Abstract
Inhibitory neurons have been shown to perform a variety of functions within brain circuits, including shaping response functions in target cells. Still, how the properties of specific inhibitory neuron classes relate to their local circuits remains unclear. To better understand the distribution and origins of orientation selectivity in inhibitory neurons expressing the calcium binding protein parvalbumin (PV) in the mouse primary visual cortex, we labeled PV+ neurons with red fluorescent protein (RFP) and targeted them for cell-attached electrophysiological recordings. PV+ neurons could be broadly tuned or sharply tuned for orientation but tended to be more broadly tuned than unlabeled neurons on average. The dendritic morphology of PV+ cells, revealed by intracellular labeling, was strongly correlated with tuning: highly tuned PV+ neurons had shorter dendrites that branched nearer to the soma and had smaller dendritic fields overall, whereas broadly tuned PV+ neurons had longer dendrites that branched farther from the soma, producing larger dendritic fields. High-speed two-photon calcium imaging of visual responses showed that the orientation preferences of highly tuned PV+ neurons resembled the preferred orientations of neighboring cells. These results suggest that the diversity of the local neighborhood and the nature of dendritic sampling may both contribute to the response selectivity of PV+ neurons.
Introduction
Inhibition has diverse roles in cortical circuits (Pouille and Scanziani, 2001; Wehr and Zador, 2003; Mariño et al., 2005; Cardin et al., 2009; Adesnik et al., 2012; Atallah et al., 2012; Lee et al., 2012; Lovett-Barron et al., 2012; Royer et al., 2012; Wilson et al., 2012). This diversity of function matches the diversity of the inhibitory population. Inhibitory neuron subclasses differ in their gene expression, axonal projection patterns, dendritic morphology, and electrophysiological properties, suggesting that specific subtypes may perform specific functions (Markram et al., 2004; Somogyi and Klausberger, 2005; Burkhalter, 2008; Isaacson and Scanziani, 2011).
Parvalbumin-positive (PV+) cells, often considered one distinct cell type, are heterogeneous across all of these dimensions. Morphologically, they include soma-targeting basket cells with small and large dendritic and axonal fields, axon-targeting chandelier cells, and dendrite-targeting multipolar bursting cells (Kisvárday and Eysel, 1993; Lund and Lewis, 1993; Han, 1994; Wang et al., 2002; Blatow et al., 2003; Krimer et al., 2005; Zaitsev et al., 2009). PV+ neurons also vary in expression of potassium channel subunits (Du et al., 1996; Chow et al., 1999), calcium binding proteins (Chow et al., 1999; Blatow et al., 2003), and other signaling molecules and receptors (Yan et al., 1998; Vruwink et al., 2001). Most PV+ neurons are fast-spiking, responding to current injections with barrages of narrow spikes; however, some PV+ neurons are regular-spiking or stuttering (Han, 1994; Blatow et al., 2003; Markram et al., 2004; Krimer et al., 2005), and PV+ basket cells include at least four electrophysiological subtypes (Helm et al., 2013).
The PV+ population is also functionally diverse; in visual cortex (V1), these cells have receptive fields that range from unselective to highly selective for orientation and other response features (Sohya et al., 2007; Liu et al., 2009; Kerlin et al., 2010; Runyan et al., 2010; Hofer et al., 2011). The origins of PV+ neuron tuning remain unresolved but might depend on the specificity of local inputs to these cells. The salt-and-pepper organization of orientation preferences in mouse V1 (Ohki and Reid, 2007), coupled with uniformly sampled inputs from the orientation map, could provide broad orientation tuning to PV+ cells, whereas narrow tuning would require precisely selected inputs. On the other hand, small homogeneities in the orientation representation (Dräger, 1975), coupled with matched dendritic arbors of a subset of PV+ neurons, could enable these neurons to achieve narrow tuning by uniform sampling of the local neighborhood alone.
We recorded from a large number of PV+ neurons and found that highly tuned PV+ neurons have smaller dendritic fields and shorter dendrites, whereas broadly tuned PV+ neurons have larger dendritic fields and longer, more tortuous dendrites. We then compared the spatial distribution of orientation preferences of the neurons surrounding PV+ neurons, finding that the orientation preferences of PV+ neurons resembled those of nearby cells, suggesting that these neurons may sample uniformly from local inputs, and the breadth of their dendritic arbors may influence their response selectivity by restricting the number and diversity of their inputs.
Materials and Methods
Mice.
Experiments were performed in mice of either sex under protocols approved by Massachusetts Institute of Technology's Animal Care and Use Committee and conformed to National Institutes of Health guidelines. Heterozygous PV-Cre knock-in driver mice, which express Cre in >90% of PV+ neurons, were backcrossed into a C57BL/6 line (Hippenmeyer et al., 2005) and used for the analysis of PV+ and PV− neurons. Thy1-GFP-S mice (Feng et al., 2000), in which a subset of superficial layer neurons are labeled, were also maintained on a C57BL/6 line and were used for the analysis of pyramidal neurons.
Viral construct and injection.
Red fluorescent protein (RFP) was expressed in PV+ neurons in V1 of PV-Cre mice by infection of an LS2L-RFP construct packaged into adeno-associated virus (serotype 2/9) as described previously (Kuhlman and Huang, 2008; Runyan et al., 2010). Six-week-old PV-Cre mice were initially anesthetized with 4% isoflurane in oxygen and maintained on 2% isoflurane. The skull was thinned along a 1 mm line at the rostral edge of V1, and the remaining skull and dura mater were carefully punctured using a glass micropipette filled with the virus. Two injections (0.25 μl of virus, injected at 10 nl/min) were made at each site, at depths of 500 and 250 μm, respectively. After each injection, the pipette was held in place for 5 min before retraction to prevent leakage.
Animal preparation.
Two to four weeks after injection, mice were anesthetized with a mixture containing fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg), supplemented with isoflurane. The eyes were protected with ophthalmic ointment during the surgery and moistened afterward with saline. A metal head plate was attached to the skull parallel to V1 using cyanoacrylate glue and dental acrylic. A 2 mm × 2 mm craniotomy was performed over V1, and the exposed area was covered with a thin layer of 2% agarose in ACSF (140 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 0.01 mm EDTA, 10 mm HEPES, 10 mm glucose, pH 7.4).
Mice were then transferred to a custom-built two-photon microscope (Majewska et al., 2000) that was powered by a Spectra Physics Mai-Tai eHP laser, passed through a Deep-See module (Spectra Physics), where the head plate was screwed into a moveable stage. To maintain anesthesia, 0.5% isoflurane in oxygen was supplied through a tube, and fentanyl/medetomidine was injected every hour. The body temperature was maintained at 37.5°C with heating pads. The recording phase of physiology experiments typically lasted for 6–12 h.
Targeted cell-attached recording.
Glass pipettes with ∼1.5 μm tip size and 3–7 MΩ resistance were filled with AlexaFluor-488 (5 μm in saline, Invitrogen) and angled into the pia above the viral injection site using a micromanipulator (Sutter, MP-285) under visual guidance with epifluorescence. Upon entering the brain, the pipette was guided toward RFP+ cells in superficial layer 2/3 of V1 under two-photon guidance. The laser was tuned to 920 nm, which allowed excitation of both the Alexa-488 and RFP fluorophores simultaneously. The pipette was targeted to RFP+ cells while applying constant positive pressure (0.2 psi), which was monitored with a digital pressure gauge (General Tools). When the pipette just touched the cell surface, positive pressure was released, and sustained negative pressure immediately applied (0.2–0.6 psi) to obtain a loose seal (Runyan et al., 2010). Cells from which well-isolated spikes were detected during the display of a drifting grating that randomly changed orientation and direction at 8 Hz were characterized further, and their orientation selectivity was assessed. Subsequently, current pulses (35 ms, 900–2000 nA) were delivered at 15 Hz for 30–60 s to fill the recorded cell through electroporation. Only single RFP+ cells that were distinctly filled during this procedure were included for analysis. In the same experiments, RFP− cells were targeted blindly. As the tip was slowly advanced through the cortex, −0.5 nA current pulses were delivered for 6.3 ms at 0.55 Hz, and the tip resistance was monitored. When the tip resistance increased substantially, positive pressure was released, and negative pressure immediately applied, as above. The location of the tip was still monitored under two-photon guidance; all RFP− cells were located within the injection site and were also filled after recording. Recordings from GFP+ pyramidal neurons in Thy1-GFP-S mice were performed similarly. Neurons in PV-Cre or Thy1 mice were targeted at depths of 100–300 μm. Recordings were made with an Axoclamp-2A amplifier (Molecular Devices) using Clampex software (Molecular Devices, v8.1) at a sampling rate of 30 kHz.
Two-photon structural imaging.
The laser was tuned to 920 nm, allowing strong excitation of GFP, RFP, and Alexa-488. Images were collected through a high performance objective lens (25× Olympus XL Plan N objective, NA = 1.05). z-stacks were collected tangentially, at 2 μm increments from the pial surface through the extent of the filled cell, at resolution of 0.4 μm/pixel, spanning 200–400 μm in x and y dimensions, using FluoView300 confocal imaging software (Olympus), on both the red (RFP+) and green (Alexa-488 dye) channels. In all filled cells, the signal on red and green channels overlapped throughout all processes, and photomultiplier tube gain was optimized to visualize the finest processes.
Two-photon calcium imaging.
For these experiments, we used a Prairie Ultima two-photon system (Prairie Technologies) driven by a Spectra Physics Mai-Tai eHP laser, passed through a Deep-See module (Spectra Physics). The excitation laser was tuned to 920 nm, and functional imaging was done through a high performance objective lens (25× Olympus XL Plan N objective, NA = 1.05).
A glass pipette filled with Oregon Green Bapta-1 AM (1.0 mm, Invitrogen) and AlexaFluor-594 (100 μm, Invitrogen) was visually guided into the brain and lowered to a depth between 100 and 200 μm below the surface, near the center of the viral injection site, and a small amount of dye was released using a picospritzer. The brain was then left undisturbed for at least 1 h before imaging.
We used a custom software system to measure changes in the calcium indicator fluorescence in response to visual stimuli, as described previously in detail (Wilson et al., 2012, 2013). Briefly, a raster scan was performed and machine vision algorithms were used to identify cell bodies. Next, an algorithm was used to identify the shortest scan path between cells, and this line scan was run at 50 Hz, with a 90% dwell time inside the cells (only 10% dwell time in neuropil), yielding high signal:noise fluorescent signals.
Visual stimulation.
Visual stimuli were displayed on a 17-inch LCD monitor placed 15 cm from the eyes. Stimuli were generated in MATLAB (Mathworks) using the PsychoPhysics Toolbox (Brainard, 1997). Square wave drifting gratings with 100% contrast were used. In targeted cell-attached recordings, spatial frequency and temporal frequency were optimized to achieve the highest firing rate for each cell: spatial frequency ranged from 0.005 to 0.05 cycles per degree (cpd), and temporal frequency ranged from 1 to 3 cycles per second (cps). Test stimuli were episodically presented, equally alternating with a blank gray screen, with 8 s cycles (4 s on, 4 s off) at each of 18 directions equally spaced by 20 degrees.
Data analysis: visual responses.
Analysis of all data was performed off-line with custom written MATLAB routines. Electrophysiological time traces were imported into MATLAB, smoothed with a Gaussian kernel, and spikes were identified by detection of events based on the derivative of the voltage traces. The spontaneous firing rate of each neuron was measured for 10 s preceding visual stimulation for each trial. A neuron was considered visually responsive if its firing rate at the preferred orientation was significantly higher than its spontaneous firing rate, determined with a t test. The spontaneous firing rate was subtracted from the response to each orientation, the mean firing rate across the entire 4 s stimulus interval. The response firing rates were then fit to a Gaussian function.
Fluorescence time traces were assigned to the appropriate cell location, to obtain a time trace sampled at 50 Hz. The response (ΔF/F) was calculated for each time point by subtracting and then dividing the fluorescence value by the mean fluorescence of the surrounding 100 time points. The maximum ΔF/F during each grating presentation was taken as the visual response (Schummers et al., 2008; Runyan et al., 2010).
The orientation selectivity index (OSI) was calculated as the vector average of responses in the preferred direction (Schummers et al., 2002). Statistical comparisons were performed using the Wilcoxon test, two-tailed t test, and the Kolmogorov-Smirnoff test and yielded indistinguishable results. Values from two-tailed t tests are reported in Results.
In the population calcium imaging datasets, the spatial arrangement of orientation preferences was assessed by calculating the local orientation scatter, or the mean difference in the preferred orientations of each cell and neighboring cells within 60, 90, 120, or 240 μm. This mean difference was compared between RFP+ cells and their neighbors and RFP− cells and their neighbors. To test the validity of the difference between the orientation mapping around RFP+ and RFP− neurons, a bootstrap procedure with 10,000 samples was used to estimate the orientation mapping in the randomized configuration, where the orientation preferences were resampled among the cells' spatial locations with replacement, and the local scatter around each cell was recalculated in each scrambled iteration (Dombeck et al., 2009).
Morphological reconstructions.
Neurons were reconstructed from tangential z-stacks that were collected in vivo, as described above, using V3D semiautomatic tracing software (Peng et al., 2010). The multi-TIF 16-bit image stacks collected with the FluoView software were converted to high-contrast 8-bit images within ImageJ software and then imported into V3D. The contrast of each image was enhanced to allow the tracing of each thin dendritic process. Both the red (RFP+) and green (Alexa-488) channels were used to follow all of the neuron's major and minor processes from the soma. Within the V3D software, stacks were carefully scrolled through to distinguish each process and to mark each insertion, branch, and tip location.
The 3D coordinates of the soma, dendritic insertion points, dendritic branches, and dendritic tips were then exported to MATLAB, where custom written software was used for the Sholl analysis (Gutierrez and Davies, 2007). The number of Sholl crossings was assessed at 20 μm increments. For each neuron, the Sholl profile was fit to a Gaussian function, from which the peak number of crossings (amplitude) and distance of the peak from the soma (displacement) were extracted. The maximal dendritic extent was calculated as the distance from the soma to the most distal dendritic tip, and the dendritic field volume was calculated by computing the convex hull of the 3D dendritic tip coordinates. The polarity of the dendritic branching was assessed as follows. The direction of each branch and tip relative to the soma in each of the three cardinal planes was measured. In each plane, a Dendritic Polarity Index (DPI) was then calculated by vector averaging the distances of the tips and branches around the soma.
RFP+ neurons from brains of PV-Cre mice were reconstructed in vitro for comparison with the in vivo reconstructions using the RFP label. The Alexa-488 dye in neurons imaged in vivo did not survive perfusion, and so the same cells were not imaged in vivo and in vitro. Coronal slices were collected at 100 μm thickness, mounted on slides, and coverslipped. z-stacks were collected at 1 μm optical slices at 4 μm per pixel on a Zeiss LSM 5 Pascal Exciter confocal microscope with the 543 laser line (RFP+ fluorophore), and neurons were reconstructed as above.
Results
Electrophysiological properties of PV+ neurons
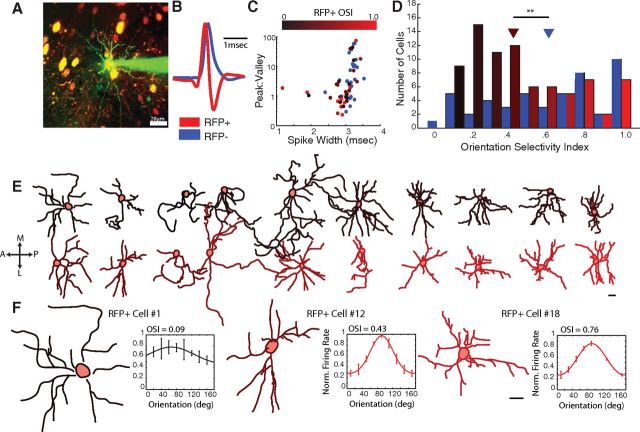
We expressed RFP in PV+ inhibitory interneurons in heterozygous PV-Cre knock-in mice (Kuhlman and Huang, 2008; Runyan et al., 2010) and used two-photon targeted cell-attached recording (Fig. 1A) to characterize 80 visually responsive RFP+ neurons and 49 visually responsive RFP− neurons in V1 of 32 PV-Cre mice. The spike shapes of the RFP+ and RFP− neurons were distinguishable (Fig. 1B,C): the RFP+ neurons had narrower spike widths (mean ± SD: RFP+, 2.9 ± 0.4 ms; RFP−, 3.1 ± 0.2 ms; p < 0.01, two-tailed two-sample t test here and below), and similar peak/valley amplitudes (RFP+, 6.7 ± 15.5; RFP−, 10.4 ± 16.5; p = 0.37) and repolarization rates (RFP+, 3.1 ± 2.3 V/s; RFP−, 2.8 ± 1.8 V/s, p = 0.61). The spontaneous firing rate was higher in the RFP+ neurons (RFP+, 1.2 ± 2.0 Hz; RFP−, 0.4 ± 0.7 Hz; p < 0.01), but the two populations had similar visually evoked firing rates (RFP+, 5.8 ± 5.9 Hz; RFP−, 5.3 ± 6.4 Hz; p = 0.67).
Figure 1.
Relating structure and function in PV+ neurons in vivo. A, The orientation selectivity of RFP+ neurons and RFP− neurons was characterized in the primary visual cortex of PV-Cre mice injected with a floxed-RFP viral construct. RFP+ neurons were targeted for recording under two-photon guidance, with a patch pipette containing green dye (Alexa-488). Each cell was then electroporated with the dye, and a z-stack was collected through the extent of the dendritic tree to enable morphological reconstruction. Scale bar, 20 μm. B, Spikes recorded from RFP+ neurons (red) and RFP− neurons (blue) were averaged and normalized by their maximum voltage. Spikes recorded from the RFP+ neurons showed the characteristic shape of fast-spiking PV+ neurons, including a strong afterhyperpolarization. C, The spike shapes of RFP+ neurons and RFP− neurons were distinguishable by the ratio of peak to valley amplitude and spike width. RFP+ neurons are color-coded by their OSI, demonstrating that spike shape in RFP+ neurons did not predict orientation selectivity (p > 0.3). D, The histogram of OSIs of RFP+ (red, shaded by OSI) and RFP− (blue) neurons reveals a wide range of orientation selectivity in both RFP+ and RFP− neurons, although RFP+ neurons tended to be more broadly tuned than RFP− neurons on average (p < 0.01). E, Twenty RFP+ neurons were reconstructed (see Materials and Methods), flattened through the z-plane (dorsal-ventral) for illustration purposes only, and color-coded by OSI as in C–D. All neurons are oriented similarly, as if in a tangential section. A, Anterior; P, posterior; M, medial; L, lateral. F, Three representative examples across the OSI range are shown, along with their orientation tuning curves. Error bars on tuning curves indicate SEM. Cell numbers indicate the cell IDs from Table 1. Scale bars: E, F, 20 μm. **p < 0.01.
Tuning properties of PV+ neurons
The diverse response features of PV+ neurons have been previously described by us (Runyan et al., 2010) and others (Liu et al., 2009; Kerlin et al., 2010; Hofer et al., 2011; Zariwala et al., 2011). We aimed in this study to carefully evaluate the orientation selectivity of a large population of these neurons and to compare their morphological features to their response selectivity. For each neuron, we measured the response to 18 drifting gratings in 20-degree increments and calculated an OSI by vector averaging the response; the OSI can range from 0 (unselective for orientation) to 1 (responsive to only one orientation). On average, the OSIs were lower in the RFP+ population (RFP+, 0.46 ± 0.28, n = 80; RFP−, 0.61 ± 0.31, n = 49; p < 0.01), although both populations included very broadly tuned and very sharply tuned cells (Fig. 1D). All recorded RFP+ cells were located between 115 and 300 μm below the pial surface (mean depth, 173 ± 70 μm), and soma depth and orientation selectivity were not correlated (r = 0.18, p = 0.13). In addition, although not all RFP+ neurons showed the narrow-spike profile typical of fast-spiking PV+ neurons (Fig. 1C), none of the measured spike shape parameters correlated significantly with OSI (spike width, r = 0.15, p = 0.37; peak/valley, r = 0.07, p = 0.77; repolarization rate, r = −0.15, p = 0.54). Thus, it is unlikely that the highly selective PV+ neurons were a depth-based subclass, such as multipolar bursting cells, a subclass of regular-spiking PV+ neurons that is located primarily at the border between layers I and II (Blatow et al., 2003) (i.e., at typical depths of 90–100 μm).
Morphology of PV+ neurons: dendritic extent, but not complexity, correlates with orientation selectivity
Differing tuning strength may indicate the existence of functional subclasses of PV+ neurons that correspond to morphological subclasses, so we reconstructed the dendritic morphology of 20 RFP+ neurons for which we had also measured orientation selectivity (Fig. 1E). After each cell's orientation tuning was characterized, it was filled with Alexa-488 (green) dye from the recording pipette, and a detailed z-stack in tangential view was collected through the extent of its dendritic tree on both the red and green channels with two-photon imaging.
The orientation selectivity of reconstructed RFP+ neurons represented the full range of broadly tuned and highly tuned OSIs we described in Figure 1D (0.09–0.87). Qualitative examination of the reconstructions suggested that the dendritic morphology of RFP+ neurons might vary systematically with orientation selectivity. The transition from larger dendritic trees to smaller dendritic trees as tuning becomes sharper is evident across the full population of reconstructed RFP+ neurons, ordered by increasing OSI (Fig. 1E) and in three representative neurons spanning the full range of tuning (Fig. 1F).
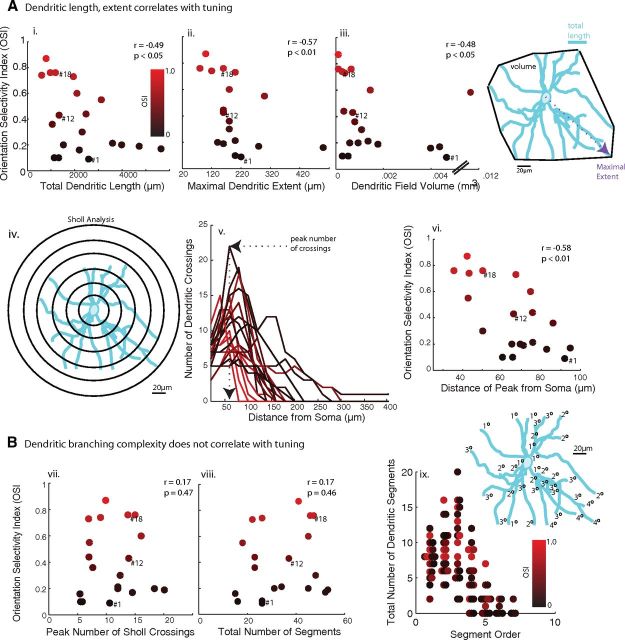
We first estimated the spatial extent of the dendritic trees by measuring four parameters for each reconstructed RFP+ neuron: (1) the total dendritic length of all of its dendritic processes, (2) the maximal dendritic extent (distance from the soma to the most distal dendritic tip), (3) the dendritic field volume (the convex volume enclosed by the dendritic tips), and (4) the distance of maximal dendritic branching from the soma (Sholl analysis). All four measures were significantly correlated with each other (p < 0.05, Spearman's rank correlation coefficient, here and below unless otherwise stated) and with orientation selectivity (Fig. 2A: dendritic length, r = −0.49, p < 0.05; maximal dendritic extent, r = −0.57, p < 0.01; dendritic field volume, r = −0.48, p < 0.05; peak branching distance, r = −0.58, p < 0.01).
Figure 2.
Dendritic extent, but not dendritic complexity, correlates with orientation selectivity of PV+ neurons. A, All four measures of dendritic extent were significantly correlated with the OSI of RFP+ neurons: i, The total dendritic length, the sum of the lengths of all dendrites; ii, the maximal dendritic extent, the distance of the most distal dendritic tip from the soma; iii, the dendritic field volume, the convex volume enclosed by the 3D coordinates of the neuron's tips; iv–vi, the distance of the Sholl peak from the soma were used to assess the spatial spread of dendritic integration by RFP+ neurons. B, Dendritic complexity, assessed by the peak number of Sholl crossings (vii; illustrated in iv, v), the total number of dendritic segments (viii), and the number of higher-order dendritic segments (ix), did not significantly correlate with OSI. Data points corresponding to the example cells of Figure 2 are indicated by their ID numbers.
Next, we assessed dendritic complexity with two parameters: (1) the peak number of branches as a function of distance from the soma (Sholl analysis) and (2) the total number of dendritic segments. The two measures of dendritic complexity correlated with one another (r = 0.8, p < 0.001), but unlike the measures of dendritic extent, neither measure correlated significantly with orientation selectivity (Fig. 2B: peak Sholl branches: r = 0.17, p = 0.47; total number of segments: r = 0.17, p = 0.46). Finally, ranking each dendritic segment by the number of branches separating it from the soma, orientation selectivity did not significantly correlate with the number of segments of low or high order (p > 0.1 for all ranks).
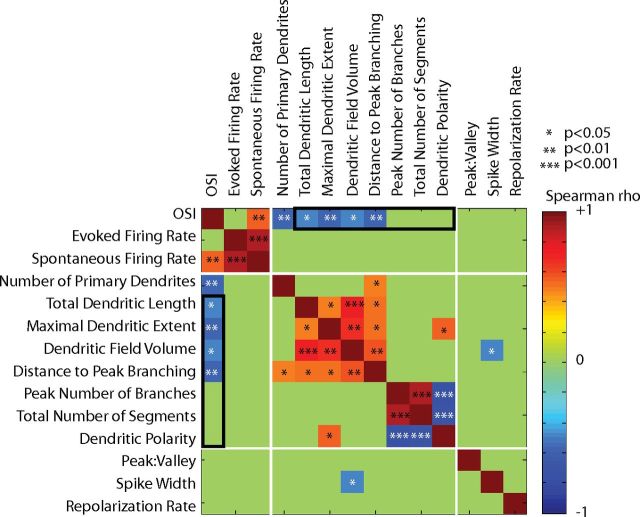
To assess whether the directionality of dendritic branching correlated with the orientation selectivity of RFP+ neurons, the angles of dendritic branches and tips from the soma were calculated in each of the three 2D planes. The polarity in each dimension was quantified by vector averaging, to obtain a DPI. The maximum DPI was not correlated to orientation selectivity, as both highly tuned and broadly tuned RFP+ neurons could have high DPIs (r = −0.17, p = 0.57). The correlations between all measured response, morphological, and electrophysiological parameters are summarized in Figure 3.
Figure 3.
Correlations between all measured response properties, morphology, and spike shape parameters in the 20 reconstructed RFP+ neurons of Figures 1 and 2. The color of each cell in the matrix corresponds to the Spearman correlation coefficient between the pair of parameters, according to the color bar at right, ranging from −1 (dark blue) to +1 (red). Coefficients with p values >0.05 have been set to 0 (green). *p < 0.05. The comparisons between OSI and morphological features described in Figure 2 are outlined in black, indicating the negative correlation between OSI and dendritic extent and lack of correlation between OSI and dendritic complexity. Three other clustered sets of related parameters show significant correlations, as expected: (1) firing rates, (2) measures of dendritic extent, and (3) measures of dendritic complexity. *p < 0.05, **p < 0.01, ***p < 0.001.
Together, these results suggest that PV+ neurons with sharper tuning extend their branches over shorter distances but maintain similar dendritic complexity than do more broadly tuned PV+ neurons, which send similarly complex processes across longer distances. All results for reconstructed RFP+ neurons are displayed in Table 1.
Table 1.
Response features and morphological measurements of reconstructed RFP+ neurons
| Cell ID | Response properties |
Morphology |
Spike shape |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OSI | Evoked firing rate (Hz) | Spontaneous firing rate (Hz) | No. of primary dendrites | Total dendritic length (mm) | Maximal dendritic extent (μm) | Dendritic field volume (mm3) | Distance of Sholl peak from the soma (μm) | Peak no. of Sholl crossings | Total no. of dendritic segments | DPI | Peak: valley | Repolarization rate | Spike width (ms) | |
| 1 | 0.09 | 5.5 | 1.7 | 7 | 2.590 | 220 | 0.00465 | 92 | 11 | 26 | 0.75 | 23.9 | 0.5 | 2.8 |
| 2 | 0.10 | 17.0 | 3.4 | 5 | 1.139 | 200 | 0.00027 | 60 | 6 | 26 | 1 | 0.9 | 1.5 | 2.6 |
| 3 | 0.10 | 7.5 | 0.8 | 5 | 1.323 | 200 | 0.00052 | 65 | 6 | 16 | 0.85 | 13.0 | 0.5 | 2.9 |
| 4 | 0.16 | 10.8 | 6.6 | 5 | 2.197 | 500 | 0.00398 | 83 | 5 | 15 | 0.82 | 1.3 | 1.6 | 1.9 |
| 5 | 0.17 | 14.3 | 4.1 | 8 | 5.646 | 280 | 0.00436 | 95 | 15 | 52 | 0.82 | 1.1 | 3.4 | 1.9 |
| 6 | 0.19 | 3.0 | 0.4 | 8 | 4.726 | 160 | 0.00177 | 71 | 20 | 53 | 0.3 | 1.2 | 1.3 | 1.1 |
| 7 | 0.20 | 5.8 | 0.7 | 8 | 1.523 | 140 | 0.00094 | 69 | 18 | 45 | 0.3 | 1.1 | 2.0 | 1.1 |
| 8 | 0.20 | 10.5 | 3.0 | 6 | 3.982 | 140 | 0.00059 | 65 | 12 | 31 | 0.53 | 0.9 | 7.2 | 3.0 |
| 9 | 0.21 | 0.8 | 0.1 | 5 | 3.597 | 200 | 0.00135 | 76 | 12 | 34 | 0.44 | 3.1 | 1.7 | 2.8 |
| 10 | 0.30 | 2.3 | 0.3 | 9 | 2.316 | 180 | 0.00108 | 50 | 12 | 48 | 0.09 | 0.3 | 3.0 | 2.8 |
| 11 | 0.36 | 2.8 | 2.2 | 7 | 1.053 | 180 | 0.00080 | 86 | 8 | 23 | 0.99 | 1.6 | 0.2 | 1.2 |
| 12 | 0.43 | 1.0 | 0.1 | 6 | 1.340 | 160 | 0.00071 | 66 | 14 | 37 | 0.22 | 1.1 | 1.8 | 1.1 |
| 13 | 0.44 | 0.5 | 0.0 | 3 | 2.479 | 160 | 0.00034 | 76 | 7 | 23 | 0.76 | 16.2 | 1.1 | 3.2 |
| 14 | 0.55 | 1.3 | 0.3 | 7 | 3.126 | 300 | 0.00120 | 43 | 7 | 18 | 0.91 | 1.4 | 2.1 | 1.9 |
| 15 | 0.60 | 0.5 | 0.0 | 7 | 2.095 | 180 | 0.00145 | 75 | 16 | 45 | 0.19 | 0.8 | 0.2 | 1.1 |
| 16 | 0.73 | 1.8 | 0.7 | 5 | 1.942 | 200 | 0.00038 | 67 | 7 | 22 | 0.59 | 1.3 | 2.2 | 1.1 |
| 17 | 0.74 | 4.5 | 0.9 | 8 | 0.606 | 120 | 0.00026 | 43 | 9 | 26 | 0.47 | 2.5 | 5.0 | 3.1 |
| 18 | 0.76 | 1.0 | 0.2 | 9 | 1.189 | 160 | 0.00064 | 50 | 14 | 47 | 0.16 | 2.5 | 2.2 | 2.7 |
| 19 | 0.76 | 2.1 | 0.0 | 11 | 0.970 | 80 | 0.00011 | 36 | 15 | 46 | 0.21 | 1.0 | 3.0 | 3.0 |
| 20 | 0.87 | 1.3 | 0.3 | 8 | 0.792 | 100 | 0.00010 | 42 | 10 | 41 | 0.63 | 17.9 | 0.2 | 3.4 |
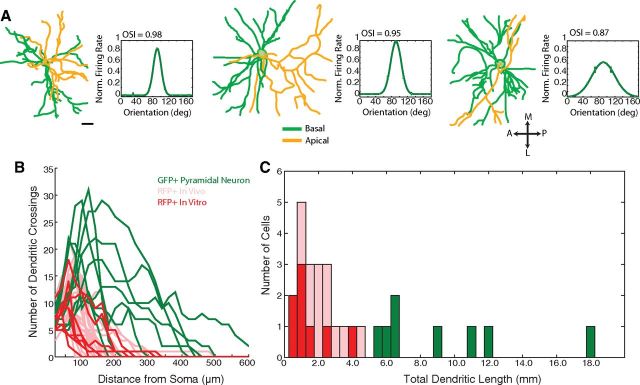
For comparison, we also reconstructed 9 GFP+ pyramidal neurons from the Thy-GFP-S mouse line (Feng et al., 2000). Example reconstructions are shown in Figure 4A, where the basal and apical processes are color-coded, and morphological and response parameters are summarized in Table 2. Furthermore, although our dendritic measurements in vivo are well within the ranges reported previously for cortical basket cells (Helmstaedter et al., 2009; Zaitsev et al., 2009), as an additional control, we performed a separate set of morphological measurements in vitro for comparison with the in vivo measurements. We sectioned V1 coronally, collected z-stacks of RFP+ neurons in vitro, and reconstructed 8 RFP+ neurons in similar detail as those imaged in vivo (Fig. 4B). The Sholl profiles of the RFP+ neurons imaged in vitro were similar to the Sholl profiles of the RFP+ neurons imaged in vivo (Fig. 4B), as were the total dendritic lengths (Fig. 4C). None of the Sholl parameters (peak number of crossings, p = 0.28; distance of peak from soma, p = 0.22; profile radius, p = 0.39; for each comparison: n = 8 RFP+ neurons imaged in vitro, n = 20 RFP+ neurons imaged in vivo) nor the total dendritic length (p = 0.30) was significantly different between the cells imaged in vivo and in vitro.
Figure 4.
Control dendritic reconstructions of pyramidal neurons in vivo, RFP+/PV+ neurons in vitro, and comparison with in vivo reconstructions of RFP+/ PV+ neurons showing similar representations of full dendritic morphologies. A, For comparison, pyramidal neurons from the Thy1-GFPS mouse line were reconstructed. Reconstructions of three example pyramidal neurons are presented as in Figure 1E–F, with apical processes colored in yellow and basal processes in green. Scale bar, 20 μm. B, Dendritic morphology is similar in RFP+ neurons reconstructed in vivo and in vitro, and distinct from GFP+ pyramidal neurons. The Sholl profiles of the RFP+ PV+ neurons imaged in vitro (bold red) and GFP+ pyramidal neurons imaged in vivo are superimposed on the in vivo RFP+ data from Figure 2 (pink). The in vitro Sholl profiles overlap extensively with the in vivo RFP+ Sholl profiles. Peak number of crossings (p = 0.28), distance of the peak from the soma (p = 0.22), and radius (p = 0.39) of the Sholl profiles were statistically similar in the in vitro and in vivo imaged cells, and distinct from pyramidal neurons (p < 0.001). C, Histogram of the total dendritic length of GFP+ pyramidal neurons (green), RFP+ neurons imaged in vitro (red), and RFP+ neurons imaged in vivo (pink). The total dendritic length was statistically similar (p = 0.30) between the two sets of RFP+ cells; however, both groups of RFP+ neurons had shorter dendrites than the GFP+ pyramidal neurons (p < 0.001). The morphological similarity between reconstructions of RFP+ neurons in vivo and in vitro suggests that in vivo imaging fully captured the dendritic arbors of these neurons.
Table 2.
Response features and morphological measurements of reconstructed GFP+ pyramidal neurons in Figure 4
| Cell ID | Response properties |
Morphology |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OSI | Evoked firing rate (Hz) | Spontaneous firing rate (Hz) | No. of primary dendrites | Total dendritic length (mm) | Maximal dendritic extent (μm) | Dendritic field volume (mm3) | Distance of Sholl peak from the soma (μm) | Peak no. of Sholl crossings | Total no. of dendritic segments | DPI | |
| 1 | 0.36 | 5.00 | 2.06 | 9 | 1.075 | 720 | 0.05443 | 171 | 17 | 59 | 0.88 |
| 2 | 0.61 | 0.75 | 0.39 | 5 | 0.891 | 500 | 0.05238 | 191 | 12 | 44 | 0.82 |
| 3 | 0.80 | 0.50 | 0.00 | 8 | 0.636 | 180 | 0.00209 | 70 | 19 | 98 | 0.98 |
| 4 | 0.85 | 0.50 | 0.22 | 5 | 0.558 | 440 | 0.01100 | 160 | 9 | 27 | 0.93 |
| 5 | 0.87 | 1.50 | 0.40 | 8 | 0.606 | 220 | 0.00814 | 111 | 29 | 101 | 0.95 |
| 6 | 0.89 | 3.75 | 0.99 | 7 | 2.032 | 420 | 0.05452 | 187 | 29 | 81 | 0.84 |
| 7 | 0.96 | 0.25 | 0.00 | 6 | 1.187 | 300 | 0.00104 | 100 | 26 | 70 | 0.69 |
| 8 | 1.00 | 1.25 | 0.30 | 9 | 0.645 | 240 | 0.00452 | 77 | 20 | 72 | 0.69 |
| 9 | 1.00 | 0.50 | 0.29 | 6 | 1.816 | 460 | 0.05987 | 208 | 17 | 52 | 0.77 |
Orientation preference maps of PV+ and PV− neurons
The correlation between sharper orientation selectivity of PV+ neurons and smaller dendritic lengths and arbor extents suggested that PV+ neurons might derive their tuning by sampling their cortical neighborhood, and sharper tuning may reflect focal sampling of locally homogeneous orientation representations. We thus examined the relationship between the tuning of PV+ neurons and surrounding neurons in the local V1 orientation map. Specifically, we used targeted, high-speed, two-photon calcium imaging (Wilson et al., 2012, 2013) to measure the orientation preferences of visually responsive RFP+/PV+ neurons and their neighbors (Fig. 5A–C). Among imaged cells, 77 of 105 RFP+/PV+ neurons and 1393 of 1601 RFP− neurons in four mice were visually responsive.
Figure 5.
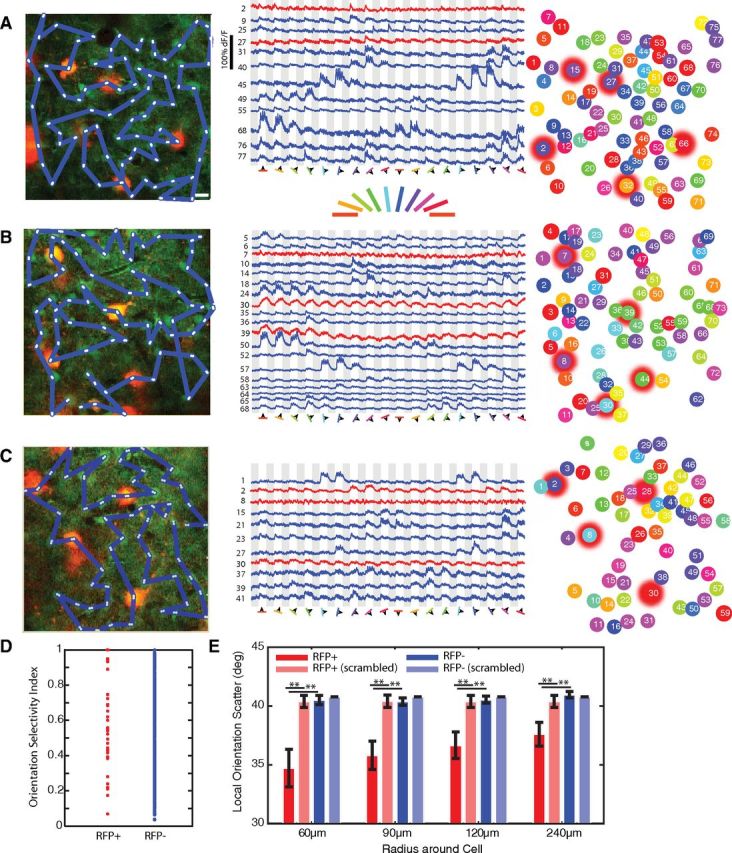
RFP+ neurons tend to share the orientation preferences of their neighbors. A–C, Left column, RFP+ neurons (red) and surrounding RFP− neurons were loaded with the calcium dye Oregon Green Bapta-1AM (green). Three representative networks are shown. The blue lines indicate the scan paths used to image calcium responses. White asterisks indicate the first and last pixels from which each cell's fluorescence response was collected within the scan path. Scale bar, 10 μm. Middle column, Example fluorescent responses to the 18 episodically presented drifting gratings (18 directions; gray represents 4 s OFF; white represents 4 s ON), from cells in each of the example networks (blue represents RFP− neuron responses; red represents RFP+ neuron responses). Right column, Each neuron is color-coded by its preferred orientation, and RFP+ neurons are indicated by red halos. A, RFP+ OSIs: cell #2, 0.23; #15, 0.84; #27, 0.80; #32, 0.29; #66, 0.48. B, RFP+ OSIs: cell #7, 0.92; #8, 0.6; #30, 0.32; #39, 0.59; #44, 0.38. C, RFP+ OSIs: cell #2, 0.74; #8, 0.31; #28, 0.51; #30, 0.35. D, The OSI spanned the full range from 0 (untuned) to 1 (highly tuned) in both RFP+ (red) and RFP− (blue) neurons, consistent with the electrophysiology measurements of Figure 1. E, The local scatter in orientation preference surrounding each neuron within expanding intercell distances (red represents RFP+ neurons; pink, RFP+ in same networks with scrambled orientation preferences; dark blue, RFP− neurons; light blue, RFP− in same networks with scrambled orientation preferences). The scatter surrounding RFP+ neurons was significantly lower than the scatter surrounding RFP− neurons (p < 0.01), or RFP+ neurons in scrambled networks (p < 0.01). Error bars indicate SEM. **p < 0.01.
The preferred orientations of cells and their neighbors across the field of view were then compared (Fig. 5A–C). We identified RFP+ cells with a range of OSIs, including high and low OSIs (Fig. 5D). The local orientation scatter surrounding each cell was calculated by measuring the difference between that cell's preferred orientation and those of the neighboring cells within 60, 90, 120, or 240 μm (Fig. 5E, pooling all RFP+ and RFP− cells from 4 animals); these distances included the maximal dendritic extent for highly tuned cells (<150 μm), and the maximal dendritic extent of most broadly tuned cells (∼250 μm; Fig. 2A). The orientation scatter surrounding both RFP+ and RFP− neurons increased with distance, being significantly greater at 240 μm than at 60 or 90 μm (p < 0.05 comparing scatter of RFP+ or RFP− neurons at 60 and 90 μm with 240 μm), reflecting the existence of local homogeneities in the orientation map that dissipate at larger distance scales.
However, the scatter surrounding RFP+ neurons was significantly lower than the scatter surrounding RFP− neurons at all distance scales (60 μm, p < 0.01; 90 μm, p < 0.01; 120 μm, p < 0.01; 240 μm, p < 0.01); that is, RFP+ neurons tended to have more similar orientation preferences to those of neighboring cells compared with RFP− neurons. To examine the validity of this difference, we scrambled the orientation preferences among all of the cells in the field of view, maintaining their spatial locations and RFP+/ RFP− identities, and again measured the scatter surrounding the RFP+ neurons. Indeed, the true scatter surrounding RFP+ neurons was less than the scatter surrounding RFP+ neurons when the orientation preferences had been resampled among the cells in the field of view (Fig. 5E; 60 μm, p < 0.01; 90 μm, p < 0.01; 120 μm, p < 0.01; 240 μm, p < 0.01). RFP− neurons, on the other hand, had similar scatter in the intact and scrambled cases at all distances examined (p > 0.2). Thus, PV+ neurons more closely resemble the orientation preferences of neighboring cells compared with RFP− neurons, and their location with the orientation map reflects local homogeneities in orientation representation that may contribute to their orientation selectivity.
Discussion
We have described the orientation selectivity of parvalbumin-expressing (PV+) inhibitory interneurons in the mouse primary visual cortex (V1), using two-photon targeted cell-attached recordings. Although PV+ neurons are less selective for orientation on average than the rest of the V1 neuronal population, the distribution of orientation selectivity in PV+ neurons includes highly tuned cells in addition to broadly tuned cells. This diversity of tuning correlates with dendritic morphology: highly tuned PV+ neurons have shorter total dendritic lengths, maximal branching closer to the soma, smaller dendritic field volumes, and shorter maximal dendritic extents compared with broadly tuned PV+ neurons. Dendritic branching complexity, however, shows no relation to tuning. Furthermore, the orientation selectivity of PV+ neurons is more similar to that of their local neighbors compared with non-PV+ neurons. Thus, the tuning of PV+ neurons may derive from sampling of local inputs coupled with their dendritic morphology: highly tuned neurons, for example, may derive their tuning by sparsely sampling a homogeneous local population with short, proximally branched, dendrites, whereas broadly tuned neurons may derive their tuning by extensively sampling a more heterogeneous population with longer, distally branched dendrites.
Several studies have now examined the orientation selectivity of inhibitory neurons in the mouse visual cortex; we and others have described the existence of both highly tuned and broadly tuned inhibitory neurons (Niell and Stryker, 2008; Ma et al., 2010; Runyan et al., 2010; Zariwala et al., 2011), whereas others have described only broadly tuned inhibitory neurons (Sohya et al., 2007; Liu et al., 2009; Kerlin et al., 2010). In the present study, we sampled a large number of PV+ inhibitory interneurons (n = 80 visually responsive PV+ neurons) to describe the distribution of orientation selectivity in this population more reliably, finding an overall more broadly tuned distribution than the rest of the V1 population, but a significant number of sharply tuned neurons as well. Differences in targeting and recording methods could account for the smaller, highly tuned, subclass missing from some studies (Hasenstaub and Callaway, 2010), as each study has used a unique combination of transgenic mice and recording techniques (Liu et al., 2009; Kerlin et al., 2010; Ma et al., 2010).
The existence of functionally diverse PV+ neurons implies that these neurons may derive their inputs in different ways. The dendritic trees of PV+ neurons were carefully and completely reconstructed in this study (Fig. 1). Fine-resolution z-stacks captured the full dorsal-ventral extent of the dendritic trees, whereas post hoc image processing to enhance the image contrast and careful manual tracing allowed each fine dendritic process to be traced through its full extent. PV+ neurons imaged in slices and reconstructed with identical methodology yielded highly similar dendritic lengths and Sholl analysis results (Fig. 4). Furthermore, the total dendritic lengths that we measured in vivo (0.9–5.6 mm; mean 2.2 mm) are similar to those described by others for inhibitory basket neurons that were imaged in vitro (Wang et al., 2002, mean 2.3 mm; Kawaguchi et al., 2006, mean 2.9 mm; Helmstaedter et al., 2009).
The finding that the orientation selectivity of PV+ neurons is correlated with the orientation distribution of cells in their local neighborhood is in agreement with recent work suggesting that inhibitory neurons receive strong local inputs from nearby cells (Bock et al., 2011; Hofer et al., 2011). Because V1 in rodents lacks the highly ordered orientation maps found in carnivores, it has been commonly assumed that orientation preferences are uniformly random across the rodent visual cortex (Ohki and Reid, 2007). However, our results in mouse V1 confirm longstanding hints that local clustering of orientation preferences exists (Dräger, 1975; Kerlin et al., 2010). Local coherence in the orientation map, combined with sparse and highly local sampling of inputs by some inhibitory neurons, could lead to high selectivity in these cells. Future studies examining the outputs of highly tuned and broadly tuned inhibitory neurons will determine whether these neurons also select their targets according to such spatial rules, or in functionally specific ways (Yoshimura and Callaway, 2005; Fino and Yuste, 2011; Packer and Yuste, 2011; Wilson et al., 2012).
Footnotes
This work was supported by National Research Service Award NS679512 (C.A.R.), National Institutes of Health Grants EY007023 and EY018041 (M.S.), and the Simons Foundation (M.S.). We thank Nathan Wilson for developing the two-photon targeted fast scan software, and the members of the M.S. laboratory for useful discussions and feedback.
The authors declare no competing financial interests.
References
- Adesnik H, Bruns W, Taniguchi H, Huang ZJ, Scanziani M. A neural circuit for spatial summation in visual cortex. Nature. 2012;490:226–231. doi: 10.1038/nature11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah BV, Bruns W, Carandini M, Scanziani M. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 2012;73:159–170. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatow M, Rozov A, Katona I, Hormuzdi SG, Meyer AH, Whittington MA, Caputi A, Monyer H. A novel network of multipolar bursting interneurons generates θ frequency oscillations in neocortex. Neuron. 2003;38:805–817. doi: 10.1016/S0896-6273(03)00300-3. [DOI] [PubMed] [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brainard DH. The psychophysics toolbox. Spat Vis. 1997;10:433–436. [PubMed] [Google Scholar]
- Burkhalter A. Many specialists for suppressing cortical excitation. Front Neurosci. 2008;2:155–167. doi: 10.3389/neuro.01.026.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Erisir A, Farb C, Nadal MS, Ozaita A, Lau D, Welker E, Rudy B. K(+) channel expression distinguishes subpopulations of parvalbumin- and somatostatin-containing neocortical interneurons. J Neurosci. 1999;19:9332–9345. doi: 10.1523/JNEUROSCI.19-21-09332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Graziano MS, Tank DW. Functional clustering of neurons in motor cortex determined by cellular resolution imaging in awake behaving mice. J Neurosci. 2009;29:13751–13760. doi: 10.1523/JNEUROSCI.2985-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dräger UC. Receptive fields of single cells and topography in mouse visual cortex. J Comp Neurol. 1975;160:269–290. doi: 10.1002/cne.901600302. [DOI] [PubMed] [Google Scholar]
- Du J, Zhang L, Weiser M, Rudy B, McBain CJ. Developmental expression and functional characterization of the potassium-channel subunit Kv3.1b in parvalbumin-containing interneurons of the rat hippocampus. J Neurosci. 1996;16:506–518. doi: 10.1523/JNEUROSCI.16-02-00506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/S0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fino E, Yuste R. Dense inhibitory connectivity in neocortex. Neuron. 2011;69:1188–1203. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez H, Davies AM. A fast and accurate procedure for deriving the Sholl profile in quantitative studies of neuronal morphology. J Neurosci Methods. 2007;163:24–30. doi: 10.1016/j.jneumeth.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Han ZS. Electrophysiological and morphological differentiation of chandelier and basket cells in the rat hippocampal formation: a study combining intracellular recording and intracellular staining with biocytin. Neurosci Res. 1994;19:101–110. doi: 10.1016/0168-0102(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Hasenstaub AR, Callaway EM. Paint it black (or red, or green): optical and genetic tools illuminate inhibitory contributions to cortical circuit function. Neuron. 2010;67:681–684. doi: 10.1016/j.neuron.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Helm J, Akgul G, Wollmuth LP. Subgroups of parvalbumin-expressing interneurons in layers 2/3 of the visual cortex. J Neurophysiol. 2013;109:1600–1613. doi: 10.1152/jn.00782.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Sakmann B, Feldmeyer D. L2/3 interneuron groups defined by multiparameter analysis of axonal projection, dendritic geometry, and electrical excitability. Cereb Cortex. 2009;19:951–962. doi: 10.1093/cercor/bhn130. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SB, Ko H, Pichler B, Vogelstein J, Ros H, Zeng H, Lein E, Lesica NA, Mrsic-Flogel TD. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat Neurosci. 2011;14:1045–1052. doi: 10.1038/nn.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Karube F, Kubota Y. Dendritic branch typing and spine expression patterns in cortical nonpyramidal cells. Cereb Cortex. 2006;16:696–711. doi: 10.1093/cercor/bhj015. [DOI] [PubMed] [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisvárday ZF, Eysel UT. Functional and structural topography of horizontal inhibitory connections in cat visual cortex. Eur J Neurosci. 1993;5:1558–1572. doi: 10.1111/j.1460-9568.1993.tb00226.x. [DOI] [PubMed] [Google Scholar]
- Krimer LS, Zaitsev AV, Czanner G, Kröner S, González-Burgos G, Povysheva NV, Iyengar S, Barrionuevo G, Lewis DA. Cluster analysis-based physiological classification and morphological properties of inhibitory neurons in layers 2–3 of monkey dorsolateral prefrontal cortex. J Neurophysiol. 2005;94:3009–3022. doi: 10.1152/jn.00156.2005. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kwan AC, Zhang S, Phoumthipphavong V, Flannery JG, Masmanidis SC, Taniguchi H, Huang ZJ, Zhang F, Boyden ES, Deisseroth K, Dan Y. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature. 2012;488:379–383. doi: 10.1038/nature11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BH, Li P, Li YT, Sun YJ, Yanagawa Y, Obata K, Zhang LI, Tao HW. Visual receptive field structure of cortical inhibitory neurons revealed by two-photon imaging guided recording. J Neurosci. 2009;29:10520–10532. doi: 10.1523/JNEUROSCI.1915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron M, Turi GF, Kaifosh P, Lee PH, Bolze F, Sun XH, Nicoud JF, Zemelman BV, Sternson SM, Losonczy A. Regulation of neuronal input transformations by tunable dendritic inhibition. Nat Neurosci. 2012;15:423–430. doi: 10.1038/nn.3024. S1–S3. [DOI] [PubMed] [Google Scholar]
- Lund JS, Lewis DA. Local circuit neurons of developing and mature macaque prefrontal cortex: Golgi and immunocytochemical characteristics. J Comp Neurol. 1993;328:282–312. doi: 10.1002/cne.903280209. [DOI] [PubMed] [Google Scholar]
- Ma WP, Liu BH, Li YT, Huang ZJ, Zhang LI, Tao HW. Visual representations by cortical somatostatin inhibitory neurons: selective but with weak and delayed responses. J Neurosci. 2010;30:14371–14379. doi: 10.1523/JNEUROSCI.3248-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska A, Yiu G, Yuste R. A custom-made two-photon microscope and deconvolution system. Pflugers Arch. 2000;441:398–408. doi: 10.1007/s004240000435. [DOI] [PubMed] [Google Scholar]
- Mariño J, Schummers J, Lyon DC, Schwabe L, Beck O, Wiesing P, Obermayer K, Sur M. Invariant computations in local cortical networks with balanced excitation and inhibition. Nat Neurosci. 2005;8:194–201. doi: 10.1038/nn1391. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki K, Reid RC. Specificity and randomness in the visual cortex. Curr Opin Neurobiol. 2007;17:401–407. doi: 10.1016/j.conb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Scanziani M. Enforcement of temporal fidelity in pyramidal cells by somatic feed-forward inhibition. Science. 2001;293:1159–1163. doi: 10.1126/science.1060342. [DOI] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, Buzsáki G. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyan CA, Schummers J, Van Wart A, Kuhlman SJ, Wilson NR, Huang ZJ, Sur M. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schummers J, Mariño J, Sur M. Synaptic integration by V1 neurons depends on location within the orientation map. Neuron. 2002;36:969–978. doi: 10.1016/S0896-6273(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Schummers J, Yu H, Sur M. Tuned responses of astrocytes and their influence on hemodynamic signals in the visual cortex. Science. 2008;320:1638–1643. doi: 10.1126/science.1156120. [DOI] [PubMed] [Google Scholar]
- Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vruwink M, Schmidt HH, Weinberg RJ, Burette A. Substance P and nitric oxide signaling in cerebral cortex: anatomical evidence for reciprocal signaling between two classes of interneurons. J Comp Neurol. 2001;441:288–301. doi: 10.1002/cne.1413. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Toledo-Rodriguez M, Wu CZ, Markram H. Anatomical, physiological, molecular and circuit properties of nest basket cells in the developing somatosensory cortex. Cereb Cortex. 2002;12:395–410. doi: 10.1093/cercor/12.4.395. [DOI] [PubMed] [Google Scholar]
- Wehr M, Zador AM. Balanced inhibition underlies tuning and sharpens spike timing in auditory cortex. Nature. 2003;426:442–446. doi: 10.1038/nature02116. [DOI] [PubMed] [Google Scholar]
- Wilson NR, Runyan CA, Wang FL, Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NR, Schummers J, Runyan CA, Yan SX, Chen RE, Deng Y, Sur M. Two-way communication with neural networks in vivo using focused light. Nat Protoc. 2013;8:1184–1203. doi: 10.1038/nprot.2013.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XX, Baram TZ, Gerth A, Schultz L, Ribak CE. Co-localization of corticotropin-releasing hormone with glutamate decarboxylase and calcium-binding proteins in infant rat neocortical interneurons. Exp Brain Res. 1998;123:334–340. doi: 10.1007/s002210050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y, Callaway EM. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat Neurosci. 2005;8:1552–1559. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- Zaitsev AV, Povysheva NV, Gonzalez-Burgos G, Rotaru D, Fish KN, Krimer LS, Lewis DA. Interneuron diversity in layers 2–3 of monkey prefrontal cortex. Cereb Cortex. 2009;19:1597–1615. doi: 10.1093/cercor/bhn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala HA, Madisen L, Ahrens KF, Bernard A, Lein ES, Jones AR, Zeng H. Visual tuning properties of genetically identified layer 2/3 neuronal types in the primary visual cortex of cre-transgenic mice. Front Syst Neurosci. 2011;4:162–116. doi: 10.3389/fnsys.2010.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]