Abstract
Zonula occludens proteins (ZO) are postsynaptic density protein-95 discs large-zonula occludens (PDZ) domain-containing proteins that play a fundamental role in the assembly of tight junctions and establishment of cell polarity. Here, we show that the second PDZ domain of ZO-1 and ZO-2 binds phosphoinositides (PtdInsP) and we identified critical residues involved in the interaction. Furthermore, peptide and PtdInsP binding of ZO PDZ2 domains are mutually exclusive. Although lipid binding does not seem to be required for plasma membrane localisation of ZO-1, phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P 2) binding to the PDZ2 domain of ZO-2 regulates ZO-2 recruitment to nuclear speckles. Knockdown of ZO-2 expression disrupts speckle morphology, indicating that ZO-2 might play an active role in formation and stabilisation of these subnuclear structures. This study shows for the first time that ZO isoforms bind PtdInsPs and offers an alternative regulatory mechanism for the formation and stabilisation of protein complexes in the nucleus.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0156-6) contains supplementary material, which is available to authorized users.
Keywords: Tight junction, Phospholipid, Post synaptic density-discs large-zonula occludens, Nucleus, Cell polarity
Introduction
Many cellular processes involve multi-protein complexes which are formed and stabilised through multiple protein–protein interactions. In recent years, protein–lipid interactions have also been shown to contribute to the targeting and stabilisation of these complexes at specific cellular locations [1]. Of particular interest are the interactions with mono- and polyphosphorylated derivatives of phosphatidylinositol (PtdIns), collectively known as phosphoinositides (PtdInsP). These lipids concentrate at the cytoplasmic face of cellular membranes and serve both a structural and a signalling role via their recruitment of proteins that contain PtdInsP binding domains [2]. They also play a role as precursors of several types of second messengers. Deregulation of the metabolisation of these lipids has been implicated in several disease processes, highlighting their physiological importance [3, 4]. Most studies on PtdInsP regulatory mechanisms have centred on cytoplasmic processes. However, PtdInsP and their biosynthetic machinery are also present in the nucleus [5].
Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P 2) is the most abundant PtdInsP that plays a pivotal role in many physiological processes, ranging from regulation of membrane trafficking to formation of stress fibres and focal adhesions, apoptosis, chemotaxis, vesicle trafficking and gene expression [6]. In addition, the products of phosphoinositide 3-kinases (PI3K), PtdIns(3)P, PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3 have been implicated in intracellular trafficking, signalling, mitogenesis and actin rearrangements [5, 7]. Recent evidence also suggests involvement of PtdInsP in the establishment of cell polarity with PtdIns(4,5)P 2 and PtdIns(3,4,5)P 3 acting respectively as key determinants of the apical and basolateral surface [8, 9].
An important protein complex required for the establishment of cell polarity in epithelial cells are the tight junctions (TJs). TJs function both as a barrier and a fence and contain both integral and peripheral membrane proteins, which are involved in the regulation of cytoskeletal organisation, establishment of polarity, and signalling to and from the nucleus [10]. One important class of such proteins are the zonula occludens proteins, ZO-1, ZO-2 and ZO-3, which are members of the membrane-associated guanylate kinase (MAGUK) protein family [11, 12]. ZOs are peripherally associated membrane proteins that interact together and anchor membrane proteins like claudins, occludin and junctional adhesion molecule (JAM) to the actin cytoskeleton, and cluster diverse kinases, phosphatases, small G proteins and nuclear and transcription factors at the TJ [13].
ZO-1 and ZO-2 also concentrate in the nucleus in response to chemical stress or mechanical injury, or when cells are cultured at sparse density [14–16]. They also contain several nuclear localisation and export signals (NLS and NES) [17]. In confluent monolayers ZO-2 is observed at TJs, while in sparse cultures it concentrates in the nucleus where it displays a speckled distribution and colocalises with the splicing factor SC35 [15]. It is also present in the nuclear matrix and associates with lamin B1 and actin [18]. The nuclear localisation of ZO-2 suggests that ZO-2 might play a role in nuclear processes, such as transcription and splicing. In this context, ZO-1 and ZO-2 have been shown to associate with proteins involved in the regulation of cell proliferation such as the transcription factors Jun, Fos, C/EBP [19], ZONAB [20] and KyoT2 [21]. Furthermore, ZO-2 has also been shown to modulate AP-1-regulated gene transcription [22].
We and others have shown that several PDZ domains bind PtdInsP, including PtdIns(4,5)P 2, thereby identifying PDZ domains as potential PtdInsP binding domains [23, 24]. Here, we report that the PDZ2 domain of ZO-1 and ZO-2 belongs to a growing family of PtdInsP binding PDZ domains. We measured the interaction of the PDZ2 domain and mutants with PtdInsPs in considerable detail by various biochemical assays. Collectively, this study provides new evidence for the existence of a subfamily of PDZ domains that are able to bind PtdInsP and offers an additional regulatory mechanism for the formation and stabilisation of protein complexes in the nucleus.
Materials and methods
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and 1-palmitoyl-2-oleoyl-sn-glycero-phosphoethanolamine (POPE) were from Avanti Polar Lipids (Alabaster, AL). (Poly)phosphoinositides were from Cayman (Ann Arbor, MI). Phospholipid concentrations were determined by a modified Bartlett analysis [25]. The Liposofast microextruder and 100-nm polycarbonate filters were from Avestin (Ottawa, Ontario). Fatty acid-free bovine serum albumin was from Bayer (Kankakee, IL). Pioneer L1 sensor chip was from Biacore (Piscataway, NJ).
Antibodies and plasmids
Rabbit and mouse anti-ZO-1/2 were from Invitrogen (Merelbeke, Belgium). Anti-GST was from Amersham. The anti-PtdIns(4,5)P 2 2C11 antibody was a kind gift of Dr. G Schiavo (Molecular Neuropathobiology, Cancer Research UK, London Research Institute, UK); a commercial anti-PtdIns (4,5)P2 IgM was obtained from Echelon. GFP-tagged human ZO-1 was a kind gift of Dr. H. Wunderli-Allenspach (Zurich, Switzerland). HA-tagged wild-type canine ZO-2 (HA-ZO-2 WT) in the CMV expression vector GW1 was a kind gift of Dr. Javier (Baylor College of Medicine, Houston, USA). Mouse ZO-2 cDNA was a gift from Dr. Tsukita (Osaka University, Japan) and was cloned in the pEGFP-C1 vector. Deletion constructs PDZ1-3 and SH3-C-terminus and the single PDZ domains and their mutants were cloned in the pEGFP-C1 and pGEX5X1 or pGEX 2TK vector.
Mutagenesis and protein expression
Mutagenesis of the PDZ2 domain of ZO-1 and -2 was performed using the Quikchange Mutagenesis kit (Stratagene). Plasmids were transformed into BL21(DE3) pLysS cells for protein expression. BL21(DE3) cells were grown at 37°C in LB medium containing 50 μg/ml ampicillin. Protein expression was induced at 37°C for 4 h with IPTG. The PDZ2 domains used for monolayer and SPR measurements were expressed as glutathione-S-transferase (GST)-fusion proteins and purified by glutathione-Sepharose affinity chromatography. Protein concentrations were determined by the method of Bradford [26] using bovine serum albumin as a standard.
Cell culture
HeLa, MDA-MB 231 and MDCK cells were maintained in Dulbecco’s modified Eagle’s medium + Glutamax (DMEM; Invitrogen), supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin and 0.1 mg/ml streptomycin.
RNAi
siRNAs specific for human ZO-2 were obtained from Eurogentec (Seraing, Belgium). The following sequences were used: sense siRNA sequence 1: 5′-GACCGCAUGUCCUACUUAAdTdT-3′, antisense sequence 1: 5′-UUAAGUAGGACAUGCGGUCdTdT-3′, sense sequence 2: 5′-GGAGAGACGUCAUCAGUAUdTdT-3′ and antisense sequence 2: 5′-AUACUGAUGACGUCUCUCCdTdT-3′. siRNA duplexes targeting canine ZO-2 were: sense sequence 1: 5′-GCAGCAGUAUUCCGACUAUdTdT-3′, antisense sequence 1: 5′-AUAGUCGGAAUACUGCUGCdTdT-3′, sense sequence 2: 5′-GCAGGAUCCGAGAAAUCUAdTdT-3′, antisense sequence 2: 5′-UAGAUUUCUCGGAUCCUGCdTdT-3′
HeLa and MDCK cells were transfected with siRNA duplexes (200 nM) at 80% confluency using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. A negative control siRNA duplex obtained from Eurogentec was used under similar conditions. Cells were harvested 48 h following transfection.
Staining and microscopy
Cultured cells were grown on glass coverslips, washed with PBS and fixed with 3% paraformaldehyde. After permeabilisation with 0.1% Triton X-100 in PBS, cells were blocked in 1% BSA in PBS and incubated at 37°C with primary antibody for 1 h or overnight at 4°C, followed by Alexa Fluor-488 goat anti-rabbit IgG (Molecular Probes, Eugene, OR) for 30 min at room temperature. Nuclei were stained using 4′-6-Diamidino-2-phenylindole (DAPI) (0.4 μg/ml) (Sigma).
For staining of plasmamembrane PtdIns(4,5)P 2 with the 2C11 antibody we adopted the protocol from Hammond et al. [27]. In short, cells were fixed for 3 h at 4°C with ice cold 4% paraformaldehyde/0.2% glutaraldehyde. Cells were rinsed three times with 50 mM NH4Cl and simultaneously permeabilised and blocked for 4 h at 4°C in 0.5% saponin, 5% normal goat serum (NGS, Sigma) and 50 mM NH4Cl in sodium glutamate buffer (NaGB). Next, cells were incubated overnight at 4°C with primary antibody solution (2C11 1/100, 0.1% saponin, and 5% NGS in NaGB). After washing with NaGB, the secondary antibody was added (Alexa Fluor-594-anti mouse IgM 1/200, 0.1% saponin and 5% NGS in NaGB) for 4 h at 4°C. Before mounting with Vectashield, cells were post-fixed for 10 min on ice and 5 min at room temperature. For staining of plasma membrane PtdIns(4,5)P 2 in MDCK cells, we used a protocol from Hammond et al. [28]. Cells were fixed for 15 min with 8% paraformaldehyde. Cells were rinsed three times with 50 mM NH4Cl and then left on ice for 2 min. Afterwards, the cells were simultaneously permeabilised and blocked for 45 min at 4°C in buffer A (137 mM NaCl, 2.7 mM KCl and 20 mM Na-PIPES, pH 6.8) supplemented with 0.5% saponin, 5% NGS and 50 mM NH4Cl. Next, cells were incubated for 1 h with primary antibody solution (2C11 1/200, 0.1% saponin, and 5% normal goat serum in buffer A). After washing with buffer A, the secondary antibody was added (Alexa Fluor-594-anti mouse IgM 1/200, 0.1% saponin and 5% NGS in buffer A) for 45 min at 4°C. Before mounting with Vectashield, cells were post-fixed for 10 min on ice and 5 min at room temperature with 2% PFA and washed three times with 50 mM NH4Cl. Co-staining of ZO-2 and PtdIns(4,5)P 2 in nuclear speckles was achieved by fixing cells with 4% PFA followed by 2C11 antibody incubation. After a brief post-fixation with PFA, cells were fixed with methanol for 10 min at −20°C, blocked with bovine serum albumin after which polyclonal anti-ZO-2 antibody was added overnight. Stained cells were examined using an Apotome Zeiss Axioplan II epifluorescence microscope. Images were captured using a cooled CCD Axiocam Camera and Axiovision 4.4 software (Zeiss, Götingen, Germany).
Western blotting
Cells were disrupted in ice-cold lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton-X100, 1 mM PMSF) and a protease inhibitor cocktail mix (Roche Diagnostics, Mannheim, Germany). Insoluble material was removed by centrifugation (20,000g for 15 min at 4°C). Western blotting was performed as described [29]. Proteins were visualised by enhanced chemiluminescence detection (ECL; Amersham Pharmacia Biotech, Buckinghamshire, UK).
Lipid-plate binding assay
Binding of GST-tagged proteins to PIP strips or GST-tagged proteins to PIP plates® (Echelon Biosciences) was done as described by the manufacturer. Briefly, GST proteins (10 nM) were incubated for 1 h at room temperature with the preblocked plates. The plate was then washed three times with PBS containing 0.1% Tween-20 and incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-GST (Amerscham Biosciences). The bound proteins were detected using the TMB Microwell Peroxidase Substrate System (Kirkegaard & Perry Laboratories). Absorbance was determined at 450 nm using a microplate reader.
Peptide synthesis, immobilisation, and binding experiments
Connexin 43 (Cx43) C-terminal peptide (NH2.PSSRASSRASSRPRPDDLEI.OH) was synthesised and purified according to established procedures [24]. The peptide was coupled to CNBr-activated Sepharose 4B (Pharmacia Biotech) at 5–7 mg/ml according to the manufacturer’s protocol. For peptide-plate assays, the peptide (1 μg) was spotted in 96-well microtiter plates (Maxisorp, Nunc) and dried at 37°C for 1 h. Blocking was done with 1% bovine serum albumin in phosphate-buffered saline (PBS) for 1 h. GST fusion proteins (1 μM) in PBS were incubated for 1 h at room temperature, and subsequently the wells were washed three times with PBS containing 0.1% Tween-20. The amount of bound protein was determined by a colorimetric reaction using HRP-conjugated anti-GST antibodies (Amersham Biosciences) and TMB substrate (Kirkegaard & Perry Laboratories).
Surface plasmon resonance (SPR) measurements
All SPR measurements were performed at 23°C using a lipid-coated L1 chip in the BIACORE X system as described previously [30]. Briefly, after washing the sensor chip surface with running buffer (20 mM HEPES, pH 7.4, 0.16 M KCl), POPC/POPE/POPS/PtdInsP (37:40:20:3) and POPC/POPE (60:40) vesicles were injected at 5 ml/min to the active surface and the control surface (typically 5–10 μl of 0.4 lipid vesicle mg/ml solution), respectively, to give the same resonance unit (RU) values (i.e. 4,000 RU), to ensure the consistent lipid coating. The level of lipid coating for both surfaces was kept low to minimise the mass transport effect and keep the total protein concentration (P 0) above the total concentration of protein binding sites on vesicles (M 0). Near-equilibrium SPR measurements were performed at the flow rate of 5 ml/min to allow sufficient time for the R values of the association phase to reach near-equilibrium values (R eq). After sensorgrams were obtained for five or more different concentrations of each protein within a tenfold range of K d, each of the sensorgrams was corrected for refractive index change by subtracting the control surface response from it. Assuming a Langmuir-type binding between the protein (P) and protein binding sites (M) on vesicles (i.e. P + M ↔ PM), R eq values were then plotted versus P 0, and the K d value was determined by a nonlinear least-squares analysis of the binding isotherm using an equation, R eq = R max/(1 + K d/P 0). Each dataset was repeated three or more times to calculate average and standard deviation values. For kinetic SPR measurements, the flow rate was maintained at 15 μl/min for both association and dissociation phases. Kinetic SPR data were collected to illustrate relative membrane affinity quantitatively and were not used for K d determination.
Isothermal titration calorimetry (ITC)
Microcalorimetric titration measurements were performed in a Microcal Omega isothermal titration calorimeter (Microcal, Northampton, MA, USA). All solutions were degassed under vacuum prior to use. In a typical experiment, 1.33 ml of 20 μM WT or mutant PDZ2 ZO-1 or ZO-2 in 20 mM HEPES, 150 mM NaCl, pH 7.5 was titrated by 20 × 15 μl injections of 200 μM Cx43 peptide. During titration, the injection syringe was rotated at 250 rpm. Time between injections was 5 min. In a blank experiment, heat evolved from dilution was measured by injecting the peptide solution into the sample cell filled with buffer only. This heat of dilution was subtracted from the peptide binding data for the PDZ2 domain. Data were integrated and fitted to an appropriate binding model using the ORIGIN software supplied by Microcal.
Results
The PDZ2 domain of ZO-1 and ZO-2 binds to phosphoinositides in vitro
ZO-1, -2 and -3 contain three N-terminal PDZ domains, followed by an SH3, a GuK domain, and a carboxy terminal end that includes an acidic domain and a proline-rich region (Fig. 1a). We performed a lipid-overlay assay using PIP strips (nitrocellulose membranes on which various phospholipids are immobilised) (Fig. 1b) for initial screening of the PDZ domains of ZO-1, -2, and -3. In this assay, the lipids are not in a membrane environment and the readout is not quantitative but it is a simple, fast and routinely used assay [31]. A GST-ZO-1 construct containing three PDZ domains (GST-ZO-1-PDZ1-3) bound non-selectively to all PtdInsPs and acidic phospholipids, including PtdIns, phosphatidylserine (PS) and phosphatidic acid (PA) (Fig. 1b). On the other hand, GST alone did not bind to any phospholipid. The PH domain of LL5α, known to non-selectively bind PtdInsPs, was used as a positive control (Fig 1b). We then subjected individual PDZ domains to the lipid-overlay assay to determine which PDZ domain contained the PtdInsP binding site. GST-ZO-1-PDZ2 bound PtdInsPs as efficiently as GST-ZO-1-PDZ1-3, while GST-ZO-1-PDZ1 and GST-ZO-1-PDZ3 showed weak to no binding (Fig 1b), indicating that the ZO-1 PDZ2 domain contained the PtdInsP binding site. Circular dichroism experiments showed that folding of the PDZ3 domain from ZO-1 and ZO-2 was very similar to the folding pattern of GST-ZO-2-PDZ2 (data not shown). Since the PDZ domains of ZO-2 are highly homologous to their counterparts in ZO-1, we also tested if the PDZ domains of ZO-2 bind PtdInsPs. As shown in Fig. 1b, the PDZ2 domain of ZO-2 also interacted with several PtdInsPs, although differences can be noticed with the ZO-1 PDZ2 domain, while PDZ1 and PDZ3 did not show significant binding. Moreover, the lack of lipid binding by a construct encompassing the Src Homology (SH3) domain up to the C-terminus (SH3-C) corroborates the notion that PtdInsP binding is restricted to the PDZ2 domain (Fig. 1b). In contrast to ZO-1 and ZO-2 PDZ domains, none of ZO-3 PDZ domains, including PDZ2, showed appreciable binding to lipids (not shown).
Fig. 1.
Structure and lipid binding properties of ZO PDZ2 domains. a Schematic overview of the domain organisation of the ZO-proteins. b Lipid-overlay assay of different domains of ZO-1 (human) and ZO-2 (mouse). Left is a schematic view of a lipid blot membrane (PIP-stripsTM). GST and GST-PH-LL5α were used as negative and positive controls, respectively. c ELISA PtdInsP binding assay. Each well of a ‘PIP specificity’ microtiter plate (Echelon Biosciences) was overlaid with GST, GST ZO-1 PDZ2, GST ZO-2 PDZ2, and GST- PH-PLC-δ1 (positive control). Data represent means ± SD (n = 3). AU Absorbance unit
We also performed a more sensitive ELISA assay on commercially available PIP plates. Again, lipids are not in a membrane environment in this assay. GST alone showed no significant affinity to any of the lipids (Fig. 1c, blue bars). We observed that both GST-ZO-1-PDZ2 and GST-ZO-2-PDZ2 bound slightly stronger to PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3 as compared to PtdIns(4,5)P 2 or PtdIns(3,5)P 2 (Fig. 1c, red and yellow bars). This indicates that GST-ZO-1-PDZ2 and GST-ZO-2-PDZ2 have some degree of lipid selectivity and also suggests that the differences between GST-PDZ2-ZO-1 and GST-PDZ2-ZO-2 observed in Fig. 1b do not reflect gross differences in lipid specificity. GST-ZO-2-PDZ1-3 showed some preference for bisphosphoinositides in this assay (Electronic Supplementary Material, ESM: Fig. S1). The pleckstrin homology (PH) domain of phospholipase Cδ (PLCδ) was used as a positive control [32], which showed stronger binding to PtdIns(4,5)P 2 than to other phosphoinositides, confirming the validity of the assay (Fig. 1c, cyan bars).
To obtain quantitative information about PtdInsP affinity and selectivity of ZO PDZ domains, we measured their binding to PtdInsP-containing vesicles coated onto the L1 sensor chip by SPR analysis. Gel-filtration chromatography showed that both GST-ZO-1-PDZ2 and GST-ZO-2-PDZ2 form dimers when the monomer concentration was as low as 100 nM (ESM: Fig. S2). Thus, these PDZ domains should exist as dimers under our experimental conditions. We first measured the binding of GST-ZO-1-PDZ2 to POPC/POPE/POPS/PtdInsP (37:40:20:3), containing each of seven PtdInsP. Similar to the ELISA assay, sensorgrams from kinetic SPR measurements (Fig. 2a) showed that the ZO-1 PDZ-2 domain showed a slight preference for PtdIns(3,4)P 2 binding, followed by PtdIns(3,4,5)P 3, PtdIns(4,5)P 2 and PtdIns(3,5)P 2. Monophosphoinositides showed weaker to no binding. A similar pattern was observed with GST-ZO-2-PDZ2 (ESM: Fig. S3A). PDZ1 and PDZ3 domains of ZO-1 and ZO-2 showed no detectable binding. To determine the lipid selectivity of the PDZ2 domains of ZO-1 and ZO-2 more quantitatively, we determined K d values for individual vesicles by equilibrium SPR analysis (K d values are listed in Table 1). The apparent affinity of the ZO-1 PDZ2 domain for POPC/POPE/POPS/PtdIns(3,4)P 2 vesicles (K d = 0.59 μM) was only twofold higher than the value obtained for the ZO-1 PDZ2 domain and POPC/POPE/POPS/PtdIns(4,5)P 2 vesicles (K d = 1.3 μM). This indicates that ZO PDZ2 domains have only a minor degree of lipid selectivity. Therefore, all our ensuing measurements were performed using PtdIns(4,5)P 2-containing vesicles because PtdIns(4,5)P 2 is a major phosphoinositide that is present in a much higher concentration than PtdIns(3,4)P 2 in mammalian cells. Of note, the latter K d value is lower (i.e. higher affinity) than that of the GST-tagged full-length syntenin-1 for the same vesicles (see Table 1), indicating that the ZO-1 PDZ-2 domain has PtdIns(4,5)P 2 affinity that is comparable to the tandem PDZ domains of syntenin 1, a well-characterised PtdIns(4,5)P 2 interacting protein [24], although the latter does not form domain swapping dimers. Consistent with our lipid overlay assay, the ZO-3 PDZ2 domain did not show appreciable binding to any phosphoinositide-containing vesicles (not shown). Overall, these results show that the ZO-1 and ZO-2 PDZ2 domains display PtdInsP binding with slight lipid selectivity, and that they have relatively high PtdInsP affinity compared with other reported lipid binding PDZ domains.
Fig. 2.
Measurement of ZO-1 PDZ2 PtdInsP binding by SPR analysis. a PtdInsP selectivity of ZO-1 PDZ determined by kinetic SPR measurements; 1 μM of protein was added to POPC/POPE/POPS/PtdInsP (37:40:20:3) vesicles containing seven different PtdInsPs. b Equilibrium SPR binding measurements between ZO-1-PDZ2 and POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles with the protein concentration varied from 0.2 to 2.5 μM. c A binding isotherm for ZO-1 PDZ2 and POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles. R eq values from Fig. 2b (average of triplicate measurements) were plotted as a function of the ZO-1-PDZ2 concentration. A solid line represents a theoretical curve constructed from R max (=1,500 ± 230) and K d (=1.3 ± 0.4 μM) values determined by nonlinear least-squares analysis of the isotherm using an equation: R eq = R max/(1 + K d/P 0); 20 mM HEPES buffer, pH 7.4, with 0.16 M KCl was used for all measurements
Table 1.
Membrane binding parameters of ZO PDZ2 domains determined from equilibrium SPR analysis
| Lipids | K d (μM)a | ||
|---|---|---|---|
| GST-ZO-1-PDZ2 | GST-ZO2-PDZ2 | GST-Syntenin1b | |
| POPC/POPE/POPS/PtdIns(3,4)P 2 (37:40:20:3) | 0.59 ± 0.13 | 1.2 ± 0.44 | NMc |
| POPC/POPE/POPS/PtdIns(3,4,5)P 3 (37:40:20:3) | 1.0 ± 0.30 | NMc | NMc |
| POPC/POPE/POPS/PtdIns(3,5)P 2 (37:40:20:3) | 1.2 ± 0.49 | NMc | NMc |
| POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) | 1.3 ± 0.30 | NMc | 2.0 ± 0.59 |
aValues represent the mean and standard deviation from triplicate measurements. All measurements were performed in 20 mM HEPES, pH 7.4, containing 0.16 M KCl
bFull-length syntenin-1 containing two PDZ domains
cNot measured
Identification of the lipid binding site in the PDZ2 domains of ZO-1 and ZO-2
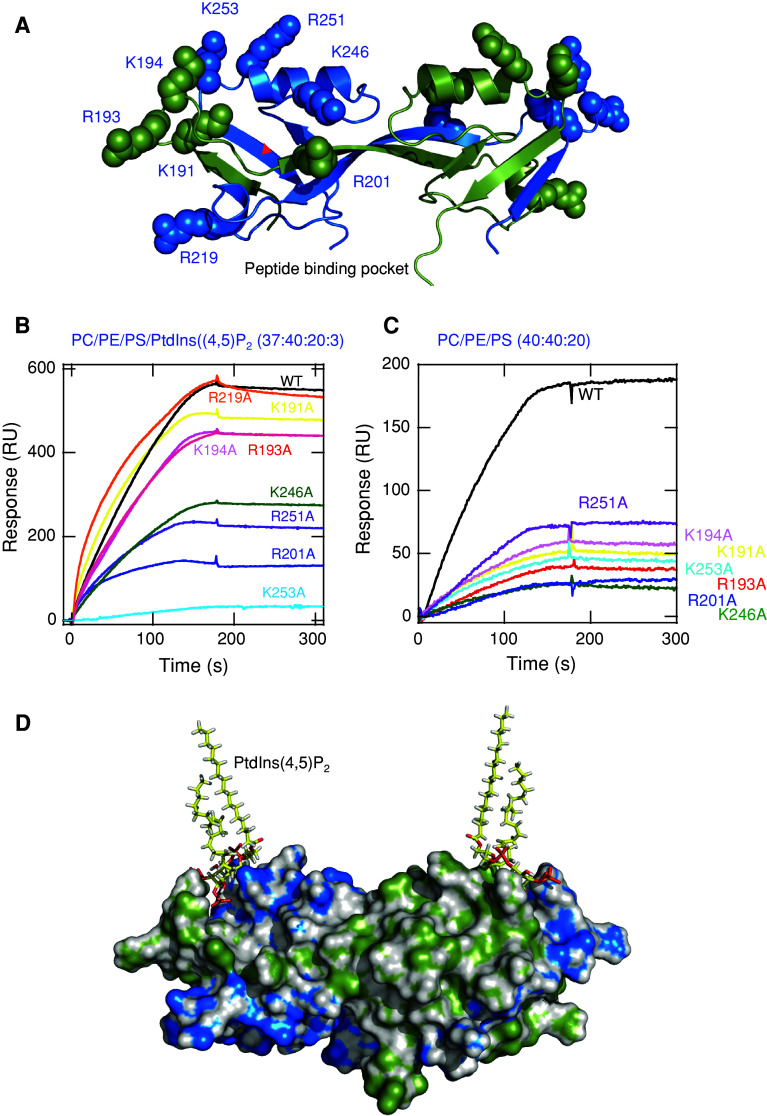
All structurally characterised PDZ domains have similar structural folds with a canonical peptide binding pocket. Recent reports indicated that PDZ domains containing basic residues on the surface surrounding the peptide binding groove can interact with anionic lipids, including PtdInsPs [23]. The PDZ2 domains of ZO-1 and ZO-2 are relatively unique among PDZ domains in that they form domain-swapping dimers [23, 33]. That is, the dimer is stabilised by extensive symmetric domain swapping of β-strands and the canonical peptide binding pocket is created by elements contributed from both monomers. As shown in Fig. 3a, the ZO-1 PDZ2 domain dimer has cationic residues surrounding the peptide binding pocket and these residues are nearly equally contributed from the two monomeric units (i.e. K191, R193, K194, and R201 from the B chain and K246, R251 and K253 from the A chain; A and B chains are arbitrarily defined in Fig. 3a). To see if these residues are involved in PtdInsP binding, we mutated them individually to Ala and measured the effects on binding to POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles by SPR analysis. Results are illustrated in Fig. 3b and also summarised in Table 2. The most dramatic effect was seen with K253A, which is followed by R201A, R251A and K246A. In fact, we could not determine the K d values for these mutants because their affinities were too low. These results suggest that R201 from B chain and K246, R251 and K253 from A chain are most directly involved in vesicle binding. Also, the mutations of K191, R193 and K194, which belong to the same B chain as R201, had modest but definite effects on vesicle binding. In contrast, the mutation of R219 that is located remotely from these residues had no effect on vesicle binding. Collectively, these results define a relatively wide lipid binding surface of the ZO-PDZ2 domains containing multiple basic residues. Similar results were obtained from mutational analysis of the ZO-2 PDZ2 domain; i.e. R302A and R347A mutations of ZO-2 PDZ2 showed similar effects to their ZO-1 PDZ2 counterparts, R201A and K246A mutations, respectively (ESM: Fig. S3B). Interestingly, R253 in ZO-1 PDZ2, which corresponds to K364 in ZO-2 PDZ2, is substituted for by Glu in the PDZ2 domain of ZO-3, and this charge-reversal substitution of a key membrane binding residue explains why the ZO-3 PDZ2 domain does not have detectable affinity for PtdInsP.
Fig. 3.
The putative PtdInsP binding site of the ZO-1 PDZ2 domain. a The structure of ZO-1 PDZ2 domain dimer. The structure (PDB id: 2RCZ) is generated by Pymol and shown in cartoon representation with basic residues surrounding the putative peptide binding groove (red arrow) shown in space-filling representation. Two monomeric units of the dimer are shown in green (A) and blue (B), respectively. A and B chains are arbitrarily defined for illustration. b Effects of mutations of basic residues on binding of the ZO-1 PDZ2 dimer to POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles were measured by kinetic SPR analysis. Protein concentrations were kept at 1 μM. K d values were determined by equilibrium SPR analysis as shown in Fig. 2b and c and listed in Table 2. c Effects of mutations of basic residues on binding of the ZO-1 PDZ2 dimer to POPC/POPE/POPS (40:40:20) vesicles measured by kinetic SPR analysis. Protein concentrations were kept at 1 μM. d A hypothetical model of the ZO-1-PDZ2- PtdIns(4,5)P 2 complex. One PtdIns(4,5)P 2 molecule is manually docked into each monomeric unit on the basis of our mutational analysis and the final conformation was determined through energy minimisation. Notice that in this model the PtdIns(4,5)P 2 head group partially blocks the entry to the putative peptide binding pocket that is outlined in red. The ZO-1 PDZ2 dimer is rotated 45° horizontally from (a) to better illustrate the potential overlap of binding sites for PtdIns(4,5)P 2 and a peptide. The approximate location of the membrane–water interface is shown as a cyan line
Table 2.
Membrane and peptide binding parameters of ZO PDZ2 domain mutants
| Proteins | K d (μM) | |
|---|---|---|
| POPC/POPE/POPS/PtdIns(4,5)P 2 a (37:40:20:3) | Connexin43 peptideb | |
| ZO-1 PDZ2 WT | 1.3 ± 0.30 | 9.9 ± 1.3 |
| ZO-1 PDZ2 K191A | NMc | NMc |
| ZO-1 PDZ2 R193A | NMc | 11 ± 2.1 |
| ZO-1 PDZ2 K194A | 2.1 ± 0.5 | 12 ± 1.6 |
| ZO-1 PDZ2 R201A | NDd | NDd |
| ZO-1 PDZ2 R219A | NMc | 11 ± 1.6 |
| ZO-1 PDZ2 K246A | 10 ± 5 | NDd |
| ZO-1 PDZ2 R251A | 15 ± 9 | 19 ± 3.4 |
| ZO-1 PDZ2 K253A | NDd | 22 ± 3.6 |
| ZO-1 PDZ2 R251AK253A | NDd | 21 ± 3.2 |
| ZO-2 PDZ2 WT | 2.6 ± 0.7 | 9.9 ± 1.2 |
| ZO-2 PDZ2 R302A | NDd | NDd |
| ZO-2 PDZ2 R347A | 20 ± 12 | NDd |
aValues represent the mean and standard deviation from triplicate SPR measurements. All measurements were performed in 20 mM HEPES, pH 7.4, containing 0.16 M KCl
bValues are representative of duplicate ITC assays. All measurements were performed in 20 mM HEPES, pH 7.4, containing 0.15 M NaCl
cNot measured
dNot detectable
Some of these residues would interact directly with the PtdInsP headgroup while others may interact non-specifically with the anionic membrane surface. To distinguish these two types of basic residues, we measured the effects of mutations on binding to POPC/POPE/POPS (40:40:20) vesicles. Notice that K253A, R201A, R251A and K246A had much lower affinities than K191A, R193A and K194A for POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles. For POPC/POPE/POPS (40:40:20) vesicles, however, all these mutants showed comparable affinities (Fig. 3c). This suggests that, although all basic residues on the putative lipid binding surface contribute to the overall binding to anionic membranes, K253, R201, R251 and K246 which are closer to the peptide binding pocket are more directly involved in PtdInsP headgroup binding than others. On the basis of these results, we performed the docking of a PtdIns(4,5)P 2 molecule to each unit of a ZO-1 PDZ2 dimer. Due to the symmetric nature of the dimer, it was postulated that the dimer would form two identical lipid binding sites. The model shown in Fig. 3d suggests that the putative PtdInsP binding site and the peptide binding site overlap to the extent that two binding processes may be mutually exclusive.
PtdIns and peptide binding are mutually exclusive
The PDZ2 domains of the ZO proteins have been shown to bind the Gap Junction protein connexin43 (Cx43). Binding involves the C-terminal peptide sequence of Cx43 (DDLEI) that fits into the hydrophobic pocket of the PDZ domain [34]. In addition, domain-swapping dimerisation of ZO-1 PDZ2 creates an extended ligand binding pocket located in PDZ dimer interface [35]. To evaluate the extent of overlap between PtdInsP binding and peptide binding of ZO PDZ2 domains, we investigated the effect of the mutations in the PtdInsP binding site on the peptide binding ability of the ZO-1 PDZ2 domain. We measured by ITC analysis the binding of wild-types and mutants of ZO-1 and -2 PDZ2 domains with a peptide, encompassing the C-terminal 20 amino acids of Cx43. The ITC analysis yielded a dissociation constant (K D) of ~9.9 μM for ZO-1 and ZO-2 PDZ2 domains, which is in good agreement with the value of 15.5 μM reported for ZO-1 PDZ2 [36], with a stoichiometry close to 1 mol peptide/mol PDZ domain (Fig. 4a; Table 2). This value is also comparable with K Ds determined for other PDZ domains [37]. As summarised in Table 2, among ZO-1 PDZ2 mutants, R201A and K246A mutants showed no significant binding to the Cx43 peptide while R251A, K253A and the double-site mutant R251A/K253A had about twofold lower peptide affinity than the wild-type. In contrast, mutations of R193 and K194, which were found to interact non-specifically with anionic membranes (see above), had little effect on peptide affinity. Likewise, the mutation of R219 that is remote from the lipid binding site had no effect on the peptide binding. Similar results were seen with ZO-2 PDZ2 mutations (summarised in Table 2). These results support the notion that there is significant overlap between the PtdInsP and peptide binding sites and that the two types of binding are mutually exclusive.
Fig. 4.
Peptide binding properties of ZO PDZ2 domains and mutants therein. a ITC titration of WT ZO PDZ2 domains with the C-terminal peptide (20 amino acids) of connexin43. Corrected raw heat data (top panel) and a binding isotherm (bottom panel) generated by plotting the area under the peak against the molar ratio of the peptide added to the PDZ domain are shown. b Data from pull-down experiments measuring the binding of GST-ZO-1 PDZ2 WT and mutants (upper panels) or GST-ZO-2 PDZ WT and mutants (lower panels) to the C-terminal peptide of connexin43 covalently bound to cyanogen bromide (CNBr)-activated Sepharose. GST-ZO-2 PDZ2 WT is also shown. The right panel shows the Coomassie staining of the same volume of aliquots from individual proteins
To further corroborate this finding, we performed two additional sets of experiments. First, we examined the effect of the mutations on the association of the GST PDZ2 domain with the Cx43 peptide by a pull-down assay, in which the Cx43 peptide was covalently coupled to CNBr-activated Sepharose (see “Materials and methods”). Following incubation with the peptide matrix, 15–20% of the input GST-ZO-1-PDZ2 WT was retained on the peptide matrix (Fig. 4b, upper left panel, compare lanes 2,6 with lanes 2,6 in right panel which represents the input) while only 5% of the input was retained in case of the R201A (lane 3) and K246A (lane 4). As expected from only a twofold difference in peptide affinity, R251A (lane 5), K253A (lane 7), and R251A/K253A (lane 8) behaved similar to the wild-type. Non-specific binding was limited to less than 5% of the total protein input, as shown for GST binding (lane 1). GST-ZO-2-PDZ2 mutants R302A and R347A also showed strongly reduced interaction with the Cx43 peptide as compared to GST-ZO-2-PDZ2 WT (Fig. 4b, lower panels).
We then performed an ELISA assay in which the Cx43 peptide immobilised on a plate was incubated with GST-ZO-2-PDZ2 WT with or without different PtdInsPs. Figure 5a shows that binding of ZO-2 PDZ2 WT to the peptide is inhibited by PtdIns(4,5)P 2, PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3 in a concentration-dependent manner and to the same extent, in contrast to PS which shows no effect. We also preincubated GST-ZO-2-PDZ2 WT with these lipids and subsequently performed a pull-down with the Cx43 peptide that was covalently bound to CNBr-activated Sepharose beads. Preincubation of 3 μg GST-ZO-2-PDZ2 WT with increasing PtdIns(4,5)P 2 concentrations (indicated at the bottom of the panels in Fig. 5b) resulted in a progressively decreasing association of the GST-ZO-2-PDZ2 with the Cx43 peptide. Likewise, PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3 inhibited association of GST-ZO-2-PDZ2 WT with the Cx43 peptide (Fig. 5b), while PS had no effect, confirming the PtdInsP specificity of the PDZ2 domain.
Fig. 5.
Lipid and peptide binding to ZO PDZ2 domains are mutually exclusive. a ELISA peptide binding assay showing that PtdInsPs compete with the Cx43 peptide for interaction with the ZO-2 PDZ2 domain. Each well of a microtiter plate was coated with 1.0 μg Cx43 peptide and incubated with 1 μM GST or GST-tagged ZO-2 PDZ2 WT with or without pre-incubation with different PtdInsPs and PS (negative control). Data shown are representative of duplicate assays, each performed in triplicate. (RCU Relative colorimetric units). b Results from a pull-down assay measuring the binding of 3 μg GST-ZO-2 PDZ2 WT to the C-terminal peptide of Cx43 covalently bound to CNBr-activated Sepharose with or without preincubation with different phospholipid liposomes at the indicated concentrations
ZO polypeptides colocalise with PtdIns(4,5)P2 at the plasma membrane
To understand the physiological roles of PtdInsP binding of ZO PDZ2 domains, we analysed the distribution of PtdIns(4,5)P 2 and ZO-1/-2 in different mammalian cells. As described above, we focused on this phospholipid because it is much more abundant in membranes of mammalian cells than PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3, while the affinity of the ZO PDZ2 domains for PtdIns(4,5)P 2, PtdIns(3,4)P 2 and PtdIns(3,4,5)P 3 is of the same order of magnitude (Table 1).
In sparsely seeded MDCK cells, endogenous ZO-1 and ZO-2 showed differential localisation. That is, ZO-1 exhibited mainly plasma membrane localisation whereas ZO-2 was also distributed in the nucleus and the cytosol, which is consistent with the previous report (Fig. 6a) [14]. In these cells, the GFP-tagged PH domain of phospholipase Cδ (PH-PLCδ) that has been used as a specific probe for PtdIns(4,5)P 2 [32] was co-localised with endogenous ZO-1 and ZO-2 in the plasma membrane, suggesting that ZO proteins might be localised in the plasma membrane due to their interaction with PtdIns(4,5)P 2 (Fig. 6b). This was further substantiated by co-staining with the 2C11 anti-PtdIns(4,5)P 2 antibody (Fig. 6c), used in earlier studies to detect PtdIns(4,5)P 2 [38, 39]. Similarly, immunostaining for PtdIns(4,5)P 2 in MDA-MB 231 breast cancer cells using the 2C11 antibody showed co-localisation with endogenous ZO-1/-2, particularly in plasma membrane extensions (Fig. 6d, e). To see if PtdIns(4,5)P 2binding is responsible for plasma membrane localisation of ZO-1 and ZO-2, we expressed GFP-tagged ZO-1 or ZO-2 wild-type and mutants in MDCK cells. Surprisingly, the R201A mutation that essentially abolished both lipid and peptide binding of the ZO-1 PDZ2 domain did not significantly affect the plasma membrane localisation of ZO-1 in MDCK cells (ESM: Fig. S4A). Also, the K253A and R251AK253A mutants, which bind the peptide with reduced affinity but show no lipid binding, were localised at the plasma membrane. Similar data were obtained with ZO-2 (ESM: Fig. S4B). These results suggest that unlike Par-3 [23], lipid binding of ZO-1/-2 is not required per sé for its plasma membrane localisation.
Fig. 6.
ZO-proteins co-localise with PtdIns(4,5)P 2. a Localisation of endogenous ZO-1 and ZO-2 in MDCK cells. Notice that ZO-1 exhibits mainly plasma membrane localisation whereas ZO-2 shows nuclear staining and cytoplasmic/membrane staining. b Endogenous ZO-1 and ZO-2 mark out areas where GFP-PH-PLCδ is localised. Nuclear ZO-2 is less clear because of focus on the plasma membrane. c Staining of MDCK cells for ZO-1/PtdIns(4,5)P 2 and ZO-2/PtdIns(4,5)P 2 with 2C11 antibody. d, e PtdIns(4,5)P 2 staining reveals co-localisation with endogenous ZO-2 d and ZO-1 e in polarised MDA-MB 231 breast cancer cells. Scale bar 10 μm
Endogenous ZO-2 targets nuclear PtdIns(4,5)P2 in speckles
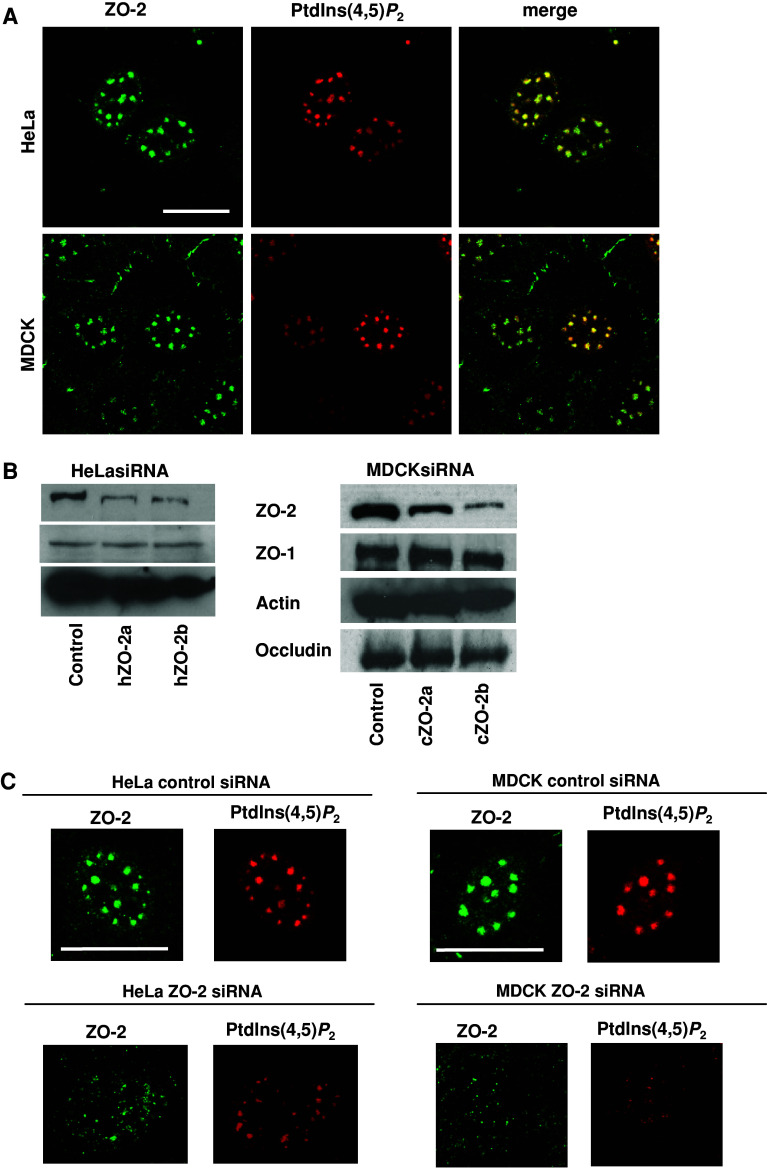
It was previously shown that ZO-2 localised to the nucleus in a cell density-dependent manner, where it displayed a speckled distribution [15, 18]. Moreover, it has been reported that nuclear PtdIns(4,5)P 2 is concentrated in speckles and nucleoli, and that the PDZ domains of syntenin-2 target these subnuclear structures due to PtdIns(4,5)P 2 interaction (34). To investigate if ZO-2 is also targeted to nuclear speckles by its ability to bind PtdIns(4,5)P 2, we studied the subcellular distribution of endogenous ZO-2. For these measurements, MDCK and HeLa cells were pre-treated with actinomycin D because transcriptional inhibitors such as actinomycin D are known to alter the spatial distribution of speckle proteins into fewer and/or abnormally large clusters [39], which helps to clearly visualise the proteins in nuclear speckles. As shown in Fig. 7a, endogenous ZO-2 showed considerable overlap with the 2C11 staining pattern suggesting that ZO-2 localises to nuclear speckles due to its ability to interact with PtdIns(4,5)P 2.
Fig. 7.
Endogenous ZO-2 localises in nuclear speckles containing PtdIns(4,5)P 2 that disperse upon ZO-2 expression knock down. a HeLa or MDCK cells treated with 1 μg/ml actinomycin D and stained for PtdIns(4,5)P 2 with 2C11 antibody. Scale bar 20 μm. b Western blot analysis of total cell extracts from HeLa cells (left panels) or MDCK cells (right panels), treated with a control siRNA or two independent species-specific siRNAs targeting ZO-2. Actin is used as a loading control. c Images of HeLa cells and MDCK cells transfected with a control siRNA or a siRNA targeting ZO-2 (h human, c canine) and treated with actinomycin D. Speckle intensity is reduced and speckles appear to have desintegrated. The nuclear PtdIns(4,5)P 2 pattern (red) was visualised using 2C11 antibody. ZO-2 staining is shown in green. Scale bar 20 μm
We then investigated the contribution of ZO-2 to nuclear speckle organisation by knocking-down endogenous ZO-2 expression in MDCK and HeLa cells. Figure 7b shows the partial knock-down of ZO-2 using two different species specific siRNA molecules, when compared to a control siRNA vector. In our hands, silencing of ZO-2 did not alter the levels of other TJ components such as occludin or ZO-1, in contrast to an earlier report [40] (Fig. 7b). Because complete knock-down of ZO-2 appeared to be toxic, we analysed cells with partial knock-down of ZO-2. In fact, ZO-2 deficient mice are embryonically lethal (unlike ZO-3−/− mice), fail to complete gastrulation and show reduced cell proliferation at E6.5 and increased apoptosis [41]. Reduction in ZO-2 expression in both cell types resulted in a dispersed PtdIns(4,5)P 2 staining pattern as observed by 2C11 staining (Fig. 7c). These results suggest that ZO-2, similar to syntenin-2 [38], could function as a scaffold in the organisation of nuclear PtdIns(4,5)P 2.
Discussion
The present work shows that the PDZ2 domains of ZO-1 and ZO-2 bind PtdInsPs with relatively high affinity, adding these PDZ domains to an increasing list of PDZ domains that interact with lipids. We and others showed previously that the tandem PDZ domains of syntenin-1 and -2 and the PDZ2 domain of PTPBL2 could bind PtdInsPs with micromolar affinity [24, 38, 42]. More recently, Wu and coworkers [23] reported that a large subset of mammalian PDZ domains, including PICK1 and another MAGUK protein Par-3, are capable of binding PtdInsPs in vitro. In general, membrane binding proteins and domains contain cationic and/or hydrophobic residues on their membrane binding surfaces [43]. Thus, PDZ domains with cationic and/or hydrophobic residues on their surfaces may have significant affinity for the membrane containing anionic lipids, including PtdInsP. Indeed, Wu et al. [23] reported that the Par-3 PDZ2 domain contained a flat surface rich in cationic residues that were shown to play critical roles in lipid binding and that other PDZ domains containing similar cationic residues also bind to anionic vesicles when measured by a vesicle pelleting assay. They also reported that the PDZ1 domain as well as the PDZ2 domain of ZO-1 and ZO-2 bound anionic vesicles; however, no quantitative data on lipid specificity and affinity were reported. We opted to rigorously characterise lipid binding of PDZ domains of ZO proteins because of the potential physiological significance of the ZO PDZ–lipid interactions. Our quantitative SPR analysis clearly shows that the isolated PDZ1 domains of ZO-1 and ZO-2 have very low affinity for anionic membranes and only PDZ2 domains of ZO proteins have relatively high PtdInsP affinity and some degree of PtdInsP selectivity. The reason for this discrepancy may derive from differences in the length of the domain constructs, different experimental conditions, or different sensitivity of measurements.
The PDZ2 domains of the ZO proteins have unique structures among PDZ domains in that they form domain-swapping dimers. However, they share a common trait with other anionic membrane binding PDZ domains including Par-3; i.e. they have a cluster of cationic residues, K194, R251 and K253 and R201 and K246, on the same surface surrounding the peptide binding pocket. In particular, K194, R251 and K253 in ZO -1 correspond to K458, R532 and K535 in Par-3, respectively, which were reported to be involved in anionic membrane binding. Unlike the Par-3 PDZ domain, however, ZO-1/-2 PDZ2 domains have modest PtdInsP selectivity. Also, clear distinction between the cationic residues of ZO-1 PDZ2 domain that are involved in PtdInsP binding and that participate in non-specific anionic membrane binding indicates the presence of a well-defined binding pocket for PtdInsPs, as illustrated in our model (see Fig. 3d).
Our results also indicate that PtdInsP binding and peptide binding of PDZ2 domains of ZO proteins are mutually exclusive because of a significant overlap of two binding sites. Indeed, binding of PtdInsPs to ZO-1 and ZO-2 PDZ2 domain reduces their interactions with a Cx43 carboxy-terminal peptide. It should be noted that Chen and co-workers [35] reported a weaker interaction between ZO-2 PDZ2 and Cx43 (K d ~ 391 μM) as compared to our data (~10 μM). This discrepancy may derive primarily from the different length of peptides used for binding studies. We determined the affinity for ZO-1 as well as ZO-2 PDZ2 in this study using the same (20aa) Cx43 peptide by ITC. The value of 391 μM was obtained by SPR using a peptide covering the last nine amino acids of Cx43. Indeed, they reported 15.5 μM Kd for ZO-1 PDZ2 when our 20aa Cx43 peptide was employed. The residues that are critical for ZO-1 PDZ2 binding to Cx43 are conserved in ZO-2 PDZ2 except for Arg367 in ZO-2 PDZ2 and Lys246 in ZO-1 PDZ2. Further, substitution of Lys246 in ZO-1 PDZ2 with an arginine residue only moderately decreases binding to Cx43 peptide (K d68 μM). Thus, our data showing that both PDZ2 domains of ZO-1 and ZO-2 have similar peptide affinity are not inconsistent with any reported data.
What are the functional consequences of this PtdInsP binding of ZO PDZ2 domains? Both endogenous ZO-1 and ZO-2 proteins are found in the plasma membrane although their overall subcellular localisation patterns differ significantly. It has been well documented that PtdIns(4,5)P 2 is the most abundant PtdInsP that is located mainly in the inner plasma membrane [44]. Unlike PtdIns(4,5)P 2, PtdIns(3,4)P 2 and PtdIns(3,4,5)P 2 are signalling lipids that are formed transiently in response to specific stimuli. Thus, their cellular concentrations are much lower (i.e. >100-fold) than that of PtdIns(4,5)P 2. This, in conjunction with the fact that ZO PDZ2 domains have much lower affinity for PtdIns(3,4)P 2 and PtdIns(3,4,5)P 2 than the specific effector domains of these lipids (e.g. PH domains), suggest that PtdIns(3,4)P 2 and PtdIns(3,4,5)P 2 binding of ZO PDZ domains may not have significant functional significance [6, 7]. Although PtdIns(4,5)P 2 is present at higher concentration than PtdIns(3,4)P 2 and PtdIns(3,4,5)P 2, there are many plasma membrane-targeting domains that have higher PtdIns(4,5)P 2 affinity than ZO PDZ2 domains and thus they may not be able to compete with other proteins for the plasma membrane pool of PtdIns(4,5)P 2. Consistent with this notion, lipid binding of ZO PDZ2 domains do not seem to play a significant role in the plasma membrane localisation of ZO proteins. This is consistent with the previous observation that the PDZ domains of ZO-1 (PDZ1-3) were not localised at the plasma membrane [45], in contrast with Par-3 for which the lipid binding activity of the PDZ domain is important for its plasma membrane localisation. Thus, the plasma membrane binding of ZO-1 and ZO-2 is more likely to be driven by protein–protein interactions [46]. As such this does not preclude a role for PtdIns(4,5)P 2 in modulating signalling complexes involving these tight junction proteins at the plasma membrane. For instance, it has been proposed that ZO-1 assembles PLCβ3 and connexin43 into a signalling complex through interaction of the third PDZ domain of ZO-1 with PLCβ3 [47] while the second PDZ domain binds to connexin43. As PtdIns(4,5)P 2 hydrolysis upon receptor activation leads to inhibition of connexin43 cell–cell based communication, these interactions allow regulation of gap junction closure by localised fluctuations in PtdIns(4,5)P 2 levels. In that study, the authors found no evidence for direct binding of PtdIns(4,5)P 2 to connexin43 but hypothesised that PtdIns(4,5)P 2 regulates junctional communication indirectly through a connexin43-associated protein. We speculate that ZO-1 itself may represent such a rheostat protein: PtdIns(4,5)P 2 interaction with the ZO-1 PDZ2 domain could modify the connexin43 regulatory tail, as PtdIns(4,5)P 2 and connexin binding are mutually exclusive, thereby shutting off gap channel function.
However, our results suggest that the PtdIns(4,5)P 2 binding activity of ZO PDZ2 domains may be important for driving subnuclear localisation of ZO proteins, ZO-2 in particular. The presence of an independent nuclear PtdInsP pool and the enzymes responsible for their metabolism is now well established [5]. Recent genetic and biochemical evidence suggests that these lipids are involved in post-transcriptional modification and chromatin-mediated gene regulation. In particular, it has been suggested that nuclear PtdIns(4,5)P 2, which is enriched in speckles and in the nucleoli [44], might be involved in the regulation of chromatin remodelling and gene transcription. Moreover, nuclear PtdIns(4,5)P 2 seems to regulate pre-mRNA splicing and controls cell growth and proliferation. Our results indicate that ZO-2 PDZ2 domain can effectively interact with these nuclear pools of PtdIns(4,5)P 2. ZO-2 is colocalised with nuclear PtdIns(4,5)P 2. Although the exact role of nuclear ZO-2 is still unknown, accumulating evidence indicates that it regulates cell growth, proliferation and apoptosis [22]. For example, Tapia et al. [48] recently reported that ZO-2 translocates to the nucleus at late G1 phase to downregulate cyclin D1 expression, and exits the nucleus during mitosis. We speculate that the transcription regulation activity of nuclear ZO-2 may be controlled by PtdInsP(4,5)P 2. Our gene knock down experiments also show that ZO-2 is involved in organising PtdIns(4,5)P 2 in nuclear speckles, as reported for syntenin-2 [38].
It has been reported that many PDZ domain-containing proteins serve as scaffolds to organise signalling complexes in the plasma membrane [49]. Our current study on the ZO PDZ2 domains and previous studies on syntenin-2 suggest that these proteins may play the same role in the nucleus and that lipid binding is critical for this role. Although the exact mechanism underlying this potentially important process and its functional consequences await further investigation, our results provide new insight into how cellular functions of PDZ domain-containing proteins can be switched by local environments that modulate the lipid binding of the PDZ domain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1. ELISA PtdInsP binding assay with PDZ domains 1-3 of ZO-2. Each well of a ‘PIP specificity’ microtiter plate (Echelon Biosciences) was overlaid with GST, GST- PH-PLC-d1 (positive control) and GST-ZO2 PDZ1-3 at a final concentration of 10 nM. Data represent means ± STDEV (n = 3). A.U. = absorbance unit (PDF 39 kb)
Supplementary Figure S2. Gel filtration chromatography of GST-ZO1-PDZ2 (red line). Molecular weight standards (blue line) include blue dextran, immunoglobin G (IgG; 150 kDa), bovine serum albumin (BSA; 67 kDa) and ovalbumin (43 kDa). 100 nM to 5 µM GST-ZO1-PDZ2 (and GST-ZO2-PDZ2) showed the same elution pattern. The estimated molecular weight of the protein was 78 kDa, which approximates that of the GST-ZO1-PDZ2 dimer (PDF 42 kb)
Supplementary material 3Supplementary Figure S3. A) PtdInsP selectivity of ZO-2 PDZ2 determined by kinetic SPR measurements. 1 µM of protein was added to POPC/POPE/POPS/PtdInsP (37:40:20:3) vesicles containing 7 different PtdInsPs. (B) Effects of mutations of basic residues on binding of the ZO-2 PDZ2 to POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles measured by kinetic SPR analysis. Protein concentrations were kept at 1 µM. Kd values were determined by equilibrium SPR analysis as shown in Figure 2B and 2C and listed in Table II. (PDF 79 kb)
Supplementary Figure S4. Expression of wild-type and mutant ZO constructs in MDCK cells. (A) Expression of various GFP-tagged ZO-1 mutants defective in lipid and/or peptide binding does not affect plasma membrane localisation of ZO-1. (B) Similar experiments for ZO-2. WT=wild type. Scale bar = 10µm. (PDF 641 kb)
Acknowledgments
We thank Dr. E. Mortier for helpful discussions during the course of this project and Dr. L. Van Troys and Dr. G. Hammond for useful advice on the lipid stainings. This work was supported by the Fund for Scientific Research-Flanders (FWO-Vlaanderen), the Concerted Actions Programme of Ghent University (GOA), the Interuniversity attraction poles (IUAP06), the Human Frontier Science Program (HFSP), a NIH grant (GM68849) (for W.C.), and the Catalyst Award from Chicago Biomedical Consortium (for W.C. and H.L.). K.M. was supported by a Postdoctoral Fellowship of the Fund for Scientific Research-Flanders (Belgium) (FWO-Vlaanderen). E.R. is supported by a fellowship from the research council of Ghent University (BOF).
Footnotes
K. Meerschaert, M. P. Tun and E. Remue contributed equally to this paper.
References
- 1.Cho W (2006) Building signaling complexes at the membrane. Sci STKE, e7 [DOI] [PubMed]
- 2.Overduin M, Cheever ML, Kutateladze TG. Signaling with phosphoinositides: better than binary. Mol Interv. 2001;1:150–159. [PubMed] [Google Scholar]
- 3.Hammond GR, Schiavo G. Polyphosphoinositol lipids: under-PPInning synaptic function in health and disease. Dev Neurobiol. 2007;67:1232–1247. doi: 10.1002/dneu.20509. [DOI] [PubMed] [Google Scholar]
- 4.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 5.Hammond G, Thomas CL, Schiavo G. Nuclear phosphoinositides and their functions. Curr Top Microbiol Immunol. 2004;282:177–206. doi: 10.1007/978-3-642-18805-3_7. [DOI] [PubMed] [Google Scholar]
- 6.Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M. Phosphoinositides: key players in cell signalling, in time and space. Cell Signal. 2001;13:377–387. doi: 10.1016/S0898-6568(01)00158-9. [DOI] [PubMed] [Google Scholar]
- 7.Toker A. Phosphoinositides and signal transduction. Cell Mol Life Sci. 2002;59:761–779. doi: 10.1007/s00018-002-8465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gassama-Diagne A, Yu W, ter Beest M, Martin-Belmonte F, Kierbel A, Engel J, Mostov K. Phosphatidylinositol-3, 4, 5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat Cell Biol. 2006;8:963–970. doi: 10.1038/ncb1461. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Belmonte F, Mostov K. Regulation of cell polarity during epithelial morphogenesis. Curr Opin Cell Biol. 2008;20:227–234. doi: 10.1016/j.ceb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Matter K, Balda MS. Epithelial tight junctions, gene expression and nucleo-junctional interplay. J Cell Sci. 2007;120:1505–1511. doi: 10.1242/jcs.005975. [DOI] [PubMed] [Google Scholar]
- 11.Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol. 2006;248:261–298. doi: 10.1016/S0074-7696(06)48005-0. [DOI] [PubMed] [Google Scholar]
- 12.Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 13.Ebnet K. Organization of multiprotein complexes at cell–cell junctions. Histochem Cell Biol. 2008;130:1–20. doi: 10.1007/s00418-008-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islas S, Vega J, Ponce L, Gonzalez-Mariscal L. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp Cell Res. 2002;274:138–148. doi: 10.1006/excr.2001.5457. [DOI] [PubMed] [Google Scholar]
- 15.Traweger A, Fuchs R, Krizbai IA, Weiger TM, Bauer HC, Bauer H. The tight junction protein ZO-2 localizes to the nucleus and interacts with the heterogeneous nuclear ribonucleoprotein scaffold attachment factor-B. J Biol Chem. 2003;278:2692–2700. doi: 10.1074/jbc.M206821200. [DOI] [PubMed] [Google Scholar]
- 16.Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell–cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Mariscal L, Ponce A, Alarcon L, Jaramillo BE. The tight junction protein ZO-2 has several functional nuclear export signals. Exp Cell Res. 2006;312:3323–3335. doi: 10.1016/j.yexcr.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Jaramillo BE, Ponce A, Moreno J, Betanzos A, Huerta M, Lopez-Bayghen E, Gonzalez-Mariscal L. Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp Cell Res. 2004;297:247–258. doi: 10.1016/j.yexcr.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Betanzos A, Huerta M, Lopez-Bayghen E, Azuara E, Amerena J, Gonzalez-Mariscal L. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp Cell Res. 2004;292:51–66. doi: 10.1016/j.yexcr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Kavanagh E, Buchert M, Tsapara A, Choquet A, Balda MS, Hollande F, Matter K. Functional interaction between the ZO-1-interacting transcription factor ZONAB/DbpA and the RNA processing factor symplekin. J Cell Sci. 2006;119:5098–5105. doi: 10.1242/jcs.03297. [DOI] [PubMed] [Google Scholar]
- 21.Huang HY, Li R, Sun Q, Wang J, Zhou P, Han H, Zhang WH. LIM protein KyoT2 interacts with human tight junction protein ZO-2-i3. Yi Chuan Xue Bao. 2002;29:953–958. [PubMed] [Google Scholar]
- 22.Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, Gonzalez-Mariscal L, Lopez-Bayghen E. Cyclin D1 is transcriptionally down-regulated by ZO-2 via an E box and the transcription factor c-Myc. Mol Biol Cell. 2007;18:4826–4836. doi: 10.1091/mbc.E07-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Feng W, Chen J, Chan LN, Huang S, Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell. 2002;9:1215–1225. doi: 10.1016/S1097-2765(02)00549-X. [DOI] [PubMed] [Google Scholar]
- 25.Kates M. Techniques of lipidology. 2. Amsterdam: Elsevier; 1986. [Google Scholar]
- 26.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Hammond GR, Dove SK, Nicol A, Pinxteren JA, Zicha D, Schiavo G. Elimination of plasma membrane phosphatidylinositol (4, 5)-bisphosphate is required for exocytosis from mast cells. J Cell Sci. 2006;119:2084–2094. doi: 10.1242/jcs.02912. [DOI] [PubMed] [Google Scholar]
- 28.Hammond GR, Schiavo G, Irvine RF (2009) Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P2. Biochem J (in press) [DOI] [PMC free article] [PubMed]
- 29.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3, 4-bisphosphate- and phosphatidylinositol 3, 4, 5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 31.Rusten TE, Stenmark H. Analyzing phosphoinositides and their interacting proteins. Nat Methods. 2006;3:251–258. doi: 10.1038/nmeth867. [DOI] [PubMed] [Google Scholar]
- 32.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fanning AS, Lye MF, Anderson JM, Lavie A. Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J Biol Chem. 2007;282:37710–37716. doi: 10.1074/jbc.M707255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giepmans BN, Verlaan I, Moolenaar WH. Connexin-43 interactions with ZO-1 and alpha- and beta-tubulin. Cell Commun Adhes. 2001;8:219–223. doi: 10.3109/15419060109080727. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Pan L, Wei Z, Zhao Y, Zhang M. Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J. 2008;27:2113–2123. doi: 10.1038/emboj.2008.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flores CE, Li X, Bennett MV, Nagy JI, Pereda AE. Interaction between connexin35 and zonula occludens-1 and its potential role in the regulation of electrical synapses. Proc Natl Acad Sci USA. 2008;105:12545–12550. doi: 10.1073/pnas.0804793105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nourry C, Grant SG, Borg JP (2003) PDZ domain proteins: plug and play! Sci STKE, RE7 [DOI] [PubMed]
- 38.Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 2005;24:2556–2565. doi: 10.1038/sj.emboj.7600722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4, 5)P 2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci. 2001;114:2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 40.Hernandez S, Chavez MB, Gonzalez-Mariscal L. ZO-2 silencing in epithelial cells perturbs the gate and fence function of tight junctions and leads to an atypical monolayer architecture. Exp Cell Res. 2007;313:1533–1547. doi: 10.1016/j.yexcr.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kachel N, Erdmann KS, Kremer W, Wolff P, Gronwald W, Heumann R, Kalbitzer HR. Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. J Mol Biol. 2003;334:143–155. doi: 10.1016/j.jmb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 43.Cho W, Stahelin RV. Membrane–protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 44.Bunce MW, Bergendahl K, Anderson RA. Nuclear PI(4, 5)P(2): a new place for an old signal. Biochim Biophys Acta. 2006;1761:560–569. doi: 10.1016/j.bbalip.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Reichert M, Muller T, Hunziker W. The PDZ domains of zonula occludens-1 induce an epithelial to mesenchymal transition of Madin–Darby canine kidney I cells. Evidence for a role of beta-catenin/Tcf/Lef signaling. J Biol Chem. 2000;275:9492–9500. doi: 10.1074/jbc.275.13.9492. [DOI] [PubMed] [Google Scholar]
- 46.Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM. The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol Biol Cell. 2007;18:721–731. doi: 10.1091/mbc.E06-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Zeijl L, Ponsioen B, Giepmans BN, Ariaens A, Postma FR, Varnai P, Balla T, Divecha N, Jalink K, Moolenaar WH. Regulation of connexin43 gap junctional communication by phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2007;177:881–891. doi: 10.1083/jcb.200610144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tapia R, Huerta M, Islas S, vila-Flores A, Lopez-Bayghen E, Weiske J, Huber O, Gonzalez-Mariscal L. Zona occludens-2 inhibits cyclin D1 expression and cell proliferation and exhibits changes in localization along the cell cycle. Mol Biol Cell. 2009;20:1102–1117. doi: 10.1091/mbc.E08-03-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. ELISA PtdInsP binding assay with PDZ domains 1-3 of ZO-2. Each well of a ‘PIP specificity’ microtiter plate (Echelon Biosciences) was overlaid with GST, GST- PH-PLC-d1 (positive control) and GST-ZO2 PDZ1-3 at a final concentration of 10 nM. Data represent means ± STDEV (n = 3). A.U. = absorbance unit (PDF 39 kb)
Supplementary Figure S2. Gel filtration chromatography of GST-ZO1-PDZ2 (red line). Molecular weight standards (blue line) include blue dextran, immunoglobin G (IgG; 150 kDa), bovine serum albumin (BSA; 67 kDa) and ovalbumin (43 kDa). 100 nM to 5 µM GST-ZO1-PDZ2 (and GST-ZO2-PDZ2) showed the same elution pattern. The estimated molecular weight of the protein was 78 kDa, which approximates that of the GST-ZO1-PDZ2 dimer (PDF 42 kb)
Supplementary material 3Supplementary Figure S3. A) PtdInsP selectivity of ZO-2 PDZ2 determined by kinetic SPR measurements. 1 µM of protein was added to POPC/POPE/POPS/PtdInsP (37:40:20:3) vesicles containing 7 different PtdInsPs. (B) Effects of mutations of basic residues on binding of the ZO-2 PDZ2 to POPC/POPE/POPS/PtdIns(4,5)P 2 (37:40:20:3) vesicles measured by kinetic SPR analysis. Protein concentrations were kept at 1 µM. Kd values were determined by equilibrium SPR analysis as shown in Figure 2B and 2C and listed in Table II. (PDF 79 kb)
Supplementary Figure S4. Expression of wild-type and mutant ZO constructs in MDCK cells. (A) Expression of various GFP-tagged ZO-1 mutants defective in lipid and/or peptide binding does not affect plasma membrane localisation of ZO-1. (B) Similar experiments for ZO-2. WT=wild type. Scale bar = 10µm. (PDF 641 kb)