Abstract
During immunoglobulin class switch recombination (CSR), activation induced cytidine deaminase (AID) induces DNA double strand breaks into transcribed, repetitive DNA elements called switch sequences. The mechanism that promotes the binding of AID specifically to switch regions remains to be elucidated. We have used a proteomic screen that employs in vivo biotinylation of AID and have identified the splicing regulator polypyrimidine tract binding protein-2 (PTBP2) as an AID interactor. Short hairpin RNA-mediated knock-down of PTBP2 in B cells led to a striking reduction in binding of AID to transcribed switch regions that resulted in marked impairment of CSR. PTBP2 is thus an effector of CSR that promotes binding of AID to switch region DNA.
In developing B cells in the bone marrow, V(D)J recombination assembles the gene segments encoding the amino-terminus variable region of the immunoglobulin heavy chain (IgH) upstream of the Cμ constant region gene segment1. The VDJ-Cμ heavy chain produced from this recombined Igh locus pairs with a similarly assembled κ or λ light chain to generate an IgM antibody molecule that is expressed on mature, naïve B cells. In secondary lymphoid organs such as the spleen and lymph nodes, the mature B cell meets antigens and undergoes Igh class switch recombination (CSR), a process by which the Cμ constant region is exchanged for one of several downstream constant region CH genes (Cγ, Cε, Cα). Thus, the B cell switches from producing IgM to one expressing a secondary antibody isotype such as IgG, IgE or IgA, each having a different effector function2.
CSR occurs between 1-12 kb long repetitive G:C-rich DNA elements termed switch (S) regions that precede each CH region2. Each of the CH gene segments is an individual transcription unit in which a cytokine-inducible promoter drives transcription through an intervening I-exon, the intronic S region and the CH gene exons2. The primary transcript is spliced and polyadenylated; however, this mature germline transcript does not have any protein coding capability2. Yet, transcription plays a major mechanistic role in CSR as mutations that inhibit germline transcription also impair CSR3. It has been proposed that transcription through the S regions generates R-loop structures in which the G-rich non-template strand is looped out as single-stranded (ss) DNA, providing an ideal substrate for AID-mediated cytidine deamination3. AID deamination of cytidines to uridines within the S regions mobilizes base-excision and mismatch repair proteins to the deaminated DNA and leads to formation of DNA double-strand breaks (DSBs)4. Ligation of DSBs between two S regions by components of the general end-joining machinery completes CSR3.
During an immune response, mature B cells in secondary lymphoid organs undergo another AID-mediated DNA alteration reaction termed somatic hypermutation (SHM)5,6. In this process, AID deamination at the variable regions of the recombined heavy and light chain genes leads to the generation B cells with increased antigen-affinity7. Thus, in B cells, the variable region genes and switch region DNA comprise the two physiological targets of AID. However, AID can mutate other transcribed genes, albeit at a significantly lower rate than variable region genes8 and induce DSBs at non-Ig regions9, 10. Such activity of AID at non-Ig regions is the major underlying cause of oncogenic mutations and translocations that are hallmarks of mature B cell lymphomas10. Elucidating the mechanism by which AID is targeted to the Ig regions is thus a major outstanding question.
It has been hypothesized that the recruitment of AID to S regions relies on the ability of AID to bind to factors that in turn can bind to regions of the Igh locus11. Multiple AID interactors have been reported, including Replication Protein A (RPA)12, Mdm2 (ref. 13) and CTNNBL1 (ref. 14). However, none of these could be classified as an AID targeting factor as mutation in these proteins or the inability of AID to interact with these proteins is not known to alter AID binding to its physiological targets. In a hunt for factors that target AID to S region DNA, we carried out a proteomic screen and have identified PTBP2 as a newly identified AID interactor that influences CSR by promoting binding of AID to S region DNA.
Results
Purification of AID complex
To purify AID complexes, we employed an in vivo biotinylation system that relies on the activity of the Escherichia coli biotin ligase BirA to biotinylate any target protein with a short sequence tag (biotag) when the two are co-expressed in a cell line (Fig. 1a). The biotinylated protein can then be affinity-purified along with its interactors using streptavidin beads15. For the in vivo biotinylation of AID, we used a previously characterized16 catalytically inactive AID (referred to as DM-AID) with two point mutations (H56R,E58Q) in the deaminase domain (Fig. 1a). The catalytically inactive AID has the potential to trap interactors that would otherwise dissociate upon deamination. For isolation of AID complexes, we used the CH12 B cell line that switches in culture from IgM to IgA upon stimulation with anti-CD40, interleukin 4 (IL-4) and transforming growth factor β (TGF-β; inducing conditions termed hereafter as CIT)17. Carrying out the purification in CH12 cells increases the opportunity to trap relevant AID complexes that are in the process of mediating CSR.
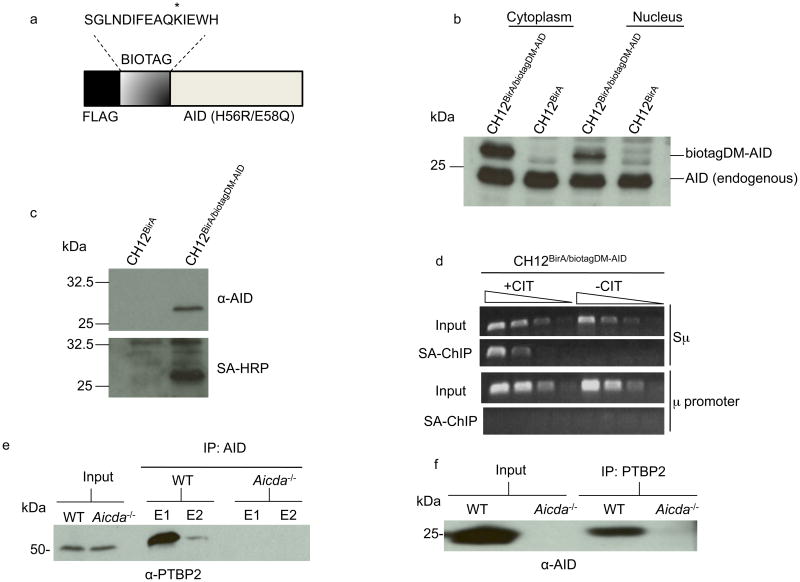
Figure 1.
AID interacts with PTBP2. (a) Schematic representation of the AID expression construct (biotagDM-AID). The lysine that is biotinylated by BirA is indicated with an asterisk. The H56R,E58Q mutation inactivates the DNA deaminase activity of AID. (b) Protein extracts derived from stimulated CH12BirA or CH12BirA/biotagDM-AID cells were analyzed on immunoblots with AID antibodies. (c) Cell extracts from CH12BirA or CH12BirA/biotagDM-AID were incubated with streptavidin-agarose beads and bound proteins analyzed by immunoblotting with AID antibodies (upper) or streptavidin-coupled to horseradish-peroxidase (SA-HRP, lower). (d) DM-AID binds to Sμ. Cross-linked DNA protein complexes from unstimulated or CIT-stimulated CH12BirA/biotagDM-AID cells were subjected to modified ChIP in which steptavidin-agarose replaced antibodies used in conventional ChIP. Three-fold dilutions of DNA bound to streptavidin agarose were analyzed by PCR for the presence of Sμ or the μ promoter. (e-f) Whole cell extracts derived from anti-CD40+IL-4-stimulated wild-type or AID-deficient mouse splenic B cells were immunoprecipitated with AID (e) or PTBP2 (f) antibodies and the immunoprecipitates were probed with anti-PTBP2 or anti-AID, respectively on immunoblots. E1 and E2 are two elutions of bound proteins. The data are representative of two independent experiments.
Cells co-expressing BirA and biotagDM-AID (designated CH12BirA/biotagDM-AID) were subcloned by serial dilution and characterized. CH12 cells that expressed BirA alone (CH12BirA) were used as negative controls. Immunoblot analysis of cytoplasmic and nuclear extracts derived from CIT-stimulated CH12BirA/biotagDM-AID and CH12BirA cells showed that the tagged AID protein, which could be distinguished from the endogenous protein due to its slower mobility on SDS gels, is not grossly over-expressed relative to endogenous AID (Fig. 1b). The tagged protein was detected in both the cytoplasm and the nucleus, with the nuclear to cytoplasmic ratio roughly similar to that observed for wild-type AID (Fig. 1b). To ensure that biotinylated biotagDM-AID could be affinity-purified, we incubated CH12BirA/biotagDM-AID extracts with streptavidin agarose. Immunoblot analysis showed that AID was retained on the streptavidin-agarose column and was biotinylated (Fig. 1c). Finally, we tested if biotagDM-AID in CH12BirA/biotagDM-AID cells could bind to S region DNA. Cross-linked DNA-protein complexes were affinity-purified through streptavidin beads and the recovered DNA analyzed by PCR (Fig. 1d). DNA from the μ-switch region (Sμ), but not from the neighboring μ-promoter (Iμ), was readily detected indicating that the biotagDM-AID protein could specifically bind S region DNA. Interestingly, biotagDM-AID, while abundantly present in unstimulated CH12 cells (Supplementary Fig. 1), required CIT stimulation for binding to S region DNA (Fig. 1d). This observation revealed a hitherto unrecognized requirement of B cell stimulation for interaction of AID with its target sequence. Overall, the proper intracellular localization of biotinylated biotagDM-AID and its binding to switch regions upon stimulation validated our use of this system to screen for AID interacting proteins.
Nuclear extracts were prepared from CIT-stimulated CH12BirA/biotagDM-AID and CH12BirA cells and subjected to streptavidin-agarose affinity-purification. A fraction of the eluted protein was analyzed by immunoblotting to confirm the presence of biotagDM-AID (Supplementary Fig. 2). The remainder was partially resolved on an SDS-gel (Supplementary Fig. 3) and proteins present in the samples were identified by LC-MS/MS mass spectrometry. The most abundant proteins detected were present in both CH12BirA/biotagDM-AID and CH12BirA samples. These comprised metabolic proteins (such as acetyl-coA carboxylase 1, pyruvate carboxylase and propionyl-coA carboxylase) that are naturally biotinylated in the cell. Among those unique to the CH12BirA/biotagDM-AID sample, thirty-eight proteins were found to be common to two independent affinity-purification–mass spectrometric analyses (Supplementary Table 1).
Approximately 30% of the potential AID interactors were proteins with undesignated functions. Several cytoskeletal and metabolic proteins and notably, DNA repair proteins, transcription factors, chromatin remodeling proteins and mRNA processing factors were detected in the complex. We sought to validate the putative interactors individually in co-immunoprecipitation assays, starting with DNA repair and mRNA processing proteins, given the intricate link between AID, CSR and RNA- or DNA-dependent reactions. We immunoprecipitated AID from activated splenic B cells (wild-type, AID-deficient or AID-deficient expressing HA-tagged AID) with AID or HA antibodies and probed the immunoprecipitate for the presence of the putative AID interactors. DNA ligase I, transportin-I or Rad50 were not detected in the AID immunoprecipitates (Supplementary Fig. 4) indicating that these proteins either represent false-positives of the mass spectrometric analysis or these interact with AID in a fashion that occluded the antibody binding site on AID during immunoprecipitation. Alternatively, since the original screen was performed using catalytically inactive AID with the objective of creating a trap for complexes, these interactions may be too transient to detect by simple co-immunoprecipitation techniques. DNA Topoisomerase-IIβ, on the other hand, was readily detected in the AID immunocomplex (Supplementary Figs. 4,5). Determining the physiological relevance of the AID-Topoisomerase-IIβ interaction in CSR will require further studies.
AID interacts with PTBP2
One molecule that was detected, albeit with only one identifiable peptide, in both sets of mass spectrometric analyses was the splicing regulator polypyrimidine tract binding protein-2 (PTBP2). The 56 kDa, primarily nuclear protein (Supplementary Fig. 6), could be readily detected in an AID complex immunoprecipitated with AID antibodies from wild-type but not AID-deficient activated splenic B cells (Fig. 1e). Conversely, immunoprecipitation of PTBP2 from activated splenic B cells revealed the presence of AID in the immunoprecipitate (Fig. 1f). The interaction between PTBP2 and AID is likely independent of RNA as recombinant his-tagged PTBP2 could bind to AID purified from 293 cells even when the proteins were treated with an excess of RNaseA (Supplementary Fig. 7). Additionally, mutation in the serine-38 phosphorylation site of AID (AID(S38A))18-20 did not abolish interaction with PTBP2 indicating that the binding is not dependent on AID phosphorylation (Supplementary Fig. 7). Finally, recombinant PTBP2 did not alter the in vitro ssDNA deaminase activity of purified AID (Supplementary Fig. 8). These results indicated that AID and PTBP2 could interact in primary B cells, that the interaction is not dependent on RNA or phosphorylation status of AID at serine-38 and that PTBP2 does not alter the ssDNA deaminase activity of AID.
PTBP2 was reported to be expressed primarily in mitotic neurons during neuronal development21. Its role in splicing was inferred from its homology to PTBP1 (also known as PTB and hnRNP I). PTBP1 is ubiquitously expressed and is one of the best-studied splicing regulators. PTBP1 and PTBP2 are generally considered repressive regulators of splicing that bind RNA with polypyrimidine tracts, block spliceosome assembly at the site and mediate exon exclusion during alternative splicing22, 23. Notably, both polypyrimidine tracts and splicing have been linked to CSR. Mutations that block splicing of germline transcripts abrogate CSR, implying that the splicing machinery or the spliced transcripts influence CSR24. Additionally, S regions are transcribed in both the sense and anti-sense orientation and the anti-sense transcripts are pyrimidine-rich25. These previous observations in conjunction with our finding that PTBP2 is a bona fide AID interactor in B cells led us to investigate the requirement of PTBP2 in CSR.
PTBP2 knock-down impairs CSR in CH12 cells
We used short-hairpin RNA (shRNA) to stably knock-down PTBP2 in CH12 cells. For this, we used the lentiviral vector pLKO-1 that transcribes the cloned shRNAs from a U6 promoter. The pLKO-1 vector also encodes a puromycin resistance gene that allowed selection of cells that had stably integrated the shRNA constructs. Two different shRNA constructs were used to knock-down PTBP2, one (PTBP2-1) directed against the coding sequence and the other (PTBP2-2) against the 3′ untranslated region (3′UTR) of PTBP2 mRNA. A “scramble” shRNA that did not target any known eukaryotic mRNA was used as a control. The puromycin-resistant cells were stimulated in culture with CIT for 72-96 h and then assayed for PTBP2 expression by immunoblotting and for CSR to IgA by flow cytometry.
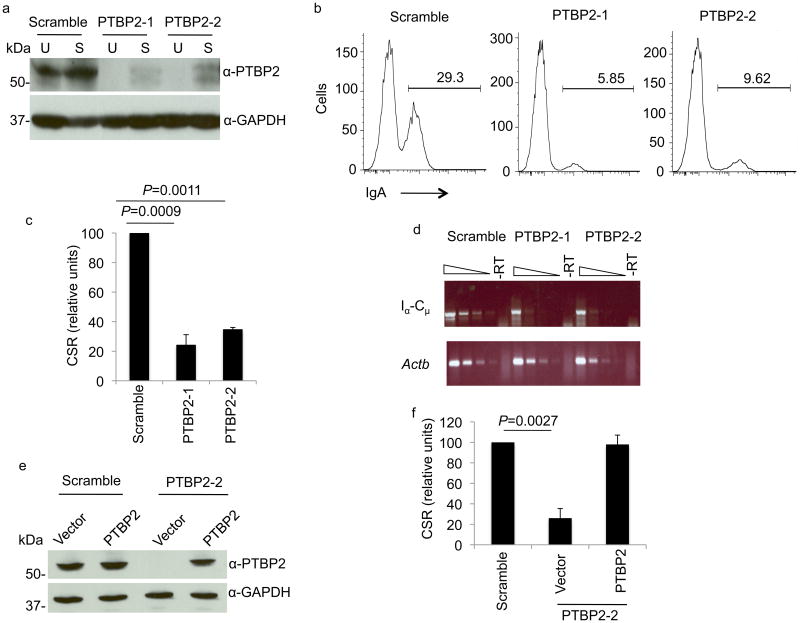
Both PTBP2-1 and PTBP2-2 led to a substantial reduction in PTBP2 protein in unstimulated and CIT-stimulated cells (Fig. 2a). When PTBP2 knock-down cells were stimulated with CIT and analyzed by flow cytometry, we observed a severe defect in CSR to IgA (Fig. 2b,c, Table 1, Supplementary Fig. 9). While on average 28% of control cells underwent CSR to IgA, only around 8% CSR was observed in PTBP2 knock-down cells (Fig. 2c, Table 1, Supplementary Fig. 9). The defect in CSR was also evident from a measure of the steady state Iα-Cμ circle transcripts that are generated from the excised DNA following recombination between Sμ and Sα (Fig. 2d). To ensure that the defect in CSR is a direct consequence of PTBP2 knock-down and not due to non-specific activity of the introduced shRNAs, we expressed PTBP2 cDNA from a lentiviral vector in CH12 cells in which the endogenous protein was knocked-down with shRNA against its 3′UTR. We observed that restoring PTBP2 expression (Fig. 2e) rescued CSR in the CH12 cells (Fig. 2f). Overall, these results suggest that PTBP2 depletion inhibits CSR.
Figure 2.
PTBP2 knock-down impairs CSR. (a) shRNAs efficiently knock-down PTBP2 expression in CH12 cells. CH12 cells were lentivirally infected with shRNAs against PTBP2 (PTBP2-1, PTBP2-2) or non-specific control (scramble). Whole cell extracts derived from unstimulated (U) or CIT- stimulated (S) cells were analyzed on immunoblots using anti-PTBP2 or control GAPDH antibodies. (b) PTBP2 knock-down or control CH12 cells were stimulated with CIT for 72 h and switching to IgA was measured by flow cytometry. A representative histogram is shown and the percentage of IgA-positive cells is indicated. (c) Quantification of CSR in PTBP2 knockdown cells. CSR in control cells was assigned an arbitrary value of 100 for each experiment. The data shows the mean of 5 independent experiments with error bars representing standard deviation from the mean. P-values were determined by the Student's t-test. (d) Reduced amounts of circle transcripts in PTBP2 knock-down cells. 3-fold dilutions of cDNA generated by reverse-transcription were amplified by PCR for Iα-Cμ or β-actin (control) transcripts. –RT represents PCR from template in which reverse-transcriptase was not added. (e) Enforced expression of PTBP2 in knock-down cells. CH12 cells expressing scrambled or PTBP2-2 shRNA were transduced with empty vector or vector harboring PTBP2 cDNA and cell extracts were analyzed by immunoblotting. (f) PTBP2 expression rescues CSR. PTBP2 knock-down cells expressing PTBP2 through a lentiviral construct were stimulated with CIT and CSR to IgA was measured by flow cytometry. CSR frequency in scramble was assigned a value of 100 in each experiment. The data represents the mean of 3 independent experiments with error bars representing standard deviation from the mean.
Table 1.
Percentage of CH12 cells that underwent CSR to IgA in individual experiments. CSR was measured by flow cytometry. Results for CH12 cells expressing scramble shRNA or two PTBP2 shRNAs (PTBP2-1, PTBP2-2) are shown. ND: not determined.
| Expt. | scramble (%CSR to IgA) | PTBP2-1 (% CSR to IgA) | PTBP2-2 (% CSR to IgA) |
|---|---|---|---|
| 1 | 29.6 | 7.6 | ND |
| 2 | 36.9 | ND | 13.3 |
| 3 | 29.3 | 5.9 | 9.6 |
| 4 | 16.7 | 5.5 | 5.5 |
| 5 | 24.8 | 5.4 | 9.0 |
| 6 | 28.1 | 5.0 | 9.3 |
Knock-down of PTBP2 did not substantially impair cell proliferation as measured by the dilution of the permanent red dye SNARF over 48 h (Supplementary Fig. 10). Since CSR is linked to cell division26, we analyzed division-dependent CSR in greater detail. Control or PTBP2 knock-down cells were stained evenly with SNARF and stimulated with CIT. After 72 h stimulation, the control cells were divided into approximately 20-percentile gates based on SNARF expression. These gates were then applied to the PTBP2 shRNA-infected cells and the percentage of IgA-positive cells within the gates was quantified. Regardless of the extent of proliferation as measured by SNARF dilution, the PTBP2 depleted cells showed a consistent reduction in CSR compared to control cells (Supplementary Fig. 11). Thus, PTBP2 knock-down does not impair B cell proliferation and in this experiment, the PTBP2-depleted cells appeared to proliferate slightly faster. We conclude that the defect in CSR in PTBP2 knock-down cells is not due to a defect in cell proliferation.
PTBP2 does not influence AID expression
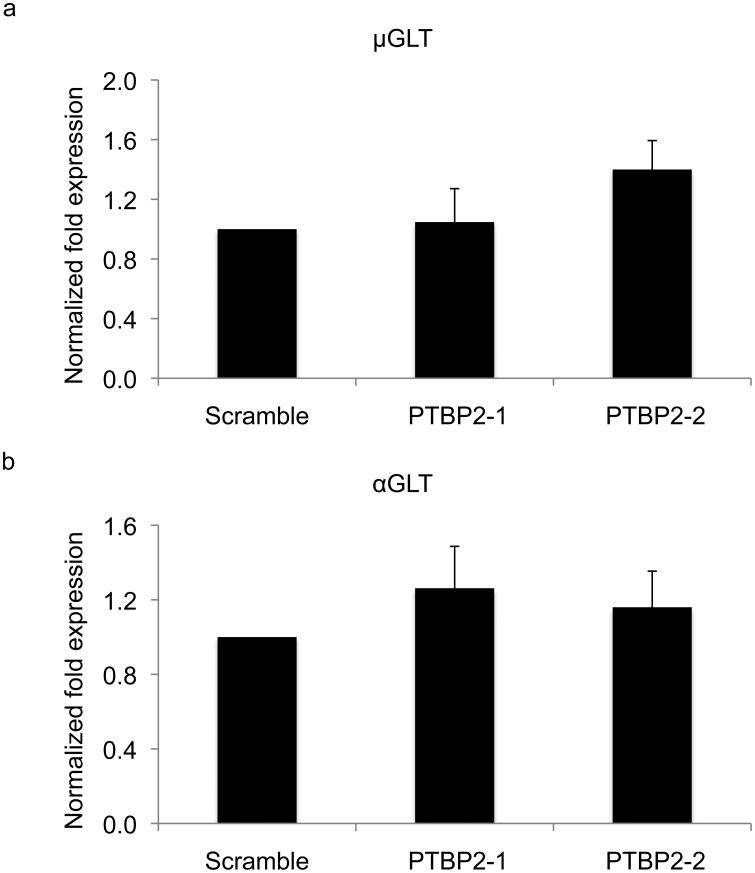
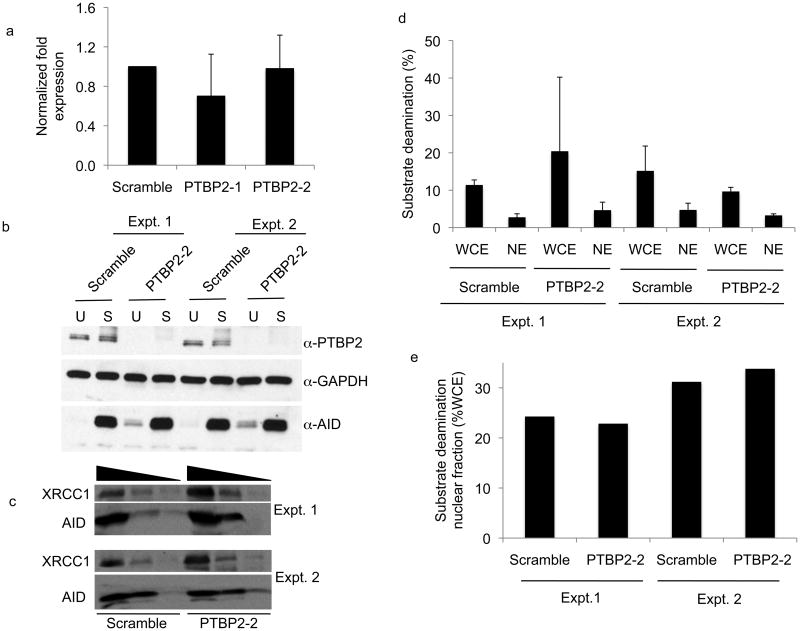
PTBP2 knock-down did not have a marked effect on the steady state abundance of μ or α germline transcripts (Fig. 3, Supplementary Fig. 12). Likewise, PTBP2 depletion did not alter the steady state abundance of AID mRNA (Fig. 4a) or total AID protein (Fig. 4b). To ensure that PTBP2 does not influence nuclear retention of AID, we analyzed AID protein abundance in the nucleus of control and PTBP2 knock-down cells by immunoblotting. While the absolute amounts of total and nuclear AID varies between experiments, the protein ratio between AID and XRCC1, a nuclear protein used as a loading control, was similar between multiple experiments (Fig. 4c, Supplementary Fig. 13, 14). To further demonstrate that PTBP2 knock-down does not alter nuclear retention of AID, we determined ssDNA cytidine deaminase activity in the nucleus as a fraction of total cellular AID activity (Fig. 4d). A low basal ssDNA cytidine activity was present in unstimulated B cells (Supplementary Fig. 15). This activity substantially increased in activated B cells upon expressing AID (Fig. 4d). Thus, ssDNA cytidine deaminase activity in activated B cells is a convenient measure of AID activity16. Consistent with earlier reports18, approximately 25-30% of total AID is nuclear in control activated B cell, which is similar to that observed in PTBP2 knock-down cells (Fig. 4e). AID activity in the nucleus was not due to contamination of cytoplasmic AID as GAPDH, a cytoplasmic protein was not detected in the nuclear fraction (Supplementary Fig. 16). Overall, we conclude that PTBP2 knock-down does not affect either the abundance or nuclear retention of AID.
Figure 3.
Germline transcription is not affected in PTBP2 knock-down cells. RNA from PTBP2 knock-down or control cells was reverse transcribed and analyzed by real-time quantitative PCR for the abundance of μ (a) and α (b) germline transcripts (GLT). Real-time data was normalized to the β-actin gene (Actb). The data represent mean of three independent experiments and error bars depict standard deviation from mean.
Figure 4.
AID expression and nuclear localization is not altered in PTBP2 knock-down cells. (a) AID mRNA expression in CIT-stimulated control or PTBP2 knock-down cells was quantified by reverse-transcription real-time quantitative PCR and normalized to β-actin. (b) Whole cell extracts (100 μg) derived from unstimulated (U) or CIT-stimulated (S) scramble or PTBP2-2 shRNA expressing cells were analyzed by immunoblots using indicated antibodies. Results from two independent knock-down experiments are shown. (c) Approximately 50 μg of nuclear protein from two independent PTBP2 knock-down experiments were analyzed by immunoblotting using AID and XRCC1 antibodies. XRCC1 served as a loading control for nuclear proteins. (d) Approximately 25 μg of whole cell (WCE) or nuclear (NE) extracts derived from two independent (Expt. 1, Expt. 2) PTBP2 knock-down or control cells were assayed for AID activity by measuring conversion of cytidine to uridine on a ssDNA substrate. Deamination was measured by the uracil release assay as described16. Deamination activity was plotted as a mean of three independent assays of each extract. Error-bars represent standard deviation from mean. (e) The fraction of nuclear AID activity was plotted as a percentage of total AID activity from the values obtained in panel (d).
PTBP2 depletion impairs binding of AID to S regions
PTBP2 is an RNA-binding protein. We therefore determined if recombinant PTBP2 could bind RNA transcribed from S region DNA. We transcribed S regions or control DNA in vitro with T7 or SP6 RNA polymerase in the presence of [α32P]-UTP. The radiolabeled RNA was incubated with immobilized recombinant his-tagged PTBP2 or his-tagged control protein Gen1 on Ni2+-agarose beads and the ability of the proteins to bind RNA was measured by the retention of radioactive counts on the beads. As predicted from the affinity of PTBP-family proteins for polypyrimidine-rich sequences, anti-sense S transcripts that are C-rich efficiently bound PTBP2 (Fig. 5). Surprisingly, RNA generated in vitro from S regions transcribed in physiological orientation displayed a 2.5-3-fold enrichment in binding to PTBP2 compared to that observed for non-S region transcripts (Fig. 5).
Figure 5.
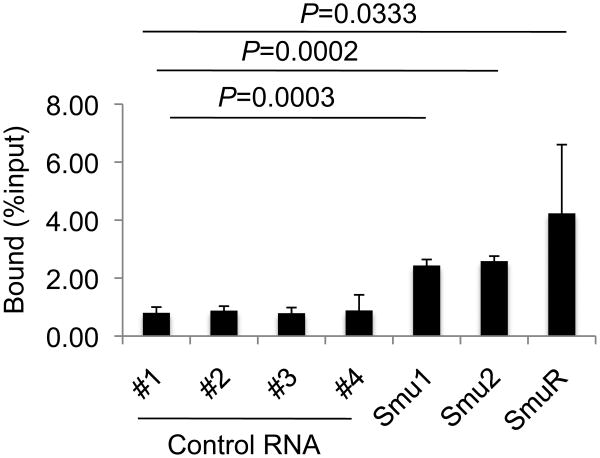
PTBP2 binds switch transcripts in vitro. His-tagged PTBP2 was immobilized on Ni2+-agarose beads and incubated with in vitro transcribed radiolabeled RNA. Beads were washed and the amount of radioactivity retained on the beads determined in a scintillation counter, subtracted from the count retained by a control protein (Gen1) and plotted as percentage of total input counts. Controls 1-4 represent transcripts derived from 4 protein-coding genes (#1, Protein kinase A Cα subunit, #2, Protein kinase A RIα subunit, #3, Ku70, and #4, AID). Smu1 and Smu2 contain, respectively, 1 kb and 3 kb Sμ RNA sequence in the sense orientation and SmuR represents a 1 kb anti-sense Sμ transcript. Values in the histogram represent the mean of three independent binding experiments and error bars represent standard deviation from mean. P-values were determined by using the Student's t-test, using RNA#1 as the non-S region control.
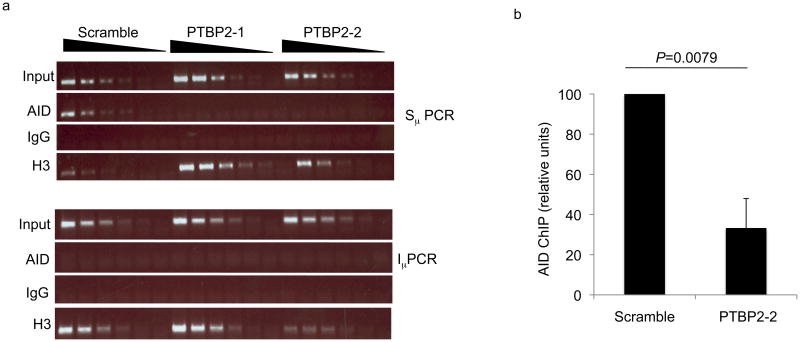
The ability of PTBP2 to bind S region transcripts prompted us to carry out chromatin immunoprecipitation (ChIP) to test if PTBP2 influences the ability of AID to bind transcribed S regions. Cross-linked DNA-protein complexes from CIT-stimulated PTBP2 knock-down or control CH12 cells were immunoprecipitated with AID antibodies and immunoprecipitated DNA was analyzed for the presence of Sμ. ChIP with histone H3 antibodies and non-specific IgG antibodies were used as positive and negative controls, respectively. AID was readily and specifically detected at Sμ, but not at the intronic μ promoter in control cells (Fig. 6). PTBP2 knock-down, on the other hand, led to a marked reduction in the amount of AID associated with Sμ (Fig. 6, Supplementary Fig. 17). These results indicate that depletion of PTBP2 significantly impairs either the recruitment or stable binding of AID to S regions.
Figure 6.
PTBP2 promotes binding of AID to S region DNA. (a) Control or PTBP2 knock-down cells were stimulated with CIT for 48 h and chromatin immunoprecipitation (ChIP) was carried out with antibodies against AID, histone H3 or IgG control. Three-fold dilutions of ChIP DNA were analyzed by PCR for the presence of Sμ or Iμ (as control). (b) The amount of Sμ in anti-AID ChIP samples was quantified by real-time PCR. The graph depicts qPCR values expressed as relative ChIP units with scrambled assigned an arbitrary value of 100. ChIP units are derived from normalizing Ct values to input and then subtracting IgG ChIP Ct values as background. Results shown are the average of three independent experiments expressed with error bars representing standard deviation from the mean. p-value was determined by using the Student's t-test.
PTBP2 influences CSR in primary B cells
Experiments described thus far investigated the ability of PTBP2 to mediate CSR specifically to IgA in the CH12 cell line. To determine if PTBP2 promotes CSR to IgG1 in primary B cells, we sought to deplete the protein from mouse splenic B cells. We transduced lentiviruses encoding PTBP2-1 and PTBP2-2 shRNAs into activated splenic B cells that were stimulated in culture to undergo CSR to IgG1 with anti-CD40 and IL-4. Unlike knock-down in CH12 cell lines where drug-selection over an extended period allowed us to generate cells that had stably integrated the shRNA constructs, use of shRNA in primary splenic B cells is complicated by their limited lifespan in culture (4-6 days). Thus, depletion of the target protein might not reach the level observed for CH12 cells before stimulation. We therefore included an additional control, an shRNA against AID, with the prediction that CSR in the AID knock-down cells would serve as an indicator of the efficiency of shRNA knockdown in this system.
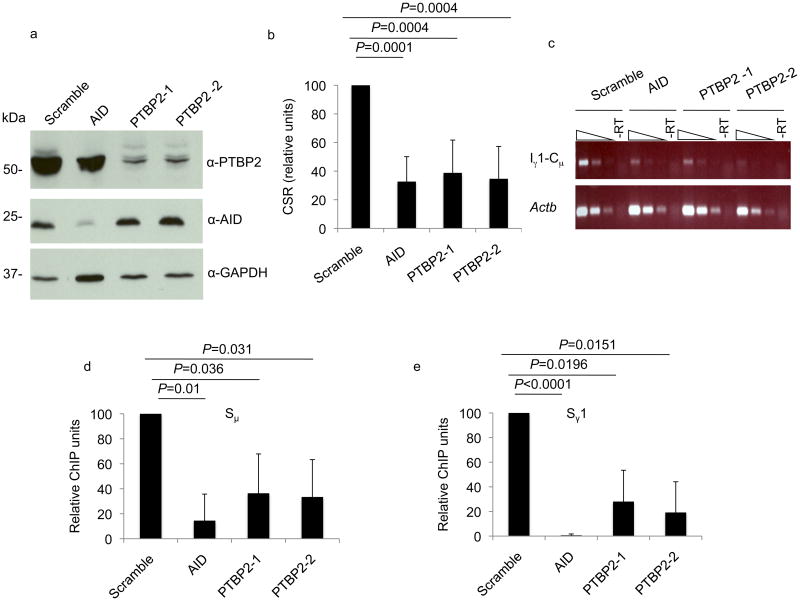
Both PTBP2 shRNAs and AID shRNA robustly depleted the target proteins in primary splenic B cells as analyzed by immunoblot (Fig. 7a). PTBP2 knock-down did not influence the amount of AID in the activated splenic B cells (Fig. 7a). As predicted from the CSR defect in CH12 cells, PTBP2 knock-down resulted in substantially impaired CSR to IgG1 as judged by both flow cytometry (Fig. 7b, Table 2, Supplementary Fig. 18) and the abundance of Iγ1-Cμ circle transcripts (Fig. 7c), with the switching frequencies approaching that observed for AID knock-down. Importantly, ChIP experiments showed that PTBP2 knock-down in primary B cells leads to a marked and significant reduction in the amount of AID associated with both Sμ and Sγ1 DNA (Fig. 7d, e). A PCR for p53 was used as a negative control for the ChIP experiment (Supplementary Fig. 19). Taken together, we conclude that the defect in CSR observed in PTBP2-depleted cells is primarily due to reduced binding of AID to S region DNA.
Figure 7.
PTBP2 knock-down impairs CSR and AID binding to activated switch regions in primary B cells. (a) Immunoblot analysis of cell extracts derived from activated splenic B cells transduced with shRNAs directed against PTBP2 or AID. (b) CSR to IgG1 was measured by flow cytometry. CSR in cells transduced with scramble shRNA was assigned a value of 100. Histogram represents mean of 7 independent experiments with error bars depicting standard deviation from the mean. p-values were determined by the Student's t-test. (c) Reduced Iγ-Cμ circle transcripts in PTBP2 knock-down cells. RNA derived from indicated cells was reverse-transcribed and 3-fold dilutions of cDNA analyzed for Iγ-Cμ circle transcripts by PCR. –RT represents PCR template in which reverse-transcriptase was omitted. (d-e) Impaired binding of AID to Sμ and Sγ1 in PTBP2 knock-down primary B cells. B cells were stimulated with anti-CD40 and IL-4 for 48 hours and then subjected to ChIP using AID or control-IgG antibodies. Levels of Sμ or Sγ1 in anti-AID ChIP samples were measured by quantitative real-time PCR. The graph depicts qPCR values expressed as relative ChIP units with scrambled assigned an arbitrary value of 100. ChIP units are derived from normalizing Ct values to input and then subtracting IgG ChIP Ct values as background. Results shown are the average of three independent experiments expressed with error bars representing standard deviation from the mean. p-values were determined by the Student's t-test.
Table 2.
Percentage of splenic B cells that underwent CSR to IgG1 in individual experiments. CSR was measured by flow cytometry. Results from activated B cells expressing scramble shRNA, two PTBP2 shRNAs (PTBP2-1, PTBP2-2) and AID shRNA are shown.
| Expt. | Scramble (%CSR to IgG1) | Scramble (%CSR to IgG1) | PTBP2-1 (% CSR to IgG1) | PTBP2-1 (% CSR to IgG1) |
|---|---|---|---|---|
| 1 | 16.7 | 6.7 | 8.2 | 8 .2 |
| 2 | 16.5 | 1.0 | 1.3 | 1.1 |
| 3 | 18.3 | 7.9 | 4.9 | 11.6 |
| 4 | 20.3 | 6.0 | 6.8 | 5.1 |
| 5 | 21.5 | 12.3 | 14.3 | 16.6 |
| 6 | 18.9 | 2.9 | 3.7 | 4.5 |
| 7 | 11.2 | 3.9 | 3.0 | 3.7 |
Discussion
We have identified PTBP2 as an AID interacting protein that plays a pivotal role in CSR. PTBP2, and its closely related family member PTBP1, are RNA-binding proteins required for alternative pre-mRNA splicing, a process critical for the generation of multiple protein isoforms from a single gene27, 28. PTBP2 was initially thought to be restricted to the brain and is therefore also known as brPTB (brain PTB) and nPTB (neuronal PTB)29-31. Subsequently, the protein was also detected in testis and at low amounts in the liver, heart, lung, skeletal muscle, and thymus30. To our knowledge, our findings are the first to describe PTBP2 expression in activated B cells.
The mechanism of CSR has been closely linked to both germline transcription and splicing. The role of splicing in this reaction is enigmatic as it is unclear whether components of the splicing machinery or the spliced transcripts themselves (or both) are required for CSR. Current models of CSR posit that transcription generates ssDNA substrates for AID in the context of R-loops32. This R-loop dependent model for CSR requires that the primary, unprocessed germline transcript stably hybridizes to the template strand. If splicing occurs co-transcriptionally and efficiently, R-loop formation would be transient. It is conceivable that PTBP2 could be recruited to S region DNA, maybe through interactions with modified histones33, and inhibit splicing, thus enhancing stability of R-loops and facilitating CSR. While the observation that the steady-state abundance of processed germline transcripts was not altered in the PTBP2 knock-down cells would argue against such a role of PTBP2, we cannot rule out transient alterations in the half-life of R-loop DNA in our studies.
The role of PTBP2 in CSR could be independent of its role in splicing. Our in vitro studies have shown that PTBP2 has the potential to bind both the sense and anti-sense S region transcripts, consistent with previous reports that the PTBP family of proteins binds a wide variety of RNA34. While transcription of S regions in the physiological orientation is essential for CSR, anti-sense transcripts encompassing S regions have also been reported25. It is therefore conceivable that PTBP2 can bind either the sense or antisense S region transcripts as they are being transcribed and recruit AID. Thus the specificity of AID for S region binding is mediated by its ability to interact with a factor, PTBP2 that in turn binds to RNA emanating from the unique S regions. Recent studies have shown that recruitment of AID to S regions could also be mediated by an RNA polymerase II associated AID interactor Spt5 (ref. 35). Thus, multiple mechanisms operate to recruit AID to S region DNA. The recruited AID could then be phosphorylated at serine residue-38 by protein kinase A, which associates with S region DNA in an AID-independent fashion36. Phosphorylated AID then binds RPA at S region DNA12, 36 and the multi-component complex thus formed could then trigger DNA deamination (by AID) and nucleation of downstream repair factors (by RPA), leading to the cascade of reactions that ultimately result in CSR.
We cannot exclude the possibility that PTBP2 influences CSR at multiple levels, including favorably altering the splicing of an unidentified protein required for AID recruitment. Additional work will surely be required to understand the precise function of PTBP2 in CSR. Likewise, the role, if any, of PTBP2 in SHM needs to be determined. Nevertheless, the results presented here suggest that PTBP2 has the potential to shepherd AID to S regions during CSR and sets the foundation for investigating how a known splicing regulator could promote AID targeting specificity.
Methods
Mice
Aicda−/− mice were a gift from T. Honjo (University of Kyoto, Japan). Wild-type BALB/c mice were from the Jackson Laboratory. All animals were maintained according to guidelines for animal welfare of the Memorial Sloan Kettering Research Animal Resource Center.
Cell lines and Plasmids
CH12 cells were obtained from T. Honjo. Stable cell lines expressing biotagDM-AID and BirA or BirA alone were generated by electroporation of the plasmids encoding each protein followed by antibiotic selection and subcloning. The biotag vector and BirA plasmids were obtained from S. Orkin (Harvard Medical School). The EF.PGK.GFP, psPAX2 and pMD2.G were obtained from Addgene.
Antibodies and Reagents
Antibodies for immunoblot were the following: anti-PTBP2 (ab57619; Abcam), anti-AID1, anti-GAPDH (6C5, Millipore). Antibodies for flow cytometry were as follows: allophycocyanin–anti-IgG1 (×56), fluorescein isothiocyanate–anti-IgA (C10-3; all from BD Pharmingen). Antibodies used for ChIP were the following: anti-AID (same as for immunoblot), anti-H3 (ab12079; Abcam) and anti–rabbit IgG (I5006; Sigma). Other reagents: Streptavidin-HRP (Invitrogen). SNARF-1 carboxylic acid, acetate, succinimidyl ester (Invitrogen).
Cell extracts and fractionation
Whole cell extracts were prepared in NP-40 lysis buffer (20 mM Tris pH 7.5, 5% glycerol, 150 mM NaCl, 5 mM β-Me, 0.5% NP-40). Cells were resuspended in lysis buffer (approximately 50 μl/1×106 cells), incubated on ice for 30 min and sonicated briefly. Lysates were centrifuged at 10,000 g and the supernatants were used as whole cell extracts. Cells were fractionated into cytoplasmic and nuclear extracts as described2.
Pull-down of AID complex with streptavidin-agarose beads
Nuclear extracts were diluted to a final salt concentration of 300 mM NaCl. Extracts were pre-cleared twice with protein-G agarose (EMD) for 2 h at 4°C. All beads were used at 1/10th the volume of the sample. Streptavidin agarose beads (Invitrogen) were washed with IP buffer (20 mM HEPES pH 7.5, 0.1% NP40, 420 mM NaCl, 0.2 mM EDTA, 10% glycerol, 0.2 mM PMSF, 1 mM DTT) and incubated with the extracts overnight. Beads were washed 7 times for 10 min in IP buffer and bound proteins were eluted by boiling for 10 min in SDS buffer (100 mM Tris-Cl pH 6.8, 4% SDS, 20% glycerol).
Mass Spectrometry
Affinity purified samples were partially resolved on SDS-gels and each lane excised as three approximately equal parts. Proteins in gel slices were digested with trypsin and the resulting polypeptide pools were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School, Boston, MA. The identified peptides were compared to a computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
Stimulation for CSR
CH12s were stimulated with anti-CD40 (1 μg/ml; HM40-3; eBioscience), IL-4 (12.5 μg/ml; R&D Systems; 404-ML) and TGF-β1 (0.1 ng/ml; R&D Systems, 240-B). Primary B cells were stimulated with anti-CD40 and IL-4 at the same concentrations.
Immunoprecipitation with AID or PTBP2 antibody
Whole cell extracts of primary B cells from wild-type or Aicda−/− mice were pre-cleared twice for one hour with protein A agarose beads and non-specific IgG. AID or PTBP2 antibody was added to each tube and rotated overnight at 4 °C. The next day protein A beads were added and the samples were rotated at 4 °C for 1 h. The beads were washed 5 times with NP40 lysis buffer and eluted with by boiling in 2× SDS buffer.
Knockdown with lentiviral shRNA
For transduction of CH12s with shRNA 10cm plates of 293T cells were transfected with 9 μg psPAX2 packaging vector, 3 μg pMD2.G envelope vector and 12μg pLKO.1 shRNA vector using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. The media was changed 24 h later to begin viral supernatant production. Another 24 h later the viral supernatant was harvested and mixed with 8 μg/ml of polybrene (Sigma). CH12 target cells were resuspended in viral supernatant at 0.25 million cells/ml and spinfected in 6-well plates at 800 g for 2 h at 25°C. Viral supernatant was then aspirated and the cells were resuspended in complete media. Puromycin (Sigma) selection (4 μg/ml) was started 24 h later. CH12s were selected in puromycin for three days (there were no live cells in the uninfected controls after 24 h of puromycin selection) before stimulation and remained in puromycin for the length of the stimulation (72 h).
For lentiviral transduction of primary B cells, 10 cm plates of 293Ts were transfected as above with 5 μg psPAX2, 5 μg pMD2.G and 10 μg pLKO.1 encoding the appropriate shRNA. The media was changed 24 h later and viral supernatant was harvested 48–72 h after that and mixed with 10 μg/ml polybrene (Sigma). On the day of viral supernatant harvest, splenic B cells were harvested and resuspended at 1 × 106 cells/ml in media containing 2 μg/ml anti-CD40. In a 12-well plate, 1 ml of B cell suspension was mixed with 1ml viral supernatant and incubated for 20 min in the hood at 25°C. Then, the plates were centrifuged for 90 min at 650 g for 1.5 h at 32 °C. The plates were carefully removed from the centrifuge and incubated with the virus overnight in the tissue culture incubator. The next day, the transduced cells were washed twice with B cell media and stimulated for CSR as described above.
Scramble shRNA in pLKO.1 was obtained from Addgene (plasmid 1864). All other shRNAs in pLKO.1 were from Sigma. PTBP2-1: 5′-CCGGGCTGTACCCTAAGG ATTGATTCTCGAGAATCAATCCTTAGGGTACAGCTTTTTG-3′; PTBP2-2: 5′-CCG GGCTGTTATCATTCCTTGGTTACTCGAGTAACCAAGGAATGATAACAGCTTTTTG-3′; AID shRNA: 5′-CCGGCATGACCTTCAAAGACTATTTCTCGAGAAATAGTC TTTGAAGGTCATGTTTTTG-3′
Rescue of knockdown in CH12 cells
To rescue the PTBP2 knockdown we used lentiviral infection of a PTBP2 expression vector in cells expressing the shRNA against the 3′UTR of PTBP2 (PTBP2-2). Cells previously transduced with PTBP2-2 and selected with puromycin were transduced with a vector encoding PTBP2 driven by the EF1α promoter (EF.PGK.GFP). Scramble or PTBP2 knockdown cells were infected with the PTBP2 expression vector or the vector control by the same protocol as that used for shRNA infection. 24 h after infection with the rescue or control construct the cells were stimulated for CSR. 72 h later the cells were analyzed for CSR by flow cytometry to assess the level of rescue.
Germline transcription and circle transcripts
Total RNA was extracted from cultured cells by using TRIzol (Invitrogen) according to manufacturer instructions. cDNA was synthesized with Superscript III (Invitrogen) by using 2-4 μg of total RNA and 50 ng of random hexamers in a 20μl reaction volume. One twentieth (GLT, γ-CTs) or one tenth (α-CTs) of the cDNA product was used as a template for reverse transcription (RT)–PCR in a 25 μl reaction volume. Primers for GLTs: (μ) ImF: 5′-CTCTGGCCCTGCTTATTGTTG-3′ and CμR: 5′-GAGACATTTGGG AAGGACTGACT-3′; (α) IαF: 5′-CCT GGCTGTTCCCCTATGAA-3′ and CαR: 5′-GAG CTGGTGGGAGTGTCAGTG-3′. Primers for CTs: CμR: 5′-AATGGTGCTGGGCAGGAA GT-3′ and IαF: 5′-CCAGGCATGGTTGAG ATAGAGATA G-3′ or Iγ1F: 5′-GGCCCTTCC AGATCTTTGAG-3′; β-actin primers: F: 5′-TGCGTGACATCAAAGAGAAG-3′ and R: 5′-CGGATGTCAACGTCACACTT-3′. Q-PCR Primers for Sμ and Sα germline transcripts are as described by Pavri et al3.
Cell Proliferation
To assess cell proliferation, PTBP2 knockdown cells or controls were stained with the permanent red dye SNARF. Knockdown or control cells were pelleted and resuspended at 2 × 107 cells/ml in staining buffer (PBS + 5% FCS pre-warmed to 37 °C). The SNARF stain was diluted in staining buffer to 18 μM. Equal volumes of cell suspension and SNARF dye were mixed to yield a final SNARF concentration of 9 μM. Cells were incubated in dye at 37 °C for 10 min and quenched with one volume of FCS at 37 °C. Quenched cells were then pelleted and washed with staining buffer. Cell-surface SNARF was detected by flow cytometry at 0, 24 and 48 and 72 h.
ChIP
ChIP was done as described4. For the biotinylated-DM-AID ChIP, SA beads were used in place of antibodies for the IP and the complexes were eluted by boiling. PCR primers: Sμ; sense: 5′-TAGTAAGCGAGGCTCTAAAAAGCAT-3′ anti-sense: 5′--AGAACAGTCCAGTGTAGGCAGTAGA-3′. Iμ promoter; sense: 5′-GCTCAGCCTGGACTTTCGGTTTGGT-3′; anti-sense: 5′-GGAGTCAAGATGGCCGATCAGAACC-3′. Sγ1; sense: 5′-TATGATGGAAAGAGGGTAGCATTCACC-3′; anti-sense: 5′-CTCCTTCCCAATCTCCCGTG-3′.
Quantitative Real-time-PCR
A Bio-Rad CFX96 Real-Time PCR Detection System was used for all assays. iQ SYBR Green Supermix (Bio-Rad) was used for real-time PCR of immunoprecipitated DNA. iScript one-step RT-PCR Kit with SYBR green (Bio-Rad) was used for quantitation of AID mRNA. Real-time PCR products were analyzed for incorporation of SYBR Green and crossing points were obtained with the CFX Manager software. Melting-curve analysis confirmed the presence of a single PCR product of the predicted size. For ChIP, ‘Relative units’ were calculated by normalization of the crossing point of ChIP with each specific antibody to the crossing point of the input DNA; the inverse of the input DNA–normalized crossing point was then calculated. The ‘IgG-preclear’ value (inverse of the input DNA–normalized crossing point) was subtracted from value obtained for ChIP with each specific antibody (inverse of the input DNA–normalized crossing point) to obtain the “relative units” for ChIP. These “relative units” were expressed as a percentage of the relative unit obtained for the scramble shRNA construct where appropriate. To quantitate differences in mRNA expression we used the delta-delta CT method. Mb1 was used as a reference gene. The primers used for real-time PCR were the same as those for conventional PCR with the addition of p53 and Mb1. p53F: 5′-CCCAGAGACTGCTGTTAAAGTAGAACC-3′; p53R: 5′-CGCCACAGCGTGGTGGTACC-3′. Mb1F: 5′-GGTACCAAGAACCGCATCATC-3′; Mb1R: 5′-AGTCAGACATATGGCAGGCAGG-3′
Supplementary Material
Acknowledgments
We thank all members of the Chaudhuri laboratory for helpful discussions and critical suggestions during the course of the study. This work was supported by grants from the Damon Runyon Cancer Research Fund, Alfred Bressler Foundation and National Institutes of Health to JC and a pre-doctoral Cancer Research Institute Fellowship to UN.
References
- 1.Jung D, Alt FW. Unraveling V(D)J recombination: Insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 4.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 6.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Goodman MF, Scharff MD, Romesberg FE. AID-initiated purposeful mutations in immunoglobulin genes. Adv Immunol. 2007;94:127–155. doi: 10.1016/S0065-2776(06)94005-X. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 9.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasqualucci L, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 11.Yang SY, Schatz DG. Targeting of AID-mediated sequence diversification by cis-acting determinants. Adv Immunol. 2007;94:109–125. doi: 10.1016/S0065-2776(06)94004-8. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 13.Macduff DA, Neuberger MS, Harris RS. MDM2 can interact with the C-terminus of AID but it is inessential for antibody diversification in DT40 B cells. Mol Immunol. 2006;43:1099–1108. doi: 10.1016/j.molimm.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Conticello SG, et al. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 15.de Boer E, et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 18.Basu U, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 19.McBride KM, et al. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci U S A. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S. Isolation of a sequence-specific RNA binding protein, polypyrimidine tract binding protein, using RNA affinity chromatography. Methods Mol Biol. 2008;488:1–8. doi: 10.1007/978-1-60327-475-3_1. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz M, Jung S, Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- 25.Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci U S A. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc Natl Acad Sci U S A. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 28.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 29.Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. Rna. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 30.Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci U S A. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markovtsov V, et al. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 33.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuong BQ, et al. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.