Abstract
In the primate visual cortex, areas V1 and V2 distribute information they receive from the retina to virtually all extrastriate cortex, parsing this information into dorsal and ventral streams. Therefore, understanding the connectivity between V1 and V2 is crucial to understand visual cortical processing. Cytochrome oxidase staining in V2 reveals a repeating pattern of pale–thick–pale–thin stripes. V1 sends parallel output pathways to distinct V2 stripes. Previous models proposed either three or two parallel V1-to-V2 pathways in macaque, but both models viewed the two pale stripes within a single stripe cycle as a single compartment. However, recent studies have suggested that the two pale stripes may be functionally distinct, and in marmosets they also differ anatomically in the laminar origin of projections they receive from V1. Here we have asked whether the two pale stripes are also anatomically distinct in macaque. We made small retrograde tracer injections in different pale stripe types. We found that while both pale stripes receive a predominant V1 input from layers 2/3, only one set of pale stripes (pale lateral) receives significant projections from layer 4B, while the other set (pale medial) receives few or no layer 4B projections. Moreover, different tracer injections in nearby pale stripe types revealed that 97–99% of layer 2/3 cells only project to a single pale stripe type. These results demonstrate that in macaque, the two pale stripes are anatomically distinct compartments, and support the notion of two distinct projection streams from V1 to the two pale stripes of V2.
Introduction
Understanding the wiring of neural circuits in the cerebral cortex is necessary to understand how cortical computations are implemented. For example, in the visual cortex, knowledge of the connectivity between areas V1 and V2 is crucial to understand how these areas reorganize and refine visual signals from the retina into dorsal and ventral processing streams for spatial and object vision, respectively (Ungerleider and Mishkin, 1982).
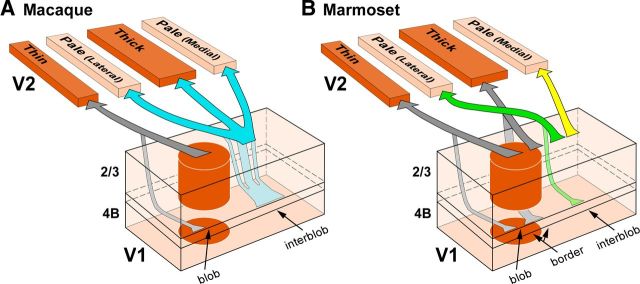
In many primate species, cytochrome oxidase (CO) staining reveals two compartments in V1 (blobs and interblobs; Horton, 1984) and three in V2 (CO-dark, thin and thick, and CO-pale stripes; Tootell et al., 1983; Wong-Riley and Carroll, 1984). V1 sends its outputs to V2 as multiple parallel pathways projecting to distinct CO stripes. A previous model proposed three parallel V1-to-V2 pathways in macaque (Livingstone and Hubel, 1984, 1987). This model was recently revised based on the anatomical demonstration of only two parallel pathways between V1 and V2, one connecting V1 blobs with V2 thin stripes, the other connecting interblobs with thick and pale stripes (Sincich and Horton, 2002a; Fig. 1A). Both models view CO-pale stripes as a single compartment. However, there are two pale stripes for each full pale–thick–pale–thin stripe cycle, and previous studies suggested that the two pale stripes are functionally and anatomically distinct. Specifically, in macaque, the two pale stripes differ in receptive field size and scatter (Roe and Ts'o, 1995), and in the selectivity for stimulus orientation as revealed by optical imaging in both owl (Xu et al., 2004) and macaque (Lim et al., 2009) monkey V2.
Figure 1.
Parallel pathways between V1 and V2 CO compartments in macaque and marmoset. Arrows indicate V1 projections to V2, with arrow thickness indicating the relative density of projections. Only projections to pale stripes are shown in color as these projections are the focus of the present study. Projections to other V2 stripe types, which were not investigated in this study, are shown in gray. A, The bipartite scheme of V1-to-V2 projections in macaque proposed by Sincich and colleagues (Sincich and Horton, 2002a; Sincich et al., 2007, 2010). According to this model, neurons throughout the depth of a blob column (predominantly in layers 2/3, and less so in layer 4B) project to thin stripes (gray arrows; sparse inputs from layers 4A and 6 also exist but are omitted in the figure), while neurons throughout the depth of an interblob column (predominantly in layers 2/3, and less so in 4B) project to both thick and pale stripes (cyan arrows; sparse inputs from layers 4A and 6 are omitted). In this model, all pale stripes receive V1 inputs from the same V1 layers. B, The quadripartite scheme of V1-to-V2 projections in marmoset proposed by Federer et al. (2009). In this model, neurons in blob columns (predominantly in layers 2/3, and less so in 4B) project to thin stripes (left pair of gray arrows; sparser inputs from 4A and 6 exist, but are omitted); neurons at the border between blobs and interblobs (predominantly and equally from layers 2/3 and 4B) project to thick stripes (right pair of gray arrows; sparser inputs from layers 4A and 6 are omitted). In this model, all pale stripes receive the strongest projection from interblob neurons in layers 2/3 (dark green and yellow arrows) and a smaller projection from layer 4A (data not shown), but while pale lateral stripes additionally receive a contribution from layer 4B (light green arrow), pale medial stripes do not receive inputs from layer 4B or 6.
Moreover, in marmoset visual cortex there are differences in the laminar origin of projections from V1 interblobs to the two pale stripes (Fig. 1B; Federer et al., 2009). Specifically, while both pale stripe types receive V1 projections from layers 2/3 and 4A, only pale stripes positioned laterally to the nearest thick stripe (here termed pale lateral) receive a small, but significant, projection from V1 layer 4B. Instead, pale stripes positioned medially to the nearest thick stripe (pale medial) receive virtually no layer 4B input. The goal of this study was to determine whether the anatomical differences in the V1 projections to the two V2 pale stripes that we found in marmosets also exist in macaque. A recent study, using larger injections of retrograde neuroanatomical tracers in macaque V2, reported no differences in the V1 layer 4B projection to the two pale stripes (Sincich et al., 2010). Here we have re-examined this issue using small retrograde tracer injections into each pale stripe of macaque V2. We found that in macaque, similar to marmoset, the two V2 pale stripes differ in the layer 4B contribution they receive from V1. Double-labeling studies further demonstrated that individual V1 cells project to either one or the other pale stripe type, but rarely to both. We conclude that the two V2 pale stripes are anatomically distinct compartments also in Old World primates, and that there exist two distinct projection streams from V1 to the pale stripes of V2 in macaque.
Materials and Methods
Animals
All experimental procedures were in accordance with protocols approved by the University of Utah Institutional Animal Care and Use Committee. The results of this study are based on quantitative analysis of 13 tracer injections (6 in pale lateral and 7 in pale medial stripes) made in five normal adult male macaque monkeys (Macaca fascicularis; Tables 1, 2). These cases were selected for analysis from a larger pool of injections (n = 65) made in eight animals for the purpose of this and other studies (criteria for selection of cases to be analyzed are described in a later section of this article). Each animal received 6–10 injections of four different tracers in dorsal area V2 of one hemisphere. Up to three injections of the same tracer were made in the same hemisphere spaced at least 5 mm apart to ensure no overlap of the resulting retrogradely labeled fields in V1. Meanwhile, injections of different tracers, instead, were regularly spaced 1.5 mm apart. The contralateral hemisphere was not injected to avoid potentially confounding label of callosal projection neurons, which are especially prominent near the vertical meridian representation at the V1/V2 border (Zeki, 1970; Van Essen et al., 1982; Kennedy et al., 1986). Injections were targeted visually just posterior to the lunate sulcus, and were made blindly with respect to stripe type, except in one case (319) in which a pale lateral stripe was identified in vivo before injecting the tracer based on intrinsic signal optical imaging of orientation maps.
Table 1.
Laminar distribution of labeled cells in V1 layers 2/3 and 4B
| CO stripe type, case number | Injection sites |
N labeled cells |
Percentage labeled V1 cells |
Transport time (d) | Figures | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tracer injected | Layers injected | Diameter (mm)a | Layer 2/3 | Layer 4B | Layer 2/3 | Layer 4B | |||
| Pale lateralb | |||||||||
| 305-647m | CTB 647 | 1–6 | 0.14 × 0.20 | 2825 | 836 | 77 | 23 | 11 | 2, 3A,B, 4A |
| 305-647l | CTB 647 | 1–6 | 0.36 × 0.17 | 2788 | 300 | 90 | 10 | 11 | 2, 3A,B, 4A |
| 305-488l | CTB 488 | 1–6 | 0.21 × 0.48 | 5071 | 328 | 94 | 6 | 11 | 2, 3A,B, 4A |
| 319 | FR | 4–6 | 0.33 × 0.30 | 84 | 26 | 77 | 23 | 3 | 4A |
| Pale medialc | |||||||||
| 305-CTBgm | CTBg | 1–6 | 0.39 × 0.51 | 1384 | 9 | 99 | 1 | 11 | 2, 3A–C, 4B |
| 305-CTBgmd | CTBg | 1–6 | 0.31 × 0.19 | 362 | 19 | 95 | 5 | 11 | 2, 3A–C, 4B |
| 305-CTBgl | CTBg | 1–6 | 0.24 × 0.51 | 582 | 1 | 100 | 0 | 11 | 2, 3A–C, 4B |
| 304-488m | CTB 488 | 1–6 | 0.37 × 0.53 | 2036 | 31 | 99 | 1 | 5 | 4B |
| 326-647l | CTB 647 | 1–6 | 0.21 × 0.67 | 1455 | 19 | 99 | 1 | 10 | 4B, 5 |
Table 2.
Number of double-labeled cells in V1 layer 2/3
| CO stripes and tracers injected |
Case number | Diameter (mm) | Interinjection distance (μm)a | Label overlap (%)b |
N single-labeled cells |
N double-labeled cells | Percentage double-labeled cells |
Figures | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pale medial (Tracer 1) | Pale lateral (Tracer 2) | Tracer 1 | Tracer 2 | Percentage of total labeled cellsc | Percentage of cells labeled with Tracer 1d | Percentage of cells labeled with Tracer 2e | |||||||
| CTBgmdf | CTB-488l | 326 | 0.39 × 0.61 | 0.25 × 0.53 | 466 | 88.6 | 1215 | 4195 | 51 | 0.93 | 4.03 | 1.22 | 5, 6A–E |
| CTB 647l | CTB-488l | 326 | 0.22 × 0.65 | 0.25 × 0.53 | 1139 | 84.9 | 1376 | 1055 | 32 | 1.28 | 2.27 | 2.94 | 5, 6F–J |
| CTBg | CTB-488 | 325 | 0.32 × 0.33 | 0.34 × 0.73 | 1262 | 92.9 | 946 | 1110 | 69 | 3.05 | 6.80 | 5.85 | |
aDistance between the lateral edge of medial-most injection site and medial edge of lateral-most injection site, measured across CO stripes.
bArea of overlap of the two labels/smallest area of single-labeled field × 100.
cCalculated as follows: N double-labeled cells * 100/(N tracer 1-labeled cells + N tracer 2-labeled cells + N double-labeled cells).
dCalculated as follows: N double-labeled cells * 100/(N cells labeled with Tracer 1 + N double-labeled cells).
eCalculated as follows: N double-labeled cells * 100/(N cells labeled with Tracer 2 + N double-labeled cells).
fThis injection straddled a thick stripe medially.
Surgery and tracer injections
Animals were preanesthetized with ketamine (25 mg/kg, i.m.), intubated with an endotracheal tube, placed in a stereotaxic apparatus, and artificially ventilated. Anesthesia was maintained with isofluorane (0.5–2%) in a mixture of 1:1 oxygen and nitrous oxide, except for case 319, in which we performed optical imaging (procedures for this case are described separately below). A craniotomy elongated parallel to the lunate sulcus was performed; small durotomies were made just caudal to the posterior edge of the lunate, and the tracers were injected. The injected retrograde tracers are listed in Tables 1 and 2. Fluororuby (FR; molecular weight 3000 and 10,000, mixed 1:1, 5% in 0.1 m PBS, pH 7.3; Invitrogen) was delivered iontophoretically using positive current in 7 s on/off cycles for 15 min and a glass micropipette of 15 μm tip diameter. All other tracers were pressure injected using glass micropipettes of 35–45 μm tip diameter and a picospritzer, in the following concentrations and volumes: gold-conjugated cholera toxin B (CTBg; List Biological Labs), 0.1% in distilled water, 90 nl; CTB-alexa-488 and CTB-alexa-647 (CTB488 and CTB647, respectively; Invitrogen), 3% in distilled water, 45–75 nl. All tracers were delivered at a cortical depth of 800 μm, except for FR, which was injected at a depth of 900 μm. These parameters typically yielded injection sites of ∼200–500 μm in diameter involving all cortical layers (but the FR injection involved only layers 4–6; Table 1). After the injections, animals were recovered from anesthesia, allowed to survive for 10–11 d (all but 2 animals; Table 1), and then killed with sodium pentobarbital (150 mg/kg, i.p.) and perfused with saline for 2–3 min, followed by 0.5% paraformaldehyde in 0.1 m phosphate buffer for 5 min.
In case 319, an injection of FR was targeted to a pale lateral stripe using intrinsic signal optical imaging of orientation maps as guidance. During imaging, anesthesia was maintained with sufentanil citrate (5–10 μg/kg/h) and the animal was paralyzed with vecuronium bromide (0.3 mg/kg/h). Surgery, chamber implantation, acquisition of optical images, and image analysis were performed as described previously (Federer et al., 2009). Visual stimuli consisted of full-field, high-contrast (100%) achromatic drifting square-wave gratings of eight different orientations and 1.0 cycles/° spatial frequency, moving back and forth at 1.5 or 2°/s in directions perpendicular to the grating orientation. After 24 h of imaging, the FR injection was targeted to a pale lateral stripe identified as a region having an orientation-preference map neighboring medially a region devoid of orientation maps (i.e., a presumed thin stripe). Unlike the other cases, this animal survived 3 additional days under anesthesia and was then perfused with 4% paraformaldehyde for 15 min.
Histology
Areas V1 and V2 were dissected away from the rest of the visual cortex by cutting through the bottom of the lunate sulcus. The V1/V2 block was then unfolded, flattened, postfixed between glass slides for 1–2 h, sunk in 30% sucrose for cryoprotection, and frozen-sectioned tangentially at 40 μm. In the optically imaged case, to avoid major tissue distortions, which could compromise accurate alignment of the images with the histological sections, no unfolding was performed; rather, the cortex was flattened gently above the imaged region and sectioned tangentially. In all cases, every third section was reacted free-floating for CO. In two cases (325, 326) injected with CTBg, CO-stained sections were mounted and coverslipped temporarily in 0.1 m phosphate buffer, and CO staining was digitized before being silver reacted to reveal CTBg. The two remaining series of sections were reacted to reveal the injected tracers. Specifically, one series of sections containing fluorescent CTB488 and CTB647 signals was mounted immediately after sectioning and coverslipped using Gel Mount (Electron Microscopy Sciences). The third series of sections was immunoreacted for CTB488 or FR as described previously (Jeffs et al., 2009). CTBg was revealed using silver intensification (Llewellyn-Smith et al., 1990) on the CO-stained sections and/or on the series of sections previously reacted for CTB488.
Data analysis
After completing all tissue staining, we first determined the location of each injection site with respect to CO stripe type, without knowledge of the resulting pattern of V1 label. This was performed as follows: CO staining was digitized at low magnification (1.25×); then, using IRtweak warping software (NCRtoolset, SCI Institute at the University of Utah; Anderson et al., 2009), 3–6 images of serial CO sections were overlaid by aligning the radial blood vessels, and merged in Adobe Photoshop. We find that the stripe pattern is much clearer in these composite images than in any single CO section, in which the stripes can be incomplete. Subsequently, V2 injection sites were outlined with camera lucida in a full series of sections; drawings from individual sections were aligned to each other using blood vessels to generate a composite injection site, and the latter was aligned to the CO image stack. From these images we selected injection sites that were confined to pale stripes that could be unambiguously classified by several investigators as either lateral or medial to the nearest thick stripe. We also used these composite reconstructions of injection sites across layers to measure the size of the injections reported in the tables. We excluded from the laminar analysis injections that straddled a neighboring stripe type or V1, and injections that produced no retrograde transport. The effective tracer uptake zone for immunoreacted CTB488 and FR was defined as the region at the injection site where no labeled cell bodies or fibers could be discerned (Ericson and Blomqvist, 1988; Luppi et al., 1990; Brandt and Apkarian, 1992; Angelucci et al., 1996). For CTBg, the tracer uptake zone was defined as the dark core seen under dark-field microscopy (Llewellyn-Smith et al., 1990). For fluorescent CTB488 and 647, the tracer uptake zone was the dense core seen under fluorescence microscopy, within which no labeled cell bodies were discernible.
Laminar analysis.
V1 label resulting from the selected tracer injections was then plotted by an investigator who had no knowledge of the stripe location of the injection sites. Retrogradely labeled cells were plotted on live images of tissue sections on all available sections (at least 1 in 3 for each tracer type) throughout the cortical thickness, using Neurolucida software (MicroBrightField). Fluorescent cells were plotted using 10–20× objectives, while CTBg and DAB-labeled cells were typically plotted under dark-field and bright-field illumination, respectively, using a 10× objective. In cases where the CTBg label was paler, or unspecific background staining was higher, cells were plotted at higher magnification (20–40×). Lack of label in layer 4B after some injections in pale medial stripes was verified by scanning the tissue sections at 40× magnification.
The laminar location of V1-labeled cells (and of tracer injection sites) was determined by aligning plots of label in single sections to adjacent CO-stained sections, using radial blood vessel profiles, and additionally by counterstaining the same sections containing the label for Nissl after plotting cell label. Layer 4B in Nissl stain appears as a pale region sandwiched between two darkly stained regions above (corresponding to layer 4A) and below (corresponding to layer 4C; see Fig. 3B,C). Laminar boundaries delineated in Nissl stain were also confirmed by alignment with adjacent CO-stained sections. In two cases, the laminar location of CTBg label was determined directly on the same sections that were also stained for CO. To quantify the laminar distribution of cell label in V1, we counted all labeled cells in layers 2/3 and 4B and estimated the percentage of labeled cells in each of these two layers as described in the Results.
Figure 3.
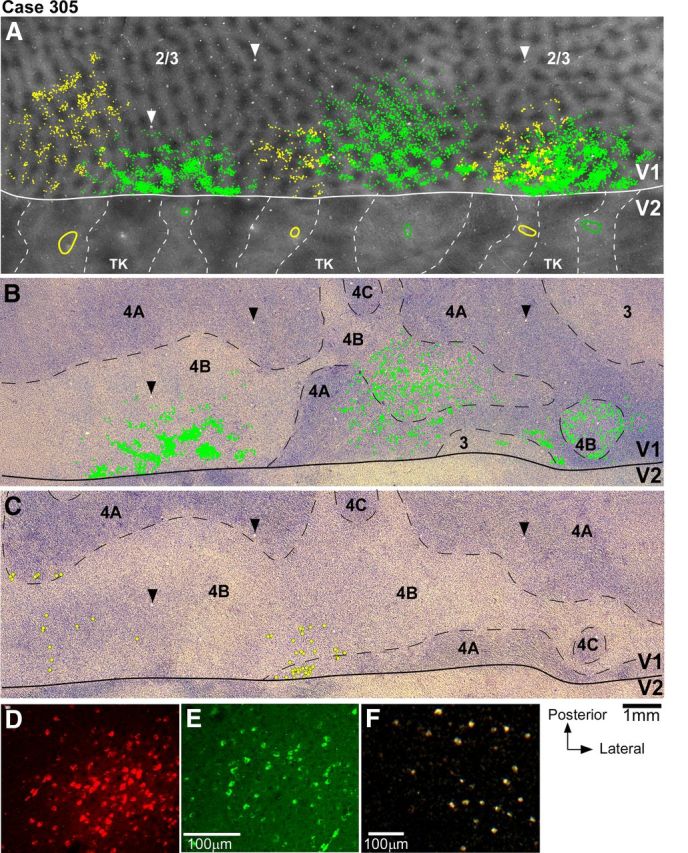
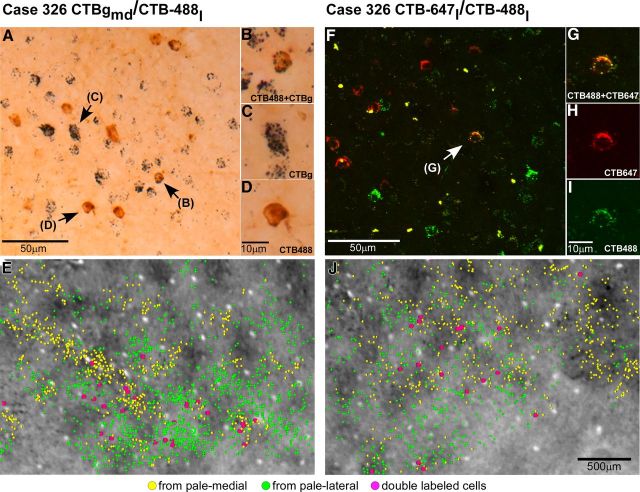
V1 projections from layers 2/3 and 4B to V2 pale lateral and pale medial stripes (case 305). A, Smaller field of view of same CO composite image as in Figure 2B, with superimposed outlines of three CTB-alexas injection sites (green ovals) in pale lateral stripes and three CTBg injection sites (yellow ovals) in pale medial stripes, and plots of the V1 cells (green and yellow dots) in layers 2/3 labeled by each respective tracer injection. Plots of V1 label are from the one tissue section containing the densest label in layers 2/3 (however, note that for purpose of analysis we plotted V1 label in a full series of sections). Arrowheads point to same blood vessel profiles as the arrowheads in B and C. B, Image of a single tissue section through layer 4B (counterstained with Nissl) that contained the densest label in this layer after injections in pale lateral stripes. Superimposed are plots of labeled cells (green dots) from this same section. Here and in C, black dashed contours delineate layer boundaries. C, Image of a Nissl-counterstained section immediately adjacent to the one in B that contained the densest label in layer 4B after injections in pale medial stripes. Yellow dots indicate the position of CTBg-labeled cells that were plotted in the same tissue section. Here cells are marked by larger dots than in A and B to facilitate detection of the very sparse label produced by the two most medial CTBg injection sites (CTBgm and CTBgmd). The most lateral CTBg injection (CTBgl) produced only one labeled cell (not in this section). D–F, High-power images of CTB647-labeled (D), CTB488-labeled (E), and CTBg-labeled (F) V1 cells, viewed under fluorescent microscopy (D–E) or dark-field illumination (F), illustrating examples of V1 label obtained in this case.
Analysis of double-labeled cells.
In two animals, paired injections of different tracers were made in nearby pale stripe types (n = 3 injection pairs; Table 2). V1 label through layers 2/3 resulting from each tracer injection was fully mapped. The V1-labeled fields resulting from each injection site were then outlined and their overlap area was measured and normalized to the area of the smallest of the two labeled fields. Within this overlap area, we counted the cells labeled with each tracer, and the cells double labeled with both tracers (Table 2), and the percentage of double-labeled cells was quantified as described in the Results. To identify cells double labeled for CTBg and CTB488, the same tissue sections were first immunoreacted to reveal CTB488 using a brown DAB reaction, and then silver reacted for CTBg. The labeled cells were plotted under direct microscopic observation and bright-field illumination using 40–64× objectives; cells were scored as double labeled if they showed cytoplasmic brown DAB product (CTB488) and black silver grains (CTBg) in the same focal plane (see Fig. 6B).
Figure 6.
Paired tracer injections in different pale stripe types reveal segregated V1 neuronal populations projecting to pale lateral and pale medial stripes. A–E, Case 326 CTBgmd/CTB-488l. A, Bright-field image of a tissue section immunoreacted for CTB488 and silver reacted for CTBg. This label was produced by the CTBg–CTB488 injection pair shown in Figure 5. Arrows point at single neurons shown at higher magnification in B–D. B–D, Higher-power images of neurons labeled with CTBg only (C), CTB488 only (D), or double labeled with both tracers (B). E, Plots of V1 layer 2/3 CTBg-labeled (yellow) and CTB488-labeled (green) cells from the section with the densest layer 2/3 label are shown superimposed to an adjacent CO-stained section. Purple dots, Double-labeled cells. F–J, Case 326 CTB-647l/CTB-488l. F, Confocal laser scanning image of a tissue section containing CTB647-labeled (red) and CTB488-labeled (green) cells. Arrow points to a double-labeled cell shown at higher power in G–I. G–I, Higher-power images of the same neuron shown under a single channel revealing a single tracer (H, I), and under both channels revealing colocalization of both tracers in the same neuron (G). J, Plots of V1 layer 2/3 CTB647-labeled (yellow) and CTB488-labeled (green) cells from the section with the densest layer 2/3 label are shown superimposed to an adjacent CO-stained section. Scale bar in J valid also for E.
Cells double labeled with fluorescent CTB488 and CTB647 were analyzed and quantified using a confocal microscope. Specifically, each 40-μm-thick tangential section containing fluorescently labeled cells in V1 layer 2/3 was scanned at 5 μm depth increments, using an Olympus FV1000 inverted laser-scanning confocal microscope with a 20× air objective; each section was scanned at both of the wavelengths needed to reveal each tracer. Two mosaic images of the label fields were generated using separate color channels for each tracer at each z-stack depth; colocalization of the two fluorescent tracers in double-labeled cells was determined by merging the two color-channel images into a single image for each z-stack depth. These image files were then imported into Neurolucida software, which enables quick scanning up and down the z-stack. The different types of labeled cells on these images were plotted using Neurolucida software. Cells labeled with either one or both tracers were identified and plotted using the single-channel and colocalization images (see Fig. 6F–I). As each laser-scanned focal plane was very narrow, we could be confident that a cell excited by two different wavelengths on an image at a single depth was indeed double labeled. Furthermore, by moving up and down the z-stack, we could identify each cell at several depths and only mark the cell once.
Results
Laminar distribution of V1 cells projecting to V2 pale stripes
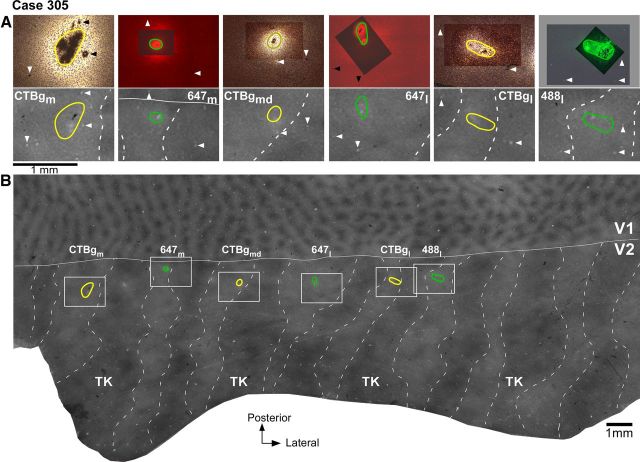
V2 in macaque is known to receive projections from V1 layers 2/3, 4A, 4B, and 5/6 (Kennedy and Bullier, 1985; Van Essen et al., 1986; Rockland, 1992). In this study, we have determined the relative contribution of each V1 layer to the two pale stripe types, by quantitatively analyzing the laminar distribution of labeled cells in V1 from tracer injections confined to either pale lateral (n = 4 injections) or pale medial (n = 5 injections) stripes (Table 1). Figure 2 shows one example case (305) in which six tracer injections landed cleanly in six consecutive pale stripes, three in pale lateral stripes and three in pale medial stripes. All six injections involved V2 layers 1–6 and the injection sites in pale lateral stripes were of similar size as the injection sites in pale medial stripes (Table 1). The resulting V1 label from these injections is shown in Figure 3. Throughout this article, the colors green and yellow are used to represent data from pale lateral and pale medial stripe injections, respectively. Qualitative observation of this case as well as of all other cases listed in Table 1 revealed that injections in either pale stripe type produced labeled cells in layers 2/3 (Fig. 3A) and 4A (Fig. 3B,C), but only injections in pale lateral stripes produced significant label in layer 4B (Fig. 3, compare B, C) and sparse label in layers 5/6 (data not shown). Consistent with previous results (Sincich et al., 2010), after injections in either pale lateral stripes or pale medial stripes, labeled neurons resided primarily in the CO-pale interblob regions of V1 (Fig. 3A).
Figure 2.
Tracer injections in V2 pale stripes (case 305). A, Top, High-power images of the tracer injection sites within the field of views boxed in B. The CTB-alexas injection sites are viewed under fluorescent microscopy, while the CTBg injection sites are viewed under dark-field microscopy. The images show the extent of the injections through the V2 cortical layers and were obtained by merging several images of sections taken at different cortical depths to demonstrate the full extent of the injection sites. To show both the injection core and the blood vessel profiles used for referencing with CO-stained sections, the latter were photographed using a longer exposure time than the former, and the two photographs were aligned in Adobe Photoshop. In the overexposed image (the one at the bottom of the stack), tracer transport around the injection core can appear as a bright glow partly hidden by the darker image of the uptake zone laying above. Note that this glow is not part of the tracer uptake zone but it is local tracer transport, as labeled cells could be discerned in this region. The tracer uptake zones are outlined in green for the injections laying in pale lateral stripes, and in yellow for the injections laying in pale medial stripes. Injection cases and tracer types are indicated in the bottom panels and correspond to those shown in B above each injection outline. Arrowheads point at the same blood vessel profiles as the arrowheads in each respective bottom panel. Bottom, Each panel shows a higher-power view of CO staining within its respective boxed region in B, with the outline of the tracer injection from the top panel superimposed. Each pair of top and bottom panels shows the same field of view in different tissue sections. B, Low-power and wide field of view of a composite CO image of unfolded and flattened V1 and V2, obtained by aligning and merging images of three tissue sections stained for CO. The image reveals V1 blob and interblob compartments in V1 and stripes (dashed contours) in V2. TK, Thick stripes; solid white contour, V1/V2 border; green and yellow ovals, outlines of tracer injection sites in pale lateral and pale medial stripes, respectively taken from the top panels in A. The regions within the white boxes are shown at higher power in the bottom panels of A. Same conventions are used in subsequent figures.
We quantified the relative contribution of V1 layers 2/3 and 4B to the two pale stripe types. We did not quantify label in layer 4A because in several cases only one in three sections was reacted for any given tracer, and since this layer is only ∼100 μm thick, we only had one section per series with label in 4A. Therefore, we could have easily missed the section(s) with the densest label in this layer. In contrast, we typically had 1.5–2 sections through layer 4B in a single, one in three, section series. While it is possible that sections with denser label through 4B may have been missed, this should have randomly affected cases in both pale stripe type groups. Quantification was performed by counting all labeled cells on a full series of sections through the full depth of layers 2/3 and 4B, and determining the percentage of labeled cells in each layer of the summed number of labeled cells in layers 2/3 and 4B. In the example case (305) shown in Figures 2 and 3, the three injections of CTB-alexas (2 CTB647, 1 CTB488) in pale lateral stripes produced 2825–5071 labeled cells in layers 2/3, and 300–836 labeled cells in layer 4B, with larger injection sites producing larger numbers of labeled cells in layers 2/3 but not necessarily in 4B (Table 1). The relative distribution of labeled cells resulting from these pale lateral stripe injections ranged between 77 and 94% in layers 2/3, and 6 and 23% in layer 4B. In contrast, the three injections of CTBg in the pale medial stripes of the same animal resulted in 362–1384 labeled cells in layers 2/3, and 1–19 labeled neurons in layer 4B, with larger injections producing greater numbers of labeled cells in layers 2/3, but not necessarily in 4B. The relative distribution of labeled cells resulting from these pale medial stripe injections ranged from 95 to 100% in layers 2/3, and 0 to 5% in layer 4B. Thus, the projection from layer 4B was proportionally much greater to the pale lateral stripes than to the pale medial stripes. However, the overall number of labeled cells (i.e., in both layers 2/3 and 4B) after pale lateral stripe injections in this case was larger than after pale medial stripe injections (Fig. 3, Table 1). This may have been caused by differences in the efficacy of transport between the two types of tracers injected in pale lateral and pale medial stripes, namely CTB-alexas and CTBg, respectively; injection sizes were similar, and tracer transport times were identical for both groups of injections (Table 1), and thus could not account for the differences in the number of labeled cells observed after pale lateral and pale medial stripe injections. We wondered whether a threshold effect in the detectability of tracers could have caused a lower percentage of cells in 4B to be labeled in cases that had an overall smaller number of labeled cells, compared with cases with larger numbers of labeled cells. Results from other cases listed in Table 1 ruled out this interpretation. Specifically, in case 319, despite the much lower number of labeled cells produced by an FR injection in a pale lateral stripe, 26 of 110 cells, i.e., 23% of cells, were located in layer 4B, versus 84 (77%) cells in layers 2/3. Moreover, injections of CTB-alexas in pale medial stripes of two additional cases (304-488m and 326-647l), which produced larger numbers of labeled cells than the CTBg injections in case 305, still produced 99% of labeled cells in layer 2/3 and only 1% in 4B. These results indicate that the proportionally more numerous projection from layer 4B to the pale lateral stripes compared with the pale medial stripes was not related to the overall number of labeled cells nor to the type of tracer injected. The data in Table 1 also suggest that the pale medial stripes receive an overall weaker projection from V1 compared with pale lateral stripes. This is because injections in pale medial stripes produced less than half the number of labeled cells produced by similar size injections of the same tracer in pale lateral stripes (e.g., compare case 304-488m with case 305-488l). Similarly, in case 326-647l, an injection of CTB647 in a pale medial stripe produced approximately half the number of V1-labeled cells produced by two smaller injections of the same tracer in pale lateral stripes (cases 305-647m and 305-647l). In fact, the CTB647 injection sites in pale lateral stripes were approximately half the size of the CTB647 injection in the pale medial stripe.
Figure 4 shows the percentage of labeled cells in layers 2/3 versus 4B for each injection case, and the population averages, grouped by stripe type. Across the population, tracer injections in pale lateral stripes labeled 84 ± 4.5% of cells in layers 2/3 and 16 ± 4.5% in layer 4B, while tracer injections in pale medial stripes labeled 98 ± 0.9% of cells in layers 2/3 and 1.7 ± 0.9% in 4B, and this difference was statistically significant (p = 0.014; Mann–Whitney U test). These results demonstrate that the two pale stripes differ with respect to the contribution they receive from V1 layer 4B, with pale lateral stripes receiving a significant contribution and pale medial stripes receiving a small or negligible contribution from this layer.
Figure 4.
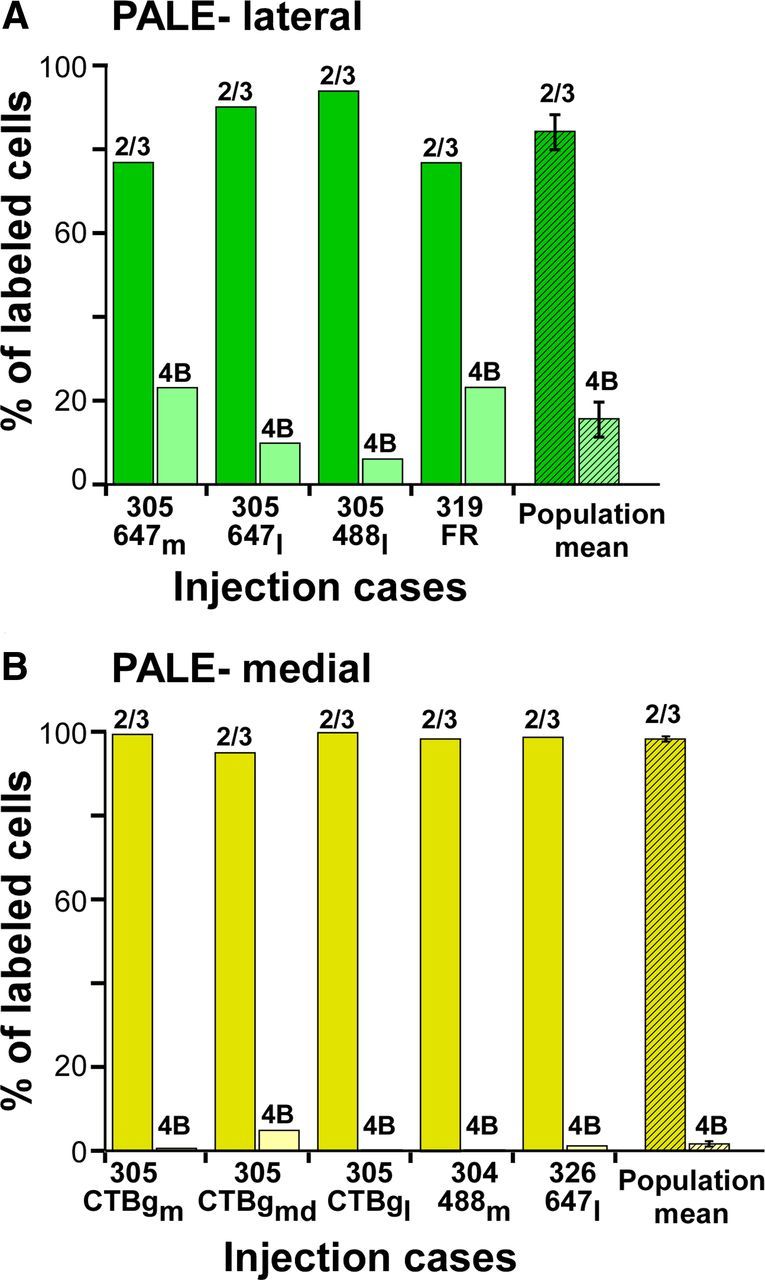
Relative contribution of layers 2/3 and 4B to pale lateral and pale medial stripes: population data. A, B, Percentage of labeled cells in V1 layers 2/3 versus 4B for each individual case (solid bars), and population means (hatched bars), resulting from injections in pale lateral (A) and pale medial (B) stripes. Error bars, SEM. Means and SEM are reported in Table 1.
Paired tracer injections in pale lateral and pale medial stripes: double-labeling study
The results above indicate differences in the layer 4B projections to the two pale stripes. However, both pale stripes receive a predominant projection from layer 2/3 cells in the interblob regions of V1. To determine whether projections from V1 layers 2/3 to the each pale stripe type arise from the same or distinct neuronal populations, we performed a double-labeling study. In two animals, paired injections of two distinguishable tracers were made in two consecutive pale stripes, one in a pale lateral stripe and one in a pale medial stripe (n = 3 injection pairs; Table 2). To ensure that the paired tracer injections encompassed several orientation-preference domains in V2, we used for this analysis slightly larger injection sizes (all but 1 measured 0.53–0.73 mm in diameter; Table 2; these measurements are not corrected for tissue shrinkage). Due to their larger size, two of these injections (CTBgmd and CTB488l) slightly straddled an adjacent thick stripe (Fig. 5). Because previous studies in both marmoset (Federer et al., 2009) and macaque (Sincich et al., 2010) found few double-labeled cells (up to 7.6% in marmoset and 13% in macaque of total labeled cells) after paired tracer injections in adjacent pale and thick stripes, we were not concerned about analyzing injections that partly straddled a thick stripe, as long as they involved a significant portion of a pale stripe.
Figure 5.
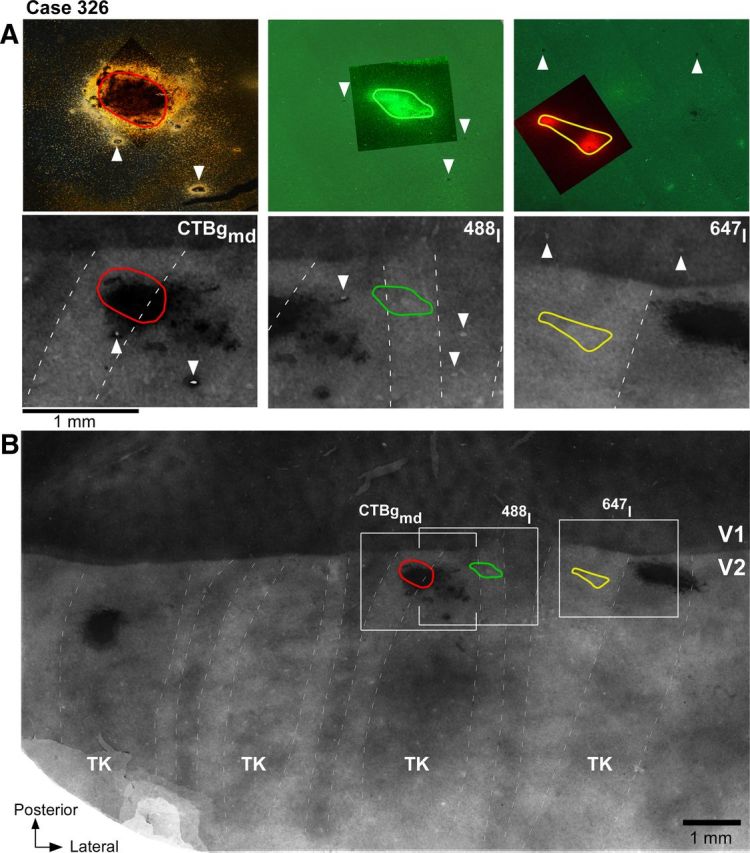
Paired tracer injections in consecutive pale stripe types (case 326). A, Top, High-power images of the three tracer injection sites within the field of views boxed in B. The CTB488 and CTB647 injection sites are viewed under fluorescent microscopy, while the CTBg injection is viewed under dark field. The images show the full extent of the injections through the cortical layers and were generated by merging images of sections at two cortical depths (from the most superficial and deepest sections that contained the injection site). The CTB647 injection appears as two separate injection sites, because we only show the uppermost and lowermost sections containing the injection, but the sections in between them also contained an injection site. Bottom, Each panel shows a higher-power view of CO staining within its respective boxed region in B, with superimposed the outline of the tracer injection from the top panel. B, Low-power and wide field of view of a composite CO image of unfolded and flattened V1 and V2 obtained by aligning and merging images of five tissue sections stained for CO. Other conventions are as in Figure 2.
Figure 5 shows one example case in which three injections landed in three consecutive pale stripes: a CTBg injection in a pale medial stripe, a CTB488 injection in a pale lateral stripe, and a CTB647 injection in a pale medial stripe. The most medial injection and the middle injection were located on each side of a thick stripe, while the middle and the most lateral injections were located on each side of a thin stripe. In one series of sections from this case, CTB488 was revealed using immunohistochemistry; the same sections were then silver reacted to reveal CTBg. An alternate series of sections from the same case was used to analyze fluorescent CTB488 and CTB647 signals. Thus, the same CTB488 injection was paired with both the CTBg and the CTB647 injections for double-labeling analysis. The CTBg–CTB488 injection pair produced fields of labeled cells that overlapped substantially (89% of the smallest field area; Table 2), indicating that the injections involved largely overlapping retinotopic regions. In addition, within the overlap area, labeled cells from each tracer injection were predominantly located in the interblobs (77% of the CTBg-labeled cells and 74% of the CTB488-labeled cells). Examples of single-labeled and double-labeled cells, and the distribution of label from each tracer within the overlap area for this case are shown in Figure 6A–E. Within the region of overlap, the CTBg injection labeled 1215 cells in layers 2/3, and the CTB488 injection labeled 4195 layer 2/3 cells in the same series of sections. Of these, only 51 cells were double labeled, i.e., 4% of the CTBg population and 1.2% of the CTB488 population, or 0.93% of the total number of labeled neurons [calculated as follows: N double-labeled cells * 100/(N cells labeled only with tracer 1 + N cells labeled only with tracer 2 + N double-labeled cells)]. The CTB647–CTB488 injection pair in case 326 also produced largely overlapped (by 85%) fields of labeled cells (79% of CTB488-labeled and 69% of the CTB647-labeled cells were located within interblobs), with 1376 CTB647-labeled and 1055 CTB488-labeled layer 2/3 cells in the overlap region. Of these, only 32 cells were double labeled, i.e., 2.3% of the CTB647 population and 2.9% of the CTB488 population, or 1.3% of the total number of labeled neurons. Examples of single-labeled and double-labeled cells, and the distribution of label from each tracer within the overlap area for this case are shown in Figure 6F–J. One additional injection pair made in a different animal (Table 2, case 325) produced 3% of double-labeled cells of the total number of labeled neurons (5.9% of the CTB488 population and 6.8% of the CTBg neuronal population). These results indicate that the great majority of layer 2/3 neurons in the V1 interblob regions project to either one or the other pale stripe type, and only rarely to both. Thus, there are largely segregated populations of output neurons from V1 to the two V2 pale stripes.
Discussion
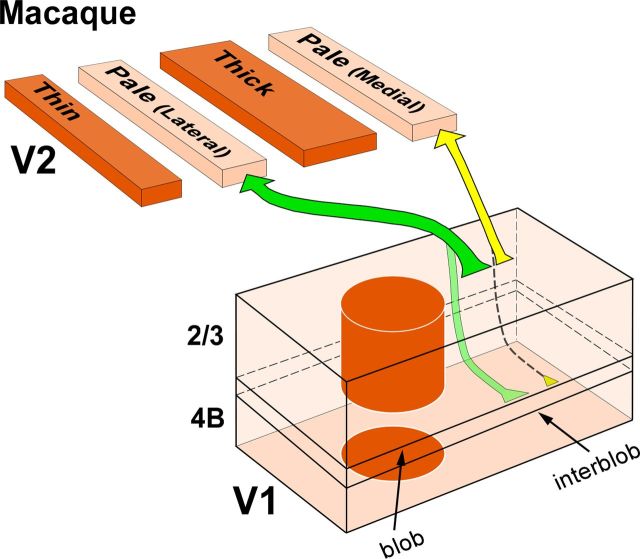
We have found that in macaque V2, the two pale stripes within a CO stripe cycle differ with respect to the projections they receive from layer 4B of V1. Pale lateral stripes receive a significant projection from layer 4B, while pale medial stripes receive few or no layer 4B projections. Furthermore, although the major projection to both pale stripe types arises from layer 2/3 in the interblob regions of V1, most individual layer 2/3 cells project only to one, but not the other, pale stripe type. These results suggest that in macaque the two pale stripes are anatomically distinct compartments, and support the existence of two distinct projection streams from V1 to the two pale stripes of V2 (Fig. 7).
Figure 7.
Two projection streams from V1 to V2 pale medial and pale lateral stripes in macaque. Our proposed model of the pathways from layers 2/3 and 4B of V1 to the two V2 pale stripe types based on the present study. Arrow thickness indicates the relative density of projections. Projections from layers 4A and 5/6 are omitted. Pale lateral stripes receive dense projections from layers 2/3 (dark green arrow), less dense projections from layers 4A (data not shown) and 4B (light green arrow), and sparse projections from layers 5/6 (data not shown). Pale medial stripes receive dense projections from layers 2/3 (thick yellow arrow), less dense projections from layer 4A (data not shown), sparse or no projections from layer 4B (dashed arrow), and no projections from layers 5/6. Pale medial stripes also receive a less robust projection than pale lateral stripes from V1 layers 2/3 (yellow arrow thinner than dark green arrow).
Previous schemes of the projections from V1 to the V2 CO stripes in macaque viewed the two pale stripes within a CO stripe cycle as a single compartment. In the original tripartite model proposed by Livingstone and Hubel (1984, 1987), pale stripes received projections exclusively from layers 2/3 in the interblobs. Instead, in the more recent bipartite scheme proposed by Sincich and Horton (2002a), pale stripes receive projections primarily from layers 2/3, but also from layers 4A, 4B, and 5/6 within an interblob column. Neither study distinguished between the two pale stripes. Here, we have found that both laminar patterns of V1 projections to pale stripes exist, but each is related to a different pale stripe type. Specifically, pale medial stripes receive projections almost exclusively from layers 2/3 (and 4A), while pale lateral stripes receive projections predominantly from layers 2/3 but also from layers 4A, 4B, and 5/6. These results resemble those of Federer et al. (2009) in marmoset visual cortex, where projections from V1 to the two pale stripes were also found to differ with respect to the layer 4B contribution they receive from V1 (Fig. 1B). However, in marmoset the projection from layer 4B of V1 to the pale medial stripes is virtually absent (representing on average 0.08% of V1 projections to pale medial stripes), while in macaque it is very sparse (on average 1.7% of V1 projections to pale medial stripes), with most pale medial stripe injections in macaque producing at least a few labeled cells in layer 4B. By comparison, pale lateral stripes in marmoset receive on average 9% of their V1 projections from layer 4B, a result that is in good agreement with our present results in macaque (mean, 16%). A recent study in macaque failed to find differences in the laminar origin of V1 projections to the two pale stripe types (Sincich et al., 2010). After injections in either pale stripe type, these authors reported that 10% of labeled cells resided in layer 4B and 87% in layers 2/3. While our population data for the pale lateral stripes is in good agreement with these authors' results (we find 16% of cells in layer 4B, vs 84% in 2/3), our results for the pale medial stripes are not consistent with those from Sincich et al. (2010). Because the validity of these experiments depends upon clearly confining tracer injections to a single stripe type, the discrepancy between our results and those of Sincich et al. (2010) may be attributable to the different size of tracer injections achieved in the two studies. Sincich et al. (2010) injected ∼140 nl volume of CTBg per injection, while we injected volumes of 90 nl for CTBg and 45–75 nl for CTB-alexas. Accordingly, in the study by Sincich et al. (2010), the tracer injection sites in pale stripes appear larger (0.6–1 mm in diameter; actual measurements of injection sizes were not reported in their study) than those in our study (0.2–0.67 mm diameter), in most cases spanning the full width of a pale stripe (Figs. 6, 9 in Sincich et al., 2010), or being displaced toward one edge of a pale stripe and spilling into an adjacent thick or thin stripe (supplementary Figs. 7, 10, 12 in Sincich et al., 2010). While an injection site spanning the full width of a stripe in principle can be desirable, in fact such large injections are more likely to involve adjacent thick or thin stripes, both of which receive projections from layer 4B (Sincich et al., 2007, 2010; Federer et al., 2009). Thus, in Sincich et al. (2010), the layer 4B labeled cells observed after pale medial stripe injections may have resulted from the large injections slightly straddling the border of an adjacent thin or thick stripe.
It has been previously shown that pale stripes in macaque receive more numerous projections from V1 compared with thick and thin stripes (Sincich and Horton, 2002b). Our results extend this finding by suggesting that the pale lateral stripes receive more robust projections from V1 than the pale medial stripes. This is because similarly sized injections of the same tracer in different pale stripe types produced significantly smaller numbers of labeled layer 2/3 neurons when located in pale medial stripes than when located in pale lateral stripes.
While the results above are strongly suggestive of two distinct pathways from V1 interblobs to V2 pale stripes, additional evidence supporting this conclusion comes from our double-labeling study, in which different tracer injections were made in nearby pale lateral and pale medial stripes. Of all labeled neurons, only 1–3% of cells were double labeled, suggesting that projections to each pale stripe type are separate, although they may arise from the same interblob regions of V1. An alternative interpretation for lack of double-labeled cells is that, although the two tracer injections were located into retinotopically matched V2 regions, they involved different functional domains. Given that a full cycle of orientations in V2 extends on average 950 μm (Vanduffel et al., 2002) and five of the six injections measured 530–730 μm, it is unlikely that these injection pairs were confined to different orientation columns in V2, as each injection would have involved at least half of an orientation hypercolumn. However, although unlikely, we cannot entirely exclude the possibility that the injections were confined to different kinds of functional domains having greater periodicity than orientation preference (Lu et al., 2010).
Segregated pathways to pale lateral and pale medial stripes suggest distinct function. Previous functional studies have indeed suggested that the two pale stripes may be functionally distinct. Optical imaging in owl monkey V2 revealed maps of high-orientation selectivity in pale medial stripes, and low-orientation selectivity in pale lateral stripes (Xu et al., 2004), a result recently confirmed in macaque monkey (Lim et al., 2009). Roe and Ts'o (1995), investigating the representation of visual space within V2 stripes, found evidence suggestive of two types of pale stripes within each stripe cycle of macaque V2, one with receptive field scatter similar to that of thin stripes, the other similar to thick stripes. Similarly, Shipp and Zeki (2002) in macaque found a tendency for pale medial stripes to have more “thin-like” receptive field properties than pale lateral stripes.
Together these functional and anatomical studies all suggest that a V2 stripe cycle contains four distinct compartments: two CO-dark and two CO-pale stripes, each with distinct function and sets of connections. Most previous studies of the response properties, functional architecture, and anatomical connectivity of the V2 stripes did not distinguish between the two pale stripes within a stripe cycle. Therefore, further studies are needed to investigate functional and anatomical differences between these two stripe types. For example, pale stripes have been previously shown to project to ventral stream area V4 (DeYoe and Van Essen, 1985; Shipp and Zeki, 1985), but whether both types of pale stripes do so remains to be demonstrated.
The laminar differences in the projections from V1 to the two pale stripes likely reflect different combinations of magno, parvo, and konio lateral geniculate inputs to the two pale stripes. In particular, the dominant layer 2/3 interblob projection, and the lack or paucity of layer 4B input to pale medial stripes suggest a dominant parvo channel between V1 and the V2 pale medial stripes (Yabuta and Callaway, 1998). Instead, the robust layer 4B contribution to pale lateral stripes may carry magno inputs to these pale stripe types. If both pale stripes indeed project to V4, then pale medial stripes would contribute predominantly parvo inputs, while pale lateral stripes would carry mixed parvo and magno inputs to V4. This magno input may be one source of the direction-selective responses found in many V4 cells (Desimone and Schein, 1987; Ferrera et al., 1994; Tolias et al., 2005) that may be used in V4 to infer object shape from motion contrast (Mysore et al., 2008; Roe et al., 2012). Since V4 contains spatially segregated maps for color, orientation (Tanigawa et al., 2010), and possibly motion direction (Li et al., 2011), an interesting question for future studies is how each pale stripe type contributes inputs to these different featural maps of V4.
Footnotes
This work was supported by National Science Foundation Grant IOS-0848106, National Institutes of Health Grant EY019743, and a grant from Research to Prevent Blindness to the Department of Ophthalmology, University of Utah. We thank Kesi Sainsbury for excellent technical assistance.
References
- Anderson JR, Jones BW, Yang JH, Shaw MV, Watt CB, Koshevoy P, Spaltenstein J, Jurrus E, UV K, Whitaker RT, Mastronarde D, Tasdizen T, Marc RE. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7:493–512. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelucci A, Clasca F, Sur M. Anterograde axonal tracing with the subunit B of cholera toxin: a highly sensitive immunohistochemical protocol for revealing fine axonal morphology in adult and neonatal brains. J Neurosci Methods. 1996;65:101–112. doi: 10.1016/0165-0270(95)00155-7. [DOI] [PubMed] [Google Scholar]
- Brandt HM, Apkarian AV. Biotin-dextran: a sensitive anterograde tracer for neuroanatomic studies in rat and monkey. J Neurosci Methods. 1992;45:35–40. doi: 10.1016/0165-0270(92)90041-B. [DOI] [PubMed] [Google Scholar]
- Desimone R, Schein SJ. Visual properties of neurons in area V4 of the macaque: sensitivity to stimulus form. J Neurophysiol. 1987;57:835–868. doi: 10.1152/jn.1987.57.3.835. [DOI] [PubMed] [Google Scholar]
- DeYoe EA, Van Essen DC. Segregation of efferent connections and receptive field properties in visual area V2 of the macaque. Nature. 1985;317:58–61. doi: 10.1038/317058a0. [DOI] [PubMed] [Google Scholar]
- Ericson H, Blomqvist A. Tracing of neuronal connections with cholera toxin subunit B: light and electron microscopic immunohistochemistry using monoclonal antibodies. J Neurosci Methods. 1988;24:225–235. doi: 10.1016/0165-0270(88)90167-7. [DOI] [PubMed] [Google Scholar]
- Federer F, Ichida JM, Jeffs J, Schiessl I, McLoughlin N, Angelucci A. Four projections streams from primate V1 to the cytochrome oxidase stripes of V2. J Neurosci. 2009;29:15455–15471. doi: 10.1523/JNEUROSCI.1648-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrera VP, Rudolph KK, Maunsell JH. Responses of neurons in the parietal and temporal visual pathways during a motion task. J Neurosci. 1994;14:6171–6186. doi: 10.1523/JNEUROSCI.14-10-06171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JC. Cytochrome oxidase patches: a new cytoarchitectonic feature of monkey visual cortex. Philos Trans R Soc Lond B. 1984;304:199–253. doi: 10.1098/rstb.1984.0021. [DOI] [PubMed] [Google Scholar]
- Jeffs J, Ichida JM, Federer F, Angelucci A. Anatomical evidence for classical and extra-classical receptive field completion across the discontinuous horizontal meridian representation of primate area V2. Cereb Cortex. 2009;19:963–981. doi: 10.1093/cercor/bhn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy H, Bullier J. A double-labeling investigation of the afferent connectivity to cortical area V1 and V2 of the macaque monkey. J Neurosci. 1985;5:2815–2830. doi: 10.1523/JNEUROSCI.05-10-02815.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy H, Dehay C, Bullier J. Organization of the callosal connections of visual areas V1 and V2 in the macaque monkey. J Comp Neurol. 1986;247:398–415. doi: 10.1002/cne.902470309. [DOI] [PubMed] [Google Scholar]
- Li P, Chen M, Han C, Zhu S, Xu H, Roe AW, Lu HD. A motion direction map in macaque V4. Soc Neurosci Abstr. 2011;37:484.03. [Google Scholar]
- Lim H, Xiao Y, Wang Y, Felleman DJ. How many V2 stripes are selective for orientation? Soc Neurosci Abstr. 2009;35:850.8. [Google Scholar]
- Livingstone MS, Hubel DH. Anatomy and physiology of a color system in the primate visual cortex. J Neurosci. 1984;4:309–356. doi: 10.1523/JNEUROSCI.04-01-00309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Hubel DH. Connections between layer 4B of area 17 and the thick cytochrome oxidase stripes of area 18 in the squirrel monkey. J Neurosci. 1987;7:3371–3377. doi: 10.1523/JNEUROSCI.07-11-03371.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn-Smith IJ, Minson JB, Wright AP, Hodgson AJ. Cholera toxin B-gold, a retrograde tracer that can be used in light and electron microscopic immunocytochemical studies. J Comp Neurol. 1990;294:179–191. doi: 10.1002/cne.902940203. [DOI] [PubMed] [Google Scholar]
- Lu HD, Chen G, Tanigawa H, Roe AW. A motion direction map in macaque V2. Neuron. 2010;68:1002–1013. doi: 10.1016/j.neuron.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppi PH, Fort P, Jouvet M. Iontophoretic application of unconjugated cholera toxin B subunit (CTb) combined with immunohistochemistry of neurochemical substances: a method for transmitter identification of retrogradely labeled neurons. Brain Res. 1990;534:209–224. doi: 10.1016/0006-8993(90)90131-T. [DOI] [PubMed] [Google Scholar]
- Mysore SG, Vogels R, Raiguel SE, Orban GA. Shape selectivity for camouflage-breaking dynamic stimuli in dorsal V4 neurons. Cereb Cortex. 2008;18:1429–4143. doi: 10.1093/cercor/bhm176. [DOI] [PubMed] [Google Scholar]
- Rockland KS. Laminar distribution of neurons projecting from area V1 to V2 in macaque and squirrel monkeys. Cereb Cortex. 1992;2:38–47. doi: 10.1093/cercor/2.1.38. [DOI] [PubMed] [Google Scholar]
- Roe AW, Ts'o DY. Visual topography in primate V2: multiple representation across functional stripes. J Neurosci. 1995;15:3689–3715. doi: 10.1523/JNEUROSCI.15-05-03689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe AW, Chelazzi L, Connor CE, Conway BR, Fujita I, Gallant JL, Lu H, Vanduffel W. Toward a unified theory of visual area V4. Neuron. 2012;74:12–29. doi: 10.1016/j.neuron.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp S, Zeki S. Segregation of pathways leading from area V2 to areas V4 and V5 of macaque monkey visual cortex. Nature. 1985;315:322–325. doi: 10.1038/315322a0. [DOI] [PubMed] [Google Scholar]
- Shipp S, Zeki S. The functional organization of area V2, I: specialization across stripes and layers. Vis Neurosci. 2002;19:187–210. doi: 10.1017/s0952523802191164. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Divided by cytochrome oxidase: a map of the projections from V1 to V2 in macaques. Science. 2002a;295:1734–1737. doi: 10.1126/science.1067902. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Horton JC. Pale cytochrome oxidase stripes in V2 receive the richest projection from macaque striate cortex. J Comp Neurol. 2002b;447:18–33. doi: 10.1002/cne.10174. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Jocson CM, Horton JC. Neurons in V1 patch columns project to V2 thin stripes. Cereb Cortex. 2007;17:935–941. doi: 10.1093/cercor/bhl004. [DOI] [PubMed] [Google Scholar]
- Sincich LC, Jocson CM, Horton JC. V1 interpatch projections to V2 thick stripes and pale stripes. J Neurosci. 2010;30:6963–6974. doi: 10.1523/JNEUROSCI.5506-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa H, Lu HD, Roe AW. Functional organization for color and orientation in macaque V4. Nat Neurosci. 2010;13:1542–1548. doi: 10.1038/nn.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias AS, Keliris GA, Smirnakis SM, Logothetis NK. Neurons in macaque area V4 acquire directional tuning after adaptation to motion stimuli. Nat Neurosci. 2005;8:591–593. doi: 10.1038/nn1446. [DOI] [PubMed] [Google Scholar]
- Tootell RB, Silverman MS, De Valois RL, Jacobs GH. Functional organization of the second cortical visual area in primates. Science. 1983;220:737–739. doi: 10.1126/science.6301017. [DOI] [PubMed] [Google Scholar]
- Ungerleider LG, Mishkin M. Two cortical visual systems. In: Ingle DJ, Mansfield JW, Goodale MA, editors. Advances in the analysis of visual behavior. Cambridge: MIT; 1982. pp. 549–586. [Google Scholar]
- Van Essen DC, Newsome WT, Bixby JL. The pattern of interhemispheric connections and its relationship to extrastriate visual areas in the macaque monkey. J Neurosci. 1982;2:265–283. doi: 10.1523/JNEUROSCI.02-03-00265.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Newsome WT, Maunsell JH, Bixby JL. The projections from striate cortex (V1) to areas V2 and V3 in the macaque monkey: asymmetries, areal boundaries, and patchy connections. J Comp Neurol. 1986;244:451–480. doi: 10.1002/cne.902440405. [DOI] [PubMed] [Google Scholar]
- Vanduffel W, Tootell RB, Schoups AA, Orban GA. The organization of orientation selectivity throughout macaque visual cortex. Cereb Cortex. 2002;12:647–662. doi: 10.1093/cercor/12.6.647. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Carroll EW. Quantitative light and electron micosciopic analysis of cytochrome oxidase-rich zones in V II prestriate cortex of the squirrel monkey. J Comp Neurol. 1984;222:18–37. doi: 10.1002/cne.902220103. [DOI] [PubMed] [Google Scholar]
- Xu X, Bosking W, Sáry G, Stefansic J, Shima D, Casagrande V. Functional organization of visual cortex in the owl monkey. J Neurosci. 2004;24:6237–6247. doi: 10.1523/JNEUROSCI.1144-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta NH, Callaway EM. Functional streams and local connections of layer 4C neurons in primary visual cortex of the macaque monkey. J Neurosci. 1998;18:9489–9499. doi: 10.1523/JNEUROSCI.18-22-09489.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeki SM. Interhemispheric connections of prestriate cortex in monkey. Brain Res. 1970;19:63–75. doi: 10.1016/0006-8993(70)90237-4. [DOI] [PubMed] [Google Scholar]