Abstract
The slow kinetics and low efficiency of reprogramming methods to generate human induced pluripotent stem cells (iPSCs) impose major limitations on their utility in biomedical applications. Here we describe a chemical approach that dramatically improves (>200 fold) the efficiency of iPSC generation from human fibroblasts, within seven days of treatment. This will provide a basis for developing safer, more efficient, non-viral methods for reprogramming human somatic cells.
Recent advances in generating human induced pluripotent stem cells (iPSCs)1–3 have raised hopes for their utility in biomedical research and clinical applications4. However, iPSC generation is still a very slow (~4 weeks) and inefficient (≤0.01%1,2) process that results in a heterogeneous population of cells. Identifying fully reprogrammed iPSCs from such a mixture is tedious, and requires specific expertise in human pluripotent cell culture.
Although the dangers of genome insertions of exogenous reprogramming factors is being overcome3, the low efficiency and slow kinetics of reprogramming continue to present a problem for ultimate applications of human iPSC. For example, genetic or epigenetic abnormalities could be enriched during the reprogramming process, where tumor suppressors may be inhibited and oncogenic pathways may be activated. Though recent studies have reported an improved efficiency of reprogramming by genetic manipulations4 in addition to the original four factors, they typically make the process even more complex, and increase the risk of genetic alterations and tumorigenicity. Thus there is still a tremendous need for a safer, easier and more efficient procedure for human iPSC generation, which would also facilitate identifying and characterizing fundamental mechanisms of reprogramming.
During four-factor (OCT4, SOX2, KLF4 & c-MYC; 4TFs hereafter) mediated reprogramming, mesenchymal type fibroblasts undergo dramatic morphological changes that result in iPSCs with distinct cell polarity, boundaries and cell-cell interactions. The cells start expressing E-cadherin, a marker for epithelial cells5, which is also highly expressed in human embryonic stem cells (hESCs). We reasoned that factors that promote the mesenchymal to epithelial transition (MET), such as TGFβ pathway antagonists, would have a direct impact on the reprogramming process. In addition, MEK-ERK pathway inhibition was previously shown to play an important role in various steps of reprogramming6,7. Furthermore, factors promoting cell survival could also be beneficial in improving reprogramming efficiency. Consequently, we focused on small molecules that can regulate these three processes and pathways, as small molecules have many advantages4,7,8 in studying biological processes and are a safer choice than genetic manipulation. Here we describe a simple chemical platform that substantially enhances generation of fully reprogrammed human iPSCs from fibroblasts through a much faster and more efficient process.
We tested known inhibitors of TGFβ receptor and MEK on 1×104 human primary fibroblasts (CRL2097 or BJ) that were retrovirally transduced with the 4TFs, for their effect on reprogramming kinetics and efficiency (see Fig. 1a for details). On day 7 (D7) post-infection, the compounds were added, individually or in combinations, and the cultures were examined for iPSCs over the next 1–3 weeks.
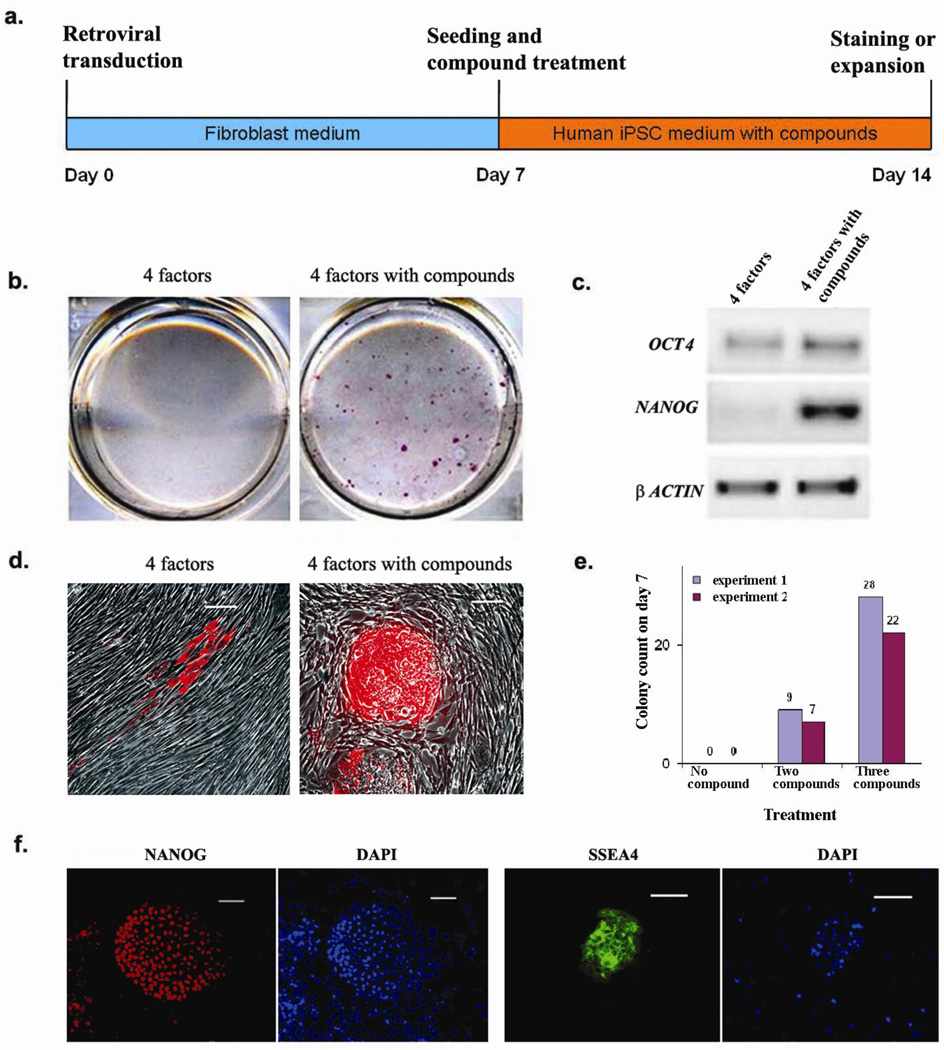
Figure 1. Compound treatment for seven days is sufficient to induce pluripotent stem cells from human fibroblasts transduced with the four reprogramming factors.
(a) Timeline for human iPSC induction using combined SB431542 and PD0325901 treatment along with 4TFs. Treatment began with cell re-seeding at day 7 after 4TF transduction and was maintained for 7 days. (b) Staining for ALP+ colonies that emerged in the untreated (left) or 2 compound-treated (right) cultures within seven days. (c) RT-PCR showing elevated endogenous mRNA expression of pluripotency markers OCT4 and NANOG in 2 compound-treated cultures. (d) Tra-1-81 staining at day 14 without (left) or with (right) 2 compound treatment. (e) The numbers of NANOG+ colonies at day 14 under different treatment conditions are plotted. (f) Typical staining for hESC-specific markers (NANOG and SSEA4) exhibited by D14 iPSCs. Scale bars, 50 µm in (d & f)
On day 7 post-treatment (D14) we observed the strongest effect in the cultures treated with a combination of ALK5 inhibitor SB431542 (2 µM) and MEK inhibitor PD0325901 (0.5 µM), which resulted in ~45 large ALP+ colonies (Fig. 1b) with characteristic hESC-like morphology, of which over 24 colonies were TRA-1-81+ (Fig. 1d), and about 6–10 colonies stained positive for SSEA4 and NANOG, a mature pluripotency factor that is not ectopically introduced (Fig. 1e and 1f). Moreover, the treated cultures showed high level expression of endogenous mRNA for the pluripotency genes (Fig. 1c). In contrast, we did not observe any NANOG+ colonies in the untreated control cultures (Fig. 1e & Supplementary Fig. 1a) or in cultures that were treated with PD0325901 alone (Supplementary Fig. 1a). However, in the cultures treated with only SB431542 we could still observe 1–2 ALP+ hESC-like colonies (Supplementary Fig. 1a). Importantly, the combined effect of both the inhibitors (Supplementary Fig. 1b & 1c), as well as the individual effect of SB431542 (data not shown) was dose dependent.
When we maintained the SB431542 plus PD0325901 treated cultures for 30 days without splitting, we obtained about 135 iPSC colonies per well (Fig. 2d), a >100 fold improvement in efficiency over the conventional method. Consistent with previous reports1, in untreated controls carrying 4TFs, we observed 1–2 iPSC colonies in addition to several granulate colonies (Fig. 2c). These granulate structures have been suggested to be partially reprogrammed colonies1. We also observed granulate colonies in the SB431542 treated cultures, which outnumbered by several fold the few hESC-like colonies. Interestingly, the number of granulate colonies was dramatically reduced in the combined SB431542 and PD0325901 treatment, which resulted in a concomitant increase in the number of hESC-like colonies. This suggested that a combined inhibition of ALK5 and MEK may guide partially reprogrammed colonies to a fully reprogrammed state and thereby improving the overall reprogramming process. Moreover, that we observed improved induction of iPSCs as early as 7 days post-treatment suggests that treatment with these small molecules not only improves the efficiency of the reprogramming process but may also accelerate its kinetics (Fig. 1a), although additional experiments are required to determine whether the reprogrammed cells at this stage indeed become fully independent of exogenous reprogramming factors earlier than in untreated cultures.
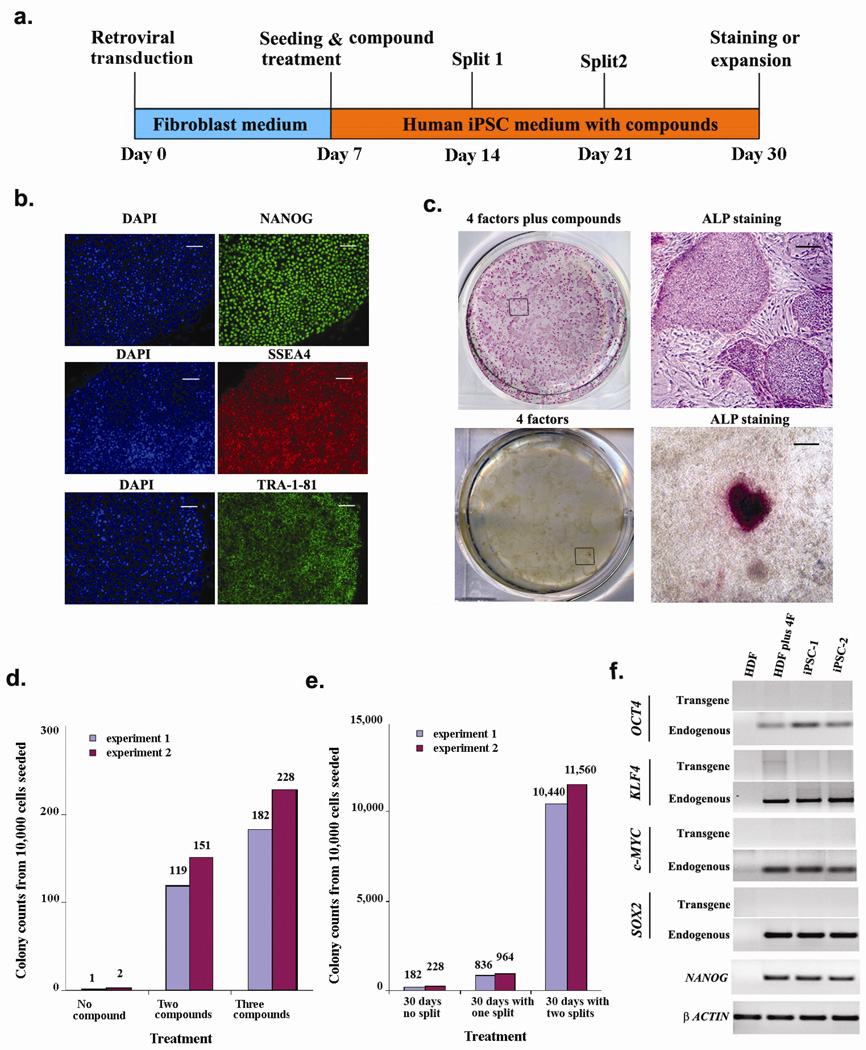
Figure 2. Prolonged compound treatment and cell passaging dramatically increased the number of reprogrammed colonies.
(a) Timeline of human iPSC induction using SB431542, PD0325901 and thiazovivin. (b) Day 30 iPSCs expressed pluripotency markers NANOG, SSEA4 and TRA-1-81. Scale bars, 50 µm (c) ALP staining of day 30 cultures with (upper panels) or without (lower panels) 3 compound treatment. Boxed areas in the left panels are enlarged in the right panels. Scale bars, 200 µm (d) Number of NANOG+ colonies on day 30 under different treatment conditions, without splitting. (e) Number of NANOG+ colonies on day 30 from 3 compound-treated cultures trypsinized as indicated. (f) RT-PCR on iPSC colonies obtained with 3 compound treatment showing reactivated expression of endogenous pluripotency markers. HDF: Human Dermal Fibroblast.
Although we could pick and expand the iPSC colonies, as in hESC cultures, splitting the cultures by trypsinization resulted in poor survival. From a recent screen performed in our laboratory we identified a novel small molecule, Thiazovivin (Supplementary Fig. 2), which dramatically improves the survival of hESCs upon trypsinization (manuscript under preparation). Addition of Thiazovivin to our cocktail of SB431542 and PD0325901 also vastly improved the survival of iPSCs after splitting by trypsinization (Fig. 2a), and meant that we could obtain a large number of reprogrammed colonies. From 10,000 cells that were originally seeded, a single 1:4 splitting on day 14 resulted in ~1,000 hESC-like colonies on day 30 (Fig. 2e), while two rounds of splitting (on day 14 and on day 21 (1:10)) resulted in ~11,000 hESC-like colonies (Fig. 2c & 2e) on day 30. These colonies showed high levels of endogenous mRNA (Fig. 2f) and protein expression (Fig. 2b & 2c) of pluripotency markers, while the expression of the four transgenes could hardly be detected (Fig. 2f). In contrast, we did not obtain any iPSC colonies from untreated or 2 compound-treated samples that were trypsinized (Supplementary table 1).
To examine whether the positive effect of Thiazovivin is solely due to survival of colonies after splitting or whether it also augments the reprogramming effect of combined SB431542 and PD0325901 treatment, we tested the 3 compound cocktail on 4TF-transduced cells that were not subjected to splitting. Interestingly, in these cultures, by day 14 we observed ~25 large colonies that were all expressing Nanog (Fig. 1e). By day 30 we observed ~205 very large NANOG+ colonies (Fig. 2d), that were also TRA-1-81+ and SSEA4+ (data not shown), which translates to a more than 200 fold improvement in efficiency over no compound treatment, and a two-fold increase over 2 compound treatment.
Two compound treatment also resulted in a larger number of alkaline phosphatase-positive colonies compared to untreated controls when the reprogramming factors were introduced using a lentiviral, rather than a retroviral system (Supplementary Fig. 3a). Furthermore, the 3 compound cocktail does not appear to influence reprogramming factor expression from retroviral vectors (Supplementary Fig. 3b–f). Together, these results suggest that the compounds do not act merely by inhibiting silencing of the retrovirally expressed reprogramming factors.
The iPSC colonies generated using the 3 compound cocktail could be readily and stably expanded for long term under conventional hESC culture conditions (over 20 passages) and they closely resemble hESCs in terms of morphology, typical pluripotency marker expression and differentiation potentials. They exhibited a normal karyotype (Supplementary Fig. 4) and could be differentiated into derivatives of all three germ layers, both in vitro (Fig. 3a & 3b) and in vivo (Fig. 3c). These results also suggest that there is no short term adverse effect associated with the much more convenient trypsinization procedure.
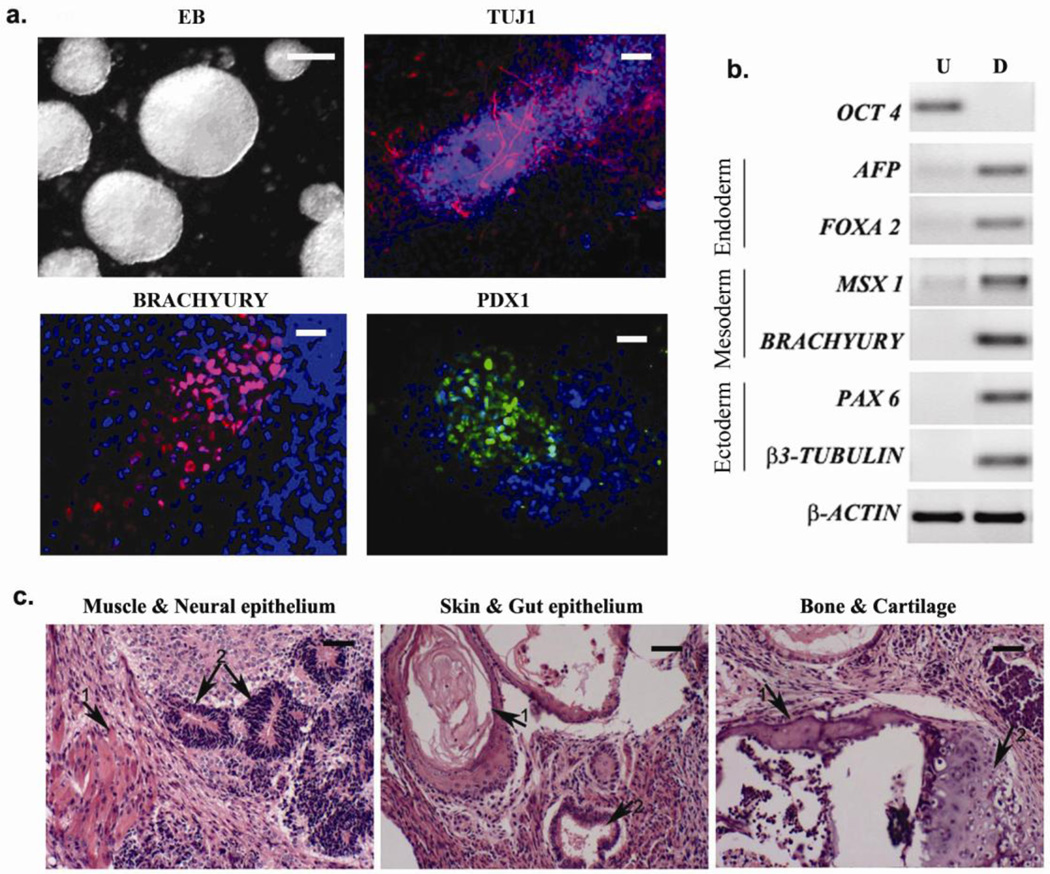
Figure 3. In vitro and in vivo differentiation of iPSCs generated with 3 compound treatment.
(a) Micrographs show embryoid bodies (EB) generated from iPSCs and in vitro differentiation into ectodermal (βIII tubulin), mesodermal (brachyury) and endodermal (PDX1) cell types. Scale bars, EB: 100 µm; others 10 µm (b) RT-PCR showing expression of representative lineage markers and the absence of OCT4 mRNA expression in differentiating cells. U- undifferentiated, D- differentiated. (c) Teratomas generated in nude mice from iPSCs (3 independent colonies tested) consist of tissues from all three germ layers. Left panel: 1- muscle, 2- neural epithelium; middle panel: 1- skin, 2- gut epithelium; right panel: 1- bone, 2- cartilage. Scale bars, 20 µm.
The demonstration that TGFβ and MEK-ERK pathway inhibition improves fibroblast reprogramming suggests critical roles for MET mechanisms in the process. Consistently, addition of TGFβ has an inhibitory effect on 4 factor-mediated reprogramming of fibroblasts (data not shown). TGFβ and its family members play important roles in self-renewal and differentiation of ESCs9. Moreover, TGFβ is a prototypical cytokine for induction of epithelial mesenchymal transition (EMT) and maintenance of the mesenchymal state10. A major end point of this signaling, in this context, is down regulation of E-cadherin11. E-cadherin has been shown to be important for the maintenance of pluripotency of ESCs and has been recently suggested to be a regulator of NANOG expression12. Therefore inhibition of TGFβ signaling, which results in de-repression of epithelial fate, could benefit the reprogramming process in multiple ways. ERK signaling also promotes EMT11, and is downstream of TGFβ in the process12. We had previously shown that the effect of reversine, a small molecule which can reprogram myoblasts to a multipotent state, is mediated in part through inhibition of MEK-ERK6. This may explain the effect observed in reprogramming when it was combined with TGFβ inhibition.
Concurrent with this study, two other studies reported an improvement in reprogramming efficiency in human cells, either by epigenetic manipulation13 or by reprogramming keratinocytes14. However, the chemical platform described here is unique, in that it modulates upstream signaling pathways and could radically improve reprogramming on a general cell type, like fibroblasts. The chemical conditions described here could provide a basic platform on which a non-viral and non-DNA based15, more efficient and safer reprogramming process could be developed, which could yield an unlimited supply of safe human iPSCs for various applications.
METHODS
Cell culture
Primary skin fibroblasts CRL2097 and BJ (neonatal foreskin) were purchased from ATCC. All cell culture media reagents were purchased from Invitrogen Corporation, CA. The cells were maintained in DMEM (10313-021) containing 10% FBS (10439-024), 1X MEM Non-Essential Amino acid (11140-050), 1X Glutamax (35050-061), 10 mM Hepes (15630-080) and 0.11mM 2-Mercaptoethanol (21985-023). Cells were passaged 1:5 using 0.05% (1X) trypsin-EDTA (25300-054).
Plasmids
The pMXs vector encoding the human cDNAs for OCT4, SOX2, c-MYC and KLF4, described before1, were obtained from ADDGENE. Mouse Slc7a1 ORF was cloned into pWPXLD (Addgene), as described previously1
Retroviral Infection and iPS Cell Generation
Lentiviruses carrying OCT4, NANOG, SOX2 & LIN28 were produced as described before2. For retrovirus production, PLAT-E packaging cells were plated at 1×106 cells/well of a 6-well plate. After 24 hours, the cells were transfected with pMXs vectors carrying OCT4, SOX2, c-MYC and KLF4 cDNAs using Fugene 6 transfection reagent (Roche) according to manufacturer’s instructions. Twenty-four hours after transfection, the medium was replaced with fresh medium and the plate was transferred to 32 °C for retrovirus production. The viruses were collected at 48 hours and 72 hours, and filtered with 0.45 µm filter before transduction.
The Slc7a1-expressing human fibroblast cells were seeded at 1×105 cells/well of a 6 well plate on the day 1. On day 2, 0.25 ml of each retroviral supernatant was added to the cells in the presence of 6 µg/ml polybrene. A second round of transduction was done on day 3. Infection efficiency was estimated by fluorescence microscopy on cells transduced in parallel with GFP or RFP gene-carrying retroviruses. Seven days after initial transduction, fibroblasts were harvested by trypsinization and re-plated at 1×104 cells/well of a 6 well plate coated with matrigel (1:50 dilution, cat 354234, BD Biosciences). For compound treatment, the cells were cultured in human reprogramming medium (DMEM/F12, 20% Knockout serum replacer, 1x MEM Non-Essential amino acid, 1x glutamax, 0.11 mM 2-Mercaptoethanol, 20 ng/ml bFGF and 1,000 U/ml LIF) and were treated with 2 µM SB431542 (Stemgent), 0.5 µM PD0325901 (Stemgent), 0.5 µM Thiazovivin, or combinations of the compounds. The media were changed every 2–3 days depending on the cell density. Seven days after compound treatment, either the plates were fixed and stained for Alkaline phosphatase (ALP) activity, or stained for protein markers, or the cultures were continued with or without indicated splitting by trypsinization till day 30. For split cultures, the cells were split (1:4) and re-plated onto irradiated CF-1 MEF feeder layer (2.5 × 105 cells/well) in each well of 6 well plate and were split (1:10) again on day 21. The cells were maintained in the same media and compound cocktail described above except for the concentrations of PD0325901 (0.5 µM for D14 and 1 µM for D21) and SB431542 (0.5–1 µM after D14). The iPSC colonies were subsequently maintained in conventional hESC media in the absence of the above compounds.
Alkaline Phosphatase Staining and Immunocytochemistry
Alkaline phosphatase staining was performed using ALP detection kit (cat no: SCR004, Chemicon) according to the product instructions. For immunocytochemistry, cells were fixed in 4% paraformaldehyde (10 min, RT), washed twice with PBS, blocked using 5% normal donkey serum (Chemicon) and 0.1% TritonX-100 (15 min, RT) and then treated with primary antibodies overnight at 4 °C. The primary antibodies used were anti-NANOG (cat no: AB9220, Chemicon, 1:1,000); anti-OCT4 (cat no: sc-5279, Santa Cruz biotech, 1:200), anti-SSEA 4 (cat no: mab4304, Chemicon, 1:500), anti-Tra-1-81 (cat 560123, BD Biosciences, 1:100), anti-Tra-1-81 (mAb 4381, Chemicon, 1:500), anti-βIII TUBULIN (cat no: MMS-435P, Covance Research Products Inc, 1:1000), anti-PDX 1 (1:500) (a kind gift from Dr. C. Wright), anti-BRACHYURY (cat No: AF2085, R&D, final concentration 0.2 µg/ml). The cells were washed twice with PBS and then treated with secondary antibodies for 1 hour at room temperature. The secondary antibodies used were Alexa fluor 488 donkey anti-rabbit or anti-mouse IgG (Invitrogen, 1:1,000) and Alexa fluor 555 donkey anti-rabbit or anti-mouse IgG (Invitrogen, 1:1,000). Nuclei were stained with 0.5 µg/ml DAPI (Sigma). Images were captured using a Nikon Eclipse TE2000-U/X-cite 120 EXFO microscope with a photometric CoolSnap HQ2 camera.
In Vitro Differentiation and Teratoma Assay
Generation of embryoid bodies and in vitro differentiation were performed as described elsewhere1. For the teratoma assay, 3–5 million cells were injected under the kidney capsule of SCID mice. Thirty one days later the tumors were excised and fixed in 4% paraformaldehyde and histologically analyzed at the TSRI histology core facility. The use of SCID mice was approved by the UCSD animal research committee.
RT-PCR
Total RNA was extracted from cells using RNeasy minikit (Qiagen). cDNAs were synthesized according to product instructions using superscript III first strand synthesis kit (Invitrogen). Two microliters of the reaction product was used for 24–28 PCR cycles using respective primers. The sequences of the primers are described elsewhere1.
Flow cytometry
For flow cytometry analysis, the cultures were mildly trypsinized and harvested from 6 well plates. The cells were washed and resuspended in FACS buffer (PBS, 2 mM EDTA, 2 mM HEPES, 1% FBS), and were analyzed on a FACS Calibur cytometer (Becton Dickinson, San Jose, CA) with the CellQuest program.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from NIH and Fate Therapeutics to SD. We thank Ronald Coleman for the assistance in the preparation of the manuscript and all the members of Ding lab for helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
TL and SD initiated the study. SD, RAn and TL made the hypothesis, designed the experiments and wrote the manuscript. TL, RAn, XY, HSH, WL conducted the experiments. SH, RAy & XL provided assistance in some of the experiments. EH generated teratomas and AH supervised EH. SD supervised the study.
References
- 1.Takahashi K, et al. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Muller LUW, Daley GQ, Williams DA. Mol. Ther. 2009;17:947–953. doi: 10.1038/mt.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng B, Ng JH, Heng JCD, Ng HH. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Hay ED. Acta Anat. (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, et al. Proc. Natl. Acad. Sci. U S A. 2007;104:10482–10487. doi: 10.1073/pnas.0704360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, et al. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Shi Y, Ding S. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 9.Watabe T, Miyazono K. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 10.Willis BC, Borok Z. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;293:L525–L534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 11.Thiery JP, Sleeman JP. Nat. Rev. Mol. Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 12.Chou YF, et al. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huangfu D, et al. Nat. Biotechnol. 2008;11:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 14.Aasen T, et al. Nat. Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H, et al. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.