Abstract
High mobility group protein 1 (HMGB1) is a non-histone nuclear protein that can activate innate immunity when in an extracellular location. As shown in in vitro studies, while polyinosinic-polycytidylic acid [poly (I:C)] and LPS, TLR3 and TLR4 ligands respectively, can induce HMGB1 release from macrophages, CpG DNA, a TLR 9 ligand, does not. Since DNA displays distinct immunostimulatory activity when transfected into cells, we investigated whether transfected DNA can induce HMGB1 release from macrophages. In these experiments, using RAW 264.7 cells as model, we show that DNA, either natural DNA or synthetic oligonucleotides, can induce HMGB1 release when used to stimulate cells with the transfection reagent Lipofectamine 2000®; release occurred irrespective of the intrinsic activity of the DNA. The induction of HMGB1 release by transfected DNA was dependent on IFN-β as shown by the inhibitory effects of an antibody. In addition, JNK activation mediated HMGB1 release induced by a transfected phosphorothioate oligonucleotide but not by transfected natural DNA. Together, these findings indicate that transfected DNA can stimulate macrophages to release HMGB1 under conditions in which free DNA is inactive and suggest a role of DNA in inducing inflammation when bound to molecules that influence its entry into cells.
Keywords: DNA, HMGB1, transfection, inflammation
1. Introduction
HMGB1 is a non-histone nuclear protein whose function depends on location. Inside the cell, HMGB1 binds DNA to modify chromatin architecture as well as mediate transcription (Czura, Wang et al. 2001; Thomas 2001). HMGB1 is a nuclear component in most cells, although it can also appear on the cell surface, shuttle between the nucleus and cytoplasm as well as exit the cell (Bianchi and Manfredi 2004; Lotze and Tracey 2005; Muller, Ronfani et al. 2004). Once outside the cell, HMGB1 can serve as a cytokine and induce a broad array of pro-inflammatory activities (Ulloa and Messmer 2006; Wang, Bloom et al. 1999). The importance of these activities has been established in animal models of inflammatory diseases such as sepsis and rheumatoid arthritis, where blocking HMG1 activity can ameliorate disease (Kokkola, Li et al. 2003; Sunden-Cullberg, Norrby-Teglund et al. 2006; Wang, Bloom et al. 1999). As such, extracellular HMGB1 can act as an alarmin to induce innate immunity or to promote inflammatory disease pathogenesis (Harris and Raucci 2006).
As shown in in vitro studies, HMGB1 can exit the cell following cell death or the activation of Toll-like receptors (TLRs) (Bell, Jiang et al. 2006; Jiang and Pisetsky 2006; Scaffidi, Misteli et al. 2002). Both events are prominent in sepsis where HMGB1 may be a target of new therapy. Thus, stimulation of either TLR4 by LPS or TLR3 by poly (I:C) can promote the extracellular translocation of HMGB1 from the nucleus, a process measured by the appearance of HMGB1 in the cell media and loss of nuclear staining by an HMGB1 antibodies (Jiang, Li et al. 2005). This translocation involves protein acetylation and phosphorylaton which alter the charge of HMGB1 to allow entrance in secretory lysosomes in the cytoplasm eventual for secretion (Bonaldi, Talamo et al. 2003; Youn and Shin 2006). With dead cells, extracellular HMGB1 results from passive release as permeability barriers break down during necrosis (Scaffidi, Misteli, and Bianchi 2002) or late apoptosis (Bell, Jiang et al. 2006).
In contrast to stimulation of TLR3 or 4, stimulation of TLR9 by CpG DNA, either bacterial DNA or CpG DNA oligonucleotides (ODN), fails to induce significant amounts of extracellular HMGB1 (Jiang, Li, Gallowitsch-Puerta, Tracey, and Pisetsky 2005). The limited expression of extracellular HMGB1 by TLR9 stimulation is notable since, under the same conditions, CpG DNA can potently induce the production of other inflammatory mediators such as TNF-α and nitric oxide. The differences in the outcome of stimulation by the various TLRs, which could be relevant to host defense in infection, may reflect different downstream signaling events. Thus, stimulation of TLR 3 and TLR4 but not TLR9 involves the TRIF pathway (Takeda and Akira 2005). Together, these findings suggest that cell activation by only certain pathways leads to HMGB1 release.
CpG DNA, either natural DNA or synthetic ODN, can stimulate cell activation through TLR9 when incubated in a free, soluble form with cells. Recent studies suggest that TLR9 is not the sole receptor for DNA, however, and that DNA can activate the immune system by non-TLR mechanisms (Ishii, Suzuki et al. 2001; Jiang, Reich et al. 2004; Stetson and Medzhitov 2006). These mechanisms, which operate when DNA is internalized into the cytoplasm of cells via transfection agents, do not depend on CpG motifs and occur with mammalian as well as bacterial DNA. Thus, mammalian DNA, which is inactive in free form because of its paucity of CpG motifs, can stimulate responses when introduced cells by a transfection reagent; transfected bacterial DNA is also active although the pattern of response differs from that of free DNA. The activity of transfected DNA most likely reflects the interaction with non-TLR cytoplasmic nucleic acid sensors that may mediate the response to nucleic acids from intracellular organisms (Bowie and Fitzgerald 2007).
Since immune activation by free DNA may differ from that of transfected DNA, we have investigated whether transfected DNA can stimulate macrophages to release HMGB1 under conditions in which free DNA is inactive. In this study, we have therefore assessed whether DNA internalized with transfection reagents can induce macrophage release of HMGB1. In results present herein, using the RAW 264.7 murine macrophage cell line as a model, we show that transfected DNA can induce HMGB1 release by a mechanism involving IFN-βand JNK. Taken together with prior findings, these results suggest that DNA can stimulate HMGB1 release from macrophages although this activity depends on the manner in which DNA contacts cells and stimulates downstream mediators.
2. Materials and Methods
2.1 Cells and reagents
RAW 264.7 cells were purchased from ATCC (Manassas, VA) and cultured in RPMI-1640 supplemented with 10% fetal bovine serum and 200 μg/ml gentamicin. Calf thymus DNA (CT DNA) and E. coli DNA (EC DNA) was purchased from Sigma-Aldrich (St. Louis, MO). Phosphodiester (Po) or phosphorothioate (Ps) oligonucleotides (ODN) were purchased from Midland Certified Reagents (Midland, TX). Compounds used in this study included a Ps CpG ODN designated 1826 (5′-TCCATGACGTTCC TGACGTT-3′); a control Ps GpC ODN (5′-TCCATGAGCTTCCTGAGCTT-3′) designated as control; a Po ODN with the same sequence as the control Ps ODN designated Po1; and a Po ODN with the same sequence as 1826 designated Po2. In addition, there were two Po ODNs with two consecutive sequences of 1826 (designated LO2) or two consecutive sequences of the control ODN (designated LO1). These ODNs were 45 bases long and included additional 5 bases at the end of the consecutive sequences. Lipofectamine® 2000 (lipofectamine) was purchase from Invitrogen (San Diego, CA) and FuGENE 6 was purchased from Roche Applied Biosciences (Indianapolis, IN). JNK inhibitor and 1400W were purchased from Alexis Corp (San Diego, CA). Recombinant IFN-β (rIFN-β), rat anti-IFN-β monoclonal antibody and IFN-β ELISA kit were purchased from R&D Systems (Minneapolis, MN).
2.2 Cell culture
RAW 264.7 cells were plated in 6-well culture plates for 2-3 hours and washed twice with Opti-MEM (Invitrogen). Cells were then stimulated with DNA or DNA-lipofectamine complexes for 20 to 24 hours and supernatants were collected for IFN-β and HMGB1 assay. DNA-liposome complexes were formed by pre-incubation of DNA and lipofectamine in Opti-MEM for 30 minutes at room temperature. For inhibition studies, anti-IFN-β antibody or a cell permeable JNK inhibitor were pre-incubated with cells for 30 minutes and cells were subsequently stimulated with DNA-lipofectamine complex for 20 hours.
2.3 Biochemical and immunochemical assays
The levels of IFN-β in supernatants were measured by ELISA and assays were performed in accordance with the manufacturer’s instructions (R&D Systems).
For Western blotting of HMGB1, supernatants were collected after 20 to 24 hours of culture and concentrated by Centricon YM-10 (Millipore, Billerica, MA). The volume of the concentrated supernatants was adjusted to 70 μl for equal loading and samples were resolved on 4-12% NuPAGE® Tris-Bis SDS polyacrylamide gel (Invitrogen). Protein was transferred to PVDF membranes (Invitrogen), blocked with 5% dry milk in TBS-Tween and blotted with either a rabbit anti-HMGB1 polyclonal antibody (BD Pharmingen, San Diego) or a mouse monoclonal anti-HMGB1 antibody (R&D Systems). The membrane was then incubated with HRP-conjugated anti-rabbit IgG followed by Super Signal® West Femto substrate (Pierce, Rockford, IL). Images were captured by exposing the membrane to a CCD camera (FluorChem8900, Alpha Innotech, San Leandro, CA).
3. Results
3.1 The induction of HMGB1 release by transfected DNA
In previous studies, we showed that CpG DNA, either bacterial DNA or a synthetic ODN, fails to induce HMGB1 release from murine macrophages despite the induction of other mediators (Jiang, Li et al. 2005). Since stimulation by DNA may vary depending upon the pathway of internalization, we have therefore investigated whether DNA, when complexed with a trasnfection reagent, can induce HMGB1 release. For this purpose, we have used the murine macrophage cell line RAW 264.7 cells a model since these cells respond well to TLR9 stimulation and release HMGB1 when stimulated with LPS or poly (I:C). In these experiments, CT DNA and EC DNA were used as natural DNA while an active and inactive form of phosphorothioate compounds were used as models for synthetic oligonucleotides. Supernatant HMGB1 level was measured by Western blotting.
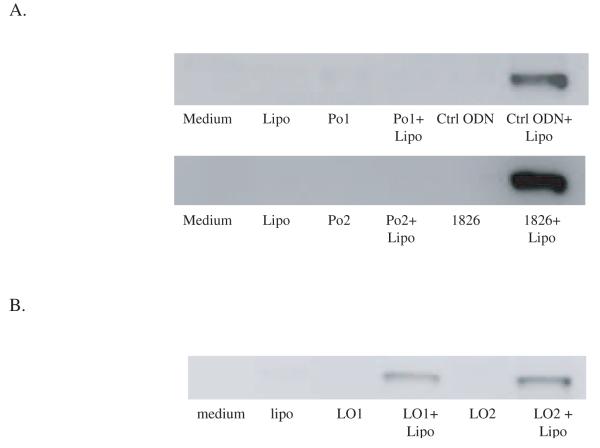
As data presented in Figure 1A show, both natural and synthetic DNA, regardless of intrinsic activity, induced the release of HMGB1 from RAW 264.7 cells when complexed with lipofectamine. For EC DNA, a limited amount of HMGB1 was observed with free DNA although the amount released was substantially greater when the DNA was transfected. Similarly, these DNA, when complexed with FuGENE 6, a nonliposome transfection reagent, also induced HMGB1 release from RAW 264.7 cells (data not shown). The induction of HMGB1 release by CT DNA was dose dependent as increasing amounts of CT DNA when mixed with a constant amount of lipofectamine produced greater amounts of HMGB1 in the media (Figure 1B). These results are consistent with previous findings and indicate the marked change in activity of DNA when it is transfected into cells.
Figure 1.
The effects of transfected DNA on the induction of HMGB1 release from RAW 264.7 cells. 10 μg E. coli DNA (EC DNA), calf thymus DNA (CT DNA), control ODN (ctrl ODN) or 1826 ODN were incubated with RAW 264.7 cells for 24 hours with or without 3 μl lipofectamine (A). CT DNA, 0, 1, 5 and 10 μg, was complexed with 3 μl lipofectamine and incubated with RAW 264.7 cells for 24 hours. Supernatants were collected and HMGB1 was detected by Western blotting.
In these experiments, we have used natural DNA which has high molecular weight. Thus, the comparison between natural and synthetic DNA involves differences of both backbone structure as well as length. To determine the properties of a synthetic Po ODN, we therefore investigated the response of two 20-base Po ODNs with the same sequence as the Ps ODNs tested before. These compounds, like the corresponding Ps ODNs, also failed to stimulate HMGB1 when incubated with cells in a free form. They differed from the Ps ODNs, however, since they did not acquire activity when transfected with lipofectamine (Figure 2A). We therefore tested the activity of a longer version of the Po ODN which has either two copies of the 1826 sequence or two copies of the control ODN sequences; these compounds were 45 bases long and had 5 additional bases at the end of the consecutive sequences. In contrast to the short Po ODNs, the 45 base compounds induced HMGB1 release when transfected into RAW 264.7 cells (Figure 2B). These results indicate that Po and Ps ODN may differ in the structural requirement for stimulation when transfected into cells.
Figure 2.
The influence of ODN length on the release of HMGB1 in cells stimulation with ODN-lipofectamine complexes. 10 μg Po ODNs with same sequences as control ODNs (Po1) or 1826 (Po2) were incubated with RAW 264.7 cells with or without 3 μl lipofectamine for 24 hours (A). 10 μg phosphodiester ODN (45 bases) with either two GpC control ODN sequence (LO1) or two 1826 sequence (LO2) were incubated with RAW 264.7 cells with or without 3 μl lipofectamine for 24 hours (B). Supernatant HMGB1 levels were measured by Western blotting.
3.2 The role of IFN-β in HMGB1 release induced by transfected DNA
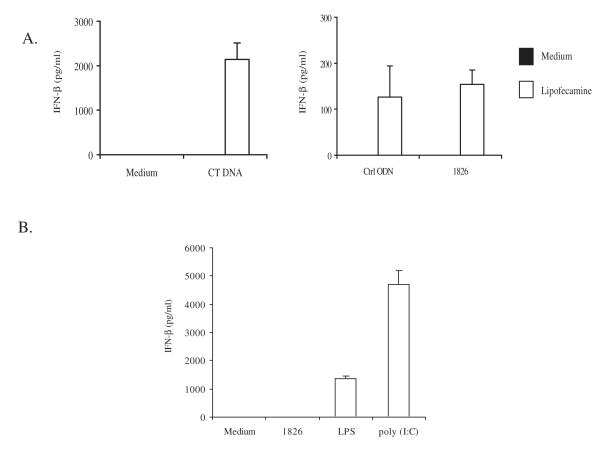
As shown by other investigators, DNA transfected into cells can induce type I IFN expression in an IRF3-dependent manner (Stetson and Medzhitov 2006). Since IFN-α can mediate HMGB1 release induced by TLR3 stimulation (Jiang and Pisetsky 2006), we therefore studied the role of IFN-β, another type I IFN, in HMGB1 release induced by transfected DNA. As shown in Figure 3A, transfection of DNA with lipofectamine induced the production of IFN-β in RAW 264.7 cells, with CT DNA showing the highest IFN-β levels. In addition, stimulation of TLR3 and TLR4 also induced IFN-β production and HMGB1 release by RAW 264.7 cells Figure 3B.
Figure 3.
The effects of transfected DNA on the induction of IFN-β production. CT DNA (10 μg), control ODN (10 μg) and 1826 ODN (10 μg) were complexed with 3 μl lipofectamine. The DNA-lipid complexes were incubated with RAW 264.7 cells for 24 hours and supernatant IFN-β levels were measured by ELISA (A). RAW 264.7 cells were stimulated with 1826 ODN (10 μg/ml), LPS (0.5 μg/ml) or poly (I:C) (0.25 μg/ml) for 20 hours and supernatant IFN-β levels were measured by ELISA (B). Each bar represents three separate experiments. Error bar depicts standard deviation.
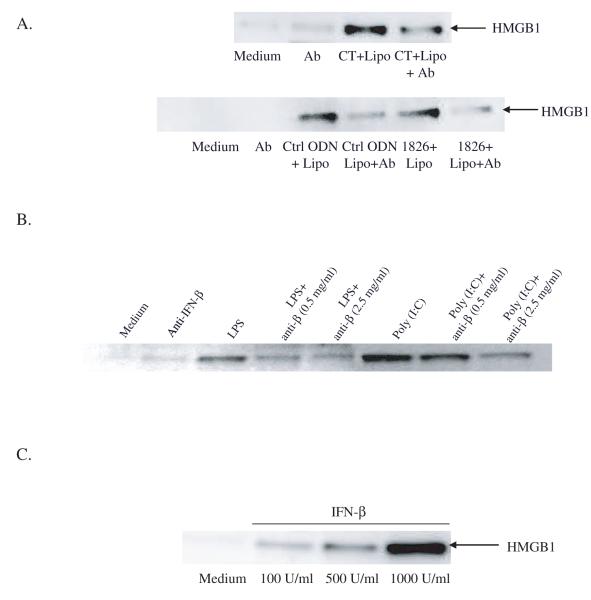
To evaluate the role of IFN-β in HMGB1 release by transfected DNA, the effects of blockade of IFN-β by an anti-IFN-β monoclonal antibody (mAb) were tested. As shown in Figure 4A, the addition of anti-IFN-β mAb to cultures of RAW 264.7 cells inhibited HMGB1 release induced by transfected DNA. Neutralizing IFN-β with a mAb also reduced HMGB1 release induced by poly (I:C) or LPS (Figure 4B), indicating a role of this cytokine in the actions of other TLRs
Figure 4.
The effect of IFN-β mediated HMGB1 release induced by transfected DNA. CT DNA, control ODN or 1826 ODN complexed with lipofectamine was incubated with RAW 264.7 cells for 24 hours in the presence of a mouse monoclonal anti-IFN-β neutralizing antibody (Ab) (A). LPS or poly (I:C) was incubated with RAW 264.7 cells for 20 hours in the presence of anti-IFN-β antibody (B). RAW 264.7 cells were also incubated with recombinant IFN-β 0, 100, 500 and 1000 U/ml for 24 hours (C). Supernatant HMGB1 levels were determined by Western blotting.
To evaluate further the effect of IFN-β in HMGB1 release, we treated RAW 264.7 cells with recombinant IFN-β and measured HMGB1 release in culture supernatants by Western blotting. As shown in Figure 4C, the addition of rIFN-β dose dependently induced HMGB1 release by RAW 264.7 cells. These results indicate a direct role of IFN-β in HMGB1 release.
3.3 The effects of JNK activation on HMGB1 release induced by transfected DNA
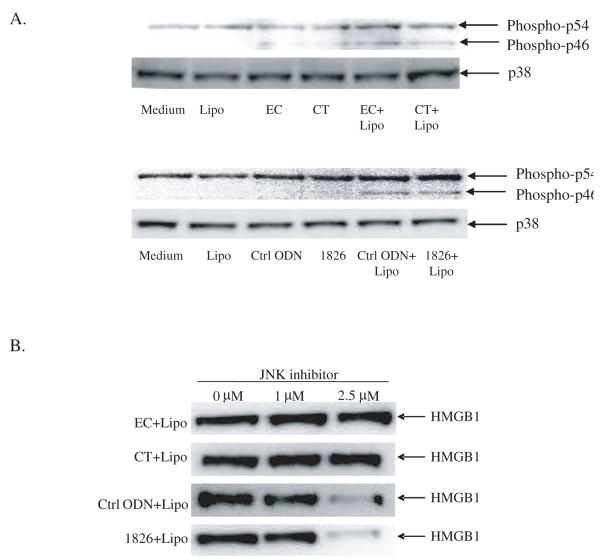
We have previously shown that transfected mammalian DNA can activate MAPKs although free DNA does not (Jiang, Reich and Pisetsky 2004). In addition, we have previously reported that JNK activation is required for HMGB1 release induced by LPS or poly (I:C). To evaluate the role of JNK in HMGB1 release induced by transfected DNA, we used a JNK inhibitor to determine whether it could block the release process. As shown in Figure 5A, with transfected DNA, JNK activation was observed with EC and CT DNA as well as the inactive and active ODN; these levels far exceeds those observed with free DNA. The effects of the JNK inhibitor differed, however, with the natural and synthetic DNA (Figure 5B). Whereas the JNK inhibitor dose dependently inhibited HMGB1 release induced by transfected CpG ODN or GpC ODN, it did not affect HMGB1 release by RAW 264.7 cell cultures transfected with CT DNA or EC DNA. These results indicate that transfected DNA can activate JNK under conditions in which free DNA is inactive, and that JNK inhibition can block HMGB1 release depending on the structure of the stimulating DNA.
Figure 5.
The effects of JNK on HMGB1 release induced by transfected DNA. DNA-liposome complexes were incubated with RAW 264.7 cells for 24 hours, washed and fresh media was added and incubated for 2 hours. Cells were then stimulated with the same DNA or DNA-liposome complex for 30 minutes and cellular protein was isolated (A). Cellular p38 levels were used as protein loading control. RAW 264.7 cells were stimulated with DNA-liposome complexes in the presence of JNK inhibitor 0, 1, 2.5 μM for 24 hours (B). Supernatant HMGB1 levels were measure by Western blotting.
4. Discussion
Results of this study provide new insights into the mechanisms of HMGB1 release and show that DNA, while unable to stimulate HMGB1 when incubated directly with RAW 264.7 cells, can nevertheless stimulate this release when transfected into cells. This release depends on the action of IFN-β which, like IFN-α, can also induce HMGB1 release. The transformation of the immune activity of DNA by transfection is consistent with previous results indicating that mammalian DNA acquires immune activity in the context of transfection reagents (Shirota, Ishii et al. 2006; Zhu, Reich et al. 2003). Together, these results suggest that transfection allows access of DNA to other, non-TLR receptors, which do not require the presence of CpG motifs.
The entry of DNA into cells and the trafficking among compartment are poorly understood, although synthetic ODN are increasingly used as immunomodulatory agents and are currently under trial for a number of clinical indications. Unlike other TLR ligands, both natural and synthetic DNA must enter cells for activity. Since TLR9 is functional in a lysosomal compartment, free DNA, when incubated with macrophages, accesses the cytoplasmic space for uptake into lysosomes where activation by TLR9 occurs (Latz, Schoenemeyer et al. 2004). Ultimately, activation depends on the content of immunostimulatory CpG motifs since even high concentrations of mammalian DNA or an non-CpG ODN fail to activate cells (Pisetsky and Reich 2000).
In contrast to the situation with free DNA, transfected DNA displays distinct structure-function relationships for stimulation, with both CpG and non-CpG DNA similarly active. Furthermore, as shown in previous studies as well as results presented herein, transfected DNA can activate certain signaling pathways (e.g., JNK) much more effectively than free DNA (Jiang, Reich and Pisetsky 2004). Transfected DNA also leads to the presence of DNA in the cytoplasm although it appears to differ from that free DNA since it leads to macrophage activation even in the absence of immunostimulatory motifs.
The activation of macrophages by transfected DNA may reflect interaction with non-TLR internal receptors that are usually inaccessible to free DNA. As shown in recent studies, cytoplasmic nucleic acids can activate a number of receptor systems which may be important in triggering innate immunity (Meylan, Tschopp et al. 2006). Such cytoplasmic nucleic acid, which may be mimicked by transfected DNA, may result from either viral or bacterial infection as well as entrance of DNA into cells in the form of immune complexes as may occur in SLE. Thus, cytoplasmic RNA can activate cells through a retinoic acid-inducible gene I/melanoma differentiation-associated gene 5 (RIG-I/MDA5) signaling pathway (Andrejeva, Childs et al. 2004; Yoneyama, Kikuchi et al. 2004). This pathway involves signaling through the adapter caspase recruitment domain adapter inducing interferon-beta (Cardif) followed by the activation of NF-κB and IRFs through the recruitment of IKK and TBK1 (Meylan, Tschopp, and Karin 2006). In addition, cytoplasmic DNA can activate immune cells to produce IFN-β through an IRF3-dependent pathway, however, the cytoplasmic receptor for this pathway is yet to be identified (Stetson and Medzhitov 2006).
Our study extends these findings by showing that transfected DNA can also induce macrophages to release HMGB1, an endogenous molecule with alarmin activity. Similar to activation by LPS or poly (I:C), this release appears to be dependent on IFN-β, a cytokine which can be induced by transfected but not free DNA. The role of IFN-β was established by showing that neutralizing IFN-β by an antibody can inhibit HMGB1 release induced by transfected DNA. It is of interest, in this regard, that a previous study showed that transfected Po ODNs can induce IFN-β production if the length of these ODNs is greater than 25 bases (Stetson and Medzhitov 2006). In our studies, Po ODNs with the same sequence and length (20 bases) as the Ps ODNs did not induce HMGB1 release even when transfected. With the length of these Po ODNs increased to 45 bases, these compounds also induced HMGB1 release when complexed with lipofectamine. These findings point to a difference in the size requirement for stimulation by transfected DNA.
In a previous study, we showed that the HMGB1 release induced by LPS and poly (I:C) depends on JNK activation (Jiang and Pisetsky 2006). JNK activation may be important to this release process since JNK activation can lead to the activation of AP-1 (Derijard, Hibi et al. 1994) and AP-1 mediates IFN-β transcription (Maniatis, Falvo et al. 1998). In our studies, while we found that transfected DNA activates JNK, the role of JNK in HMGB1 release differ depending on DNA backbone structure. As such, HMGB1 release induced by transfected Ps ODNs appeared dependent on JNK. On the other hand, inhibition of JNK did not prevent HMGB1 release induced by a natural DNA. These findings are consistent with a large body of data indicating that Po and Ps DNA may utilize different signaling transduction pathways despite a common interaction with TLR9.
While these studies involve transfected DNA, the mechanism we postulate may nevertheless occur during infection or inflammatory disease. During infection, internal nucleic acids most likely arise from intracellular viruses or microorganisms. During inflammation, the internal nucleic acid may result from immune complexes or complexes of DNA with DNA binding molecules that functionally “transfect” DNA into cells. Among these molecules, HMGB1 itself has transfecting properties and can promote DNA entry into cells (Bottger, Vogel et al. 1988; Kato, Nakanishi et al. 1991). In the setting of cell death or inflammation, extracellular DNA-HMGB1 complexes may form from nuclear material extruded or released from dead and dying cells; activated cells may also release these complexes. In addition, such complexes may form because of a high concentration of extracellular DNA and HMGB1.
Whatever their origin, such complexes may allow access of DNA to internal nucleic acid receptors which lead to a cycle of cytokine production and HMGB1 release that further intensifies inflammation by expanding the array of pro-inflammatory mediators released. As shown in studies on immune complexes in SLE, HMGB1 may also affect activation because of triggering of other receptors (i.e. RAGE) as well as its to interact with TLR9 in the lysosomal compartment in way that determine stimulation by CpG DNA (Ivanov, Dragoi et al. 2007; Tian, Avalos et al. 2007). The interaction of DNA and HMGB1 in inflammation may therefore be multifaceted. Future studies are in progress to determine whether DNA-HMGB1 complexes can act similarly as transfected DNA and to identify agents that can block this distinct pathway of DNA stimulation.
Acknowledgement
This work was supported by the Lupus Research Institute, VA Medical Research Service. W.J. was supported by a NIH training grant T32 AI 007217.
Abbreviations
- Cardif
caspase recruitment domain adapter inducing interferon-beta
- CT DNA
calf thymus DNA
- EC DNA
Escherichia coli DNA
- HMGB1
high mobility group protein 1
- IFN-β
interferon beta
- IKK
IκB kinase
- IRF
interferon regulatory factor
- LPS
lipopolysaccharide
- MDA
melanoma-differentiation-associated gene
- ODN
oligonucleotide
- poly (I:C)
polyinosinic-polycytidylic acid
- Po
phosphordiester
- Ps
phosphorothioate
- RAGE
receptor for advanced glycation end products
- RIG
retinoic acid-inducible gene
- TBK
Tank-binding kinase
- TLR
Toll-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell CW, Jiang W, Reich CF, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am. J. Physiol Cell Physiol. 2006;291:C1318–C1325. doi: 10.1152/ajpcell.00616.2005. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi ME, Manfredi A. Chromatin and cell death. Biochim. Biophys. Acta. 2004;1677:181–186. doi: 10.1016/j.bbaexp.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottger M, Vogel F, Platzer M, Kiessling U, Grade K, Strauss M. Condensation of vector DNA by the chromosomal protein HMG1 results in efficient transfection. Biochim. Biophys. Acta. 1988;950:221–228. doi: 10.1016/0167-4781(88)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Bowie AG, Fitzgerald KA. RIG-I: tri-ing to discriminate between self and non-self RNA. Trends Immunol. 2007;28:147–150. doi: 10.1016/j.it.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Czura CJ, Wang H, Tracey KJ. Dual roles for HMGB1: DNA binding and cytokine. J.Endotoxin.Res. 2001;7:315–321. doi: 10.1177/09680519010070041401. [DOI] [PubMed] [Google Scholar]
- 8.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 9.Harris HE, Raucci A. Alarmin(g) news about danger: workshop on innate danger signals and HMGB1. EMBO Rep. 2006;7:774–778. doi: 10.1038/sj.embor.7400759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii KJ, Suzuki K, Coban C, Takeshita F, Itoh Y, Matoba H, Kohn LD, Klinman DM. Genomic DNA released by dying cells induces the maturation of APCs. J. Immunol. 2001;167:2602–2607. doi: 10.4049/jimmunol.167.5.2602. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov S, Dragoi AM, Wang X, Dallacosta C, Louten J, Musco G, Sitia G, Yap GS, Wan Y, Biron CA, Bianchi ME, Wang H, Chu WM. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007 doi: 10.1182/blood-2006-09-044776. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W, Pisetsky DS. The role of IFN-alpha and nitric oxide in the release of HMGB1 by RAW 264.7 cells stimulated with polyinosinic-polycytidylic acid and lipopolysaccharide. J. Immunol. 2006;177:3337–3343. doi: 10.4049/jimmunol.177.5.3337. [DOI] [PubMed] [Google Scholar]
- 13.Jiang W, Li J, Gallowitsch-Puerta M, Tracey KJ, Pisetsky DS. The effects of CpG DNA on HMGB1 release by murine macrophage cell lines. J. Leukoc. Biol. 2005;78:930–936. doi: 10.1189/jlb.0405208. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W, Reich CF, III, Pisetsky DS. Mechanisms of activation of the RAW264.7 macrophage cell line by transfected mammalian DNA. Cell Immunol. 2004;229:31–40. doi: 10.1016/j.cellimm.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Kato K, Nakanishi M, Kaneda Y, Uchida T, Okada Y. Expression of hepatitis B virus surface antigen in adult rat liver. Co-introduction of DNA and nuclear protein by a simplified liposome method. J. Biol. Chem. 1991;266:3361–3364. [PubMed] [Google Scholar]
- 16.Kokkola R, Li J, Sundberg E, Aveberger AC, Palmblad K, Yang H, Tracey KJ, Andersson U, Harris HE. Successful treatment of collagen-induced arthritis in mice and rats by targeting extracellular high mobility group box chromosomal protein 1 activity. Arthritis Rheum. 2003;48:2052–2058. doi: 10.1002/art.11161. [DOI] [PubMed] [Google Scholar]
- 17.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 18.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 20.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 21.Muller S, Ronfani L, Bianchi ME. Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J. Intern. Med. 2004;255:332–343. doi: 10.1111/j.1365-2796.2003.01296.x. [DOI] [PubMed] [Google Scholar]
- 22.Pisetsky DS, Reich CF. Inhibition of murine macrophage IL-12 production by natural and synthetic DNA. Clin. Immunol. 2000;96:198–204. doi: 10.1006/clim.2000.4897. [DOI] [PubMed] [Google Scholar]
- 23.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 24.Shirota H, Ishii KJ, Takakuwa H, Klinman DM. Contribution of interferon-beta to the immune activation induced by double-stranded DNA. Immunology. 2006;118:302–310. doi: 10.1111/j.1365-2567.2006.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Sunden-Cullberg J, Norrby-Teglund A, Treutiger CJ. The role of high mobility group box-1 protein in severe sepsis. Curr. Opin. Infect. Dis. 2006;19:231–236. doi: 10.1097/01.qco.0000224816.96986.67. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K, Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 28.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 29.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 30.Ulloa L, Messmer D. High-mobility group box 1 (HMGB1) protein: friend and foe. Cytokine Growth Factor Rev. 2006;17:189–201. doi: 10.1016/j.cytogfr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 33.Youn JH, Shin JS. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 34.Zhu FG, Reich CF, Pisetsky DS. Effect of cytofectins on the immune response of murine macrophages to mammalian DNA. Immunology. 2003;109:255–262. doi: 10.1046/j.1365-2567.2003.01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]