Abstract
A CotA Multicopper Oxidase (MCO) from Bacillus pumilus, previously identified as a laccase, has been studied and characterized as a new bacterial Bilirubin Oxidase (BOD). The 59kDa protein containing four coppers, was successfully over-expressed in Escherichia coli and purified to homogeneity in one step. This 509 amino-acid enzyme, having 67% and 26% sequence identity with CotA from Bacillus subtilis and BOD from Myrothecium verrucaria, respectively, shows higher turnover activity towards bilirubin compared to other bacterial MCOs. The current density for O2 reduction, when immobilized in a redox hydrogel, is only 12% smaller than the current obtained with Trachyderma tsunodae BOD. Under continuous electrocatalysis, an electrode modified with the new BOD is more stable, and has a higher tolerance towards NaCl, than a T. tsunodae BOD modified electrode. This makes BOD from B. pumilus an attractive new candidate for application in biofuel cells and biosensors.
Keywords: Biofuel cells, Bilirubin Oxidase, Oxygen reduction, Bacillus pumilus, osmium polymer
1. Introduction
Enzymatic miniature membrane-less glucose/O2 Biofuel Cells (BFCs) are of particular interest because they may power implanted medical devices, in the near future. (Barton et al. 2004; Heller 2004) Despite the continued improvement of the performance of enzyme-based biofuel cells, the power output and lifetime are still not sufficient for direct applications. (Osman et al. 2011) In most of the BFCs, these limitations are generally attributed to the cathode (Gallaway and Barton 2008). At the cathode, two enzymes in particular have been used for bioelectrocatalysis: laccases and bilirubin oxidases (BOD). Both enzymes belong to the multicopper oxidase (MCO) family which utilize a minimum of four copper ions to catalyze the four-electron reduction of O2 to water, without release of H2O2.(Solomon et al. 1996) The copper ions of these enzymes are classified into three types depending on their optical and magnetic properties. Type 1 (T1) copper has an intense Cys to Cu(II) charge transfer absorption band around 600 nm. This copper center accepts electrons from the electron-donating substrate, e.g. phenols or metal ions, and relays these to the O2 reduction site. This site is a tri-nuclear cluster (TNC), consisting of an EPR active type 2 (T2) or “normal” copper ion, and a type 3 (T3) pair of copper ions, antiferromagnetically coupled via a bridging hydroxide, with a characteristic 330 nm absorption shoulder.
High potential laccases, eg Trametes versicolor, are characterized by high T1 Cu redox potentials and high catalytic activities at low pH (4-5). They have proven to be efficient electrocatalysts for the electroreduction of O2, (Gallaway et al. 2008; Hussein et al. 2011; Mano 2008; Rengaraj et al. 2011a; Rengaraj et al. 2011b; Rincón et al. 2011) although poor stability in the presence of chloride and/or at neutral pH, limit their utilization, especially in implantable devices, even though different strategies are developed to overcome both issues. (Beyl et al. 2011; Kavanagh et al. 2008) More recently, bilirubin oxidases have attracted attention because, unlike laccases, they show high activity at neutral pH and high tolerance towards NaCl. In addition to more traditional laccase substrates such as syringaldazine (SGZ), 2,6-dimetoxyphenol (DMP) or 2,2’-azino-di-[3-ethylbenzthiazoline-6-sulphonic acid] (ABTS), BODs also catalyze the oxidation of bilirubin to biliverdin which can be used for the diagnosis of jaundice and hyperbilirubinemia. (Doumas et al. 1999; Kirihigashi et al. 2000; Kosaka et al. 1987; Kurosakaa et al. 1998) Fungal BOD from Myrothecium verrucaria (Guo et al. 1991) and Trachyderma tsunodae (Hiromi et al. 1992) have been widely used for the elaboration of biocathodes, either immobilized in redox polymer or directly connected to electrode surfaces, mainly because of their commercial availability. (Gupta et al. 2011; Mano et al. 2003; Ramirez et al. 2008; Tsujimura et al. 2004; Wang et al. 2012; Zloczewska et al. 2011) These enzymes, however, have significant limitations including low turnover activity (M. verrucaria) and instability (T. tsudonae), which have motivated the search for new BODs with improved properties.
Recently, Reiss and co-workers identified a new CotA enzyme from Bacillus pumilus and classified it as a laccase (Reiss et al. 2011). In their study, however, the spectroscopic properties and bilirubin oxidation were not investigated.
We now present the spectroscopic properties of the new CotA enzyme from Bacillus pumilus, which are consistent with features observed, in general, for MCOs. Also, the activity towards bilirubin, allows us to assign this enzyme as a bilirubin oxidase. Furthermore, biochemical and biophysical evaluations show that this enzyme is an excellent candidate for the elaboration of cathodes for biofuel cells operating under physiological conditions, due to their high electrocatalytic currents at neutral pH, thermostability, and tolerance towards NaCl.
2. Materials and methods
Chemicals
IPTG (Isopropyl β-D-1-thiogalactopyranoside) was purchased from Euromedex (Souffelweyersheim, France) and all other chemicals were of analytical grade or higher and purchased from Sigma (Sigma-Aldrich, Saint-Louis, MO). Restriction enzymes were obtained from New England Biolabs (Ipswich, MA). Oligonucleotides were provided by Genecust (Genecust Europe, Luxembourg). All solutions were made with deionised water passed through a AQ 10 Milli-Q purification system from Millipore (Molsheim, France). AKTA purifier 10 UV 900, HisPrep FF 16/10 column, containing Ni Sepharose 6 Fast Flow medium, were from GE Healthcare Bio-Sciences AB (Uppsala, Sweden). Amicon Ultra ultrafiltration columns were from Millipore (Molsheim, France). Polymerase chain reactions were carried out in an automated thermal cycler (VWR). DNA was introduced into E. coli by electroporation using a Eppendorf Eporator system (Eppendorf, France).
UV- visible and Electron paramagnetic resonance
Spectroscopic UV-visible measurements were performed on a Cary 100 system from Varian, Inc (Palo Alto, CA), equipped with a peltier thermostable multicell holder. X-band EPR spectra were obtained at 77K using a Bruker EMX spectrometer equipped with an ER051QR microwave bridge and a dual mode ER4116DM cavity. Purified enzyme was added to a 4mm Wilmad EPR tube, and frozen in liquid nitrogen. The following parameters were applied: 9.65GHz microwave frequency, 10mW microwave power, 10 G modulation amplitude, 100kHz modulation frequency, 328ms time constant, and 82ms conversion time.
Resonance raman
Resonance Raman spectra were obtained in a ~135° backscattering configuration using a Coherent I90C-K Kr+ CW ion laser. The 568.2 nm laser line with an incident power of 25 mW was used as the excitation source. Scattered light was dispersed through a Spex 1877 CP triple monochromator with 1200, 1800, and 2400 grooves/mm holographic gratings and detected with an Andor Newton charge-coupled device (CCD) detector cooled to −80 °C. Samples were contained in a 4 mm NMR tube immersed in a liquid nitrogen finger dewar for measurements. The Raman energy scale was calibrated using Na2SO4 and citric acid. Background spectra were obtained using ground, activated charcoal at 77 K in an equivalent NMR tube. Frequencies are accurate to within 2 cm−1.
Electrochemical experiments
Measurements were performed using a bipotentiostat (CH Instruments, model CHI 842B, Austin, TX, USA) with a dedicated computer. A platinum spiral wire was used as counter electrode and all potentials were referred to a Ag/AgCl (3 M NaCl) electrode (BAS, West Lafayette, IN). All electrochemical measurements were performed in a water-jacket electrochemical cell in 100 mM sodium phosphate buffer (PB) at pH 7.2 at 37°C. The 3 mm diameter glassy carbon electrodes were rotated using a Pine Instruments Rotator and the temperature was controlled by an isothermal circulator (Lab Companion, FR).
Preparation of the modified electrodes
Modified electrodes were prepared as previously described. (Mano et al. 2003) The deposition solution for Bacillus pumilus electrodes consisted of l.26 μL of polymer (PAA-PVI-{Os(4,4′-dichloro-2,2′-bipyridine)2Cl}+/2+ at 10mg/mL, 1μL of Tris H2SO4 50mM buffer at pH7.5, l.24 μL of BOD from B. pumilus at 5mg/mL in the same buffer, and 0.75 μL of Poly(ethylene glycol) (400) diglycidyl ether at 2mg/mL. 1.48μL were deposited on the 3mm diameter glassy carbon electrodes for a total loading of 100μg/cm−2. For Trachyderma tsunodae electrode, the proportions were 1.25μL redox polymer, 1.2μL water, 0.49μL BOD at 5mg/mL and 0.6μL PEGDGE at 2mg/mL, from which 1.55μL were deposited on the electrode to obtain a total loading of 100μg/cm−2. After deposition, the electrodes were cured, protected from dust, during 18 hours in an oven at 25°C.
Enzyme assay and kinetic measurements
The enzyme activity was determined spectrophotometrically at 37°C by following the oxidation of ABTS at 420nm (ε420nm = 36 mM−1cm−1) in a Mcllvaine's citrate-phosphate buffer pH 3.2. Syringaldazine at 22μM (SGZ, ε530nm = 64 mM−1cm−1) and 2,6-dimethoxyphenol at 1mM (DMP, ε468nm = 14.8 mM−1cm−1) were used to measure protein activity at pH 6.2 and 6.8, respectively, in Mcllvaine's citrate-phosphate buffers. Unconjugated bilirubin and conjugated bilirubin were used to assay the bilirubin activity at 37°C and were measured spectrophotometrically at 450nm and 440nm (ε450nm = 32 mM−1cm−1 and ε440nm = 25 mM−1cm−1) in sodium phosphate 50mM pH 7.2 and in a Mcllvaine's citrate-phosphate 0.1M buffer pH 4.8, respectively. Due to low solubility at low pH, the oxidation of unconjugated bilirubin can only be measured for pH >7. One unit was defined as the amount of enzyme that oxidized 1μmol of substrate per minute. Km and kcat were determined in the 2.5μM-5mM concentration range for ABTS, 2.5μM-300μM for SGZ, 2.5μM-4mM for DMP and 2.5μM-150μM for conjugated bilirubin by fitting the data with the simple Michealis-Menten model by non-linear regression.
Mcllvaine's citrate-phosphate 0.1M buffers at pH 3-7.5 and Tris-H2SO4 50mM at pH above 7.5 were used to study the influence of pH on substrate oxidation by the enzyme. Mcllvaine's citrate-phosphate 0.1M buffer at pH 4 was used to study the influence of temperature on ABTS oxidation by the enzyme. The effect of NaCl on the enzyme activity in solution was studied at 25°C in the presence of 0.05mM SGZ in Mcllvaine's citrate-phosphate 0.1M buffer pH 7. The thermostability of the enzymes was investigated by incubating the proteins for different times in the absence of substrate in a Mcllvaine's citrate/phosphate 0.1M buffer pH 7 at 80°C and/or 37°C, followed by activity measurement, determined with 1mM of ABTS at 37°C in a Mcllvaine's citrate/phosphate 0.1M buffer pH 4 at 420nm.
Other details such as strains, plasmids, culture conditions, cloning of the genes, expression and purification of the protein can be found in supporting information.
3. Results
3.1 Expression and purification
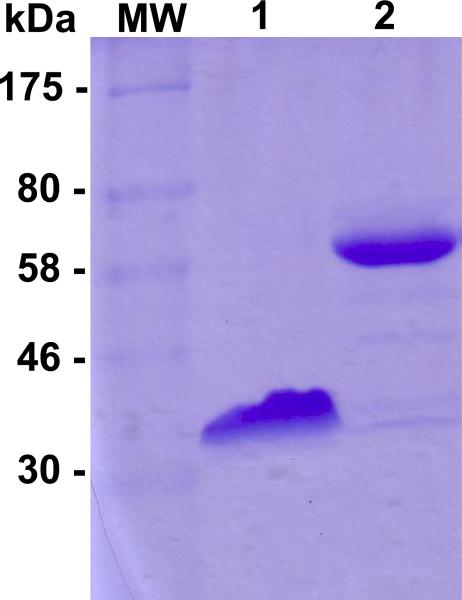
CotA from B. pumilus was identified by a similar approach to that of Reiss et. al (Fig. 1 in supp information). The CotA gene from B. pumilus was inserted into E.coli, over-expressed, and purified in a single step, yielding 15 mg of protein per batch. Figure 1 shows the migration of the purified protein in SDS-PAGE before heat treatment (line 1) and after 30 minutes at 100°C (line 2). Denaturing SDS-PAGE unfolds the enzyme and allows for estimation of the purity and the molecular weight of the enzyme. SDS is a strong anionic detergent which generally disrupts the structure of the protein in combination with a reducing agent such as 2-β-mercaptoethanol leading to the formation of a linear polypeptide chain. Thus, in SDS-PAGE, the migration relates to the overall number of amino acids in the protein. Without heat denaturation, only one band is observed corresponding to an apparent molecular weight of 35kDa. After 30 minutes of boiling, one band at 60-65kDa corresponding to the complete unfolded protein, is observed. The molecular weight of the purified enzyme was measured by mass spectrometry and was found to be 61005.9Da, corresponding to the protein truncated at a methionine at the N-terminus during over-expression in E. coli. The Cu loading, determined by the biquinoline method and by atomic absorption spectroscopy, was 3.8 Cu/enzyme, consistent with close to full occupancy of the four Cu ions. (Felsenfeld 1960)
Figure 1.
Blue Coomassie-stained SDS-PAGE gel (12%) of purified CotA from Bacillus pumilus. Lane 1: without boiling, Lane 2: with 30min of boiling.
3.2 Spectroscopic characterization
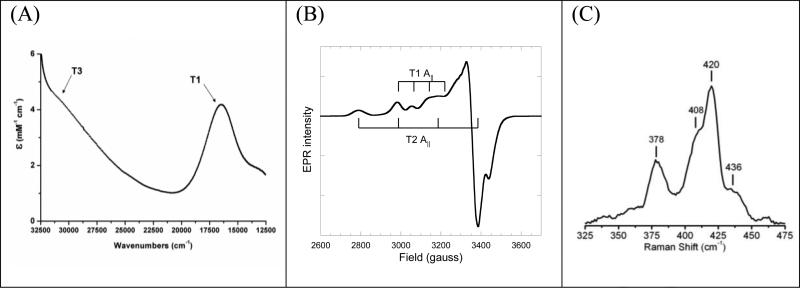
The purified enzyme from B. pumilus has the deep blue color generally observed for multicopper oxidases,consistent with the UV-visible (UVvis) spectrum of the protein, showing an intense absorption band at ~600nm (~16,700cm−1) originating from a Cys-S to T1 Cu(II) charge transfer transition (Fig. 2A). Additionally, in the UVvis spectrum a broad shoulder is observed at ~330nm (30,300cm−1), diagnostic of the antiferromagnetically coupled T3 binuclear Cu(II) pair. The absorbance ratio between 607nm and 278nm is 17 and constant for all purification batches.
Figure 2.
UV-visible (A), X-band EPR (B), and resonance raman (C) spectra of purified BOD from Bacillus pumilus
The EPR spectrum (Fig. 2B) of the purified enzyme, has contributions from two paramagnetic centers (g∥ = 2.230/ A∥ = 78×10−4cm−1, and g∥ = 2.248/ A∥ = 195×10−4cm−1), consistent with T1 and T2 Cu(II)'s, respectively (Solomon et al. 1996). Spin integration of the EPR spectrum revealed 1.8-1.9 spin/molecule, as expected based on Cu loading. The resonance Raman spectrum of the purified enzyme obtained using laser excitation at 568.2 nm, probing the strength of the Cys-S to T1 Cu(II) bond, is shown in Figure 2C. The overall appearance of the spectrum and the calculated effective vibrational frequency (obtained from the intensity weighted peak energies), 〈νCu-S〉 = 410 cm−1, are similar to previously published results for CotA from B. subtilis and BOD from M. verrucaria (Chen et al. 2010; Shimizu et al. 2003).
3.3 Biochemical and biophysical properties
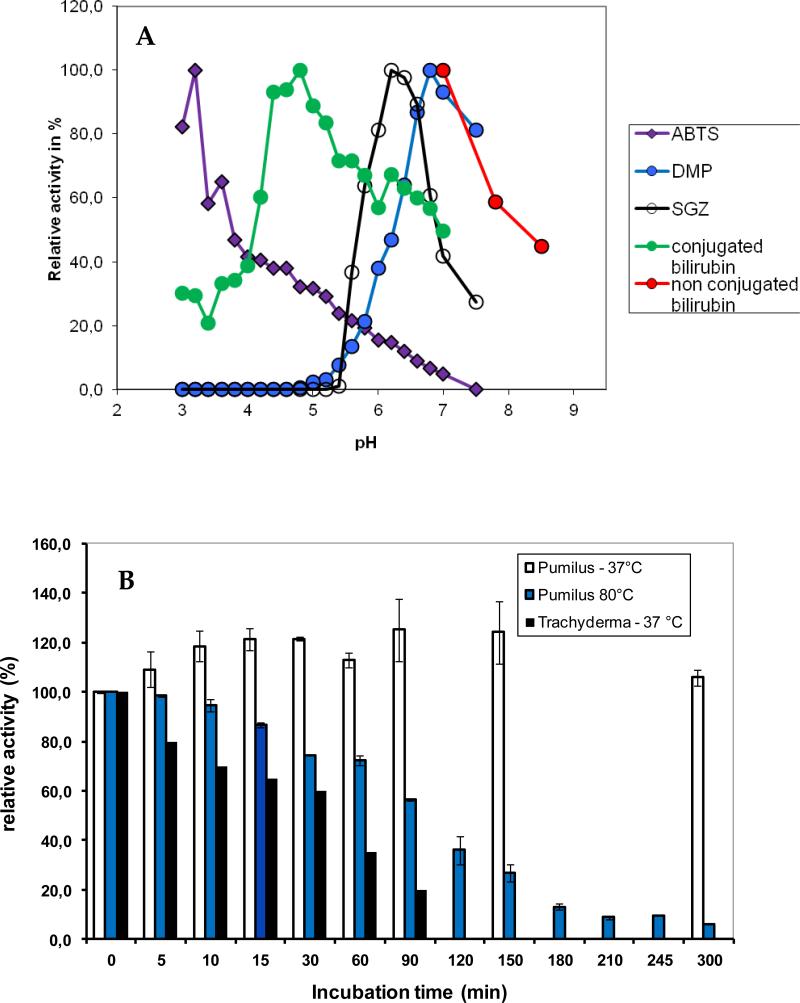
Enzyme activity was investigated using the laccase substrates ABTS, SGZ and 2,6 DMP. For each substrate, the optimal pH was determined in a Mcllvaine's citrate-phosphate 0.1M buffer with the pH varying between 2.6 and 7.5, or in Tris-H2SO4 50mM buffer for pH above 7.5 (Fig. 3). The kinetic parameters, kcat and KM, determined at 37°C and optimal pH are: 391s−1 and 31.7μM for ABTS; 116s−1 and 45μM for SGZ and 57s−1 and 822μM for 2,6-DMP.
Figure 3.
A) Dependence of enzyme activity on pH for the purified BOD from Bacillus pumilus. The activity was measured at 37°C using 1mM ABTS (purple symbols), 1mM DMP (blue squares), 0.5mM SGZ (empty circles), 100μM conjugated bilirubin (green circles) and 30μM unconjugated bilirubin (red circles) in 0.1M phosphate-citrate buffer. B) Thermostability of the purified BOD Bacillus pumilus incubated either at 37°C or 80°C in a 0.1M citrate-phosphate buffer pH 7 and of BOD from Trachyderma tsunodae incubated at 37°C in a 0.1M citrate-phosphate buffer pH 7. The enzyme activity was measured at 37°C in a 0.1M citrate-phosphate buffer pH 4 in the presence of 1mM ABTS.
In addition, activity towards conjugated and unconjugated bilirubin was evaluated. Both forms were investigated because bilirubin exists in serum either in the free unconjugated form, or in the conjugated form. (Kirihigashi et al. 2000) Activity was observed with both substrates, with kinetic constants of kcat =66.8s−1 and KM = 35.1μM and kcat =70s−1 and KM = 22μM for conjugated and unconjugated bilirubin, respectively, obtained at the optimal pH's (Fig. 3A) and 37°C. Based on this, we now classify CotA from B. pumilus as a BOD.
To evaluate the thermal deactivation of the purified BOD in solution, we measured the activity towards ABTS of enzyme pre-incubated at 37°C and 80°C, respectively, in Mcllvaine's citrate-phosphate 0.1M buffer pH 7 (Fig. 3B). For comparison, deactivation at 37°C for Trachyderma tsunodae was also evaluated. Control experiments with enzyme kept in the same buffer at 4°C, showed no decrease in catalytic activity after pre-incubation for 300min. As seen in Figure 3B, no activity decrease was observed for B. pumilus after preincubation at 37°C for 300min, whereas T. tsudonae lost more than 50% activity after 90min. At 80°C, B. pumilus activity had decreased by less than 50% at 90min.
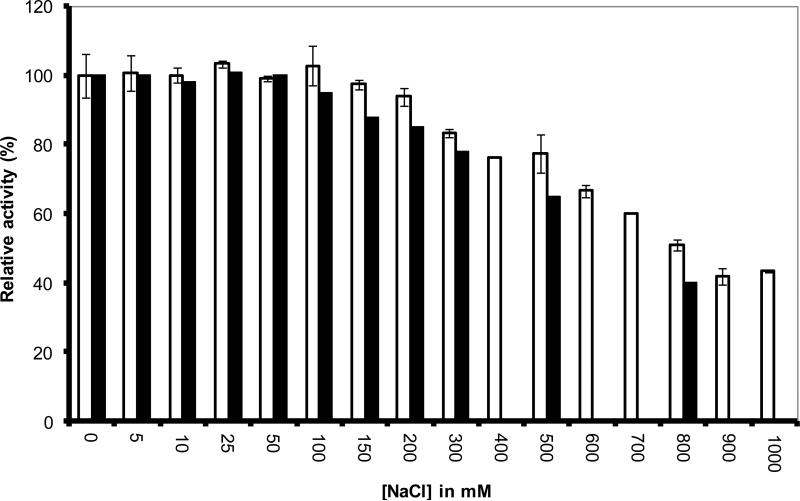
Because some MCOs are inhibited by Cl−, we evaluated the enzyme activity dependence on NaCl concentration, in the presence of 0.1mM SGZ. As seen in Figure 4, both B. pumilus and T. tsudonae showed relative high tolerance towards NaCl at concentrations up to 100mM, with inhibition at higher concentrations being more pronounced for T. tsudonae BOD.
Figure 4.
Dependence of enzyme activity on NaCl concentration for purified BOD from Bacillus pumilus (white) and BOD from Trachyderma tsunodae (black). The enzyme activity was measured at 25°C in a 0.1M citrate-phosphate buffer pH 7 in the presence of 0.05mM SGZ.
3.4 Electrochemical Properties
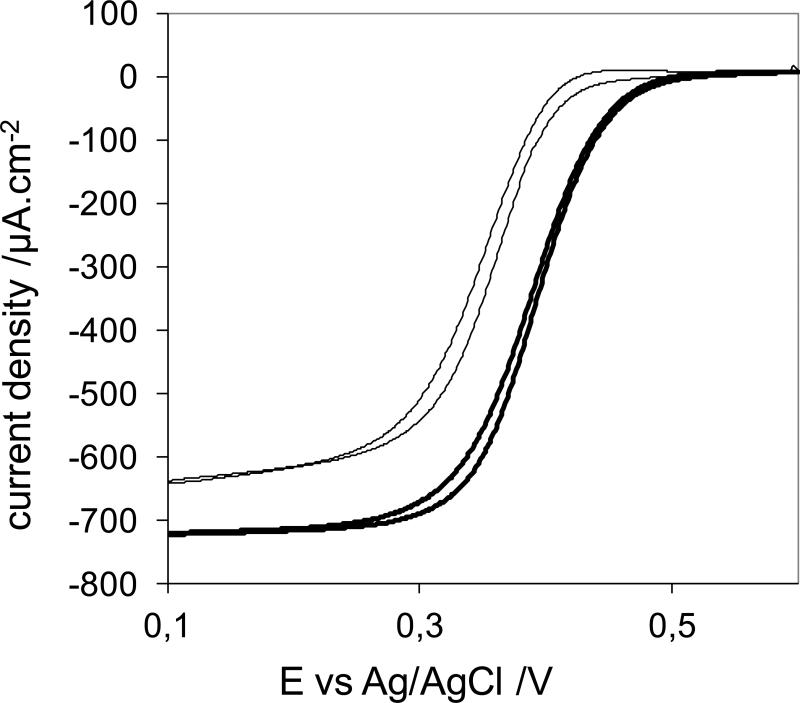
Due to the interest in utilizing MCOs in biofuel cells and biosensors, we investigated the electrochemical properties of the new BOD and compared it to those of BOD from T. tsudonae. Electrodes were prepared as previously reported (Heller 1992), where Os-redox polymers provided the electrical contact between enzyme and electrode. Figure 6 shows catalytic currents for O2 reduction on electrodes modified with B. pumilus (thin line) or T. tsunodae (thick line) BOD, respectively, each at their optimum enzyme/polymer/cross-linker ratio and same loading. As observed, the onset for O2 reduction was +0.42V for the B. pumilus modified electrode, only 0.08V lower than for T. tsunodae. At +0.1V, the current density for the electrode modified with B. pumilus was 640μAcm−2 vs. 720μA.cm−2 for T. tsunodae. At +0.35V, these values were 300μA cm−2 and 600μA.cm−2, respectively.
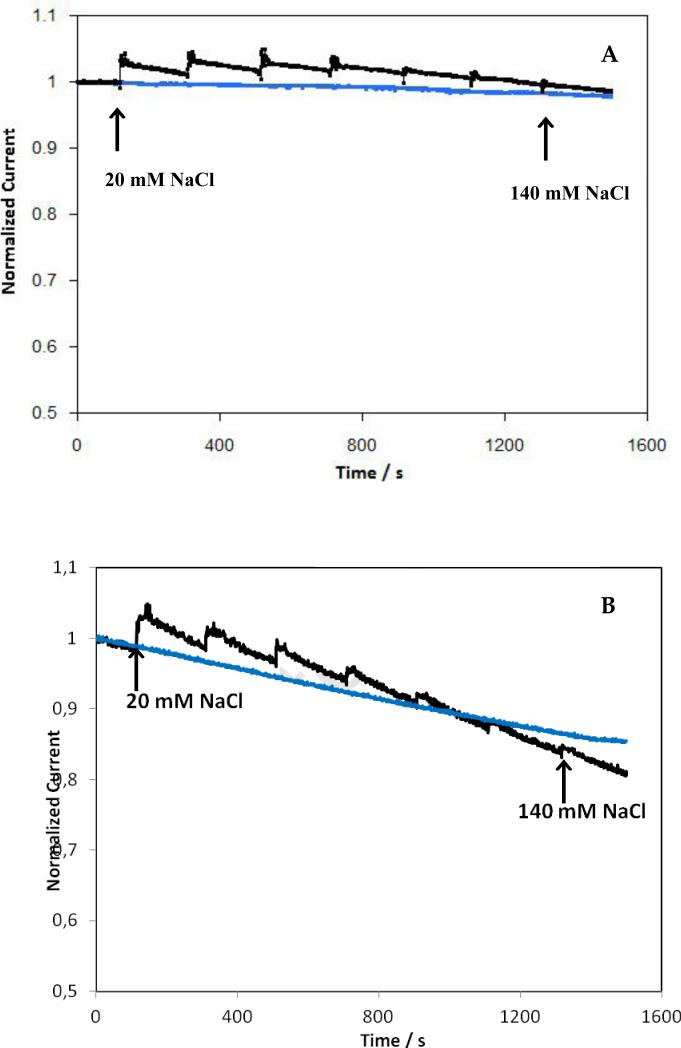
Figure 6.
Dependence of current density on time for electrodes modified with BOD from B. pumilus (A) or T. tsunodae (B) in absence (blue curves) or presence of successive additions of Na Cl (black curves). The arrows indicate the total concentration of NaCl in the cell. Electrodes modified as in Figure 4. 100mM phosphate buffer pH7.2, under O2, 37°C, 500rpm, measured at +0.1V vs Ag/AgCl.
In Figure 6 the current density as a function of time for electrodes modified with BOD from B. pumilus (A) or T. tsunodae (B) in the absence (blue lines) or presence of successive additions of NaCl (black lines), is depicted. Experiments were performed at + 0.1 V and 500 rpm in a 100 mM phosphate buffer pH 7.2 at 37° C. After 50mins of continuous operation, in the absence of chloride, the current density declined by ~2 and ~15% for the B. pumilus and T. tsunodae modified electrodes, respectively (Fig. 6, top and bottom blue traces). With subsequent NaCl additions, an increase in catalytic current was initially observed for both BODs. This behaviour is similar to that observed for laccase from Melanocarpus albomyces; explained by a possible Donnan potential effect, due to the high local concentration of ions present in the film (Kavanagh et al. 2008). The long term loss of current, due to NaCl inhibition, was evaluated by subtracting the blue trace from the black (subtracting the temperature dependent deactivation). The current density of the electrode modified with B. pumilus BOD, was almost independent of the concentration of chloride, while it declined by 6% in the presence of 140 mM Cl− for the electrode modified with T. tsunodae.
4. Discussion
High thermal stability, low sensitivity to chloride ions, and high activity at pH 7 are three of the majors conditions required for the elaboration of an efficient enzymatic cathode. To this end, BODs are attractive candidates for enzymatic cathodes in biofuel cells, because unlike laccases, they possess most of these characteristics. Earlier, Reiss et al. identified a new CotA enzyme from B. pumilus and characterized it as a bacterial laccase. (Reiss et al. 2011) We have now shown that due to its high catalytic activity with bilirubin as substrate, this enzyme should be classified as a bilirubin oxidase.
Here, we cloned the B. pumilus gene in a pET21 vector to produce the enzyme with a 6Xhis tag at its C-terminus and its over-expression in E. coli was optimized to facilitate purification under native conditions in one step. The spectroscopic features of the new BOD generally resemble those of the other MCOs including the BODs from B. subtilis and M. verrucaria (Chen et al. 2010; Shimizu et al. 2003). The features of the T1 Cu, including an intense LMCT transition in the absorption spectrum, a small parallel hyperfine splitting in the EPR spectrum, and a vibrational frequency average of 410 cm−1 in rR, are all properties of the highly covalent Cys-S – Cu bond found in T1 sites (Solomon et al. 2004). Furthermore, the 410cm−1 〈νCu-S〉 reflects the 2xHis, 1xCys, and 1xMet overall ligand set, relevant for the blue copper center of this new BOD. With respect to the T2 Cu(II), its EPR g and A parallel values are almost identical to those of B. subtilis BOD, whereas small deviations from values reported in M. verrucaria BOD, are observed (Chen et al. 2010; Shimizu et al. 2003). These results correlate with the sequence homology of the three MCOs (supp. Fig 1).
The optimal pHs, determined in this study, for oxidation of ABTS, SGZ and DMP are similar to those of other bacterial (Koschorreck et al. 2008) and fungal BODs (Pakhadnia et al. 2009). The ratios of kcat/KM's suggest, that this protein is efficient in the oxidation of ABTS substrate but less so in the oxidation of phenolic substrates. A three-dimensional structure of the T1 copper region as well as molecular modelling would be helpful in understanding these differences in substrate affinities. To this end, protein crystallization is under way.
Catalytic activity towards conjugated and unconjugated bilirubin confirmed that this enzyme should be classified as a bilirubin oxidase, the only sub-class of MCOs showing this reactivity. The kcat/Km ratio (3200 mM−1 s−1) is similar to the ratio for BOD from B. subtilis (3400 mM−1 s−1) and higher than that obtained for the BOD from M. verrucaria, expressed in native fungus (980mM−1 s−1) or in Pichia pastoris (2000 mM−1 s−1). In addition, the low KM (22μM) makes it a potential candidate for determining serum bilirubin levels in clinical samples. In addition, this bacterial BOD also shows high activity towards unconjugated bilirubin with a specific activity higher than that of B. subtilis (52U/mg versus 10U/mg).
To evaluate the electrochemical efficiency of the new BOD from B. pumilus (figure 5, thin line), we compared it to BOD from T. tsunodae (figure 5, thick line), usually considered the most efficient BOD for the reduction of O2 to water. (Mano et al. 2003) The optimal composition of the new enzyme electrode was determined in a series of experiments in which the weight percentages of the enzyme and of the redox polymer varied (data not shown). The crosslinker (PEGDGE) was fixed at 7.4 wt %, and the total loading of the three components was kept at 100 μg/cm2. The resulting optimal catalyst was composed of 30.5 wt % BOD from B. pumilus, 62.1 wt % polymer and 7.4 wt % PEDGE at 100 μg/cm2 total loading. The effect of the total loading on the kinetic limit of the catalytic current was not optimized because the optimum would vary with the ratios of the components. The observed difference in onset potentials of B. pumilus and T. tsudonae of ~80mV, observed in Figure 5, is related to the difference in axial ligation of the T1 copper. BOD from B. pumilus has a weakly coordinating methionine residue in this position, whereas in T. tsunodae, the corresponding residue is a non-coordinating phenylalanine (Supporting Information, Fig. 1). This difference has previously been shown to correspond to a ~100mV difference in the redox potential of the T1 Cu, consistent with our result. The current obtained at +100mV is only 12% smaller for the new BOD compared to that of T. tsunodae. This difference is related to the difference in T1 potentials, which determines the driving force, and thereby electron transfer rate, for electrons from the redox polymer to the enzyme. Mutations of the T1 residues are currently being pursued in order to increase the redox potential of B. pumilus BOD.
Figure 5.
O2 reduction for electrodes made with the optimal composition of catalyst, with BOD from B. pumilus (30.5 wt% BOD, 62.1 wt% redox polymer, 7.4 wt% PEGDGE, thin line) and BOD from T. tsunodae (15.4 wt% BOD, 77.2 wt% redox polymer PAA-PVI-{Os(4,4′-dichloro-2,2′-bipyridine)2Cl}+/2+, 7.4 wt% Poly(ethylene glycol) (400) diglycidyl ether, thick line), as evaluated by Cyclic voltammetry at 5mV/s under 1 atm O2, 100 mM phosphate buffer pH 7.2 at 37° C, 500 rpm.
In addition to high current density, the BOD must be thermostable at neutral pH, and in the presence of NaCl to be used as an efficient enzyme cathode. CotA proteins usually display the highest thermostability among MCOs, with half-lifes of >100min at 80°C, and it has been hypothesized that it was related to a more compact structure of the protein and/or to coppers deeply buried inside the enzymes. (Hilden et al. 2009). This hypothesis is confirmed by the SDS-PAGE that shows that 30 minutes of boiling is necessary to totally unfold the new BOD (Fig. 1). In addition, the time dependence of the current density at 37 °C for the modified electrodes (figure 6, blue traces), demonstrates that the new BOD is more stable at physiological temperatures than T. tsunodae under continuous operation, in agreement with the thermostability experiments performed in homogenous solution (Fig. 3B). These experiments demonstrate that this new BOD is a promising enzyme for the elaboration of biofuel cells. No attempt was made to increase the stability of the immobilized enzymes by employing, for examples, nanotube fibers instead of bare glassy carbon electrodes. (Gao et al. 2010)
Finally, the new BOD showed a higher tolerance towards Cl− compared to T. tsudonae, when the enzymes were immobilized in redox polymers. Our electrochemical experiments (Fig 6, black traces) are in agreement with those obtained in homogeneous solution (Fig. 4), demonstrating that this new BOD, in contrast to many laccases, is almost insensitive to chloride.
5. Conclusion
A CotA enzyme from Bacillus pumilus, previously identified as a laccase, has been studied and characterized as a new bacterial Bilirubin Oxidase. Because of its high thermal stability, its low sensitivity to chloride ions, and its high activity at pH 7, this new BOD is an excellent candidate for the elaboration of efficient biofuel cells and bilirubin biosensors operating under physiological conditions.
Supplementary Material
Highlights.
We identified a new bacterial Bilirubin oxidase from Bacillus Pumilus
We present a full characterization of this enzyme in solution and on modified electrodes
The current for O2 reduction is 68% higher than for Myrothecium Verrucaria
The modified electrode is 400 % more stable under continuous operation than other BOD
Acknowledgment
This work was supported by a European Young Investigator Award (EURYI), la Région Aquitaine, a France-Stanford grant, and by National Institutes of Health Grant DK-31450.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barton SC, Gallaway J, Atanassov P. Chem. Rev. 2004;104:4867–4886. doi: 10.1021/cr020719k. [DOI] [PubMed] [Google Scholar]
- Beyl Y, Guschin DA, Shleev S, Schuhmann W. Electrochem. Comm. 2011;13:474–476. [Google Scholar]
- Chen Z, Durao P, Silva CS, Pereira MM, Todorovic S, Hildebrandt P, Bento I, Lindley PF, Martins LO. Dalton Trans. 2010;39(11):2875–2882. doi: 10.1039/b922734b. [DOI] [PubMed] [Google Scholar]
- Doumas BT, Yein f., Perry B, Jendrzejczak B, Kessner A. Clin. Chem. 1999;45(8):1255–1260. [PubMed] [Google Scholar]
- Felsenfeld G. Arch Biochem Biophys. 1960;87:247–251. doi: 10.1016/0003-9861(60)90168-5. [DOI] [PubMed] [Google Scholar]
- Gallaway J, Barton SC. J Am Chem Soc. 2008;130:8527–8536. doi: 10.1021/ja0781543. [DOI] [PubMed] [Google Scholar]
- Gallaway J, Wheeldon I, Rincon R, Atanassov P, Banta S, Barton SC. Biosens Bioelectron. 2008:1229–1235. doi: 10.1016/j.bios.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Gao F, Viry L, Maugey M, Poulin P, Mano N. Nat Commun. 2010;1(1) doi: 10.1038/ncomms1000. 10.1038/ncomms1000. [DOI] [PubMed] [Google Scholar]
- Guo J, Liang XX, Mo PS, Li GX. Appl Biochem Biotechnol. 1991;31(2):135–143. doi: 10.1007/BF02921784. [DOI] [PubMed] [Google Scholar]
- Gupta S, Lau C, Rajendran A, Colon F, Branch B, Ivnitski D, Atanassov P. Electrochem. Comm. 2011;13:247–249. [Google Scholar]
- Heller A. J Phys. Chem.B. 1992;96:3579–3587. [Google Scholar]
- Heller A. Phys. Chem. Chem. Phys. 2004;6:209–216. [Google Scholar]
- Hilden K, Hakala TK, Lundell T. Biotechnol Lett. 2009;31(8):1117–1128. doi: 10.1007/s10529-009-9998-0. [DOI] [PubMed] [Google Scholar]
- Hiromi K, Yamaguchi S, Sugiura Y, Iwamoto H, Hirose J. Biosci. Biotech. Biochem. 1992;56:1349–1350. [Google Scholar]
- Hussein L, Rubenwolf S, von Stetten F, Urban G, Zengerle R, Krueger M, Kerzenmacher S. Biosens Bioelectron. 2011;26:4133–4138. doi: 10.1016/j.bios.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Kavanagh P, Jenkins P, Leech D. Electrochem. Comm. 2008;10:970–972. [Google Scholar]
- Kirihigashi K, Tatsumi N, Hino M, Yamane T, Ohta K. Osaka City Med J. 2000;46(1):55–70. [PubMed] [Google Scholar]
- Kosaka A, Yamamoto A, Morishita Y, Nakane K. Clin. Biochem. 1987;20:451–458. doi: 10.1016/0009-9120(87)90014-2. [DOI] [PubMed] [Google Scholar]
- Koschorreck K, Richter SM, Ene AB, Roduner E, Schmid RD, Urlacher VB. Appl Microbiol Biotechnol. 2008;79(2):217–224. doi: 10.1007/s00253-008-1417-2. [DOI] [PubMed] [Google Scholar]
- Kurosakaa K, Senbaa S, Tsubotab T, Kondoa H. Clinica Chimica Acta. 1998;269:125–136. doi: 10.1016/s0009-8981(97)00194-0. [DOI] [PubMed] [Google Scholar]
- Mano N. Chem Commun (Camb) 2008:2221–2223. doi: 10.1039/b801786g. [DOI] [PubMed] [Google Scholar]
- Mano N, Fernandez JL, Kim Y, Shin W, Bard AJ, Heller A. J. Am. Chem. Soc. 2003;125:15290–15291. doi: 10.1021/ja038285d. [DOI] [PubMed] [Google Scholar]
- Osman MH, Shah AA, Walsh FC. Biosens Bioelectron. 2011;26:3087–3102. doi: 10.1016/j.bios.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Pakhadnia YG, Malinouski NI, Lapko AG. Biochemistry (Mosc) 2009;74(9):1027–1034. doi: 10.1134/s0006297909090119. [DOI] [PubMed] [Google Scholar]
- Ramirez P, Mano N, Andreu R, Ruzgas T, Heller A, Gorton L, Shleev S. Biochimica et Biophysica Acta. 2008;1777:1364–1369. doi: 10.1016/j.bbabio.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Reiss R, Ihssen J, Thony-Meyer L. BMC Biotechnol. 2011;11(1):9. doi: 10.1186/1472-6750-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengaraj S, Kavanagh P, Leech D. Biosens Bioelectron. 2011a;30:294–299. doi: 10.1016/j.bios.2011.09.032. [DOI] [PubMed] [Google Scholar]
- Rengaraj S, Mani V, Kavanagh P, Rusling J, Leech D. Chem. Commun. 2011b;47:11861–11863. doi: 10.1039/c1cc15002b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón RC, Lau C, Luckarift HR, Garcia KE, Adkins E, Johnson GR, Atanassov P. Biosens Bioelectron. 2011;27:132–136. doi: 10.1016/j.bios.2011.06.029. [DOI] [PubMed] [Google Scholar]
- Shimizu A, Samejima T, Hirota S, Yamaguchi S, Sakurai N, Sakurai T. J Biochem. 2003;133(6):767–772. doi: 10.1093/jb/mvg098. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Sundaram UM, Machonkin TE. Chem. Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Szilagyi RK, DeBeer George S, Basumallick L. Chem Rev. 2004;104(2):419–458. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- Tsujimura S, Nakagawa T, kano K, Ikeda T. Electrochemistry. 2004;72(6):437–439. [Google Scholar]
- Wang X, Falk MC, Ortiz R, Matsumura H, Boback J, Ludwig R, Bergelin M, Gorton L, Shleev S. Biosens Bioelectron. 2012;31:219–225. doi: 10.1016/j.bios.2011.10.020. [DOI] [PubMed] [Google Scholar]
- Zloczewska A, Jonsson-Niedziolka M, Rogalski J, Opallo M. Electrochim. Acta. 2011;56:3947–3953. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.