Background: Forkhead box O (FOXO) transcription factors affect life span and age-related diseases.
Results: Evolutionarily conserved role for RALA in stress-induced assembly of a JIP1 scaffold complex to ensure FOXO activity.
Conclusion: RALA regulates formation of a JIP1 scaffold complex to propagate JNK signaling toward FOXO4 in response to ROS.
Significance: JIP1 is important for ROS-induced signaling from RALA to FOXO.
Keywords: C. elegans, MAP Kinases (MAPKs), Reactive Oxygen Species (ROS), Scaffold Proteins, Signal Transduction, Small GTPases, Transcription Factors
Abstract
FOXO (forkhead box O) transcription factors are tumor suppressors and increase the life spans of model organisms. Cellular stress, in particular oxidative stress caused by an increase in levels of reactive oxygen species (ROS), activates FOXOs through JNK-mediated phosphorylation. Importantly, JNK regulation of FOXO is evolutionarily conserved. Here we identified the pathway that mediates ROS-induced JNK-dependent FOXO regulation. Following increased ROS, RALA is activated by the exchange factor RLF (RalGDS-like factor), which is in complex with JIP1 (C-Jun-amino-terminal-interacting protein 1) and JNK. Active RALA consequently regulates assembly and activation of MLK3, MKK4, and JNK onto the JIP1 scaffold. Furthermore, regulation of FOXO by RALA and JIP1 is conserved in C. elegans, where both ral-1 and jip-1 depletion impairs heat shock-induced nuclear translocation of the FOXO orthologue DAF16.
Introduction
The transcription factors of the forkhead box O (FOXO) class are involved in a variety of cellular processes like cell cycle regulation, apoptosis, and metabolism (reviewed in Refs. 1 and 2). In model organisms, FOXO activity affects life span as well as age-related diseases like cancer and diabetes (reviewed in Ref. 3). Multiple upstream pathways regulate FOXO activity through post-translational modifications that influence nucleocytoplasmic shuttling of FOXO (4). FOXOs are negatively regulated by the PI3K/PKB (or AKT) pathway. Activation of PI3K/PKB induces phosphorylation and nuclear exclusion of FOXO, thereby inhibiting FOXO transcriptional activity. In addition, signaling induced by ROS2 relocates FOXO to the nucleus and thereby potentiates its activation (reviewed in Ref. 4). Previously, we described the importance of JNK-dependent phosphorylation of FOXOs in this response toward ROS (5). Both the insulin/PI3K/PKB signaling and ROS/JNK signaling pathways toward FOXO have been conserved through evolution in species as diverse as Caenorhabditis elegans (6), Drosophila melanogaster (7), and Hydra vulgaris (8). In agreement, JNK activity extends life span in C. elegans (6) and D. melanogaster (7), and this requires the FOXO homologue DAF-16.
The JNK signaling pathway is regulated in time and space by the formation of signaling modules. These signaling modules typically consist of a scaffold protein that interacts with proteins that relay the extracellular signal from kinase to kinase, ultimately resulting in phosphorylation of JNK (reviewed in Refs. 9 and 10). Examples of scaffold proteins that are able to interact with members of the JNK pathway are Filamin (11), β-Arrestin (12), p130Cas (13), and JIP1, 2, 3, and 4 (e.g., as reviewed in Ref. 10). Several extracellular signals can regulate the assembly of different scaffold complexes. For example, the assembly of the Filamin signaling complex containing TRAF2 and MKK4 is induced by TNFα (11), and in hippocampal neurons, it has been shown that JIP1 is required for stress-induced JNK activation (14). Whereas a scaffolding molecule ensures specificity in terms of the use of upstream regulators of JNK, a recruitment signal is often needed to specify the location of the signaling complex. Small GTPases are known to function as a recruitment signal. For example, Ras-GTP induces dephosphorylation of the MAPK scaffold KSR (kinase suppressor of Ras-1), which results in its translocation from the cytosol to the membrane. There it associates with Raf and downstream effector proteins, such as MEK and ERK, facilitating sequential kinase reactions (15). RAL is a Ras-like small GTPase that cycles between an active GTP-bound and an inactive GDP-bound state. The two RAL isoforms, RALA and RALB, share 85% sequence homology. Several signals result in RAL activation, including growth factor stimulation (EGF and insulin) (16) and Ca2+ mobilization (17). RAL is involved in several physiological and pathophysiological cellular processes, such as exocytosis (18), endocytosis (19, 20), cellular transformation (21–24), and transcriptional regulation (25–27). RAL regulates activation of several transcription factors including c-Jun (25), NF-κB (28), Zonab (29), and FOXO4 (26); however, the molecular mechanism whereby RAL regulates transcriptional activity is largely unexplored. Overexpression of a constitutive active RAL exchange factor or increased ROS levels result in JNK-dependent phosphorylation and activation of FOXO4 (5). This suggests a pathway whereby RALA regulates JNK to modulate FOXO transcriptional activity. However, how RALA mediates specificity toward JNK-mediated FOXO regulation is poorly understood. Because small GTPases are reported to recruit signaling complexes to confer specificity to signaling, we here addressed the role of JNK scaffold proteins in the regulation of JNK by RALA and the signaling toward FOXO4. We show that activation of RALA results in the assembly of an active JIP1 scaffold complex consisting of MLK3, MKK4, and JNK. Furthermore, formation of this complex by RALA is necessary for proper JNK signaling toward FOXO4 and subsequent FOXO4 transcriptional activity. Finally, we show the conservation of RAL/JIP1-dependent FOXO regulation in C. elegans.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HEK293T and NIH3T3 cells were maintained in DMEM (Cambrex Bioscience, Verviers, Belgium), supplemented with 10% fetal bovine serum, Penicillin/streptomycin, and 0.05% l-glutamine. The cells were transfected using FuGENE 6 (Roche Applied Science), according to the manufacturer's instructions. The experiments were carried out 48 h after transfection. siRNAs for Scrambled, RALA, MLK3, or JIP1 were transfected using either Oligofectamine (Invitrogen) or INTERFERin (Polyplus Transfection) according to the manufacturer's instructions; experiments were analyzed 72 h after transfection. The cells were treated with 200 μm H2O2 or 5 μg/μl anisomycin for the indicated time points or left untreated.
Plasmids and siRNA Oligonucleotides
MKK4 and dnMKK4 in pCMV-FLAG were kindly provided by Dr. T. Katada, University of Tokyo (30, 31). JIP1 in pCDNA3-HA was kindly provided by Dr. T. Tsuzuki. pCDNA3-MLK3 was kindly provided by J. Gutkind (32). HA-MLK3 mutants were obtained by conventional cloning; more information is available upon request. The following plasmids have been described before: JNK1 in pMT2-HA (33), RALA, RLF, RFLCAAX, RLFΔcatCAAX (deletion of amino acids 212–327) in pMT2-HA (34), RALA in pCDNA3-Myc, GLOFLAG3-FLAG-FOXO4 (35), 6× FOXO DNA-binding element (DBE) firefly luciferase, and TK-Renilla-luciferase (36). All of the siRNAs used were synthesized by Dharmacon. The target sequence for the scrambled siRNA is 5′-UAGCGACUAAACACAUCAAUU-3′. The target sequences for RALA (37) and MLK3 (38) were described previously and for knockdown of JIP1 a predesigned ON-TARGET plus SMARTpool was used.
Antibodies
The following antibodies were purchased: phospho-JNK (Thr-183/Tyr-185), phospho-cjun (Ser-63), phospho-MKK4 (Ser-257/Thr-261), phospho-MLK3 (Thr-277/Ser-281), RALA and MLK3 antibodies from Cell Signaling, anti-JIP1 (Santa Cruz, B-7), anti-JNK1 (BIOSOURCE), anti-α-tubulin (Calbiochem), and anti-FLAG-M2 (Sigma). Anti-HA (12CA5), anti-Myc (9E10), and anti-phospho-FOXO4 (Thr-223/Ser-226) were described before (39, 40).
Immunoprecipitation and Western Blot
For immunoprecipitation, the cells were lysed in buffer containing 50 mm Tris-HCl (pH 7.5), 1% Nonidet P-40, 5 mm EDTA, 100 mm NaCl, protease, and phosphatase inhibitors. The cell lysates were centrifuged for 10 min at 14,000 rpm at 4 °C, and 5% of the supernatant was used as input material. Immunoprecipitation of the proteins of interest was done with protein agarose beads, coupled to the protein or tag specific antibody, for 2 h at 4 °C. The beads were washed with lysis buffer and resuspended in Laemmli sample buffer. The samples were separated on SDS-polyacrylamide gels and transferred to PVDF membrane. Western blot analysis was performed on an Odyssey scanner (LI-COR Biosciences) using fluorescently labeled secondary antibodies (see Fig. 5A, FLAG blot) or by enhanced chemiluminescence (all other figures). Experiments were performed at least two to five times, of which a representative figure is shown. The blots were quantified with ImageJ (see Fig. 2, E and F).
FIGURE 5.
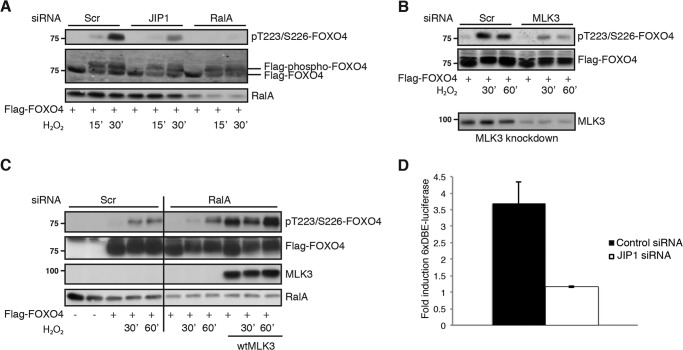
The assembly of a functional JIP1 scaffold complex by RALA ensures proper JNK signaling toward FOXO4. A, knockdown of RALA or JIP1 inhibits FOXO4 phosphorylation on JNK sites (Thr-223/Ser-226). FLAG-FOXO4 was immunoprecipitated from NIH3T3 cells with anti-FLAG antibody, and Western blots were analyzed for phosphorylation of Thr-223/Ser-226 on FOXO4, FLAG (total FOXO4), and RALA. B, MLK3 knockdown inhibits ROS-induced FOXO4 phosphorylation on JNK sites. The experiment was performed as described for A. Western blots were analyzed for phosphorylation of Thr-223/Ser-226 on FOXO4 and FLAG (total FOXO4). Efficiency of MLK3 siRNA is shown below on overexpressed MLK3, because of the bad quality of MLK3 antibody for detecting endogenous MLK3. C, knockdown of RALA inhibits JNK-induced FOXO4 phosphorylation and MLK3 overexpression rescues this inhibition. Cell lysates were examined for MLK3, RALA, and phosphorylation of FOXO4 on the JNK sites (Thr-223/Ser-226), total FOXO4 levels remained equal. wtMLK3, wild type MLK3. D, JIP1 knockdown inhibits FOXO4 transcriptional activity. NIH3T3 cells were transfected with FLAG-FOXO4, tk-Renilla-luciferase, 6×DBE luciferase, and siRNA against JIP1 or a scrambled control sequence. The graph shows fold induction of FOXO4 over GFP. The data are represented as means ± S.E. Scr, Scrambled.
FIGURE 2.
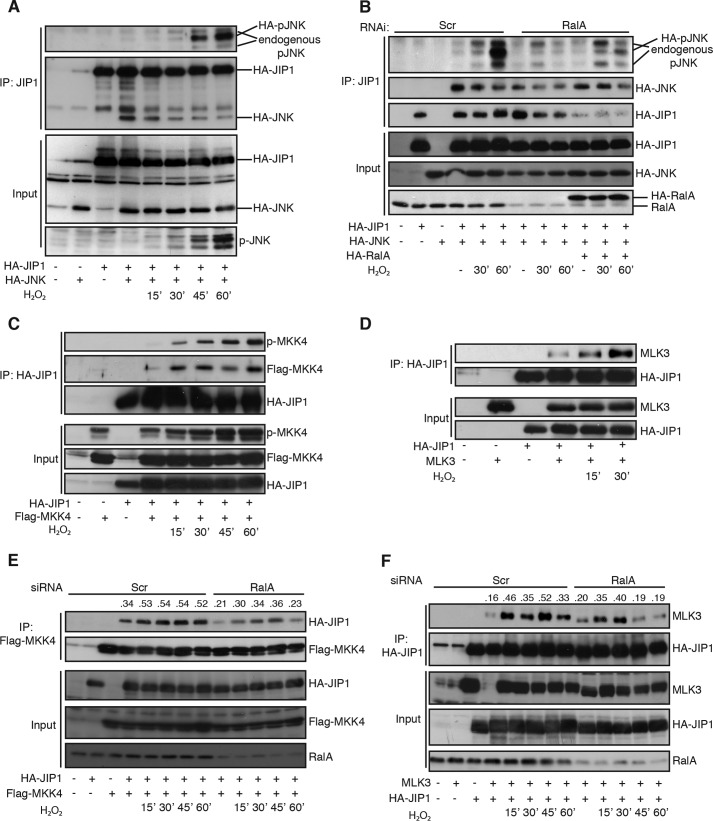
RALA regulates assembly of a functional JIP1 scaffold complex consisting of MLK3-MKK4 and JNK. A, ROS-induced activation of co-immunoprecipitated JNK with JIP1. HA-JIP1 was immunoprecipitated with an anti-JIP1 antibody. Western blots were probed for pJNK (Thr-183/Tyr-185) and HA (JNK and JIP1). B, ectopically expressed RALA rescues inhibition of JNK activation on JIP1 upon RALA knockdown. HA-JIP1 was immunoprecipitated with anti-JIP1 antibody, and Western blots were analyzed for pJNK (Thr-183/Tyr-185), HA (JNK, RALA, and JIP1), and endogenous RALA. C, ROS-induced binding and activation of MKK4 on JIP1. The experiment was carried out as in Fig. 2A. D, ROS-induced interaction of MLK3 to JIP1. The experiment was performed as described for A. E and F, RALA mediates ROS-induced interaction of MKK4 (E) and MLK3 (F) to JIP1. FLAG-MKK4 (E) or HA-JIP1 (F) was immunoprecipitated with anti-FLAG or HA antibody, and Western blots were analyzed for pJNK (Thr-183/Tyr-185), HA (JIP1), FLAG (MKK4), MLK3, and RALA. The numbers indicate ratio of bound JIP1 over immunoprecipitated MKK4 (E) and bound MLK3 over immunoprecipitated JIP1 (F). Scr, Scrambled; IP, immunoprecipitation.
GST Pulldown
Purification of RAL and loading with GppNHp (a nonhydrolyzable GTP) or GDP was done as described previously for Rap (41). Recombinant GST-RALA-GppNHp and GST-RALA-GDP were precoupled to glutathione-agarose beads for 10 min at 4 °C in GST buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, 5% glycerol, and 5 mm DTT) and washed three times with RAL buffer (10% glycerol, 1% Nonidet P-40, 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 2.5 mm MgCl2, protease, and phosphatase inhibitors). The cells were lysed in RAL buffer and centrifuged for 10 min at 4 °C, and lysates containing HA-JIP1 or MLK3 were incubated with the precoupled beads for 5 min at 4 °C. The beads were washed with RAL buffer and resuspended in Laemmli sample buffer, followed by Western blotting.
Luciferase Reporter Assay
Luciferase-based measurement of FOXO activity was performed as described before (5). Briefly, NIH3T3 cells were transfected with siRNA against JIP1 or scrambled siRNA and the 6xDBE-luciferase reporter construct, together with TK-Renilla-luciferase as internal control for transfection. Luciferase activity was analyzed with a dual luciferase assay system (MicroLumat Plus LB 96V; Berthold Technologies), according to the manufacturer's instructions.
Quantitative RT-PCR
RNA isolation from NIH3T3 cells was performed using the RNeasy mini kit from Qiagen, and cDNA was created using iScript cDNA synthesis kit from Bio-Rad. IQ SybrGreen (Bio-Rad) real time quantitative PCR was performed according to the manufacturer's instructions. The relative abundance JIP1 mRNA was corrected using PGBD mRNA. The following primers were designed to detect JIP1 mRNA: forward primer, 5′-CGTTCCTCCAGTGCTGAGTC-3′; and reverse primer, 5′-CCGAGGCACAAACCTGAATA-3′. For detection of PBGD mRNA (used as housekeeping gene), the following primers were designed: forward primer, 5′-GCCTACCATACTACCTCCTGGCT-3′; and reverse primer, 5′-AAGACAACAGCATCACAAGGGTT-3′.
C. elegans
TJ356 (Pdaf-16::daf-16-gfp; rol-6) (42) and VC20318 (nonsense mutation in jip-1 coding exon) strains were obtained from the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (Grant P40 OD010440) and maintained under standard conditions. RNAi against ral-1 was kindly provided by F. Zwartkruis and is described previously (43). TJ356 was crossed with VC20318 to obtain mutant jip-1 DAF16::GFP strains. We obtained multiple positive lines, of which we used three for the translocation experiments. Crossings were checked for mutation in the jip-1 gene by PCR and subsequent sequencing with the following primers: forward primer, 5′-ATCAGCGGAAAATCCAGAAA-3′; and reverse primer, 5′-CAATGGAGTCCTCGTCGATT-3′. TJ356 and jip-1 mutant worms were synchronized by bleaching, and L1 larvae were transferred to RNAi plates containing Escherichia coli expressing either control RNAi (L4440) or ral-1 RNAi (43) or standard agar plates with E. coli. Worms were grown until they reached adulthood (indicated by presence of eggs on the plates) and subsequently exposed to heat shock (33 °C). Translocation of DAF16::GFP was scored double blind to be nuclear, cytoplasmic or intermediate at least every hour in at least 25 worms per condition using a NIKON SMZ1500 dissection microscope equipped with an epifluorescence setup. Kinase assays using worm lysates were performed as follows: protein lysates of at least 40 worms per condition were incubated with GST-Jun (44) for 30 min following affinity purification of GST-Jun using glutathione beads. After washing three times with RAL buffer without MgCl2 (10% glycerol, 1% Nonidet P-40, 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, protease and phosphatase inhibitors) and two times with kinase buffer without ATP (25 mm Tri-HCl, pH 7.5, 20 mm MgCl2, 2 mm DTT), the beads were incubated with kinase buffer (25 mm Tri-HCl, pH 7.5, 20 mm MgCl2, 2 mm DTT, 100 μm ATP) for 30 min at 30 °C. The kinase reaction was stopped by adding 5× concentrated Laemmli sample buffer and heated for 5 min at 95 °C. The samples were analyzed by SDS-PAGE, followed by autoradiography. Subsequently, the Western blots were analyzed with a phospho-c-Jun (Ser-63) antibody.
RESULTS
The Scaffold Protein JIP1 Is Important for RALA-induced JNK Activation
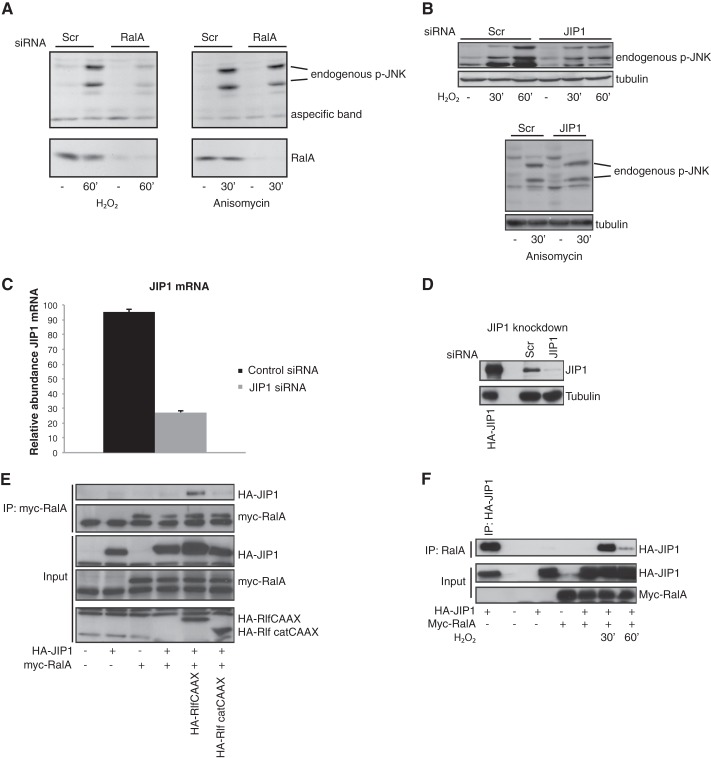
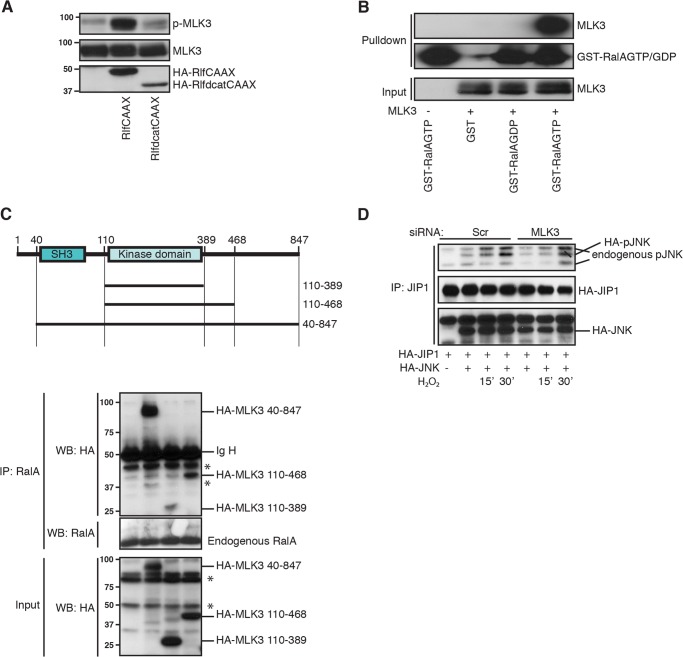
Scaffold proteins are important in the assembly of a functional JNK module and can be regulated by small GTPases. Therefore we examined whether scaffold proteins can provide the cross-talk between RALA and JNK. We concentrated on JIP1 (JNK interacting protein 1), because JIP1 has been described to selectively bind to JNK and not to other MAP kinases like p38 and ERK (45), and we have no evidence that RALA participates in H2O2-induced p38 activation (data not shown and Ref. 5). Furthermore, JIP1 is important in ROS-induced JNK activation (14). Similar to RALA depletion (Fig. 1A), siRNA-mediated knockdown of JIP1 inhibited ROS-induced JNK activation (Fig. 1B). Both RALA and JIP1 are required specifically for ROS-induced JNK activation, because anisomycin-induced JNK activation was not affected by knockdown of RALA or JIP1 (Fig. 1, A and B). The efficiency of the JIP1 siRNA oligonucleotides in NIH3T3 cells was confirmed both by quantitative PCR and Western blot (Fig. 1, C and D). In agreement with the possibility that RALA regulates JIP1, we observe binding of RALA to JIP1. Importantly, this interaction depends on RALA activity. To activate RALA, we used expression of RLFCAAX, an active mutant of the RalGEF RLF, in which the Ras-binding domain of RLF has been replaced by a CAAX membrane localization motif, resulting in a constitutively active RalGEF (34). RLFCAAX expression induces binding of RALA to JIP1, whereas expression of the inactive mutant (RLFΔcatCAAX), carrying a deletion of the catalytic domain does not (Fig. 1E). In addition, H2O2 treatment of cells (which activates RALA as we previously published (5)) also induced the interaction of RALA and JIP1 (Fig. 1F). Hence, activation of RALA results in binding of RALA to the JIP1 complex.
FIGURE 1.
The small GTPase RALA regulates JNK activity via the scaffold protein JIP1. A, knockdown of RALA inhibits ROS-induced JNK activation but not anisomycin-induced JNK activation. Phosphorylation of JNK (Thr-183/Tyr-185) and knockdown of RALA are shown. B, knockdown of JIP1 results in inhibition of ROS-induced JNK activation but not of anisomycin-induced JNK activation. The experiment was carried out as described for A. C, knockdown of JIP1 was verified with quantitative PCR. Relative abundance of JIP1 over PBGD is shown. The data are represented as means ± S.E. D, and by Western blot (D). Overexpressed HA-JIP1 is shown in the first lane to control for the position of JIP1. E, JIP1 binds to RLFCAAX-induced active RALA. Myc-RALA was immunoprecipitated, and Western blots were analyzed for HA (JIP1, RLFCAAX, and RLFΔcatCAAX) and Myc (RALA). F, ROS-induced interaction of RALA with JIP1. Myc-RALA was immunoprecipitated, and Western blots were analyzed for HA (JIP1) and Myc (RALA). Scr, Scrambled; IP, immunoprecipitation.
ROS Induced Assembly of the JNK Signaling Cascade on the Scaffold Protein JIP1
Next we looked into the interaction between JNK and JIP1. As shown in Fig. 2A, the interaction of JNK with JIP1, as assessed by co-immunoprecipitation was already evident under basal conditions, and treatment of cells with H2O2 did not significantly affect binding of JNK to JIP1. However, increased ROS resulted in time-dependent activation of the JNK pool co-immunoprecipitated with JIP1 (Fig. 2A). Consequently, we investigated the role of RALA in activation of JNK on JIP1. Knockdown of endogenous RALA inhibits activation of JNK in complex with JIP1. In addition, we could rescue this inhibition by expression of HA-RALA (Fig. 2B). From these data we conclude that the JNK pool interacting with JIP1 is regulated by RALA, following increased ROS.
Scaffold proteins can direct activity of MAP kinases (like JNK) by assembly of the complete MAP kinase cascade complex. Herein, a MAP kinase kinase kinase (MAP(3)K) phosphorylates and activates a MAP kinase kinase (MAP(2)K). The activated MAP(2)K subsequently phosphorylates and activates a MAP kinase (MAPK) (reviewed in Refs. 9, 10, and 46). Components of the JNK cascade are MAP(3)Ks, such as members of the MEK kinase and mixed lineage kinases and MAP(2)K members like MKK4 and MKK7. To determine which MAP(2)K is important in signaling from RALA to JNK via JIP1, we analyzed binding of the MAP(2)K MKK4 to JIP1 following H2O2 treatment. Binding between JIP1 and MKK4 is rapidly induced upon H2O2 treatment (Fig. 2C). Furthermore, MKK4 activation (measured with a phospho-antibody against Ser-257/Thr-261) was also rapidly induced and increased over a longer time period compared with total MKK4. This suggests not only that H2O2 signaling mediates the binding of MKK4 to JIP1 but also results in activation of MKK4 over time (Fig. 2C). We verified the involvement of MKK4 in ROS-induced JNK activation by ectopic expression of either dominant-negative MKK4 (dnMKK4) or wild type MKK4 (wtMKK4). dnMKK4 was able to block the H2O2-induced JNK activation, whereas wtMKK4 could not (data not shown). Because both MKK4 and MKK7 are described to be important for full JNK activation (47, 48), and MKK7 is described to bind JIP1 (45), we tested the involvement of RALA in regulation of MKK7 binding to JIP1. In agreement with the literature, we found that MKK7 bound to JIP1 (data not shown); however, in contrast to MKK4, the interaction appeared not regulated by RALA. This does not exclude a role for MKK7, but at least suggests MKK4 to be the main mediator of ROS/RALA signaling toward JNK via the JIP1 scaffold complex.
Next, we tested MAP(3)Ks (MLK3 and ASK1), known to be important in JNK signaling, for interaction with JIP1 following increased ROS. As shown in Fig. 2D, we observed increased binding of MLK3 upon H2O2 treatment, suggesting that MLK3 might be important for regulation of RALA-mediated JNK activation. Taken together, upon increased ROS, a MAPK module consisting of MLK3, MKK4, and JNK is assembled onto the JIP1 scaffold protein and subsequently activated.
Subsequently we determined the role of RALA in the assembly and activation of the above outlined ROS-induced JIP1-JNK complex. As shown in Fig. 2B, knockdown of RALA had no effect on JNK binding to JIP1 but reduced the activation of JNK on JIP1. In contrast, upon RALA knockdown we observed a decrease in ROS-induced binding of MKK4 (Fig. 2E) and MLK3 (Fig. 2F) to JIP1. From these data we conclude that the small GTPase RALA regulates binding of both MLK3 and MKK4 to JIP1, which subsequently results in activation of JNK on JIP1. Hence, RALA-mediated JNK activation is accomplished through the regulation of a MLK3/MMK4/JNK cascade formed onto the JIP1 scaffold complex in response to ROS, Whether the regulation of this cascade proceeds through an orderly and sequential binding of the individual components or whether this involves the formation of a complex in which all components are bound simultaneously remains to be determined.
The RALGEF RLF Is in Complex with JIP1 to Regulate RALA-mediated JNK Activation
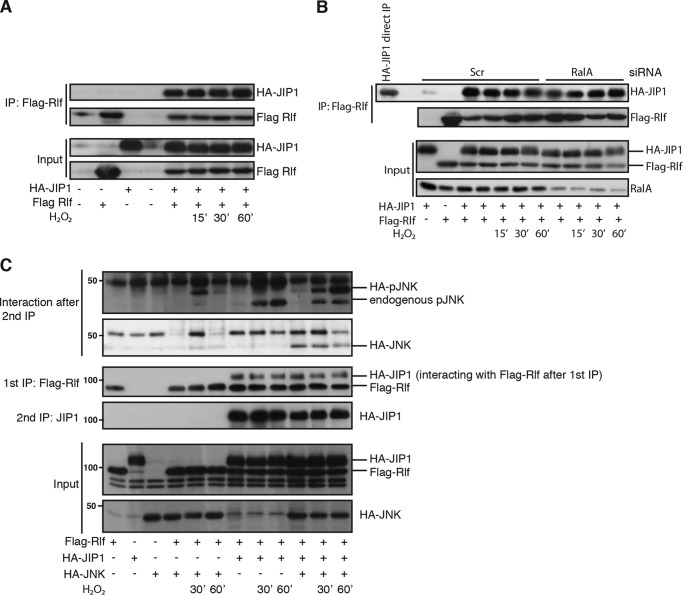
Activation of small GTPases such as RAL usually proceeds through activation of guanine nucleotide exchange factors (GEFs). We observed that assembly of an active JIP1-JNK complex following increased ROS requires RALA. Furthermore, RALA activation by ROS or expression of RLFCAAX induced the binding of RALA to JIP1. Therefore, we tested the involvement of the RALGEF, RLF (34), in mediating JIP1-JNK complex assembly induced by RALA. Indeed, RLF interacts with JIP1 however, similar to JNK, this interaction was independent of ROS (Fig. 3A). Because our experiments indicate that activated RALA binds to JIP1 and GEFs such as RLF bind to inactive GTPases, we tested whether the presence of RALA was required for the interaction of RLF to JIP1. Upon RALA knockdown, RLF is still bound to JIP1, indicating that this interaction, similar to JNK binding to JIP1, occurs independent of RALA (Fig. 3B). Simultaneous binding of RLF and JNK to JIP1 prior to RALA activation might allow rapid and directed activation of RALA toward JNK. To examine whether RLF is indeed present in one complex together with JIP1 and JNK, we performed a sequential pulldown. First FLAG-RLF was immunoprecipitated, and this precipitate of RLF and associated proteins were eluted from the beads and subsequently immunoprecipitated for JIP1. The JIP1 pulldown was assayed for the presence and activity of JNK. As shown in Fig. 3C, immunoprecipitation of RLF resulted in a pulldown of JNK bound to JIP1, and in addition we observed activation of JNK upon H2O2 treatment in this trimeric RLF-JIP1-JNK complex. Next to the binding of HA-pJNK to JIP1, we were also able to detect endogenous pJNK in the RLF-JIP1 complex (Fig. 3C).
FIGURE 3.
The RALA GEF RLF is in complex with JIP1 and mediates RALA-induced JNK activation. A, interaction of RLF with JIP1 is independent of ROS. FLAG-RLF was immunoprecipitated, and Western blots were analyzed for HA (JIP1) and FLAG (RLF). B, interaction of RLF with JIP1 is independent of RALA. The experiment was performed as described for A. C, RLF is in one complex with JIP1 and JNK. FLAG-RLF was immunoprecipitated with an anti-FLAG antibody and eluted from the beads with FLAG peptide. The eluate was subsequently used to immunoprecipitate HA-JIP1 with anti-JIP1 antibody. Western blots were analyzed for pJNK (Thr-183/Tyr-185), HA (JIP1 and JNK), and FLAG (RLF). Scr, Scrambled; IP, immunoprecipitation.
Activation of RAL Is Necessary to Activate MLK3
Small GTPases are well known to directly activate kinases; a typical example is the Ras-Raf pathway, where Ras activates Raf-1 (49–51). Therefore we tested whether RALA was able to directly activate MLK3 or whether this may require additional components. For this purpose, we analyzed activation of MLK3 using a phospho-specific MLK3 antibody, recognizing the phosphorylation sites in its activation loop (Thr-277/Ser-281). Ectopic expression of RLFCAAX, but not the inactive version RLFΔcatCAAX, was sufficient to induce autophosphorylation of MLK3 (Fig. 4A). Effector proteins of small GTPases such as Raf-1 bind preferentially to the active GTP-bound form of these GTPases (49). Therefore, we performed a pulldown with GST-RALA, loaded either with GDP or GTP, to see whether MLK3 prefers binding to either the GDP- or GTP-bound form of RALA. As shown in Fig. 4B, MLK3 preferentially binds RALGTP, suggesting that MLK3 is an effector protein of RALA. To map the region where RALA is binding to MLK3, we constructed several mutants (Fig. 4C, top panel). A mutant of MLK3 that contains only the kinase domain can still interact with endogenous RALA (Fig. 4C), suggesting that RALA interacts with the kinase domain of MLK3. Finally, we investigated the importance of MLK3 in the assembly of an active JIP1-JNK complex after increased ROS. Upon H2O2 treatment, JNK is activated on JIP1, and this activation is delayed in presence of MLK3 RNAi (Fig. 4D).
FIGURE 4.
MLK3 is important downstream of RALA in activation of the JIP1-JNK complex. A, activation of RALA results in phosphorylation of MLK3 in its activation loop. HEK293T cells were analyzed for phosphorylation on Thr-277/Ser-281. B, MLK3 specifically binds to GTP-bound RALA. HEK293T cells were transfected with the indicated constructs, and the GST pulldown was performed as described under “Experimental Procedures.” Western blots were probed for MLK3 and GST. C, RALA interacts with the kinase domain of MLK3. HA-tagged MLK3 mutants, as depicted in the upper panel, were overexpressed, and endogenous RALA was immunoprecipitated. Western blots were analyzed for HA and RALA. Asterisks indicate background bands. D, knockdown of MLK3 inhibits ROS-induced activation of JNK on JIP1. HA-JIP1 was immunoprecipitated with JIP1 antibody. Western blots were analyzed for pJNK (Thr-183/Tyr-185) and HA (JIP1 and JNK). Scr, Scrambled; IP, immunoprecipitation; WB, Western blot.
RALA-JIP1-JNK Complex Assembly Is Necessary for ROS-induced FOXO4 Regulation
As described previously, the forkhead box O transcription factor FOXO4 is phosphorylated and activated by JNK in a ROS- and RAL-dependent manner (5). To determine the relevance of the above described RALA-JIP1-JNK complex toward FOXO4, we performed knockdown of several components of the identified JNK signaling pathway and analyzed JNK-dependent phosphorylation of FOXO4 with a phosphospecific antibody against two of the JNK sites; phospho-FOXO4-Thr-223/Ser-226 (52). Knockdown of both RALA and JIP1 resulted in decreased H2O2-induced FOXO4 phosphorylation (Fig. 5A). In addition, knockdown of MLK3 also decreased FOXO4 phosphorylation (Fig. 5B). Furthermore, overexpression of MLK3 could rescue the decrease in FOXO4 phosphorylation after RALA RNAi (Fig. 5C), showing the ability of MLK3 to act downstream of RALA to regulate JNK-mediated FOXO4 activation. In addition, as expected from the effects on FOXO4 phosphorylation, JIP1 is also required for full FOXO4 activity, as shown by a luciferase reporter (Fig. 5D). FOXO4 overexpression results in increased transcriptional activity measured by 6×DBE-luciferase activity, whereas knockdown of JIP1 inhibits FOXO4 transcriptional activity.
Conserved Role for RALA and JIP1 in Stress-induced DAF16 Nuclear Localization
In both D. melanogaster and C. elegans, activation of JNK results in nuclear localization of dFOXO/DAF16 and increased life span (6, 7). Based on this notion, we questioned whether FOXO regulation by RALA observed in mammalian cells (5) is evolutionarily conserved in C. elegans. Therefore, we used a C. elegans strain expressing DAF-16::GFP (TJ356). This strain has been shown to respond to environmental stresses, such as heat shock, by displaying nuclear translocation of DAF-16::GFP (42). RNAi-mediated knockdown of ral-1 resulted in a delayed heat shock-induced DAF16 nuclear localization compared with control RNAi (L4440) (Fig. 6A). To confirm that in C. elegans heat shock regulates JNK activity and this may involve ral-1, we analyzed JNK activity by performing a GST-Jun pulldown kinase assay (44) on worm lysates subjected to heat shock with control or ral-1 RNAi. Heat shock indeed induces JNK activity (53), and importantly this induction occurred in a ral-1-dependent manner (Fig. 6B), confirming the role of ral-1 in regulation of JNK signaling upon heat shock. Because we demonstrate the importance of JIP1 in the assembly of a complete signaling complex to mediate JNK activity and regulation of FOXO4 activity in mammalian cells, we questioned whether JIP1-mediated FOXO regulation is also conserved. To this end we crossed the DAF16::GFP line with a jip-1 mutant line. Subsequently we tested three jip-1 mutant lines for heat shock-induced translocation of DAF16. All three jip-1 mutant strains showed delayed nuclear translocation upon heat shock, similar to what we observed with ral-1 siRNA. A representative example is shown in Fig. 6C. Apparently, the evolutionarily conserved regulation of FOXO by JNK observed in C. elegans proceeds through a signaling pathway that consists of the small GTPase RAL and the JNK scaffold JIP1. These data show not only that the well known regulation of FOXO by PI3K/PKB (AKT) signaling is evolutionarily conserved, but also that the RALA/JIP1/JNK axis in regulation of the FOXO transcription factor is conserved.
FIGURE 6.
Conserved regulation of FOXO by RALA/JIP1 in C. elegans. A, C. elegans ral-1 is important in stress-induced nuclear DAF16 translocation. TJ356 (DAF16::GFP) were synchronized and fed with control (L4440) RNAi or ral-1 RNAi. After they reached adulthood, the worms were exposed to heat stress (33 °C) and counted for the presence of nuclear, intermediate, or cytoplasmic DAF16::GFP at indicated time points. The upper panel shows representative pictures of cytoplasmic, intermediate, and nuclear DAF16 localization. Intermediate and nuclear translocation was pooled (and scored as nuclear) to perform Fisher's exact test. ***, p = 0.0002. B, heat shock-induced JNK activation is mediated by ral-1. TJ356 worms were exposed to heat stress and subsequently lysed to perform a GST-c-Jun pulldown. c-Jun phosphorylation was detected by both anti-phospho-c-Jun (Ser-63) and autoradiography (γ-32P); Coomassie Blue staining of the gel is shown as loading control. C, jip-1 is important for heat shock-induced nuclear DAF16 translocation. TJ356 and jip-1 mutant worms were synchronized, and after they reached adulthood, the worms were exposed to heat stress (33 °C) and counted for the presence of nuclear, intermediate, or cytoplasmic DAF16::GFP at the indicated time points. Intermediate and nuclear translocation was pooled (and scored as nuclear) to perform Fisher's exact test. p values are 0.0002 for 120 min and <0.0001 for 180 min of heat shock (according to Fisher's exact test). Cntrl, control.
DISCUSSION
Numerous signaling pathways have been shown to modulate FOXO activity. Two main pathways, the PI3K/PKB (AKT) pathway and JNK, appear evolutionarily conserved to tightly regulate FOXO activity. In the presence of growth factors, the insulin pathway through PI3K/PKB negatively regulates FOXOs, whereas FOXOs are activated by JNK following oxidative stress. Previously we identified a role for the small GTPase RALA in JNK regulation of FOXO4. However, how RALA regulates JNK-mediated FOXO activation was unclear; therefore we analyzed the pathway that specifies RALA induced JNK activation following increased cellular oxidative stress. We show that upon increased ROS, RALA mediates JIP1-dependent activation of JNK, which results in phosphorylation, and activation of FOXO4. RALA regulates the assembly of a MAPK cascade consisting of MLK3, MKK4, and JNK on the scaffold protein JIP1. Under basal conditions, JIP1 is in complex with RLF and JNK, possibly to allow rapid activation of JNK by RALA. Upon increased ROS, the small GTPase RALA is recruited to JIP1 and activated by RLF; this subsequently results in the recruitment and activation of MLK3. Because GTP-bound RALA, but not GDP-bound RALA, binds to MLK3, these results suggest MLK3 to be a potential novel RAL effector. Following MLK3 activation, MKK4 is recruited to JIP1 and activated; this subsequently results in phosphorylation and activation of JNK. Active JNK subsequently phosphorylates and thereby activates FOXO4 (depicted in the model in Fig. 7). Finally we show that RALA/JIP1-mediated FOXO regulation is conserved in C. elegans. Although it is not entirely clear how heat stress results in an increase in oxidizing conditions, it has been shown that heat stress results in JNK-dependent nuclear shuttling of DAF16 (6). We indeed observed that in C. elegans, heat shock-induced DAF-16 nuclear translocation as well as JNK activation was reduced following RNAi against ral-1. Combined, these results and our data clearly suggest RALA/JNK/DAF-16 regulation to be conserved.
FIGURE 7.
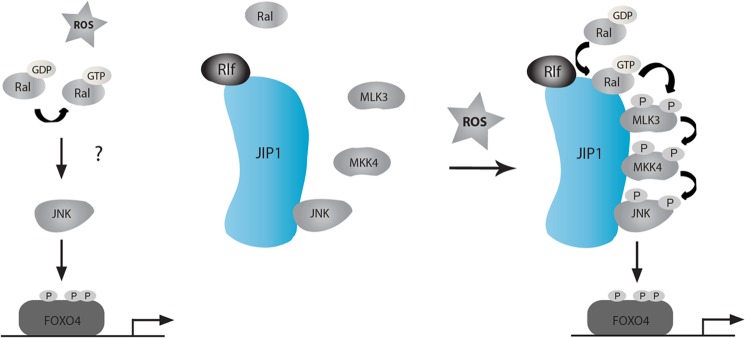
Model for RALA dependent FOXO4 regulation. Upon ROS signaling, RALA is activated by RLF on JIP1. Subsequently, the binding and activation of both MLK3 and MKK4 on JIP1 is mediated by RALA, which ultimately results in activation of JNK. Finally, activated JNK can phosphorylate and activate FOXO4. For further details see text.
How Does ROS Regulate the JIP1 Scaffold Complex?
We identified the RAL exchange factor RLF and RALA as the components most proximal to ROS. However, the question still remains how ROS activates RLF/RALA. It has been described that GTPases can be sensitive to cysteine oxidation and that this might regulate their activity (reviewed in Refs. 54 and 55). Redox-mediated cysteine modification has been suggested to collaborate with and/or enhance the action of guanine nucleotide exchange factors in the activation of small GTPases. Such a mechanism was proposed for Ras and Rho family members and could explain activation of RALA by ROS. Another possibility is that ROS induce modifications on either JIP1 or RLF, which results in relocalization of the complex to the membrane where RALA is present and becomes activated. Finally, it is possible that JIP1 binding to RLF inhibits RLF function and that ROS-induced modifications might relieve this inhibition. However, we tested whether the presence of recombinant JIP1 affected RLF exchange activity in vitro and did not observe a change in activity (data not shown). We are currently further exploring these possibilities.
In our study we identified MLK3 as the MAPKKK important for JIP1-mediated JNK activation following increase in ROS. Also for MLK3, redox-mediated cysteine oxidation has been described. Cysteine oxidation of MLK3 results in its dimerization and subsequent activation (56). However, we found that RALA can activate MLK3 independent of ROS (Fig. 4A). In agreement, others also demonstrated direct activation of MLK3 by small GTPases (Rac and CDC42) (32). This suggests that MLK3 can be activated by several mechanisms, either via cysteine oxidation or by binding of small GTPases. Whether MLK3, next to RALA-mediated activation, is also regulated by cysteine oxidation in the JIP1 scaffold complex, remains to be investigated.
Potential Mechanism of MLK3 Activation by RALA
As described by others, the small GTPase Cdc42 can activate MLK3 through binding and membrane targeting (57, 58). Binding of Cdc42 was mapped to the SH3 region of MLK3, and it was suggested that the binding of Cdc42GTP to MLK3 induces release of its SH3-mediated autoinhibition and subsequently promotes dimerization and (auto) phosphorylation within its activation loop. The mechanism whereby RALA mediates MLK3 activation differs from Cdc42, as we mapped the interaction of RALA to the kinase domain of MLK3. Furthermore, our results show that RALA, in addition to activation of MLK3, also regulates binding of MLK3 to JIP1. Thus, alternative to the mechanism of Cdc42 activation, RALA binding to the MLK3 kinase domain may facilitate binding of the SH3 domain of MLK3 to JIP1 and thereby relieve autoinhibition indirectly. This and other possibilities as to how RALA is able to regulate MLK3 activity by binding to its kinase domain are currently under investigation.
RALA in D. melanogaster and C. elegans
Whereas we demonstrate a clear role of RALA in the activation of JNK in both mammalian cells and the nematode C. elegans, others have shown in D. melanogaster that RALA plays an inhibitory role toward JNK (59, 60). An important difference between these developmental studies and ours is that we address a postdevelopmental role of RALA. Importantly, the outcome of JNK signaling is highly context-dependent, either resulting in pro-apoptotic or anti-apoptotic activity. It might be that during development, JNK activity is differently regulated compared with postdevelopment. In addition, whereas we used siRNA against RALA, these studies used a dominant-negative version of RAL (RALN28) to inhibit RAL signaling. Surprisingly, in D. melanogaster ectopic expression of dominant-negative RALN28 induces in part similar phenotypes as ectopic expression of dominant-active RALV20 (60). We indeed observe no difference between siRNA-mediated inhibition of RAL (Fig. 1A) and ectopic expression of RALN28 (5). However, we were not able to observe activation of JNK when expressing the dominant active version of mammalian RAL (RALV23). RALV23, being a GTP-bound form of RAL, does not bind to RALGEFs and therefore potentially does not bind to JIP1. We show here that RAL activation needs to occur at the JIP1 scaffold to activate JNK, and therefore RALV23 is probably unable to activate JNK.
At present little is known concerning the role of RAL in C. elegans development and/or adult life. A recent study indicates a role for RAL in determining vulval pattern formation during development, and this was attributed to a role for RAL in antagonizing RAS-RAF signaling (61). In addition, a synthetic lethal screen for loss of RAP-1 in C. elegans identified RAL as being essential for survival of RAP-1 null worms (43). In our study we describe an important role for RALA in mediating JNK signaling in C. elegans.
Activating versus Inhibiting Signals toward FOXO
JNK-mediated FOXO phosphorylation is emerging as an important positive regulatory pathway opposing the negative regulation by PI3K/PKB signaling. Interestingly, JIP1 has been described to play an important role in PKB regulation; however current literature is unclear, or maybe even contradictory, as to whether JIP1 acts as a negative or positive regulator of PKB (62–64). More importantly there is extensive cross-talk between JNK and PKB signaling. For example, PKB has been described to negatively regulate several components of the MAPK signaling complex described here (65–67). Whereas JNK is able to negatively regulate PKB, for example through direct phosphorylation and subsequent inhibition of IRS1 (68). Surprisingly, RALGDS, a RAL exchange factor similar to RLF, is found to recruit PDK1 to PKB via JIP1 and thereby enhances PDK1-mediated PKB phosphorylation and subsequent activation (69). However, this function of RALGDS does not require its ability to activate RAL. The interplay between PKB and JNK signaling is clearly complex and requires further detailed and directed studies. Nevertheless, it supports the idea that exchange factors for RAL such as RLF and RALGDS are upstream components of pathways leading to JNK versus PKB, respectively, and that JIP1 can be of importance in determining the control of negative and positive signaling pathways toward FOXO4.
The cellular response to ROS is tightly controlled and ranges from proliferation to cell cycle arrest and apoptosis, dependent on ROS level, cell type, and developmental stage. FOXO plays a central role in this response at the level of transcription and is regulated in multiple ways through ROS-mediated post-translational modifications (70). FOXO activity is probably the most important determinant of organismal life span, and this has been suggested to be dependent on its regulation of the cellular response to ROS (71). The pathway we describe here from ROS to FOXO activity illustrates how a specific ROS signal could be fine-tuned to carefully adjust FOXO activity and regulate the appropriate response. Like FOXO, JNK and recently also MKK4 (72) have been implicated in aging, which suggests that the RAL/RLF/JIP-1/MLK3/MKK4/JNK signaling cascade toward FOXO delineated here could be important for life span determination.
Acknowledgments
We thank Sanne Weijzen for initiating the project, Geert Kops for critically reading the manuscript, and Marije Rensen for technical assistance.
This work was supported by grants from the Dutch Cancer Society, the Center for Biomedical Genetics, and the Cancer Genomics Center.
- ROS
- reactive oxygen species
- GEF
- guanine nucleotide exchange factors
- RLF
- RalGDS-like factor
- KSR
- kinase suppressor of Ras-1.
REFERENCES
- 1. van der Horst A., Burgering B. (2007) Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 8, 440–450 [DOI] [PubMed] [Google Scholar]
- 2. Eijkelenboom A., Burgering B. M. (2013) FOXOs. Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14, 83–97 [DOI] [PubMed] [Google Scholar]
- 3. Dansen T. B., Burgering B. M. (2008) Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 [DOI] [PubMed] [Google Scholar]
- 4. Salih D. A., Brunet A. (2008) FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 20, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23, 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh S. W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R. J., Tissenbaum H. A. (2005) JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. U.S.A. 102, 4494–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang M. C., Bohmann D., Jasper H. (2005) JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121, 115–125 [DOI] [PubMed] [Google Scholar]
- 8. Bridge D., Theofiles A. G., Holler R. L., Marcinkevicius E., Steele R. E., Martínez D. E. (2010) FoxO and stress responses in the cnidarian Hydra vulgaris. PLoS ONE 5, e11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morrison D. K., Davis R. J. (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 10. Dhanasekaran D. N., Kashef K., Lee C. M., Xu H., Reddy E. P. (2007) Scaffold proteins of MAP-kinase modules. Oncogene 26, 3185–3202 [DOI] [PubMed] [Google Scholar]
- 11. Marti A., Luo Z., Cunningham C., Ohta Y., Hartwig J., Stossel T. P., Kyriakis J. M., Avruch J. (1997) Actin-binding protein-280 binds the stress-activated protein kinase (SAPK) activator SEK-1 and is required for tumor necrosis factor-α activation of SAPK in melanoma cells. J. Biol. Chem. 272, 2620–2628 [DOI] [PubMed] [Google Scholar]
- 12. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) β-Arrestin 2. A receptor-regulated MAPK scaffold for the activation of JNK3. Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 13. Girardin S. E., Yaniv M. (2001) A direct interaction between JNK1 and CrkII is critical for Rac1-induced JNK activation. EMBO J. 20, 3437–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whitmarsh A. J., Kuan C. Y., Kennedy N. J., Kelkar N., Haydar T. F., Mordes J. P., Appel M., Rossini A. A., Jones S. N., Flavell R. A., Rakic P., Davis R. J. (2001) Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev. 15, 2421–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 16. Wolthuis R. M., Zwartkruis F., Moen T. C., Bos J. L. (1998) Ras-dependent activation of the small GTPase Ral. Curr. Biol. 8, 471–474 [DOI] [PubMed] [Google Scholar]
- 17. Hofer F., Berdeaux R., Martin G. S. (1998) Ras-independent activation of Ral by a Ca(2+)-dependent pathway. Curr. Biol. 8, 839–842 [DOI] [PubMed] [Google Scholar]
- 18. Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. (2002) The exocyst is a Ral effector complex. Nat. Cell Biol. 4, 66–72 [DOI] [PubMed] [Google Scholar]
- 19. Nakashima S., Morinaka K., Koyama S., Ikeda M., Kishida M., Okawa K., Iwamatsu A., Kishida S., Kikuchi A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 18, 3629–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jullien-Flores V., Mahé Y., Mirey G., Leprince C., Meunier-Bisceuil B., Sorkin A., Camonis J. H. (2000) RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex. Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 113, 2837–2844 [DOI] [PubMed] [Google Scholar]
- 21. Urano T., Emkey R., Feig L. A. (1996) Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 15, 810–816 [PMC free article] [PubMed] [Google Scholar]
- 22. White M. A., Vale T., Camonis J. H., Schaefer E., Wigler M. H. (1996) A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J. Biol. Chem. 271, 16439–16442 [DOI] [PubMed] [Google Scholar]
- 23. Rangarajan A., Hong S. J., Gifford A., Weinberg R. A. (2004) Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6, 171–183 [DOI] [PubMed] [Google Scholar]
- 24. Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. (2005) Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7, 533–545 [DOI] [PubMed] [Google Scholar]
- 25. de Ruiter N. D., Wolthuis R. M., van Dam H., Burgering B. M., Bos J. L. (2000) Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol. Cell Biol. 20, 8480–8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Ruiter N. D., Burgering B. M., Bos J. L. (2001) Regulation of the forkhead transcription factor AFX by Ral-dependent phosphorylation of threonines 447 and 451. Mol. Cell Biol. 21, 8225–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ouwens D. M., de Ruiter N. D., van der Zon G. C., Carter A. P., Schouten J., van der Burgt C., Kooistra K., Bos J. L., Maassen J. A., van Dam H. (2002) Growth factors can activate ATF2 via a two-step mechanism. Phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 21, 3782–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henry D. O., Moskalenko S. A., Kaur K. J., Fu M., Pestell R. G., Camonis J. H., White M. A. (2000) Ral GTPases contribute to regulation of cyclin D1 through activation of NF-κB. Mol. Cell Biol. 20, 8084–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frankel P., Aronheim A., Kavanagh E., Balda M. S., Matter K., Bunney T. D., Marshall C. J. (2005) RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 24, 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kishimoto H., Nakagawa K., Watanabe T., Kitagawa D., Momose H., Seo J., Nishitai G., Shimizu N., Ohata S., Tanemura S., Asaka S., Goto T., Fukushi H., Yoshida H., Suzuki A., Sasaki T., Wada T., Penninger J. M., Nishina H., Katada T. (2003) Different properties of SEK1 and MKK7 in dual phosphorylation of stress-induced activated protein kinase SAPK/JNK in embryonic stem cells. J. Biol. Chem. 278, 16595–16601 [DOI] [PubMed] [Google Scholar]
- 31. Wada T., Nakagawa K., Watanabe T., Nishitai G., Seo J., Kishimoto H., Kitagawa D., Sasaki T., Penninger J. M., Nishina H., Katada T. (2001) Impaired synergistic activation of stress-activated protein kinase SAPK/JNK in mouse embryonic stem cells lacking SEK1/MKK4. Different contribution of SEK2/MKK7 isoforms to the synergistic activation. J. Biol. Chem. 276, 30892–30897 [DOI] [PubMed] [Google Scholar]
- 32. Teramoto H., Coso O. A., Miyata H., Igishi T., Miki T., Gutkind J. S. (1996) Signaling from the small GTP-binding proteins Rac1 and Cdc42 to the c-Jun N-terminal kinase/stress-activated protein kinase pathway. A role for mixed lineage kinase 3/protein-tyrosine kinase 1, a novel member of the mixed lineage kinase family. J. Biol. Chem. 271, 27225–27228 [DOI] [PubMed] [Google Scholar]
- 33. de Groot R. P., van Dijk T. B., Caldenhoven E., Coffer P. J., Raaijmakers J. A., Lammers J. W., Koenderman L. (1997) Activation of 12-O-tetradecanoylphorbol-13-acetate response element- and dyad symmetry element-dependent transcription by interleukin-5 is mediated by Jun N-terminal kinase/stress-activated protein kinase kinases. J. Biol. Chem. 272, 2319–2325 [DOI] [PubMed] [Google Scholar]
- 34. Wolthuis R. M., de Ruiter N. D., Cool R. H., Bos J. L. (1997) Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 16, 6748–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van der Horst A., Tertoolen L. G., de Vries-Smits L. M., Frye R. A., Medema R. H., Burgering B. M. (2004) FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 279, 28873–28879 [DOI] [PubMed] [Google Scholar]
- 36. Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W., Coffer P. J., Huang T. T., Bos J. L., Medema R. H., Burgering B. M. (2002) Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419, 316–321 [DOI] [PubMed] [Google Scholar]
- 37. Chien Y., White M. A. (2003) RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 4, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chadee D. N., Kyriakis J. M. (2004) MLK3 is required for mitogen activation of B-Raf, ERK and cell proliferation. Nat. Cell Biol. 6, 770–776 [DOI] [PubMed] [Google Scholar]
- 39. Medema R. H., Kops G. J., Bos J. L., Burgering B. M. (2000) AFX-like forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782–787 [DOI] [PubMed] [Google Scholar]
- 40. de Keizer P. L., Packer L. M., Szypowska A. A., Riedl-Polderman P. E., van den Broek N. J., de Bruin A., Dansen T. B., Marais R., Brenkman A. B., Burgering B. M. (2010) Activation of forkhead box O transcription factors by oncogenic BRAF promotes p21cip1-dependent senescence. Cancer Res. 70, 8526–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rehmann H. (2006) Characterization of the activation of the Rap-specific exchange factor Epac by cyclic nucleotides. Methods Enzymol. 407, 159–173 [DOI] [PubMed] [Google Scholar]
- 42. Henderson S. T., Johnson T. E. (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980 [DOI] [PubMed] [Google Scholar]
- 43. Frische E. W., Pellis-van Berkel W., van Haaften G., Cuppen E., Plasterk R. H., Tijsterman M., Bos J. L., Zwartkruis F. J. (2007) RAP-1 and the RAL-1/exocyst pathway coordinate hypodermal cell organization in Caenorhabditis elegans. EMBO J. 26, 5083–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hibi M., Lin A., Smeal T., Minden A., Karin M. (1993) Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7, 2135–2148 [DOI] [PubMed] [Google Scholar]
- 45. Whitmarsh A. J., Cavanagh J., Tournier C., Yasuda J., Davis R. J. (1998) A mammalian scaffold complex that selectively mediates MAP kinase activation. Science 281, 1671–1674 [DOI] [PubMed] [Google Scholar]
- 46. van Drogen F., Peter M. (2002) MAP kinase cascades. Scaffolding signal specificity. Curr. Biol. 12, R53–55 [DOI] [PubMed] [Google Scholar]
- 47. Lawler S., Fleming Y., Goedert M., Cohen P. (1998) Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr. Biol. 8, 1387–1390 [DOI] [PubMed] [Google Scholar]
- 48. Fleming Y., Armstrong C. G., Morrice N., Paterson A., Goedert M., Cohen P. (2000) Synergistic activation of stress-activated protein kinase 1/c-Jun N-terminal kinase (SAPK1/JNK) isoforms by mitogen-activated protein kinase kinase 4 (MKK4) and MKK7. Biochem. J. 352, 145–154 [PMC free article] [PubMed] [Google Scholar]
- 49. Koide H., Satoh T., Nakafuku M., Kaziro Y. (1993) GTP-dependent association of Raf-1 with Ha-Ras. Identification of Raf as a target downstream of Ras in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 90, 8683–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stokoe D., Macdonald S. G., Cadwallader K., Symons M., Hancock J. F. (1994) Activation of Raf as a result of recruitment to the plasma membrane. Science 264, 1463–1467 [DOI] [PubMed] [Google Scholar]
- 51. Leevers S. J., Paterson H. F., Marshall C. J. (1994) Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 369, 411–414 [DOI] [PubMed] [Google Scholar]
- 52. de Keizer P. L., Szypowska A. A., Riedl-Polderman P. E., van den Broek N. J., de Bruin A., Dansen T. B., Marais R., Brenkman A. B., Burgering B. M. (2010) Activation of FOXO transcription factors by oncogenic BRAF promotes p21cip1-dependent senescence. Cancer Res. 70, 8526–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Minden A., Lin A., Smeal T., Dérijard B., Cobb M., Davis R., Karin M. (1994) c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol. Cell Biol. 14, 6683–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitchell L., Hobbs G. A., Aghajanian A., Campbell S. L. (2013) Redox regulation of Ras and Rho GTPases. Mechanism and function. Antioxid. Redox Signal. 18, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heo J. (2011) Redox control of GTPases. From molecular mechanisms to functional significance in health and disease. Antioxid. Redox Signal. 14, 689–724 [DOI] [PubMed] [Google Scholar]
- 56. Hu S. Q., Ye J. S., Zong Y. Y., Sun C. C., Liu D. H., Wu Y. P., Song T., Zhang G. Y. (2012) S-Nitrosylation of mixed lineage kinase 3 contributes to its activation after cerebral ischemia. J. Biol. Chem. 287, 2364–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Du Y., Böck B. C., Schachter K. A., Chao M., Gallo K. A. (2005) Cdc42 induces activation loop phosphorylation and membrane targeting of mixed lineage kinase 3. J. Biol. Chem. 280, 42984–42993 [DOI] [PubMed] [Google Scholar]
- 58. Böck B. C., Vacratsis P. O., Qamirani E., Gallo K. A. (2000) Cdc42-induced activation of the mixed-lineage kinase SPRK in vivo. Requirement of the Cdc42/Rac interactive binding motif and changes in phosphorylation. J. Biol. Chem. 275, 14231–14241 [DOI] [PubMed] [Google Scholar]
- 59. Balakireva M., Rossé C., Langevin J., Chien Y. C., Gho M., Gonzy-Treboul G., Voegeling-Lemaire S., Aresta S., Lepesant J. A., Bellaiche Y., White M., Camonis J. (2006) The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol. Cell Biol. 26, 8953–8963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sawamoto K., Winge P., Koyama S., Hirota Y., Yamada C., Miyao S., Yoshikawa S., Jin M. H., Kikuchi A., Okano H. (1999) The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH2-terminal kinase pathway. J. Cell Biol. 146, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zand T. P., Reiner D. J., Der C. J. (2011) Ras effector switching promotes divergent cell fates in C. elegans vulval patterning. Dev. Cell 20, 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim A. H., Sasaki T., Chao M. V. (2003) JNK-interacting protein 1 promotes Akt1 activation. J. Biol. Chem. 278, 29830–29836 [DOI] [PubMed] [Google Scholar]
- 63. Kim A. H., Yano H., Cho H., Meyer D., Monks B., Margolis B., Birnbaum M. J., Chao M. V. (2002) Akt1 regulates a JNK scaffold during excitotoxic apoptosis. Neuron 35, 697–709 [DOI] [PubMed] [Google Scholar]
- 64. Song J. J., Lee Y. J. (2005) Cross-talk between JIP3 and JIP1 during glucose deprivation. SEK1-JNK2 and Akt1 act as mediators. J. Biol. Chem. 280, 26845–26855 [DOI] [PubMed] [Google Scholar]
- 65. Barthwal M. K., Sathyanarayana P., Kundu C. N., Rana B., Pradeep A., Sharma C., Woodgett J. R., Rana A. (2003) Negative regulation of mixed lineage kinase 3 by protein kinase B/AKT leads to cell survival. J. Biol. Chem. 278, 3897–3902 [DOI] [PubMed] [Google Scholar]
- 66. Figueroa C., Tarras S., Taylor J., Vojtek A. B. (2003) Akt2 negatively regulates assembly of the POSH-MLK-JNK signaling complex. J. Biol. Chem. 278, 47922–47927 [DOI] [PubMed] [Google Scholar]
- 67. Park H.-S., Kim M.-S., Huh S.-H., Park J., Chung J., Kang S. S., Choi E.-J. (2002) Akt (protein kinase B) negatively regulates SEK1 by means of protein phosphorylation. J. Biol. Chem. 277, 2573–2578 [DOI] [PubMed] [Google Scholar]
- 68. Aguirre V., Uchida T., Yenush L., Davis R., White M. F. (2000) The c-Jun NH2-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser307. J. Biol. Chem. 275, 9047–9054 [DOI] [PubMed] [Google Scholar]
- 69. Hao Y., Wong R., Feig L. A. (2008) RalGDS couples growth factor signaling to Akt activation. Mol. Cell Biol. 28, 2851–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de Keizer P. L., Burgering B. M., Dansen T. B. (2011) Forkhead box O as a sensor, mediator, and regulator of redox signaling. Antioxid. Redox Signal. 14, 1093–1106 [DOI] [PubMed] [Google Scholar]
- 71. Honda Y., Honda S. (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13, 1385–1393 [PubMed] [Google Scholar]
- 72. Marasa B. S., Srikantan S., Masuda K., Abdelmohsen K., Kuwano Y., Yang X., Martindale J. L., Rinker-Schaeffer C. W., Gorospe M. (2009) Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci. Signal. 2, ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]