Background: Many bacterial pathogens, such as Bacillus anthracis, increase cAMP in monocytes, leading to disruption of immune responses.
Results: In human monocytes, cAMP up-regulates transducin-like enhancer of split (TLE) and activates Notch signaling.
Conclusion: Our findings demonstrate novel signaling mechanisms used by cAMP to enhance Notch signaling.
Significance: This work delineates how cAMP modifies a signaling pathway critical to innate immune responses during infection.
Keywords: Bacterial Pathogenesis, Bacterial Toxins, Cyclic AMP (cAMP), Monocytes, Notch Pathway
Abstract
In cells of the innate immune system, pathological increases in intracellular cAMP attenuate immune responses and contribute to infections by bacteria such as Bacillus anthracis. In this work, cAMP from B. anthracis edema toxin (ET) is found to activate the Notch signaling pathway in both mouse macrophages and human monocytes. ET as well as a cell-permeable activator of PKA induce Notch target genes (HES1, HEY1, IL2RA, and IL7R) and are able to significantly enhance the induction of these Notch target genes by a Toll-like receptor ligand. Elevated cAMP also resulted in increased levels of Groucho/transducin-like enhancer of Split (TLE) and led to increased amounts of a transcriptional repressor complex consisting of TLE and the Notch target Hes1. To address the mechanism used by ET to activate Notch signaling, components of Notch signaling were examined, and results revealed that ET increased levels of recombinant recognition sequence binding protein at the Jκ site (RBP-J), a DNA binding protein and principal transcriptional regulator of Notch signaling. Overexpression studies indicated that RBP-J was sufficient to activate Notch signaling and potentiate LPS-induced Notch signaling. Further examination of the mechanism used by ET to activate Notch signaling revealed that C/EBP β, a transcription factor activated by cAMP, helped activate Notch signaling and up-regulated RBP-J. These studies demonstrate that cAMP activates Notch signaling and increases the expression of TLE, which could be an important mechanism utilized by cAMP to suppress immune responses.
Introduction
Many pathogenic bacteria (e.g. Bacillus anthracis, Mycobacterium tuberculosis, Bordetella pertussis) have evolved strategies that increase cAMP levels in host cells (1). When cAMP is increased in cells of the innate immune system, such as macrophages, the ability of these cells to respond to bacteria is dampened, and this creates conditions that are advantageous to the bacterial pathogen. An important example of a pathogen that utilizes such a system is B. anthracis, which increases cAMP within host cells through the activities of edema toxin (ET)2 (2). ET is a secreted binary toxin composed of edema factor (EF) and protective antigen (PA). PA mediates the entry of EF into the cytoplasm of host cells, where EF functions as a potent adenylate cyclase (2, 3). A recent study demonstrated that toxin-mediated inactivation of myeloid cells (neutrophils and macrophages) is essential for the onset of anthrax disease (4). Therefore, studying how ET alters signaling pathways in these types of cells is critical for understanding the early stages of this disease.
Although much is known about how B. anthracis as well as other pathogens elevate cAMP within host cells, much less is known about how cAMP modifies signaling programs within macrophages and, ultimately, attenuates immune responses. Many of the signaling activities of cAMP are initiated by the activation of PKA and the basic region leucine zipper transcription factors CREB and C/EBP β (5–7). These cAMP effectors are used ubiquitously by cells, but how these proteins feed into other signaling pathways and eventually affect macrophages phenotypes is not completely understood. In an effort to identify and understand signaling pathways impacted by cAMP in macrophages, we evaluated a panel of gene reporters in macrophages and found that the Notch signaling pathway was modified by ET. We were particularly interested in this change in Notch signaling in macrophages because ET was found recently to disrupt Notch signaling in Drosophila melanogaster and human endothelial cells through a mechanism involving inhibition of endocytic recycling by the Rab11-dependent exocyst system (8).
The Notch signaling pathway is used ubiquitously, with critical roles in processes such as embryogenesis and lymphopoiesis (9, 10). To activate Notch signaling, one of four Notch receptors (Notch 1–4) binds Notch ligands such as Jagged1, Jagged2, Delta-like ligand 1 (DLL1), DLL3, and DLL4. These interactions between receptors and ligands lead to the proteolytic cleavage of the Notch receptor and the release of the Notch intracellular domain. When released, the Notch intracellular domain localizes to the nucleus and binds to the DNA binding protein, the recombinant recognition sequence binding protein at the Jκ site (RBP-J). Binding between the Notch intracellular domain and RBP-J facilitates the formation of a transcriptional activation complex that promotes the expression of Notch target genes (11). Of the Notch target genes, the most important and best characterized are the families of basic helix-loop-helix proteins, hairy and enhancer of split (Hes) and hairy and enhancer of split with YRPW motif (Hey) (12). These DNA binding proteins function as transcriptional repressors and are the primary effectors of Notch signaling.
Recently, Notch signaling has emerged as a critical signaling pathway in macrophages (13–17). In these types of cells, Notch signaling combines with Toll-like receptor (TLR) signaling to induce Notch target genes such as Hes and Hey (13). A striking outcome of Notch signaling in macrophages is that Hes and Hey function as part of a feedback loop that attenuates the production of cytokines (13). In the following studies, cAMP from ET is found to induce Notch signaling and potentiate the induction of Notch target genes by TLR ligands. Further studies reveal mechanisms used by ET to activate Notch signaling and to enhance the formation of the Hes transcription repressor complex.
EXPERIMENTAL PROCEDURES
Recombinant Proteins and Other Reagents
PA and EF were purchased from List Biological Laboratories (Campbell, CA). The membrane-permeable activator of PKA, N6-monobutyryladenosine-3′,5′-cyclic monophosphate (6-MB-cAMP), was obtained from Biolog (Bremen, Germany). LPS (catalog no. L4391) was purchased from Sigma-Aldrich (St. Louis, MO).
Maintenance and Use of Cell Lines
Cell lines used in these studies were purchased from the ATCC. THP-1 cells were grown in RPMI 1640 medium (Invitrogen) with 10% fetal bovine serum (ATCC) supplemented with 50 μm 2-mercaptoethanol. RAW 264.7, HEK-293, and L-929 cells were grown in DMEM (ATCC) containing 10% fetal bovine serum (ATCC). L-929 cell were used to produce conditioned medium necessary for the growth of BMDMs. HEK-293 cells were used to produce retrovirus particles. All of these cell lines were used between passages 5 and 20.
Isolation and Culture of BMDM
C57BL/6J mice were purchased from The Jackson Laboratories (Bar Harbor, ME) and handled in accordance with campus IACUC guidelines. BMDMs were cultured from bone marrow isolated from mouse femurs. Isolated bone marrow was added to macrophage culture medium (DMEM with 10% FBS and 30% L-929 conditioned medium) and plated on bacterial-grade Petri dishes. Adherent bone marrow cells were discarded after 24 h, and non-adherent bone marrow cells were maintained in culture for 6 days until a monolayer of macrophages was observed. The resulting macrophages were then plated in macrophage culture medium at a density of 5 × 105 cells/well of a 6-well tissue culture-treated plate. After maintaining the culture overnight, the macrophages were subjected to the experimental conditions.
Isolation of Human Monocytes
In accordance with the guideline of the University of Oklahoma Health Sciences Center Institutional Review Board, buffy coats derived from human whole blood were obtained from the Oklahoma Blood Institute. Buffy coats were diluted in complete RPMI 1640 medium (10% FBS plus penicillin/streptomycin), and peripheral blood mononuclear cells were then isolated using density gradient centrifugation utilizing Histopaque 1077 (Sigma-Aldrich). Monocytes were then isolated from the peripheral blood mononuclear cells with a monocyte isolation kit purchased from Miltenyi Biotec (Auburn, CA). This kit utilizes a negative selection method in which all non-monocytes are labeled magnetically and separated from the monocytes. The isolated monocytes were diluted into complete RPMI 1640 medium at a concentration of 1.0 × 106 cells/ml and then subjected to the conditions detailed in the figure legends.
Reverse Transcriptase qPCR Analysis
Isolated RNA was converted to cDNA by performing reverse transcription reactions using SuperScript III (Invitrogen). The qPCR analysis was performed by combining the cDNA with a SYBR Green PCR master mix (SABiosciences) and gene-specific primers. An Applied Biosystems 7500 real-time PCR system was used to perform the amplification reactions, and relative changes in levels of the mRNA of the gene of interest were compared with the levels of Actb mRNA using the 2−ΔΔCt method.
Measurement of Notch Activity in RAW 264.7 Cells by Gene Reporter
To use a gene reporter to monitor Notch activity, RAW 264.7 cells were transduced with lentivirus particles (SABioscience) containing a luciferase gene linked to a minimal promoter plus RBP-J binding elements (RAW-Notch). A gene reporter that contained only the minimal promoter was also generated and used as a control (RAW-Neg). To transduce RAW 264.7 cells, lentiviral particles (multiplicity of infection of ∼10) were added to cells with 8 μg/ml Polybrene and then incubated for 24 h at 32 °C. After this incubation, stable cells were isolated by selection with 3.5 μg/ml of puromycin. These stable cells were then subjected to experimental conditions outlined in the figure legends. Luciferase expression was quantified using the luciferase assay system (Promega) followed by measurement of the luminescence signal with a Victor3 (PerkinElmer Life Sciences) plate reader. Luciferase expression in RAW-Neg was used to normalize for changes to the minimal promoter that are not dependent on the Notch signal. This procedure was also utilized to perform the screen presented in the supplemental data.
Immunoprecipitation of TLE
To immunoprecipitate TLE, THP-1 cells were lysed by incubating for 5 min in lysis buffer (20 mm HEPES (pH 7.9), 350 mm NaCl, 30 mm MgCl2, 10% glycerol, 0.5% Nonidet P-40, 200 μm DTT, and protease inhibitor mixture), passing the lysates through a 22-gauge needle 10 times, and then centrifuging the lysates at 18,000 × g for 5 min. From this resulting lysate, 500 μg of total protein was diluted into a binding buffer (20 mm HEPES (pH 7.9), 30 mm MgCl2, 10% glycerol, 0.2% Nonidet P-40, 200 μm DTT, and protease inhibitor mixture) to give a final volume of 300 μl. One microgram of pan-TLE antibodies (Santa Cruz Biotechnology, catalog no. sc-13373) or control IgG was added to each immunoprecipitation mixture. This mixture was incubated for 2 h at 4 °C. Then, protein G-conjugated magnetic beads were added, and the incubation was continued for an additional 30 min. After removing the input protein, the magnetic beads were washed three times in binding buffer, and the immunoprecipitated proteins were subsequently eluted. The resulting eluted proteins were then detected by immunoblot analysis.
Expression of RBP-J by Retrovirus Transduction
PCR was used to amplify Rbpj from mouse cDNA and to fuse a FLAG tag to its N terminus. This PCR product was cloned into the pENTR/D-TOPO vector (Invitrogen) and subsequently transferred into expression vectors via a recombination reaction using LR Clonase (Invitrogen). The retrovirus expression vectors used to express RBP-J in these experiments were pQCXP and pQCXIB (Addgene plasmids 17386 and 17487, originating from E. Campeau (18)).
To express RBP-J by retrovirus, the RBP-J expression plasmid along with murine leukemia virus gag/pol and vesicular stomatitis virus G expression plasmids were each cotransfected into HEK-293 cells using the calcium phosphate transfection method. Forty-eight hours after transfection, retroviral particles that were released into the medium were harvested and used to transduce RAW 264.7 cells. To infect the RAW 264.7 cells, the retrovirus particles plus 8 μg/ml Polybrene were added to cells plated at 5.0 × 104 cells/well in a 12-well plate. The plate containing the cell and virus mixture were then centrifuged for 2 h at 1200 × g at 25 °C and placed in the tissue culture incubator overnight. Virus was then removed and replaced with fresh medium containing 3.5 μg/ml of puromycin. The cells remained under puromycin selection for 3–4 days to kill non-transduced cells. The cells were then used for experiments or continued to be cultured in the presence of puromycin. For experiments using Notch reporter cells (RAW-Notch), blasticidin was used as the selection antibiotic.
Immunoblot Analysis
After subjecting cells to experimental conditions, total proteins were extracted by removing culture medium and then adding lysis buffer chilled to 4 °C containing 1% SDS, 50 mm Tris (pH 7.4), 5 mm EDTA, and a protease inhibitor mixture (Sigma, catalog no. P8340). The cells were incubated in this lysis buffer on ice for 15 min, passed through a 22-gauge needle 10 times, and finally centrifuged for 5 min at 20,000 × g. To perform an immunoblot analysis, these proteins extracts (10 μg/well) were combined with sample buffer (62.5 mm Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.001% bromphenol blue) and heated at 95 °C for 7 min. Proteins were then separated using 10–12% SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane by electroblotting. The membrane was blocked with 5% nonfat milk in a wash buffer consisting of 20 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 0.1% Tween 20. The membranes were then probed with primary antibodies that recognize the following proteins: Hes1 (Millipore, catalog no. ab15470), Jagged1 (Santa Cruz Biotechnology, catalog no. sc-6011), GAPDH (Abcam, catalog no. ab8245), GSK-3β (Cell Signaling Technology, catalog no. 9315), pan-TLE (Cell Signaling Technology, catalog no. 4681), C/EBP β (Santa Cruz Biotechnology, catalog no. sc-896), RBP-J (Cell Signaling Technology, catalog no. 5442), and RBP-J (Abcam, catalog no. ab25949). The membrane was then washed and incubated with a secondary antibody conjugated to horseradish peroxidase. After another round of washes, the blots were developed with an enhanced chemiluminescent protein development system (GE Healthcare) and exposed to film. Densitometry analysis of immunoblot bands was carried out on digitized images using ImageJ 1.37V software (Wayne Rasband, National Institutes of Health).
siRNA Transfection
RAW 264.7 cells were transfected with siRNA using the transfection reagent HiPerFect (Qiagen). To begin the transfection procedure, 3.0 × 105 cells were plated into a well of a 12-well plate in 400 μl of DMEM with 10% FBS. Immediately after plating the cells, the transfection mixture containing 12 μl of HiPerFect and 100 nm siRNA was prepared in 400 μl of Opti-MEM. Following a 10-min incubation, the transfection mixture was added to the cells and incubated for 6 h. After this incubation, the transfection mixture was removed and replaced with fresh DMEM containing 10% FBS. The siRNAs against C/EBP β (Santa Cruz Biotechnology, catalog no. sc-29862) contain three target-specific siRNAs. As a negative control, cells were transfected with a scrambled sequence not targeting any known gene (Santa Cruz Biotechnology, catalog no. sc-37007).
ELISA
Cell supernatants were collected from RAW 264.7 cells. IL-6 and IL-10 levels were measured using ELISA Ready-set-go! kits from eBioscience, Inc. (San Diego, CA). TNF-α levels were quantified using a Instant ELISA kit from eBioscience, Inc.
RESULTS
Increased cAMP in Macrophages/Monocytes Activates Notch-induced Gene Expression
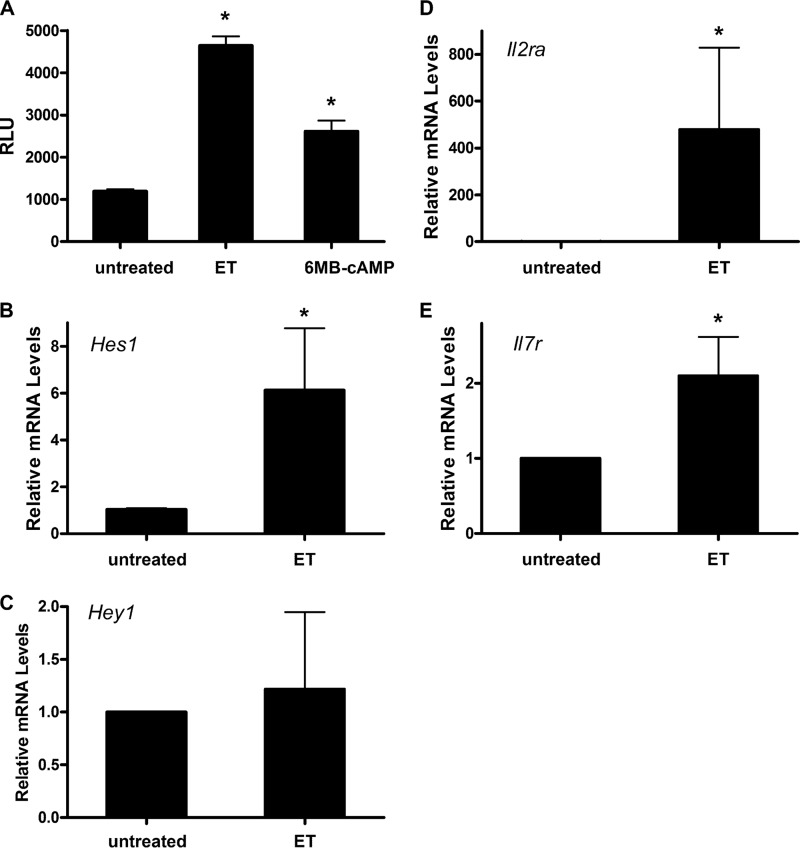
In an effort to identify and understand signaling pathways impacted by cAMP generated by ET, we screened a selection of RAW 264.7 cell lines stably transduced with luciferase-based gene reporters (supplemental Fig. S1). Out of this screen, the Notch pathway was found to be activated by exposures to either ET or 6-MB-cAMP, a membrane-permeable analog of cAMP (Fig. 1A). This cAMP analog is a specific agonist of PKA and demonstrates that activation of this pathway can be mediated through PKA. To support the Notch reporter data, mouse BMDMs were examined to determine whether ET exposures induce Notch target genes such as Hes1, Hey1, Il2ra, and Il7r. As shown by RT-qPCR, with the exception of Hey1, each of these target genes were up-regulated after exposure to ET (Fig. 1, B–E). Collectively, these results demonstrate that cAMP from ET activates Notch signaling in mouse macrophages.
FIGURE 1.
ET activates a Notch gene reporter and induces Notch target genes in mouse macrophages. A, as described under “Experimental Procedures,” RAW 264.7 cells containing a Notch luciferase gene reporter were created. Luciferase levels were then determined in the Notch reporter cell line (RAW-Notch) after a 7-h exposure to 10 nm ET (10 nm EF plus 10 nm PA) or 1 mm 6MB-cAMP. The values in the bar graph represent normalized average relative luciferase units (RLU) from three independent experiments. B–E, BMDMs were cultured as described under “Experimental Procedures.” These cells were exposed for 6 h to 10 nm ET (10 nm EF and 10 nm PA), and RT-qPCR was used to measure levels of Hes1, Hey1, Il2ra, and Il7r. Bar graphs are representative of three independent experiments. For the BMDMs, each experiment used a different culture generated from a different mouse. Error bars indicate mean ± S.D. *, p < 0.05.
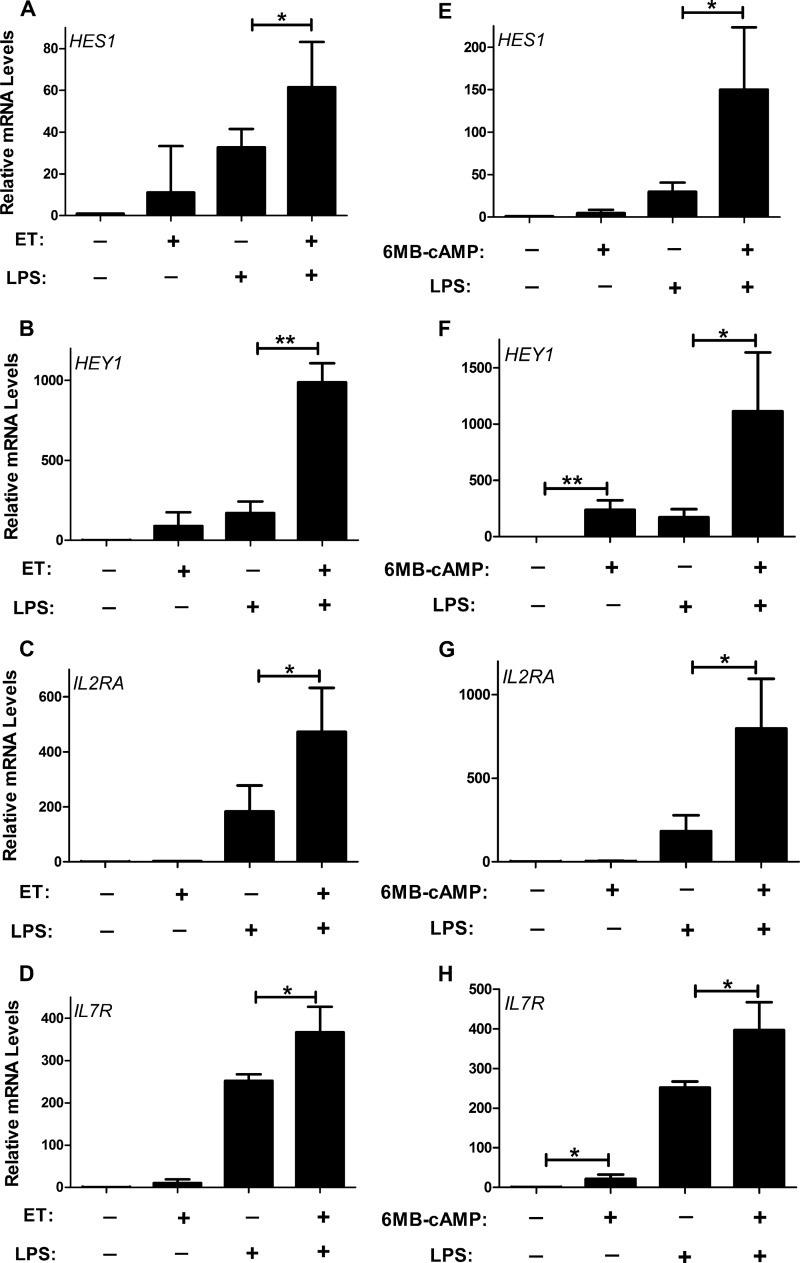
Next, human monocytes were isolated from donor blood, and the ability of cAMP to impact Notch responsive genes was examined in these cells. As shown by RT-qPCR, HES1, HEY1, and IL7r were induced by ET or 6-MB-cAMP in these cells (Fig. 2). Recently, human macrophages were observed to activate the Notch signaling pathway when stimulated by TLR agonists (13). Therefore, we examined human monocytes to determine how cAMP modulates this signaling pathway when monocytes are stimulated with the TLR agonist LPS. As shown in Fig. 2, A–D, LPS induces the production of Notch-inducible genes (HES1, HEY1, IL2RA, and IL7R), which is in agreement with previous observations by Hu et al. (13). Interestingly, exposing human monocytes to a combination of LPS and ET led to a significant increase in these Notch-responsive genes above levels observed in human monocytes treated with only LPS. For HEY1, this combination of LPS and ET led to 6-fold more HEY1 transcript than observed in human monocytes exposed to only LPS, which suggests a synergistic effect. We also determined whether 6-MB-cAMP had a similar effect on LPS stimulated monocytes. As shown in Fig. 2, E–H, combining LPS and 6-MB-cAMP also triggered an increase in these Notch target genes above levels obtained in human monocytes exposed to only LPS. Together, these results reveal that cAMP can induce Notch target genes in human monocytes and, importantly, that cAMP potentiates the LPS stimulation of the Notch signaling pathway.
FIGURE 2.
Increased cAMP induces Notch target genes in human monocytes. Human monocytes were isolated as described under “Experimental Procedures.” These cells were exposed for 6 h to one of the following or a combination of the following: 10 nm ET (10 nm EF and 10 nm PA), 1 mm 6MB-cAMP, or 100 ng/ml LPS. RT-qPCR was then used to quantify transcript levels of HES1, HEY1, IL2RA, and IL7R. Bar graphs are representative of three independent experiments. Each experiment used monocytes obtained from a different donor. Error bars indicate mean ± S.D. *, p < 0.06; **, p < 0.005.
Increased cAMP Elevates TLE and Increases Interactions between TLE and Hes1
The activation of the Notch pathway results in the up-regulation of the family of DNA binding transcription repressors Hey and Hes. In a previous study, these transcription repressors were found to help control macrophage responses to TLR agonists by attenuating the production of certain cytokines (13). One established mechanism utilized by Hes to repress transcription is to bind and recruit a family of transcriptional corepressor proteins, TLE1–4 (19–21). Because repressor activities of Hes are critical in macrophages, we sought to determine whether ET impacts TLE and possibly alters interactions between TLE and Hes1. To begin to address this, levels of the TLE1 transcript were first examined in human monocytes. As shown by RT-qPCR, both ET and 6MB-cAMP were able to induce TLE1 transcripts (Fig. 3A). A similar effect was also observed for ET and 6MB-cAMP when immunoblotting using pan-TLE antibodies was performed with extracts from the human monocyte cell line THP-1 (Fig. 3, B and C). Next, we determined whether increases in TLE levels and the activation of the Notch pathway ultimately correlate with an increase in the formation of a complex containing Hes1 and TLE. TLE was immunoprecipitated from extracts taken from THP-1 cells, and then interactions with Hes1 were determined by immunoblot analysis. As shown in Fig. 3D, this coimmunoprecipitation experiment revealed that the addition of ET or 6MB-cAMP to THP-1 cells led to an increase in binding between TLE and Hes1. These results demonstrate that cAMP is able to stimulate the formation of a transcription repressor complex consisting of Hes1 and TLE.
FIGURE 3.
ET and 6MB-cAMP elevate TLE and trigger increased interactions between TLE and Hes1. A, RT-qPCR was utilized to quantify transcript levels of TLE1 in human monocytes. These cells were exposed for 6 h to 10 nm ET (10 nm EF and 10 nm PA) or 1 mm 6MB-cAMP. Bar graphs are representative of three independent experiments. Each experiment used monocytes obtained from a different donor. B, THP-1 cells were exposed to 10 nm ET (10 nm EF and 10 nm PA) or 1 mm 6MB-cAMP, and immunoblot analyses were performed with antibodies recognizing pan-TLE and antibodies against GSK-3β (loading control). C, the bar graph represents the densitometry analysis of three pan-TLE immunoblots. D, immunoprecipitation (IP) was performed using protein extracts taken from THP-1 cells exposed for 4.5 h to 10 nm ET (10 nm EF and 10 nm PA) or for 3 h to 500 μm 6MB-cAMP. Immunoprecipitation was carried out with control antibodies or pan-TLE antibodies, and the subsequent immunoblot was probed with antibodies against Hes1. Error bars indicate mean ± S.D. *, p < 0.05.
Elevated cAMP in Monocytes Increases Levels of RBP-J
To begin to understand the mechanism used by ET to activate the Notch pathway, the expression of components of the Notch signaling pathway were analyzed in human monocytes after cAMP was elevated. When Notch ligands were analyzed, ET and 6MB-cAMP stimulated an increase in transcripts for DLL1 and JAG1, but these transcripts only reached statistical significance during exposures to 6MB-cAMP (Fig. 4, A and B). Although DLL1 and JAG1 are likely important for the cAMP-mediated activation of Notch signaling, this high degree of variability suggests that other factors may be used by cAMP to activate Notch signaling. DLL3 and DLL4 were also examined but were not detected after exposure to ET or 6MB-cAMP. Notch receptors (NOTCH1 and NOTCH2) were analyzed at the mRNA level and were not found to increase (data not shown), which suggests that the induction of the receptors is not important for cAMP-mediated Notch signaling.
FIGURE 4.
Expression level of Notch signaling components in human monocytes after exposure to ET or 6MB-cAMP. A–C, human monocytes were isolated and exposed for 6 h to 10 nm ET (10 nm EF and 10 nm PA) or 1 mm 6MB-cAMP. RT-qPCR was then used to quantify transcript levels in three independent experiments. Each experiment used monocytes obtained from a different donor. D, human monocytes were exposed to 10 nm ET (10 nm EF and 10 nm PA) for 6 h, and immunoblot analyses were performed with antibodies recognizing RBP-J and antibodies against GSK-3β (loading control). E, the bar graph represents the densitometry analysis of three RBP-J immunoblots. Error bars indicate mean ± S.D. *, p < 0.05; **, indicates p < 0.005.
In addition to analyzing the Notch ligands and receptors, levels of the primary transcriptional regulator of Notch signaling, RBP-J, were assessed. As shown in Fig. 4C, human monocytes exposed to ET or 6-MB-cAMP had a significant increase in levels of the RBPJ transcript. Analysis of RBP-J by immunoblotting also revealed that RBP-J was increased when human monocytes were exposed to ET (Fig. 4, D and E). These data indicate that RBP-J is increased significantly in human monocytes exposed to cAMP, suggesting RBP-J may be a critical factor that mediates the activation of Notch signaling by cAMP.
Overexpression of RBP-J Activates Notch Signaling
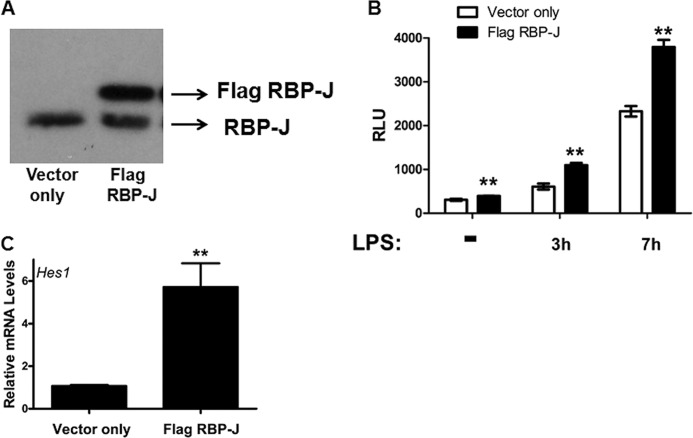
Because RBP-J is up-regulated by cAMP, we hypothesized that increased levels of RBP-J may help drive Notch signaling. To examine this possibility, RBP-J was overexpressed in RAW 264.7 cells by transducing these cells with a retrovirus containing a FLAG-tagged form of RBP-J. As shown by immunoblot analysis, transduced cells had high levels of the FLAG-tagged form of RBP-J in addition to endogenous RBP-J (Fig. 5A). To measure Notch signaling activity, we next utilized a RAW 264.7 cell line possessing a luciferase-based Notch reporter. As shown in Fig. 5B, the reporter cells overexpressing RBP-J stimulated slightly more luciferase production than cells transduced with the negative control retrovirus. As previously discussed, the Notch pathway is stimulated by LPS in macrophages (13). Therefore, we next sought to determine whether the overexpression of RBP-J could enhance the LPS activation of Notch signaling. As shown in Fig. 5B, the LPS induction of the Notch signaling pathway was significantly higher in macrophages with RBP-J elevated by retrovirus transduction. Hence, elevated RBP-J is able to increase Notch signaling in LPS-stimulated macrophages.
FIGURE 5.
Increased RBP-J triggers the activation of Notch signaling. A, RAW 264.7 cells were transduced with retrovirus-containing vector only control or FLAG-tagged RBP-J as described under “Experimental Procedures.” To demonstrate expression of FLAG-tagged RBP-J, immunoblotting was performed with antibodies against RBP-J. B, RAW-Notch and RAW-Neg reporter cells were subjected to retrovirus transduction with vector-only control or FLAG-tagged RBP-J. After transduction and selection, the reporter cells were exposed to 100 ng/ml of LPS, and a luciferase assay was performed. The values from this assay represent normalized average relative luciferase units (RLU) from three independent experiments. C, RT-qPCR was used to measure transcript levels of Hes1 in RAW 264.7 cells that were transduced with retrovirus. Bar graphs are representative of four independent experiments. Error bars indicate mean ± S.D. **, p < 0.005.
Next, we sought to determine whether the overexpression of RBP-J could lead to increased levels of the Notch target gene, Hes1. As shown by RT-qPCR, RAW 264.7 cells overexpressing RBP-J also possessed increased levels of the Hes1 transcript when compared with levels in control-transduced cells (Fig. 5C). These data suggest that elevated RBP-J is sufficient to activate the Notch signaling pathway and could be an important mechanism for increasing Notch signaling in macrophages.
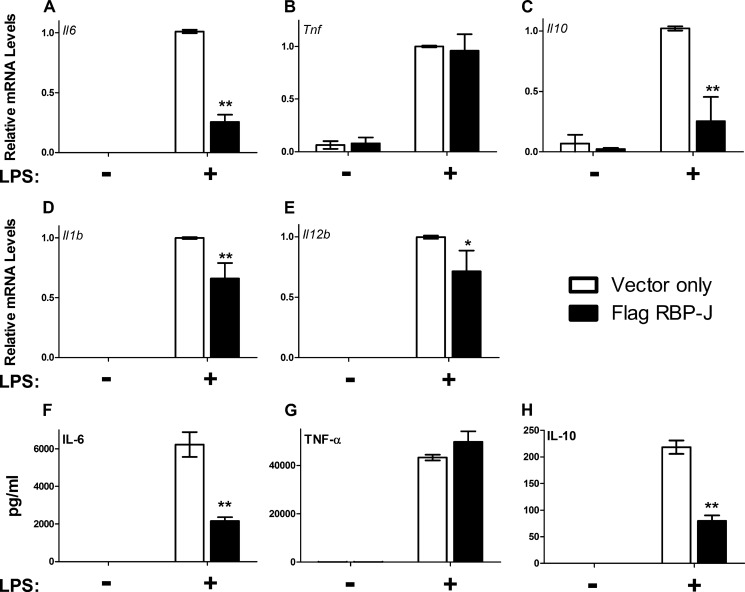
Because increased levels of Hes1 are predicted to repress genes that contain N- or E-box DNA sequences within their promoters such as cytokines (22, 23), we next determined whether the LPS-meditated induction of cytokines could be suppressed in macrophages overexpressing RBP-J. Therefore, IL-6, IL-12, IL-1β, IL-10, and TNF-α were examined to determine whether cytokine suppression was triggered by overexpressing RBP-J. As shown by RT-qPCR, Il6, Il12b, Il1b, and Il10 transcripts were decreased in LPS-stimulated macrophages overexpressing RBP-J (Fig. 6). The secretion of IL-6 and IL-10 proteins was examined by ELISA, and the results also demonstrated that overexpressing RBP-J suppressed these cytokines (Fig. 6). IL-1β and IL-12 secretion was examined but was not detected by ELISA under these conditions (data not shown). In contrast to these cytokines, TNF-α levels were not altered significantly in LPS-exposed macrophages overexpressing RBP-J when examined by RT-qPCR or ELISA (Fig. 6). These data suggest that activating Notch signaling through RBP-J overexpression can lead to the suppression of cytokines.
FIGURE 6.
Increased RBP-J reduces cytokine production. RAW 264.7 cells were transduced with retrovirus-containing vector only control or FLAG-tagged RBP-J as detailed under “Experimental Procedures.” Cells expressing high levels of RBP-J were selected and used for these experiments. After transduction and selection, these cells were exposed to 100 ng/ml LPS for 6 h. A–E, RT-qPCR was used to quantify transcript levels of Il6, TNF, Il10, Il1b, and Il12b. F–H, the levels of IL-6, TNF-α, and IL-10 secreted into the culture medium were measured by ELISA. IL-1β and IL-12 were not detected in the culture medium by ELISA. Bar graphs are representative of a minimum of three independent experiments. Error bars indicate mean ± S.D. *, p < 0.05; **, p < 0.005.
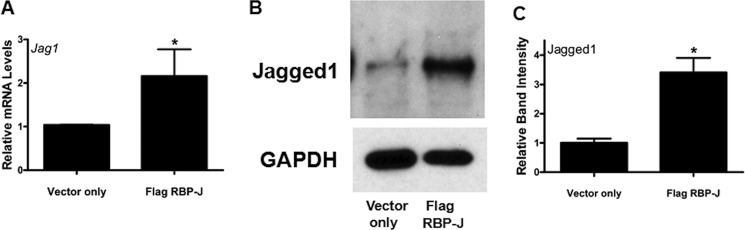
Elevated RBP-J Increases Jagged1
A recent study demonstrated that the Notch ligand Jagged1 is a RBP-J target gene that can be induced by Notch signaling and can thus function as part of an autoamplification loop (14). Therefore, we sought to determine whether the overexpression of RBP-J can induce Jagged1, which could in part account for the ability of elevated RBP-J to induce Notch signaling. As shown by RT-qPCR and immunoblot analyses, overexpressing RBP-J in macrophages resulted in increased levels of Jagged1 when compared with control transduced cells (Fig. 7). These data demonstrate that macrophages overexpressing RBP-J can induce a Notch target gene that can possibly further amplify Notch signaling.
FIGURE 7.
Elevated RBP-J increases levels of Jagged1. RAW 264.7 cells were transduced with retrovirus-containing vector only control or FLAG-tagged RBP-J as described under “Experimental Procedures.” A, RT-qPCR was used to measure transcript levels of Jag1. Bar graphs are representative of four independent experiments. B, immunoblot analysis was performed with antibodies against Jagged1 and GAPDH (loading control). C, the bar graph represents the densitometry analysis of four Jagged1 immunoblots. Error bars indicate mean ± S.D. *, p < 0.05.
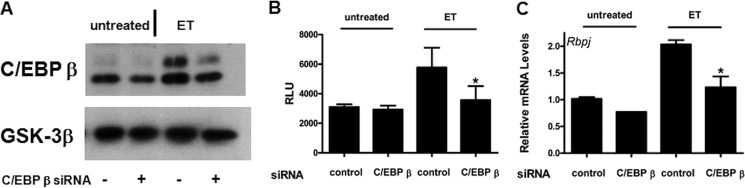
C/EBP β Activates Notch Signaling and Up-regulates RBP-J
Next, experiments were conducted to gain a better understanding of how cAMP signaling is linked to the activation of Notch signaling. We demonstrated previously that C/EBP β is critically involved in regulating much of the gene expression activities of ET (6). Therefore, C/EBP β was hypothesized to be involved in regulating Notch activation. To address this possibility, levels of cellular C/EBP β were reduced using siRNA (Fig. 8A), and then the activation of the Notch pathway was measured in Notch reporter cells. As shown in Fig. 8B, silencing C/EBP β inhibited the activation of the Notch reporter by ET suggesting C/EBP β was necessary for the induction of Notch signaling. Because RBP-J is elevated in the human monocytes and this increase is able to help drive Notch signaling, we determined whether RBP-J was regulated by C/EBP β. As shown by RT-qPCR, siRNA targeting C/EBP β repressed the induction of Rbpj by ET (Fig. 8C), which suggests that RBP-J induction is through a C/EBP β-dependent mechanism.
FIGURE 8.
Silencing C/EBP β inhibits the ability of cAMP to activate Notch signaling and up-regulated RBP-J. RAW 264.7 cells were transfected with siRNA directed against C/EBP β or negative control siRNA. A, an immunoblot analysis was performed demonstrating that siRNA against C/EBP β reduces levels of C/EBP β after exposure to ET (10 nm EF and 10 nm PA for 6 h). B, RAW-Notch and RAW-Neg reporter cells were transfected with siRNA and then exposed to ET (10 nm EF and 10 nm PA for 6 h). A luciferase assay was performed next, and the values from this assay are presented as normalized average relative luciferase units (RLU) from three independent experiments. C, RT-qPCR was used to measure transcript levels of Rbpj in RAW 264.7 cells that were transfected with siRNA. Error bars indicate mean ± S.D. *, p < 0.05.
DISCUSSION
The ability of pathogens such as B. anthracis to increase cAMP levels in macrophages is a key step in disease progression. By increasing cAMP in macrophages, pathogens are able to suppress immune responses by disrupting functions such as phagocytosis, intracellular killing, and induction of proinflammatory cytokines (24, 25). A major goal of this research is to understand mechanisms of cell signaling that are impacted by cAMP and can lead to attenuated immune responses. In these studies, cAMP from ET is found to activate Notch signaling and potentiate the induction of Notch target genes by TLR ligands (Figs. 1 and 2). This observation is especially interesting considering that ET was recently found to inhibit Notch signaling in a D. melanogaster model system and in human endothelial cells (8). These contrasting findings highlight the connection between Notch and cAMP signaling and demonstrate that the mechanisms connecting these two pathways differ by cell type. In terms of human disease, these observations support the following model. During the early stages of anthrax, ET activates Notch signaling in macrophages, potentially resulting in the suppression of immune responses and allowing B. anthracis to grow to high levels. Then, in the late stages of anthrax, ET helps triggers vascular dysfunction, possibly by inhibiting Notch signaling in endothelial cells.
In macrophages, the Notch signaling pathway has become recognized as a key signaling pathway that combines with TLR signaling to regulate many macrophage activities (13–16). One important result of the activation of Notch signaling is the up-regulation of Hes and Hey. These DNA binding proteins are transcriptional repressors that target genes that contain N- or E-box DNA sequences within their promoters (12). Recent studies have demonstrated that Hes and Hey are part of a feedback loop that represses the TLR induction of cytokines such as IL-12 and IL-6 (13). Many other cytokines and other NF-κB-regulated genes also possess N- or E- box DNA binding sites and could be potential targets of Hes and Hey (22, 23). In addition to activating the Notch signaling pathway, cAMP is able to increase levels of TLE, which is a transcriptional corepressor recruited to gene promoters by DNA binding proteins such as Hes (19–21, 26). The transcription repressor complex resulting from TLE and Hes1 is important for mediating many of the Notch signaling effects and was observed recently to play a role in the ability of zymosan to suppress IL-12 production in dendritic cells (27). Results from these studies indicate that cAMP induces the formation of the TLE/Hes1 complex, which likely results from the ability of cAMP to activate the Notch pathway and increase levels of TLE.
This ability of cAMP to both up-regulate TLE and potentiate TLR-induced Notch signaling could shift the balance of the TLR/Notch signaling pathway. This shift could make the Hes-directed feedback loop more effective and could lead to a more substantial suppression of TLR-induced cytokines. This is in contrast to macrophages exposed to IFN-γ, a cytokine that augments TLR responses. In macrophages treated with IFN-γ, Hu et al. (13) demonstrated that the TLR-induced activation of Notch target genes was inhibited and that the Hes and Hey feedback loops were thus disrupted. Therefore, Notch signaling represents a critical target for ET and a key element for controlling the strength of immune responses.
To address the mechanism used by cAMP to activate Notch signaling, levels of components of Notch signaling were examined. Interestingly, RBP-J was found to be up-regulated by cAMP in these studies (Fig. 4). RBP-J is the primary transcription regulator of Notch target genes. In the absence of Notch signaling, this DNA-binding protein interacts with a transcriptional repressor complex. However, after Notch signaling is activated and RBP-J binds the Notch intracellular domain, this repressor complex is replaced with an activator complex that promotes gene transcription. Because RBP-J is up-regulated by cAMP, experiments were designed to determine how this cAMP-mediated increase in RBP-J may impact Notch signaling. Therefore, RBP-J was overexpressed by retrovirus transduction, and Notch signaling was examined. The overexpression of RBP-J resulted in the activation of Notch signaling, as shown by using a Notch reporter as well as by examining levels of Hes1 (Fig. 5). These data suggest that RBP-J is a limiting factor in macrophages and that increases in RBP-J are sufficient to increase Notch signaling. This also suggests that a low level of Notch activity is being maintained continually in these macrophages. The impact of overexpressed RBP-J was also examined when TLR signaling was activated in macrophages. Under these conditions, overexpressed RBP-J was able to amplify LPS-induced Notch signaling in a manner similar to how ET impacts TLR-induced Notch signaling. Therefore, increased RBP-J is capable of intensifying TLR-activated Notch signaling and could possibly be a mechanism used by ET to enhance TLR-activated Notch signaling. The RBP-J-mediated activation of Notch also correlated with the suppression of LPS-induced IL-6, IL-12, IL-1β, and IL-10. IL-6 and IL-12 are cytokines that have been shown previously to be repressed by Hes and Hey (13). In our studies, TNF-α was not changed, which agrees with prior data that show that TNF-α is not very sensitive to Hes or Hey (13). These studies suggest that increases in RBP-J could be one important mechanism used by cAMP to control cytokine production.
The ability of increased levels of RBP-J to activate Notch signaling could result in part from an auto-amplification loop. Recent studies have demonstrated that Notch ligands such as Jagged1 and DLL4 have RBP-J binding sites within their promoters and can be induced by Notch signaling (14, 28). Therefore, Notch signals can potentially up-regulate Notch ligands and further amplify this signaling pathway. As shown in Fig. 7, this idea is supported by the observation that Jagged1 is up-regulated in macrophages that overexpress RBP-J. Hence, this autoamplification loop could in some measure account for the activation of Notch in macrophages by increased levels of RBP-J.
To further address the mechanism utilized by ET to activate Notch signaling, cAMP effectors were examined to determine how these proteins impact Notch signaling. PKA was found to be an important mediator of cAMP-dependent activation of Notch signaling because a PKA-specific agonist, 6-MB-cAMP, was found to be a potent activator of Notch signaling. Additionally, C/EBP β was examined to determine whether it was necessary for the activation of Notch signaling by ET. C/EBP β is part of the basic region leucine zipper family of transcription factors and is essential for regulating many ET activities (6). In these studies, Notch activation by ET was found to depend on C/EBP β because silencing C/EBP β with siRNA could inhibit the ET-mediated induction of Notch signaling. Furthermore, reducing C/EBP β by siRNA inhibited the ability of ET to induce RBP-J, which further supports the idea that RBP-J levels are important for controlling cAMP-induced Notch signaling.
In conclusion, ET-generated cAMP activates Notch signaling and potentiates the induction of Notch target genes by TLR ligands. This activation of Notch signaling depends on the ability of cAMP to up-regulate RBP-J through a mechanism involving PKA and C/EBP β. This ability of cAMP to enhance Notch signaling and up-regulate TLE could possibly strengthen the repressor activities of Hes, leading to attenuated immune responses.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant U19 AI 062629 (to J. D. B.).

This article contains supplemental Fig. S1.
- ET
- edema toxin
- EF
- edema factor
- PA
- protective antigen
- RBP-J
- recombinant recognition sequence binding protein at the Jκ site
- TLE
- Groucho/transducin-like enhancer of split
- TLR
- Toll-like receptor
- 6-MB-cAMP
- N6-monobutyryladenosine-3′,5′-cyclic monophosphate
- BMDM
- bone-marrow-derived macrophage
- qPCR
- quantitative PCR.
REFERENCES
- 1. McDonough K. A., Rodriguez A. (2012) The myriad roles of cyclic AMP in microbial pathogens. From signal to sword. Nat. Rev. Microbiol. 10, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leppla S. H. (1982) Anthrax toxin edema factor. A bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. 79, 3162–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Young J. A., Collier R. J. (2007) Anthrax toxin. Receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem. 76, 243–265 [DOI] [PubMed] [Google Scholar]
- 4. Liu S., Miller-Randolph S., Crown D., Moayeri M., Sastalla I., Okugawa S., Leppla S. H. (2010) Anthrax toxin targeting of myeloid cells through the CMG2 receptor is essential for establishment of Bacillus anthracis infections in mice. Cell Host Microbe 8, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim C., Wilcox-Adelman S., Sano Y., Tang W.-J., Collier R. J., Park J. M. (2008) Antiinflammatory cAMP signaling and cell migration genes co-opted by the anthrax bacillus. Proc. Natl. Acad. Sci. 105, 6150–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larabee J. L., Shakir S. M., Hightower L., Ballard J. D. (2011) Adenomatous polyposis coli protein associates with C/EBP beta and increases Bacillus anthracis edema toxin-stimulated gene expression in macrophages. J. Biol. Chem. 286, 19364–19372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Larabee J. L., Maldonado-Arocho F. J., Pacheco S., France B., DeGiusti K., Shakir S. M., Bradley K. A., Ballard J. D. (2011) Glycogen synthase kinase 3 activation is important for anthrax edema toxin-induced dendritic cell maturation and anthrax toxin receptor 2 expression in macrophages. Infect. Immun. 79, 3302–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guichard A., McGillivray S. M., Cruz-Moreno B., van Sorge N. M., Nizet V., Bier E. (2010) Anthrax toxins cooperatively inhibit endocytic recycling by the Rab11/Sec15 exocyst. Nature 467, 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maillard I., Adler S. H., Pear W. S. (2003) Notch and the immune system. Immunity 19, 781–791 [DOI] [PubMed] [Google Scholar]
- 10. Lewis J., Hanisch A., Holder M. (2009) Notch signaling, the segmentation clock, and the patterning of vertebrate somites. J. Biol. 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bray S. J. (2006) Notch signalling. A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 12. Fischer A., Gessler M. (2007) Delta, Notch, and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 35, 4583–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu X., Chung A. Y., Wu I., Foldi J., Chen J., Ji J. D., Tateya T., Kang Y. J., Han J., Gessler M., Kageyama R., Ivashkiv L. B. (2008) Integrated regulation of Toll-like receptor responses by Notch and interferon-γ pathways. Immunity 29, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Foldi J., Chung A. Y., Xu H., Zhu J., Outtz H. H., Kitajewski J., Li Y., Hu X., Ivashkiv L. B. (2010) Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J. Immunol. 185, 5023–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu H., Zhu J., Smith S., Foldi J., Zhao B., Chung A. Y., Outtz H., Kitajewski J., Shi C., Weber S., Saftig P., Li Y., Ozato K., Blobel C. P., Ivashkiv L. B., Hu X. (2012) Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 13, 642–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Q., Wang C., Liu Z., Liu X., Han C., Cao X., Li N. (2012) Notch signal suppresses Toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J. Biol. Chem. 287, 6208–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palaga T., Buranaruk C., Rengpipat S., Fauq A. H., Golde T. E., Kaufmann S. H., Osborne B. A. (2008) Notch signaling is activated by TLR stimulation and regulates macrophage functions. Eur. J. Immunol. 38, 174–183 [DOI] [PubMed] [Google Scholar]
- 18. Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grbavec D., Stifani S. (1996) Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223, 701–705 [DOI] [PubMed] [Google Scholar]
- 20. Chen G., Courey A. J. (2000) Groucho/TLE family proteins and transcriptional repression. Gene 249, 1–16 [DOI] [PubMed] [Google Scholar]
- 21. Grbavec D., Lo R., Liu Y., Stifani S. (1998) Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258, 339–349 [DOI] [PubMed] [Google Scholar]
- 22. Sharif M. N., Sosic D., Rothlin C. V., Kelly E., Lemke G., Olson E. N., Ivashkiv L. B. (2006) Twist mediates suppression of inflammation by type I IFNs and Axl. J. Exp. Med. 203, 1891–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Šoši D., Richardson J. A., Yu K., Ornitz D. M., Olson E. N. (2003) Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell 112, 169–180 [DOI] [PubMed] [Google Scholar]
- 24. Peters-Golden M. (2009) Putting on the Brakes. Cyclic AMP as a multipronged controller of macrophage function. Sci. Signal. 2, pe37. [DOI] [PubMed] [Google Scholar]
- 25. Serezani C. H., Ballinger M. N., Aronoff D. M., Peters-Golden M. (2008) Cyclic AMP. Master regulator of innate immune cell function. Am. J. Respir. Cell Mol. Biol. 39, 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tetsuka T., Uranishi H., Imai H., Ono T., Sonta S., Takahashi N., Asamitsu K., Okamoto T. (2000) Inhibition of nuclear factor-κB-mediated Transcription by association with the amino-terminal enhancer of Split, a Groucho-related protein lacking WD40 repeats. J. Biol. Chem. 275, 4383–4390 [DOI] [PubMed] [Google Scholar]
- 27. Alvarez Y., Municio C., Hugo E., Zhu J., Alonso S., Hu X., Fernández N., Sánchez Crespo M. (2011) Notch- and transducin-like enhancer of split (TLE)-dependent histone deacetylation explain interleukin 12 (IL-12) p70 inhibition by zymosan. J. Biol. Chem. 286, 16583–16595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caolo V., van den Akker N. M., Verbruggen S., Donners M. M., Swennen G., Schulten H., Waltenberger J., Post M. J., Molin D. G. (2010) Feed-forward signaling by membrane-bound ligand receptor circuit. The case of Notch Delta-like 4 ligand in endothelial cells. J. Biol. Chem. 285, 40681–40689 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.