Background: Based on their sequences, the closely related yeast Hpa2 and Hpa3 proteins appear to be acetyltransferases.
Results: Hpa2 and Hpa3 acetylate histones, polyamines, and some small basic proteins.
Conclusion: Although Hpa2 and Hpa3 are very similar in sequence, they have somewhat different substrate preferences.
Significance: This is the first biochemical characterization of these two acetyltransferases.
Keywords: Histone Acetylase, Histone Modification, Mass Spectrometry (MS), Polyamines, Translation, Acetyltransferase
Abstract
Based on their sequences, the Saccharomyces cerevisiae Hpa2 and Hpa3 proteins are annotated as two closely related members of the Gcn5 acetyltransferase family. Here, we describe the biochemical characterization of Hpa2 and Hpa3 as bona fide acetyltransferases with different substrate specificities. Mutational and MALDI-TOF analyses showed that Hpa3 translation initiates primarily from Met-19 rather than the annotated start site, Met-1, with a minor product starting at Met-27. When expressed in Escherichia coli and assayed in vitro, Hpa2 and Hpa3 (from Met-19) acetylated histones and polyamines. Whereas Hpa2 acetylated histones H3 and H4 (at H3 Lys-14, H4 Lys-5, and H4 Lys-12), Hpa3 acetylated only histone H4 (at Lys-8). Additionally, Hpa2, but not Hpa3, acetylated certain small basic proteins. Hpa3, but not Hpa2, has been reported to acetylate d-amino acids, and we present results consistent with that. Overexpression of Hpa2 or Hpa3 is toxic to yeast cells. However, their deletions do not show any standard phenotypic defects. These results suggest that Hpa2 and Hpa3 are similar but distinct acetyltransferases that might have overlapping roles with other known acetyltransferases in vivo in acetylating histones and other small proteins.
Introduction
Acetyltransferases catalyze the transfer of an acetyl group from acetyl-CoA to an acceptor residue on proteins or small molecules and have wide-ranging roles in gene regulation, antibiotic inactivation, transcriptional silencing, and cell cycle progression (1, 2). The best characterized enzymes in this group, the histone acetyltransferases (HATs),3 acetylate the ϵ-amino group of lysines on histone N-terminal tails and core regions and bring about changes in chromatin structure and dynamics, often resulting in transcriptional activation (3).
Many well known HATs, such as Gcn5, Hat1, and PCAF (p300/CBP-associated factor), belong to the GNAT (Gcn5-related N-acetyltransferases) family with clear sequence and structural similarity, but they acetylate different residues on different histones (3). These HATs are often present in multisubunit complexes in vivo, such as Gcn5 in SAGA and ADA complexes, Hat1 in the HAT-B complex, etc. In yeast, the SAGA complex has well defined roles in transcriptional activation at promoters and also has roles in transcription elongation and mRNA export (4). In the context of the SAGA complex, Gcn5 is able to acetylate histones and nucleosomes primarily on H3 Lys-14. Hat1 in the HAT-B complex acetylates free histone H4 at Lys-5 and Lys-12 and is thought to be the primary acetyltransferase for newly synthesized histones (5). NuA4 and NuA3 are two other yeast HAT complexes. They contain the MYST family acetyltransferases Esa1 and Sas3 as the catalytic subunits, acetylating histones H4 and H3, respectively (6, 7). The multiplicity of HATs and their overlapping specificities indicate built-in redundancy to maintain proper in vivo function.
Hpa2 and Hpa3 are two closely related members of the GNAT superfamily, with nearly 50% identity and 68% similarity over 156 amino acids (3). Both proteins share significant sequence and structural similarity in their catalytic and acetyl-CoA-binding regions (motifs A and D) with other members of the GNAT superfamily (8). The Hpa2 crystal structure shows that motif C is structurally conserved within the superfamily even though there is very limited sequence conservation. Sedimentation analysis shows that Hpa2 forms a dimer in solution and can tetramerize in the presence of acetyl-CoA (8). The crystal structure of the oligomer reveals that two Hpa2 dimers are held together by interaction between the bound acetyl-CoA molecules (8).
Despite their sequence homology, Hpa3, but not Hpa2, has been shown to acetylate a wide variety of d-amino acids in vitro (9). This activity is required in vivo to prevent toxicity when d-amino acids are taken up by the cell (10). Uptake of exogenous d-amino acids through the Gap1 permease is inhibited by the presence of l-amino acids and rich nitrogen sources such as ammonium ions (11). However, in limiting nitrogen medium, even a low concentration of d-amino acids is toxic to cells presumably by being misincorporated into proteins. Acetylation of d-amino acids by Hpa3 combined with the action of d-tyrosyl-tRNATyr deacylase to recycle mislabeled d-aminoacyl-tRNAs allows the cells to export the d-amino acids out of the cell and overcome their toxicity (10, 12).
Our goal in this study was to understand the substrate specificity of Hpa2 and Hpa3. Our earlier attempts to understand the HAT activity profile of these proteins led us to hypothesize that only Hpa2 can acetylate histones (8). However, as we show in this study, the natural form of Hpa3 can acetylate histones but with a different substrate specificity than Hpa2. Our biochemical analysis also shows that Hpa2 and Hpa3 are unique acetyltransferases, with a wide range of substrates in vitro, including histones, polyamines, and some non-histone chromatin proteins.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The following strains were used in this study: W303-1a (MATa ade2 ura3 leu2 his3 trp1 can1), VSY48 (MATα ura3 can1, a segregant from a cross of W303-1a with AN1 MATα lys1), VSY49 (MATα can1, a prototroph generated by integrating pRS306 at the ura3 locus of VSY48), VSY50 (MATα ura3 can1 hpa3Δ::URA3), VSY51 (MATα ura3 can1 hpa2Δ::URA3), and VSY52 (MATα ura3 can1 hpa3Δ::kanMX4). Yeast cells were grown at 30 °C in yeast/peptone/dextrose or synthetic complete medium plus appropriate supplements or in minimal synthetic defined medium for prototrophs (13). Standard yeast manipulations were performed as described (14).
Plasmids
pVS7 was generated by cloning the HPA3 upstream region (nucleotides −420 to 1) and ORF in frame with a C-terminal GST tag in pRS426. pMS04 was generated by replacing the GST tag with the His6-HA-protease 3C-protein A (ProtA) ZZ domain in pVS7. pVS46 (M1R), pVS47 (M19R), pVS48 (M27R), and pVS49 (M19R/M27R) were generated by site-directed mutagenesis of the HPA3 ORF in pMS04.
To overexpress Hpa2 and Hpa3 in yeast, the HPA2 and HPA3 ORFs were cloned into the p423 TEF expression vector (15) at BamHI and XhoI sites, generating pLBS68 and pLBS71, respectively. pLBS71 expresses Hpa3 amino acids (aa) 19–179. pSTT103 and pJL1 express LexA fusions to Hpa2 and Hpa3(19–179) that were generated by inserting the ORFs as EcoRI-SalI fragments into the two-hybrid vector pSTT91 (16). HPA2 and HPA3 (expressing aa 19–179) ORFs were cloned as EcoRI-SalI fragments in frame with the GAL4 activation domain in pGAD424 (17) to generate pSTT105 and pJL2, respectively.
Plasmids for expressing Hpa2 and Hpa3 proteins in Escherichia coli were constructed as follows. pSTT97 was constructed by inserting the HPA2 ORF into the pET28c expression vector as a BamHI-XhoI fragment. pSTT104 was made by cloning the HPA3 ORF (aa 1–179) into the BamHI-XhoI sites of pET28c. pLBS61 was made by insertion of the HPA3 ORF (aa 19–179) into the NdeI and XhoI sites of pET28a. Plasmids pLBS53, pLBS51, pLBS55, and pLBS57 were constructed by inserting the ORFs of NHP6a, NHP6b, HMO1, and ABF2 into the NcoI and XhoI sites of pET28b. The HMO2 ORF was inserted into the NdeI and XhoI sites of pET28a to form plasmid pLBS56.
Protein Expression and Purification in E. coli
Hpa2 and Hpa3 proteins were expressed in E. coli BL21(DE3) cells after induction with 1 mm isopropyl β-d-thiogalactopyranoside at room temperature for 5 h. The protein was purified using His·Bind affinity resin (Novagen) according to the manufacturer's instructions. Purified Hpa2 protein was dialyzed against 50 mm Tris-HCl (pH 8.0) and frozen at −80 °C by adding glycerol to a final concentration of 10%. Purified Hpa3 protein was directly frozen at −80 °C by adding glycerol to a final concentration of 10% without being dialyzed. Nhp6a, Nhp6b, Hmo1, and Abf2 were also cloned into pET28 and expressed and purified as described above.
Hpa3 Purification from Yeast
VSY48 expressing pMS04 was grown overnight at 30 °C in 1 liter of synthetic defined medium to A600 = 2. His-HA-ProtA-tagged Hpa3 was purified according to established protocols (18). Briefly, the cell pellet was lysed by bead beating, and ProtA-tagged Hpa3 was recovered by overnight incubation with IgG-Sepharose beads, followed by washing three times with 25 mm Tris-HCl (pH 8) and 0.1% Nonidet P-40 and once with 50 mm Tris-HCl (pH 7), 150 mm NaCl, 1 mm EDTA, and 1 mm DTT. His-HA-tagged Hpa3 was cleaved from the IgG-Sepharose beads by incubation with GST-protease 3C (GE Healthcare), followed by incubation with glutathione-Sepharose beads for 2 h. The supernatant containing lysed His-HA-Hpa3 was further purified through a nickel-nitrilotriacetic acid column and quantitated on a 15% SDS-polyacrylamide gel with BSA protein standards.
Mass Spectrometry Characterization
Hpa3 was analyzed on a Voyager-DE STR MALDI-TOF mass spectrometer system (Applied Biosystems) operated in the linear mode. Samples were dissolved in a 50% solution of acetonitrile and 0.1% TFA containing sinapinic acid (10 mg/ml) and then dried on the sample plate. The accelerating voltage was set to 25 kV with a delay of 300 ns. The mass scale (m/z 1000–25,000) was calibrated with myoglobin (1 pm/μl), and ∼200 laser shots were used to produce each spectrum.
SDS-PAGE and Western Blot Analysis
For the mutational analysis of Hpa3 start sites, the strains were grown to A600 = 1, lysed by bead beating according to standard protocols, and electrophoresed on a 20-cm 15% SDS-polyacrylamide gel in a Sturdier apparatus (Hoefer Inc.) at 250 V for 3–4 h. The gel was transferred onto a PVDF membrane using the Bio-Rad wet transfer system. The membrane was divided at the 50-kDa mark. The lower part was probed with a mouse anti-HA monoclonal antibody, and the upper part with a rat anti-tubulin antibody.
HAT Assays
HAT assays were carried out in a 25-μl total reaction volume with 37.5 mm Tris-HCl (pH 7.9), 1 mm DTT, 0.1 mm EDTA, 0.25 μCi of [3H]acetyl-CoA (5.60 Ci/mmol; catalog no. TRK688, Amersham Biosciences), 0.5–1 μg of recombinant Hpa2 or Hpa3, and 10 μg of chicken histones purified from erythrocytes or ∼1 μg of recombinant non-histone proteins as substrates. Reactions were incubated at 30 °C for 30 min and then spotted onto Whatman P-81 cation exchange paper and dried in a hot air oven. The dried papers were washed three times for 5 min each with 50 mm sodium carbonate (pH 9.0) and once with acetone. Radioactivity retained on the paper was quantified in a liquid scintillation counter.
Polyamine Acetyltransferase Assays
Assays were carried out in a 50-μl total reaction volume containing 3 mm polyamines (putrescine, spermidine, or spermine; Sigma), 100 mm Tris-HCl (pH 7.9), 0.5 μCi of [3H]acetyl-CoA (5.60 Ci/mmol), and 0.5–1 μg of recombinant Hpa2 or Hpa3. Reactions were incubated at 30 °C for 30 min, and 10 μl of 1 m hydroxylamine hydrochloride was then added to terminate the reaction. The reactions were boiled for 3 min, and the precipitate was removed by centrifugation at 12,000 × g for 2 min. 40 μl of supernatant was spotted onto Whatman P-81 paper and dried. The dried papers were washed once with tap water, three times with distilled water for 2 min each, and finally once with 100% ethanol to remove unbound [3H]acetyl-CoA. The filters were dried in a hot air oven, and radioactivity retained on the filters was quantified in a liquid scintillation counter.
SDS-PAGE of Acetylated Histones
The HAT reactions were performed as described above, and instead of spotting the reactions on P-81 paper, the reactions were terminated by the addition of 5 μl of 5× SDS loading buffer. Samples were heated to 100 °C for 5 min and resolved on a 15% SDS-polyacrylamide gel. The gel was stained with Coomassie Blue, destained, saturated with EN3HANCE (PerkinElmer Life Sciences), dried under vacuum, and subjected to autoradiography.
Amino Acid Sequence Analysis of Acetylated Histones
HAT assays were done as described above using the recombinant Hpa2 and Hpa3 proteins and either the H3 or H4 peptide as substrate. Four reactions were pooled, applied to ProSorb membranes (Applied Biosystems), and processed for protein sequencing according to the manufacturer's instructions. The samples were subjected to Edman degradation. Eluates from each cycle were collected, dried, resuspended in 50 mm sodium acetate (pH 5.2), and counted in a liquid scintillation counter.
Two-hybrid Analysis
The LexA-Hpa2 and LexA-Hpa3 fusions were tested for two-hybrid interactions with Gal4 activation domain-Hpa2 or Gal4 activation domain-Hpa3 in strain L40 as described previously (19).
RESULTS
Hpa2, but Not Hpa3(1–179), Acetylates Histones
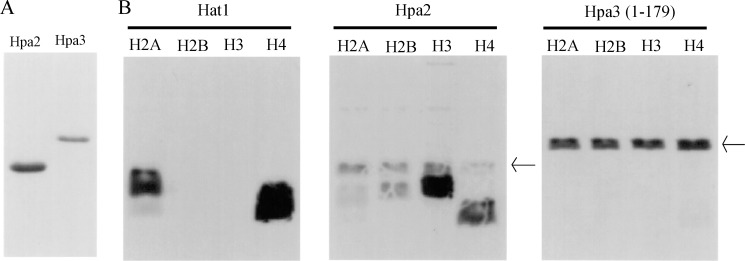
To analyze the biochemical properties of Hpa2 and Hpa3, we purified the full-length proteins as C-terminally His-tagged fusions from E. coli (Fig. 1A) and assayed them for HAT activity in vitro using Hat1 as a positive control. As shown in Fig. 1B, Hpa2 showed strong activity with purified histone H3 and somewhat less with histone H4. In contrast, full-length Hpa3 showed no acetylation activity against histones but had significant autoacetylation activity.
FIGURE 1.
A, full-length Hpa2 and Hpa3, as annotated in the Saccharomyces Genome Database, were expressed and purified from E. coli BL21(DE3) cells and subjected to SDS-PAGE. The Coomassie Blue-stained gel is shown. B, Hpa2, but not Hpa3, can acetylate histones H3 and H4. Recombinant Hpa2, Hpa3, and Hat1 were assayed for HAT activity against recombinant purified individual histones in the presence of [3H]acetyl-CoA, followed by SDS-PAGE and autoradiography. The arrows show the positions of autoacetylated Hpa2 and Hpa3.
Hpa3 Translation Initiates from Met-19 in Saccharomyces cerevisiae
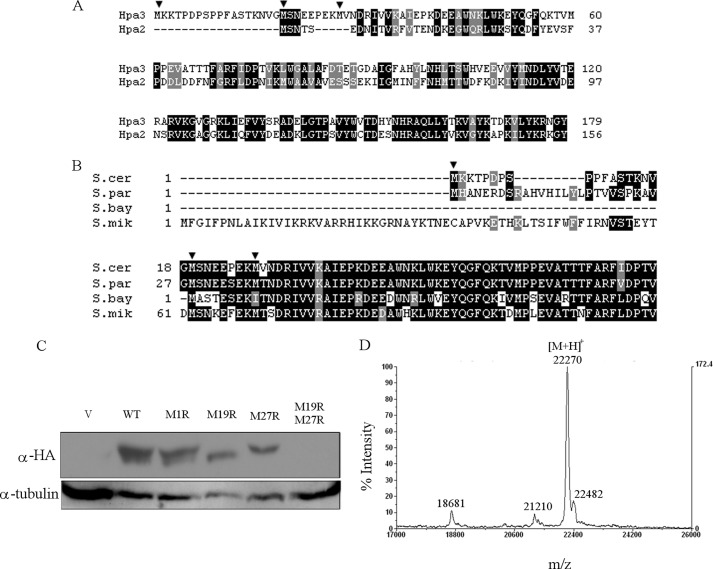
The disparity in the acetylation abilities of Hpa2 and Hpa3, despite their 50% sequence identity, led us to reexamine the sequence alignment between Hpa2 and Hpa3. Hpa3 protein has three in-frame methionines within the first 27 aa (Met-1, Met-19, and Met-27 of the ORF). It was apparent that Hpa2 aligns with Hpa3 starting at Met-19 (Fig. 2A). Significantly, Hpa3 protein alignment begins at Met-19 in four closely related Saccharomyces species (Fig. 2B). This led us to hypothesize that Met-19 is the in vivo translational start site for Hpa3 in S. cerevisiae.
FIGURE 2.
Hpa3 is produced from the second methionine in the annotated sequence. A, sequence alignment of Hpa2 and Hpa3 showing their similarity. Identical residues are shown in black boxes, and similar residues are shown in gray boxes. The three in-frame methionines of Hpa3 at positions 1, 19, and 27 are indicated with arrowheads. B, sequence alignment of Hpa3 from S. cerevisiae (S.cer), Saccharomyces paradoxus (S.par), Saccharomyces bayanus (S.bay), and Saccharomyces mikatae (S.mik) shows that the proteins have no homology before Met-19 of S. cerevisiae Hpa3. Identical and similar residues are shaded as described for A. C, Hpa3 is expressed primarily from the second methionine, Met-19. Strain W303-1a carrying the vector pRS426 (V), wild-type pMS04 (His-HA-ProtA-tagged WT Hpa3), or mutant pVS46–49 (M1R, M19R, M27R, or M19R/M27R) was assayed for Hpa3 protein levels using anti-HA antibody. Anti-tubulin antibodies were used to measure tubulin levels as a loading control. D, MALDI-TOF analysis confirmed that Hpa3 in yeast is produced from the second methionine in vivo. Wild-type Hpa3 purified from strain VSY48 containing pMS04 was subjected to MALDI-TOF analysis. Shown is the intensity profile against mass/charge ratio. The major and minor peaks are shown as protonated molecular ions.
We used mutational analysis and performed MALDI-TOF measurements on the purified protein to confirm this hypothesis. The ORF of HPA3 was expressed from its natural promoter and cloned in frame with a His6-HA-ProtA tag in a 2μ plasmid to facilitate the monitoring and purification of the Hpa3 protein. Site-directed mutagenesis in this plasmid background was used to generate mutants of all three methionines either singly or in combination (M1R, M19R, M27R, and M19R/M27R). The wild-type and mutant plasmids were used to express the proteins in yeast. Protein extracts were separated on a 20-cm 15% SDS-polyacrylamide gel and analyzed by Western blotting with anti-HA antibody. As shown in Fig. 2C, the first annotated start site, Met-1, was not used, as mutating it did not affect Hpa3 production. Both the wild-type protein and the M1R mutant produced a doublet that differed in size by ∼0.8 kDa. The top band appears to be due to Met-19 because it is missing in the M19R mutant. The bottom band appears to be due to Met-27 because it is missing in the M27R mutant (Fig. 2C). Consistent with this interpretation, the M19R/M27R double mutant did not produce any Hpa3.
To confirm the start of the Hpa3 protein from Met-19, the His-HA-ProtA-tagged wild-type Hpa3 protein was purified from yeast and subjected to MALDI-TOF analysis. Taking into account the appropriate modifications such as methionine cleavage and Nα-acetylation, the predicted molecular masses of proteins expressed from this plasmid starting at Met-1, Met-19, and Met-27 are 24,232.54, 22,259.16, and 21,272.15 Da, respectively. In the MALDI-TOF analysis, the predominant peak seen with a mass of 22,270 Da corresponds closely to the expected molecular mass of Hpa3 beginning at Met-19 (Fig. 2D). The minor peaks could represent alternative forms of Hpa3, but because they were not reproducible in two different preparations of Hpa3, they are unlikely to be significant products.
Taken together, the results from the MALDI-TOF and mutational analyses strongly suggest that Met-19 is the preferred in vivo translational start site for Hpa3. Henceforth, protein 19–179 is referred to as Hpa3.
Hpa2 and Hpa3 Are Similar but Distinct HATs
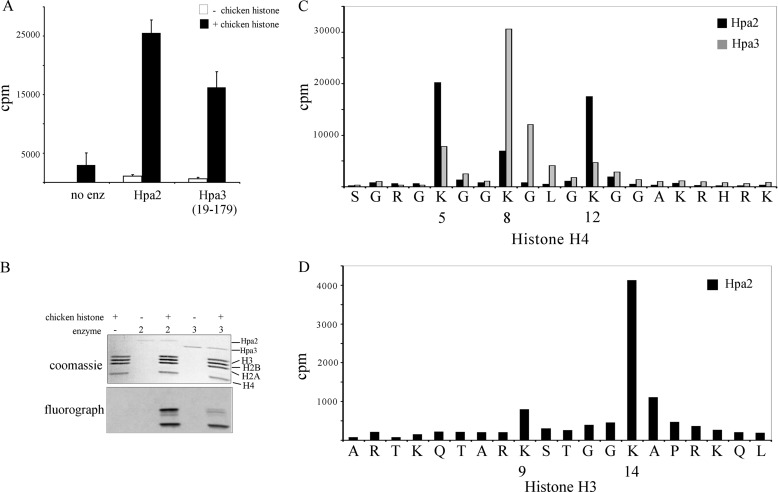
The newly identified form of Hpa3 (aa 19–179) was utilized in histone acetylation assays against chicken histones. As shown in Fig. 3A, unlike Hpa3(1–179), this form of Hpa3 possesses strong histone acetylation activity like Hpa2. Both Hpa2 and Hpa3 could not acetylate nucleosomes in vitro (data not shown).
FIGURE 3.
Hpa2 and Hpa3 have HAT activity. A, recombinant Hpa2 and Hpa3(19–179) show HAT activity against chicken histones in the presence of [3H]acetyl-CoA. The results shown are the means ± S.D. of three independent experiments. B, Hpa2 can acetylate histones H3 and H4, whereas Hpa3 can acetylate only histone H4. Reaction products from HAT assays performed as described for A were electrophoresed on two 15% SDS-polyacrylamide gels and subject to autoradiography and Coomassie Blue staining. The results from a representative experiment are shown. C, Hpa2 and Hpa3 have different HAT profiles. HAT assays were performed with histone H4 peptide 1–20 in the presence of Hpa2 or Hpa3, and the reaction products were subjected to sequencing by Edman degradation, followed by scintillation counting to identify residues that carry the [3H]acetyl group. D, same as in C except histone H3 peptide 1–20 was used as a substrate.
Hpa2 and Hpa3 have different histone acetylation profiles. Hpa3 acetylated only H4, whereas Hpa2 acetylated both H3 and H4 (Figs. 1B and 3B). Sequence analysis of the acetylated histones offered further evidence that Hpa2 and Hpa3 have different substrate specificities. Hpa2 preferred to acetylate H4 Lys-5 and Lys-12 over Lys-8 (Fig. 3C). In contrast, Hpa3 preferentially acetylated H4 Lys-8 over H4 Lys-5 and Lys-12. Hpa2 also acetylated H3 Lys-14 (Fig. 3D), whereas Hpa3 had little or no activity against H3 (Fig. 3B).
Hpa2 and Hpa3 Possess Polyamine Acetyltransferase Activity
Polyamines such as putrescine, spermine, and spermidine are small cationic molecules that bind to DNA and chromatin to perform essential but as yet unknown functions in cells (20). We have previously identified a member of the GNAT superfamily, Paa1, as a polyamine acetyltransferase (21). Paa1 acetylates the free amino groups on many polyamines but does not acetylate histones (21). However, some mammalian acetyltransferases have both histone and polyamine acetyltransferase activities (22).
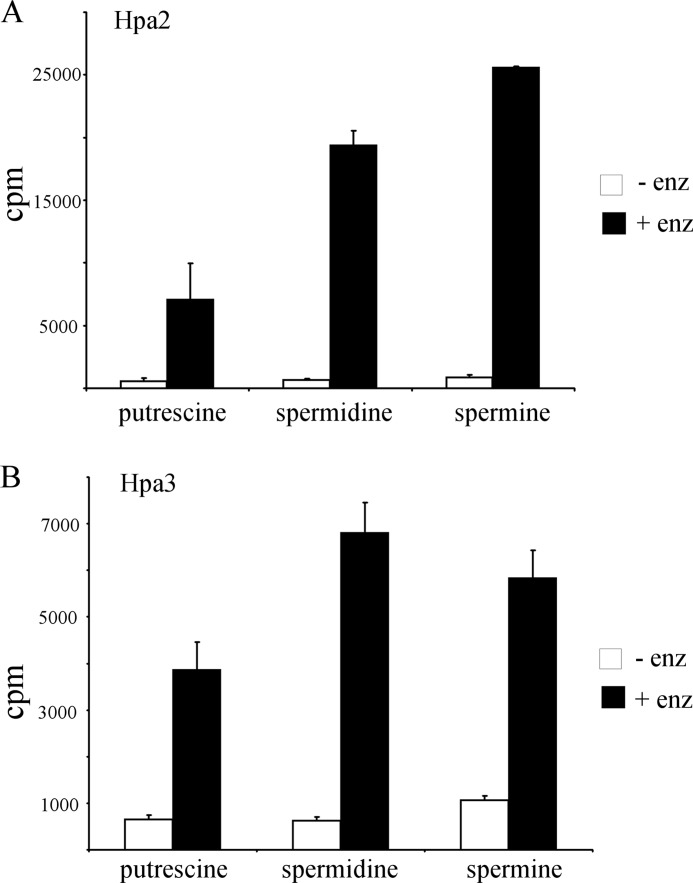
We tested whether Hpa2 and Hpa3 are able to acetylate polyamines by subjecting putrescine, spermidine, and spermine to acetyltransferase assays. As shown in Fig. 4 (A and B), both Hpa2 and Hpa3 were able to acetylate all three polyamines, with a preference for spermine and spermidine over putrescine.
FIGURE 4.
Hpa2 and Hpa3 can acetylate polyamines. Putrescine, spermine, and spermidine were subjected to acetylation in the presence or absence of Hpa2 (A) and Hpa3 (B). The results shown are the means ± S.D. of three independent experiments. enz, enzyme.
Hpa2, but Not Hpa3, Can Acetylate Some Small Basic Proteins
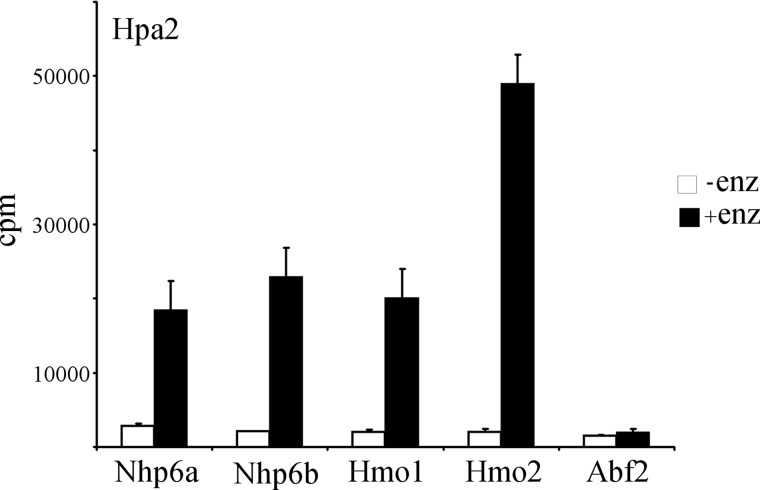
Many members of the HMG family are known to be acetylated by p300/CBP, Gcn5, and Hat1 HATs (3). For instance, the S. cerevisiae Spt2 protein, an HMG1 homolog, is acetylated by Gcn5 (23). We tested the ability of Hpa2 and Hpa3 to acetylate the following yeast HMG-like proteins: Nhp6a, Nhp6b, Hmo1, Hmo2, and Abf2. As shown in Fig. 5, Hpa2 was able to acetylate all of the nuclear HMG proteins tested, with a preference for Hmo2. However, Abf2, a mitochondrial member of the HMG family, was not acetylated. Hpa3 did not acetylate any of the HMG proteins tested (data not shown).
FIGURE 5.
Hpa2 can acetylate nuclear non-histone chromatin proteins. Hpa2 was assayed for its ability to acetylate nuclear (Nhp6a, Nhp6b, Hmo1, and Hmo2) and mitochondrial (Abf2) members of the HMG family in the presence of [3H]acetyl-CoA. The results shown are the means ± S.D. of three independent experiments. enz, enzyme.
Hpa2 and Hpa3 Self-associate but Do Not Interact with Each Other
Sedimentation and crystal structure analyses of Hpa2 clearly show that Hpa2 is dimeric in solution and tetramerizes in the unit crystal (8). To assess the self- and heterodimerization abilities of Hpa2 and Hpa3, we carried out two-hybrid analysis of Hpa2 and Hpa3 as fusions to LexA and Gal4 activation domains. Both Hpa2 and Hpa3 proteins showed significant self-association but did not heterodimerize with each other in vivo (Table 1).
TABLE 1.
Two-hybrid analysis of Hpa2-Hpa3 interaction
| Two-hybrid interaction | GAD-Hpa2 | GAD-Hpa3 |
|---|---|---|
| LexA-Hpa2 | ++++ | − |
| LexA-Hpa3 | − | ++++ |
Hpa3 and d-Amino Acid Detoxification
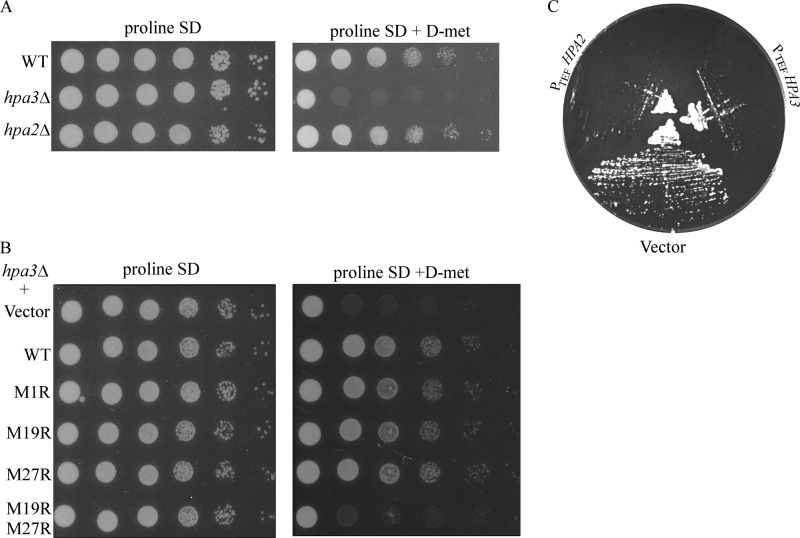
Hpa3, but not Hpa2, has been shown to be important for d-amino acid detoxification through its ability to acetylate d-amino acids (10). As d-amino acid uptake is inhibited by the presence of l-amino acids in regular media, we generated prototrophic hpa3Δ and hpa2Δ strains and assessed them for their ability to grow in synthetic medium in the presence of 0.1 mm d-methionine, as described under “Experimental Procedures.” We confirmed that hpa3Δ mutants were unable to grow in the presence of d-methionine. In contrast, the growth of hpa2Δ mutants was not affected by d-methionine (Fig. 6A). The hpa3Δ growth defect could be rescued by all of the plasmid-borne single Hpa3 missense mutants (M1R, M19R, and M27R) (Fig. 6B), suggesting that the Hpa3 protein produced from either Met-19 or Met-27 is capable of acetylating d-amino acids. As expected, the Hpa3 M19R/M27R mutant, which did not produce any Hpa3 protein, was not able to rescue the d-methionine sensitivity of the hpa3Δ strain.
FIGURE 6.
A, Hpa3, but not Hpa2, is required to overcome d-amino acid toxicity. Prototrophic strains VSY49 (WT), VSY50 (hpa3Δ), and VSY51 (hpa2Δ) were assayed for their ability to grow in the presence of 0.1 mm d-methionine in synthetic defined medium (SD) containing proline as the major nitrogen source. The plates were incubated at 30 °C for 2 days and photographed. B, strain VSY52 (hpa3Δ) was transformed with plasmid pRS426 (vector), pMS04 (WT Hpa3), pVS46 (Hpa3 M1R), pVS47 (M19R), pVS48 (M27R), or pVS49 (M19R/M27R) and assayed for d-amino acid toxicity as described for A. The plates were incubated at 30 °C for 2 days and photographed. C, overexpression of Hpa2 and Hpa3 is toxic to yeast. Strain W303-1a containing the overexpression plasmid pLBS68 (PTEF HPA2) or pLBS71 (PTEF HPA3) or pRS423 TEF as the vector control was assayed for growth on His− synthetic complete medium following incubation at 30 °C for 2 days.
Lack of Hpa2 and Hpa3 Does Not Have Any Phenotypic Consequence under Normal Conditions
To understand other in vivo functions of Hpa2 and Hpa3, we tested hpa2Δ, hpa3Δ, and hpa2Δhpa3Δ mutants for phenotypic defects. We did not observe any defects in growth at 30 or 37 °C, sporulation, growth in the presence of genotoxic agents such as hydroxyurea or methyl methanesulfonate, recovery from stationary phase, or transcriptional silencing (data not shown). The only growth defect noted for hpa2Δ (but not hpa3Δ) mutants was minor 6-azauracil sensitivity (data not shown).
We also tested whether overexpression of Hpa2 and Hpa3 has any phenotypic consequences. When overexpressed from the strong constitutive PTEF promoter, HPA2 and HPA3 ORFs were toxic to the growth of yeast cells (Fig. 6C).
DISCUSSION
Hpa2 and Hpa3 are members of the GNAT family of acetyltransferases, a group of proteins with a wide variety of substrates ranging from small molecules such as antibiotics and amino acids to proteins (3). Although Hpa3 has been shown to be a d-amino acid acetyltransferase that detoxifies d-amino acids, the exact in vivo role of Hpa2 is not yet clear (10). In this study, we have shown that both Hpa2 and Hpa3 can acetylate histones and other proteins with distinct substrate specificities.
Histone Acetylation by Hpa2 and Hpa3
Acetylation of histone N-terminal tails is a well known modulator of chromatin structure and gene regulation. Hpa2 preferred to acetylate H3 Lys-14 and H4 Lys-5 and Lys-12 (Fig. 3, B and C). These sites are also the preferred targets of Gcn5 and the Hat1-containing HAT-B complex, respectively (3, 5). In contrast, Hpa3 preferred to acetylate H4 Lys-8 (Fig. 3, B and C), which is also targeted by the Esa1-containing NuA4 complex in vivo (6). The redundant nature of these acetyltransferases could explain the lack of phenotypes seen for hpa2Δ or hpa3Δ mutants. Because hat1 mutants do not have obvious growth defects (24), we checked whether hpa2Δhat1Δ, hpa3Δhat1Δ, or hpa2Δhpa3Δhat1Δ had growth defects and found that they did not (data not shown). These results suggest that Hpa2 and Hpa3 do not acetylate H4 Lys-5 and Lys-12 in vivo or that there are other overlapping HATs that can compensate for these deletions. Consistent with these possibilities, histones purified from hpa2Δ or hpa3Δ mutants did not show any changes in their acetylation status when electrophoresed on Triton/acid/urea gels (data not shown). Additionally, overexpression of Hpa2 and Hpa3 was toxic to yeast cells (Fig. 6C), suggesting that hyperacetylation of their target protein(s) is detrimental to growth. It would be interesting to know whether the weak 6-azauracil sensitivity seen for hpa2Δ mutants is indicative of transcription defects in vivo.
Other Substrates for Hpa2 and Hpa3
It is possible that histones are not the primary in vivo targets of Hpa2 and Hpa3. These proteins are unusual in that they can also acetylate polyamines; none of the well characterized yeast acetyltransferases have been shown to have both histone and polyamine acetyltransferase activities. Both Hpa2 and Hpa3 were able to acetylate the polyamines spermine, spermidine, and putrescine in vitro (Fig. 4), similar to Paa1, the primary polyamine acetyltransferase in yeast (21). Acetylation of polyamines is thought to be necessary for their breakdown or export from cells (25). Unlike Paa1, overexpression of Hpa2 or Hpa3 did not cause additional growth defects on media lacking pantothenate (data not shown). Moreover, the hpa2Δhpa3Δpaa1Δ mutant was no more sensitive to hydroxyurea than the paa1Δ mutant (data not shown), suggesting that polyamine acetylation might not be the primary role for Hpa2 and Hpa3 in vivo. Many years ago, Libby (22) isolated calf liver N-acetyltransferases, which have molecular masses similar to those of Hpa2 and Hpa3 and are capable of acetylating both histones and polyamines; they might be mammalian homologs of Hpa2 and Hpa3.
Prior experiments and recent whole genome acetylation profiles have revealed a wide variety of non-histone proteins that are acetylated at internal lysine residues, including non-histone chromatin proteins such as the HMG family and transcriptional activators such as p53 (3, 26, 27). In keeping with these findings, we found that Hpa2, but not Hpa3, was able to acetylate many small non-histone proteins belonging to the HMG family, with a special preference for Hmo2. It is interesting that Hpa2 activity on these small basic proteins does not correlate with their pI or the percentage of lysine residues, as Hmo2 was the least basic of the four nuclear proteins tested (pI of 8.06 and 12% Lys versus an average pI of 9.7 and 14% Lys).
Hpa2 and Hpa3 Have Different Substrate Specificities
Although Hpa2 and Hpa3 are very similar proteins, with 50% identity and 68% similarity, they have different substrate specificities. Hpa3, but not Hpa2, can detoxify d-amino acids, whereas Hpa2 alone acetylates small non-histone chromatin proteins (Figs. 5 and 6A). This difference in substrate specificity is presumably due to amino acids not conserved between the two proteins.
As mentioned above, Hpa2 forms a dimer in solution and can tetramerize in the presence of the cofactor acetyl-CoA. The crystal structure of Hpa2 shows that an extensive dimer interface is formed in large part by projections of strand β3-turn β3/β4-strand β4 and strand β7 into the adjacent monomer (8). A comparison of the primary sequences showed that only 7 of the 15 residues in the first projection (and only two of the nine residues in turn β3/β4) are identical between Hpa2 and Hpa3. This difference in the dimer interface could explain why Hpa2 and Hpa3 do not interact with each other while maintaining the ability to self-interact (Table 1).
Is d-Amino Acid Acetylation the Primary Role for Hpa3?
As mentioned above, Hpa3 has a clearly defined role in detoxifying d-amino acids by acetylation (10). As d-amino acids are taken into yeast cells only under low nitrogen conditions, it was intriguing to note that under normal high nitrogen conditions, the Hpa3 protein is present at ∼1000 copies/cell (28). The toxicity associated with overexpression of HPA3 cannot be explained by hyperacetylation of d-amino acids, as these experiments were carried out under high nitrogen conditions (Fig. 6C). The toxicity could be due to inappropriate acetylation of histones or polyamines, or perhaps other proteins.
Hpa3 Translation Initiates Downstream of the Annotated Start Site
Our experiments clearly show that the Hpa3 protein is produced from Met-19, a methionine downstream of the annotated start site, Met-1 (Fig. 2, C and D), in agreement with the primary sequence homology between Hpa3 and its orthologs (Fig. 2, A and B). Transcript analysis also suggests that Hpa3 must be translated from the second or third methionine (Met-19 and Met-27). The three in-frame methionines are at chromosome V coordinates 26667, 26721, and 26745. Based on the presence of an intact G-cap, Hpa3 mRNA transcripts begin at coordinates 26679 and 26717 (29). Therefore, both transcripts begin downstream of the first methionine, Met-1, precluding translation from that methionine.
It was interesting to note that recombinant Hpa3 protein produced from the first methionine did not have HAT activity, whereas the correct version did (Figs. 1B and 3A), suggesting that the extra N-terminal residues in the recombinant protein interfere with either substrate binding or the catalytic activity on histones. However, these extra N-terminal residues do not affect detoxification and hence acetylation of d-amino acids (9), presumably because the small molecule substrate can bind to the active site, whereas larger substrates such as histones cannot.
This work was supported by National Institutes of Health Grant R01 GM55641 (to R. S.).
- HAT
- histone acetyltransferase
- ProtA
- protein A
- aa
- amino acid(s).
REFERENCES
- 1. Carrozza M. J., Utley R. T., Workman J. L., Côté J. (2003) The diverse functions of histone acetyltransferase complexes. Trends Genet. 19, 321–329 [DOI] [PubMed] [Google Scholar]
- 2. Davies J. (1994) Inactivation of antibiotics and the dissemination of resistance genes. Science 264, 375–382 [DOI] [PubMed] [Google Scholar]
- 3. Sterner D. E., Berger S. L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baker S. P., Grant P. A. (2007) The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26, 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parthun M. R. (2007) Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328 [DOI] [PubMed] [Google Scholar]
- 6. Doyon Y., Côté J. (2004) The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14, 147–154 [DOI] [PubMed] [Google Scholar]
- 7. John S., Howe L., Tafrov S. T., Grant P. A., Sternglanz R., Workman J. L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14, 1196–1208 [PMC free article] [PubMed] [Google Scholar]
- 8. Angus-Hill M. L., Dutnall R. N., Tafrov S. T., Sternglanz R., Ramakrishnan V. (1999) Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol. 294, 1311–1325 [DOI] [PubMed] [Google Scholar]
- 9. Yow G. Y., Uo T., Yoshimura T., Esaki N. (2004) D-Amino acid N-acetyltransferase of Saccharomyces cerevisiae: a close homologue of histone acetyltransferase Hpa2p acting exclusively on free d-amino acids. Arch. Microbiol. 182, 396–403 [DOI] [PubMed] [Google Scholar]
- 10. Yow G. Y., Uo T., Yoshimura T., Esaki N. (2006) Physiological role of d-amino acid-N-acetyltransferase of Saccharomyces cerevisiae: detoxification of d-amino acids. Arch. Microbiol. 185, 39–46 [DOI] [PubMed] [Google Scholar]
- 11. Rytka J. (1975) Positive selection of general amino acid permease mutants in Saccharomyces cerevisiae. J. Bacteriol. 121, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soutourina J., Blanquet S., Plateau P. (2000) d-tyrosyl-tRNATyr metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 275, 11626–11630 [DOI] [PubMed] [Google Scholar]
- 13. Ausubel F. M. (1988) Current Protocols in Molecular Biology, Wiley-Interscience, New York [Google Scholar]
- 14. Guthrie C., Fink G. R. (1991) Guide to Yeast Genetics and Molecular Biology, Academic Press, San Diego [Google Scholar]
- 15. Mumberg D., Müller R., Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 16. Sutton A., Heller R. C., Landry J., Choy J. S., Sirko A., Sternglanz R. (2001) A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21, 3514–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartel P. L., Chien C., Sternglanz R., Fields S. (1993) Using the two-hybrid system to detect protein-protein interactions. in Cellular Interactions in Development: A Practical Approach (Hartley D. A., ed) p. xviii, IRL Press, Oxford [Google Scholar]
- 18. Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., Wise K. J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., Dumont M. E., Phizicky E. M., Snyder M., Grayhack E. J. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chien C. T., Bartel P. L., Sternglanz R., Fields S. (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. U.S.A. 88, 9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tabor C. W., Tabor H. (1984) Polyamines. Annu. Rev. Biochem. 53, 749–790 [DOI] [PubMed] [Google Scholar]
- 21. Liu B., Sutton A., Sternglanz R. (2005) A yeast polyamine acetyltransferase. J. Biol. Chem. 280, 16659–16664 [DOI] [PubMed] [Google Scholar]
- 22. Libby P. R. (1978) Calf liver nuclear N-acetyltransferases. Purification and properties of two enzymes with both spermidine acetyltransferase and histone acetyltransferase activities. J. Biol. Chem. 253, 233–237 [PubMed] [Google Scholar]
- 23. Pollard K. J., Peterson C. L. (1997) Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell Biol. 17, 6212–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kleff S., Andrulis E. D., Anderson C. W., Sternglanz R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270, 24674–24677 [DOI] [PubMed] [Google Scholar]
- 25. Casero R. A., Jr., Pegg A. E. (1993) Spermidine/spermine N1-acetyltransferase–the turning point in polyamine metabolism. FASEB J. 7, 653–661 [PubMed] [Google Scholar]
- 26. Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., Hake S. B. (2009) Proteome-wide prediction of acetylation substrates. Proc. Natl. Acad. Sci. U.S.A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuan H., Marmorstein R. (2013) Histone acetyltransferases: rising ancient counterparts to protein kinases. Biopolymers 99, 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 29. Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T. (2006) A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. U.S.A. 103, 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]