Abstract
Splicing of mRNA precursors was discovered over 30 years ago. It is one of the most complex steps in gene expression and therefore must be tightly controlled to ensure that splicing occurs efficiently and accurately. Splicing takes place in a large complex, the spliceosome, which contains approximately 200 proteins and five small RNAs (U snRNAs). Since its discovery, much work has been done to elucidate the pathway of the chemical reaction as well as the proteins and RNAs involved in catalysis. A variety of studies have established the potential for U2 and U6 snRNAs to play a role in splicing catalysis, raising the possibility that the spliceosome is a ribozyme. If correct, this would point to the spliceosomal proteins playing a supporting role during splicing. On the other hand, it may be that proteins contribute more directly to the spliceosomal active site, with the highly evolutionarily conserved Prp8 protein being an excellent candidate. This review will concentrate on recent work on splicing catalysis, and elucidating possible roles proteins play in this process.
Introduction
The splicing reaction entails the removal of introns and the joining of exons by two consecutive transesterification reactions (reviewed in refs. 1–6). In order to remove introns precisely, the spliceosome recognizes a number of sequences in the mRNA precursor (pre-mRNA), including the 5′ splice site, the 3′ splice site and the branch site (see Figure 1A). In the first step, the 2′OH group of a residue in the branchsite, usually an adenosine (the branchpoint), bulged from an RNA-RNA duplex created by base pairing with U2 snRNA, is activated to attack the phosphodiester bond at the 5′ splice site, resulting in the formation of a branched, or lariat, intermediate containing a 2′ to 5′ phosphodiester bond, and release of the 5′ exon (see Figure 1B). The second step involves a nucleophilic attack by the 3′OH of the 5′ exon on the phosphodiester bond at the 3′ splice site, resulting in ligation of the two exons and excision of the intron lariat.
Figure 1. Pathway of pre-mRNA splicing.
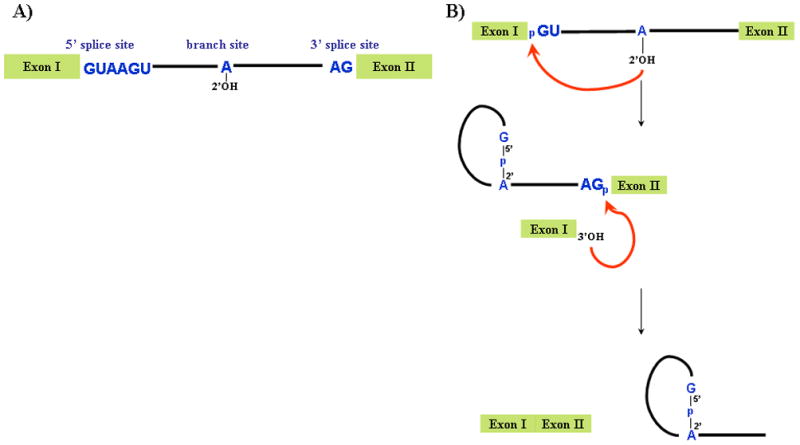
A) Schematic of pre-mRNA highlighting the sequences required for splicing. B) The two steps of the splicing reaction, with the 2′OH of the branched adenosine, are shown. The red arrow indicates the two nucleophilic attacks required for splicing. In the first step, the 2′OH of the branched adenosine attacks the 5′ splice site resulting in a free exon I and the intron lariat-exon II intermediate. In the second step, exon I attacks the 3′ splice site, resulting in ligated exons and an excised intron lariat.
Both genetic and biochemical studies have shown extensive interactions between the pre-mRNA and the different U snRNAs (reviewed in ref. 2). The first step, before the spliceosome is activated, involves recognition of the 5′ splice site and branchsite by U1 and U2 snRNP, respectively, forming the A complex. This is followed by the addition of the so-called U4.U6/U5 tri-snRNP to form B complex. In order to form the catalytically active C complex, U4 and U6 snRNAs, extensively base paired in the tri-snRNP, are unwound, U1 snRNA is displaced by U6 at the 5′ splice site, and U2 and U6 become extensively base-paired (see Figure 2). The multiple steps required before assembling a catalytically active spliceosome allows the system to check itself to ensure that the proper splicing signals are recognized and that splicing occurs accurately. The spliceosome is now primed for the first step of splicing, after which more rearrangements are required before the second step can take place7. These include a change in the conformation of U2 snRNA8, 9, a perturbation in the U2-branch site interaction10, and tethering of the 5′ and 3′ splice sites by U5 snRNA11.
Figure 2. U2 and U6 snRNA are extensively base-paired in the catalytically active spliceosome.
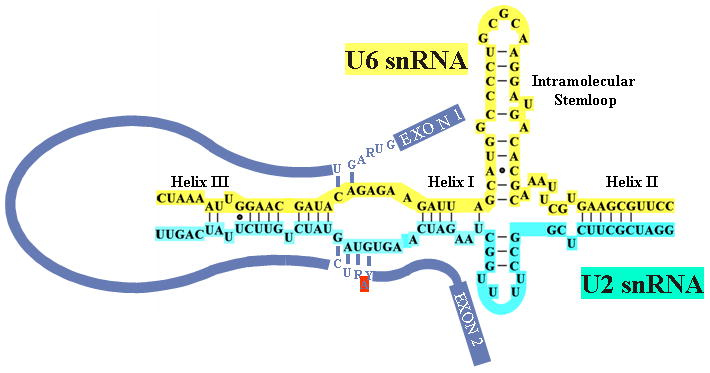
The proposed base-pairing interactions between the central domains of human U2 (turquoise) and U6 (yellow) snRNAs are shown, along with the pre-mRNA (blue). The interactions between the 5′ splice site and U6 snRNA and the interaction between the branch site and U2 snRNA are depicted with the bulged adenosine branch point indicated in red. The base-paired helices I, II, and III and the intramolecular stem-loop of U6 are shown.
The spliceosome contains approximately 200 proteins12, 13. These include proteins required for the first step, the second step or both steps. Included in the multitude of proteins are proteins that constitute the snRNPs (snRNP proteins) as well as so-called non snRNP proteins. One of the critical questions regarding splicing catalysis is whether the reaction is catalyzed by RNA or proteins, or both. Considerable evidence exists that the spliceosome is in fact a ribozyme, in which U snRNAs perform the chemistry of the reaction. However, whether or not splicing is an RNA catalyzed reaction, there is no question that many proteins are needed to assemble the catalytically active spliceosome as well as to orchestrate the many rearrangements required during splicing.
This review will focus on some of the recent work trying to answer the question of whether the spliceosome is a ribozyme, identifying the proteins involved in the various steps of spliceosome assembly and the reaction, and how these proteins are regulated during the course of the splicing reaction.
Role of U snRNAs in splicing catalysis: Is the spliceosome a ribozyme?
One of the longstanding questions in the splicing field is if the spliceosome is a ribozyme14, 15. Is the reaction catalyzed by the U snRNAs, by one or more of the many proteins associated with the spliceosome, or by a combination of the two? The similarities between pre-mRNA splicing and self-splicing of group II introns have led many to believe that splicing is an RNA catalyzed reaction (e.g., refs. 3, 16). The two steps of the splicing reactions share the same stereochemistry as well as other similarities in their structure and conservation of catalytically important domains16, 17. If the spliceosome is in essence a ribozyme, then which of the U snRNAs is responsible for catalysis? Since U1 and U4 snRNAs are removed from the spliceosome before catalysis, it is safe to assume that they are not part of the active core required for splicing. The conserved loop of U5 snRNA has been shown to be important to correctly position the exons for the second step of splicing18, 19, although it has been shown to be dispensable for both steps of splicing in mammals20. This would lead to the possibility that U2 and U6 snRNAs form at least part of the catalytic core of the spliceosome. Mutagenesis data have shown that there are two domains in U6 that are crucial for catalysis, the evolutionarily conserved ACAGAGA box21 and the AGC triad22. Another clue that U6 is involved in catalysis is its ability to bind a divalent cation at a position (U74) that has been shown to be important for catalysis23, 24. Furthermore, structural studies of a fragment of the yeast U2-U6 duplex revealed that it contains a group-II intron like domain and contains a four-fold junction that has the potential to juxtapose catalytically crucial regions of the snRNAs25.
A number of studies have provided evidence that splicing may indeed be RNA catalyzed. In one, in vitro selection was performed on variants of U6 snRNA and several ribozymes were selected that had 2′,5′-branch forming activity26. A significant step towards answering this question was taken when it was shown that in vitro transcribed U2 and U6 sRNAs are able to form a duplex in vitro under splicing conditions, and that secondary and tertiary RNA-RNA interactions that had previously been identified by genetic tools were present in this duplex27. When this duplex was formed and a short RNA oligonucleotide containing the branch site consensus (Br oligo; see Figure 3A) added in the presence of Mg++, the U2-U6 duplex catalyzed a reaction that led to the formation of a novel RNA, RNA X 28, 29. RNA X was shown to contain a covalent linkage of the branch oligo with U6, by an unusual phosphotriester bond linking the 2′OH of the bulged adenosine of the branch site and a phosphate in the catalytically important AGC triad of U6 (see Figure 3B). Specifically, this region of U6 was previously shown to function as a traditional branch site acceptor in nematode cell extracts30. The phosphotriester bond is related to the phosphodiester bond found in the intron lariat intermediate of splicing, but with the difference that in RNA X the 5′ end of the branch acceptor (U6) is not displaced, and the leaving group is water. It was shown that conserved regions of U6 that are known to be crucial for splicing, namely the AGC triad and ACAGAGA box, were necessary for RNA X formation.
Figure 3. The protein-free RNA based splicing system.
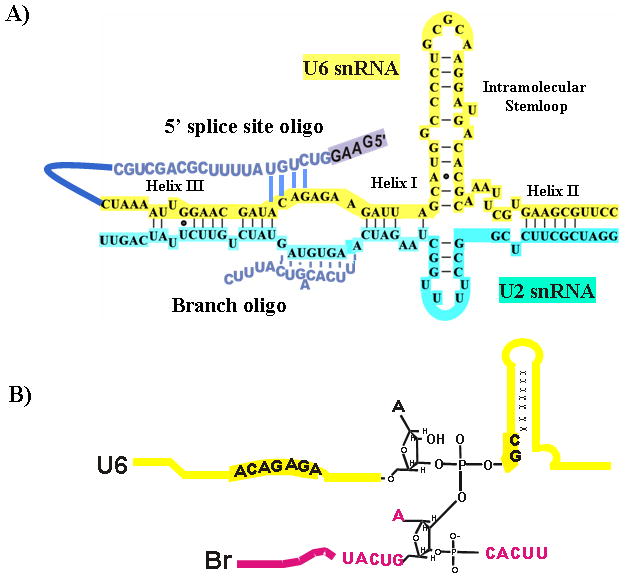
A) Base-pairing interactions in the in vitro-assembled complex of human U2-U6 snRNAs and a branchsite consensus-containing RNA oligonucleotide (Br). A 5′ splice site oligo (blue) that was linked to U6 for use in the RNA Y system is shown with the proposed exon boxed. B) Proposed chemistry of RNA X. RNA X was shown to contain a covalent linkage of the branch oligo (shown in pink) with U6 (shown in yellow), by an unusual phosphotriester bond linking the 2′OH of the bulged adenosine of the branch site and a phosphate in the catalytically important AGC triad of U6.
More recently, a second reaction related to the first step of splicing was shown to be catalyzed by the U2-U6 duplex31. A short RNA sequence containing a consensus 5′ splice site was linked to U6 (5′SS-U6). This RNA was then annealed to U2 and the complex incubated with Br oligo (see Figure 3A). After incubation a new RNA was produced in a Mg++ dependent manner. This RNA, RNA Y, contains full-length Br oligo and 5′SS-U6. However, the 5′ end of the 5′ splice site is not present in RNA Y, suggesting it has been removed during formation of RNA Y, as is the 5′ exon during the first step of the splicing reaction. The linkage between U6 and Br appears to be at or near the putative bulged adenosine. Therefore, it is apparent that the splicing reaction can be catalyzed by RNA. However, the protein-free splicing-related reactions are very slow and inefficient, leading to the conclusion that (a) co-factor(s) are required for splicing of pre-mRNA to take place in the cell.
Role of spliceosomal proteins: Prp8 and splicing catalysis
The most likely candidate protein to function in splicing catalysis is Prp8, the largest (2413 and 2335 amino acids in yeast and human, respectively) and most highly conserved protein (62% identity between yeast and human) in the spliceosome32. Due to its location at the heart of the spliceosome and its extensive interactions with crucial sequences required for splicing Prp8 has been proposed to be a master regulator of the splicing reaction14, 33. The sequence of Prp8 does not shed any light on its function in splicing. While it contains a putative RNA recognition domain, containing RNP-1 and RNP-2 sequences33, and a Jab1/MPN domain, a possible ubiquitin binding domain34, 35, no other known domains have been identified. Prp8 has been shown to interact with the 5′ splice site both within the exon36, 37 and at the canonical GU dinucleotide38. Prp8 also can be cross-linked to the 3′ splice site37 and the branch site39. In addition to its interactions with the pre-mRNA at these important sequences, Prp8 interacts with several U snRNAs, including cross-linking to U540 and U641. It has also been shown to genetically interact with U4 and U6 snRNAs and to govern their unwinding during activation of the spliceosome in yeast42. Prp8 has been implicated in controlling a number of protein splicing factors, including the helicase Brr2, which is required for both splicing catalysis and spliceosome disassembly43, 44, and Snu114, which regulates Brr2 activity45.
More recently, Prp8 has been shown to be involved in the transition between the two steps of splicing. Work by Query and Konarska has suggested a two-stage model in which there is a major reconfiguration of the spliceosome between the two steps of splicing, with Prp8 playing a central role. The first line of evidence came from examining the interaction between the 5′ splice site and U6 snRNA. Hyper-stabilizing the base-pairing between the two RNAs led to inhibition of the second step, indicating this interaction must be disrupted between the two steps of splicing46. Furthermore, weakening the interaction between the two RNAs enhanced the second step of splicing of a mutant substrate defective in the second step of splicing. A second line of evidence came from a mutational analysis of Prp8. A search for Prp8 alleles uncovered a group of mutants that improve the second step of splicing by inhibiting the first step47, which led the authors to investigate if an opposite class exists. Indeed, they identified Prp8 mutants that promote the first step by inhibiting the second step48. Putting this data together the authors suggested a model in which the active spliceosome has two conformations. The first conformation favors the first step of splicing while inhibiting the second step. After the first step is completed, the spliceosome is rearranged to a conformation that promotes the second step while inhibiting the first step. Recently, it was shown that under certain salt conditions both steps of splicing can be reversed49. This lends support to the model proposed by Query and Konarska in that while under normal conditions the forward reactions are favored, the spliceosome can still perform the reverse reactions. If the model is correct it will be of great interest to find the factors required to ensure that the splicing reaction favors moving forward over going backwards. However, the ability to go back and forth between the two steps may allow for a proof-reading mechanism in the event that the spliceosome makes a mistake.
A major effort has been made to identify a catalytic activity of Prp8. Since the protein interacts with all the critical RNA elements required for splicing and appears to be involved in the transition between the two steps of splicing, there is little doubt that it is an indispensable protein for the reaction. Three independent groups recently solved the crystal structure of a central domain of both the human and yeast Prp850–52, which has furthered our understanding of the role Prp8 plays in splicing. The groups all crystallized a similar domain, which contains the region that cross-links with the 5′ splice site of the pre-mRNA. The structure is similar to a mitten containing a palm region, fingers and a flexible thumb region which consists of a β-finger. The most striking feature is the protruding thumb or β-finger, which is reminiscent of many ribosomal proteins52, and likely interacts with other proteins or RNA similar to the protrusions of ribosomal proteins. Interestingly, Prp8 mutations that effect the equilibrium between the two steps of splicing48 are located in the β-finger. The fragment of Prp8 adopts a fold similar to that found in the catalytic core of RNase H53, although lacking key catalytic residues. Proteins, including Prp8, that contain this RNAse H fold all contain a similar five-stranded β-sheet and three corresponding α-helices surrounding the sheet. Pena et al. (2008)50 derived a model for the binding of the 5′ splice site RNA with Prp8 by comparison to the RNase H-substrate complex. Significantly, the model agrees with both genetic and biochemical data in regard to the interactions of Prp8 and the pre-mRNA.
The RNase H fold seems ideal for facilitating the RNA rearrangements that are proposed to be governed by Prp8. Interestingly, while Yang et al. (2008)52 found the domain of Prp8 only binds RNA weakly, Ritchie et al. (2008)51 found that Prp8 had a higher affinity for duplexes and a 5–10 fold greater affinity for an RNA containing a four-helix assembly resembling the fold of U2 and U6 snRNAs in the catalytic core25. Luhrmann and colleagues also identified residues within the RNase H fold that may be involved in the catalysis of splicing by potentially assisting RNA residues in coordinating a catalytic metal ion50. However, Yang et al. (2008)52 provided evidence that at least one of these residues is important for Prp8 stability, and perhaps not catalysis. Indeed, attempts by all three groups failed to show any ability of Prp8 to bind divalent metal ions. Further work is required to determine whether the U2-U6 duplex works in concert with Prp8 to catalyze splicing.
Identification of proteins in the catalytically active spliceosome
Ever since splicing was discovered work has been done to identify the protein factors needed for assembly of a functional spliceosome. The last ten years have seen numerous attempts to identify all the splicing factors by mass spectrometry. The first studies, using heterogeneous populations of spliceosomes purified at different stages of splicing identified over 200 proteins12, 13. Recently, Luhrmann and colleagues purified and identified proteins from the B and C complexes54 by using a bi-molecular system first established by Anderson and Moore (1997)55, which enables the study of the second step of splicing. Strikingly, the purified C complexes were fully active for the second step of splicing. By comparing the proteins identified in the different spliceosome, preparations, Bessnov et al. (2008)54 provided new insight into which proteins associate with the spliceosome at different stages. Supporting previous work, they found that SF3a and SF3b, while strongly associated with B complex, were under-represented in C complexes, and more importantly were completely absent form salt-treated C complexes. SF3a and SF3b are known to be required for U2 snRNA association with the branch site. This raises the possibility that the U2 snRNA-branchpoint interaction is not required for the second step of splicing. Indeed, recent work in yeast10 showed that in the presence of a mutated 3′ splice site U2 can be used as a 3′ splice site, suggesting that its interaction with the branchpoint is disrupted during the transition from the first to second step.
One of the more interesting results from the above analysis54 was the identification of the few proteins found when C complex was purified under high salt conditions. Besides a number of U5 snRNP associated proteins, including Prp8, the vast majority of the proteins were from the PRP19 complex and PRP19 related factors. First discovered in 199456, the PRP19 complex was later shown to be required for the stable association of U5 and U6 with the spliceosome after U4 dissociates57. The highly stable association of the PRP19 complex with the C complex reflects its requirement for U5 and U6 stabilization. Furthermore, the number of PRP19 associated factors may indicate a supporting role for these proteins in stabilizing C complex. The ability to purify spliceosomes at different stages will hopefully lead to a clearer picture of the requirements for splicing progression. The purified spliceosomes can also be used for structural studies as well as potentially x-ray crystallography experiments that will help elucidate the architecture of the spliceosome and further our understanding of how the splicing reaction works.
Protein modifications and splicing
As with most complex processes in the cell, post-translational protein modifications play important roles in splicing. Several examples indicate that phosphorylation and dephosphorylation play significant roles in modulating transitions in the spliceosome during catalysis. Recently, Luhrmann and colleagues showed that the protein kinase SRPK2 is associated with the U4.U6/U5 tri-snRNP and is required for B complex formation58. They further showed that the kinase is needed to phosphorylate PRP28, a U5 snRNP component that is required for the dissociation of U1 from the 5′ splice site59, and that this phosphorylation is required for the protein’s stable association with the tri-snRNP and for tri-snRNP integration into complex B. Another study showed that dephosphorylation is required for the second step of splicing60. The phosphatases PP1 and PP2A were found to be required for the second step, to associate with the spliceosome, and to dephosphorylate the U2 and U5 snRNP components SAP155 and U5-116 kDa, respectively. This was proposed to facilitate essential structural rearrangements in the spliceosome during the transition from the first to the second step. These and other studies indicate that phosphorylation plays a significant role in splicing by controlling the activity of several important spliceosomal proteins.
The role for phosphorylation at different steps of splicing leads to the question of whether other post-translational modifications are involved in splicing. A recent study investigated the possible role of ubiquitin in splicing61. Ubiquitination of proteins is known to be involved in many cellular processes, and besides targeting proteins to the proteasome has been shown to be able to affect a protein’s function62. Using a ubiquitin mutant that can enter the conjugation pathway but is unable to function, Bellare et al. (2008)61 found that ubiquitin is required for splicing. The mutant ubiquitin inhibited splicing by reducing the levels of tri-snRNP. Further investigation showed that ubiquitin is required to repress U4/U6 unwinding. Since it was previously shown that Prp8 binds ubiquitin via its Jab1/Mpn domain63, the authors examined whether Prp8 is involved, and found that Prp8 is ubiquinated within purified tri-snRNPs. It has been suggested that Prp8 can stabilize various tertiary RNA interactions in the spliceosome as well as facilitate formation of the catalytic core14, and ubiquitin may play a role in stabilizing the snRNPs. Ubiquitin binding has been proposed to modulate intermolecular protein interactions as well as alter protein function64. Bellare et al. (2008)61 propose that the ubiquitination of Prp8 alters its ability to activate the helicase Brr2. It is possible that the ubiquitin status of Prp8 mediates how Prp8 interacts with other splicing factors in stabilizing the catalytic core.
Summary
The splicing reaction takes place in a large complicated machine that requires regulation in order for splicing to proceed correctly. The reaction is catalyzed by RNA or perhaps by RNA with the help of proteins, in all likelihood U2 and U6 snRNA with the aid of Prp8. While the two reactions are chemically simple, due to its importance splicing requires close to 200 proteins to ensure that it proceeds with exquisite accuracy, and can be effectively regulated and coupled with other cellular processes. These proteins are needed to form a catalytically active spliceosome and for the rearrangements required for the conversion from the first step to the second. Post-translational modifications of various splicing factors play an important role in splicing regulation. The further purification of spliceosomes stalled at specific stages will allow for the identification of the components needed for each step as well as allow for structural studies to understand how the splicing reaction proceeds. The protein-free RNA based systems already established28, 31 can be complemented with different proteins to determine their role in splicing catalysis. The further development of minimal RNA-based systems that recapitulate the second step or both steps of splicing will allow for the study of the individual contribution of protein factors to catalysis.
Acknowledgments
We would like to thank Saba Valadkhan (Case Western University) for help with the figures. Work done in the lab of J.L.M. was supported by the NIH.
References
- 1.Moore M, Querry CC, Sharp PA. In: The RNA world. Gesteland RF, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1993. pp. 303–357. [Google Scholar]
- 2.Nilsen TW. In: RNA structure and function. Simon R, Grunberg-Manago M, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1998. pp. 279–307. [Google Scholar]
- 3.Burge CB, Tuschl TH, Sharp PA. In: The RNA World, second edition. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 525–560. [Google Scholar]
- 4.Brow DA. Annual Review Genetics. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen TW. Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 6.Matlin AJ, Moore MJ. Adv Exp Med Biol. 2007;623:14–35. doi: 10.1007/978-0-387-77374-2_2. [DOI] [PubMed] [Google Scholar]
- 7.Umen JG, Guthrie C. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 8.Hilliker AK, Mefford MA, Staley JP. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perriman RJ, Ares M., Jr Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DJ, Query CC, Konarska MM. Mol Cell. 2007;26:883–890. doi: 10.1016/j.molcel.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman AJ. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappsilber J, Ryder U, Lamond AI, Mann M. Genome Research. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Licklider LJ, Gygi SP, Reed R. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 14.Collins CA, Guthrie C. Nat Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 15.Valadkhan S. Biol Chem. 2007;388:693–697. doi: 10.1515/BC.2007.080. [DOI] [PubMed] [Google Scholar]
- 16.Dayie KT, Padgett RA. RNA. 2008;14:1697–1703. doi: 10.1261/rna.1154408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel F, Ferat JL. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 18.O’Keefe RT, Newman AJ. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Keefe RT, Norman C, Newman AJ. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 20.Segault V, Will CL, Polycarpou-Schwarz M, Mattaj IW, Branlant C, Luhrmann R. Mol Cell Biol. 1999;19:2782–2790. doi: 10.1128/mcb.19.4.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesser CF, Guthrie C. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 22.Hilliker AK, Staley JP. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Nat Struct Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 24.Yean SL, Wuenschell G, Termini J, Lin RJ. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sashital DG, Cornilescu G, Butcher SE. Nat Struct Mol Biol. 2004;11:1237–1242. doi: 10.1038/nsmb863. [DOI] [PubMed] [Google Scholar]
- 26.Tuschl T, Sharp PA, Bartel DP. RNA. 2001;7:29–43. doi: 10.1017/s1355838201001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valadkhan S, Manley JL. RNA. 2000;6:206–219. doi: 10.1017/s1355838200992197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valadkhan S, Manley JL. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 29.Valadkhan S, Manley JL. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu YT, Maroney PA, Nilsen TW. Cell. 1993;75:1049–1059. doi: 10.1016/0092-8674(93)90315-h. [DOI] [PubMed] [Google Scholar]
- 31.Valadkhan S, Mohammadi A, Wachtel C, Manley JL. RNA. 2007;13:2300–2311. doi: 10.1261/rna.626207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodges PE, Jackson SP, Brown JD, Beggs JD. Yeast. 1995;11:337–342. doi: 10.1002/yea.320110406. [DOI] [PubMed] [Google Scholar]
- 33.Grainger RJ, Beggs JD. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC. Mol Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Shen J, Guarnieri MT, Heroux A, Yang K, Zhao R. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siatecka M, Reyes JL, Konarska MM. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teigelkamp S, Newman AJ, Beggs JD. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes JL, Gustafson EH, Luo HR, Moore M, Konarska MM. RNA. 1999;5:167–179. doi: 10.1017/s1355838299981785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins CA, Guthrie C. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal VP, Verdone L, Mayes AE, Beggs JD. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn AN, Li Z, Brow DA. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 43.Raghunathan PL, Guthrie C. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 44.Laggerbauer B, Achsel T, Luhrmann R. Proc Natl Acad Sci U S A. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Small EC, Leggett SR, Winans AA, Staley JP. Mol Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konarska MM, Vilardell J, Query CC. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Querry CC, Konarska MM. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Query CC, Konarska MM. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 49.Tseng CK, Cheng SC. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- 50.Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC. EMBO J. 2008;27:2929–2940. doi: 10.1038/emboj.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM. Nat Struct Mol Biol. 2008;15:1199–1205. doi: 10.1038/nsmb.1505. [DOI] [PubMed] [Google Scholar]
- 52.Yang K, Zhang L, Xu T, Heroux A, Zhao R. Proc Natl Acad Sci U S A. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- 55.Anderson K, Moore MJ. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 56.Tarn WY, Lee KR, Cheng SC. Proc Natl Acad Sci U S A. 1993;90:10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan SP, Kao DI, Tsai WY, Cheng SC. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 58.Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Nat Struct Mol Biol. 2008;15:435–443.0. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 59.Staley JP, Guthrie C. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 60.Shi Y, Reddy B, Manley JL. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 61.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. Nat Struct Mol Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 63.Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schnell JD, Hicke L. J Biol Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]


