Abstract
BACKGROUND
Cutaneous squamous-cell carcinomas and keratoacanthomas are common findings in patients treated with BRAF inhibitors.
METHODS
We performed a molecular analysis to identify oncogenic mutations (HRAS, KRAS, NRAS, CDKN2A, and TP53) in the lesions from patients treated with the BRAF inhibitor vemurafenib. An analysis of an independent validation set and functional studies with BRAF inhibitors in the presence of the prevalent RAS mutation was also performed.
RESULTS
Among 21 tumor samples, 13 had RAS mutations (12 in HRAS). In a validation set of 14 samples, 8 had RAS mutations (4 in HRAS). Thus, 60% (21 of 35) of the specimens harbored RAS mutations, the most prevalent being HRAS Q61L. Increased proliferation of HRAS Q61L–mutant cell lines exposed to vemurafenib was associated with mitogen-activated protein kinase (MAPK)–pathway signaling and activation of ERK-mediated transcription. In a mouse model of HRAS Q61L–mediated skin carcinogenesis, the vemurafenib analogue PLX4720 was not an initiator or a promoter of carcinogenesis but accelerated growth of the lesions harboring HRAS mutations, and this growth was blocked by concomitant treatment with a MEK inhibitor.
CONCLUSIONS
Mutations in RAS, particularly HRAS, are frequent in cutaneous squamous-cell carcinomas and keratoacanthomas that develop in patients treated with vemurafenib. The molecular mechanism is consistent with the paradoxical activation of MAPK signaling and leads to accelerated growth of these lesions. (Funded by Hoffmann–La Roche and others; ClinicalTrials.gov numbers, NCT00405587, NCT00949702, NCT01001299, and NCT01006980.)
The T→A transversion at position 1799 of BRAF (BRAF V600E) is present in approximately 50% of patients with metastatic melanoma.1,2 BRAF V600E induces constitutive signaling through the mitogen-activated protein kinase (MAPK) pathway, stimulating cancer-cell proliferation and survival.2 The clinical development of inhibitors of oncogenic BRAF, termed type I BRAF inhibitors, which block the active conformation of the BRAF kinase, has led to a high rate of objective tumor responses and improvement in overall survival, as compared with standard chemotherapy.3–5 However, nonmelanoma skin cancers — well-differentiated cutaneous squamous-cell carcinomas and keratoacanthomas — have developed in approximately 15 to 30% of patients treated with type I BRAF inhibitors such as vemurafenib and dabrafenib (GSK-2118436).3,4,6
The antitumor activity of vemurafenib against BRAF V600E–mutant cells in cell cultures, animal models, and humans is associated with the inhibition of oncogenic MAPK signaling, as evidenced by the inhibition of phosphorylated ERK (pERK), a downstream effector of BRAF that is active when phosphorylated.3,7–11 However, BRAF inhibitors induce the opposite effect — that is, increasing pERK in cell lines with wild-type BRAF that harbor upstream pathway activation such as oncogenic RAS or up-regulated receptor tyrosine kinases.12–14 This RAF inhibitor–dependent activation of MAPK signaling in BRAF wild-type cells is termed paradoxical MAPK-pathway activation15 and is driven by the formation of RAF dimers that lead to signaling through CRAF and consequently MAPK-pathway hyperactivation.12–14
Studies modeling cutaneous squamous-cell carcinomas and keratoacanthomas in mice suggest that these tumors develop from a multistep process whereby an initial carcinogenic event (carcinogenesis inducer), driven by a chemical carcinogen or ultraviolet-light exposure, is followed by a tumor-promoting event.16 The initiating event in the commonly used two-stage skin carcinogenesis model is an oncogenic driver mutation in RAS, preferentially in HRAS.17,18 In humans, sporadic, well-differentiated cutaneous squamous-cell carcinomas and keratoacanthomas harbor HRAS mutations at a frequency of 3 to 30%,19,20 which is less frequent than in the mouse model. In some of these lesions in humans, receptor tyrosine kinases such as the epidermal growth factor receptor (EGFR)19 are hyperactive, which would also activate RAS and consequently MAPK signaling. Other reported oncogenic events in these lesions in humans include frequent mutations or deletions in TP5321,22 and the cell-cycle control gene CDKN2A.
METHODS
PATIENTS AND LESION SAMPLES
Patients participated in the vemurafenib phase 1 dose-escalation study (ClinicalTrials.gov number, NCT00405587), the phase 2 study (NCT00949702), the phase 3 study (NCT01006980), or the drug–drug interaction study (NCT01001299). All patients had BRAF V600–mutant metastatic melanoma and received 720 or 960 mg of vemurafenib orally twice a day. Patients provided written informed consent for molecular analyses of the skin-cancer lesions excised during dermatologic examinations while they were in the study.
MOLECULAR ANALYSES OF TUMOR SPECIMENS
DNA extracted from the tumor specimens was sequenced for HRAS (exons 1 and 2), NRAS (exons 1 and 2), KRAS (exons 1 and 2), and CDKN2A (exon 2) with the use of polymerase-chain-reaction (PCR) amplification. (For primers, see Tables 1 and 2 in the Supplementary Appendix, available with the full text of this article at NEJM.org.) This was followed by Sanger sequencing23 (see the Methods section in the Supplementary Appendix). Single-base substitutions or deletions in TP53 exons 2 through 11 were analyzed with the use of an investigational AmpliChip p53 Test (Roche Molecular Systems), according to the manufacturer’s instructions. ERK phosphorylation was assessed by means of immunohistochemical analysis.
CELLULAR ANALYSES OF THE INTERACTION BETWEEN MUTANT HRAS AND BRAF INHIBITORS
The HRAS-mutant B9 murine cutaneous squamous-cell carcinoma cell line was seeded in soft agar with vemurafenib, with the analogue tool compound PLX4720 (both from Plexxikon), or with dimethyl-sulfoxide (Sigma-Aldrich) vehicle control, for studies of anchorage-independent clonal growth, as described previously.11 A431 human squamous-cell carcinoma cells (ATCC) either were transfected with empty vector or a plasmid carrying HRAS Q61L with the use of Fugene 6 (Roche Molecular Systems), according to the manufacturer’s instructions, or were stably transduced with a control or HRAS Q61L lentiviral vector, as described previously.24 Cells were analyzed for proliferation after vemurafenib exposure by means of cell-viability counts or MTT or MTS assays, as described previously.9,11,25 NIH3T3 cells (ATCC) were transfected with empty vector or an HRAS Q61L plasmid with the use of Fugene 6 and analyzed for colony formation in a soft agar.11 Western blotting was performed as described previously.9,11,25 At least two independent experiments were performed in triplicate with the use of each model.
ANALYSIS OF GENE EXPRESSION
B9 cells were plated in dimethylsulfoxide control or 1 μM of vemurafenib or PLX4720 and incubated for 16 hours. Cells were harvested, total RNA was isolated (RNeasy Mini Kit, Qiagen), and gene expression was measured with the use of Affymetrix Mouse 420 2.0 array chips, according to the manufacturer’s instructions. Vemurafenib and PLX4720 response genes were identified as those that changed by a factor of more than 2 (up-regulated) or less than 0.5 (down-regulated) relative to controls. The gene patterns of the B9 cells were compared with MAPK-pathway output genes from five human melanoma cell lines,7 and differential gene expression was confirmed by PCR assay.
STUDIES IN MICE
The procedures in animals were performed in accordance with local animal ethics committees. The two-stage skin carcinogenesis procedures were essentially those that have been described previously,16,26 with six animals per group. The BRAF inhibitor PLX4720 and the MEK inhibitor PD184352 were synthesized at the Institute of Cancer Research and delivered by means of oral gavage (25 mg per kilogram of body weight per day) in 200 μl of di-methylsulfoxide in water (1:19 solution).
STUDY OVERSIGHT
Data generated and collected by the study investigators were analyzed by the senior academic and industry authors, who vouch for the completeness and accuracy of the analyses and reported results. The clinical protocol summaries for the four trials are available at NEJM.org.
STATISTICAL ANALYSIS
Results of mutation testing in the initial 21 specimens of cutaneous squamous-cell carcinoma lesions of the keratoadenoma subtype and in 14 independent validation specimens are presented as mutation frequencies with 95% confidence intervals. Statistical analysis of gene-expression profiling was performed with the use of the z-test and chi-square analysis with Yates’ correction. Analysis of the cell-line culture experiments was performed with the use of Student’s t-test and a two-way analysis of variance with a Bonferroni post-test analysis. In the studies of mice, analysis of variance was performed with the use of the Kruskal–Wallis test.
RESULTS
CHARACTERISTICS OF THE PATIENTS AND LESIONS
As part of the dermatopathological review of vemurafenib-treated patients, 21 centrally confirmed samples of cutaneous squamous-cell carcinoma or keratoacanthoma obtained from 11 patients with metastatic melanoma were analyzed (Table 3A in the Supplementary Appendix). A validation set of 14 samples from 12 vemurafenib-treated patients was subsequently analyzed to confirm the high frequency of RAS mutations (Table 3B in the Supplementary Appendix). Overall, the mean time to diagnosis of the first cutaneous squamous-cell carcinoma or keratoacanthoma in the combined series was 10 weeks, with the earliest lesion appearing at 3 weeks (Table 4 in the Supplementary Appendix). The mean age at diagnosis was 60 years (range, 44 to 84). Eighteen of the 23 patients (78%) had a history and clinical signs of chronically sun-damaged skin, and 4 (17%) had a history of cutaneous squamous-cell carcinomas or keratoacanthomas. The lesions were widely distributed, with 34% on the head and neck areas, 23% on the torso, and 43% on the extremities. Twenty-two lesions (63%) were characterized as keratoacanthomas (Fig. 1, and Fig. 1 in the Supplementary Appendix), and 13 (37%) were cutaneous squamous-cell carcinomas.
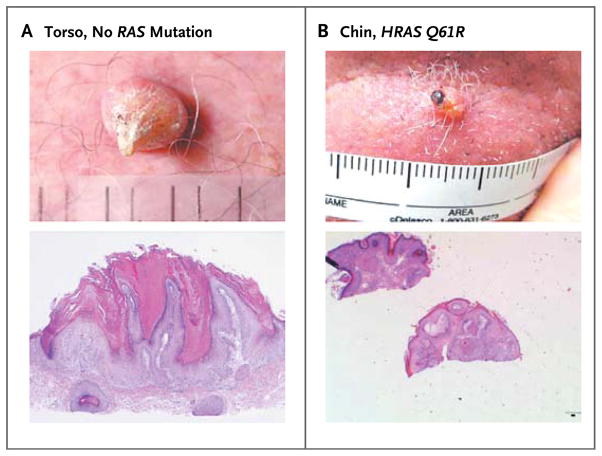
Figure 1. Cutaneous Squamous-Cell Carcinomas or Keratoacanthomas in Patients Treated with Vemurafenib.
Representative photographs and photomicrographs of nonmelanoma skin lesions in vemurafenib-treated patients are shown. The upper image in Panel A shows a lesion with the clinical features of keratoacanthoma, noted on day 98 after the patient had begun taking vemurafenib at a dose of 960 mg twice daily, in the centrally analyzed initial series. The lower image in Panel A is a low-power view of a section of a lesion obtained from the skin of the torso of the same patient with no RAS mutation, reported as squamous-cell carcinoma of the keratoacanthoma subtype (hematoxylin and eosin). Panel B shows the clinical appearance (upper image) and histopathological appearance (lower image, hematoxylin and eosin) of a keratoacanthoma from the chin of a patient with HRAS Q61R in the validation series.
GENE MUTATIONS AND ERK ACTIVATION IN LESION SPECIMENS
In the initial cohort of 21 centrally analyzed samples, there were 14 mutations in hotspot codons (12, 13, and 61) of different RAS genes (HRAS, NRAS, or KRAS) in 13 specimens (62%; 95% confidence interval [CI], 38 to 82) (Fig. 1C and 1D and Table 3A in the Supplementary Appendix). The most prevalent HRAS substitutions occurred at codon 61 (8 of 14), with fewer at codon 12 (4 of 14) and codon 13 (2 of 14). Two of 18 samples had TP53 mutations that change amino acids in the p53 protein (Table 3A in the Supplementary Appendix). One sample had an intronic mutation in TP53, and for 3 samples (all from Patient 4), no results were obtained. No mutations were identified in exon 2 of CDKN2A. ERK phosphorylation was assessed in 10 cutaneous squamous-cell carcinomas or keratoacanthomas and the surrounding normal epithelia and was found to be higher in these lesions than in the normal epidermis (Fig. 2A and 2B in the Supplementary Appendix).
In the validation set, RAS mutations were noted in 8 of 14 samples (57%; 95% CI, 29 to 82): 4 in HRAS and 4 in KRAS (Table 3B in the Supplementary Appendix). In the 4 samples with sufficient surrounding normal skin to perform RAS mutational analyses, no mutations were detected (Table 3B and Fig. 2C and 2D in the Supplementary Appendix). Staining for pERK, which was performed in 3 of the 4 samples, was more extensive in the cells of the lesions than in those of the surrounding normal skin (Fig. 2). Thus, RAS mutations were noted in 60% of the combined series, and HRAS Q61L was the most frequent mutation, so oncogenic HRAS was selected for pre-clinical mechanistic investigations.
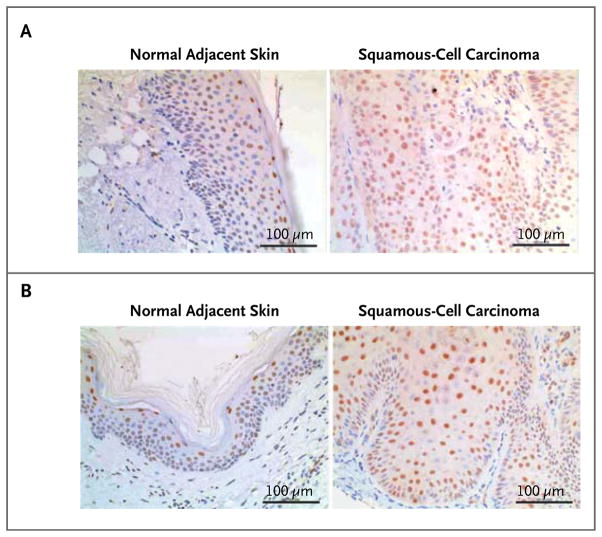
Figure 2. Examples of the Mutation and MAPK-Signaling Analysis in Cutaneous Squamous-Cell Carcinomas or Keratoacanthomas.
Immunohistochemical staining for pERK (brown) of samples of normal adjacent skin (left) and of cutaneous squamous-cell carcinomas (right) is shown from two patients in the validation set who were treated with vemurafenib. In these patients, both of whom had KRAS G12 mutations, the lesions were diagnosed as well-differentiated squamous-cell carcinomas that appeared 87 days (Panel A) and 51 days (Panel B) after vemurafenib therapy was begun.
EFFECTS OF BRAF INHIBITORS ON CELLS HARBORING HRAS MUTATIONS
Paradoxical Increase in MAPK Signaling and Proliferation of Cells Harboring Mutated HRAS
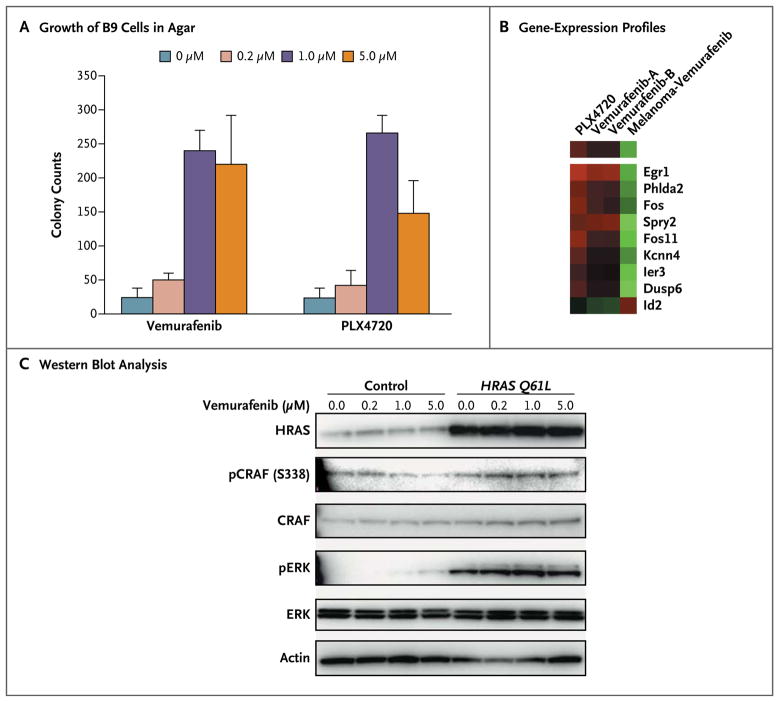
To investigate the effects of BRAF inhibitors on HRAS mutations in cutaneous squamous-cell carcinomas, we used the murine cell line B9, which harbors the HRAS Q61L mutation.27 Exposure of B9 cells to vemurafenib or its analogue PLX4720 stimulated cell proliferation in soft agar (Fig. 3A). To analyze the underlying mechanism, we compared gene-expression profiles of B9 cells before and after they were exposed to vemurafenib (two independent replicates) or PLX4720. An assessment that included a total of 306 gene probes (representing 250 genes) suggested a shared trend in treatment-induced changes in gene expression that included well-characterized MAPK-pathway genes (Spry2, Dusp6, Fos, Fos11, and Egr1) (P<0.001 by z-test of the three data sets) (Fig. 3A in the Supplementary Appendix). Supervised analysis of these changes in the B9 cells exposed to vemurafenib and PLX4720 revealed a change by a factor of two in nine murine homologues of genes that were differentially expressed when BRAF was inhibited in human melanoma cells expressing the BRAF V600E mutation (P<0.001 by z-test).7 Because of the different cell lineages in these two studies, this overlap is significant (P<0.001 by chi-square analysis with Yates’ correction, two-tailed test). For these nine genes, the change in B9 cells was the opposite of the change in the gene signature observed in the human BRAF V600E melanoma cells (Fig. 3B) (see Fig. 3B in the Supplementary Appendix for confirmation with the use of the reverse-transcriptase PCR assay), suggesting that the proliferation stimulated by the BRAF inhibitor results from paradoxically increased MAPK-pathway transcriptional effects in cells with mutant HRAS.
Figure 3. Mechanistic Studies Showing Paradoxical MAPK Activation in Cell Lines.
Panel A shows stimulation of B9 cell growth in soft agar after exposure to vemurafenib or PLX4720. Panel B shows differential expression of selected MAPK-regulated output genes in B9 cells after exposure to vemurafenib or PLX4720 overnight, as compared with gene expression of five human melanoma cell lines used as a reference.7 Red indicates higher expression and green indicates lower expression than untreated cells. Panel C shows the results of Western blot analysis of NIH3T3 cells transfected with a control vector or with mutated HRAS Q61L and treated with different concentrations of vemurafenib.
To further investigate the functional role of the most prevalent mutation, HRAS Q61L was expressed in the human squamous-cell carcinoma cell line A431, which already harbors an amplified EGFR but has wild-type RAS. Exposure to vemurafenib induced modest proliferation in A431 cells transfected with a control plasmid vector. In A431 cells expressing HRAS Q61L with the use of plasmid transfection, 1 to 3 μM of vemurafenib stimulated cell proliferation, whereas higher concentrations decreased proliferation (Fig. 4A and 4B in the Supplementary Appendix). Similar data were obtained with the expression of HRAS Q61L in A431 cells with the use of lentiviral transduction (Fig. 4C in the Supplementary Appendix). Western blot analysis showed a dose-dependent increase in pERK that was most notable at 24 hours, with a minimal effect on the parallel phosphatidylinositol 3-kinase (PI3K)–AKT signaling pathway (Fig. 5A in the Supplementary Appendix).
Since amplification of EGFR in A431 cells may interfere with the effects of vemurafenib on cells expressing oncogenic HRAS, we studied the interaction between HRAS Q61L and vemurafenib in NIH3T3 cells, an immortalized fibroblast cell line with wild-type RAS and without EGFR amplification. Cells transfected with the use of a control vector did not form colonies; when cells were transfected with HRAS Q61L, the size of the colonies increased in a dose-dependent fashion (Fig. 5B in the Supplementary Appendix), with an increase in pERK, after exposure to vemurafenib (Fig. 3C).
These three in vitro models show that vemurafenib stimulates proliferation in cells with mutated HRAS Q61L through paradoxical activation of the MAPK pathway, evidenced by increased ERK phosphorylation and increased expression of ERK-regulated genes.
PLX4720 and Reduced Tumor Latency in a Mouse Model of Skin Carcinogenesis
To model the in vivo effects of BRAF inhibition in cutaneous squamous-cell carcinomas and keratoacanthomas, we used the two-stage skin carcinogenesis mouse model in which topical application of the carcinogen 7,12-dimethylbenz-(a)anthracene (DMBA) induces HRAS Q61L mutations in mouse keratinocytes.17 Subsequent application of the tumor promoter 12-O-tetradecanoyl-phorbol-13-acetate (TPA) then induces these lesions.17
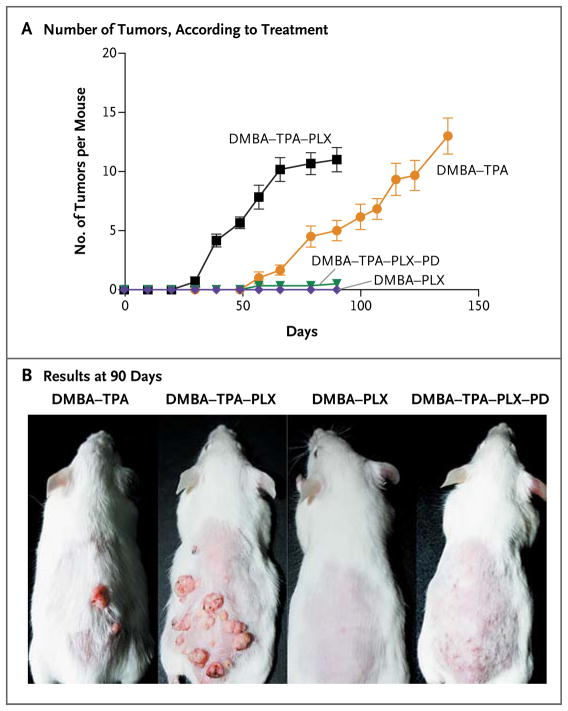
In mice that received DMBA and TPA along with PLX4720, the appearance of these lesions was markedly accelerated, as compared with the mice given DMBA and TPA alone (Fig. 4). The addition of PLX4720 did not increase the number of lesions induced by DMBA and TPA, but it reduced tumor latency by 45% and reduced the interval between the initial development of lesions and the maximal tumor burden by 35% (P = 0.002 by the Kruskal–Wallis test). The lesions arising in the mice given DMBA and TPA alone and in those given DMBA, TPA, and PLX4720 were clinically and histologically similar, consistent with keratoacanthomas and well-differentiated invasive cutaneous squamous-cell carcinomas, and the lesions in both groups of mice had the HRAS Q61L mutation (Fig. 6 in the Supplementary Appendix). Mice given the combination of DMBA and PLX4720 alone had no visible or palpable tumors (Fig. 4). For these studies, the analogue compound was used in preference to vemurafenib because it had excellent oral bioavailability in mice, whereas the available formulation of vemurafenib had poor bioavailability when delivered by either the oral or the intravenous route (data not shown).
Figure 4. Acceleration of Growth of Nonmelanoma Skin Tumors in Mice with BRAF Inhibition.
Panel A shows the number of palpable tumors arising over time in mice treated with the carcinogen 7,12-dimethylbenz(a)anthracene (DMBA), the tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA), the BRAF inhibitor PLX4720 (PLX) (25 mg per kilogram of body weight per day), and the MEK inhibitor PD184352 (PD) (25 mg per kilogram per day). Each cohort consisted of six mice, and the mean number of tumors per mouse is shown. I bars represent standard deviations. Panel B shows the results at 90 days after mice were treated with four different regimens.
Suppression of Tumor Development by a MEK Inhibitor
PDV cells are DMBA-transformed mouse keratinocytes that express HRAS Q61L.28 PLX4720 induced pERK and cellular proliferation in PDV cells, and the MEK inhibitor PD184352 blocked pERK activation by PLX4720 (Fig. 7A and 7B in the Supplementary Appendix). In the mouse model, PD184352 administration suppressed tumor development by 91% in mice given DMBA, TPA, and PLX4720 (Fig. 4) but did not mediate tumor remission in mice with established tumors (Fig. 7C in the Supplementary Appendix).
DISCUSSION
RAS was the first oncogene discovered in which point mutations led to cellular transformation.29 Although RAS mutations alone typically result in cellular senescence, in conjunction with other events that alter control of the cell cycle and apoptosis, they induce cellular transformation.30 Mutant RAS functions as a driver oncogene in approximately one third of human cancers.31 The prevalence of RAS mutations in sporadic cases of cutaneous squamous-cell carcinomas or keratoacanthomas is not known for sure but reportedly ranges between 3 and 30%.19,20 Our data indicate that RAS mutations are present in approximately 60% of cases in patients treated with vemurafenib, suggesting that preexisting mutations may confer a predisposition to the development of squamous-cell carcinomas or keratoacanthomas. Results of research on the paradoxical activation of MAPK by RAF inhibitors predicts that upstream oncogenic events, either activating mutations in RAS or mutations or amplifications in receptor tyrosine kinases that strongly elevate levels of the RAS–guanosine triphosphate complex in the absence of a BRAF V600E mutation, would potentiate signaling through the MAPK pathway.12–14 Our functional studies showing HRAS-primed activation of the MAPK pathway in models of squamous-cell carcinoma treated with BRAF inhibitors provide evidence that the toxicity related to BRAF inhibition may arise from paradoxical MAPK-pathway activation. Recent studies have shown that vemurafenib resistance can be mediated by receptor tyrosine kinases such as the platelet-derived growth factor and insulin-like growth factor 1 receptors.32,33 Preexisting amplification of the EGFR gene in the A431 cell-line model also resulted in paradoxical MAPK-pathway signaling in functional assays, although at a lower level than that driven by oncogenic HRAS. These data from in vitro models suggest that similar mutations or amplifications of receptor tyrosine kinases may account for the development of cutaneous squamous-cell carcinomas and keratoacanthomas in the 40% of samples in our combined series in which no RAS mutations were found.
The timing of the appearance of these lesions after vemurafenib treatment is decidedly different from that of secondary cancers associated with cytotoxic chemotherapy. In the case of vemurafenib, the lesions tend to appear within the first few weeks after the start of therapy, whereas cancers that are due to the genotoxic effects of chemotherapy develop years after exposure. The specificity of vemurafenib for a limited number of kinases,8,10 along with our finding that RAS mutations occur frequently in lesions arising preferentially in sun-damaged skin, suggests that vemurafenib may not have direct carcinogenic effects but instead may potentiate preexisting initiating oncogenic events.
In the skin carcinogenesis model, the BRAF inhibitor PLX4720 drove paradoxical activation of the MAPK pathway and proliferation of HRAS Q61L-transformed keratinocytes, with decreased latency and accelerated growth of cutaneous squamous-cell carcinomas and keratoacanthomas. PLX4720 was not itself a true tumor promoter because it could not substitute for TPA. Instead, PLX4720 accelerated the growth of preexisting RAS-mutant lesions. Taken together with the clinical observations and functional analyses, our data provide circumstantial evidence to suggest that vemurafenib does not initiate tumorigenesis but rather accelerates the progression of preexisting subclinical cancerous lesions with strong upstream MAPK-signaling potential. These findings explain why the lesions generally develop early after vemurafenib treatment and only in a subset of patients.
In conclusion, our data provide a molecular mechanism for the development of clinical toxicity that is the opposite of what would be expected from a targeted oncogene inhibitor. This mechanism accounts for the development of cutaneous squamous-cell carcinomas and keratoacanthomas, notably of the skin, but it is not clear whether it is relevant to the development of squamous-cell carcinomas in other organs. Our findings support the caution against investigating single-agent type I BRAF inhibitors in patients with cancers driven by RAS or by activated receptor tyrosine kinases. The discovery that the development of these lesions is driven by RAS and by MAPK in patients receiving BRAF inhibitors, as well as the effects noted in the animal model, point to the usefulness of combining a BRAF inhibitor with a MEK inhibitor to prevent this toxic effect34 and make way for the clinical development of a new generation of BRAF inhibitors selected to avoid paradoxical MAPK-pathway activation.
Supplementary Material
Acknowledgments
Supported in part by Hoffmann–La Roche and Plexxikon (to Drs. Su, Trunzer, and Bollag); the Seaver Institute, the Louise Belley and Richard Schnarr Fund, the Fred L. Hartley Family Foundation, the Wesley Coyle Memorial Fund, the Ruby Family Foundation, the Albert Stroberg and Betsy Patterson Fund, the Jonsson Cancer Center Foundation, and the Caltech–UCLA Joint Center for Translational Medicine (to Drs. Lo and Ribas); the European Organisation for Research and Treatment of Cancer Melanoma Group, Cancer Research U.K. (CRUK) (C107/A10433 and CRUK-A8274), the American Institute for Cancer Research (09-0773), and the Institute of Cancer Research (to Dr. Marais); and Breakthrough Breast Cancer and the 2010 CRUK Future Leaders Prize in Cancer Research (to Dr. Reis-Filho).
We thank the patients and study investigators for participating in the two studies; the Association for International Cancer Research; Carla Duymelinck, Suzanne Cheng, Jennifer Narozny, Louise Cockey, Astrid Koehler, and Michael Stumm for assisting with organizing and conducting the molecular analyses of patient specimens; Dr. Alain Balmain (University of California, San Francisco) for providing the B9 cells; Dr. Miguel Quintanilla (Madrid) for providing PDV cells; Dr. Kay Savage, Eric Ward, and Annette Lane (Institute of Cancer Research), and Ngan Doan (University of California, Los Angeles), for assisting with histology and immunohistochemistry; and Alice Smith and Vidya Pawar (Institute of Cancer Research) for assisting with DNA sequencing.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribas A, Kim K, Schuchter L, et al. BRIM-2: an open-label, multicenter Phase II study of RG7204 (PLX4032) in previously treated patients with BRAF V600E mutation-positive metastatic melanoma. J Clin Oncol. 2011;29(Suppl):8509. abstract. [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kefford R, Arkenau H, Brown MP, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J Clin Oncol. 2010;28(Suppl):611s. abstract. [Google Scholar]
- 7.Joseph EW, Pratilas CA, Poulikakos PI, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107:14903–8. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Søndergaard JN, Nazarian R, Wang Q, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Higgins B, Kolinsky K, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70:5518–27. doi: 10.1158/0008-5472.CAN-10-0646. [Erratum, Cancer Res 2010;70:9527.] [DOI] [PubMed] [Google Scholar]
- 12.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 14.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall-Jackson CA, Eyers PA, Cohen P, et al. Paradoxical activation of Raf by a novel Raf inhibitor. Chem Biol. 1999;6:559–68. doi: 10.1016/s1074-5521(99)80088-x. [DOI] [PubMed] [Google Scholar]
- 16.Abel EL, Angel JM, Kiguchi K, Di-Giovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4:1350–62. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Haras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 18.Balmain A, Ramsden M, Bowden GT, Smith J. Activation of the mouse cellular Harveyras gene in chemically induced benign skin papillomas. Nature. 1984;307:658–60. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- 19.Uribe P, Gonzalez S. Epidermal growth factor receptor (EGFR) and squamous cell carcinoma of the skin: molecular bases for EGFR-targeted therapy. Pathol Res Pract. 2011;207:337–42. doi: 10.1016/j.prp.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Oberholzer PA, Kee D, Dziunycz P, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2011 Nov 7; doi: 10.1200/JCO.2011.36.7680. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344:975–83. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 22.White AC, Tran K, Khuu J, et al. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108:7425–30. doi: 10.1073/pnas.1012670108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koya RC, Kasahara N, Pullarkat V, Levine AM, Stripecke R. Transduction of acute myeloid leukemia cells with third generation self-inactivating lentiviral vectors expressing CD80 and GM-CSF: effects on proliferation, differentiation, and stimulation of allogeneic and autologous anti-leukemia immune responses. Leukemia. 2002;16:1645–54. doi: 10.1038/sj.leu.2402582. [DOI] [PubMed] [Google Scholar]
- 25.Niehr F, von Euw E, Attar N, et al. Combination therapy with vemurafenib (PLX4032/RG7204) and metformin in melanoma cell lines with distinct driver mutations. J Transl Med. 2011;9:76. doi: 10.1186/1479-5876-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-García A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–26. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 27.Burns PA, Jack A, Neilson F, Haddow S, Balmain A. Transformation of mouse skin endothelial cells in vivo by direct application of plasmid DNA encoding the human T24 H-ras oncogene. Oncogene. 1991;6:1973–8. [PubMed] [Google Scholar]
- 28.Quintanilla M, Haddow S, Jonas D, Jaffe D, Bowden GT, Balmain A. Comparison of ras activation during epidermal carcinogenesis in vitro and in vivo. Carcinogenesis. 1991;12:1875–81. doi: 10.1093/carcin/12.10.1875. [DOI] [PubMed] [Google Scholar]
- 29.Sukumar S, Notario V, Martin-Zanca D, Barbacid M. Induction of mammary carcinomas in rats by nitrosomethylurea involves malignant activation of H-ras-1 locus by single point mutations. Nature. 1983;306:658–61. doi: 10.1038/306658a0. [DOI] [PubMed] [Google Scholar]
- 30.Collado M, Gil J, Efeyan A, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 31.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [Erratum, Nat Rev Cancer 2003;3:708.] [DOI] [PubMed] [Google Scholar]
- 32.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Infante JR, Falchook GS, Lawrence DP, et al. Phase I/II study to assess safety, pharmacokinetics, and efficacy of the oral MEK 1/2 inhibitor GSK1120212 (GSK212) dosed in combination with the oral BRAF inhibitor GSK2118436 (GSK436) J Clin Oncol. 2011;29(Suppl) abstract. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.