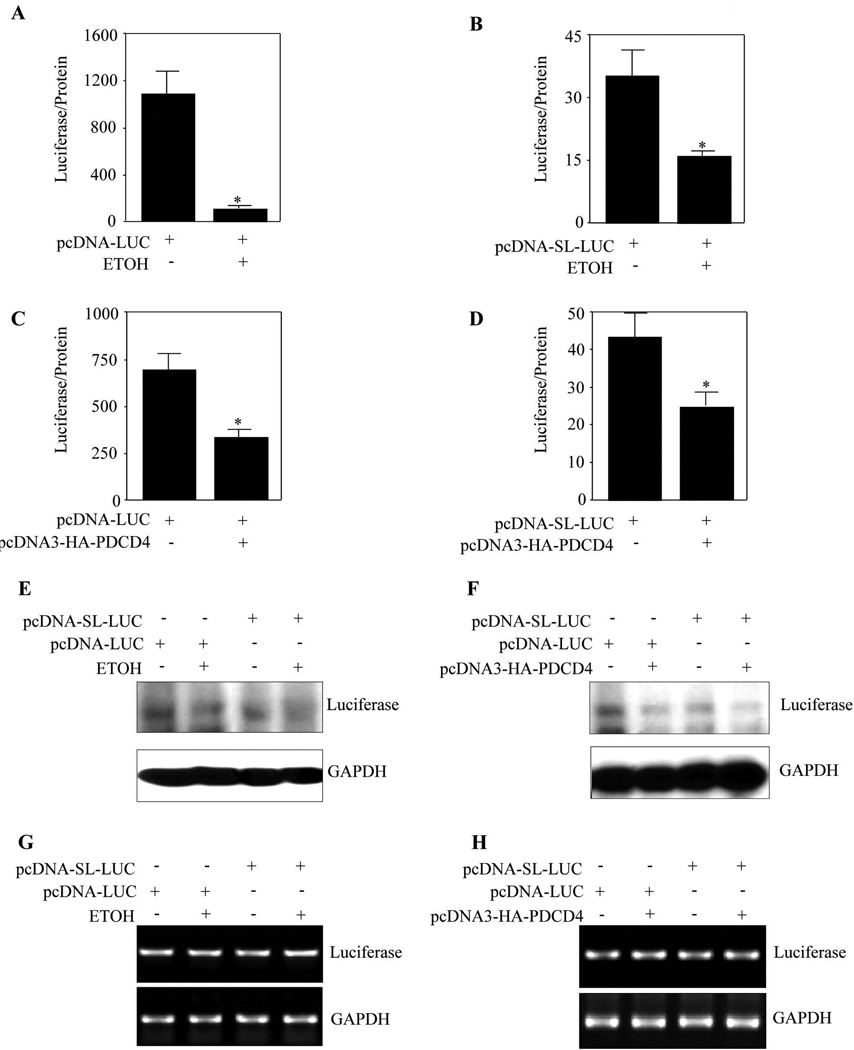
Having shown that ETOH induces eIF4A sequestration by PDCD4 and subsequently affects eIF4A association with methyl cap, we next determined if the reporter RNAs in question are synthesized and if this is translated into protein products. To address this, we examined the effect of ETOH on reporter activity of unstructured luciferase construct (pcDNA-Luc) or a structured construct with a 24 bp stem loop and 97 nucleotides downstream of the cap of mRNA (pcDNA-SL-Luc) driving luciferase gene in the "in vivo translation assay" (Yang et al., 2004). This classical assay involving stem-loop based luciferase construct is to evaluate the “translation in vivo” wherein the endogenous eIF4A would unwind the stem-loop under “normal translation” allowing the translation of luciferase mRNA to protein which is reflected in a relatively high luciferase readout. On the contrary, when translation process is hampered due to inactivation of eIF4A, the stem-loop remains unwound resulting in less efficient translation of luciferase mRNA that can be observed from a relatively decreased luminescence output. Pause and Sonenberg (1992) reported that helicase activity of eIF4A is critical for efficient translation of mRNAs with secondary structure in 5’ UTR and any interference of eIF4A helicase activity by PDCD4 suppresses translation (Yang et al., 2003a). Thus the use of a hairpin structured (stem loop) reporter construct in this assay facilitates the reliable measurement of translational efficiency in intact cells. Further the relative increase/decrease in luciferase readout in this assay could serve as an indirect measure of eIF4A’s helicase activity (Fig. s1). Translation of stem-loop luciferase only ranged between 3% and 6% as efficient as that of non-stem-loop luciferase transcript (col. 1 of Fig. 5A vs 5B; Fig. 5C vs 5D). In response to ETOH, PCNs transfected with non-structured luciferase construct displayed a 10 fold reduction in translation (Fig. 5A). In addition, a significant (p<0.05) 3-fold reduction in the translation of luciferase gene in pcDNA- SL-Luc transfected neurons was observed on ETOH treatment (Fig 5B). Further, Western blotting confirmed the ETOH-induced relative decrease in luciferase protein in both structured and non-structured transfected PCNs (Fig. 5E) substantiating the repressed translation of luciferase mRNA constructs (Fig. 5A and 5B). As ETOH induced translation could be a result of inhibition of eIF4A helicase activity by PDCD4, overexpression of PDCD4 would be expected to produce an equivalent effect as that of ETOH. To test this, pcDNA3-HA-PDCD4 was co-transfected with non-stem-loop or stem-loop luciferase mRNA constructs in PCNs and the translation efficiency was determined by luciferase activity. As expected, PDCD4 overexpression blocked translation of both the unstructured and structured mRNA constructs significantly (p<0.05) by about 2.5 and 2 fold respectively (Fig. 5C and 5D). Similarly, immunoblotting using the same lysates confirmed that PDCD4 overexpression significantly inhibited the luciferase protein level (Fig. 5F). RT-PCR analysis showed that neither ETOH treatment nor PDCD4 overexpression resulted in luciferase mRNA changes in PCNs overexpressed with stem-loop or non-stem loop constructs (Fig. 5G, 5H) suggesting the changes are only due to translation. These results suggest that ETOH induced PDCD4 can inhibit in vivo translation in cerebral cortical neurons.
Figure 5. ETOH and transient overexpression of PDCD4 inhibits in vivo translation in PCNs.
(A) PCNs were transfected with non-structured mRNA luciferase construct using Fugene HD. 24 h post transfection the cells were treated with or without ETOH (4mg/ml) for additional 24 h. Luciferase activity was measured following normalization to protein levels. (B) PCNs were transfected with structured mRNA luciferase construct using Fugene HD. 24 h following transfection, the cells were treated with or without ETOH (4mg/ml) for additional 24 h. Luciferase activity was determined and the activity was expressed as Luciferase/protein. (C) PCNs were co-transfected with non-stem loop mRNA luciferase construct and PDCD4 expression construct using Fugene HD. 48 h post transfection the cells were processed for determination of luciferase activity. The results were plotted as luciferase/protein values. (D) PCNs were co-transfected with stem-loop mRNA luciferase construct and PDCD4 expression construct using Fugene HD. 48 h following transfection the luciferase activity was determined and the activity was normalized to protein levels. (E) Equal amount of protein samples from 5A, 5B were resolved in SDS-PAGE electrophoresis and immunoblotted for luciferase protein and GAPDH expression. (F) Protein lysates from 5C, 5D were analyzed for luciferase protein and GAPDH expression by immunoblotting. (G) PCNs were transfected and treated as in 5A and 5B. At the end of the experimental period, one step RT-PCR analysis was performed for luciferase mRNA and GAPDH mRNA expression. (H) Total RNA from neurons transfected as in 5C and 5D were subjected for one step RT-PCR analysis with primers specific for luciferase and GAPDH. In Panels A–D, Student’s t test was performed to determine the significance of treatment. * - P<0.05 compared with untreated control (mean ± s.e.m, n=6).
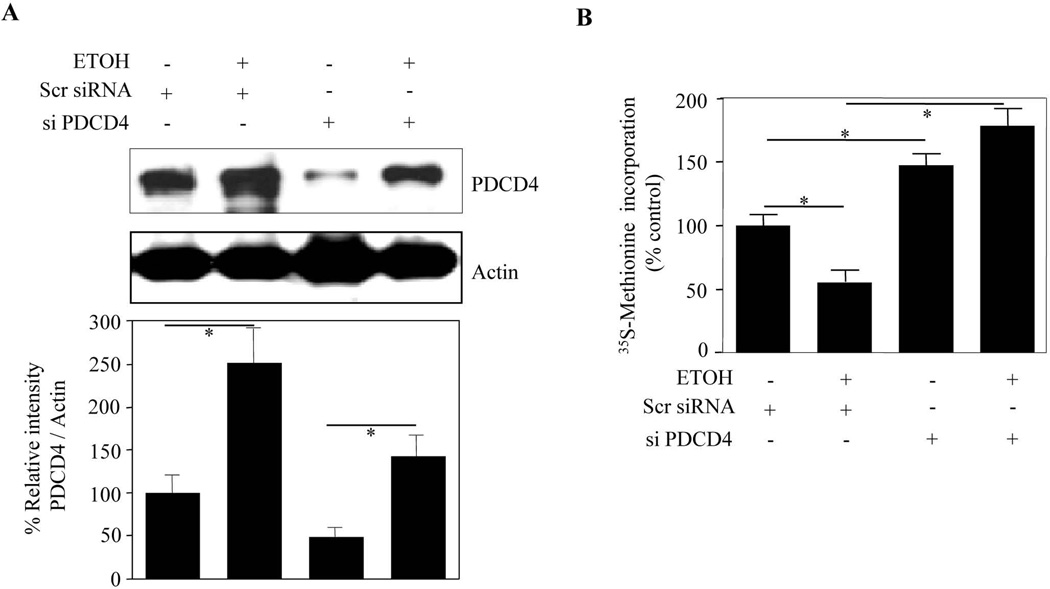
PDCD4 silencing mitigates ETOH-induced inhibition of protein synthesis
In order to confirm that the observed effect of ETOH-induced inhibition of protein synthesis is PDCD4 dependent, we carried out PDCD4 silencing strategy in PCNs. Targeted inhibition of PDCD4 by siRNA significantly (p<0.05) reduced PDCD4 abundance by about 60% when compared with scramble siRNA (Fig. 6A; col. 1 vs col. 3). This manipulation further blocked ETOH-induced PDCD4 expression significantly (p<0.05). Using this siRNA strategy, we next assessed the effect of PDCD4 silencing on the ETOH-induced reduction of protein synthesis. When untreated PCNs were exposed to ETOH, [35S]-methionine incorporation into proteins was reduced significantly by 45% (Fig. 6B). PDCD4 knockdown by itself enhanced the basal protein synthesis by 40% (p<0.01). Furthermore, this measurement was more than tripled (3.5 fold, p<0.001) in ETOH-treated PDCD4 depleted neurons when compared to neurons exposed to ETOH alone (col. 2 vs col. 4; Fig. 6B). As translation and polysome assembly involving PDCD4-eIF4A dependent regulation has a stronger preference for mRNA subset that contain secondary structures in 5’UTRs, we did not rule out that PDCD4 silencing controlled the ETOH-induced protein alterations only partially. However, these observations demonstrate a definite functional relevance of ETOH-induced PDCD4 upregulation in neurons, the silencing of which mitigates ETOH-induced impairment in protein synthesis.
Figure 6. RNAi mediated PDCD4 downregulation reversed ethanol induced suppression of protein translation in PCNs.
(A) PCNs were transfected with either non-targeting scramble siRNA or smart pool mix of four siPDCD4 using siPORT amine. 24 h post transfection of scrambled siRNA or siPDCD4 the cells were treated with or without ETOH (4mg/ml) for additional 24 h. Protein extracts were then immunoblot analysed with anti-PDCD4 and anti-actin. (B) PCNs were transfected and treated as in 6A and 2h before the termination of the experiment the cells were labeled with 10 µCi/ml [35S] methionine and processed for [35S Methionine] incorporation into proteins. The amount of [35S] radioactivity in the TCA precipitating material was measured and the percentage incorporation of [35S] radioactivity over control was represented. One way ANOVA was performed to determine the significance of treatment. * - represents P<0.05 (mean ± s.e.m, n=6).
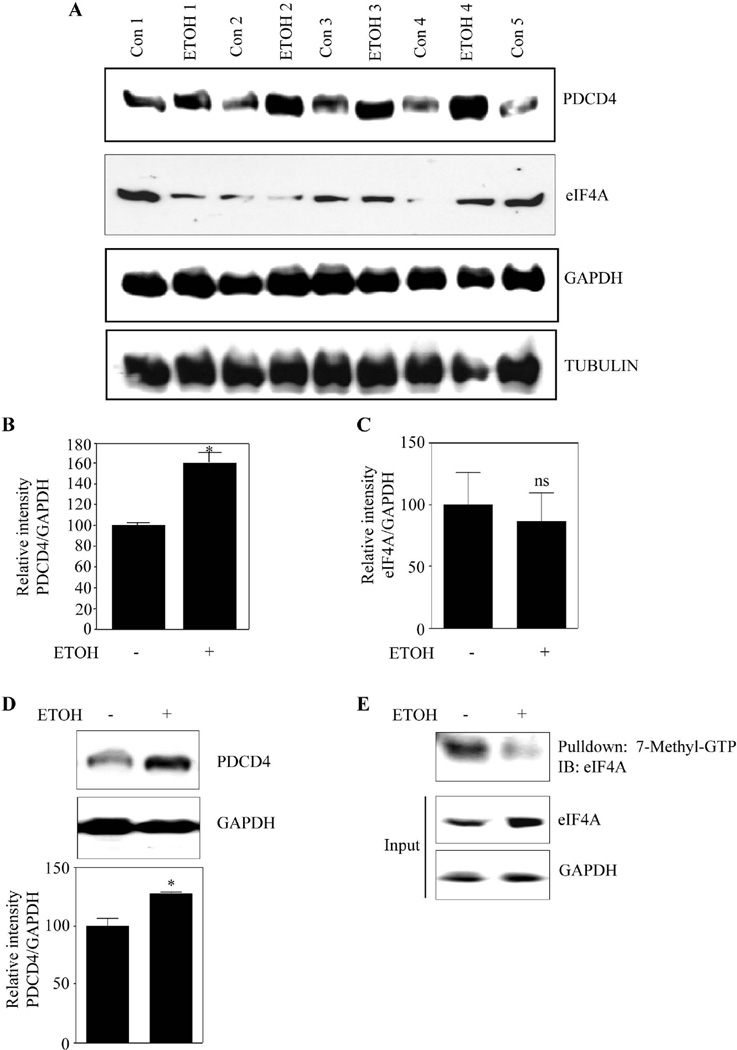
In utero ETOH exposure induces PDCD4 protein expression in brain cortex and rest of the brain regions of embryos
To assess whether the ETOH-induced PDCD4 changes observed in primary neuronal cultures, reflect in vivo setting, we assessed PDCD4 protein levels in an in utero binge model. Similar to in vitro observations, in utero exposure to ETOH resulted in a significant increase of PDCD4 protein expression by 60% in brain cerebral cortices of embryos when compared to isocaloric dextrose received groups (Fig. 7A and 7B). As eIF4A level and its activity can dictate the translation process, we checked whether alcohol influences eIF4A levels. eIF4A levels were unaltered in ETOH-exposed fetal brain cortices (Fig. 7A and 7C). Thus, in the current binge alcohol model, significant repression of protein synthesis in fetal brain cortices might occur in a PDCD4 dependent manner. We also observed a 25% increase in PDCD4 in "rest of the brain" from fetuses of ETOH group (Fig. 7D), suggesting that PDCD4 is a player in ETOH-induced inhibition of protein synthesis in other brain regions. Further, binge alcohol treatment of pregnant dams reduced eIF4A association with methyl cap in fetal brain cortices (Fig. 7E). This data is consistent with our in vitro observations.
Figure 7. Prenatal ethanol exposure increases PDCD4 expression in fetal brain cortex and rest of the brain.
(A) Pregnant rats (Sprague-Dawley) at embryonic day 16 (E16) were administered ETOH (4g/kg body weight) or isocaloric dextrose by gastric intubation at 12 h intervals for two days. At E18 brain cortex from embryos were dissected and processed for PDCD4, eIF4A, GAPDH, Tubulin protein expression by immunoblotting (n=6). (B) This panel illustrates the densitometric scanning ratio of PDCD4/GAPDH intensities. Student’s t test was performed to determine the significance of treatment. * P<0.05 compared with isocaloric dextrose administered animals (mean ± s.e.m, n=6). (C) Densitometric scanning ratio of eIF4A/ GAPDH intensities. Statistical analysis was determined by Student’s t test. ns - represent not significant compared with untreated controls (mean ± s.e.m, n=3). (D) Equal amount of protein lysates from rest of the brain devoid of cortex was separated by SDS-PAGE electrophoresis and immunoblotted with PDCD4 and GAPDH. Lower panel show the densitrometric analysis of PDCD4/GAPDH band intensities. Statistical analysis using Student’s t test indicates * P<0.05 when compared with isocaloric dextrose administered animals. (E) 225µg of cortical brain lysates obtained from 7A is processed for methyl cap analysis as in Fig. 4A and a representative immunoblot is given.
Discussion
We now report a novel molecular mechanism involving PDCD4 by which ETOH suppresses protein synthesis in PCNs and in intact fetal cerebral cortex. Ontogenic development of brain begins early in gestation and continues into the postnatal period in humans, nonhuman primates, rodents and others. Multiple studies documented that maternal alcohol consumption during the critical periods of fetal brain development disrupt brain architecture and function in children that can persist throughout life (Archibald et al., 2001; Bookstein et al., 2002; Riley et al., 2004; Sowell et al., 2002). The literature to-date has documented that gestational alcohol interferes with protein synthesis in various brain regions, including cerebral cortex, however there is a paucity of data on the mechanism(s) underlying this phenomenon (Bonner et al., 2003; Rawat 1985).
In this study, exposure of PCNs to ETOH resulted in reduction of general protein synthesis by about 40% (Fig. 1A). ETOH could impair protein translation by impacting on several key points viz. (a) suppressing mTOR/S6K levels (Hong-Brown et al., 2010; Zargar et al., 2011), (b) increasing eIF2α phosphorylation (Chen et al., 2006; Karinch et al., 2008), and/or (c) decreasing levels of elongation factors, eEF1A and eEF2 (Vary et al., 2002). Additionally, this negative effect of ETOH on activity of mTORC1 and protein synthesis could be physiologically counterbalanced by mTORC2 (Hong-Brown et al., 2011). However, our data indicate that the p70S6K-4EBP1 and eIF2α based mechanisms appear not to significantly contribute in ETOH induced protein translation impairment in neurons. Thus, based on different alcohol treatments and models, it is evident that the translation process is controlled by diverse factors. This emphasizes the need for identifying new molecular candidates to target specific translational checkpoints. PDCD4, a tumor suppressor gene with RNA binding property acts as a repressor of protein translation in non-neuronal systems (Yang et al., 2003b; Zargar et al., 2011). To our knowledge there have been no reports documenting the expression of PDCD4 protein and its message in developing brain. In this study, we show that PDCD4 mRNA and protein are expressed in developing rat brain and cultured PCNs (Fig. 1B and 1C) suggesting a role for such an endogenous repressor of protein synthesis during normal brain development. Additionally, the current study demonstrates ETOH-related dose and time-dependent enhancements of expression of PDCD4 in PCNs (Fig. 2A and 2B). Further, the PDCD4 downregulation experiment demonstrates a causal connection between the ETOH-related decrease in protein synthesis and the increased PDCD4. An important significance of these responses to ETOH resides in the potential impact of inhibited protein synthesis on the dynamic developing brain. Even if these PDCD4-related events are periodic and reverse on withdrawal of ETOH, there are critical windows of vulnerability of the various brain areas where damage could be inflicted that would ultimately be reflected in structural anomalies of the mature brain. These are the sort of apparent heterogeneous, brain-wide adverse responses presented in Fetal Alcohol Syndrome/Fetal Alcohol Spectrum Disorders.
Our data show that ETOH enhanced the cytoplasmic presence of PDCD4 in PCNs (Fig. 3A), and interactions of PDCD4 with initiation factors, eIF4A, and eIF4G in cytoplasm are implicated in suppression of protein translation (Göke et al., 2002; Kang et al., 2002; Yang et al., 2003a). ETOH also increased PDCD4 localization in the nucleus (Fig. 3A) suggesting that (a) neither the nuclear export nor the import of PDCD4 is impaired (b) ETOH augments the expression of PDCD4 in both compartments rather than altering the distribution between them. This is likely due to a comparable “volume transmission” phenomenon which brain cells utilize to transmit substances such as ions, transmitters, peptides, neurosteriods etc (Zoli et al., 1999). Studies by others have also found PDCD4 to be localized to cell nucleus on overexpression (Schlichter et al., 2001) and interfering with prosurvival events (Yang et al., 2006). However, the role for nuclear increases in neurons is obscure at present.
Since eIF4A unwinds the complex secondary structure at 5’ cap of mRNA and facilitates the scanning of ribosomes, inhibition of eIF4A activity may directly correlate with inhibition of overall translation (Miron and Sonenberg 2001; Thornton et al., 2003). Our studies demonstrated that ETOH strikingly reduced the interaction of eIF4A to cap analog (Fig. 4A; Fig. 7E) suggesting the unavailability of eIF4A to initiate translation. Notably, the clear increase in eIF4A associated with PDCD4 in methyl cap-eIF4A depleted supernatants from ETOH exposed neurons (Fig. 4B) indicates that eIF4A is in a locked state with PDCD4. Thus, the classic cap-dependent translation suppression induced by ETOH in neurons that is mediated by sequestration of eIF4A, agrees with previous findings (Göke et al., 2002; Yang et al., 2003a). It has been proposed that DExH/D family of proteins (x - any amino acid) can reorganize the structure of ribonucleoprotein assemblies that play an important role in normal development of cerebral cortex (Jankowsky et al., 2001; Thyagarajan and Szaro 2008). Thus, by hampering the activity of eIF4A, a member of DEAD-box RNA helicase, PDCD4 can affect structural rearrangement of RNPs thereby affecting neuronal development besides translation during ETOH exposure.
The translation assays demonstrated that both ETOH and enforced PDCD4 overexpression inhibited translation of non-stem loop and stem-loop luciferase mRNA constructs in PCNs (Fig.5A–5D). This suggests that increased PDCD4 can hijack and interfere with eIF4A-methyl cap association thus suppressing translation. Increases in PDCD4 inhibited translation of non-structured mRNA more efficiently than that of structured mRNA. This finding is at odds with the observations of Yang et al. (2004), in that PDCD4 has a greater preference for inhibiting translation of structured luciferase construct than the non-structured ones. It is possible that when the role for PDCD4 is limited towards an eIF4A-dependent mechanism as in the case of non-stem loop mRNA translation (Fig. 5A, 5C), it can generate a strong alternative repression via inhibiting eIF4E (Jiang et al., 2010) in a cell type specific manner. Different stimuli alter the availability and activity of other accessory factors such as eIF4E and eIF2 affecting the initiation of cap-dependent translational process (Van Der Kelen et al., 2009). Notably, ETOH represses mRNA expression of eIF4E and MAPK which phosphorylates eIF4E in mouse neurons (Gutala et al., 2004). Thus, our results suggest, that in the setting where unwinding is required (as in stem-loop construct translation), a PDCD4-eIF4A based mechanism could be favored. To the contrary, when complex structure is not involved, a PDCD4-eIF4E dependent mechanism could be preferred, with either case repressing translation.
Blockade of PDCD4 may result in efficient translation in non-neuronal models (Yang et al., 2003b). Our results suggest that as a parallel process, down regulation of PDCD4 could result in restoration of protein synthesis and further ETOH induced suppression of protein synthesis can be restored (Fig. 6B). As the translation process is controlled at several points which involve different translation factors, we cannot discount other mechanisms that could have an impact on ETOH-induced protein synthesis aberrations. Although PDCD4 checks the growth of cancer cells by suppressing translation, to our knowledge, this is the first report of PDCD4 upregulation eliciting an adverse outcome in fetal brain in a well-established rat alcohol binge model (Fig. 7A; 7B and 7D). Importantly, PDCD4 acts as a dominant endogenous inhibitor of eIF4A as one molecule of PDCD4 binds to and inactivates two molecules of eIF4A (Chang et al., 2009). Although both phosphorylation status and abundance can dictate the functionality of PDCD4, the latter is sufficient to regulate mRNA translation (Zargar et al., 2011). Thus, in the context of gestational alcohol exposure in rats, our data implicate PDCD4 abundance as an inhibitor of protein synthesis in developing fetal cerebral cortex and cortical neurons (Fig. 8). As during reduced cap-dependent translation in stress conditions such as hypoxia, ER stress or serum deprivation, IRES based selective mRNA translation occurs (Holcik and Sonenberg, 2005). Therefore, it will be interesting and important in the future to understand the role of alcohol responsive IRES phenomenon in regards to PDCD4 in neurons and developing brain. PDCD4 has been very recently shown to repress IRES mediated translation (Liwak et al., 2012).
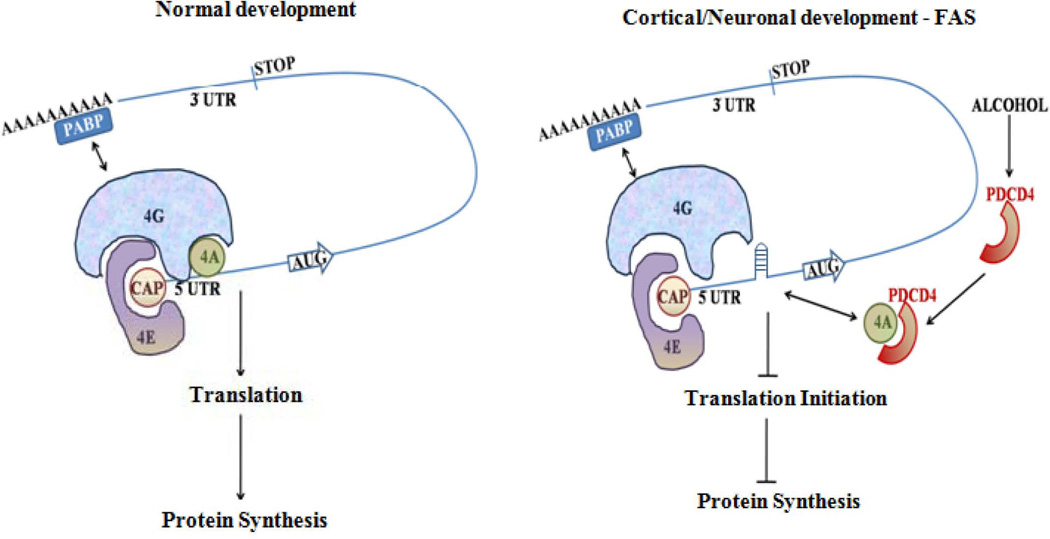
Figure 8. Schematic showing role of PDCD4 in FAS model.
During normal development, when PDCD4 expression is maintained at physiological level, eIF4A is available to bind to cap structure and unwinds the mRNA enabling normal translational process. In the event of ETOH abuse during pregnancy, increase in endogenous PDCD4 levels could sequester eIF4A thus hindering the association of eIF4A to the cap structure. Thus the complex structure of mRNA 5’ UTR is left unresolved leading to hampered scanning of ribosomes, inhibiting translation initiation.
Conclusions
This data, for the first time illustrates that alcohol induced dysregulation of protein synthesis is mediated by PDCD4. Furthermore, an ETOH-induced PDCD4 upregulation appears to influence the translation of selective mRNA subsets which is evident from the upregulation of other factors involved in alcohol-induced neurotoxic phenomena (Narasimhan et al., 2011; Xu et al., 2008). Thus, our current report suggests that targeting multiple translational checkpoints instead of independent regulatory points would be a promising pharmacological intervention strategy in FAS-related translation impairment.
Supplementary Material
Acknowledgements
This work was supported by RO1 AA010114 (to G.I.H). SM is supported by grants awarded by the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development (1I01BX000975) and by NIH (RO1AI043279). We thank Dr. Michele Pagano, NYUMC, New York for PDCD4 plasmid.
Abbreviations
- ETOH
ethanol
- PCNs
primary cortical neurons
- PDCD4
programmed cell death protein 4
- eIF4A
eukaryotic initiation factor 4A
- DIV
days in vitro
- FAS
fetal alcohol syndrome
- EWD
ethanol withdrawal
- EtBr
ethidium bromide
- UTR
untranslated region
References
- Archibald SL, Fennema-Notestine C, Gamst A, Riley EP, Mattson SN, Jernigan TL. Brain dysmorphology in individuals with severe prenatal alcohol exposure. Dev Med Child Neurol. 2001;43:148–154. [PubMed] [Google Scholar]
- Böhm M, Sawicka K, Siebrasse JP, Brehmer-Fastnacht A, Peters R, Klempnauer KH. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene. 2003;22:4905–4910. doi: 10.1038/sj.onc.1206710. [DOI] [PubMed] [Google Scholar]
- Bonner AB, Dalwai S, Marway JS, Preedy VR. Acute exposure to the nutritional toxin alcohol reduces brain protein synthesis in vivo. Metabolism. 2003;52:389–396. doi: 10.1053/meta.2003.50009. [DOI] [PubMed] [Google Scholar]
- Bookstein FL, Streissguth AP, Sampson PD, Connor PD, Barr HM. Corpus callosum shape and neuropsychological deficits in adult males with heavy fetal alcohol exposure. Neuroimage. 2002;15:233–251. doi: 10.1006/nimg.2001.0977. [DOI] [PubMed] [Google Scholar]
- Chang JH, Cho YH, Sohn SY, Choi JM, Kim A, Kim YC, Jang SK, Cho Y. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci U S A. 2009;106:3148–3153. doi: 10.1073/pnas.0808275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ma C, Bower KA, Ke Z, Luo J. Interaction between RAX and PKR modulates the effect of ethanol on protein synthesis and survival of neurons. J Biol Chem. 2006;281:15909–15915. doi: 10.1074/jbc.M600612200. [DOI] [PubMed] [Google Scholar]
- Dong J, Yan D, Chen SY. Stabilization of Nrf2 protein by D3T provides protection against ethanol-induced apoptosis in PC12 cells. PLoS One. 2011;6(2):e16845. doi: 10.1371/journal.pone.0016845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Göke A, Göke R, Knolle A, Trusheim H, Schmidt H, Wilmen A, Carmody R, Göke B, Chen YH. DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem Biophys Res Commun. 2002;297:78–82. doi: 10.1016/s0006-291x(02)02129-0. [DOI] [PubMed] [Google Scholar]
- Göke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1) Am J Physiol Cell Physiol. 2004;287:C1541–C1546. doi: 10.1152/ajpcell.00025.2004. [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- Gutala R, Wang J, Kadapakkam S, Hwang Y, Ticku M, Li MD. Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. Alcohol Clin Exp Res. 2004;28:1779–1788. doi: 10.1097/01.alc.0000148117.17707.b4. [DOI] [PubMed] [Google Scholar]
- Henderson GI, Devi BG, Perez A, Schenker S. In utero ethanol exposure elicits oxidative stress in the rat fetus. Alcohol Clin Exp Res. 1995;19:714–720. doi: 10.1111/j.1530-0277.1995.tb01572.x. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nature Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hong-Brown LQ, Brown CR, Kazi AA, Huber DS, Pruznak AM, Lang CH. Alcohol and PRAS40 knockdown decrease mTOR activity and protein synthesis via AMPK signaling and changes in mTORC1 interaction. J Cell Biochem. 2010;109:1172–1184. doi: 10.1002/jcb.22496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong-Brown LQ, Brown CR, Navaratnarajah M, Huber DS, Lang CH. Alcohol-induced modulation of rictor and mTORC2 activity in C2C12 myoblasts. Alcohol Clin Exp Res. 2011;35:1445–1453. doi: 10.1111/j.1530-0277.2011.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Zhang SH, Han GQ, Qin CY. Interaction of Pdcd4 with eIF4E inhibits the metastatic potential of hepatocellular carcinoma. Biomed Pharmacother. 2010;64:424–429. doi: 10.1016/j.biopha.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Ahn HS, Lee JY, Matsuhashi S, Park WY. Up-regulation of PDCD4 in senescent human diploid fibroblasts. Biochem Biophys Res Commun. 2002;293:617–621. doi: 10.1016/S0006-291X(02)00264-4. [DOI] [PubMed] [Google Scholar]
- Karinch AM, Martin JH, Vary TC. Acute and chronic ethanol consumption differentially impact pathways limiting hepatic protein synthesis. Am J Physiol Endocrinol Metab. 2008;295:E3–E9. doi: 10.1152/ajpendo.00026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann E, Deverm TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J Biol Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Lankat-Buttgereit B, Göke R. The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell. 2009;101:309–317. doi: 10.1042/BC20080191. [DOI] [PubMed] [Google Scholar]
- Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- Liwak U, Thakor N, Jordan LE, Roy R, Lewis SM, Pardo O, Seckl M, Holcik M. Tumour Suppressor PDCD4 Represses IRES-Mediated Translation of Anti-Apoptotic Proteins and is Regulated by S6 Kinase 2. Mol Cell Biol. 2012 doi: 10.1128/MCB.06317-11. Mar 19: Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahimainathan L, Das F, Venkatesan B, Choudhury GG. Mesangial cell hypertrophy by high glucose is mediated by downregulation of the tumor suppressor PTEN. Diabetes. 2006;55:2115–2125. doi: 10.2337/db05-1326. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Riley EP, Delis DC, Stern C, Jones KL. Verbal learning and memory in children with fetal alcohol syndrome. Alcohol Clin Exp Res. 1996;20:810–816. doi: 10.1111/j.1530-0277.1996.tb05256.x. [DOI] [PubMed] [Google Scholar]
- Miron M, Sonenberg N. Regulation of translation via TOR signaling: insights from Drosophila melanogaster. J Nutr. 2001;131:2988S–2993S. doi: 10.1093/jn/131.11.2988S. [DOI] [PubMed] [Google Scholar]
- Morley SJ, Coldwell MJ, Clemens MJ. Initiation factor modifications in the preapoptotic phase. Cell Death Differ. 2005;12:571–584. doi: 10.1038/sj.cdd.4401591. [DOI] [PubMed] [Google Scholar]
- Narasimhan M, Mahimainathan L, Rathinam ML, Riar AK, Henderson GI. Overexpression of nrf2 protects cerebral cortical neurons from ethanol-induced apoptotic death. Mol Pharmacol. 2011;80:988–999. doi: 10.1124/mol.111.073262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney JW, Tenkova T, Dikranian K, Qin YQ, Labruyere J, Ikonomidou C. Ethanol-induced apoptotic neurodegeneration in the developing C57BL/6 mouse brain. Brain Res Dev Brain Res. 2002;133:115–126. doi: 10.1016/s0165-3806(02)00279-1. [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Akar U, Steiner M, Zorrilla-Calancha I, Tirado-Gomez M, Colburn N, Danilenko M, Kornblau S, Berestein GL. Programmed cell death-4 tumor suppressor protein contributes to retinoic acid-induced terminal granulocytic differentiation of human myeloid leukemia cells. Mol Cancer Res. 2007;5:95–108. doi: 10.1158/1541-7786.MCR-06-0125. [DOI] [PubMed] [Google Scholar]
- Palamarchuk A, Efanov A, Maximov V, Aqeilan RI, Croce CM, Pekarsky Y. Akt phosphorylates and regulates Pdcd4 tumor suppressor protein. Cancer Res. 2005;65:11282–11286. doi: 10.1158/0008-5472.CAN-05-3469. [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. Current advances in local protein synthesis and synaptic plasticity. J Neurosci. 2006;26:7147–7150. doi: 10.1523/JNEUROSCI.1797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Watts LT, Maffi SK, Chen J, Schenker S, Henderson G. Ethanol-induced oxidative stress precedes mitochondrially mediated apoptotic death of cultured fetal cortical neurons. J Neurosci Res. 2003;74:577–588. doi: 10.1002/jnr.10767. [DOI] [PubMed] [Google Scholar]
- Rathinam ML, Watts LT, Stark AA, Mahimainathan L, Stewart J, Schenker S, Henderson GI. Astrocyte control of fetal cortical neuron glutathione homeostasis: up-regulation by ethanol. J Neurochem. 2006;96:1289–1300. doi: 10.1111/j.1471-4159.2006.03674.x. [DOI] [PubMed] [Google Scholar]
- Rawat AK. Nucleic acid and protein synthesis inhibition in developing brain by ethanol in the absence of hypothermia. Neurobehav Toxicol Teratol. 1985;7:161–166. [PubMed] [Google Scholar]
- Riley EP, McGee CL, Sowell ER. Teratogenic effects of alcohol: a decade of brain imaging. Am J Med Genet C Semin Med Genet. 2004;127C:35–41. doi: 10.1002/ajmg.c.30014. [DOI] [PubMed] [Google Scholar]
- Roebuck TM, Mattson SN, Riley EP. Behavioral and psychosocial profiles of alcohol-exposed children. Alcohol Clin Exp Res. 1999;23:1070–1076. [PubMed] [Google Scholar]
- Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci USA. 1997;94(21):11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichter U, Burk O, Worpenberg S, Klempnauer KH. The chicken Pdcd4 gene is regulated by v-Myb. Oncogene. 2001;20:231–239. doi: 10.1038/sj.onc.1204071. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Mattson SN, Tessner KD, Jernigan TL, Riley EP, Toga AW. Regional brain shape abnormalities persist into adolescence after heavy prenatal alcohol exposure. Cereb Cortex. 2002;12:856–865. doi: 10.1093/cercor/12.8.856. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Masters SE, Bagnall MW, Carew TJ. Molecular mechanisms underlying a unique intermediate phase of memory in aplysia. Neuron. 2001;31:143–154. doi: 10.1016/s0896-6273(01)00342-7. [DOI] [PubMed] [Google Scholar]
- Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J Mol Med. 2003;81:536–548. doi: 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res. 2008;1189:33–42. doi: 10.1016/j.brainres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Van Der Kelen K, Beyaert R, Inzé D, De Veylder L. Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol. 2009;44:143–168. doi: 10.1080/10409230902882090. [DOI] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci USA. 2002;99:1639–1644. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary TC, Nairn AC, Deiter G, Lang CH. Differential effects of alcohol consumption on eukaryotic elongation factors in heart, skeletal muscle and liver. Alcohol Clin Exp Res. 2002;26:1794–1802. [PubMed] [Google Scholar]
- Xu Y, Tang Y, Li Y. Effect of folic acid on prenatal alcohol-induced modification of brain proteome in mice. Br J Nutr. 2008;99:455–461. doi: 10.1017/S0007114507812074. [DOI] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003a;23:26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003b;22:3712–3720. doi: 10.1038/sj.onc.1206433. [DOI] [PubMed] [Google Scholar]
- Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol. 2004;24:3894–3906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, Tan TH, Colburn NH. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga H, Matsuhashi S, Fujiyama C, Masaki Z. Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int. 1999;49:1067–1077. doi: 10.1046/j.1440-1827.1999.00995.x. [DOI] [PubMed] [Google Scholar]
- Zargar S, Moreira TS, Samimi-Seisan H, Jeganathan S, Kakade D, Islam N, Campbell J, Adegoke OA. Skeletal muscle protein synthesis and the abundance of the mRNA translation initiation repressor PDCD4 are inversely regulated by fasting and refeeding in rats. Am J Physiol Endocrinol Metab. 2011;300:E986–E992. doi: 10.1152/ajpendo.00642.2010. [DOI] [PubMed] [Google Scholar]
- Zoli M, Jansson A, Syková E, Agnati LF, Fuxe K. Volume transmission in the CNS and its relevance for neuropsychopharmacology. Trends Pharmacol Sci. 1999;20:142–150. doi: 10.1016/s0165-6147(99)01343-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.