Background: Opportune expression of MafA is essential for the development and function of pancreatic β cells.
Results: Onecut1 markedly decreased MafA gene expression through the Foxa2-binding cis-element on the MafA gene enhancer region.
Conclusion: Onecut1 functions as a negative regulator of MafA gene expression.
Significance: Onecut1 is likely involved in MafA gene expression in embryonic and diabetic β cells.
Keywords: Cell Biology, Diabetes, Insulin Synthesis, Islet, Transcription Factors
Abstract
The transcription factor MafA is a key regulator of insulin gene expression and maturation of islet β cells. Despite its importance, the regulatory mechanism of MafA gene expression is still unclear. To identify the transcriptional regulators of MafA, we examined various transcription factors, which are potentially involved in β cell differentiation. An adenovirus-mediated overexpression study clearly demonstrated that Onecut1 suppresses the promoter activity of MafA through the Foxa2-binding cis-element on the MafA enhancer region (named area A). However, ChIP analysis showed that Foxa2 but not Onecut1 could directly bind to area A. Furthermore, overexpression of Onecut1 inhibited the binding of Foxa2 onto area A upon ChIP analysis. Importantly, insertion of a mutation in the Foxa2-binding site of area A significantly decreased the promoter activity of MafA. These findings suggest that Onecut1 suppresses MafA gene expression through the Foxa2-binding site. In the mouse pancreas, MafA expression was first detected at the latest stage of β cell differentiation and was scarcely observed in Onecut1-positive cells during pancreas development. In addition, Onecut1 expression was significantly increased in the islets of diabetic db/db mice, whereas MafA expression was markedly decreased. The improved glucose levels of db/db mice with insulin injections significantly reduced Onecut1 expression and rescued the reduction of MafA expression. These in vivo experiments also suggest that Onecut1 is a negative regulator of MafA gene expression. This study implicates the novel role of Onecut1 in the control of normal β cell differentiation and its involvement in β cell dysfunction under diabetic conditions by suppressing MafA gene expression.
Introduction
Insulin is the critical hormone for the regulation of blood glucose levels and is synthesized exclusively in β cells of the islets of Langerhans. The molecular mechanisms that control insulin gene transcription are well characterized (1–7). MafA was identified as a strong trans-activating factor of the insulin gene (8–10). Compared with other insulin transcription factors, such as Pdx1 and Neurod1, which are expressed early in pancreas development, MafA expression in mice is detected at embryonic day E13.5, the late stage of pancreas development. In addition, MafA can be detected only in insulin-positive cells during the pre- and postnatal period, although both Pdx1 and Neurod1 are expressed in various cell types in the pancreas (11–16). These findings suggested that MafA is one of the key regulators of insulin gene expression, and its own expression is also precisely regulated by other transcription factors involved in pancreas and islet development. In fact, MafA knock-out mice displayed decreased insulin gene expression and impaired glucose-stimulated insulin secretion resulting in the development of diabetes (14). Furthermore, the expression of MafA in the early embryonic pancreas inhibits proliferation of pancreatic progenitors and reduces both pancreatic size and the number of endocrine cells (16). All of these reports indicated that timely expression of MafA is essential for the development and normal function of pancreatic β cells.
Many transcription factors, including Pdx1 and Neurod1, are known to be involved in the development of the pancreas (12–21). Among them, Onecut1 (formerly known as HNF6), a homeodomain-containing transcription factor, is an important regulator of pancreatic endocrine development (17–23), as shown with Onecut1 null mice, in which endocrine differentiation was disturbed during embryogenesis (22). Onecut1 is expressed early in pancreatogenesis in all endodermally derived cells and activates expression of the transcription factors Pdx1 (22) and Ngn3 (23), but its expression rapidly disappears as pancreatic progenitors are specified to the endocrine lineage (22–24). Prolonged expression of Onecut1 in the pancreatic endocrine cell lineage using a transgenic strategy showed disrupted islet morphogenesis in which the numbers of β cells were relatively decreased (24). These results and its expression pattern indicate that Onecut1 is a critical regulator for the initiation of endocrine specification.
Foxa2 (Forkhead box protein A2; formerly known as HNF3β) is one of the key regulators of endodermal cell lineage development (25–27). The expression of Foxa2 mRNA is first detected at E6.5 in the node at the anterior end of the primitive streak in all three germ layers, and in the developing pancreas, the expression of Foxa2 is restricted in the endocrine cells and absent in the ductal epithelium (27). In the islet β cells, Foxa2 binds to the conserved cis-acting elements of the MafA and Pdx1 genes, and inserting of a mutation in its binding site reduced the transcription activity of these genes (28, 29). These findings suggest that Foxa2 positively regulates MafA and Pdx1 gene expression and plays a key role in β cell function.
In this study, we found that Onecut1 suppresses MafA gene expression by negatively regulating the Foxa2-binding cis-element of the MafA gene. Consistent with our in vitro results, immunostaining experiments showed that Onecut1-positive cells scarcely expressed MafA during pancreas development. Interestingly, Onecut1 gene expression was markedly increased in the islet cells under diabetic conditions, whereas MafA expression is suppressed in contrast to Onecut1. These findings suggest a novel role of Onecut1 in the regulation of normal β cell differentiation and function through MafA gene expression.
EXPERIMENTAL PROCEDURES
Preparation of Expression Plasmids and Reporter Analysis
The whole enhancer/promoter sequence of the mouse MafA gene, including −10,427 to +22 bp from the MafA-coding sequence (a kind gift from Dr. Roland Stein) was inserted upstream of the firefly luciferase coding sequence in the PGL4 basic reporter plasmid (Promega). Using the whole MafA reporter plasmid as a template, individual reporter plasmids, including fragments of the MafA enhancer/promoter region (area A, −8152 to −7780 bp; area B, −6160 to −5634 bp; area C, −2291 to −1747 bp; area D, −878 to −641 bp; and area E, −249 to −1 bp), were constructed. For the insertion of mutations into area A, the QuikChange mutagenesis kit (Stratagene) was utilized. All sequences were confirmed by DNA sequencing.
The coding sequence of each transcription factor (Foxa2, Onecut1, Pdx1, Hb9, Ptf1a, Sox9, Ngn3, Neurod1, Insm1, and Hes1) was cloned into the BamHI and SacI sites of pcDNA3.1 (Invitrogen). Dual-Luciferase® reporter assays were performed 48 and 60 h after transfection, according to the manufacturer's protocol (Promega). The normalized firefly luciferase data by TK-Renilla luciferase (phRL-TK) were statistically analyzed by the two-tailed t test.
Preparation of Adenoviruses and Sample Isolation
Recombinant adenoviruses expressing Foxa2 (Ad-Foxa2), Onecut1 (Ad-Onecut1), Hb9 (Ad-Hb9), and Ptf1a (Ad-Ptf1a) were prepared as described previously (30). An adenovirus expressing only GFP (Ad-GFP) was also prepared as a control. MIN6 cells were treated with each adenovirus for 60 h, followed by isolation of nuclear proteins and total RNA.
Western Blotting Analysis
Western blotting analyses were performed using goat β-actin antibody (Santa Cruz Biotechnology), rabbit MafA antibody (Bethyl), goat anti-Foxa2 antibody, rabbit anti-Onecut1 antibody (Santa Cruz Biotechnology), and rabbit anti-Pdx1 antiserum (31).
Real Time RT-PCR
One microgram of total RNA was reverse-transcribed and used for real time PCR analysis. Primer sets for mouse MafA (numbering relative to ATG, forward 757TTCAGCAAGGAGGAGGTCAT and reverse 973CCGCCAACTTCTCGTATTTC; 217 bp) and mouse β-actin (forward 778GCTCTTTTCCAGCCTTCCTT and reverse 945CTTCTGCATCCTGTCAGCAA; 168 bp) were utilized to quantify each factor.
Electrophoretic Mobility Shift Assay
The gel-shift assay was performed with DIG Gel shift kit, 2nd generation (Roche Applied Science). DNA probes used in this assay were as follows (mutated bases are underlined): area A-1, GGCTCCACTCAGCCTTGTTTAGGGAGAAAA, area A-1 mutated competitor, GGCTCCACTCAGCCTTGCGTGGGAGAAAA; area A-2, CTTTCTGTAAACATTTTACAGCTCTCTGCG, area A-2 mutated competitor, CTTTCTGTACGCATTTTACAGCTCTCTGCG; area A-3, TATCATTTTATTGTCATATTTCACGGCCG, area A-3 mutated competitor, TATCATTTTATTGTCATAGCTCACGGCCG. According to the manufacturer's instructions, the binding reactions were performed with 6 μg of nuclear extract from MIN6 cells and 30 fmol of digoxigenin-labeled probes. Competition analysis was performed with a molar excess of unlabeled competitor to labeled probe (250-fold). Antibody supershift analyses were performed with 0.4 μg of goat anti-Foxa2 antibody (Santa Cruz Biotechnology) or rabbit anti-Onecut1 antibody (Santa Cruz Biotechnology) preincubated with extract protein for 15 min prior to the addition of the DNA probe. The protein-DNA probe complexes were electrophoresed on a 6% nondenaturing polyacrylamide gel (acrylamide/bisacrylamide ratio of 29:1) in 0.5 × TBE buffer. The gel was electroblotted onto a positively charged nylon membrane (Roche Applied Science). The blotted membranes were cross-linked with UV (1200 × 100 μJ/cm2). The digoxigenin-labeled oligonucleotides were visualized by an enzyme immunoassay using alkaline phosphatase-conjugated anti-digoxigenin antibody and the chemiluminescent substrate CSPD (disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate).
Immunoprecipitation Assay
Co-immunoprecipitations were performed using the co-immunoprecipitation kit (Pierce). According to the manufacturer's instructions, 10 μg of goat anti-Foxa2 antibody (Santa Cruz Biotechnology) or goat anti-Onecut1 antibody (MafA gene) was coupled with 10 μg of MIN6 nuclear extract with or without adenoviral Onecut1 overexpression. Immune complexes were eluted from the resin, and samples were analyzed by SDS-PAGE followed by immunoblotting analysis using rabbit anti-Foxa2 antibody and rabbit anti-Onecut1 antibody.
Immunohistochemistry
Pancreas tissues were fixed overnight in 4% paraformaldehyde at 4 °C. Embedded samples in paraffin were sliced into 4-μm sections and mounted on glass slides. Double immunofluorescence staining was performed using rabbit or goat anti-MafA at 1:500 dilution, goat anti-Foxa2 at 1:200 dilution (Santa Cruz Biotechnology), and rabbit anti-Onecut1 antibody at 1:200 dilution (Santa Cruz Biotechnology). Secondary antibodies were donkey anti-rabbit and anti-goat antibodies at 1:200 dilution (Jackson ImmunoResearch). Improved sensitivity of goat anti-MafA antibody was achieved by using the avidin-biotin complex. Fluorescent images were captured on a confocal microscope.
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP analysis was performed as described previously with some modifications (32). MIN6 cells were preincubated with each adenovirus for 60 h. The cells were then formaldehyde cross-linked for 10 min, followed by quenching in 125 mm glycine. Cells were resuspended in lysis buffer and sonicated to obtain 500–1000-bp DNA fragments. The protein-DNA complexes were isolated by incubation with 100 μg of Dynal beads and 10 μg of the following antibodies: goat anti-Foxa2 antibody (Santa Cruz Biotechnology), rabbit anti-Onecut1 antibody (Santa Cruz Biotechnology), and normal rabbit IgG and normal goat IgG. Real time PCR was performed with purified immunoprecipitated DNA as a template using the primers specific for area A-2 (forward GAGGGCTGATTTAATTAGAAAG and reverse CGTTACGGCCGTGAAATATGA).
Knockdown Experiments
shRNA-expressing lentiviruses were utilized for the knockdown study according to the manufacturer's instructions. MIN6 cells were infected with lentivirus containing control or Foxa2-targeting shRNA (shFoxa2, CGCAGCTACACACACGCCA).
After selecting shRNA-integrated MIN6 cells in the presence of 2 μg/ml puromycin (Sigma) for 3 days, cells were reseeded and cultured in complete medium. Two days later, total RNA was isolated and subjected to quantitative RT-PCR experiments.
Statistical Analysis
Data are expressed as means ± S.D. Statistical analysis was performed using the two-tailed Student's t test. A value of p < 0.05 was considered to be statistically significant.
RESULTS
Excessive Expression of Onecut1 or Foxa2 Reduces MafA Gene Expression
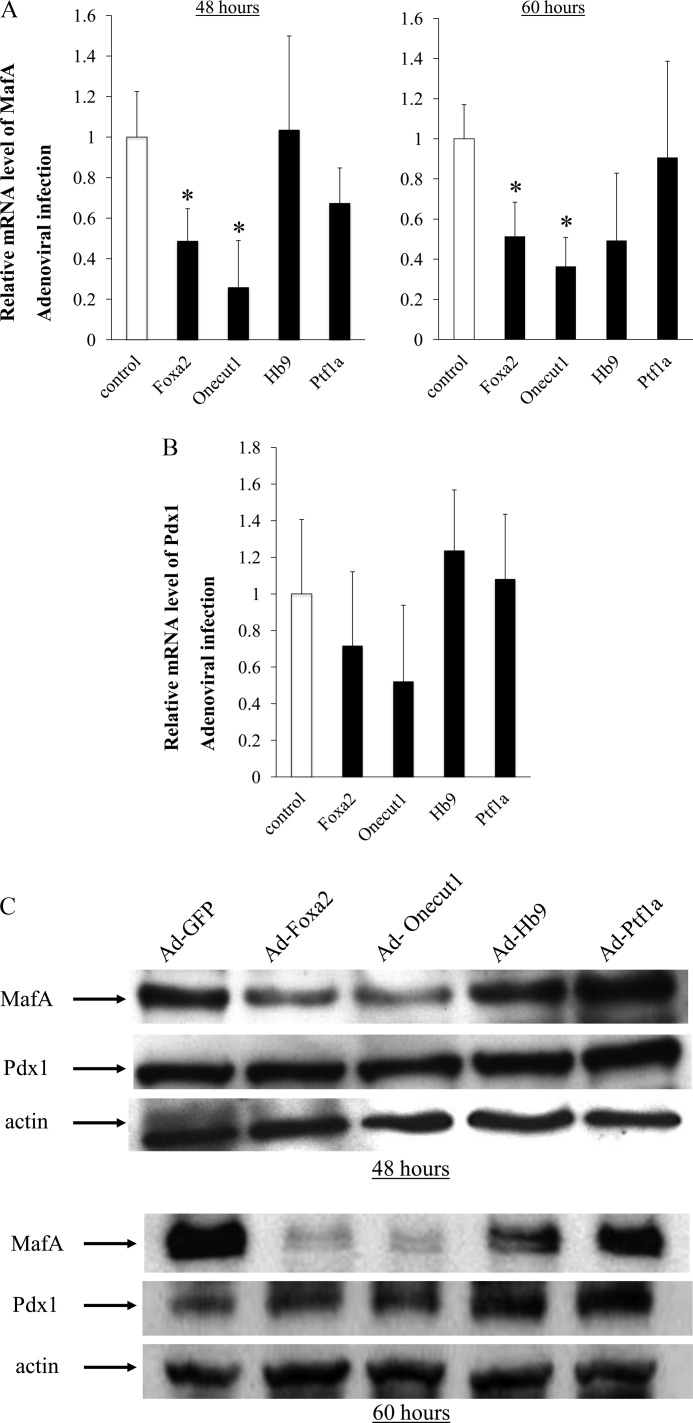
We first evaluated various transcription factors involved in pancreas development as candidates for the transcriptional regulator of MafA gene expression. To determine whether these transcription factors regulate MafA promoter activity, reporter gene analysis was performed by co-transfecting a MafA enhancer/promoter reporter plasmid and each expression vector for the candidate transcription factors into MIN6 cells. As shown in Fig. 1, the overexpression of Ptf1a, Onecut1, Hb9, or Foxa2 significantly reduced MafA promoter activity. This result suggests that these four transcription factors negatively regulate MafA gene expression. To examine whether these transcription factors actually influence endogenous MafA gene expression, we infected MIN6 cells with adenoviruses expressing Foxa2 Oncut1, Hb9 or Ptf1a. As shown in Fig. 2, A and B, overexpression of Onecut1 or Foxa2 significantly reduced MafA mRNA levels within 48 h after adenovirus infection, although the mRNA level of another key insulin transcription factor, Pdx1, was not significantly affected by these factors. Consistent with these results, Western blot analysis showed that overexpression of Foxa2 or Onecut1 dramatically reduced MafA protein levels (Fig. 2C). However, Pdx1 was not influenced as much by Foxa2 or Onecut1. These results demonstrate that overexpression of Foxa2 or Onecut1 exclusively reduces endogenous MafA expression.
FIGURE 1.
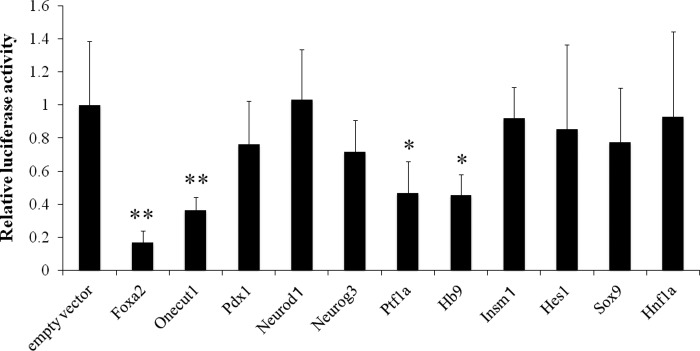
Suppressive effect of transcription factors on MafA promoter activity. A reporter plasmid, including the mouse MafA promoter/enhancer region (−10.5 to −1 kbp from the coding sequence) upstream of the firefly luciferase coding sequence, was co-transfected into MIN6 cells together with the expression plasmids for various transcription factors. The activities of firefly luciferase were normalized with phRL-TK-driven Renilla luciferase activity. Data are expressed as mean ± S.D. with the basal MafA promoter activity being arbitrarily set at 1 (n ≧5). *, p < 0.05; **, p < 0.01 versus control empty vector.
FIGURE 2.
Foxa2 and Onecut1 suppress the expression of MafA. Total RNA and nuclear protein were isolated from MIN6 cells at 48 or 60 h after Ad-Foxa2, Ad-Onecut1, Ad-Hb9, or Ad-Ptf1a infection. Real time PCR analysis was performed using 1 μg of total RNA to evaluate the amount of mRNA of MafA (A), Pdx1 (B), and the β-actin control. Each MafA mRNA level was normalized with that of β-actin. Data are presented as relative amounts ± S.D., with the ratio of MafA mRNA level with control Ad-GFP treatment being arbitrarily set at 1 (n = 4). *, p < 0.05. C, Western blotting was performed with 10 μg of nuclear protein isolated from MIN6 cells that were incubated with each adenovirus or control Ad-GFP for 48 or 60 h.
Onecut1 and Foxa2 Regulate MafA Promoter Activity through Area A
To determine the cis-element responsible for regulation of the MafA gene by Foxa2 and Onecut1, we constructed a reporter plasmid, including −10427 to +22 bp of the mouse MafA enhancer/promoter region, which shows maximum activity for the MafA promoter in β cells (29). Within the MafA enhancer region on the reporter plasmid, there are 2 and 20 consensus binding sites of Onecut1 and Foxa2, respectively (data not shown).
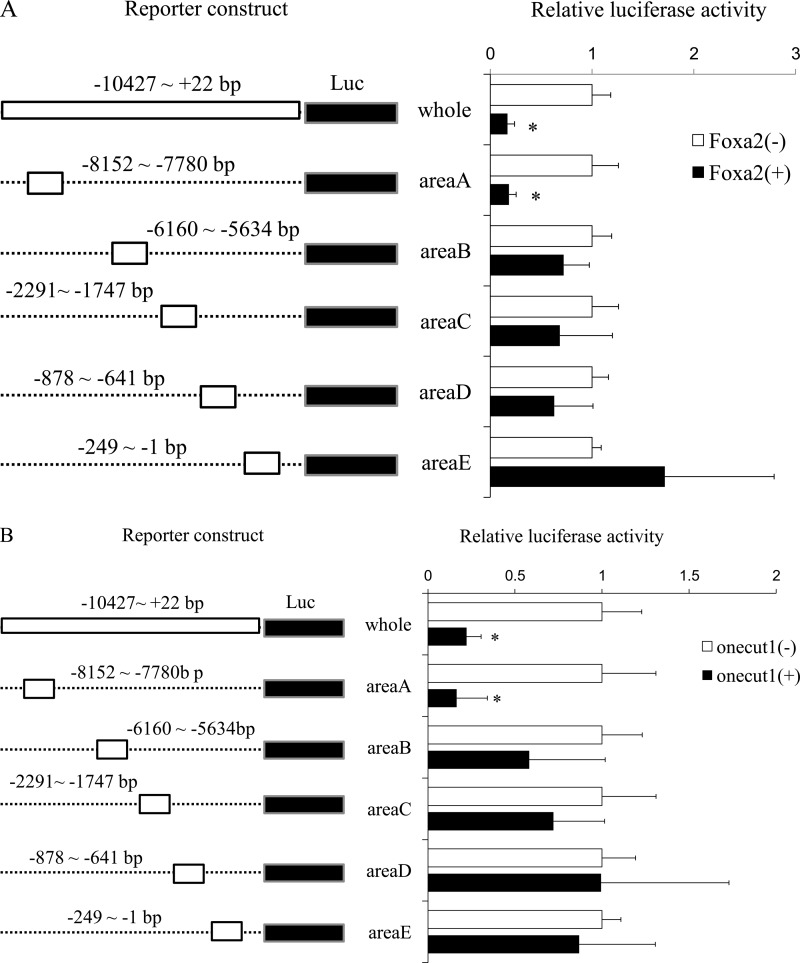
Next, we constructed reporter plasmids, including several of these consensus sites, which we named areas A to E. As shown in Fig. 3, the expression of Foxa2 or Onecut1 significantly reduced the reporter activity driven by the MafA area A enhancer region, as well as the whole length of the MafA enhancer region. These results suggest that Onecut1 and Foxa2 regulate MafA gene expression through area A.
FIGURE 3.
Foxa2 and Onecut1 regulate MafA promoter activity through area A. Reporter plasmids containing various lengths MafA promoter fragments (area A to E) were co-transfected with a Foxa2- (A) or Onecut1 (B)-expressing plasmid into MIN6 cells. Sixty hours after transfection, cells were harvested, and luciferase assays were performed. Activity levels of MafA promoter-driven firefly luciferase were normalized with phRL-TK-driven Renilla luciferase (Luc) activity. *, p < 0.05.
Foxa2 Binds to Area A of the MafA Enhancer Region
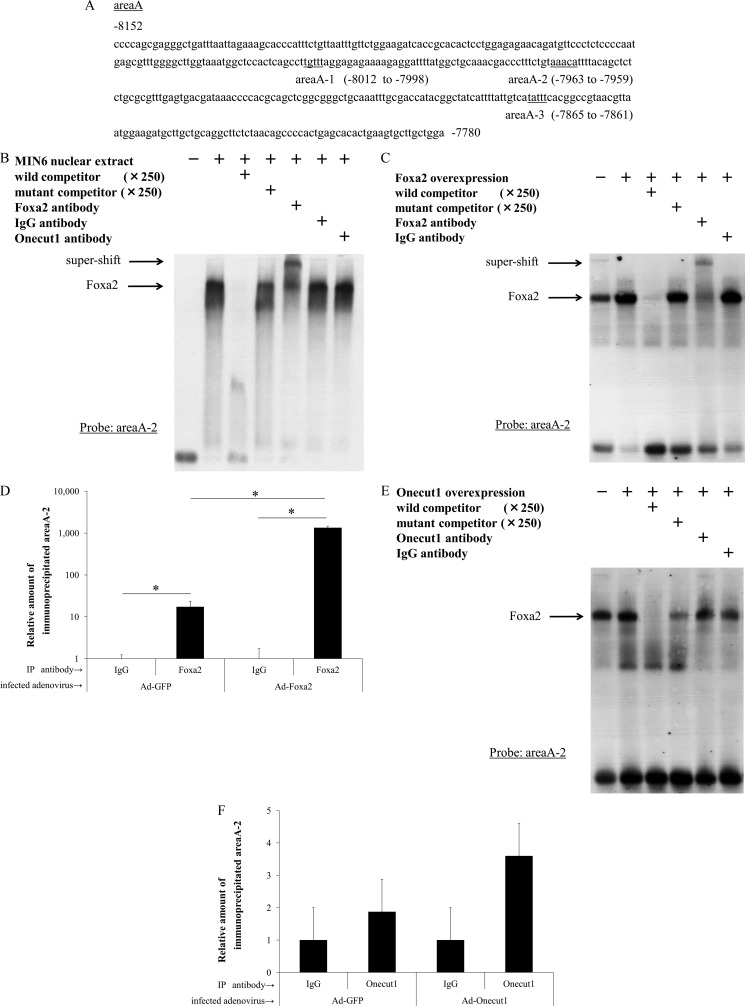
Area A is a highly conserved region between rodent and human, and this region includes three Foxa2 consensus binding sites, which we named area A-1 to A-3 (Fig. 4A). It is notable that there are no Onecut1 consensus binding sites within area A. Gel-shift analysis showed that endogenous and overexpressed Foxa2 could directly bind to area A-2 (Fig. 4, B and C) and that area A-2-binding protein was mainly Foxa2 as indicated by supershift analysis, although it did not bind to area A-1 or A-3 (data not shown). The ChIP with an anti-Foxa2 antibody also demonstrated that Foxa2 directly binds to area A in vivo (Fig. 4D). Unlike Foxa2, protein expression of Onecut1 was not detectable in MIN6 cells by Western blotting (data not shown). This is consistent with in vivo observations that Onecut1 expression is absent in pancreatic cells producing endocrine hormones. Thus, we first overexpressed Onecut1 in MIN6 cells by the adenoviral delivery system, and we performed gel-shift analysis using nuclear extracts from these cells, as well as ChIP analysis with an anti-Onecut1 antibody (Fig. 4, E and F). Both experiments clearly showed that Onecut1 did not bind to area A-2, although Onecut1 seems to act as a negative regulator.
FIGURE 4.
FoxA2 directly binds to area A-2 in vivo. A, area A (−8152 to −7780 bp) of the MafA gene enhancer region is illustrated. The mutation designed in the Foxa2 binding consensus within area A is underlined. A potential Foxa2-binding cis-element within area A-2 was used as a probe in gel-shift binding assays with MIN6 nuclear extract (B), nuclear extract from Foxa2- (C) or Onecut1 (E)-overexpressed MIN6 cells. The specificity of protein-DNA (area A-2) complex formation was determined by competition with a 250-fold excess of unlabeled wild-type competitor (lane 3), mutant competitor (lane 4), or specific antibodies as indicated. D and F, ChIP analysis was performed with MIN6 cells preincubated with Ad-Foxa2, Ad-Onecut1, or control Ad-GFP for 60 h. Formaldehyde cross-linked chromatin from MIN6 cells was incubated with antibodies specific to Foxa2, Onecut1, or control IgG antibody. The amount of immunoprecipitated DNA was analyzed by real time PCR using primers specific to area A-2. Data are presented as relative amounts ± S.D., with the level of immunoprecipitated DNA by nonspecific IgG arbitrarily set at 1 (n = 4). *, p < 0.05.
Onecut1 Inhibits Foxa2 Binding to the MafA Promoter
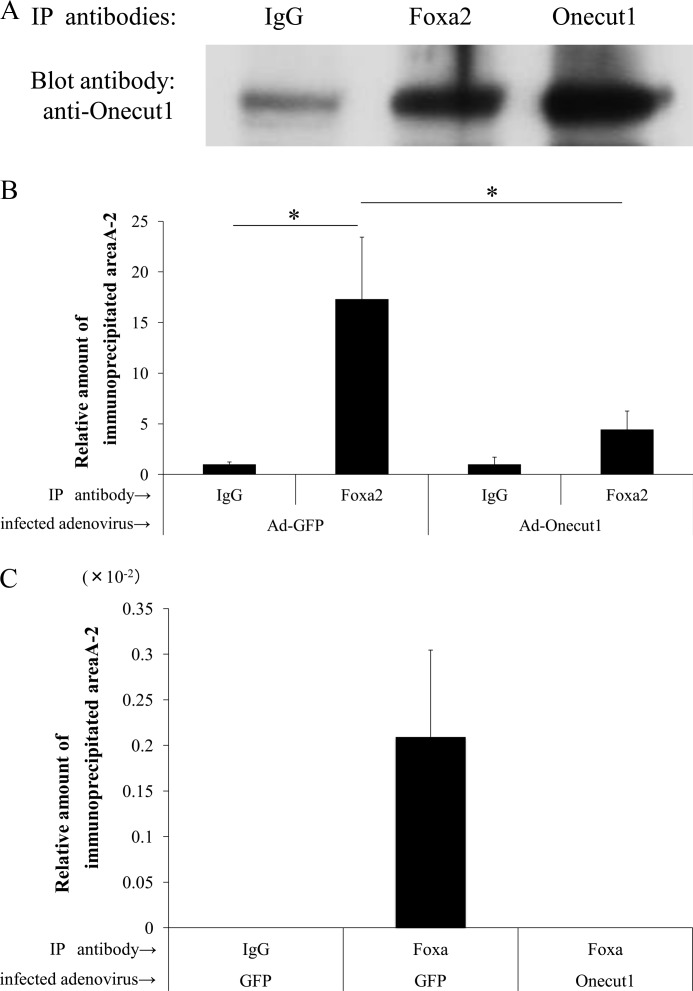
Although overexpression of Onecut1 suppresses MafA gene expression through area A (Fig. 3), it did not show any direct binding to this area of the MafA promoter on gel-shift analysis. These results led us to the hypothesis that Onecut1 suppresses MafA gene expression through the Foxa2-binding cis-element on area A-2. To examine if Onecut1 affects Foxa2 binding, an immunoprecipitation assay was performed using nuclear extracts of Onecut1-overexpressed MIN6 cells. It was clearly demonstrated that the Foxa2 antibody pulls down the Onecut1 protein, which indicates the interaction of Foxa2 and Onecut1 (Fig. 5A). To evaluate the effects of Onecut1 on Foxa2 activity, ChIP was performed. As shown in Fig. 5B, Onecut1 significantly attenuated the binding of endogenous Foxa2 to area A in MIN6 cells. Furthermore, the results using islet cells also showed Onecut1 overexpression in islet cells decreased the pull downed amount of DNA fragments by Foxa2 antibody to undetectable levels. These results suggest that Onecut1 directly suppresses the effect of endogenous Foxa2 on MafA gene expression.
FIGURE 5.
Onecut1 interacts with Foxa2 and inhibits binding ability of Foxa2 to MafA promoter in vivo. A, 10 μg of goat anti-Foxa2, goat anti-Onecut1, or control goat IgG was coupled with 10 μg of MIN6 nuclear extract with adenoviral Onecut1 overexpression. Immune complexes were eluted from the resin and applied to SDS-PAGE followed by immunoblot analysis using a rabbit anti-Onecut1 antibody. ChIP analysis was performed with MIN6 (B) and mouse primary cultured islet (C) cells preincubated with Ad-Onecut1 or control Ad-GFP for 60 h. Formaldehyde cross-linked chromatin from MIN6 cells was incubated with antibodies specific to Foxa2 or a control IgG antibody. Immunoprecipitated (IP) DNA was quantified by real time PCR using primers specific to area A-2. Data are presented as relative amounts ± S.E., with the ratio of immunoprecipitated DNA level by nonspecific IgG arbitrarily set at 1 (n = 4) in B, or DNA level from input sample arbitrarily set at 1 (n = 4) in C. *, p < 0.05.
Appropriate Amount of Foxa2 Activates MafA Gene Expression
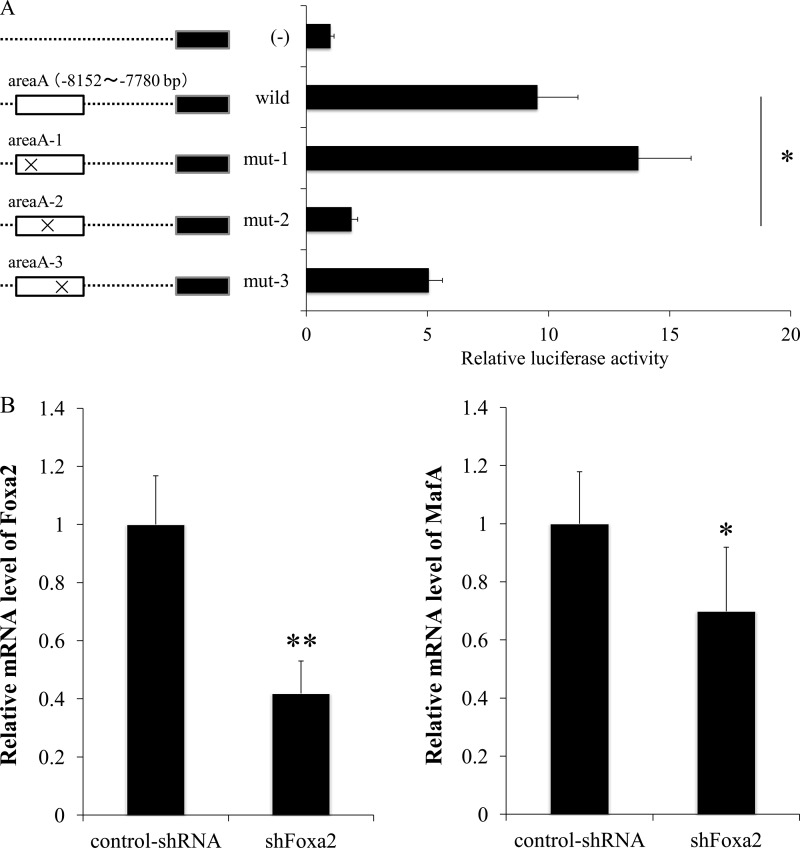
Because Onecut1 suppressed MafA gene expression and simultaneously reduced the binding activity of Foxa2 to area A on the enhancer region of the MafA gene, we examined in detail the effect of Foxa2 on MafA gene expression. For this purpose, we performed reporter gene analyses using luciferase reporter plasmids containing mutated Foxa2-binding sites of area A on MafA enhancer region (Fig. 6A). Insertion of a mutation within area A-2 significantly decreased MafA promoter activity, whereas mutation of area A-1 and area A-3 showed no effects on that activity. These results demonstrate that the area A-2 cis-element is critical for MafA gene activation, which is consistent with a previous report (29). To gain further insight into the role of Foxa2 on MafA gene expression, Foxa2 was knocked down using the lentivirus-mediated shRNA expression system. As shown in Fig. 6B, the mRNA level of Foxa2 was reduced down to 40% using this system, and MafA gene expression was significantly decreased. These results suggest that an appropriate amount of Foxa2 promotes MafA gene expression through its binding to the area A-2 cis-element of the MafA gene.
FIGURE 6.
Area A-2 is a positive regulatory cis-element for MafA gene expression. A, reporter gene analysis was performed with various reporter plasmids, including mutated area A as indicated in Fig. 4A. Sixty hours after the transfection of MIN6 cells, luciferase assays were performed. Activity levels of area A-driven firefly luciferase were normalized with phRL-TK-driven Renilla luciferase activity. *, p < 0.01. B, MIN6 cells were transfected with lentivirus containing control shRNA or Foxa2 shRNAs. FoxA2 (left panel) and MafA (right panel) mRNA levels were determined by real time PCR and are shown as mean ± S.D. *, p < 0.05; **, p < 0.01.
Onecut1 Can Be a Negative Regulator of MafA in Vivo
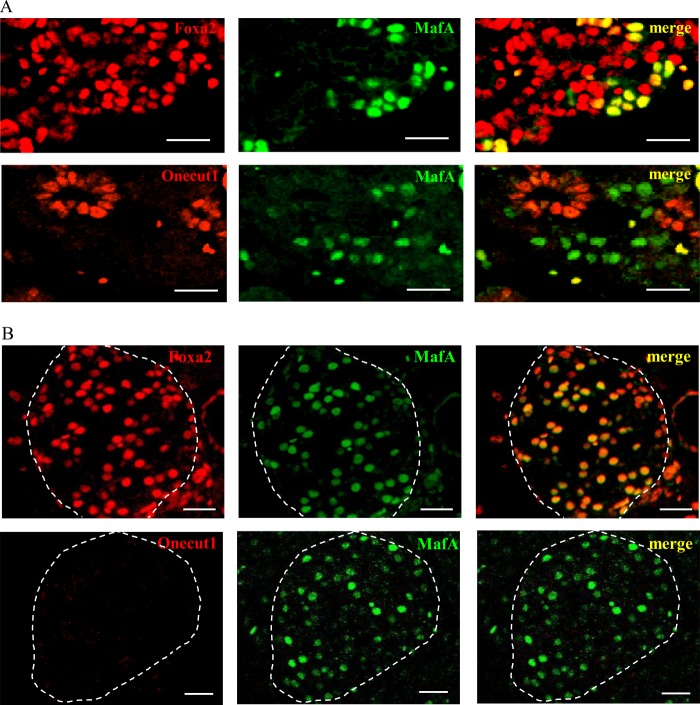
To examine the physiological relevance of Onecut1 or Foxa2 on MafA expression in vivo, immunohistochemical analysis was performed. Because MafA expression is first detected at the beginning of the principal phase of insulin-producing cell emergence, namely E13.5, and a large amount of MafA expression is observed at E16.5, the expression patterns of these transcription factors were examined at E16.5. As shown in Fig. 7A, MafA expression was observed in the embryonic pancreas within Foxa2-positive cells. In contrast, MafA expression was not observed in most Onecut1-positive cells. In the adult pancreas, Foxa2 and MafA continue to be co-expressed, although Onecut1 expression was scarcely detected in islet cells under normal conditions (Fig. 7B). These results imply that the disappearance of Onecut1 is required for initiating MafA gene expression in the embryonic pancreas.
FIGURE 7.
MafA expression can be detected only in Onecut1 negative cells. Pancreatic sections from mouse embryo at E16.5 (A) and mature mouse at 7 weeks of age (B) were immunostained with Foxa2 or Onecut1 (red) and MafA (green) antibodies. Islets are outlined with dotted line. Scale bars, 20 μm.
Increased Expression of Onecut1 under Diabetic Conditions
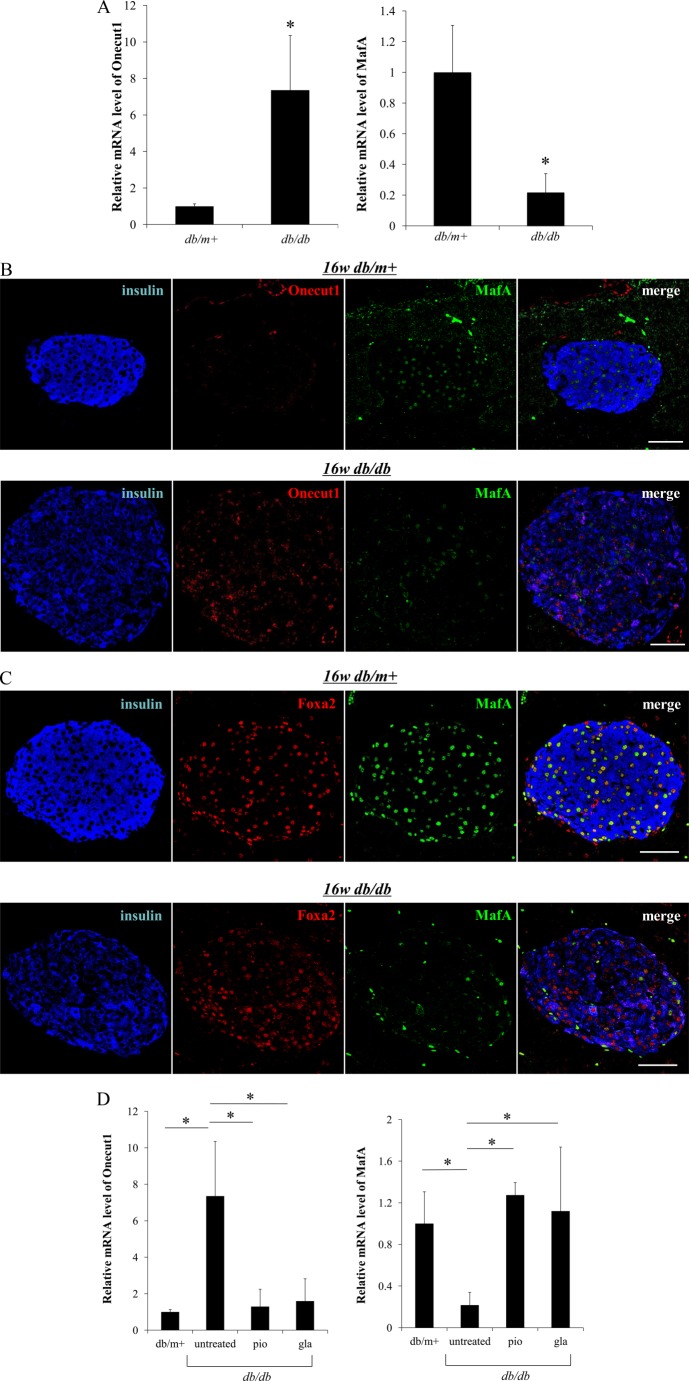
As another experiment, we evaluated the expression pattern of those factors in the pancreas of db/db type 2 diabetic model mice, as MafA expression is markedly decreased under diabetic conditions (33–35). Intriguingly, Onecut1 expression was dramatically increased in these islets under diabetic conditions (Fig. 8A), and MafA expression was not detected in most Onecut1-positive cells, as observed in the embryonic pancreas (Fig. 8B). With respect to Foxa2, its expression appeared to be decreased under diabetic conditions, with the expression not being as much as MafA (Fig. 8C). As we showed previously, improvement of blood glucose levels preserves the expression of MafA in islet β cells (35). Thus, to determine whether improvement of glycemic control in db/db mice could modulate Onecut1 expression, we administered insulin (glargine) or thiazolidine (pioglitazone) into db/db mice as reported previously (35). As shown in Fig. 8D, improvement of glycemic control with insulin or thiazolidine administration cancelled the increase in expression of Onecut1 and the suppression of MafA. These results suggest that Onecut1 induced under diabetic conditions is involved in the reduction of MafA, which contributes to the β cell dysfunction.
FIGURE 8.
Onecut1 expression is increased under diabetic conditions. A, mRNA expression levels of Onecut1 and MafA in pancreatic islets of diabetic C57B/KsJ-db/db (db/db) mice and normal C57B/KsJ-db/misty (db/m+) mice were quantified at the age of 16 weeks. The levels of mRNA were normalized with β-actin and shown as a change from db/m+ mice. Results are means ± S.D. *, p < 0.05. B, pancreatic sections from db/db and db/m+ mice at the age of 16 weeks were immunostained with insulin (blue), Onecut1 (red), and MafA (green) antibodies. To enhance MafA staining detected with goat MafA antibody, biotin and streptavidin binding was utilized in these figures. C, pancreatic sections from db/db and db/m+ mice at the age of 16 weeks were immunostained with insulin (blue), Foxa2 (red), and MafA (green) antibodies. Scale bars, 50 μm. D, 8-week-old db/db mice were divided into three groups. Each group was fed a diet containing no medicine or 0.02% pioglitazone (pio) (equivalent to 50.9 mg kg−1 day−1) or insulin glargine (gla) injected subcutaneously daily from 8 to 16 weeks of age. At 16 weeks of age, mRNA levels of Onecut1 (left) and MafA (right) in the islets from each group were quantified by real time PCR and normalized with that of β-actin and shown as a change from db/m+ mice. Results are means ± S.D. *, p < 0.05 versus untreated. (n = 4–6).
DISCUSSION
MafA plays a critical role in islet β cell function, especially in insulin biosynthesis and secretion. Consistent with its importance in islet β cells, MafA is expressed only in insulin-producing cells during pancreas development that are destined to populate the islet β cells, which is an expression pattern not found for any other characterized transcription factors. Such a specific expression pattern of MafA led to the idea that MafA gene expression is elaborately regulated by the transcription factors associated with development of the pancreas. Thus, in this study, we examined whether MafA gene expression is regulated by such transcription factors. Among many of these factors, we showed that Onecut1 directly suppresses transcription of the MafA gene. Our finding is consistent with the report using transgenic mice in which Onecut1 is overexpressed in postnatal islets, resulting in β cell dysfunction accompanied by suppressed expression of MafA (36). The physiological expression pattern of Onecut1, during pancreas development, also supports the association between MafA and Onecut1 as MafA expression was detected mostly in Onecut1 negative cells (Fig. 7). Considering that Onecut1 is essential for early endocrine differentiation, its rapid down-regulation is necessary for specification to the endocrine lineage (23, 24), and that MafA is a critical factor for the final differentiation of islet β cells (16), it is surmised that Onecut1 is required for inhibiting MafA gene expression to maintain a pool of immature endocrine cells until the proper developmental period. However, another critical insulin transcription factor, Pdx1, tends to be but is not significantly suppressed by Onecut1, although Pdx1 is one of the target genes of Foxa2. These differences between MafA and Pdx1 imply the existence of an elaborate mechanism on transcriptional regulation for each factor.
Intriguingly, we identified that Onecut1 affects MafA gene promoter activity through area A-2 located within area A, which was reported as a critical enhancer region to control whole MafA gene expression (29). Our detailed experiments indicate that there is no binding site of Onecut1 within area A-2, which is the critical enhancer region on the MafA gene, although Onecut1 inhibits MafA gene promoter activity through this area. As an important observation, area A-2 includes a Foxa2-binding consensus sequence (29), and adding a mutation into this consensus sequence results in a significant decrease of MafA gene promoter activity (Fig. 6A). These findings suggest that Onecut1 decreases the expression of MafA via inhibiting positive regulators that bind to area A-2, such as Foxa2, during embryonic stage and under diabetic conditions (Fig. 9). In contrast to Onecut1, ChIP analysis demonstrated that Foxa2 directly binds to area A (Fig. 4C), and an appropriate amount of Foxa2 appears to increase the transcription of the MafA gene, as shown in the results of Foxa2 knockdown analysis (Fig. 6B). Furthermore, the protein pulldown assay proved the binding of Onecut1 and Foxa2 in vivo. All of these results support the idea that Onecut1 decreases the activity of Foxa2, and consequently, MafA gene expression is suppressed by Onecut1. In fact, protein and the mRNA level of Foxa2 were suppressed under diabetic conditions (data not shown), but the decline of MafA mRNA was more remarkable (Fig. 8A). These results may suggest there is post-translational regulation of Foxa2 on MafA gene expression under diabetic conditions. Confusingly, the knockdown of Foxa2 in MIN6 cells decreased the expression of MafA (Fig. 6B), which is apparently the unexpected result from the decreased expression of MafA by overexpression of Foxa2 (Fig. 1). These results indicate the importance of maintaining appropriate amounts of Foxa2 to activate MafA gene promoter activity, and they may suggest the existence of another factor, which facilitates area A-2 in cooperation with Foxa2. Specifically, normal activation of Foxa2 can be interrupted by the excessive expression of Foxa2. DNA-dependent protein kinase, which has been extensively characterized as a key participant in DNA repair pathways, is reported to interact with Foxa2 and positively modulate its transcriptional potential (37). Although our ChIP analysis using a DNA-dependent protein kinase antibody failed to prove that DNA-dependent protein kinase and Foxa2 form a complex on area A (data not shown), it is still plausible that the biphasic function of Foxa2 on MafA is due to the existence of Foxa2 regulators.
FIGURE 9.
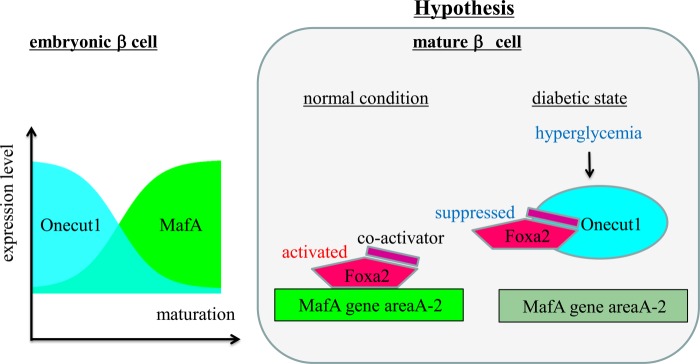
Converse expression pattern of Onecut1 to MafA and a possible explanation how Onecut1 suppresses the gene expression of MafA. The expression pattern of Onecut1 is contrary to that of MafA in embryonic pancreas and mature islets. We showed that Onecut1 decreases the expression of MafA via inhibiting positive regulators that bind to area A-2, such as Foxa2. Although the mechanism for biphasic function of Foxa2 on MafA remains to be elucidated, it might be explained by the existence of co-activator of Foxa2. Excessive amount of Foxa2 may occupy the co-activator and inhibit the adequate activation of Foxa2 on MafA gene area A-2.
The expression level of MafA in islet β cells is markedly decreased under diabetic conditions, and improved glucose tolerance with some anti-diabetic medications preserved the expression of MafA and β cell function in diabetic mice (35). These findings suggest the pathophysiological importance of MafA under diabetic conditions, as it is critical for β cell function. In this study, we showed that Onecut1, which is not or is negligibly expressed in mature islet β cells, is increased markedly in those cells of diabetic db/db mice. Furthermore, improved glycemic control attenuated the appearance of Onecut1, which is opposite MafA (Fig. 8D). These results may provide evidence that Onecut1 suppresses the expression of MafA not only in the developing pancreas but also in mature islets under specific conditions such as diabetes. Although it still remains to be uncovered as to how the expression of Onecut1 is induced under diabetic conditions, examining its function in islet β cells may provide new insights for understanding β cell dysfunction.
In conclusion, this study implicates a novel role of Onecut1 in the control of normal β cell differentiation and its involvement in β cell dysfunction under diabetic conditions by suppressing MafA gene expression.
Acknowledgments
We thank Chikayo Yokogawa and Miku Miyawaki for secretarial assistance.
Footnotes
This work was supported by Juvenile Diabetes Research Foundation Career Development Award 2-2005-946 (to T. M.) and KAKENHI Grant 10379258 (to T. M.).
REFERENCES
- 1. Edlund T., Walker M. D., Barr P. J., Rutter W. J. (1985) Cell-specific expression of the rat insulin gene: evidence for role of two distinct 5′-flanking elements. Science 230, 912–916 [DOI] [PubMed] [Google Scholar]
- 2. Karlsson O., Edlund T., Moss J. B., Rutter W. J., Walker M. D. (1987) A mutational analysis of the insulin gene transcription control region: expression in beta cells is dependent on two related sequences within the enhancer. Proc. Natl. Acad. Sci. U.S.A. 84, 8819–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crowe D. T., Tsai M.-J. (1989) Mutagenesis of the rat insulin II 5′-flanking region defines sequences important for expression in HIT cells. Mol. Cell. Biol. 9, 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whelan J., Poon D., Weil P. A., Stein R. (1989) Pancreatic beta-cell-type-specific expression of the rat insulin II gene is controlled by positive and negative cellular transcriptional elements. Mol. Cell. Biol. 9, 3253–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohlsson H., Karlsson K., Edlund T. (1993) IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12, 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peshavaria M., Gamer L., Henderson E., Teitelman G., Wright C. V., Stein R. (1994) XIHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol. Endocrinol. 8, 806–816 [DOI] [PubMed] [Google Scholar]
- 7. Petersen H. V., Serup P., Leonard J., Michelsen B. K., Madsen O. D. (1994) Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc. Natl. Acad. Sci. U.S.A. 91, 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olbrot M., Rud J., Moss L. G., Sharma A. (2002) Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. U.S.A. 99, 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kataoka K., Han S. I., Shioda S., Hirai M., Nishizawa M., Handa H. (2002) MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 277, 49903–49910 [DOI] [PubMed] [Google Scholar]
- 10. Matsuoka T. A., Zhao L., Artner I., Jarrett H. W., Friedman D., Means A., Stein R. (2003) Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol. Cell. Biol. 23, 6049–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuoka T. A., Artner I., Henderson E., Means A., Sander M., Stein R. (2004) The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. U.S.A. 101, 2930–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 13. Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., Wright C. V. (1996) PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122, 983–995 [DOI] [PubMed] [Google Scholar]
- 14. Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., Kudo T., Engel J. D., Yamamoto M., Takahashi S. (2005) MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25, 4969–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naya F. J., Stellrecht C. M., Tsai M. J. (1995) Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 9, 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishimura W., Bonner-Weir S., Sharma A. (2009) Expression of MafA in pancreatic progenitors is detrimental for pancreatic development. Dev. Biol. 333, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landry C., Clotman F., Hioki T., Oda H., Picard J. J., Lemaigre F. P., Rousseau G. G. (1997) HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver-enriched transcription factors. Dev. Biol. 192, 247–257 [DOI] [PubMed] [Google Scholar]
- 18. Rausa F., Samadani U., Ye H., Lim L., Fletcher C. F., Jenkins N. A., Copeland N. G., Costa R. H. (1997) The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3β in the developing murine liver and pancreas. Dev. Biol. 192, 228–246 [DOI] [PubMed] [Google Scholar]
- 19. Jacquemin P., Durviaux S. M., Jensen J., Godfraind C., Gradwohl G., Guillemot F., Madsen O. D., Carmeliet P., Dewerchin M., Collen D., Rousseau G. G., Lemaigre F. P. (2000) Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 20, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gradwohl G., Dierich A., LeMeur M., Guillemot F. (2000) Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 97, 1607–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Villasenor A., Chong D. C., Cleaver O. (2008) Biphasic Ngn3 expression in the developing pancreas. Dev. Dyn. 237, 3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacquemin P., Lemaigre F. P., Rousseau G. G. (2003) The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev. Biol. 258, 105–116 [DOI] [PubMed] [Google Scholar]
- 23. Zhang H., Ables E. T., Pope C. F., Washington M. K., Hipkens S., Means A. L., Path G., Seufert J., Costa R. H., Leiter A. B., Magnuson M. A., Gannon M. (2009) Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech. Dev. 126, 958–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tweedie E., Artner I., Crawford L., Poffenberger G., Thorens B., Stein R., Powers A. C., Gannon M. (2006) Maintenance of hepatic nuclear factor 6 in postnatal islets impairs terminal differentiation and function of beta-cells. Diabetes 55, 3264–3270 [DOI] [PubMed] [Google Scholar]
- 25. Ang S. L., Rossant J. (1994) HNF-3β is essential for node and notochord formation in mouse development. Cell 78, 561–574 [DOI] [PubMed] [Google Scholar]
- 26. Monaghan A. P., Kaestner K. H., Grau E., Schütz G. (1993) Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm, and neuroectoderm. Development 119, 567–578 [DOI] [PubMed] [Google Scholar]
- 27. Wu K. L., Gannon M., Peshavaria M., Offield M. F., Henderson E., Ray M., Marks A., Gamer L. W., Wright C. V., Stein R. (1997) Hepatocyte nuclear factor 3β is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 17, 6002–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerrish K., Van Velkinburgh J. C., Stein R. (2004) Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3β sites. Mol. Endocrinol. 18, 533–54814701942 [Google Scholar]
- 29. Raum J. C., Gerrish K., Artner I., Henderson E., Guo M., Sussel L., Schisler J. C., Newgard C. B., Stein R. (2006) FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific MafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol. Cell. Biol. 26, 5735–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuoka T. A., Kaneto H., Stein R., Miyatsuka T., Kawamori D., Henderson E., Kojima I., Matsuhisa M., Hori M., Yamasaki Y. (2007) MafA regulates expression of genes important to islet beta-cell function. Mol. Endocrinol. 21, 2764–2774 [DOI] [PubMed] [Google Scholar]
- 31. Kaneto H., Kajimoto Y., Miyagawa J., Matsuoka T., Fujitani Y., Umayahara Y., Hanafusa T., Matsuzawa Y., Yamasaki Y., Hori M. (1999) Beneficial effects of antioxidants in diabetes: possible protection of pancreatic beta-cells against glucose toxicity. Diabetes 48, 2398–2406 [DOI] [PubMed] [Google Scholar]
- 32. Lee T. I., Johnstone S. E., Young R. A. (2006) Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaneto H., Matsuoka T. A., Nakatani Y., Miyatsuka T., Matsuhisa M., Hori M., Yamasaki Y. (2005) A crucial role of MafA as a novel therapeutic target for diabetes. J. Biol. Chem. 280, 15047–15052 [DOI] [PubMed] [Google Scholar]
- 34. Matsuoka T. A., Kaneto H., Miyatsuka T., Yamamoto T., Yamamoto K., Kato K., Shimomura I., Stein R., Matsuhisa M. (2010) Regulation of MafA expression in pancreatic beta-cells in db/db mice with diabetes. Diabetes 59, 1709–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawashima S., Matsuoka T. A., Kaneto H., Tochino Y., Kato K., Yamamoto K., Yamamoto T., Matsuhisa M., Shimomura I. (2011) Effect of alogliptin, pioglitazone, and glargine on pancreatic β-cells in diabetic db/db mice. Biochem. Biophys. Res. Commun. 404, 534–540 [DOI] [PubMed] [Google Scholar]
- 36. Gannon M., Ray M. K., Van Zee K., Rausa F., Costa R. H., Wright C. V. (2000) Persistent expression of HNF6 in islet endocrine cells causes disrupted islet architecture and loss of beta cell function. Development 127, 2883–2895 [DOI] [PubMed] [Google Scholar]
- 37. Nock A., Ascano J. M., Jones T., Barrero M. J., Sugiyama N., Tomita M., Ishihama Y., Malik S. (2009) Identification of DNA-dependent protein kinase as a cofactor for the forkhead transcription factor FoxA2. J. Biol. Chem. 284, 19915–19926 [DOI] [PMC free article] [PubMed] [Google Scholar]