Abstract
Purpose
The purpose of this preclinical study was to determine the effectiveness of RAF265, a multi-kinase inhibitor, for treatment of human metastatic melanoma and to characterize traits associated with drug response.
Experimental Design
Advanced metastatic melanoma tumors from 34 patients were orthotopically implanted to nude mice. Tumors that grew in mice (17 of 34) were evaluated for response to RAF265 (40 mg/kg, every day) over 30 days. The relation between patient characteristics, gene mutation profile, global gene expression profile, and RAF265 effects on tumor growth, mitogen-activated protein/extracellular signal-regulated kinase (MEK)/extracellular signal-regulated kinase (ERK) phosphorylation, proliferation, and apoptosis markers was evaluated.
Results
Nine of the 17 tumors that successfully implanted (53%) were mutant BRAF (BRAFV600E/K), whereas eight of 17 (47%) tumors were BRAF wild type (BRAFWT). Tumor implants from 7 of 17 patients (41%) responded to RAF265 treatment with more than 50% reduction in tumor growth. Five of the 7 (71%) responders were BRAFWT, of which 1 carried c-KITL576P and another N-RASQ61R mutation, while only 2 (29%) of the responding tumors were BRAFV600E/K. Gene expression microarray data from nonimplanted tumors revealed that responders exhibited enriched expression of genes involved in cell growth, proliferation, development, cell signaling, gene expression, and cancer pathways. Although response to RAF265 did not correlate with pERK1/2 reduction, RAF265 responders did exhibit reduced pMEK1, reduced proliferation based upon reduced Ki-67, cyclin D1 and polo-like kinase1 levels, and induction of the apoptosis mediator BCL2-like 11.
Conclusions
Orthotopic implants of patient tumors in mice may predict prognosis and treatment response for melanoma patients. A subpopulation of human melanoma tumors responds to RAF265 and can be characterized by gene mutation and gene expression profiles.
Introduction
For the past 2 decades, the incidence of cutaneous melanoma has steadily increased. In 2010, 68,130 new cases with 8,700 deaths were estimated to have occurred in the United States (1, 2). Identification of effective strategies for the treatment of melanoma remains an urgent need. Recently, immunotherapy with anti-CTLA4 antibody has shown improved overall survival when compared with a peptide vaccine for patients with refractory advanced melanoma (3). Moreover, genetic changes are linked to the intrinsic molecular defects that are identified to be "causal" for tumor development and as a result, therapeutic strategies often target oncogenes (4–6). Approximately 43% to 50% of melanoma patients exhibit somatic mutation in the BRAF gene and another 20% exhibit mutation in NRAS (www.sanger.ac.uk/genetics/CGP/cosmic/). Indeed, recently this has been successfully shown in melanoma through clinical trials whereby highly specific inhibitors targeting the V600E BRAF mutation [Vemurafenib (Zelboraf) and GSK2118436] substantially reduced tumor burden in patients with melanoma harboring this mutation (7). Indeed, BRAF-targeted therapy has been approved by the U.S. Food and Drug Administration as standard of care for metastatic melanoma patients with proven BRAF mutation (7–11). Given the specificity of such small-molecule inhibitors, identification of genetically defined patient subgroups is critical to gain better outcomes and avoid druginduced adverse effects (12, 13). Moreover, resistance that results after vemurafenib treatment can result from activation of c-RAF, suggesting that combined therapy with an inhibitor that targets multiple kinases, like RAF265, or a mitogen-activated protein (MAP)/extracellular signal-regulated kinase (ERK; MEK) inhibitor may be more effective.
RAF265 is an orally bioavailable small molecule with preclinical antitumor activity that currently is being tested in phase I clinical trials. Much like sorafenib, in vitro kinase assays show RAF265 inhibits the activities of several intracellular kinases, including BRAF(V600E), BRAF(wild type), c-RAF, VEGF receptor 2 (VEGFR2), platelet-derived growth factor receptor (PDGFR), colony-stimulating factor (CSF) 1R, RET and c-KIT, SRC, STE20, and others with IC50 ranging from less than 20 to more than 100 nmol/L. However, in cell-based assays, RAF265 is most potent for BRAFV600E, and VEGFR2, but less active for PDGFRB and c-KIT (14, 15; and Stuart and colleagues, submitted manuscript]. RAF265 inhibited BRAF-mediated downstream activation of ERK, which was conceived as the major underlying mechanism for the growth inhibition of human colorectal carcinoma in an orthotopic transplant tumor model (16). The efficacy of RAF265 in treating human melanoma is under evaluation, though the ongoing melanoma phase I clinical trials are based upon cell line xenograft studies (15, 17). Because melanoma cells possess multiple mechanisms to invade, metastasize, and resist therapies, the multiple-targeting agents like RAF265 may inhibit the pathways critical for tumor and induce tumor regression. Because limited data are available about responsiveness to RAF265, we wished to examine response to this drug in a preclinical setting that evaluates the response of melanoma tumors taken directly from the patient, where genetic markers and gene expression profiles which may predict response to the drug are determined. The response to RAF265 seemed effective in more than 70% of BRAF wild-type melanomas. In addition, analysis of the global gene expression profile of human melanoma tumor samples revealed differential expression of genes known to be relevant to cell cycle, apoptosis, cell–cell adhesion, epithelial–mesenchymal transition, and drug resistance in RAF265 responders compared with nonresponders. Using this information, it may be possible to predict which melanoma patients will respond to RAF265.
Materials and Methods
Chemical agent and antibodies
A detailed list of reagents and antibodies is found in the Supplementary Methods section.
Patient characteristics
Thirty-four patients with advanced melanoma underwent surgical resection of regional lymph node or distant metastases between February 2007 and August 2009. A single patient (V30) had a tumor obtained from a locally advanced primary of the heel. All patients gave informed consent to participate in an Institutional Review Board–approved melanoma and cutaneous malignancy tissue repository. Immediately after resection of the tumor, the sample was divided, and fresh tissue was placed in medium for subcutaneous implantation into BALB/C nu/Foxn1 athymic nude mice for the evaluation of tumor response to treatment. Other samples were fixed in paraformaldehyde, flash frozen for signaling, or processed in RNAlater for gene expression microarray experiments. The remainder of the specimen was sent to pathology for standard histologic analyses. The demographic features, pathology, and treatment of patients included in the study are listed in Supplementary Table S1. Tumor thickness and Clark level varied widely, and many patients had melanoma metastatic to a lymph node from an unidentified primary site. Most patients received single or multiple immunologic, radiation or chemotherapies before enrollment in the study. All patients were followed prospectively and recurrence and death were recorded.
Orthotopic tumor implantation model
Melanoma tissue biopsy was obtained from patients undergoing surgical resection at Vanderbilt University Medical Center. Tissues were implanted into BALB/C nu/Foxn1 athymic nude mice and drug response was evaluated according to the guidelines of the Vanderbilt Institutional Animal Care and Use Committee. Detailed protocols are located in Supplementary Methods.
Isolation of total RNA from melanoma tissue
At resection, 20 to 30 mg aliquots of melanoma tissues from patients were immediately placed into cryovials containing 1 mL of RNAlater Solution. Sample cryovials were placed at room temperature for 24 hours then stored at −70°C until RNA extraction by RNeasy Fibrous Tissue Mini Kit (QIAGEN) with DNase I digestion, following the manufacturer’s protocol.
Genome-wide expression profiling of melanoma
Gene expression profiling was performed in the Vanderbilt Functional Genomics Shared Resource. Detailed protocols are available in Supplementary Methods.
Accession number
The microarray data were submitted to the Gene Expression Omnibus (GEO) repository according to Minimum Information About a Microarray Experiment (MIAME) criteria. The GEO accession number is: GSE30812.
Western blot analysis
Western blots were carried out as described in Supplementary Methods.
SNaPshot genotyping assay
The SNaPshot mutational profiling method is characterized by multiplexed PCR, multiplexed single-base primer extension, followed by capillary electrophoresis. This assay was designed to detect 43 point mutations in 6 genes (Supplementary Table S2). SNaPshot analysis was conducted as previously described (18). Briefly, PCR primers were pooled to amplify the target DNA, and PCR was carried out with the following conditions: 95°C (8 minutes), followed by 40 cycles of 95°C (20 seconds), 58°C (30 seconds), and 72°C (1 minute), and then a final extension of 72°C (3 minutes). PCR primers used for this assay were previously described (19). Next, PAGE-purified primers were pooled together and multiplex extension reactions were carried out with the following conditions: 96°C (30 seconds), followed by 25 cycles of 96°C (10 seconds), 50°C (5 seconds), and 60°C (30 seconds). Single-base extension primers were previously described (19). Extension products were applied to capillary electrophoresis in an ABI 3730 analyzer and the data were interpreted with ABI GeneMapper software (version 4.0). Human male genomic DNA (Promega) was used as a wild-type control.
Direct dideoxynucleotide-based sequencing
All mutations detected by SNaPshot genotyping assay were confirmed with direct sequencing. Exons with mutations were amplified using a HotStarTaq Master Mix Kit (QIAGEN) and M13-tagged gene-specific primers. The following PCR conditions were used: 96°C (15 minutes), followed by 40 cycles of 95°C (30 seconds), 60°C (30 seconds), 72°C (60 seconds), and a final extension step of 72°C (10 minutes). Excess primers and nucleotides were digested with ExoSAP (USB). Sequencing reactions were carried out with Applied Biosystems version 3.1 Big Dye Terminator chemistry and analyzed on an Applied Biosystems 3730XL Sequencer. All sequence chromatograms were read in both forward and reverse directions.
Tissue microarray analysis
Melanoma tissues were collected and fixed in 4% paraformaldehyde solution for 24 hours at room temperature and embedded in paraffin blocks, and tissue microarrays were prepared according to standard procedures described in detail in Supplementary Materials and Methods. Immunohistochemistry (IHC) scores were made in reference to positive and negative areas in the cores of tissue controls in which +0.5 indicated low staining over approximately 50% of the cells, +1 indicated a low level of staining over approximately 80% of the cells, +2 indicated moderate staining over approximately 80% of the cells, and +3 indicated strong staining over approximately 80% of the cells. Tissue microarray (TMA) slides were evaluated and scored by Sara Kantrow, a licensed and board certified dermatopathologist, or by Kelli Boyd, and by at least one other scientist, each of which was blind to treatment protocol.
Statistical analyses
Statistical significance was analyzed with Student t test, Bernard exact test, χ2, and/or ANOVA.
Results
Patient profile
Among the tissue biopsies or surgical resections from 34 enrolled patients, tumor tissues from 17 patients were successfully grown in nude mice. In this 17 patient cohort, 13 were male and 4 were female. The mean survival time of this cohort from the time the patient was diagnosed was 30 months (range 6–85 months) at the time this study ended. Melanoma lesions were located on trunk and lower limbs. Tumor ulceration was present in 7 patients. Most patients underwent prior single or combined treatments of immunotherapy, radiation therapy, and chemotherapy (Table 1 and Supplementary Table S1).
Table 1.
Patient characteristics
| Total patient population |
Patient tumors in RAF265 regime |
|
|---|---|---|
| Gender | ||
| Male/female | 25/9 | 13/4 |
| Survival (mo) | ||
| Mean (range) | 34.4 (3–174) | 30 (6–85) |
| Site of primary | ||
| Head/neck | 3 | 1 |
| Trunk | 12 | 5 |
| Upper limbs | 6 | 0 |
| Lower Limbs | 4 | 4 |
| Finger | 1 | 0 |
| Toe | 1 | 1 |
| Unknown | 7 | 7 |
| Ulceration | ||
| Yes | 15 | 7 |
| No | 9 | 3 |
| Unknown | 10 | 7 |
| Combined treatments | ||
| Immunotherapy | 18 | 8 |
| Radiation therapy | 23 | 11 |
| Chemotherapy | 15 | 5 |
NOTE: The clinical information of 34 enrolled patients was summarized. The mean survival time was calculated based on the follow-up data before the end of 2010.
Tumor responses to RAF265
Of the 34 patients studied, we had sufficient tumor implant growth for evaluation of response to RAF265 for 17 patients (Fig. 1). (Tumor tissue from patient V9 and V26 also grew, but are excluded because the number of mice evaluated did not allow fully a powered analysis of drug response). The mean survival time from the time of diagnosis for those patients whose tumors grew in the mouse model was 28 months as compared with survival for 50 months from diagnosis for those patients whose tumors did not grow sufficiently in mice for evaluation in the study, suggesting the tumors that can be successfully transplanted are the most aggressive. The gene expression microarray analysis of representative tumors that grew in mice compared with tumor taken directly from the patient was 90% concordant, indicating that tumor implants in mice are representative of patient tumor (data not shown).
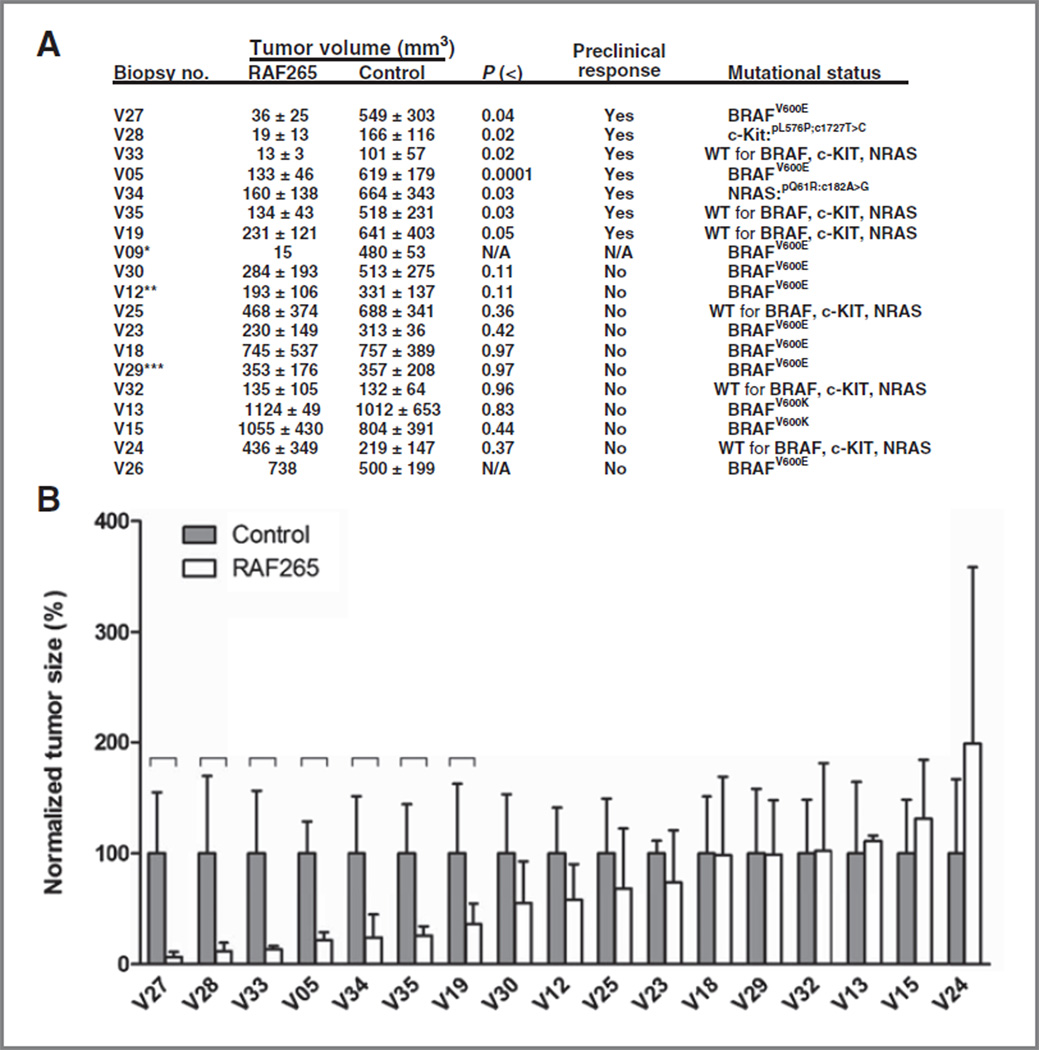
Figure 1.
A, orthotopic transplant tumor response to RAF265. The 17 human melanoma biopsy tissues were successfully grown on nude mice and the mean ± SD (biological repeats, n = 5) of measured tumor volumes of RAF265 treatment and control groups were shown. Seven melanoma biopsy tumor-derived orthotopic implants showed significant growth inhibition illustrated by unpaired Student t test analysis. The definition of treatment response in this study: When compared with control, more than 50% reduction of tumor volume is achieved by RAF265 at the end of 30-day treatment period. Mutational status (BRAF, cKIT, and NRAS) is noted in the right hand column. Annotations are as follows: *, mice carrying xenografts of patient V09 had only 1 mouse in the RAF265 treatment group and 4 in the control group, making statistical analysis of response not applicable (N/A); **, patient V12 received 800mg of the old formulation of PLX4032 which was inadequate and the patient progressed; ***, patient V29 was initially treated with 1600 mg of an older formulation of PLX4032, then switched to the newer formula and allowed to escalate to 360 and then to 720 mg/kg. After an initial response, patient V29 progressed at 15 months, and the PLX4032-resistant tumor was biopsied and used in the tumor implant study. B, graphical comparison of orthotopic transplant tumor size after 15- to 30-day treatment with RAF265. The tumor sizes on RAF265-treated nude mice were measured, and the difference of tumor volume between treatment and control groups was analyzed by unpaired Student t test (P <0.05). The figure shows normalized tumor sizes (mean ± SD, n = 5) in RAF265 treatment and control groups.
The tumor volumes at treatment day 30 (or the time point upon which mice had to be euthanized due to tumor size or other AAALAC criteria) were compared to evaluate RAF265 response, defined as more than 50% decrease of tumor volume relative to control. Significant drug responses in transplanted tumors occurred in 7 different patients (samples V27, V28, V33, V05, V34, V35, and V19), which accounted for about 41% of all tumors evaluated (Fig. 1A and B, Supplementary Fig. S3). Partial responses were observed in tumors from patients V12, V30, V25, and V24 (P = 0.11, 0.11, 0.35, and 0.42, respectively).
Melanoma mutational profile and ERK inactivation by RAF265
To further investigate whether RAF265-induced response in human melanoma tissues correlates with melanomarelevant genomic alterations, SNaPshot mutational profiling designed to survey 43 common somatic point mutations in 6 genes (BRAF, NRAS, GNAQ, GNA11, and CTNNB1) was carried out. There were no detectable mutations inCTNNB1,GNAQ, andGNA11, while 1 sample had a cKITL576P mutation and another had an NRASP61R mutation (Fig. 1A and Supplementary Table S2). About BRAF gene alterations, 8 of 17 samples (47%) were wild type for BRAF, whereas 7 of 17 samples had the V600E mutation (41%) and 2 of 17 the V600K mutation (12%). No tumors had mutations in more than one of these genes. To clarify whether the mutation profile in tumors would be faithfully maintained after being implanted and grown in nude mice, transplant tumor tissues were also subjected to SNaPshot analysis. The mutational profile between each orthotopic transplant and its human tumor counterpart that had not been transplanted to the mouse were identical (data not shown). Genotyping of 7 of the tumors that did not grow as an implant revealed that 3 were BRAFV600E mutant, 2 had NRASP61R, and 2 were BRAFWT with no mutation in NRAS or cKIT. Thus of the 24 tumors genotyped, 50% had BRAF mutation, 12.5% had NRAS mutation, and 4% had cKIT mutation.
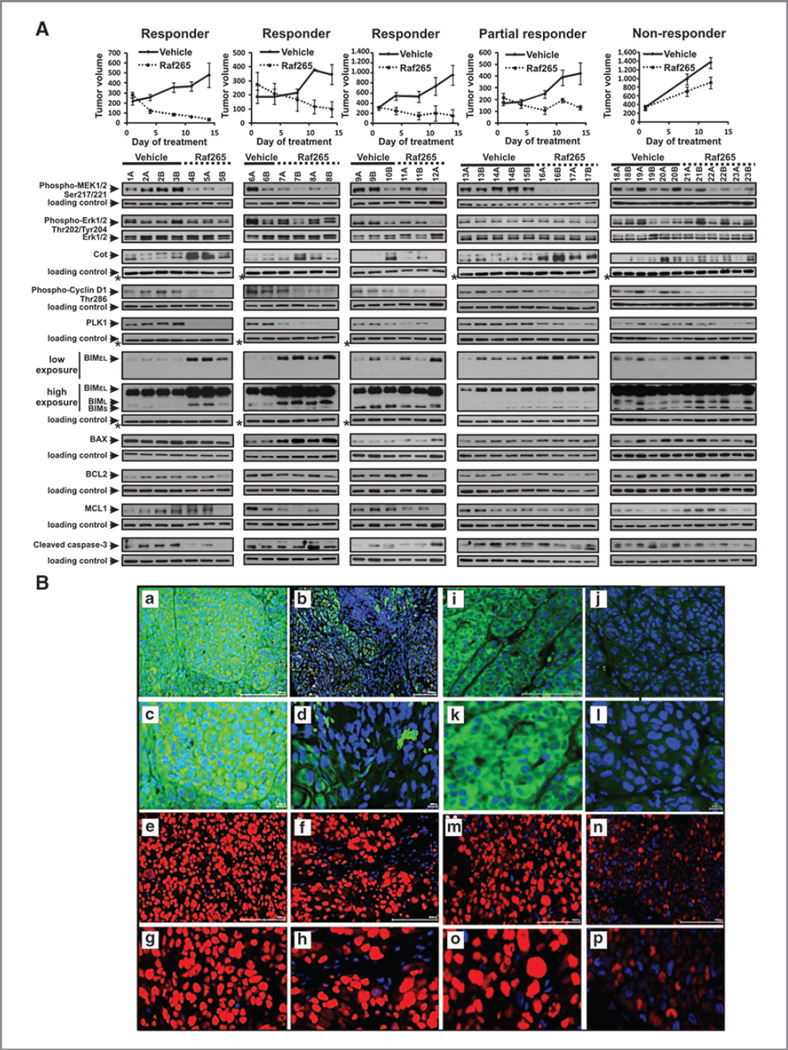
As RAF265 was previously shown to inhibit the growth of tumors with BRAFV600E mutation and part of the underlying mechanism was related to inhibition of ERK (16), we then asked 2 questions: (i) whether the responding tumors in this study had BRAFV600E mutation, and (ii) whether RAF265-induced inhibition of melanoma growth involved inhibition of pERK. By analyzing data from SNaPshot mutational profiling, we noticed that most responding tumors (5 of 7, 71%) were BRAFWT (Table 2) and most of the BRAFV600E mutant tumors (77%) were RAF265 nonresponders (Table 2). Immunohistochemical staining of TMAs revealed no significant difference between ERK phosphorylation status of RAF265 responding and nonresponding human tumors before implantation, or between implanted tumors treated with RAF265 compared with vehicle control (Supplementary Fig. S1A, S1B, and S1C (unpaired Student t test, P = 0.2296). These results suggest that ERK inactivation was not achieved by RAF265 at 40 mg/kg once daily, in this orthotopic implant model at the time point evaluated. The results also suggest that the antitumor effect of RAF265 on human melanoma may depend on mechanisms independent of the RAF/ERK signaling pathway. Western blot analysis of representative RAF265 responding tumors also revealed no inhibition of pERK1/2 using 2 different antibodies that gave the same result. Interestingly however, there was substantial inhibition of phospho-MEK in 4 out of 4 responding tumors analyzed by Western blot (Fig. 2A). TMA analysis of phosphorylated MEK (pMEK)1 staining showed reduction of pMEK in 4 of 7 RAF265 responders and 1 partial responder (V30; Fig. 2B). The differences between TMA and Western blot analysis are likely due to the technical difficulty of successfully preserving and capturing phosphoprotein staining in TMAs. The inhibition of pMEK observed in Western blot of freshly extracted tumors treated with RAF265 was accompanied by reduction in phospho-cyclin D1 (Thr286) and polo-like kinase1 (PLK1) indicating an inhibition of cell-cycle progression in response to RAF265. Although we cannot rule out the possibility that a subpopulation of responding cells may have exhibited inhibition of ERK phosphorylation at an earlier time point, the data suggest that the tumors have developed an alternate mechanism for activation or ERK1/2.Arecent paper from the Garraway laboratory (20) showed that the MAP3K COT is capable of directly phosphorylating ERK1/2 in melanoma cells. We did observe COT expression in 5 out of 5 melanoma tumor explants by Western blot analysis and COT was elevated in response to RAF265 in three fourth responders, suggesting that COT might directly phosphorylate ERK in those tumors in which pMEK was reduced in response to RAF265 (Fig. 2A).
Table 2.
Drug response associates with BRAF genotype
| BRAF/wild type |
BRAF/V600E,K |
||
|---|---|---|---|
| Sample | Response | Sample | Response |
| V28 | Yes | V27 | Yes |
| V33 | Yes | V05 | Yes |
| V34 | Yes | V30 | No |
| V35 | Yes | V12 | No |
| V19 | Yes | V23 | No |
| V25 | No | V18 | No |
| V32 | No | V29 | No |
| V24 | No | V13 | No |
| V15 | No | ||
| % of response: | 62.5 (5/8)* | 22.2 (2/9) | |
NOTE: Tumor samples were separated based on their BRAF genotype and RAF265-induced drug responses were compared. An increased response with marginal statistical difference was indicated in tumors of wild-type BRAF (*, P = 0.0605, Barnard exact test).
Figure 2.
A, Western blot analysis of lysates from 5 selected melanoma implants reveals that RAF265 treatment downregulates level of phosphorylated MEK without affecting ERK1/2 phosphorylation. It also reduces phospho-cyclin D1 and PLK1 levels, and elevates BIM1 and COT as compared with vehicle-treated controls in tumors responsive to the drug treatment. Top, tumor growth charts for 3 representative Raf265-responsive melanoma implants, 1 partial responder, and 1 nonresponder tumor during the 14-day period of daily treatment with Raf265 or vehicle control. Bottom, Western blot analysis of phospho-MEK, phospho-ERKand total ERK,Cot, phospho-Cyclin D1, PLK, BIM, BAX, BCL2, MCL1, and cleaved caspase-3 levels in lysates extracted from Raf265 and vehicle-treated melanoma tumors shown above. Western blot with β-actin–specific antibody was used as a loading control everywhere except for cases indicated with * in which GAPDH levels were analyzed to ensure equal loading. B, photomicrographs showing immunohistochemical localization of pMEK1 (green) and cyclin D1 (red) in tissue cores from TMAs assembled from tumors implanted into mice and treated with RAF265 or vehicle control. Nuclei are stained with DAPI and appear blue. a, pMEK1 staining of tumor core from V27 treated with vehicle control. Magnification, ×20. b, pMEK staining of tumor core from V27 treated with RAF265, ×20 magnification. c, pMEK1 staining of tumor core from V27 treated with vehicle control, ×40 magnification. d, pMEK staining of tumor core from V27 treated with RAF265, ×40 magnification. e, cyclin D1 staining of tumor core from V27 treated with vehicle control, ×20 magnification. f, cyclin D1 staining of tumor core from V27 treated with RAF265, ×20 magnification. g, cyclin D1 staining of tumor core from V27 treated with vehicle control, ×40 magnification. h, cyclin D1 staining of tumor core from V27 treated with RAF265, ×40 magnification. i, pMEK1 staining of tumor core from V05 treated with vehicle control, ×20 magnification. j, pMEK staining of tumor core from V05 treated with RAF265, ×20 magnification. k, pMEK1 staining of tumor core from V05 treated with vehicle control, ×40 magnification. l, pMEK staining of tumor core from V05 treated with RAF265, ×40 magnification. m, cyclin D1 staining of tumor core from V05 treated with vehicle control, ×20 magnification. n, cyclin D1 staining of tumor core from V05 treated with RAF265, ×20 magnification. o, cyclin D1 staining of tumor core from V27 treated with vehicle control, ×40 magnification. p, cyclin D1 staining of tumor core from V05 treated with RAF265, ×40 magnification.
RAF265 inhibits proliferation or enhances apoptosis in responding tumors
Proliferation and apoptosis in tumor implants were evaluated by Ki-67 and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining. Of the tumor implants responding to RAF265, those from patient V27, V28, V19, and V35 showed a statistically significant reduction (P≤0.05) in Ki-67 staining in the tumor implants on mice treated with RAF265 (40 mg/kg every day; Supplementary Fig. S2A). Tumor implants from patient V33 showed a reduction in Ki-67, but the mean reduction did not reach statistical significance (P = 0.08). Of these tumors with reduction in Ki-67 after RAF265 treatment, one exhibited the BRAFV600E mutation (V27), one had a cKITI576P mutation (V28) and 3 did not show mutation in any of the genes evaluated in the SNaPshot analysis (V19, V33, V35). V34 carried the NRASQ61R mutation and did not exhibit reduced Ki-67 staining with RAF265 treatment. In addition, 2 tumors that had a partial response to RAF265 (~30% reduction in tumor volume) exhibited a statistically significant reduction in Ki-67 staining in response to RAF265 treatment (V23 and V25). Moreover, TMA analysis of total cyclin D1 revealed differences between control and treated tumors for V19, V18, V28, V12, V27, V25, and V05 in which there was 54%, 42%, 46%, 39%, 36%, 33%, and 22% reduction, respectively (Fig. 2B). Western blot analysis for phospho-cyclin D1 on representative responding tumors (V27, V34, and V35) revealed strong reduction in phosphocyclin D1 in response to RAF265, again suggesting that RAF265 inhibits cell-cycle progression in responding tumors (Fig. 2A). In summary, approximately 70% of the RAF265-responding tumors showed a reduction in Ki-67 and/or cyclin D1 that correlated with the inhibition of tumor growth in response to this drug (Supplementary Fig. S2C).
Tumors from both vehicle control and RAF265-treated mice had significant levels of apoptosis, based upon TUNEL staining. However, only V19 and V25 showed strong increase in TUNEL staining in the RAF265-treated group. (Supplementary Fig. S2B and S2C), and V34 and V23 showed small increases in TUNEL staining compared with vehicle control. Upregulation of BCL2-like 11 (BIM) expression is often coincident with induction of apoptosis, because it can bind to and inhibit BCL-2 proteins and also bind and activate BAX (21). To obtain additional insight into the ability of RAF265 to induce apoptosis in melanoma tumors, Western blot analysis for BIM and for cleaved caspase 3 was conducted on representative tumors. We observed that treatment with RAF265 induced BIMEL, BIML, and BIMS isoforms (22) in representative RAF265-responsive tumors (V27, V35), while the nonresponding V29 tumor showed minimal change in BIM expression (Fig. 2A). V34 had an NRAS mutation and did not consistently exhibit induction of BIM in response to RAF265. Because BCL-2, BCL-XL, and MCL-1 bind to proapoptotic proteins like BAK, and BAX (23) to prevent apoptosis, we compared the levels of proapoptotic (BAX) and antiapoptotic proteins (BCL-2 and MCL1) and BIM in tumors from RAF265 responders and non-responders treated with RAF265 or vehicle control (Fig. 2A). Although BIM expression was induced in response to RAF265, we did not observe induction of BAX, BCL-2, or MCL1 in response to RAF265, though significant levels of these proteins were detected. V34 exhibited a reduction in MCL1 in RAF265-treated tumors. Moreover, we did not detect increases in cleaved caspase 3 in response to the RAF265 in the 4 responding tumors analyzed by Western blot (Fig. 2A). These data support the data from the TMA analysis in which we did not observe increased TUNEL staining in response to RAF265 for RAF265 responders, with the exception of V19, and one partial responder, V25 (Supplementary Fig. S2B). Altogether, these data suggest that sufficient BCL2 and MCL1 may be present in melanoma tumors to override the potential pro-apoptotic effect of enhanced BIM.
Genome-wide expression profiling
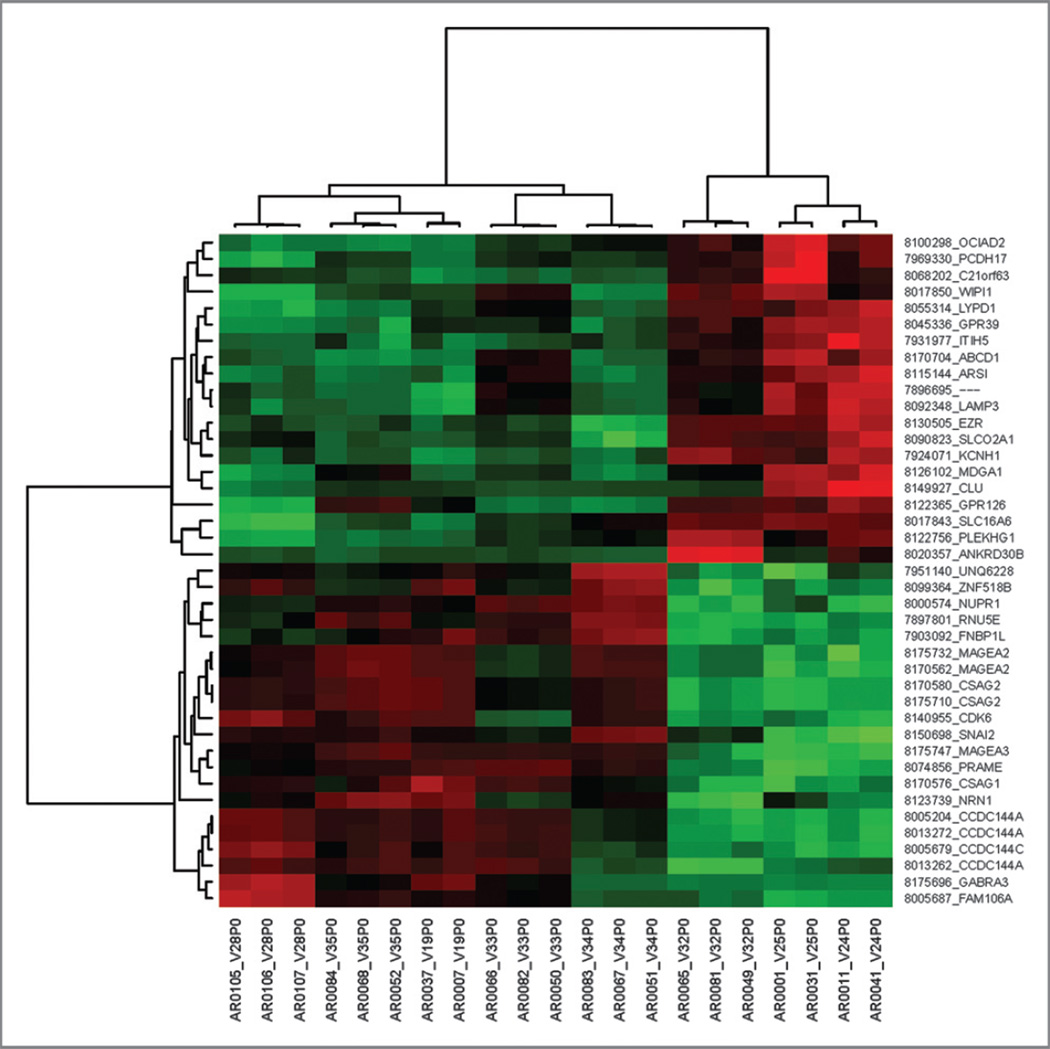
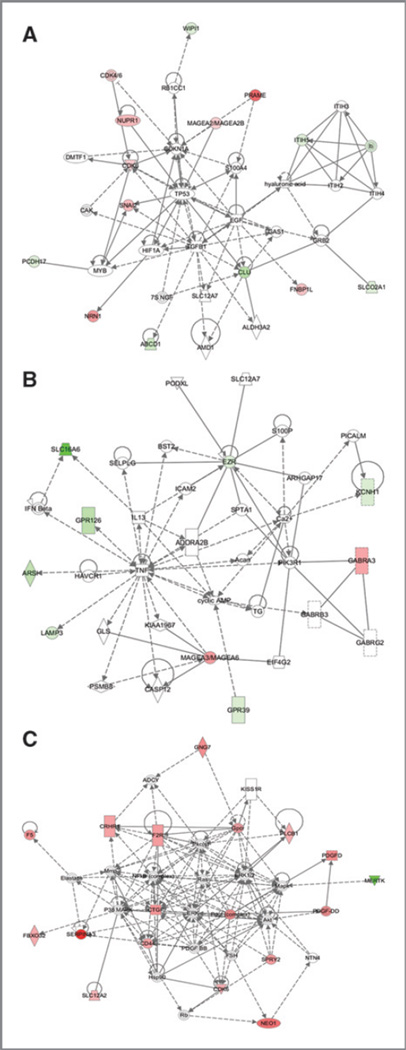
Because the RAF265-mediated tumor response occurred mainly among BRAFWT samples (Table 2), we directed our focus on human melanomas harboring wild-type BRAF. To identify the possible intrinsic molecular characteristics associated with RAF265-induced drug response, we conducted a global gene expression profiling assay by Affymetrix Human Gene ST 1.0 Array on human melanoma samples harboring wild-type BRAF. We sorted the data and carried out unsupervised clustering on the 28,869 interrogated genes, which allowed separation of wild-type BRAF human melanoma samples into 2 major categories, with 5 responder tumors (V28, V35, V19, V33, and V34) in one group and 3 non-responders (V32, V25, and V24) in the other group (Tables 2 and 3 and Fig. 3). Several genes that were expressed at a higher level in RAF265-responding BRAFWT tumors were in the cancer antigen family (CSAG2, MAGEA3, MAGEA2, PRAME, and CSAG1) while others are involved in cell cycle (CDK6, SNAI2). BRAFWT RAF265 responsive tumors also exhibited a lower level of expression of genes involved in transport (SLC16A6, KCNH1, SLCO2A1, ABCD1),metabolism (CLU, ARS1), or signaling (GPR126, GPR39, PLEKHG1). Of interest, based upon Ingenuity Analysis, the primary pathway that linked the differentially expressed genes in the RAF265 responders versus nonresponders were involved in cell development and growth regulation (Fig. 4A). The cell signaling pathways that linked differentially expressed genes in BRAFWT responders compared with BRAFWT non-responders showed hubs around TNF, PI3KR1, and MAGEA3/MAGEA6 (Fig. 4B). Gene Set Enrichment Analysis (GSEA) analyses were also conducted on the gene expression data and many of the genes identified as enriched in GSEA were significantly differentially expressed in the RMA/limma analysis (in Table 3–5 in which ** beside the gene name designates those genes identified as differentially expressed by both analyses). Moreover, the GSEA analysis BRAFWT tumors that did not respond to RAF265 showed enrichment of genes in the MAPK/RAS pathway, cancer, cell death, growth, and proliferation (Supplementary Table S4 and Fig. S6A).
Table 3.
Differentially expressed genes in BRAF WT melanoma tumors responding to RAF265 compared with those not responding to Raf265
| Gene ID | Description | Fold | Functional group P value | |
|---|---|---|---|---|
| CSAG2** | Cancer/testis antigen family 24, member 2 | 9.97 | Melanoma antigen | 2.37E-.06 |
| CCDC144A | Coiled-coil domain containing 144A | 7.21 | Unknown | 9.68E-.05 |
| PRAME** | Preferentially expressed antigen in melanoma | 7.21 | Melanoma antigen | 1.35E-.06 |
| NRN1** | Neuritin 1 | 4.87 | Melanoma protein | 0.083 |
| MAGEA3 | Melanoma antigen family A, member3 | 4.73 | Melanoma antigen | 9.44E-.05 |
| GABRA3 | γ-aminobutyric acid (GABA) A receptor, α3 | 4.30 | Signal transduction | 0.083 |
| CCDC144C | Coiled-coil domain containing 144C | 3.69 | Unknown | 0.032 |
| CSAG1 | Chondrosarcoma associated gene 1 | 3.40 | Melanoma antigen | 0.083 |
| NUPR1 | Nuclear protein 1 | 3.29 | Apoptosis | 0.004 |
| SNAI2 | Protein snail homolog 2 | 3.21 | EMT | 0.012 |
| RNU5E | U5E small nuclear RNA | 3.14 | Unknown | 0.003 |
| UNQ6228 | Hypothetical gene | 3.04 | Unknown | 0.083 |
| FNBP1L** | Formin binding protein 1-like | 2.93 | Lipid binding | 0.081 |
| FAM106A | Family with sequence similarity 106, member A | 2.53 | Unknown | 0.06 |
| ZNF518B | Zinc finger protein 518B | 2.51 | Transcription | 0.083 |
| CDK6 | Cyclin-dependent kinase 6 | 2.43 | Cell cycle | 0.044 |
| MAGEA2 | Melanoma antigen family A. member2 | 2.29 | Melanoma antigen | 0.026 |
| ITIH5** | Inter-α (globulin) inhibitor H5 | −2.18 | Metabolism | 0.083 |
| SLCO2A1** | Solute carrier organic anion transporter family, member 2A1 | −2.20 | Solute carrier transport | 0.026 |
| PCDH17** | Protocadherin 17 | −2.24 | Cell adhesion | 0.054 |
| WIPI1** | WD repeat domain, phosphoinositide interacting 1 | −2.38 | Autophagy | 0.053 |
| EZR | Ezrin | −2.40 | Cell adhesion | 0.006 |
| KCNH1 | Potassium voltage-gated channel, subfamily H (eag-related), member 1 |
−2.63 | Membrane protein | 0.006 |
| LAMP3 | Lysosomal-associated membrane protein 3 | −2.65 | Metastasis | 0.083 |
| GPR39 | G protein-coupled receptor 39 | −2.71 | Signal transduction | 0.032 |
| C21orf63 | Chromosome 21 open reading frame 63 | −2.75 | Membrane protein | 0.007 |
| PLEKHG1 | Pleckstrin homology domain containing, family G (with Gef domain) member 1 |
−3.27 | Signal transduction | 0.006 |
| ABCD1 | ATP-binding cassette, sub-family D (ALD), member 1 | −3.54 | Metabolism | 0.014 |
| ANKRD30B | Ankyrin repeat domain 30B | −3.54 | Unknown | 0.091 |
| MDGA1 | MAM domain containing glycosylphosphatidylinositol anchor 1 | −3.96 | Cell adhesion | 0.039 |
| ARSI | Arylsulfatase family, member l | −4.46 | Metabolism | 0.032 |
| GPR126** | G protein-coupled receptor 126 | −4.73 | Signal transduction | 0.039 |
| LYPD1 | LY6/PLAUR domain containing 1 | −5.07 | Membrane protein | 0.003 |
| CLU** | Clusterin | −5.08 | Metabolism | 0.039 |
| OCIAD2 | OCIA domain containing 2 | −5.44 | Unknown | 0.004 |
| SLC16A6** | Solute carrier family 16, member 6 (monocarboxylic acid transporter 7 |
−14.77 | Solute carrier transport | 0.002 |
NOTE: **, indicates those genes also detected as differentially expressed between responders and nonresponders by GSEA. The genes were chosen based on at least 2-fold differential expression, with less than 10%false discovery rate, between responder and nonresponder tumors. Fold change was noted to have a positive value when the mean expression is greater in the responder compared with nonresponder and has a negative value if the mean expression in the responder is lower than in the nonresponders.
Figure 3.
Heatmap of differentially expressed genes and their patterns. Unsupervised Clustering categorized wild typeBRAFhuman melanoma into two groups which was in agreement of the separation based on RAF265-induced responsiveness. The lines on the left are used to group the similar gene expression pattern together. These lines separate into two large groups based on whether the gene expression is up-regulated or down-regulated. Further, into each group, there are subgroups, each having genes sharing even more similar patterns as identified by the unsupervised clustering software. There are seven levels of gene expression patterns shown on the left. The top lines of the figure are used to group samples of RNA based on the similarity of gene expression patterns shown on the right side of the figure. These RNA samples were from primary patient tumors so they were labeled P0, for example V28P0 means passage 0 primary melanoma tumor from patient V28. Gene expression of three RNA sample repeats from each patient, except V19, 25, and 24 which only have two repeats, were included. The pseudo-color red indicates the up-regulation while green color for down-regulation of gene expression in the tumor RNA samples.
Figure 4.
Pathway analysis of differentially expressed genes in RAF265 responding versus nonresponding tumors. Ingenuity software was utilized to recognize pathways connecting genes that were differentially expressed in RAF265 responding versus nonresponding patients. A, Pathway analysis of genes involved in cell development and growth regulation that are differentially expressed in BRAFWT responding tumors to RAF265 versus those not responding to RAF265. B, Pathway analysis of genes involved in cell signaling, nucleotide metabolism, and small molecule biochemistry that are differentially expressed in BRAFWT tumors responding to RAF265 versus those not responding to RAF265. C, Pathway analysis of genes involved in cell signaling interactions that are differentially expressed in RAF265 responding versus nonresponding tumors, irrespective of BRAF mutation status.
Table 5.
Differentially expressed genes in melanoma tumors responding to RAF265 compared with those expressed in melanoma tumors not responding to RAF265
| Gene symbol | Description |
P value responder/ nonresponder |
Fold change/RAF265 responder/nonresponder |
|---|---|---|---|
| SERPINA3** | Serpin peptidase inhibitor clade A | 0.037 | 5.77 |
| CNTN1** | Contactin 1 | 0.018 | 5.04 |
| SLC25A27 | Solute carrier family25 member27 | 0.012 | 4.78 |
| MPPED2** | Metallophosphoesterase domain containing 2 | 0.024 | 4.66 |
| ANO4 | Anoctamin 4 | 0.007 | 4.07 |
| RBMY2EP | RNA-binding motif protein, Y-linked, 2E | 0.012 | 3.94 |
| TMEM47** | Transmembrane protein 47 | 0.005 | 3.91 |
| ST8SIA6 | ST8α-N-acetyl-neuraminide-α2,5-sialy-transferase | 0.023 | 3.87 |
| NEO1** | Neogenin homolog 1 | 0.013 | 3.84 |
| GPC3** | Glypican3 | 0.006 | 3.50 |
| SLC26A2** | Solute carrier family 26 member 2 | 0.034 | 3.42 |
| TXNDC13** | Thioredoxin domain containing 13 | 0.015 | 3.15 |
| PDGFD** | Platelet-derived growth factor D | 0.011 | 3.14 |
| ETNK2 | Ethanolamine kinase 2 | 0.032 | 3.12 |
| GNG7 | Guanine nucleotide binding protein gamma 7 | 0.001 | 3.09 |
| AMD1 | Adenosylmethionine decarbosylase 1 | 0.011 | 3.07 |
| ZNF320 | Zinc finger binding protein 320 | 0.011 | 2.99 |
| ANGPTL7 | Angiopoietin-like 7 | 0.0002 | 2.98 |
| NPAL3 | NPA-like domain containing 3 | 0.029 | 2.97 |
| F5 | Coagulation factor V | 0.015 | 2.95 |
| CTSF** | Cathepsin F | 0.0002 | 2.91 |
| CTGF** | Connective tissue growth factor | 0.028 | 2.90 |
| RAPGEF4 | Rap guanine nucleotide exchange factor | 0.011 | 2.88 |
| ABCA5 | ATP-binding cassette,A5 | 0.013 | 2.77 |
| RPL15 | Ribosomal protein L15 | 0.023 | 2.75 |
| GLT8D4 | Glycosyltransferase 8 domain containing 4 | 0.013 | 2.74 |
| WDR17 | WD repeat domain 17 | 0.039 | 2.74 |
| F2R* | Coagulation factor 2 receptor | 0.039 | 2.70 |
| SPRY2** | Sprouty homolog 2 | 0.039 | 2.70 |
| SCRG1** | Scrapie responsive protein 1 | 0.015 | 2.64 |
| MBNL2 | Muscleblind-like 2 | 0.034 | 2.63 |
| PCMTD2 | Protein-D-aspartate-O-methyltransferase | 0.023 | 2.62 |
| KCNJ10** | Potassium inwardly rectifying channel J10 | 0.024 | 2.57 |
| CRHR1** | Corticotropin releasing hormone receptor 1 | 0.026 | 2.56 |
| CD44 | CD44 molecule (Indian Blood Group) | 0.006 | 2.49 |
| CD59 | Complement regulatory protein | 0.038 | 2.46 |
| C5orf26 | Chromosome 5 open reading frame 26 | 0.019 | 2.42 |
| NR1D2 | Nuclear receptor subfamily1 D2 | 0.011 | 2.42 |
| GAPDHS | Glyceraldehyde-3-phosphate dehydrogenase | 0.032 | 2.39 |
| SNX7 | Sorting nexin 7 | 0.030 | 2.35 |
| FBXO32 | F-box protein 32 | 0.042 | 2.34 |
| FOXR2 | Forkhead box R2 | 0.037 | 2.31 |
| SPATA13 | Spermatogenesis associated 13 | 0.037 | 2.29 |
| SLITRK6 | SLIT, NTRK-like family member 6 | 0.039 | 2.28 |
| ARID5B | AT-rich interactive domain 5B | 0.013 | 2.28 |
| TAPT1 | Transmembrane anterior/posterior transformation | 0.032 | 2.27 |
| MCTP1 | Multiple C2 domains, transmembrane 1 | 0.031 | 2.27 |
| MRAP2 | Melanocortin 2 receptor accessory protein 2 | 0.033 | 2.25 |
| CDK6** | Cyclin-dependent kinase 6 | 0.046 | 2.24 |
| PDZRN3 | PDZ domain containing ring finger 3 | 0.036 | 2.24 |
| C20orf30 | Chromosome 20 ORF 30 | 0.033 | 2.23 |
| ZCCHC14 | Zinc finger CCHC domain containing 14 | 0.013 | 2.21 |
| RPL22 | Ribosomal protein L22 | 0.019 | 2.20 |
| TSC22D1 | TSC22 domain family member 1 | 0.001 | 2.19 |
| PLCB1** | Phospholipase C beta 1 | 0.032 | 2.18 |
| CYP39A1 | Cytochrome P450 family 39,A1 | 0.013 | 2.15 |
| CAMLG | Calcium-modulating ligand | 0.014 | 2.13 |
| AIG1 | Androgen induced 1 | 0.037 | 2.11 |
| AQP7P1 | Aquaporin 7 pseudogene 1 | 0.025 | 2.10 |
| ACSS2 | Actyl-CoA synthetase short-chain 2 | 0.006 | 2.09 |
| HPS5 | Hermansky-Pudlak syndrome 5 | 0.006 | 2.08 |
| CDC42EP3** | CDC42 effector protein 3 | 0.021 | 2.08 |
| ANKRD10 | Ankyrin repeat domain 10 | 0.004 | 2.08 |
| ZNF844 | Zinc finger protein 844 | 0.015 | 2.07 |
| PIGA** | Phosphatidylinositol glycan anchor biosynthesis A | 0.035 | 2.07 |
| AMFR | Autocrine motility factor receptor | 0.043 | 2.06 |
| FAM172A | Family with sequence similarity 172A | 0.027 | 2.04 |
| ENDOD1** | Endonuclease domain containing 1 | 0.037 | 2.04 |
| TBC1D19 | TBC1 domain family member 19 | 0.011 | 2.03 |
| SLC12A2** | Solute carrier family 12A2 | 0.039 | 2.03 |
| JMY** | Junction-mediating and regulatory protein | 0.037 | 2.01 |
| FLJ26056 | Hypothetical protein LOC375127 | 0.048 | 0.50 |
| LOC728212 | Hypothetical LOC 728212 | 0.038 | 0.48 |
| KNTC1** | Kinetochore associated 1 | 0.012 | 0.47 |
| DKFZP434L187 | Hypothetical LOC 26082 | 0.031 | 0.46 |
| WDHD1** | WD repeat & HMG-box DNA Binding Protein1 | 0.006 | 0.46 |
| MERTK** | c-mer proto-oncogene tyrosine kinase | 0.001 | 0.46 |
| DKFZP434B061 | DKFZP434B061 protein | 0.038 | 0.45 |
| NOSTRIN | Nitric oxide synthase trafficker | 0.032 | 0.38 |
NOTE. **, indicates those genes also detected as differentially expressed between responders and nonresponders by GSEA.
Using the same procedures described above, we analyzed the genes that were differentially expressed in BRAFV600E/K RAF265-responding versus nonresponding tumor implants. There were only 2 responders in this group (Table 2). Only 2 genes were identified to be differentially expressed between BRAFV600E/K RAF265 responders and nonresponders: integrinA10 (ITGA10), 8.6-fold elevated in responders, P <0.006); and calcium-dependent secretion activator (CADPS), 3.9-fold elevated in responders, P <0.02 (Table 4). These genes are involved in cell adhesion and vesicle exocytosis, respectively.
Table 4.
Differentially expressed genes in BRAF mutant melanoma tumors responding to RAF265 compared with those not responding to Raf265
| Gene symbol |
Description | P | Fold change |
|---|---|---|---|
| ITGA10 | Integrin α-10 calcium dependent |
0.006 | 8.6 |
| CADPS | Secretion activator1 | 0.019 | 3.9 |
| CPNE5 | Copine V -Ca–dependent phospholipids-binding protein 5 |
0.100 | 3.0 |
| SERINC5 | Serine incorporator 5 | 0.172 | 2.2 |
| NUP188 | Nucleoporin 188 | 0.166 | −2.0 |
Comparison of the genes differentially expressed in responders versus nonresponders, regardless of the BRAF mutation status, NUPR1 was most significantly upregulated (>9-fold) in responders (adjusted P = 0.045). NUPR1 is a nuclear protein and a transcriptional regulator that is structurally similar to high-mobility group family transcriptional regulators involved in stress response and cancer progression. NUPR1 regulates cell cycle, apoptosis, autophagy, access to chromatin, and is required for regulation of TGFβ responses (24). Using less stringent criteria (Hochberg’s step-up method) for data analysis, we identified 36 more genes with more than 2-fold differential expression (P <0.05) between responder and nonresponder tumors (Table 5). RAF265 responder tumors exhibited enhanced expression of genes the protein products of which are involved in cell growth (PDGFD, CTGF, SPRY2), transport (SLC25A27, SLC26A2, ABCA5, KCNJ10), metabolism (SERPINA3, MPPED2, TXNDC13, AMD1, CTSF, RPGGEF4, GLT8D4, PCMTD2), among other processes. Ingenuity pathway analysis placed a number of differentially expressed genes in the cell–cell signaling interaction pathway (NEO1, CDK6, SPRY2, FBXO32, PDGFD, CTGF, CD44, SERPINA3, SLC12A2, F5, CRHR1, F2R, GNG7, PLCB1, GPCR, MERTK; Fig. 4C). Additional pathway analyses for Raf265 responders versus nonresponders revealed connections that are shown in Supplementary Fig. S4A–C. Interestingly, many of these pathways indirectly intersect the ERK and NF-κB pathways. GSEA analysis revealed significant differences in expression of many of the same genes, as indicated by the ** in Table 5. Moreover, RAF265 nonresponders showed enrichment in genes involved in theMAPKpathway (MAP3K11) and elevated expression of HRAS and genes involved in cell cycle and cell proliferation (Supplementary Table S4 and Fig. S6B and S6C). A subset of these genes differentially expressed may prove to be clinically useful to predict responders versus nonresponders.
Discussion
The RAS-RAF-MEK-ERK signal transduction has been shown as one of the critical pathways for melanoma tumorigenesis, and inactivation of ERK through inhibition of BRAF is dependent on the mutation status of RAS and BRAF (11, 25–27). In this study, we observed a 41% (7 of 17) response rate for orthotopic implants of human melanoma after treatment with RAF265. Earlier phase II trials showed at most 1 of 34 patients clinically respond to Sorafenib alone (28). Although the numbers are small, preliminary reports suggest that RAF265 has a higher response rate than sorafenib in melanoma (17). Phase I clinical trials have shown some clinical antitumor activity for RAF265 and the clinical activity was not limited to BRAFV600E-mutated tumors. Phase I trials of RAF265 defined the MTD of oral RAF265 at 48 mg, and clinical activity was observed in melanoma patients with wild type and mutated BRAF (17). There is an ongoing phase 1b trial combining an MEK inhibitor with RAF265 in melanoma (NCT01352273).
In our study, based upon IHC and Western blot, we did not observe RAF265 inhibition of ERK1/2 phosphorylation in the BRAFV600E/K or BRAFWT tumors at the dose applied and the end point examined. Moreover, the ERK activation status in either nontransplant or transplant melanoma samples did not correlate with tumor response to RAF265. As has been reported with other RAF kinase inhibitors, RAF265 may not potently inhibit the ERK pathway in cells expressing wild-type BRAF (12–13, 26, 29, 30). However, Western blot and IHC analyses indicate reduction in pMEK1/2 in some RAF265-responding tumors. Although it is not surprising that the tumor cells develop RAF-independent mechanisms to activate ERK in BRAF wild-type melanoma implants in response to RAF265, the reduction in MEK activation coincident with no change in phospho-ERK1/2 is intriguing. We observed that the MAP3K COT that can directly phosphorylate ERK1/2 (20) is expressed in a representative set of the melanoma tumor implants analyzed herein, suggesting that COT may substitute for pMEK to phosphorylate ERK when MEK is inhibited. Alternatively, one might speculate that MAPK phosphatases like DUSP might be down-regulated by RAF265, resulting in retention of a lower level of ERK1/2 phosphorylation (31).
It has been previously shown using cell lines that RAF265 alone does not induce caspase3 activity or PARP cleavage, but it does reduce Cyclin D1 expression, in keeping with its ability to induce cell-cycle arrest (14, 30). Here, we observed a major decrease in phospho-Cyclin D1 and in PLK1 in response to RAF265 treatment of 4 of 4 responding orthotopic implants of human melanoma tumors based upon Western blot (Fig. 2). We also saw decreased total Cyclin D1 in TMA analysis for 4 of 7 responders and 2 of 3 partial responders, suggesting that cell-cycle arrest is a major mechanism of action of RAF265 in melanoma. However, in 2 RAF265 responders and 2 partial responders, there was some increase in TUNEL staining, indicating that induction of apoptosis can also contribute to RAF265 inhibition of melanoma growth.
Our study showed little overlap between the differentially expressed genes in our RAF265-responding versus nonresponding melanoma tumors with other signatures reported associated with BRAF-ERK signaling effects (32–34). The gene expression profile we identified in association with RAF265 nonresponsive tumors is quite different from that identified by Dry (35) to be associated with resistance to inhibitors of MEK, with the following exceptions: SPRY2, CD44, and CLU from the Dry analysis were also in the RMA/limma statistical analysis, while the GSEA analysis of BRAFWT responders detected ERBB1, TRIB1, SERPINB1 (down in resistant cells in the Dry study), while when the comparison was made between RAF265 responsive and nonresponsive tumors regardless of BRAF mutation status TYR, TRIO, CLU, FZD2, FGFR1, and COL5A1 from the Dry study were also shown to be enriched (Tables 3 and 5 and Supplementary Table S4). Results from these experiments are consistent with RAF265 having significant antitumor activity through ERK-dependent and ERK-independent pathways. Indeed, we observed that tumors that respond to RAF265, regardless of BRAF mutation status, exhibited a 3-fold higher basal expression of PDGFD which regulates cell cycle and VEGF and Notch-1 (36; Table 5), as well as elevated expression of genes involved in glycolysis/gluconeogenesis and energy metabolism, carbon fixation, intracellular transport, and signal transduction, while the nonresponding tumors exhibited upregulation of genes involved in nucleic acid synthesis, cell cycle, growth regulation (Supplementary Table S4). Though we did not conduct gene expression microarray analysis posttreatment, future studies will interrogate changes in gene expression profiles after RAF265 treatment.
Here, we report the BRAFWT human melanoma tumors that responded to RAF265 exhibited higher basal expression of genes encoding proteins that regulate (i) transcription of genes involved in cell cycle and apoptosis (NUPR1), (ii) initiation of EMT (SNAI2), (iii) promotion of cell cycle (CDK6), and (iv) elevated expression clusters of genes highly expressed in melanoma that regulate proteins involved in cell cycle [CTA family genes including MAGE3, MAGE2 (37, 38)]. CDK6 plays an important role for cell-cycle progression and G1–S transition, cell differentiation, apoptosis, tumor development, and metastasis (39, 40). SNAI2 is one of the critical regulators in early phase of EMT (41), in which it is involved in the transcriptional suppression of tumor cell E-cadherin and Puma, as well as the induction of Mt4-mmp (42–44). Both MAGEA2 and MAGEA3 function as suppressors of p53 to promote tumor progression (45–47), and MAGEA3 can downregulate p21 and promote Rb phosphorylation (48), whereas PRAME may downregulate genes that modulate apoptosis (49). Thus, our data indicate that the RAF265-responsive BRAFWT tumors exhibit higher basal expression of genes involved in cell-cycle progression and EMT than do BRAFWT tumors nonresponsive to RAF265. GSEA analysis of the gene expression data also showed that there is enrichment of genes involved in cell cycle and cell proliferation in RAF265-responsive tumors whereas RAF265-nonresponsive tumors (both BRAF mutant and WT) showed enrichment in genes involved in the MAPK pathway (MAP3K11), HRAS, and genes involved in cell cycle and cell proliferation.
The genes exhibiting reduced expression (before treatment with drug) in BRAFWT tumors identified as RAF265 responsive are associated with cell adhesion, migration, metabolism, biogenesis, solute carrier transport, and autophagy. The functions of protocadherin 17 (PDH17) and EZR are clearly related to cell adhesion and migration. (50–53). Because EZRIN and ABCD1 are both involved in multidrug resistance in melanoma (54), lower levels of expression may result in enhanced sensitivity to RAF265. SLC16A6 (MCT7) mRNA is reduced nearly 15-fold and is involved in the transport of lactate, pyruvate, and other monocarboxylates, and reduced expression could result in reduction of lactate-fueled ATP production impacting tumor cell survival in the glucose-deficient tumor microenvironment (55), Last, CLU encodes a multifunction protein clusterin, which is involved in cell death, cell cycle, apoptosis, cell proliferation, cell survival, cell differentiation (56). Altogether, the downregulation of genes involved in cell adhesion, multidrug resistance, and apoptosis in RAF265-responsive melanomas provides important insight into the mechanism for RAF265 inhibition of tumor growth in BRAFWT melanoma. In conclusion, RAF265 induces substantial growth inhibition of a subgroup of human melanoma tumors in an orthotopic implant tumor model, which closely represents the clinical setting. In this study, the antitumor effect of this small-molecule inhibitor gave a higher response rate in BRAFWT melanoma than in those melanoma tumors harboring BRAFV600E/K mutations. Drug treatment resulted in inhibition of expression of phospho-cyclin D1; total cyclin D1 and PLK1, upregulation of BIM1 and inhibition of cell proliferation based upon Ki-67 staining. In addition to RAF, RAF265 also targets tyrosine kinase receptors, such as VEGFR, PDGFR, c-KIT-mediated pathways. We show here that tumors that can respond to RAF265 exhibited higher basal expression of 2 growth factors, PDGFD and angiopoietin-like 7, both of which can affect angiogenesis. However, at the times analyzed here, we did not observe decreases in CD31 or factor VIII staining in RAF265-responsive tumor implants compared with nonresponsive tumor implants (Supplementary Fig. S5), though we cannot rule out the possibility that at earlier time points there may have been changes in angiogenesis or that there may be changes in the integrity of the vascular endothelium with RAF265 treatment. Examination of the drug response of orthotopic implants of human tumors on nude mice combined with analysis of global gene expression profiles clearly suggest the likelihood that pathways in addition to the RAS-RAF-MAPK pathway are involved in the responsiveness of melanoma tumors to RAF265. Furthermore, the resultant gene expression signature provides information related to the possible working molecular mechanisms for RAF265 in melanoma and also provides important information that may prove to be helpful for patient candidate selection for RAF265 treatment. It will be interesting to further study the gene expression pattern of patients treated with RAF265 in the clinic to determine whether the responders exhibit a similar gene expression profile.
Supplementary Material
Translational Relevance.
Analysis of response of patient tumors orthotopically implanted in mice provides a useful model for determining the potential therapeutic value of a drug for treatment of genetically identified patient subgroups. Our results show that RAF265 may provide a useful therapy for a subset of melanoma patients who are not eligible for treatment with therapeutics directed specifically to patients with BRAFV600 mutations. The gene expression signature of patients that respond to RAF265 provides information that may be helpful for selecting patients for RAF265 treatment.
Acknowledgments
The authors thank Donna J. Hicks and Donald Hucks for their assistance in running the SNaPshot mutational analysis of the samples; Susan Opleiear and Blake King, Division of Surgical Oncology, Department of Surgery, Vanderbilt Medical Center, for their assistance in experimental sample collection and processing; Anthony L. Frazier, Human Tissue Acquisition and Pathology Shared Resource, Vanderbilt-Ingram Cancer Center, for the assistance on tissue microarray preparation; Melissa Downing also from the VUMC Immunohistochemistry core for immunostaining staining of the TMAs; Joseph T. Roland, Ph.D. for assistance with the Ariol scanning of the TMAs; and Linda W. Horton for helpful assistance in managing the animal study protocols for this study.
The SNaPshot genotyping assay was conducted in the Vanderbilt Innovative Translational Research Shared Resource supported by the Vanderbilt-Ingram Cancer Center and the TJ Martell Foundation. Sequence validation of the mutations was done in the Vanderbilt Sequencing Core Facilities (CA68485).
Footnotes
Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.National Cancer Institute N, DHHS. Cancer trends progress report-2009/2010 update. 2010. [Google Scholar]
- 2.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan N, Telang S, Miller D, Chesney J. Novel immunotherapeutic agents and small molecule antagonists of signalling kinases for the treatment of metastatic melanoma. Drugs. 2011;71:1233–1250. doi: 10.2165/11591380-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 5.Janne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty KT, Fisher DE. New strategies in metastatic melanoma: oncogene-defined taxonomy leads to therapeutic advances. Clin Cancer Res. 2011;17:4922–4928. doi: 10.1158/1078-0432.CCR-10-2612. [DOI] [PubMed] [Google Scholar]
- 7.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolchok JD. Vemurafemib approved! Good news for melanoma patients. Medscape News Today: Hematology-Oncology. 2011 [Google Scholar]
- 9.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 12.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuart D, Aardalen KM, Lorenzana EG, Salagsang RD, Venetsanakos E, Tan N, et al. Characterization of a novel Raf kinase inhibitor that causes target dependent tumor regression in human melanoma xeno-grafts expressing mutant B-Raf. Proc Amer Assoc Cancer Res. 2006;47:4856. [Google Scholar]
- 15.Tseng JR, Stuart D, Aardalen K, Kaplan A, Aziz N, Hughes NP, et al. Use of DNA microarray and small animal positron emission tomography in preclinical drug evaluation of RAF265, a novel B-Raf/VEGFR-2 inhibitor. Neoplasia. 2011;13:266–275. doi: 10.1593/neo.101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramurthy S, Subramanian S, Aikawa M, Amiri P, Costales A, Dove J, et al. Design and synthesis of orally bioavailable benzimidazoles as Raf kinase inhibitors. J Med Chem. 2008;51:7049–7052. doi: 10.1021/jm801050k. [DOI] [PubMed] [Google Scholar]
- 17.Sharfman WH, Hodi FS, Lawrence DP, Flaherty KT, Amaravadi RK, Kim KB, et al. Results from the first-in-human (FIH) phase I study of the oral RAF inhibitor RAF265 administered daily to patients with advanced cutaneous melanoma. J Clin Oncol. 2011;29(Suppl 15):8508. [Google Scholar]
- 18.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fohn L, Su Z, Dahlman KB, Dias-Santagata D, Duke M, Hicks D, et al. A rapid sensitive test for detection of 43 mutations in 6 genes with relevance to targeted therapy in melanoma. Plos One. 2012 In press. [Google Scholar]
- 20.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118:3651–3659. doi: 10.1172/JCI35437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang CC, Lai F, Tay KH, Croft A, Rizos H, Becker TM, et al. Apoptosis of human melanoma cells induced by inhibition of B-RAFV600E involves preferential splicing of bimS. Cell Death Dis. 2010;1:e69. doi: 10.1038/cddis.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams JM, Cory S. Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol. 2007;19:488–496. doi: 10.1016/j.coi.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cano CE, Hamidi T, Sandi MJ, Iovanna JL. Nupr1: the Swiss-knife of cancer. J Cell Physiol. 2011;226:1439–1443. doi: 10.1002/jcp.22324. [DOI] [PubMed] [Google Scholar]
- 25.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan FM, Mastrangelo MJ, Aplin AE. The wrath of RAFs: rogue behavior of B-RAF kinase inhibitors. J Invest Dermatol. 2010;130:2669–2671. doi: 10.1038/jid.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen T, Ahmad T, Flaherty KT, Gore M, Kaye S, Marais R, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dittmer A, Fuchs A, Oerlecke I, Leyh B, Kaiser S, Martens JW, et al. Mesenchymal stem cells and carcinoma-associated fibroblasts sensitize breast cancer cells in 3D cultures to kinase inhibitors. Int J Oncol. 2011;39:689–696. doi: 10.3892/ijo.2011.1073. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Shen Q, Labow M, Gaither LA. Protein kinase D3 sensitizes RAF inhibitor RAF265 in melanoma cells by preventing reactivation of MAPK signaling. Cancer Res. 2011;71:4280–4291. doi: 10.1158/0008-5472.CAN-10-3761. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Kobayashi S, Borczuk AC, Leidner RS, Laframboise T, Levine AD, et al. Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis. 2010;31:577–586. doi: 10.1093/carcin/bgq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 33.Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 34.Pavey S, Johansson P, Packer L, Taylor J, Stark M, Pollock PM, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23:4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 35.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Kong D, Banerjee S, Li Y, Adsay NV, Abbruzzese J, et al. Down-regulation of platelet-derived growth factor-D inhibits cell growth and angiogenesis through inactivation of Notch-1 and nuclear factor-kappaB signaling. Cancer Res. 2007;67:11377–11385. doi: 10.1158/0008-5472.CAN-07-2803. [DOI] [PubMed] [Google Scholar]
- 37.Vasseur S, Hoffmeister A, Garcia S, Bagnis C, Dagorn JC, Iovanna JL. p8 is critical for tumour development induced by rasV12 mutated protein and E1A oncogene. EMBO Rep. 2002;3:165–170. doi: 10.1093/embo-reports/kvf023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang WG, Davies G, Martin TA, Kynaston H, Mason MD, Fodstad O. Com-1/p8 acts as a putative tumour suppressor in prostate cancer. Int J Mol Med. 2006;18:981–986. [PubMed] [Google Scholar]
- 39.Meyer CA, Jacobs HW, Datar SA, Du W, Edgar BA, Lehner CF. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 2000;19:4533–4542. doi: 10.1093/emboj/19.17.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimoto T, Anderson K, Jacobsen SE, Nishikawa SI, Nerlov C. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26:2361–2370. doi: 10.1038/sj.emboj.7601675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh M. Comparative genomics on SNAI1, SNAI2, and SNAI3 ortho-logs. Oncol Rep. 2005;14:1083–1086. [PubMed] [Google Scholar]
- 42.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 43.Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, et al. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11:1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gjerstorff MF, Kock K, Nielsen O, Ditzel HJ. MAGE-A1, GAGE and NYESO-1 cancer/testis antigen expression during human gonadal development. Hum Reprod. 2007;22:953–960. doi: 10.1093/humrep/del494. [DOI] [PubMed] [Google Scholar]
- 46.Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, et al. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A. 2006;103:11160–11165. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 48.Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68:8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 49.Steinbach D, Pfaffendorf N, Wittig S, Gruhn B. PRAME expression is not associated with down-regulation of retinoic acid signaling in primary acute myeloid leukemia. Cancer Genet Cytogenet. 2007;177:51–54. doi: 10.1016/j.cancergencyto.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, et al. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31:1027–1036. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 51.Mielgo A, Brondani V, Landmann L, Glaser-Ruhm A, Erb P, Stupack D, et al. The CD44 standard/ezrin complex regulates Fas-mediated apoptosis in Jurkat cells. Apoptosis. 2007;12:2051–2061. doi: 10.1007/s10495-007-0115-3. [DOI] [PubMed] [Google Scholar]
- 52.Hiscox S, Jiang WG. Ezrin regulates cell-cell and cell-matrix adhesion, a possible role with E-cadherin/beta-catenin. J Cell Sci. 1999;112(Pt 18):3081–3090. doi: 10.1242/jcs.112.18.3081. [DOI] [PubMed] [Google Scholar]
- 53.Endo K, Kondo S, Shackleford J, Horikawa T, Kitagawa N, Yoshizaki T, et al. Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene. 2009;28:1725–1735. doi: 10.1038/onc.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 55.Draoui N, Feron O. Lactate shuttles at a glance: from physiological paradigms to anti-cancer treatments. Dis Model Mech. 2011;4:727–732. doi: 10.1242/dmm.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.