Abstract
The incorporation of potentially catalytic groups into DNA is of interest for the in vitro selection of novel deoxyribozymes. We have devised synthetic routes to a series of three C7 modified 7-deaza-dATP derivatives with pendant aminopropyl, Z-aminopropenyl and aminopropynyl side chains. These modified triphosphates have been tested as substrates for Taq polymerase during PCR. All the modifications are tolerated by this enzyme, with the aminopropynyl side chain giving the best result. Most protein enzymes have more than one type of catalytic group located in their active site. By using C5-imidazolyl-modified dUTPs together with 3-(aminopropynyl)-7-deaza-dATP in place of the natural nucleotides dTTP and dATP, we have demonstrated the simultaneous incorporation of both amino and imidazolyl moieties into a DNA molecule during PCR. The PCR product containing the four natural bases was fully digested by XbaI, while PCR products containing the modified 7-deaza-dATP analogues were not cleaved. Direct evidence for the simultaneous incorporation during PCR of an imidazole-modified dUTP and an amino-modified 7-deaza-dATP has been obtained using mass spectrometry.
INTRODUCTION
In vitro selection techniques have been used to select nucleic acid catalysts for a variety of reactions which include several phosphoryl transfer reactions (1–5), amide bond formation (6–9) and a Diels–Alder reaction (10). The technique is essentially a combinatorial chemistry methodology in which functional nucleic acids are isolated from random libraries of single-stranded DNA or RNA. However, most ribozymes and deoxyribozymes isolated in this manner do not approach the turnover numbers normally associated with protein catalysts.
Recently there has been interest in the potential of adding extra protein-like functionality to nucleic acids by incorporation of modified triphosphates during PCR amplification, in the case of selection for DNA catalysts, and during transcription in the RNA case. In these experiments, a modified nucleoside triphosphate completely replaces its unmodified counterpart. A variety of functional groups have been conjugated to the nucleoside triphosphates, but particular attention has been paid to the provision of primary amino and imidazolyl residues as these mimic the lysine and histidine residues which are often intimately involved in catalysis in protein enzymes.
Initial work centred on the use of modified ribonucleoside triphosphates that can be used for the selection of RNA catalysts. (8,10–16). However, the greater chemical and enzymatic stability of DNA compared to RNA makes it a more attractive scaffold for the generation of functional catalysts. Thus, there has been recent interest in the preparation and evaluation of modified 2′-deoxynucleoside triphosphates. Sakthivel and Barbas (17) have described a range of conjugates of 5-(E-3-aminopropenyl)-2′-deoxyuridine triphosphate which have been demonstrated to be substrates for DNA polymerases typically used in PCR. The triphosphate of 5-(3-aminopropynyl)-2′-deoxyuridine is a substrate for DNA polymerases (18). We have compared the substrate properties of a number of 5-modified-2′−deoxyuridine triphosphates and found that 5-(3-aminopropynyl)dUTP and 5-(E-3-aminopropenyl)dUTP and their imidazole-4-acetic acid and urocanic acid-modified conjugates were substrates in a PCR using Taq polymerase (6). However, C5-amino-modified dUTPs with alkane or Z-alkene linkers and their corresponding conjugates were not substrates.
In contrast to the modified triphosphates of 2′-deoxyuridine, little attention has been paid to the synthesis and evaluation of the substrate properties of modified purine 2′-deoxynucleoside triphosphates in the context of SELEX experiments. Indeed, only a single report of an 8-modified dATP derivative has appeared in the literature (19). Here we describe the synthesis of three amino-modified 7-deazaadenosine derivatives, with linker arms of differing flexibility and assess these analogues as substrates during PCR.
Typically the active sites of protein enzymes contain more than one type of amino acid. Imidazole residues and amino groups are a particular combination seen in the active sites of enzymes catalysing such diverse reactions as RNA cleavage (RNase) and the aldol reaction (Class I aldolases). To ensure that faithful amplification during the selection procedure takes place and that primary structure determination of the resultant catalyst is possible, a single nucleoside triphosphate must replace a single natural counterpart. Thus, in order to introduce two functionalities, such as an imidazole and an amino group into DNA, two of the natural nucleotides must be replaced with modified triphosphates. Hélène and co-workers (19) have demonstrated the simultaneous incorporation of 5-(E-3-aminopropenyl)-2′-deoxyuridine triphosphate (replacing dTTP) and 8-(2-(4-imidazolyl)ethylamino)-2′-deoxyadenosine (replacing dATP) using modified T7 DNA polymerase (Sequenase). However, the analogues were incorporated with reduced efficiency compared to their natural counterparts and no report was made of the simultaneous incorporation of both during PCR (19). Therefore, in any SELEX strategy using this combination of analogues, a PCR amplification step with normal nucleotides would have to follow selection and then the modifications would require re-introduction by a Sequenase-mediated DNA polymerisation. A more attractive strategy is a direct PCR amplification using nucleotide analogues. Here we report the direct PCR-mediated synthesis of DNA containing both imidazolyl and amino functionalities as the result of the simultaneous incorporation of modified 2′-deoxyuridine and 7-deaza-2′-deoxyadenosine triphosphates by Taq polymerase.
MATERIALS AND METHODS
Chemicals and reagents
AnalaR glacial acetic acid and tetra-sodium pyrophosphate decahydrate were from BDH. Chemicals/reagents from Aldrich: Anhydrous DMF, phosphoryl chloride (phosphorus oxychloride), proton sponge® (1,8-bisdimethylamino naphthalene), tri-n-butylamine (99+%), triethylamine (99%), trimethylphosphate (99+%), 3 Å molecular sieves. The following resins were used: Dowex® 50Wx8 in the H+ form (100–200 mesh), Dowex® AG1X8 from BDH, Sephadex®–DEAE A-25, ion exchange resin (40–120 micron) from Aldrich. Triethylamine was refluxed and distilled from potassium hydroxide pellets before use. Acetonitrile was dried by reflux and distillation from calcium hydride.
1H NMR (250.134 MHz) and 31P NMR spectra (101.256 MHz) were obtained on a Bruker AC250 and are given in p.p.m. relative to external standards of tetramethyl silane and 85% phosphoric acid, respectively.
4-Chloro-5-iodo-7H-pyrrolo[2,3-d]pyrimidine (3). 4-Chloropyrrolo[2,3-d]pyrimidine (20) (2.50 g, 16.3 mmol) was stirred with N-iodosuccinimide (4.10 g, 18.3 mmol) in dry N,N-dimethylformamide (DMF) (160 ml) at room temperature for 3 days. The solution was then evaporated, and the residue partitioned between ethyl acetate (500 ml) and saturated aqueous sodium bicarbonate solution (50 ml). The organic layer was dried (MgSO4), evaporated and the pure product obtained as a white solid (4.10 g, 14.7 mmol, 90%) following silica gel column chromatography (6% MeOH/CH2Cl2). The material gave identical analytical data to that obtained from reaction with N-iodosuccinimide in CH2Cl2 (21).
4-Chloro-7-(2-deoxy-3,5-di-O-(4-toluoyl)-β-D-erythro-pentofuranosyl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidine (4). 4-Chloro-5-iodopyrrolo[2,3-d]pyrimidine (3) (21) (300 mg, 1.07 mmol) and was suspended in dry acetonitrile (25 ml) and stirred at room temperature for 30 min, with sodium hydride (60% in oil, 48 mg, 1.2 mmol) under argon. After this time, a clear solution had formed and 1-chloro-2-deoxy-3,5-di-O-p-toluoyl-α-d-erythro-pentofuranose (22) (583 mg, 1.50 mmol) was added in three portions over 20 min. The solution was then stirred overnight, evaporated and the resulting oil partitioned between dichloromethane (300 ml) and water (50 ml). The organic layer was dried (MgSO4), evaporated and the crude product purified by silica gel column chromatography (ethyl acetate/hexane 85:15, v/v) to give the desired product as a white foam. Recrystallisation from isopropanol gave white needles (477 mg, 0.76 mmol, 71%), which had identical analytical data to that described for 4 from alternative synthetic routes (23,24).
4-Amino-7-(2-deoxy-β-d-erythro-pentofuranosyl)-5-iodo-7H-pyrrolo[2,3-d]pyrimidine (5). Compound 4 (1.20 g, 1.89 mmol) was heated in saturated ammonia in methanol (60 ml) at 65°C for 4 days in a glass pressure bottle. The solution was then cooled to room temperature and the solvent evaporated. The crude product was adsorbed onto silica and purified by column chromatography (10% MeOH/CH2Cl2) to give a white foam (627 mg, 1.67 mmol, 88%) which gave identical analytical data to that described for 5 when alternative ammonolysis strategies are employed (23–25).
4-Amino-7-(2-deoxy-β-d-erythro-pentofuranosyl)-5-[3-(trifluoroacetamido)-Z-prop-1-enyl]-7H-pyrrolo[2,3-d]pyrimidine (7). Compound 6 (26) (70 mg, 0.175 mmol) was dissolved in anhydrous methanol (8 ml) and cooled to –78°C. Nickel (II) chloride (45.8 mg, 0.19 mmol) was then added as a solution in anhydrous methanol (2 ml). Sodium borohydride (7.3 mg, 0.19 mmol) was then added portionwise over 2 min. The reaction mixture was then stirred at –78°C for 30 min and allowed to warm slowly to room temperature until the solution turned black, whereupon silica (100 mg) was added immediately and the reaction mixture concentrated. Purification by silica chromatography (5% MeOH in ethyl acetate) gave the desired compound as a white foam (40.6 mg, 0.102 mmol, 58%).
Rf (ethyl acetate/MeOH, 9/1), 0.2; δH([2H6]-DMSO) 9.78 (1 H, t, NH), 8.11 (1 H, s, 2-H), 7.46 (1 H, s, 6-H), 6.91 (3 H, m, CH, NH2), 6.57 (1 H, dd, 1′-H, J 6.1/7.94), 5.58 (1 H, m, CH), 5.31 (1 H, Br s, OH), 4.4 (1 H, Br s, 3′-H), 4.12 (2 H, m, CH2), 3.85 (1 H, dd, 4′-H, J 4.27/6.71), 3.57 (2 H, m, 2×5′-H), 2.57 (1 H, m, 2′a-H), 2.21 (1 H, m, 2′b-H). ). δC([2H6]-DMSO) 157.8 (C4), 151.3 (C2), 150.4 (C7a), 126.7 (CH Alkene), 122.8 (CH Alkene), 121.9 (C6), 111.3 (C5), 101.8 (C4a), 87.9 (C′4), 83.7 (C′1), 71.4 (C′3), 62.4 (C′5) (Note: signals due to CF3CO are not assigned due to their very low intensities). FAB+ MS m/z 402 [8% (M+H)+]. Calc. 402.138914, found 402.137726.
4-Amino-7-(2-deoxy-β-d-erythro-pentofuranosyl)-5-[3-(trifluoroacetamido)propyl]-7H-pyrrolo[2,3-d]pyrimidine (8). Compound 6 (26) (50 mg, 0.125 mmol) was dissolved in anhydrous methanol (20 ml) and hydrogenated for 1 h over PtO2. Silica TLC (10% MeOH in dichloromethane) indicated the reaction to be complete and the mixture was then filtered through celite and chromatographed on silica gel (14% MeOH/CH2Cl2) to give the product as a white foam (50 mg, 0.125 mmol, 99% yield).
Rf (CH2Cl2/MeOH, 9/1), 0.23; δH([2H6]-DMSO) 9.62 (1 H, t, NH), 8.20 (1 H, s, 2-H), 7.30 (1 H, s, 8-H), 6.79 (2 H, m, NH2), 6.66 (1 H, dd, 1′-H, J 5.79/8.24), 5.42 (1 H, d, OH), 5.26 (1 H, Br s, OH), 4.49 (1 H, Br s, 3′-H), 3.96 (1 H, m, 4′-H), 3.68 (2 H, m, 2×5′-H), 3.39 (2 H, m, CH2), 2.93 (2 H, m, CH2), 2.63 (1 H, m, 2′a-H), 2.29 (1 H, m, 2′b-H), 1.95 (2 H, m, CH2). δC([2H6]-DMSO) 158.0 (C4), 151.8 (C2), 150.9 (C7a), 119.3 (C6), 114.7 (C5), 102.5 (C4a), 87.6 (C′4), 83.2 (C′1), 71.6 (C′3), 62.6 (C′5), 29.9 (CH2), 23.6 (CH2) (Note: signals due to CF3CO are not assigned due to their very low intensities, C′2 and CH2 are hidden by DMSO). FAB+ MS m/z 404 [100% (M+H)+]. Calc. 404.154564, found 404.151950.
Nucleoside triphosphates (9–11). [The nucleoside triphosphates 12 and 13 were synthesised as described (6,17)]. A typical reaction is shown below. Modified nucleoside 6 (100 mg, 0.25 mmol) was dried overnight over P2O5 and then dissolved in anhydrous trimethylphosphate (0.6 ml) under argon. Proton sponge (64 mg, 0.30 mmol, also dried overnight over P2O5) was added to the solution and dissolved with gentle heating. The reaction mixture was then cooled to 0°C and freshly distilled POCl3 (35 µl, 0.33 mmol) was added dropwise to the stirred solution via a syringe. The reaction mixture was stirred for 2 h, removed from the ice bath and a well vortexed mixture of 4 equivalents, bis tri-n-butylammonium pyrophosphate (2.0 ml of a 0.5 M solution in DMF) and tri-n-butylamine (0.22 ml), was added in one portion. The reaction mixture was stirred at room temperature for 15 min, and then 20 ml 0.1 M triethylammonium bicarbonate solution (TEAB) was added. The reaction mixture was then stirred at room temperature for a further 1 h. Concentrated aqueous ammonia solution (20 ml) was then added to the reaction mixture, which was stirred at room temperature overnight. The solution was concentrated to remove the ammonia and the resultant solution purified by ion-exchange chromatography on DEAE–Sephadex A-25 using a linear gradient of TEAB (0.05–0.70 M pH 8, 2 l each). The triphosphates 9–11 eluted at the concentrations of TEAB buffer indicated below. Further purification was by preparative reversed phase HPLC on an SB-C18 Zorbax 250 × 21.2 mm column (Anachem) using the following gradient: buffer A (0.1 M TEAB pH 7.5), buffer B (0.1 M TEAB pH 7.5, 30% acetonitrile), using the gradient t = 0 min 5% B, t = 5 min 5% B, t = 25 min 60% B. Yields were as follows: 6 (0.25 mmol) gave 295 OD280nm of 9; 7 (0.10 mmol) gave 21 OD280nm of 10; 8 (0.10 mmol) gave 190 OD277nm of 11.
31P NMR (D2O) 9 –5.9 (d, γP), –10.6 (d, αP), –21.6 (t, βP); 10 –5.9 (d, γP), –10.5 (d, αP), –21.3 (t, βP); 11 –5.6 (d, γP), –10.6 (d, αP), –21.5 (t, βP), ES-MS 9 542 (100% [M–H]–); 10 544 (38% [M–H]–); 11 546 (100% [M–H]–), retention time on DEAE–Sephadex 9 0.35–0.45 mM TEAB; 10 0.45–0.55 mM TEAB; 11 0.35–0.40 mM TEAB, retention time on RP-HPLC 9 20.9 min; 10 21.2 min; 11, 21.0 min, λmax 9 280 nM; 10 280 nM; 11 277 nM.
PCR amplification
A typical PCR contained between 100 pg and 10 ng of DNA template [pUC19 (Pharmacia) or pLitmus28 (New England Biolabs)], 0.5 µM each of the forward and reverse primers, 1 µl Taq polymerase (1–5 U; Promega), 5 µl 10× magnesium-free polymerase buffer (Promega), 2 mM MgCl2, 100 µM unmodified dNTPs [control reactions dATP, dCTP, dGTP and dTTP (MBI Fermentas); reactions containing modified C7-dATP triphosphates dTTP, dCTP, dGTP only; reactions containing modified dUTP triphosphates dATP, dCTP, dGTP only; reactions containing modified C7-dATP and modified dUTP triphosphates dCTP, dGTP only], 500 µM modified dUTP, 500 µM modified C7-dATP or 500 µM each modified dUTP and modified C7-dATP (omitted in control reactions) in a 50 µl reaction. For all reactions, a hot start (3 min at 95°C, addition of MgCl2, followed by a further 3 min at 95°C) was used, followed by 30 cycles of amplification (1 min at 95°C, 1 min at 52°C, 2 min at 72°C) and a final incubation for 10 min at 72°C. The primer sequences used were as follows for a 98mer product using pUC19 template: forward primer-1 d(GTA AAA CGA CGG CCA GT); reverse primer d(AAC AGC TAT GAC CAT GA). A shorter PCR product from pUC19 (62mer), was formed using the above reverse primer and the following forward primer: forward primer-2 d(GGG GAT CCT CTA GAG TCG). The PCR products were separated by denaturing 12% (100mer), 14% (62mer) PAGE containing 7 M urea. The products were visualised following ethidium bromide staining by transillumination and the image recorded with a Ultra-Violet Products CCD camera and the Labworks analysis software.
Restriction enzyme digest of PCR products
PCR products formed using modified C7-dATP derivatives and control reactions were ethanol precipitated and digested according to the manufacturer’s protocols. The DNA was incubated with XbaI (Promega) in 6 mM Tris–HCl pH 7.9, 150 mM NaCl, 1 mM DTT for 1.5 h at 37°C for DNA containing natural nucleotides and for 1.5 h at 37°C and then overnight at room temperature for DNA containing modified nucleotides. Alternatively the DNA was incubated with SmaI (Promega) in 10 mM Tris–HCl pH 7.0, 7 mM MgCl2, 50 mM KCl, 1 mM DTT for 1.5 h at 25°C for DNA containing natural nucleotides and for 1.5 h at 25°C and then overnight at room temperature for DNA containing modified nucleotides. Restriction digests were visualised by gel electrophoresis as described above.
Mass spectrometry of PCR products
Shorter PCR products (62/63mer) were obtained from pUC19 using forward primer-2 and biotinylated reverse primer. The products of 50 × 100 µl PCRs (each using 200 pg DNA template and 5 U enzyme) were combined, extracted by phenol/chloroform/ether, desalted on NAP-10 columns (Pharmacia) and then precipitated with 10 vol ethanol from 0.75 M ammonium acetate solution. DNA precipitates were dissolved in 200 µl capture buffer (20 mM Tris–HCl pH 7.5, 2 M NaCl, 1 mm EDTA) and captured on magnetic streptavidin beads (Sigma) which were previously washed three times with the same buffer. Beads were shaken at 25°C for 15 min, washed three times with capture buffer (20 mM Tris–HCl pH 7.5, 2 M NaCl, 1 mM EDTA) and eluted by 3 × 200 µl 0.2 M NaOH. Eluates were combined, neutralised by addition of 0.5 vol 7.5 M ammonium acetate, desalted on NAP-10 column and desalted by microdialysis (Millipore microcon YM-10). Single-stranded PCR products were dissolved in water (5–10 µl) and used for mass spectrometry.
Mass spectrometry was performed using 1 µl of a solution containing single-stranded PCR product (approximate concentration 20 µM) mixed in a 1:1 ratio of matrix (3-hydroxypicolinic acid 25 mg/ml) on a Bruker REFLEX III MALDI-TOF instrument. Dowex 50 W (NH4+) beads were used to desalt the oligomers prior to analysis. A standard of trypsinogen (23982 [M+H]+ and 11991 [M+2H]2+) was used to calibrate the instrument. Modified 62 nt fragment d(GGG GA*T CC CTA* GA*G TCG A*CC U*GC A*GG CA*U* GCA* A*GC U*U*G GCG U*A*A* U*CA* U*GG U*CA* U*A*G CU*G U*U*), U* = using modified dUTP 13 in PCR; A* = using modified 7-deazadATP 9 in PCR. Calc. MW (62mer) = 22433, found 22480 (difference from calculated MW: +47, 0.2%). (Note: for the unmodified sequence, Taq polymerase adds an extra dA to the 3′-end (6). The 63 nt sequence has a mass of 19494). Mass spectrum is available as Supplementary Material.
RESULTS AND DISCUSSION
Design and synthesis of modified triphosphates
Primary amino groups from lysine residues and imidazolyl groups from histidine residues play both important structural and catalytic roles in protein enzymes. In contrast, nucleic acids lack the functional group repertoire normally associated with proteins. To mimic the role of lysine and histidine residues, it is necessary to incorporate two separate 2′-deoxyribonucleoside triphosphates bearing either a primary amino group or an imidazolyl residue during PCR. In the pyrimidine series, C5 is the choice for attachment of the required functionality (6,17,18). The most attractive position for modification of the purine nucleobase appears to be at the 7-position, located in double helical DNA in the major groove. Seela and Thomas (27) studied the stability of duplexes containing 7-substituted 7-deaza-2′-deoxyadenosines and concluded that the addition of functionality to this position has a stabilising effect. In contrast, the addition of functionality at C8 is destabilising in duplex DNA (28) and is known to alter the conformation of the nucleobase with respect to the sugar from anti to syn. Furthermore, it is known that 8-substituted 7-deazaguanines within alternating CG sequences can promote the B→Z transition (29). Thus, the addition of pendant groups at the 7-position should not interfere with duplex formation during PCR. Modification of a naturally occurring purine nucleoside at N7 would give rise to a positively charged nucleosidic derivative, with a highly labile glycosidic bond. To circumvent this problem, we have therefore chosen to modify the 7-deaza derivative of 2′-deoxyadenosine.
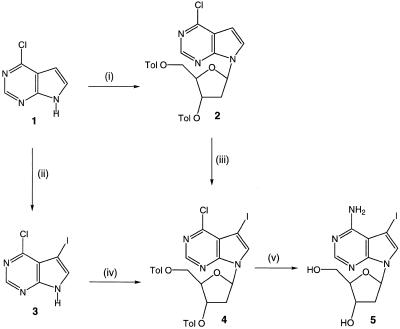
In previous work, we found that the flexibility of the linker arm between the amine functionality and the C5-position of dUTP strongly affects the substrate properties of the analogues for Taq polymerase. We assumed that the same trend could not be extended to 7-deaza-dATP analogues and therefore synthesised such modified nucleotides bearing alkyne, alkene and alkane linkers. The propargylamino modified 7-deaza-2′-deoxyadenosine, 6, was synthesised from 7-deaza-7-iodo-2′-deoxyadenosine, 5, by a palladium-catalysed coupling reaction according to the method of Seela and Zulauf (26) (Figs 1 and 2). The iodo compound 5 was prepared from 4 by reaction with methanolic ammonia at 65°C for 4 days in 88% yield (Fig. 1). We found this to give a superior yield to the ammonolyses at elevated temperatures which have been reported previously (23–25). Compound 4 was synthesised from 1 (30) either via 2 (23) or 3 (24). Compound 3 was formed from 1 in 90% yield when the iodination solvent was changed from dichloromethane (21) to DMF. Of the two literature routes to 4, we found the latter method, i.e. glycosylation of 3, to be the most efficient. In this case, however, we used the sodium salt glycosylation methodology (20) in preference to the procedure described by Seela and Zulauf (24), which afforded 4 in good yield (71%) after column chromatography and recrystallisation.
Figure 1.
The synthesis of 7-deaza-7-iodo-2′-deoxyadenosine. (i) NaH, CH3CN, then 1-chloro-2-deoxy-3,5-di-O-p-toluoyl-α-d-erythro-pentofuranose, room temperature, 71% (20); (ii) N-iodosuccinimide, DMF, room temperature, 3 days, 90%; (iii) N-iodosuccinimide, DMF, room temperature, 71%; (iv) NaH, CH3CN, then 1-chloro-2-deoxy-3,5-di-O-p-toluoyl-α-d-erythro-pentofuranose room temperature, 71%; (v) NH3/MeOH, 70°C, 4 days, 88%.
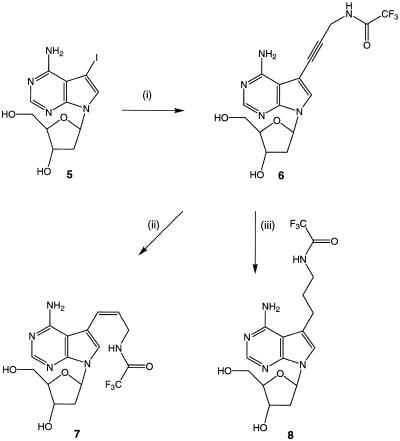
Figure 2.
The synthesis of protected C7-amino-modified 7-deaza-2′-deoxyadenosines from 5-iodo-2′-deoxyadenosine. (i) N-propynyltrifluoracetamide, Pd(PPh3)4, CuI, Et3N, DMF, 60% yield; (ii) NiCl2/NaBH4, MeOH, –78°C, 58% yield; (iii) PtO2, H2, MeOH, 98%.
The Z-alkene, 7, was obtained from 6 in a 58% yield using the NiCl2/NaBH4 system that we have previously employed for the synthesis of 5-(Z)-[3-(trifluoroacetamido)propenyl]-2′-deoxyuridine (31) (Fig. 2). Although reduction of the alkyne nucleoside 6 to the corresponding alkane, 8, could be achieved in 50% yield using the same reducing agent, we found that catalytic hydrogenation using a PtO2 catalyst was essentially quantitative.
We were unable to prepare the E-alkene amino analogue of 7 from 5. The palladium-catalysed coupling reaction previously employed in the synthesis of 5-(E)-[3-(trifluoroacetamido)propenyl]-2′-deoxyuridine (17) which uses sodium tetrachloropalladate in aqueous buffered DMF solution afforded unidentified polar material. Likewise, the method of Seela and Zulauf (24) employing Pd(Ph3P)4 in the Heck reaction, used previously for the synthesis of the methyl acrylate derivative of 7-deaza-2′-deoxyadenosine was also unsuccessful. In this case, the starting material was returned unchanged after the reaction. Methyl acrylate and other electron-deficient alkenes are known to be the most reactive alkenes in the Heck reaction. We therefore assume that 3-(trifluoroacetamido)propene is not sufficiently reactive to result in an efficient Heck reaction with 5.
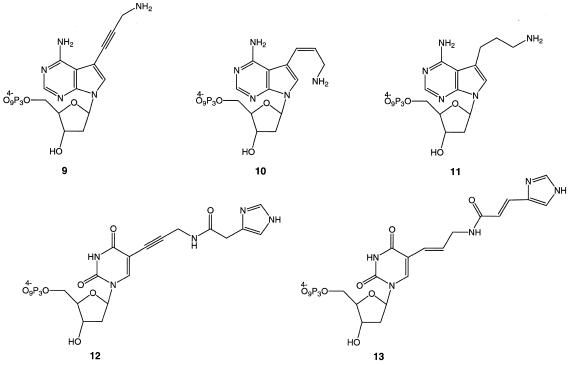
The nucleosides 6–8 were converted to the corresponding triphosphates, 9–11, by adapting the method of Kovacz and Ötvös (32) (Fig. 3). The imidazole-conjugated modified 2′-deoxyuridine triphosphates 12 and 13 were prepared according to the literature (6,17).
Figure 3.
The C5-modified triphosphates employed in this study. The triphosphates were prepared according as described in Materials and Methods and Lee et al. (6).
PCR
In order to be of use for in vitro selection procedures, the modified deoxynucleoside triphosphates are required to be substrates during PCR. Thus, the triphosphates must be good substrates for thermostable DNA polymerases and the modification must not interfere with the fidelity of Watson–Crick base pairing, so that multiple rounds of PCR can take place. Furthermore, in order that the sequence of the final selected DNA can be determined using conventional techniques and to allow faithful copying of the template DNA, the modified triphosphate must completely replace its unmodified counterpart during PCR.
We have studied the incorporation of the C7-modified triphosphates, which replace dATP during PCR mediated by Taq polymerase, using pUC19 template DNA with the appropriate primers to yield 62 or 98 nt products. The results of the substrate studies with the modified triphosphates are summarised in Table 1. All three modified C7-dATP derivatives 9–11 produce 98 nt PCR products in good yields that are comparable to those obtained using the natural nucleotides (Fig. 4). However, the alkyne 9, gives the highest yield of product. In previous studies in the C5-modified pyrimidine series, we observed that the triphosphates of 5-[3-aminopropynyl]-2′-deoxyuridine and 5-(E)-[3-aminopropenyl]-2′-deoxyuridine are both substrates for Taq polymerase whilst the related cis-alkene and alkane are not (6). Interestingly, the substrate properties of amino-modified C7-dATP analogues appear to be much less sensitive to the stereochemistry of the amino linker. Of the three C7-dATP derivatives 9–11, synthetic yields of 9 and 11 and their respective precursors 6 and 8 are greatest and as such, their use is to be preferred. Since in vitro selection is essentially an empirical procedure, it is difficult to predict the best way of displaying an amino function to achieve any desired structural diversity or catalytic function. Thus, all three modified C7-dATP analogues deserve investigation during selection experiments.
Table 1. Summary of substrate properties of the nucleotide analogues.
| Triphosphate analogue | Length of product | |
| |
62 nt |
98 nt |
|
9 (alkyne amino
C7-dATP) |
+++ |
+++ |
|
10 (Z-alkene
amino C7-dATP) |
no. exp |
++ |
|
11 (alkane amino
C7-dATP) |
no. exp |
++ |
|
9 and 12 |
+ |
– |
| 9 and 13 | ++ | + |
In each case, the indicated nucleotide was used as a replacement for dATP of dATP and dTTP during PCR. Results are shown for synthesis of PCR products of lengths of 62–98 nt. Details of conditions are described in Materials and Methods. The absence of PCR product is indicated by –. ′+′ indicates that full-length PCR product was obtained (the number of ′+′ signs is proportional to the efficiency/yield of PCR product). ‘No exp’ indicates that no PCR experiment was made.
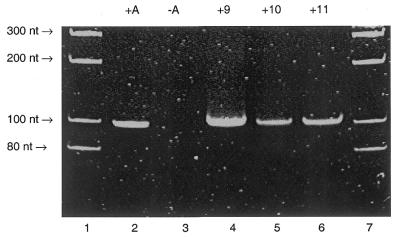
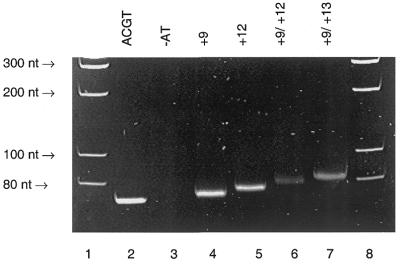
Figure 4.
PAGE gel image of the ethidium bromide-stained and UV-visualised 98 nt PCR fragment derived from pUC19 and the C7-amino-modified 7-deaza-dATP analogues 9–11. Lane 1, molecular weight markers; lane 2, a PCR containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP; lane 3, a PCR containing dTTP, dCTP and dGTP does not result in the formation of any product; lane 4, a PCR containing dTTP, dCTP, dGTP and 9; lane 5, a PCR containing dTTP, dCTP, dGTP and 10; lane 6, a PCR dTTP, dCTP, dGTP and 11; lane 7, molecular weight markers. All triphosphates 9–11 (Fig. 3) are substrates for Taq polymerase during PCR.
In order to produce DNA with two different modifications during PCR, the amplification must be performed using two different modified triphosphates in place of their natural counterparts. In previous work we have studied the substrate properties of C5-imidazole-modified dUTPs, and found compounds 12 and 13 (Fig. 3) to be the best substrates for Taq polymerase (6). Consequently, we investigated whether full-length PCR products could be formed during a reaction containing the amino alkyne C7-dATP derivative 9 in place of dATP and either 12 or 13 in place of dTTP (Fig. 5). DNA produced from such a PCR contains both primary amino and imidazolyl modifications and when used in in vitro selection protocols has the potential to deliver catalysts which mimic the active sites of protein enzymes such as RNase A. In both cases we found that PCR products of 62 nt in length were formed, although the yield of PCR product is slightly higher when 9 and 13 are used. In contrast, only 9 and 13 allowed the formation of 98 nt product (Table 1).
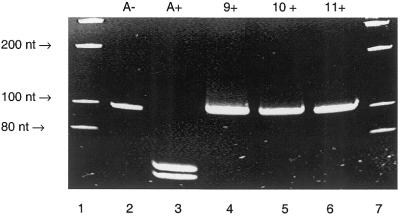
Figure 5.
PAGE gel image of the ethidium bromide-stained and UV-visualised 62 nt PCR fragment derived from pUC19 and the C7-amino-modified 7-deaza-dATP analogues 9–11 and dUTP imidazole conjugates 12 and 13. Lane 1, molecular weight markers; lane 2, a PCR containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP; lane 3, a PCR containing dCTP and dGTP does not result in the formation of any product; lane 4, a PCR containing dTTP, dCTP, dGTP and 9; lane 5, a PCR containing dATP, dCTP, dGTP and 12; lane 6, a PCR dCTP, dGTP, 9 and 12; lane 7, a PCR dCTP, dGTP, 9 and 13; lane 8, molecular weight markers. Full-length 62 nt PCR products were formed using 9 and 12, and 9 and 13 in place of dATP and dUTP, respectively, using Taq polymerase.
To our knowledge, this is the first report of the PCR-mediated synthesis of DNA bearing pendant groups displaying both imidazolyl and amino functionalities. The incorporation of two such analogues replacing dTTP and dATP necessitates Watson–Crick base pairing of the two modified nucleosides, which is surprisingly well accommodated by Taq polymerase. However, it may be advantageous to investigate the substrate properties of combinations of analogues for which mutual base pairing during the PCR is not a pre-requisite. To this end we are currently investigating routes to the corresponding C5-modified dCTP analogues.
Characterisation of modified PCR products
In order to confirm the presence of the modifications, we investigated the cleavage of the 98 nt PCR products using the restriction enzyme XbaI (T^CTAGA, ^ represents the site of hydrolysis). We and others (6,17) have demonstrated previously that replacement of thymidines with C5-amino-and imidazolyl-modified 2′-deoxyuridines leads to inhibition of restriction of the resulting DNA by XbaI. Consistent with this, incorporation of any of the C7-dATP analogues 9–11 is also inhibitory to cleavage by XbaI (Fig. 6). To our surprise, cleavage of the same PCR products by SmaI (CCC^GGG, ^ represents the site of hydrolysis) was also inhibited, whilst the same DNA sequence containing the natural nucleotides was completely hydrolysed (data not shown). It is noteworthy that the SmaI site in the 98 nt PCR product is flanked at the 5′-end with dTA and at the 3′-end with dGA, where A represents a site of modification. Under the conditions of restriction digest the amino groups of the modification would be expected to be largely protonated. Furthermore, the modifications introduce both steric bulk into the major groove and a heterocyclic base with differing electronic properties due to the replacement of N7 by a methine moiety. Although this modification is not displayed directly in the recognition sequence it is likely that the restriction endonuclease also interacts with the flanking DNA sequence and that the changes introduced are inhibitory to this. These findings are currently under further investigation.
Figure 6.
PAGE gel image of the ethidium bromide-stained and UV-visualised PCR fragments derived from pUC19 and the C7-amino-modified 7-deaza-dATP analogues 9–11 (Fig. 4) following incubation with XbaI. Lane 1, molecular weight markers; lane 2, control 98 nt PCR product containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP in the absence of XbaI; lane 3, the 98 nt PCR product containing all four natural triphosphates, dATP, dCTP, dGTP and dTTP following incubation with XbaI; lane 4, the 98 nt PCR product containing dTTP, dCTP, dGTP and 9 following incubation with XbaI; lane 5, the 98 nt PCR product containing dTTP, dCTP, dGTP and 10 following incubation with XbaI; lane 6, the 98 nt PCR product containing dTTP, dCTP, dGTP and 11 following incubation with XbaI; lane 7, molecular weight markers. Incorporation of analogues 9–11 into recognition site for XbaI (T^CTAGA) results in inhibition of cleavage.
The lack of digestion by XbaI of the PCR product obtained using a mixture of dCTP, dGTP and analogues 9 and 13 is consistent with the simultaneous incorporation of both analogues (data not shown). The large decrease in mobility of this product during PAGE relative to the PCR product containing the four natural bases (Fig. 5) is also consistent with the incorporation of both 9 and 13, since the substituents of these analogues are likely to be substantially protonated, thereby decreasing the overall negative charge of the DNA. In order to provide direct evidence for the simultaneous incorporation of both 9 and 13, we used MALDI-TOF mass spectrometry as described previously (6). Thus, we prepared a PCR product of 62 nt in length and biotinylated on one strand and determined the mass of the non-biotinylated strand as we have described previously (6). The observed mass was within 0.2% of the calculated mass and 3.0 kDa greater than that of the corresponding PCR product containing the four natural bases (Fig. S1). This demonstrates unambiguously that both analogues are indeed incorporated during PCR.
CONCLUSIONS
We have demonstrated that the incorporation of C7-dATP derivatives bearing either propynyl amino, cis-propenyl amino or propyl amino functionalities is possible in PCRs containing the modified nucleotide and dCTP, dGTP and dTTP. Surprisingly, in contrast to the case of C5-amino-modified dUTP derivatives, the substrate properties of the amino-modified C7-dATP derivatives are not dramatically affected by the stereochemistry of the linker arm. The resultant DNA possesses primary amino groups which would provide additional structural diversity and potential catalytic function when the DNA is subjected to in vitro selection experiments. To further enhance the structural diversity and catalytic potential of DNA we have also demonstrated the simultaneous incorporation of imidazolyl and amino groups during PCR mediated by Taq polymerase using modified C7-dATP and C5-modified dUTP in place of their natural counterparts.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Prof. David M. Perrin, University of British Columbia, for sharing information prior to publication. We are grateful to Dr Brian Taylor and Ms Sue Bradshaw for obtaining NMR spectra and we thank Ms Elaine Frary for her expert technical assistance. This work was supported by the Leverhulme Trust F/118/AQ (N.M., T.G. and A.S.), the BBSRC grant no. 50B09442 (S.E.L.), The Wellcome Trust grant ref no. 053924 (A.S.), EPSRC grant no. GR/L85749 (MALDI-TOF) and Royal Society equipment grants 18048 and 21084. J.A.G. is a BBSRC Advanced Research Fellow.
References
- 1.Breaker R.R. and Joyce,G.F. (1995) A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol., 2, 655–660. [DOI] [PubMed] [Google Scholar]
- 2.Cuenoud B. and Szostak,J.W. (1995) A DNA metalloenzyme with DNA-ligase activity. Nature, 375, 611–614. [DOI] [PubMed] [Google Scholar]
- 3.Lorsch J.R. and Szostak,J.W. (1994) In-vitro evolution of new ribozymes with polynucleotide kinase-activity. Nature, 371, 31–36. [DOI] [PubMed] [Google Scholar]
- 4.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santoro S.W., Joyce,G.F., Sakthivel,K., Gramatikova,S. and Barbas,C.F. (2000) RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc., 122, 2433–2439. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.E., Sidorov,A., Gourlain,T., Mignet,N., Thorpe,S.J., Dickman,M.J., Hornby,D.P., Grasby,J.A. and Williams,D.M. (2001) Enhancing the catalytic repertoire of nucleic acids: a study of linker length and rigidity. Nucleic Acids Res., 29, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B.L. and Cech,T.R. (1997) Peptide bond formation by in vitro selected ribozymes. Nature, 390, 96–100. [DOI] [PubMed] [Google Scholar]
- 8.Wiegand T.W., Janssen,R.C. and Eaton,B.E. (1997) Selection of RNA amide synthases. Chem. Biol., 4, 675–683. [DOI] [PubMed] [Google Scholar]
- 9.Lohse P.A. and Szostak,J.W. (1996) Ribozyme-catalysed amino-acid transfer reactions. Nature, 381, 442–444. [DOI] [PubMed] [Google Scholar]
- 10.Tarasow T.M., Tarasow,S.L. and Eaton,B.E. (1997) RNA-catalysed carbon–carbon bond formation. Nature, 389, 54–57. [DOI] [PubMed] [Google Scholar]
- 11.Tarasow T.M., Tarasow,S.L., Tu,C., Kellogg,E. and Eaton,B.E. (1999) Characteristics of an RNA Diels-Alderase active site. J. Am. Chem. Soc., 121, 3614–3617. [Google Scholar]
- 12.Dewey T.M., Zyzniewski,M.C. and Eaton,B.E. (1996) RNA world: functional diversity in a nucleoside by carboxyamidation of uridine. Nucl. Nucl., 15, 1611–1617. [Google Scholar]
- 13.Dewey T.M., Mundt,A.A., Crouch,G.J., Zyzniewski,M.C. and Eaton,B.E. (1995) New uridine derivatives for systematic evolution of RNA ligands by exponential enrichment. J. Am. Chem. Soc., 117, 8474–8475. [Google Scholar]
- 14.Tu C., Keane,C. and Eaton,B.E. (1995) Palladium catalysis in the synthesis of 8-position modified adenosine, 2′-deoxyadenosine and guanosine. Nucl. Nucl., 14, 1631–1638. [Google Scholar]
- 15.Vaish N.K., Fraley,A.W., Szostak,J.W. and McLaughlin,L.W. (2000) Expanding the structural and functional diversity of RNA: analog uridine triphosphates as candidates for in vitro selection of nucleic acids. Nucleic Acids Res., 28, 3316–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matulic-Adamic J., Daniher,A.T., Karpeisky,A., Haeberli,P., Sweedler,D. and Beigelman,L. (2000) Nucleoside triphosphates for in vitro selection of new catalytic RNAs. Bioorg. Med. Chem. Lett., 10, 1299–1302. [DOI] [PubMed] [Google Scholar]
- 17.Sakthivel K. and Barbas,C.F. (1998) Expanding the potential of DNA for binding and catalysis: highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem. Int. Ed. Engl., 37, 2872–2875. [DOI] [PubMed] [Google Scholar]
- 18.Battersby T.R., Ang,D.N., Burgstaller,P., Jurczyk,S.C., Bowser,M.T., Buchanan,D.D., Kennedy,R.T. and Benner,S.A. (1999) Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc., 121, 9781–9789. [DOI] [PubMed] [Google Scholar]
- 19.Perrin D.M., Garestier,T. and Hélène,C. (1999) Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucl. Nucl., 18, 377–391. [DOI] [PubMed] [Google Scholar]
- 20.Kazimierczuk Z., Cottam,H.B., Revankar,G.R. and Robins,R.K. (1984) The synthesis of 2′-deoxytubercidin, 2′-deoxyadenosine and related 2′-deoxynucleosides via a novel direct stereospecific sodium salt glycosylation procedure. J. Am. Chem. Soc., 106, 6379–6382. [Google Scholar]
- 21.Pudlo J.S., Nassiri,M.R., Kern,E.R., Wotring,L.L., Drach,J.C. and Townsend,L.B. (1990) Synthesis, antiproliferative and antiviral activity of certain 4-substituted and 4,5-disubstituted 7-[(1,3-dihydro-2-propoxy)methyl]pyrrolo[2,3-d]pyrimidines. J. Med. Chem., 33, 1984–1992. [DOI] [PubMed] [Google Scholar]
- 22.Hoffer M. (1960) α–Thymidin. Chem. Ber., 93, 2777–2780. [Google Scholar]
- 23.Buhr C.A., Wagner,R.W., Grant,E. and Froehler,B.C. (1996) Oligonucleotides containing C7 propyne analogs of 7-deaza-2′-deoxyguanosine and 7-deaza-2′-deoxyadenosine. Nucleic Acids Res., 24, 2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seela F. and Zulauf,M. (1996) Palladium-catalysed cross coupling of 7-iodo-2′-deoxytubercidin with terminal alkynes. Synthesis, 726–730. [Google Scholar]
- 25.Seela F., Zulauf,M., Rosemeyer,H. and Reuter,H. (1996) The DNA-stablising nucleoside 7-iodo-2′-deoxytubercidin: its structure in the solid state and in solution. J. Chem. Soc. Perkin Trans. II, 2373–2376. [Google Scholar]
- 26.Seela F. and Zulauf,M. (1999) Oligonucleotides containing 7-deazaadenines: the influence of the 7-substituent chain length and charge on the duplex stability. Helv. Chim. Acta, 82, 1878–1898. [Google Scholar]
- 27.Seela F. and Thomas,H. (1995) Duplex stabilisation of DNA: oligonucleotides containing 7-substituted 7-deazaadenines. Helv. Chim. Acta, 78, 94–108. [Google Scholar]
- 28.Kanaya E., Howard,F.B., Frazier,J. and Miles,H.T. (1987) Poly(2-amino-8-methyldeoxyadenylinic acid)—contrasting effects in deoxynucleotides and ribopolynucleotides of 2-amino and 8-methyl substituents. Biochemistry, 26, 7159–7165. [DOI] [PubMed] [Google Scholar]
- 29.Seela F. and Chen,Y.M. (1996) Oligonucleotides containing 7- or 8-methyl-7-deazaguanine: steric requirements of major groove substituents on the DNA structure. J. Chem. Soc. Chem. Commun., 2263–2264. [Google Scholar]
- 30.Davoll J. (1960) Pyrrolo[2,3-d]pyrimidines. J. Chem. Soc. Perkin Trans. II, 131–138. [Google Scholar]
- 31.Lee S.E., Vyle,J.S., Williams,D.M. and Grasby,J.A. (2000) Novel syntheses of (Z)-alkene and alkane base-modified nucleosides. Tetrahedron Lett., 41, 267–270. [Google Scholar]
- 32.Kovacz T. and Ötvös,L. (1988) Simple synthesis of 5-vinyl- and 5-ethynyl-2′-deoxyuridine-5′-triphosphates. Tetrahedron Lett., 29, 4525–4528. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.